Abstract
ESR1 mutations were recently found to be an important mechanism of endocrine resistance in ER-positive (ER + ) metastatic breast cancer. To determine the clinicopathological features driving the emergence of the ESR1 mutations we studied plasma cfDNA and detailed clinical data collected from patients with metastatic breast cancer. Droplet Digital PCR was performed for the detection of the most common ESR1 mutations and PIK3CA mutations. Among the patients with ER + /HER2- disease, ESR1 mutations were detected in 30% of the patients. There were no associations between the pathological features of the primary disease or time to distant recurrence and the emergence of ESR1 mutations in metastatic disease. The prevalence of the ESR1 mutations was significantly associated with prior treatment with an aromatase inhibitor in the adjuvant or metastatic setting. The prevalence of the ESR1 mutations was also positively associated with prior fulvestrant treatment. Conversely, the prevalence of ESR1 mutations was lower after treatment with a CDK4/6 inhibitor. There were no significant associations between specific systemic treatments and the prevalence of PIK3CA mutations. These results support the evolution of the ESR1 mutations under the selective pressure of treatment with aromatase inhibitors in the adjuvant and metastatic settings and have important implications in the optimization of adjuvant and metastatic treatment in ER + breast cancer.
Genetics: Drug treatment spurs new resistance mutations
Treatment with aromatase inhibitors, a class of drugs that suppress the synthesis of estrogen, can drive the evolution of mutations in the estrogen receptor gene ESR1, leading to tumor resistance against hormone therapies. To better understand the emergence of ESR1 mutations, Rinath Jeselsohn from the Dana-Farber Cancer Institute in Boston, Massachusetts, USA, and coworkers tested tumor DNA contained within blood samples from 155 women with metastatic breast cancer. They found ESR1 mutations rarely in women with any molecular subtype of cancer other than estrogen receptor-positive disease. Nothing about the primary tumor predicted who would develop ESR1 mutations; however, treatment with an aromatase inhibitor was associated with mutations arising. The findings highlight the need to develop therapeutic regimens that reduce the selective pressure for ESR1 mutations and/or target these mutations directly.
Introduction
The ESR1 ligand-binding domain (LBD) mutations were unveiled in recent years as an important mechanism of acquired endocrine resistance that evolves under the selective pressure of endocrine treatments. These mutations are rarely found in primary estrogen receptor-positive (ER + ) breast cancers but have a high prevalence in metastatic disease and lead to constitutive ligand independent activity.1–3 The most prevalent mutations as detected in a number of studies are the Y537S and D538G mutations. The third most common mutation is the E380Q mutation, also located in the LBD.4
Liquid biopsies detecting circulating tumor DNA (cfDNA) are emerging as a useful non-invasive tool for serial monitoring of genomic alterations in patients with metastatic cancer. Multiple studies have now shown that the ESR1 LBD mutations can be successfully detected in the plasma of patients with metastatic ER + breast cancer.5,6 Patients with ER + metastatic breast cancer who received an aromatase inhibitor (AI) in the metastatic setting compared to AI naive patients had a higher prevalence of cfDNA ESR1 mutations.7 Moreover, patients with metastatic ER + breast cancer with detectable cfDNA ESR1 mutations had decreased progression free survival on subsequent treatment with an aromatase AI.6
In this study, we sought to comprehensively study the associations between the emergence of the ESR1 mutations in cfDNA, clinicopathological features, and treatments in the adjuvant and metastatic settings. We prospectively collected plasma samples from patients with metastatic breast cancer from a single institution and tested for the most common ESR1 mutations using droplet digital PCR (ddPCR). We also tested for the most common PIK3CA mutations, as PIK3CA mutations have been reported to be an early event in ER + breast cancer and are found in more than 30% of ER + primary treatment naive breast cancers. The frequency of PIK3CA mutations do not change under the pressure of endocrine treatments or the development of endocrine resistance and metastatic disease.2,8
Results
Patient and sample characteristics
We prospectively collected 155 plasma samples from patients with metastatic breast cancer enrolled on this biospecimen collection protocol. Median age at initial breast cancer diagnosis was 46 years, (range 29–81 years). Subtype distribution was as follows: ER + /HER2-, n = 113 (73%); HER2 + (either ER + or ER-), n = 25 (16%); ER and PR-negative/HER2-negative (triple-negative breast cancer, TNBC), n = 17 (11%). Results of clinical ER, PR, and HER2 testing on an archival metastatic biopsy specimen were available in 147 (95%) of the 155 patients included in this study.
Patient and sample characteristics of the ER + /HER2- cohort are shown in Table 1. Among patients who presented with stage I–III disease, median disease-free interval was 58.9 months (range 0–368.8). Among the 113 patients presenting with hormone receptor-positive/HER2-negative breast cancer, the primary tumor was ER and PR-positive in 92 (81.4%) of patients; 15 (13.3%) patients had ER-positive/PR-negative disease. Approximately one-fifth of the patients (n = 21) presented with de novo metastatic disease. Among patients who presented with stage I–III ER and/or PR-positive disease, 84% received adjuvant endocrine therapy, and median disease-free interval was 67.4 months (range 6.1–312.1). Contemporaneous research metastatic biopsies were available for 23 of the 113 patients.
Table 1.
