Abstract
MicroRNAs (miRNAs) have been recognized as key regulators of tumorigenesis and progression. Serum miR-302c-3p expression is prominently deregulated in HCV-related hepatocellular carcinoma (HCC). However, the expression of miR-302c-3p and its functional role in HBV-related HCC are rarely investigated. In this study, we found that the expression levels of miR-302c-3p were prominently down-regulated in HCC tissues compared to matched tumor-adjacent tissues. Moreover, miR-302c-3p under-expression was detected in HCC cell lines compared to a normal hepatic cell line LO2. Low miR-302c-3p expression was positively correlated with multiple tumor nodes, venous infiltration and advanced TNM tumor stage of HCC patients. Notably, our follow up data and TCGA data demonstrated that low miR-302c-3p expression predicted a poor survival of HCC patients. Functionally, miR-302c-3p overexpression inhibited migration and invasion of MHCC97H cells in vitro. Additionally, miR-302c-3p knockdown showed an opposite effect on these metastatic behaviors of HepG2 cells. MiR-302c-3p negatively regulated tumor necrosis factor receptor associated factor 4 (TRAF4) abundance by directly targeting 3'-UTR of TRAF4 mRNA. The expression of TRAF4 was up-regulated in HCC tissues. The level of TRAF4 mRNA was inversely correlated with miR-302c-3p expression in HCC specimens. Mechanistically, miR-302c-3p restrained AKT-mediated epithelial-mesenchymal transition (EMT) in HCC cells. Importantly, TRAF4 restoration reversed the inhibitory effect of miR-302c-3p on AKT-induced EMT and HCC cell metastasis. MK2206, an AKT inhibitor, inhibited miR-302c-3p knockdown-induced EMT in HepG2 cells. In summary, these results indicate that miR-302c-3p exhibits a tumor suppressive role in HCC by targeting TRAF4. Inhibition of miR-302c-3p/TRAF4 axis may serve as a therapeutic target for HCC.
Keywords: miR-302c-3p; TRAF4; hepatocellular carcionoma; EMT, metastasis
Introduction
Hepatocellular carcinoma (HCC), the fifth most common cancer, exhibits extremely high morbidity and mortality rates worldwide 1. In 2012, more than approximately 700,000 died of HCC, which accounts for 9.1% in all cancers 2. Tumor recurrence and metastasis are the main causes for the poor survival rate of patients with advanced stage HCC 3. Therefore, identification of novel biomarkers for early stage detection and novel potential therapeutic targets for HCC is desirable and urgently needed.
MicroRNAs (miRNAs), as a family of small non-coding RNAs (ncRNAs), are approximately 22 nucleotides in length, which act as post-transcriptional regulators by binding to the 3'-untranslated regions (UTRs) of their target mRNAs 4. Recent studies have shown the association between miRNAs and HCC progression. For example, miR-187-3p, miR-542-3p, miR-1296 etc. function as tumor suppressors by targeting different genes 5-7, while miR-324-3p, miR-1468 and miR-519a act as oncogenes in HCC 8-10 and miR-367 promotes cell proliferation and metastasis during the progression of human HCC 11. MiR-302c-3p has been recognized as a cancer-related miRNA in previous studies. MiR-302c-3p is identified a potent estrogen receptor-alpha (ERα) regulating miRNA and inhibits estrogen-induced cell growth of breast cancer 12, 13. Down-regulation of miR-302c-3p leads to the resistance of tumor cells to natural killer (NK) cells via up-regulation of UL-16 binding protein 2 (ULBP2) and MHC class I chain-related A and B (MICA/B) 14. Furthermore, miR-302c-3p acts as a tumor suppressor in glioma via inhibiting proliferation and invasion of cancer cells 15. MiR-302c-3p/interleukin-8 (IL8) axis plays an essential role in receptor for activated C-kinase 1 (RACK1)-mediated metastasis of gastric cancer 16. Meanwhile, miR-302c-3p suppresses HCC growth via inhibiting endothelial cell-mediated angiogenesis 17. Recently, serum miR-302c-3p expression is prominently deregulated in hepatitis C virus (HCV)-related HCC 18. However, the expression of miR-302c-3p and its functional role in hepatitis B virus (HBV)-related HCC are rarely investigated.
In the current study, we were aimed to investigate the expression and clinical significance of miR-302c-3p in HCC, and disclosed the effects of miR-302c-3p on migration and invasion of HCC cells. Down-regulation of miR-302c-3p was observed in HCC tissues. Tumor necrosis factor receptor associated factor 4 (TRAF4) was recognized as a novel target of miR-302c-3p and it possibly mediated the tumor suppressive role of miR-302c-3p in HCC.