Characteristics of the ER + /HER2-cohort at time of diagnosis of primary disease
| Characteristic | All patients (113) | ESR1 mutant (34) | ESR1 WT (79) | p-values | PIK3CA mutant (36) | PIK3CA WT (77) | p-values |
|---|---|---|---|---|---|---|---|
| Median age at random assignment, years (IQR) | 57.1 (48.7, 65.1) | 57.1 (52.8, 68.1) | 56.7 (46.7, 64.6) | 0.16 | 57.9 (49.1, 63.4) | 56.7 (48.4, 65.1) | 0.61 |
| Tumor grade | |||||||
| I | 12 (10.6%) | 6 (17.6%) | 6 (7.6%) | 0.19 | 6 (16.7%) | 6 (7.8%) | 0.19 |
| II | 50 (44.2%) | 15 (44.1%) | 35 (44.3%) | 15 (41.7%) | 35 (45.4%) | ||
| III | 43(38.1%) | 10 (29.4%) | 33 (41.8%) | 13 (36.1%) | 30 (39.0%) | ||
| Unknown | 8 (7.1%) | 3 (8.8%) | 5 (6.3%) | 2 (5.5%) | 6 (7.8%) | ||
| Stage | |||||||
| 0 | 1 (0.9%) | 0 (0%) | 1 (1.3%) | 0.022 | 0 (0%) | 1 (1.3%) | 0.48 |
| I | 23 (20.4%) | 13 (38.2%) | 10 (12.7%) | 10 (27.8%) | 13 (16.9%) | ||
| II | 52 (46.0%) | 14 (41.2%) | 38 (48.1%) | 15 (41.7%) | 37 (48.0%) | ||
| III | 10 (8.8%) | 4 (11.8%) | 6 (7.6%) | 5 (13.9%) | 5 (6.5%) | ||
| IV | 21 (18.6%) | 3 (8.8%) | 18 (22.8%) | 6 (16.7%) | 15 (19.5%) | ||
| Unknown | 6 (5.3%) | 0 (0%) | 6 (7.6%) | 0 (0%) | 6 (7.8%) | ||
| PR status-primary | |||||||
| Positive | 92 (81.4%) | 27 (79.4%) | 65 (82.3%) | 0.77 | 30 (83.3%) | 62 (80.5%) | 0.38 |
| Negative | 15 (13.3%) | 5 (14.7%) | 10 (12.7%) | 3 (8.3%) | 12 (15.6%) | ||
| Unkown | 6 (5.3%) | 2 (5.9%) | 4 (5.1%) | 3 (8.3%) | 3 (3.9%) | ||
ESR1 and PIK3CA mutations detected in cfDNA of patients with metastatic breast cancer are highly concordant with metastatic tumor samples
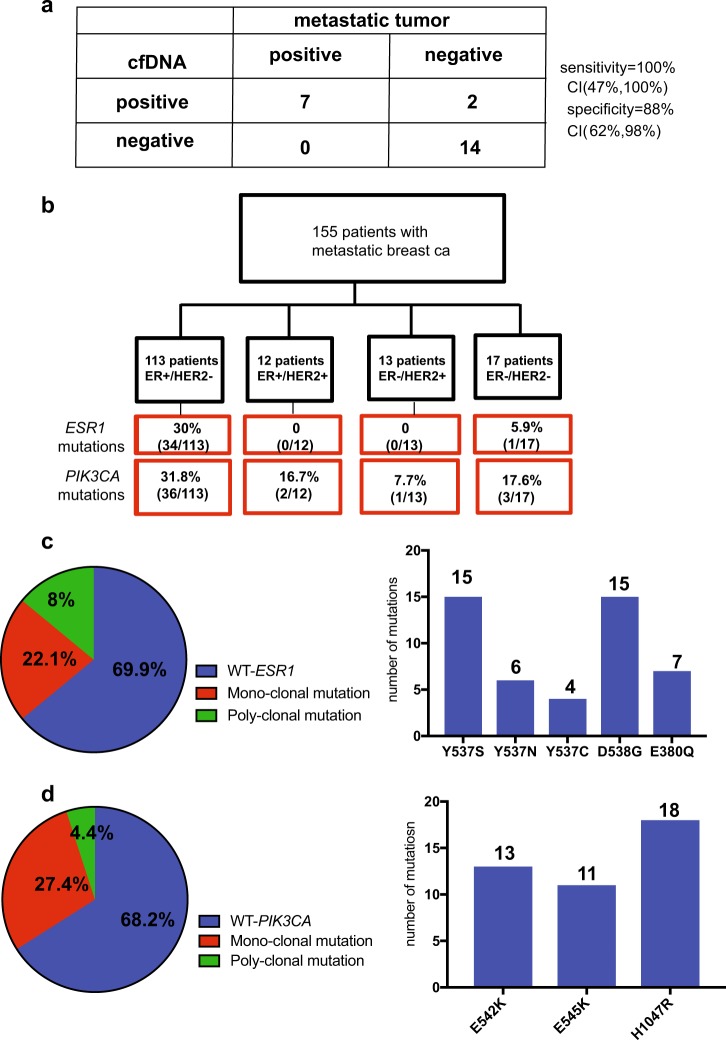
We developed a highly sensitive assay using droplet digital PCR (ddPCR) for the detection of the most common ESR1 (Y537S, D538G, E380Q, Y537N, Y537C) and PIK3CA mutations (H1047R, E542K, E545K). To examine the sensitivity and specificity of ESR1 mutant detection in cfDNA compared to detection in tissue biopsies both tested by ddPCR, we tested for the ESR1 mutations in a subset of 23 patients from whom contemporaneous metastatic tumor biopsies were available. Seven ESR1 mutations were found in the tissue samples and all were detected by the cfDNA analysis. There were two mutations detected by the cfDNA analysis that were not detected in the tumor samples, which likely reflects the ability of the cfDNA test to capture information from genetically heterogeneous metastatic samples. Overall, the plasma and metastatic tumor samples were highly concordant with 100% sensitivity and 88% specificity for the plasma ESR1 cfDNA assay compared to testing metastatic tissue samples applying ddPCR (Fig. 1a). High concordance between cfDNA ESR1 mutations and metastatic tissues, was seen in previous studies.6,9
Fig. 1.
Prevalence of cfDNA ESR1 and PIK3CA mutations in patients with metastatic breast cancer. a The sensitivity and specificity of mutation detection in cfDNA compared to paired metastatic tissue samples. b REMARK diagram. c Prevalence of ESR1 mutations in cfDNA among ER + /HER2-metastatic patients. d Prevalence of PIK3CA mutations in cfDNA among ER + /HER2- metastatic patients
ESR1 and PIK3CA mutations are frequently detected in cfDNA of patients with metastatic breast cancer
Given the concordance between ddPCR in blood vs. tumor specimens, we next proceeded to evaluate the frequencies of ESR1 mutations and PIK3CA mutations in the entire cohort (n = 155). We found 35 (22.6%) and 42 (27%) patients to have at least 1 ESR1 mutation and 1 PIK3CA mutation, respectively. All patients with an ESR1 mutation had ER + disease, with the exception of one patient with TNBC (Fig. 1b). This patient was diagnosed with TNBC based on pathology of the primary and a metastatic lesion. The cfDNA ESR1 mutation was confirmed in replicate assays. This patient did not have a history of ER + breast cancer and this mutation likely reflects a sub-clone of ER + cells that is a rare event as in previous studies the ESR1 recurrent mutations were not detected in TNBC metastatic tissue samples.2 In the subgroup of patients with ER + /HER2-metastatic breast cancer, 30% were found to have an ESR1 mutation and 26% of these patients had more than one ESR1 mutation suggesting polyclonal disease (Fig. 1c). Similar to previous reports, the most common mutations were the Y537S (15 or 31.9%) and D538G (15 or 31.9%) mutations followed by the E380Q mutation (7 or 14.9%). In contrast to the ESR1 mutations, the PIK3CA mutations were not significantly more prevalent in ER + /HER2-disease compared to other breast cancer subtypes and 14% of the PIK3CA mutant samples had more than one PIK3CA mutation (Fig. 1d). The presence of ESR1 and PIK3CA mutations were not independent of each other. Among the 113 patients with ER + disease, patients without a detectable PIK3CA mutation were less likely to have an ESR1 mutation (21% of the patients with WT PIK3CA had a ESR1 mutation, whereas 50% of the patients with a PIK3CA mutation also had an ESR1 mutation, p-value = 0.002).
The E380Q mutation confers ligand independent cell growth and relative resistance to tamoxifen and fulvestrant
The three most commonly reported ER LBD mutations are the D538G, Y537S, and the E380Q mutations; our data confirm these findings. The D538G and Y537S mutations are both within the C-terminal helix within helix 12 and their structure has been solved by crystallography studies.10,11 In addition, a number of studies have investigated the functional roles of these mutations.12 The structure of the E380Q mutation has not been solved. However, since the E380 residue is in the vicinity of helix 12, it is highly plausible that this mutation has similar functional characteristics. To date, only a limited number of studies have investigated the functional consequences of the E380Q mutation and the data are conflicting. In one study, a PDX model harboring this mutation did not display E2 independent tumor growth, whereas overexpression of the E380Q mutation in MCF7 cells resulted in E2 independent transcriptional activity and proliferation, albeit to a lesser extent compared to cells overexpressing the Y537S mutation.4,13
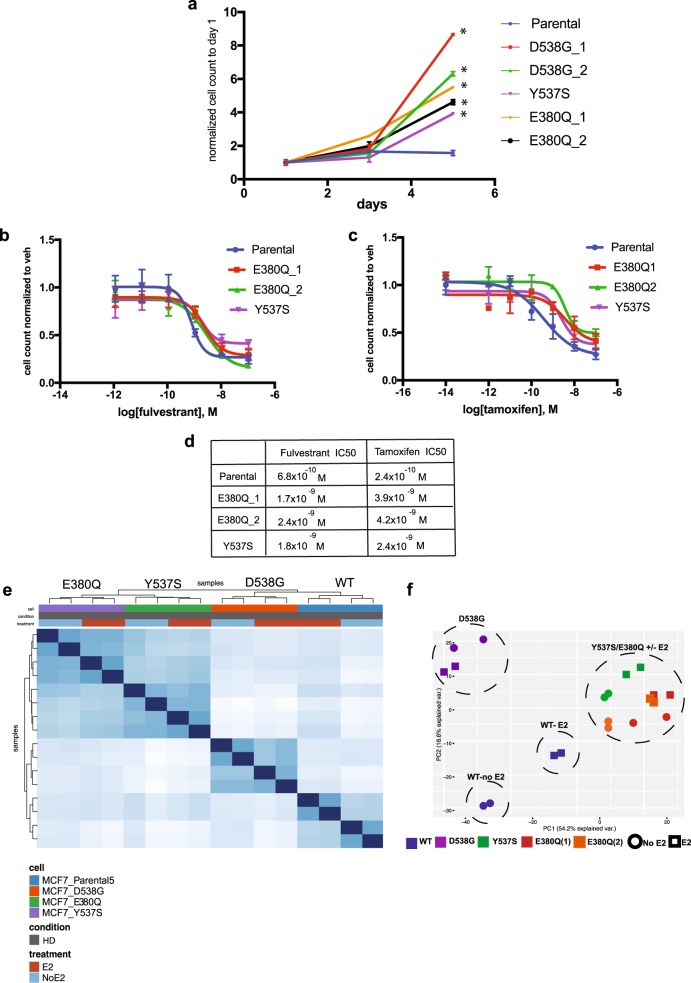
To study the functional roles of the E380Q mutation, we generated and characterized single allele E380Q, Y537S, and D538G knock-in MCF7 cells. Similar to the Y537S and D538G knocked-in MCF7 cells, the E380Q knocked-in cells displayed a significant growth advantage in hormone-depleted (HD) conditions when compared to the MCF7 WT ESR1 parental cells (Fig. 2a). In addition, the E380Q knocked-in cells were relatively resistant to tamoxifen and fulvestrant similar to the Y537S mutant cells (Fig. 2b–d). To study the global transcriptional effects of the E380Q mutation, we performed RNA-seq in the knock-in cell lines. Pairwise correlation analysis of the RNA-seq in HD and HD + Estradiol (E2) clustered the E380Q and Y537S mutant cells together and distinctly separated from the D538G mutant and the WT-ER cells (Fig. 2e). Similarly, in a principal components analysis (Fig. 2f), the principal component 1 (PC1) is driven by the ESR1 allele with the E380Q allele clustering with Y537S and the WT and D538G alleles were in distinct clusters. The medium conditions resolved along PC2 and distinctly segregated the WT cells in HD conditions from HD + E2 conditions. This distinct separation between the HD and HD + E2 conditions was not seen in the mutant knock-in cells. These results indicate that the E380Q mutation, similar to the Y537S and D538G mutation, exhibits constitutive transcriptional activity, ligand independent cell proliferation, and relative resistance to tamoxifen and fulvestrant. Previous studies have demonstrated that the Y537S and D538G mutations confer distinct transcriptional programs.14,15 Here, we show that the E380Q mutant transcriptional program resembles the transcriptional program of the Y537S mutant and both are different from the D538G mutant. Taken together, these results support the inclusion of the E380Q ESR1 allele as an important resistance mutation.
Fig. 2.
The E380Q mutation confers ligand independent cell growth and relative resistance to tamoxifen and fulvestrant. a Cell growth of the E380Q (clone 1 and 2), Y537S and D538G (clone 1 and 2) knocked-in MCF7 cells and parental MCF7 cells in hormone-depleted (HD) conditions. Error bars represent S.D., n = 3. *p-value < 0.001. b Dose–response curves of the MCF7 parental cells, E380Q (clones 1 and 2) and Y537S cells with fulvestrant treatment on day 5. Error bars represent S.D., n = 3. c Dose–response curves of the MCF7 parental cells and E380Q cells (clones 1 and 2) with tamoxifen treatment on day 5. Error bars represent S.D., n = 3. d IC50 values for fulvestrant and tamoxifen in MCF7 parental cells and E380Q knocked-in MCF7 cells (clones 1 and 2). e Pairwise correlation analysis of the RNA-seq in HD and HD + Estradiol (E2) conditions. f Principal component analysis of the transcriptomes of ESR1 WT, D538G, Y537S, and E380Q mutant cells
ESR1 and not PIK3CA mutations are associated with presenting clinicopathological features and sites of metastasis in ER + /HER2-metastatic disease
To examine the pathological and clinical features associated with cfDNA ESR1 and PIK3CA mutations in the 113 ER + /HER-cases, we looked at differences in the clinical features of the breast cancer between patients who had and did not have detectable ESR1 or PIK3CA mutations in cfDNA. There were no significant associations between the pathological features of the primary tumor and cfDNA detection of ESR1 or PIK3CA mutations. Features examined included tumor grade, stage (I, II, or III) and PR status (Table 1). In contrast, there was a significant positive association between PR expression in the metastasis and ESR1 mutations (p-value = 0.006), but not PIK3CA mutations (p-value = 0.22), potentially indicative of active ER transcription in the presence of the ESR1 mutations (Table 2).
Table 2.