Materials and methods
Clinical samples
Eighty pairs of HCC and adjacent normal tissues were obtained from the Zhejiang Provincial People's Hospital. All samples were confirmed by experienced pathologists and were conserved in liquid nitrogen for further analysis. All patients did not receive therapies including radiotherapy, chemotherapy and radiofrequency ablation prior to surgery. The study was approved by Ethic Committee of the Zhejiang Provincial People's Hospital and written informed consent was signed by all patients. The clinicopathologic parameters of patients were shown in Table 1.
Table 1.
Clinicopathological correlation analysis of miR-302c-3p expression in hepatocellular carcinoma
| Clinicopathologic features | n | miR-302c-3p expression | P | ||
|---|---|---|---|---|---|
| Low level (n=40) | High level (n=40) | ||||
| Age (year) | ≤50 | 32 | 14 | 18 | 0.361 |
| >50 | 48 | 26 | 22 | ||
| Sex | Male | 64 | 30 | 34 | 0.264 |
| Female | 16 | 10 | 6 | ||
| HBV infection | No | 17 | 6 | 9 | 0.390 |
| Yes | 63 | 34 | 31 | ||
| Serum AFP level (ng/mL) | ≤20 | 22 | 8 | 14 | 0.133 |
| >20 | 58 | 32 | 26 | ||
| Tumor size (cm) | ≤5 | 20 | 8 | 12 | 0.302 |
| >5 | 60 | 32 | 28 | ||
| No. of tumor nodules | 1 | 68 | 30 | 38 | 0.012* |
| ≥2 | 12 | 10 | 2 | ||
| Cirrhosis | Absent | 28 | 16 | 12 | 0.348 |
| Present | 52 | 24 | 28 | ||
| Venous infiltration | Absent | 40 | 15 | 25 | 0.025* |
| Present | 40 | 25 | 15 | ||
| Edmondson-Steiner grading | I+II | 60 | 28 | 32 | 0.302 |
| III+IV | 20 | 12 | 8 | ||
| TNM tumor stage | I+II | 64 | 27 | 37 | 0.005* |
| III+IV | 16 | 13 | 3 | ||
HBV, hepatitis B virus; AFP, alpha-fetoprotein; TNM, tumor-node-metastasis. *Statistically significant.
Cell culture, transfection and reagent
The human HCC cell lines (HepG2, SMMC-7721, Huh7, Hep3B and MHCC97H) and normal hepatic cell line (LO2) were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA, USA) added with 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA) and 1% penicillin/streptomycin (HyClone, Logan, UT, USA). All cell lines were incubated in a humidified atmosphere with 5% CO2 at 37°C.
The miR-302c-3p mimics, miR-302c-3p inhibitors and their corresponding negative control were purchased from GeneCopoeia (Guangzhou, China). TRAF4 expression plasmid (pcDNA3.1-TRAF4) was constructed and purchased from Ribobio (Guangzhou, China). Cells transfection was performed by using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. An AKT inhibitor, MK-2206 (1μM, Selleck Chemicals, Houston, TX, USA) was used to treat HCC cells for 12h.
RNA extraction and quantitative real-time PCR (qRT-PCR)
The total RNA from HCC cells and tissues was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. cDNA was synthesized by TaqMan miRNA reverse transcription (Applied Biosystems, Foster City, CA, USA) and a PrimeScript Reverse Transcriptase kit (Takara, Dalian, China). The relative expression of miR-302c-3p and TRAF4 mRNA were quantified using miRNA-specific TaqMan miRNA Assay Kit (Applied Biosystems) and the SYBR Premix Ex Taq™ Kit (Takara, Shiga, Japan) in the Applied Biosystems 7500 Sequence Detection system. The relative expression of miR-302c-3p and TRAF4 mRNA were normalized by U6 small nuclear RNA and GAPDH, respectively. The primers of miR-302c-3p, U6, TRAF4 and GAPDH were designed and synthesized by Sangon Biotech (Shanghai, China).
Western blotting
Western blotting was performed according to the protocol described previously 19. The protein concentration was measured using spectrophotometer (BIO-RAD, Hercules, CA), then equal protein was separated by SDS-PAGE and transferred to PVDF membranes. Subsequently, the PVDF membranes were probed with antibody against TRAF4 (Abcam, Cambridge, MA, USA), p-AKT (Ser473, Cell Signaling Technology, Beverly, MA, USA), AKT (Cell Signaling Technology), E-cadherin (Cell Signaling Technology), Vimentin (Cell Signaling Technology), Slug (Cell Signaling Technology) and GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA), then probed with HRP-conjugated secondary antibodies (#7074 and #7076, Cell Signaling Technology). The western blot was detected with enhanced chemiluminescence regent (Thermo Scientific, Waltham, MA, USA).
Luciferase reporter assay
The sequence of TRAF4 3'-UTR containing the putative miR-302c-3p binding region was amplified from human genomic DNA. Then the sequence was cloned into pGL3 luciferase reporter vector (Promega, Madison, WI, USA). The potential miR-302c-3p binding sites were mutated by the Quick-change site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). The wild type (wt) TRAF4 3'-UTR vector or mutant (mt) TRAF4 3'-UTR vector and miR-302c-3p mimics or miR-302c-3p inhibitors were co-transfected into HepG2 cells by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) under luminometer (Berthold Detection System, Pforzheim, Germany), and luciferase activity was normalized to Renilla activity.
Cell migration and invasion assay
1 × 104 indicated HCC cells were seeded into the upper chamber coated with or without Matrigel (BD Bioscience, San Jose, CA, USA) and added DMEM without FBS. Then, the chamber was put into the cell culture plate containing DMEM supplemented with 10% FBS and incubated at 37°C for 24 hours. subsequently, the cells inside the upper chamber were carefully removed with cotton swabs. Migrated and invaded cells were fixed with 1% paraformaldehyde for 10 min and subsequently stained by hematoxylin for 5 min. The migratory and invasive cells were finally examined and counted under microscope.
Statistical analysis
All data were shown as mean ± standard deviation (SD) and analyzed by using GraphPad Prism software version 5.0 (San Diego, CA, USA). Statistical analysis was calculated by Chi-squared test, Student's t-test, ANOVA, Pearson correlation analysis, Kaplan-Meier method and Log-rank test. P-value < 0.05 was considered as statistical significance. Each experiment was repeated three times.
Results
The expression of miR-302c-3p in HCC specimens
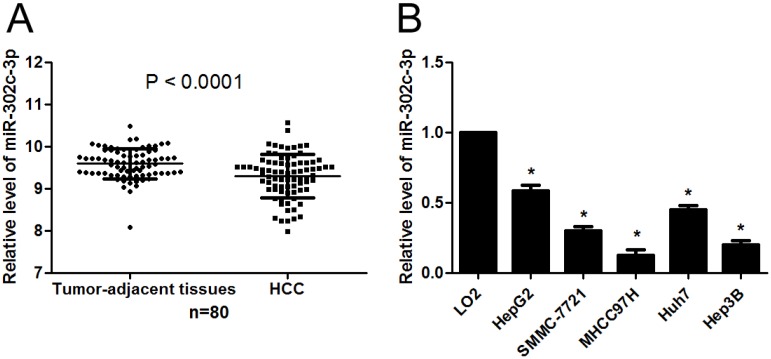
First, the expression of miR-302c-3p between HCC and tumor-adjacent tissues was determined by qRT-PCR. Our data showed that the expression of miR-302c-3p in HCC was obviously lower than that in tumor-adjacent tissues (P<0.0001, Figure 1A). Furthermore, the levels of miR-302c-3p in different HCC cell lines were detected. In accordance, low miR-302c-3p level was observed in all HCC cell lines compared to normal hepatic cell line LO2 (P<0.05, respectively, Figure 1B). These data reveal an under-expression of miR-302c in HCC.
Figure 1.
miR-302c-3p expression is down-regulated in HCC. (A) The expression of miR-302c-3p in HCC tissues was significantly lower than that in matched tumor-adjacent tissues. n=80, P<0.0001 by Student's t-test. (B) The expression differences of miR-302c-3p between HCC cell lines (HepG2, SMMC-7721, MHCC97H, Huh7 and Hep3B) and normal hepatic cell line (LO2). n= three repeats with similar results, *P<0.05 by one-way ANOVA.