Patient characteristics of ER + /HER2-cohort after diagnosis of metastatic disease
| Characteristic | All patients (113) | ESR1 mutant (34) | ESR1 WT (79) | p-values | PIK3CA mutant (36) | PIK3CA WT (77) | p-values |
|---|---|---|---|---|---|---|---|
| PR metastatic | |||||||
| Positive | 47 (41.6%) | 22 (64.7%) | 25 (31.6%) | 0.006 | 19 (52.8%) | 28 (36.4%) | 0.22 |
| Negative | 59 (52.2%) | 12 (35.3%) | 47 (59.5%) | 17 (47.2%) | 42 (54.5%) | ||
| Unknown | 7 (6.2%) | 0 (0%) | 7 (8.9%) | 0 (0%) | 7 (9.1%) | ||
| Presentation of metastatic disease | |||||||
| Relapsed | 86 (76.1%) | 31 (91.2%) | 55 (69.6%) | 0.016 | 29 (80.6%) | 57 (74.0%) | 0.5 |
| De novo | 27 (23.9%) | 3 (8.8%) | 24 (30.4%) | 7 (19.4%) | 20 (26.0%) | ||
| Site of mets | |||||||
| Liver | |||||||
| Yes | 74 (65.5%) | 30 (88.2%) | 44 (55.7%) | 0.001 | 26 (72.2%) | 48 (62.3%) | 0.4 |
| No | 38 (33.6%) | 4 (11.8%) | 34 (43.0%) | 10 (27.8%) | 28 (36.4%) | ||
| Bone | |||||||
| Yes | 87 (77.0%) | 33 (97.1%) | 54 (68.4%) | < 0.001 | 32 (88.9%) | 55 (71.4%) | 0.056 |
| No | 25 (22.1%) | 1 (2.9%) | 24 (30.4%) | 4 (11.1%) | 21 (27.3%) | ||
| Lung | |||||||
| Yes | 38 (33.6%) | 13 (38.2%) | 24 (30.4%) | 0.51 | 12 (33.3%) | 25 (32.5%) | 1.00 |
| No | 75 (66.4%) | 21 (61.8%) | 54 (68.4%) | 24 (66.7%) | 51 (66.2%) | ||
| Visceral disease | |||||||
| Yes | 69 (61.1%) | 25 (73.5%) | 44 (55.7%) | 0.2 | 26 (72.2%) | 43 (55.8%) | 0.21 |
| No | 40 (35.4%) | 9 (26.5%) | 31 (39.2%) | 10 (27.8%) | 30 (39.0%) | ||
| Unknown | 4 (3.5%) | 0 (0%) | 4 (5.1%) | 0 (0%) | 4 (5.2%) | ||
| Status of metastatic disease | |||||||
| New diagnosis of mets | 35 (31.0%) | 3 (8.8%) | 32 (40.5%) | < 0.001 | 9 (25.0%) | 26 (33.8%) | 0.002 |
| Stable disease | 14 (12.4%) | 1 (2.9%) | 13 (16.5%) | 0 (0%) | 14(18.2%) | ||
| Progressive disease | 63 (55.8%) | 30 (88.2%) | 33 (41.8%) | 27 (75.0%) | 36 (46.8%) | ||
| Unknown | 1 (0.9%) | 0 (0%) | 1 (1.3%) | 0 | 1 | ||
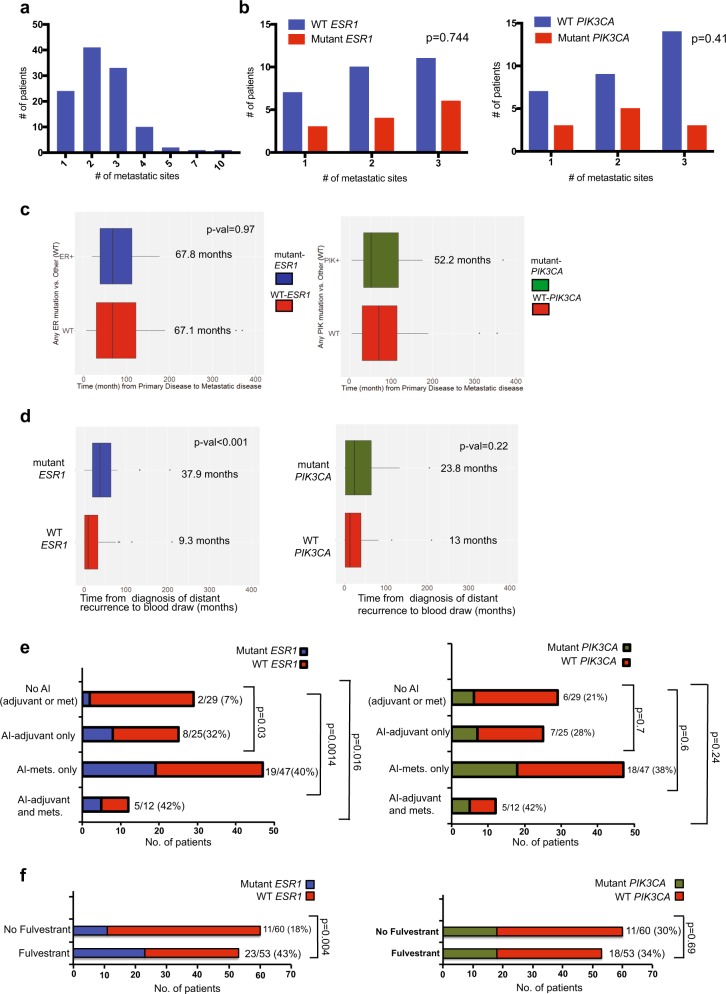
While there was no association between visceral vs. bone disease only or the number of metastatic sites and the presence of cfDNA ESR1 or PIK3CA mutations, patients with liver and bone metastases were more likely to have detectable cfDNA ESR1 but not PIK3CA mutations (p-value = 0.001 for liver and p-value < 0.001 for bone metastases) (Fig. 3a-3b and Table 2). We have detected an association between the D538G mutations and liver metastases in previous studies.15 In this study, the association between the ESR1 mutations and liver metastases is seen when including all the allelic LBD mutations tested here and not specifically the D538G mutation. This is possibly due to the fact that cfDNA is capturing mutations from multiple metastatic sites and not specifically the liver. The mechanism of this organotropism remains to be elucidated.
Fig. 3.
Associations between clinical features and ESR1 mutations. a The distribution of the patients based on number of metastatic sites for all 113 ER + /HER2-patients. b Associations between the number of metastatic sites and the prevalence of ESR1 or PIK3CA mutations. c Interval time from diagnosis of primary disease and distant recurrence in patients with and without ESR1 mutations and in patients with and without PIK3CA mutations. Center line is median, box limits are the upper and lower quartiles, whiskers are the 1.5 x inter-quartiles and the points are the outliers. d Interval time from diagnosis of distant recurrence to blood draw in patients with and without ESR1 mutations and in patients with and without PIK3CA mutations. Center line is median, box limits are the upper and lower quartiles, whiskers are the 1.5 x inter-quartiles and the points are the outliers. e Associations between aromatase inhibitor (AI) treatment and the prevalence of ESR1 and PIK3CA mutations. f Associations between fulvestrant treatment and the prevalence of ESR1 and PIK3CA mutations
ESR1 mutation frequency varies according to disease status, timing of blood draw and emerge after aromatase inhibitor treatment in the adjuvant and metastatic settings
In this study, a convenience blood sample was collected after study enrollment at any point in a patient’s metastatic disease course. We thus evaluated the relationship between the timing of blood collection relative to disease status and endocrine therapy, and the frequency of ESR1 and PIK3CA alterations. The clinical status of the metastatic disease at the time of the plasma collection was strongly linked to the detection of ESR1 and PIK3CA mutations in cfDNA (Table 2). Patients who had stable disease or a new diagnosis of metastatic disease at the time of the blood draw were less likely to have cfDNA ESR1 (11.7%) or PIK3CA (25%) mutations compared to patients with progressive disease (88.2% for ESR1 mutations and 75% for PIK3CA mutations, respectively) at the time of the blood draw. This finding was seen for mutations in both genes and is likely reflective of less shedding of cfDNA at the time of stable disease or new diagnosis of metastases, as was shown for other mutations and cancer types.16–18
The disease-free interval defined as the time from the diagnosis of primary disease to the development of distant recurrence did not significantly differ in patients who had or did not have detectable cfDNA ESR1 or PIK3CA mutations (Fig. 3c). Conversely, cfDNA ESR1 mutation frequency positively correlated with the duration of time elapsed from the diagnosis of distant recurrence to the plasma collection (average time 9.3 months for WT ESR1 and 37.9 months for mutant ESR1, p-value < 0.001) (Fig. 3d). In addition, patients who had relapsed metastatic disease were more likely to have detectable cfDNA ESR1 mutations (p-value = 0.016 for ESR1), compared to patients who presented with de novo metastatic breast cancer (Table 2). These two associations were not seen with cfDNA PIK3CA mutations and may be the consequence of more prolonged selection by endocrine therapies.
To test this hypothesis, we next investigated the impact of systemic treatments given prior to the blood sample collection on the prevalence of ESR1 and PIK3CA mutations in cfDNA in the ER + /HER2-cohort. Chemotherapy treatment in the setting of neoadjuvant/adjuvant treatment for early-stage disease or metastatic disease was not associated with the prevalence of ESR1 or PIK3CA mutations in cfDNA (Table 3). The majority of the patients were exposed to treatment with an AI (n = 84) either in the adjuvant, metastatic setting or both prior to the blood draw. Patients who received an AI at any time compared to patients who did not receive AI treatment were more likely to have detectable ESR1 mutations in cfDNA (Fig. 3e). We next examined whether the timing of AI treatment influenced the prevalence of the ESR1 and PIK3CA mutations. We observed high rates of ESR1 mutations among patients with metastatic breast cancer, irrespective of whether AI exposure was in the adjuvant setting only (32%, p-value = 0.03), metastatic setting only (40%, p-value = 0.0014), or in both the adjuvant and metastatic settings (42%, p-value = 0.016). There was no significant difference in the prevalence of ESR1 mutations in patients who received AI treatment in the adjuvant setting only vs. patients who received AI treatment for metastatic disease only (p-value = 0.62). Notably, given the proposed role of the PI3K pathway in mediating endocrine resistance, there was no association between AI exposure in the adjuvant, metastatic or both and PIK3CA mutations (p-value > 0.05 for all treatment settings), supporting a selection effect of AI treatment unique to ESR1 mutations. We next studied the association of fulvestrant treatment for metastatic disease and ESR1 and PIK3CA mutations. Similar to AI treatment, we observed an increase in the prevalence of the ESR1 mutations in patients who received fulvestrant compared to those who did not (p-value = 0.004). Again, this association was not seen for the PIK3CA mutations (p = 0.69) (Fig. 3f). We also tested the associations between tamoxifen treatment and the ESR1 mutations. Overall 49 patients received either single agent adjuvant tamoxifen or sequential treatment with tamoxifen followed by an aromatase inhibitor. To look at the effect of adjuvant tamoxifen treatment on the emergence of the ESR1 mutations, we selected patients that received adjuvant tamoxifen and no endocrine treatment (AI or fulvestarnt) in the metastatic setting prior to the blood draw. Among these patients, six patients received sequential adjuvant tamoxifen followed by an AI and one patient out of these patients was found to have an ESR1 mutation. There were nine patients that received adjuvant tamoxifen only and of these patients, one patient was found to have an ESR1 mutation. There was no significant positive or negative associations between adjuvant tamoxifen treatment (adjuvant tamoxifen/adjuvant tamoxifen followed by an AI) and the emergence of the ESR1 mutations (Supplementary Fig. S1). This analysis was limited because of the small numbers of patients and should be interpreted with caution. Nonetheless, these results demonstrate that the ESR1 mutations are found in a small number of patients that received prior endocrine treatment with tamoxifen alone, albeit in small numbers. In the metastatic setting, 20 patients received tamoxifen treatment prior to the blood draw. We did not detect a significant association between the tamoxifen treatment for metastatic disease and the prevalence of the ESR1 mutations. Collectively, these results support the clonal evolution unique to the ESR1 mutations under the selective pressure of AI treatment in the adjuvant and metastatic settings, and fulvestrant treatment.
Table 3.
Associations between treatments and mutations in ER + /HER2-cohort
| Treatment | All patients (113) | ESR1 mutant (34) | ESR1 WT (79) | p-values | PIK3CA mutant (36) | PIK3CA WT (77) | p-values |
|---|---|---|---|---|---|---|---|
| Neo/adj chemotherapy | |||||||
| Yes | 68 (60.2%) | 24 (70.6%) | 44 (55.7%) | 0.15 | 20 (55.6%) | 48 (62.3%) | 0.54 |
| No | 45 (39.8%) | 10 (29.4%) | 35 (44.3%) | 16 (44.4%) | 29 (37.7%) | ||
| Chemotherapy in metastatic disease | |||||||
| Yes | 61 (54.0%) | 23 (67.6%) | 38 (48.1%) | 0.067 | 19 (52.8%) | 42 (54.5%) | 1.00 |
| No | 52 (46.0%) | 11 (32.4%) | 41 (51.9%) | 17 (47.2%) | 35 (45.5%) | ||
| Palbociclib | |||||||
| Yes | 23 (20.4%) | 2 (5.9%) | 21 (26.6%) | 0.011 | 5 (13.9%) | 18 (23.4%) | 0.32 |
| No | 90 (79.6%) | 32 (94.1%) | 58 (73.4%) | 31 (86.1%) | 59 (76.6%) | ||
| Everolimus | |||||||
| Yes | 16 (14.2%) | 7 (20.6%) | 9 (11.4%) | 0.24 | 4 (11.1%) | 12 (15.6%) | 0.77 |
| No | 97 (85.8%) | 27 (79.4%) | 70 (88.6%) | 32 (88.9%) | 65 (84.4%) | ||
ESR1 alterations in the setting of novel targeted agents
A subset of patients in this study received a CDK4/6 inhibitor (n = 23) or everolimus (n = 16) prior to collection of the blood specimen for cfDNA. Interestingly, patients who received palbociclib for metastatic disease were less likely to have detectable ESR1 mutations in cfDNA but this was not the case for PIK3CA mutations (p-value = 0.01) (Table 3). In contrast, there was no association between everolimus treatment in metastatic disease and the prevalence of ESR1 or PIK3CA mutations in cfDNA (Table 3). These results suggest that palbociclib treatment may be effective in inhibiting tumors harboring the ESR1 mutations, which is in line with previous reports demonstrating that the tumors harboring the ESR1 mutation are sensitive to palbociclib.19
Discussion
Previous studies have shown that the ESR1 endocrine resistance mutations are rarely detected in primary treatment naive tumors and evolve predominantly in metastatic disease under the selective pressure of endocrine treatment. cfDNA analysis using ddPCR is emerging as a non-invasive highly sensitive test that can capture the heterogeneity of the mutational landscape from multiple metastatic sites. In this study, we used cfDNA to comprehensively study the clinicopathological features and treatments associated with the evolution of the ESR1 mutations. Since these mutations drive endocrine therapy resistance and confer worse outcomes in metastatic disease,20 understanding these associations is clinically important.
In line with previous studies, we observed a strong link between treatment with aromatase inhibitors and the evolution of the ESR1 mutations. Importantly, in our study the prevalence of the ESR1 mutations increased equally after AI treatment in the adjuvant setting, metastatic setting or both. The finding that the ESR1 mutations emerge during adjuvant AI treatment differs from an earlier study and has important clinical implications.6 This finding may explain why adjuvant AI in comparison to adjuvant tamoxifen reduces the risk of distant recurrence, but in most studies has not been shown to significantly improve overall survival.21 This finding also highlights the importance of improving adjuvant endocrine treatments to reduce the clonal selection of the ESR1 mutations, particularly for patients at high risk of recurrence.
In a previous study, the emergence of the ESR1 mutations was seen with AI treatment in the metastatic setting only.6 The reason for this discrepancy between our study and the study by Schiavon et al. is not clear. This may be due to the overall small number of patients in both studies. Another possible explanation for the discrepancy is differences between the two study centers in the rates of adherence to adjuvant aromatase inhibitors. Non-adherence to adjuvant aromatase inhibitors is substantial and in the range of 30%,22,23 which could account for the discordant results. Nonetheless, a larger study to resolve this inconsistency is needed.
Notably, we did not find significant correlations between the detection of cfDNA ESR1 mutations in metastatic disease and clinical or pathological features at the time of initial presentation of early-stage ER + disease. In addition, chemotherapy treatment, either adjuvant or for metastatic diseases, did not influence the prevalence of the ESR1 mutations. The emergence of the ESR1 mutations was also not linked to the timing of distant recurrence (i.e., early vs. late recurrences). Conversely, the likelihood of detecting ESR1 cfDNA mutation was increased the longer the duration of ER + metastatic disease. This finding underscores the concept that ESR1 mutations occur under the selective pressure of endocrine therapy, and highlight the need for improved treatments targeting these ER mutations both to circumvent tumor resistance and effectively treat it when it occurs.
The PIK3CA mutations are relatively common in ER + breast cancers and are an early mutational event in the development of breast cancer.24 Previous reports have indicated that the prevalence of the PIK3CA mutations in primary and metastatic tumors is comparable and these mutations likely do not evolve under the selective pressure of current treatments.2 In this study, we analyzed the most common PIK3CA mutations in addition to the ESR1 mutations. Despite the putative role of the PI3K pathway in mediating endocrine resistance, we did not observe any enrichment of PIK3CA mutations in patients according to prior endocrine therapy exposure. The strength of our results compared to existing data using metastatic tumor specimens (which are often collected at or near the time of metastatic recurrence) is the span of timing of blood sample collection, with 55% of samples collected ≥ 12 months from the date of metastatic recurrence, suggesting that even with long-term endocrine selection, PIK3CA mutations are not selected.
Finally, we report on an intriguing association between the use of CDK4/6 inhibitors and reduced emergence of ESR1 mutations, though our observations are based on small numbers of patients. Of note, we recently showed in preclinical studies that breast cancers harboring the ESR1 mutations remain sensitive to palbociclib.15 Overall survival data from multiple large, randomized, phase 3 studies evaluating the role of CDK4/6 inhibitors in ER-positive metastatic breast cancer are not yet mature. However, if an OS advantage is eventually demonstrated, our data would suggest a potential mechanism for this, i.e., suppression of emerging ESR1 mutations that may be associated with resistance to future lines of endocrine therapy, and which would support their preferential use in the first-line setting. In addition, our data would support ongoing trials evaluating the role of CDK4/6 inhibitors in the adjuvant setting (NCT01864746, NCT02513394, and NCT03078751).
Our study has a number of limitations. Because of the relatively small number of patients, we analyzed the different ESR1 mutations together. However, a number of studies showed that the ESR1 mutations have allele-specific functional characteristics14,15 In addition, the preclinical studies characterizing the E380Q mutation included cell clones derived from one cell line. Lastly, because of the relatively small number of patients we could not perform a meaningful multivariate analysis and, therefore, we could not test for confounding factors. Despite these limitations, our study illuminates the significance of the E380Q mutation, the high prevalence of the ESR1 mutations in metastatic ER + breast cancer, the fidelity of cfDNA testing for the ESR1 mutations and the unique evolution of the ESR1 mutations under the selective pressure of AI treatment in the adjuvant and metastatic settings.
Methods
Patients and samples
Patients presenting to the breast oncology clinic at Dana-Farber Cancer Institute were approached for participation in an IRB-approved, prospective protocol, which included clinical data collection, a research biopsy, serial blood collection, and permission for genomic analysis. The Dana-Farber/Harvard Cancer Center (DF/HCC) institutional review board (IRB) approved the protocol (DF/HCC IRB #05-246). The protocol was later amended to include blood collection for ddPCR; hence, though nearly all patients enrolled in the protocol underwent a biopsy as part of the study, only in a subset of patients was the research biopsy contemporaneous with the blood sample collected for ddPCR. Patients provided written informed consent prior to the initiation of any study procedures.
Blood samples for ddPCR were collected from 10/2014 through 9/2016. Pathology of the primary tumors for all patients was reviewed at the Dana Farber Cancer Institute as part of routine clinical care. ER, progesterone receptor (PR), and human epidermal growth receptor 2 (HER2) status were assessed based on the ASCO/CAP guidelines.25,26 Breast cancer subtypes (ER + /HER2-, ER + /HER2 + , ER-/HER2 + and triple-negative) were assigned based on the receptor status of the primary tumors.
Circulating-free DNA extraction
Whole venous blood (6–10 mL) was collected from patients into EDTA lavender capped vacutainer tubes (Becton and Dickinson). Blood was processed within 2 h of collection. Whole blood was centrifuged for 10 min at 1200 × g and the plasma supernatant was further cleared by centrifugation for 10 min at 3000 × g. Cleared plasma was stored in cryostat tubes at –80 °C until use. Cell-free DNA was isolated using the QIAmp Circulating Nucleic Acid Kit (Qiagen) according to the manufacturer’s protocol. DNA was eluted in AVE buffer (100 μL) and stored at –80 °C until use.
Droplet digital PCR materials
Droplet digital PCR reagents were ordered from Bio-Rad. Primer/probe mix for ESR1 and PIK3CA mutations were custom-made by Life Technologies. The allele-specific MGB probes are labeled with either VIC or FAM at the 5' end and a nonfluorescent quencher (NFQ) at the 3' end.
The sequences used are:
ESR1 E380Q: forward primer, 5'-TGGATTTGACCCTCCATGATCAG-3',
reverse primer, 5'-AGACGAGACCAATCATCAGGATCT-3';
probe sequences: 5'-VIC-CCACCTTCTAGAATGTG-MGB-NFQ-3',
5'-FAM-CCACCTTCTACAATGTG-MGB-NFQ-3'.
ESR1 Y537C: forward primer, 5'-CAGCATGAAGTGCAAGAACGT-3',
reverse primer, 5'-TGGGCGTCCAGCATCTC-3';
probe sequences: 5'-VIC-TGCCCCTCTATGACCTG-MGB-NFQ-3',
5'-FAM- CCCCTCTGTGACCTG-MGB-NFQ-3'.
ESR1 Y537N: forward primer, 5'-CTGTACAGCATGAAGTGCAAGAAC-3',
reverse primer, 5'-TGGGCGTCCAGCATCTC-3';
probe sequences: 5'-VIC-TGGTGCCCCTCTATGAC-MGB-NFQ-3',
5'-FAM-TGCCCCTCAATGAC-MGB-NFQ-3'.
ESR1 Y537S: forward primer, 5'-CAGCATGAAGTGCAAGAACGT-3',
reverse primer, 5'-TGGGCGTCCAGCATCTC-3';
probe sequences: 5'-VIC-CCCCTCTATGACCTGC-MGB-NFQ-3',
5'-FAM-CCCTCTCTGACCTGC-MGB-NFQ-3'.
ESR1 D538G: forward primer, 5'-CAGCATGAAGTGCAAGAACGT-3',
reverse primer, 5'-TGGGCGTCCAGCATCTC-3';
probe sequences: 5'-VIC-CCCCTCTATGACCTGCT-MGB-NFQ-3',
5'-FAM-CCCTCTATGGCCTGCT-MGB-NFQ-3'.
PIK3CA E542K: forward primer, 5'-GGGAAAATGACAAAGAACAGCTCAA-3',
reverse primer, 5'-CTGTGACTCCATAGAAAATCTTTCTCCT-3';
probe sequences: 5'-VIC-CCTCTCTCTGAAATCA-MGB-NFQ-3',
5'-FAM-CCTCTCTCTAAAATCA-MGB-NFQ-3'.
PIK3CA E545K: forward primer, 5'-TCAAAGCAATTTCTACACGAGATCCT-3',
reverse primer, 5'-CTGTGACTCCATAGAAAATCTTTCTC-3';
probe sequences: 5'-VIC-CTCTCTGAAATCACTGAGCAG-MGB-NFQ-3',
5'- FAM-CTCTGAAATCACTAAGCAG-MGB-NFQ-3'.
PIK3CA H1047R: forward primer, 5'-GCAAGAGGCTTTGGAGTATTTCATG-3',
reverse primer, 5'-GCTGTTTAATTGTGTGGAAGATCCAA-3';
probe sequences: 5'-VIC-CCACCATGATGTGCATC-MGB-NFQ-3',
5'-FAM-CACCATGACGTGCATC-MGB-NFQ-3'.
ESR1 mutations
Standard curves were prepared using ESR1 mutant and wild-type plasmids ranging from 1000 copies/μL to 25 copies/μL. Mutation-specific TaqMan probes/primers (Life Technologies) were used in the PCR reaction, which had the following cycling conditions: 95 °C x 10 min (1 cycle), 40 cycles of 94 °C x 30 s and 56 °C x 1 min, and 10 °C hold.
PIK3CA mutations
Genomic DNA was extracted from the following cell lines: T84 (E542K heterozygous), MCF7 (E545K heterozygous), and HCT116 (H1047R heterozygous) using DNeasy Blood and Tissue Kit (Qiagen). Standard curves were prepared using genomic DNA from each of these cell lines with concentrations ranging from 1000 genome copies/μL to 10 genome copies/μL. Mutation-specific TaqMan probes/primers (Life Technologies) were used in the PCR reaction, which had the following cycling conditions: 95 °C x 10 min (1 cycle), 40 cycles of 94 °C x 30 s and 59 °C x 1 min, followed by 10 °C hold.
Droplet digital PCR workflow
ddPCR was performed as previously described.27 Briefly, TaqMan PCR reaction mixtures were assembled from a 2 × ddPCR Mastermix (Bio-Rad) and custom 40x TaqMan probes/primers made specific for each assay. Twenty-five microliters of assembled ddPCR reaction mixture, which include either 5 μL of cfDNA sample or water as no template control was loaded into wells of a 96-well PCR plate. The heat-sealed PCR plate was subsequently loaded onto the Automated Droplet Generator (Bio-Rad). After droplet generation, the new 96-well PCR plate was heat-sealed, placed on a conventional thermal cycler, and amplified to the end-point. After PCR, the 96-well PCR plate was read on the QX100 droplet reader (Bio-Rad). Analysis of the ddPCR data was performed with QuantaSoft analysis software (Bio-Rad) that accompanied the droplet reader.
Cell lines
MCF7 cells were purchased from ATCC. The cells were authenticated and regularly tested for mycoplasma contamination. The cells were maintained in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S). For hormone-depleted (HD) conditions, cells were kept in phenol-red free medium supplemented with 10% heat-inactivated charcoal-stripped (CS)-FBS and 1% P/S. All cells were incubated at 37 °C in 5% CO2.
For the cell proliferation assays the cells were plated in 24-well plates (2.5 × 104/well). The cells were trypsinized and collected at the noted time points. The number of viable cells was determined by Trypan blue exclusion staining and manually counted with a hemocytometer using independent triplicates.
Generation of the ESR1 mutant knocked-in cells
The generation of the ESR1 Y537S and D538G knock-in mutant cells in MCF7 cells was described previously.14 The ESR1 E380Q knock-in cells in MCF7 cells were generated using a similar protocol. The TALENs were designed to target intron 4 and the TALEN recognition sequences were:
XTN1: (bold = TAL binding sites, non-bold = cut region)
5' TGCTCCTAACTTGCTCTtggacaggtaagtgacctGGCTGTAGCTTAGGAGTA 3'
XTN2: (bold = TAL binding sites, non-bold = cut region)
5' TAACTTGCTCTTGGACAGgtaagtgacctggctGTAGCTTAGGAGTAGCA 3'
The sequences were synthesized and cloned into the SQT281 vector (Transposagen) that includes a Fok1 nuclease. For the homologous recombination we used a donor vector that contained the targeting constructs of piggyBac transposon and the a puromycin-thymidine kinase selection cassette flanked by about 500 bp of ESR1 genomic sequence with the coding changes (GAA > CAA) in the 3' end matching the exon 5 coding region. The MCF7 cells were transiently transfected with the TALEN vector and donor vector using Lipofectamine 2000 (Invitrogen). After puromycin selection, clones with the desired mutation were detected by Sanger sequencing and transiently transfected with transposase expression plasmids (Transposagen) for removal of the selection cassettes followed by gancyclovir treatment for negative selection. We confirmed the mutations by Sanger sequencing and RNA-sequencing.
RNA-seq
Total RNA was isolated using an RNeasy Mini Kit (Qiagen). For all cells and conditions samples were done in triplicates. RNA-seq libraries were made using the TruSeq RNA Sample Preparation Kit (Illumina) adapted for use on the Sciclone (Perkin-Elmer) liquid handler. Samples were sequenced on an Illumina Nextseq500. Alignment to the hg19 human genome was done using STAR v2.5.128 followed by Transcript assembly using cufflinks v2.2.129 and quality control steps were done using STAR v2.5.1 and RseQC v2.6.230.
Statistical analysis
Descriptive statistics were used to describe the disease and treatment characteristics of patients with and without an ESR1 mutation or a PIK3CA mutation. The association between the emergence of the ESR1 mutations in cfDNA, clinicopathological features and treatments at the time of the early-stage and metastatic disease was assessed using Fisher’s exact test. A Wilcoxon rank sum test was conducted to test association between time of disease development and mutation status. The sensitivity and specificity of mutation detection in plasma and metastatic tumor samples were calculated with 95% confidence interval. Analyses were performed using R version 3.3.1. (www.r-project.org). The Student’s t-test was used for the analysis of the cell line proliferation studies. The IC50 was calculated using GraphPad PRISM.
Data availability
All RNA-seq data was uploaded to GEO, accession number GSE112243. All other data are available from the corresponding author upon reasonable request.
Electronic supplementary material
Acknowledgements
We thank the patients for participating in this study. This work was supported in part by the Breast Cancer Alliance Exceptional Project Award (to R.J.), the NIH K08 CA191058-03 (to R.J.), Claudia Adams Barr Program for Innovative Cancer Research (to R.J.), NIH Breast Cancer SPORE Career Development Award (to R.J.), Friends of Dana Farber Cancer Institute Award (to R.J and I.E.K), Susan Komen Leadership Grant (to M.B.), the Breast Cancer Research Foundation (to N.U.L. and E.P.W.), Pan-Mass Challenge (to Dana-Farber Cancer Institute Breast Oncology Center), Fashion Footwear Association of New York (to Dana-Farber Cancer Institute Breast Oncology Center), National Comprehensive Cancer Network Oncology Research Program-Pfizer Independent Grants for Learning and Change (to N.U.L.), National Cancer Institute Specialized Program of Research Excellence (SPORE) Grant 1P50CA168504 (to Dana-Farber/Harvard Cancer Center) and Expect Miracles (to C.P.P. and Y.K.).
Author contributions
Y.K., P.K, P.A.J., C.P.J., N.U.L., I.E.K., E.P.W., M.B., and R.J. were responsible for the conception and design. Y.K., B.S., M.P., M.C., G.B., M.E.H., N.W., N.U.L., and R.J. contributed to data acquisition. J.A., M.C., and W.T.B. performed the biostatistical and computational analyses. Y.K., N.U.L., M.B., and R.J. wrote the manuscript with the input from all authors. All authors approved the final manuscript.
Competing interests
Dr. Pasi A. Jänne has received consulting fees from AstraZeneca, Boehringer Ingelheim, Pfizer, Merrimack Pharmaceuticals, Roche/Genentech, Chugai Pharmaceuticals, ACEA Biosciences, Ignyta, LOXO Oncology, Ariad Pharmaceuticals, Eli Lilly and Company and Araxes Pharmaceuticals; sponsored research funding from Astellas Pharmaceuticals, AstraZeneca, Daiichi Sankyo, PUMA and Eli Lilly and Company and receives post-marketing royalties on DFCI owned intellectual property on EGFR mutations licensed to Lab Corp. Dr. Cloud P. Paweletz has received honoraria from AstraZeneca, Clovis Oncology, and Bio-RAD and is a SAB member of DropWorks. Drs. Cloud P. Paweletz, Yanan Kuang, and Pasi A. Jänne are inventors on a pending patent related to non-invasive genotyping of cfDNA. The other authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Toy W, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 2013;45:1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeselsohn R, et al. Emergence of constitutively active estrogen receptor-α mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin. Cancer Res. 2014;20:1757–1767. doi: 10.1158/1078-0432.CCR-13-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson DR, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 2013;45:1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toy W, et al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 2017;7:277–287. doi: 10.1158/2159-8290.CD-15-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu D, et al. ESR1 mutations in circulating plasma tumor DNA from metastatic breast cancer patients. Clin. Cancer Res. 2016;22:993–999. doi: 10.1158/1078-0432.CCR-15-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schiavon G, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci. Transl. Med. 2015;7:313ra182. doi: 10.1126/scitranslmed.aac7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fribbens C, et al. Tracking evolution of aromatase inhibitor resistance with circulating tumour DNA analysis in metastatic breast cancer. Ann. Oncol. 2018;29:145–153. doi: 10.1093/annonc/mdx483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Network T. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spoerke JM, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat. Commun. 2016;7:11579. doi: 10.1038/ncomms11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nettles KW, et al. NFκB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses. Nat. Chem. Biol. 2008;4:241–247. doi: 10.1038/nchembio.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fanning SW, et al. Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation. Elife. 2016;5:e12792. doi: 10.7554/eLife.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeselsohn R, et al. ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer. Nat. Rev. Clin. Oncol. 2015;12:573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4:1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bahreini A, et al. Mutation site and context dependent effects of ESR1 mutation in genome-edited breast cancer cell models. Breast Cancer Res. 2017;19:60. doi: 10.1186/s13058-017-0851-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeselsohn R, et al. Allele-specific chromatin recruitment and therapeutic vulnerabilities of ESR1 activating mutations. Cancer Cell. 2018;33:173–186.e5. doi: 10.1016/j.ccell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ono A, et al. Circulating tumor DNA analysis for liver cancers and its usefulness as a liquid biopsy. Cell Mol. Gastroenterol. Hepatol. 2015;1:516–534. doi: 10.1016/j.jcmgh.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan YT, et al. Clinical relevance of cell-free DNA in gastrointestinal tract malignancy. Oncotarget. 2017;8:3009–3017. doi: 10.18632/oncotarget.13821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray ES, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget. 2015;6:42008–42018. doi: 10.18632/oncotarget.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fribbens C, et al. Plasma ESR1 Mutations and the treatment of estrogen receptor-Positive advanced breast cancer. J. Clin. Oncol. 2016;34:2961–2968. doi: 10.1200/JCO.2016.67.3061. [DOI] [PubMed] [Google Scholar]
- 20.Chandarlapaty S, et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer. JAMA Oncol. 2016;2:1310–1315. doi: 10.1001/jamaoncol.2016.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dowsett M, et al. Meta-analysis of breast cancer outcomes in adjuvant trials of aromatase inhibitors versus tamoxifen. J. Clin. Oncol. 2010;28:509–518. doi: 10.1200/JCO.2009.23.1274. [DOI] [PubMed] [Google Scholar]
- 22.Partridge AH, et al. Adherence to initial adjuvant anastrozole therapy among women with early-stage breast cancer. J. Clin. Oncol. 2008;26:556–562. doi: 10.1200/JCO.2007.11.5451. [DOI] [PubMed] [Google Scholar]
- 23.Hershman DL, et al. Early discontinuation and nonadherence to adjuvant hormonal therapy in a cohort of 8,769 early-stage breast cancer patients. J. Clin. Oncol. 2010;28:4120–4128. doi: 10.1200/JCO.2009.25.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller TW, Rexer BN, Garrett JT, Arteaga CL. Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res. 2011;13:224. doi: 10.1186/bcr3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond MEH, Hayes DF, Wolff AC, Mangu PB, Temin S. American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Oncol. Pract. 2010;6:195–197. doi: 10.1200/JOP.777003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolff AC, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Update. J. Clin. Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 27.Oxnard GR, et al. Noninvasive detection of response and resistance in egfrmutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin. Cancer Res. 2014;20:1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts andisoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, et al. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data was uploaded to GEO, accession number GSE112243. All other data are available from the corresponding author upon reasonable request.