The clinical significance of miR-302c-3p in HCC patients
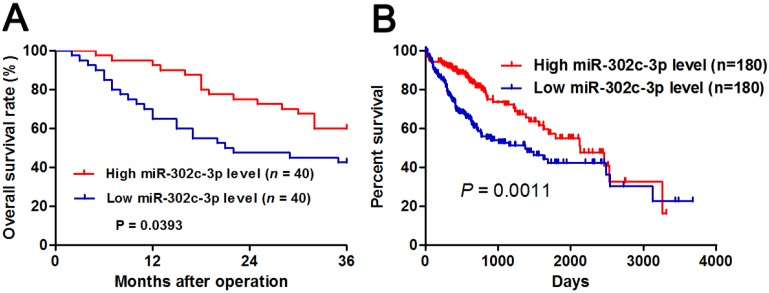
Different subgroups (low/high miR-302c-3p expression) were divided according to the median level of miR-302c-3p in the cohort. Clinicopathological correlation analysis revealed that low miR-302c-3p level was correlated with multiple tumor nodes, venous infiltration and advanced TNM tumor stage (P < 0.05, respectively, Table 1). Notably, Kaplan-Meier plots indicated that HCC patients with low miR-302c-3p level had a significant shorter 3-year overall survival compared to cases with high miR-302c-3p expression (P=0.0393, Figure 2A). Meanwhile, The Cancer Genome Atlas (TCGA) data also showed that low miR-302c-3p level indicated a poor prognosis of HCC patients (P=0.0011, Figure 2B). These data suggest that miR-302c-3p is a potential prognostic biomarker for HCC patients.
Figure 2.
The prognostic significance of miR-302c-3p in HCC patients. (A) Different subgroups (low/high miR-302c-3p expression) were divided according to the median of miR-302c-3p in our cohort. HCC patients with low miR-302c-3p level had a significant shorter overall survival compared to those with high miR-302c-3p level. P<0.05 by Log-rank test. (B) TCGA data showed that low miR-302c-3p level predicted an obvious poor prognosis of HCC patients. P<0.05 by Log-rank test.
MiR-302c-3p inhibits migration and invasion of HCC cells
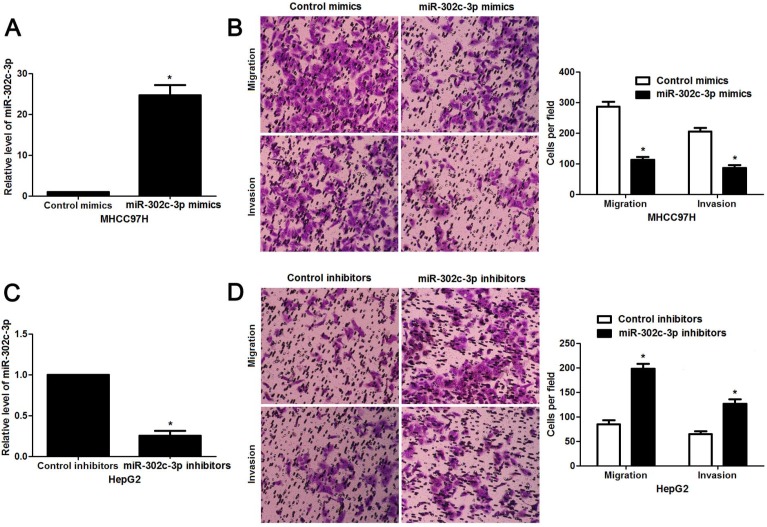
Since our clinical data revealed the correlation between miR-302c-3p expression and metastatic features of HCC, the regulatory effect of miR-302c-3p on migration and invasion of HCC cells was determined. MHCC97H cells with low miR-302c-3p level and HepG2 cells with high miR-302c-3p level were used for gain- and loss-of-function experiments, respectively. Further experiments were performed in MHCC97H cells after miR-302c-3p overexpression (P < 0.05, Figure 3A). Transwell assays showed that miR-302c-3p overexpression remarkably decreased the migration and invasion capacities of MHCC97H cells (P < 0.05, respectively, Figure 3B). Furthermore, miR-302c-3p knockdown prominently facilitated migration and invasion of HepG2 cell (P < 0.05, respectively, Figure 3C and 3D). Thus, miR-302c-3p functions as a tumor suppressor by suppressing migration and invasion of HCC cells.
Figure 3.
miR-302c-3p inhibits migration and invasion of HCC cells. (A) MHCC97H cells that were transfected with miR-302c-3p mimics and control mimics, respectively, were subjected to qRT-PCR for miR-302c-3p expression. n= three repeats with similar results, *P<0.05 by Student's t-test. (B) Transwell assays indicated that miR-302c-3p overexpression suppressed the migration and invasion of MHCC97H cells. n= three repeats with similar results, *P<0.05 by Student's t-test. (C) HepG2 cells that were transfected with miR-302c-3p inhibitors and control inhibitors, respectively, were detected by qRT-PCR for miR-302c-3p expression. n= three repeats with similar results, *P<0.05 by Student's t-test. (D) The migration and invasion capacities of HepG2 cells was enhanced by miR-302c-3p knockdown. n= three repeats with similar results, *P<0.05 by Student's t-test.
miR-302c-3p regulates TRAF4 abundance by directly targeting its 3'UTR in HCC cells
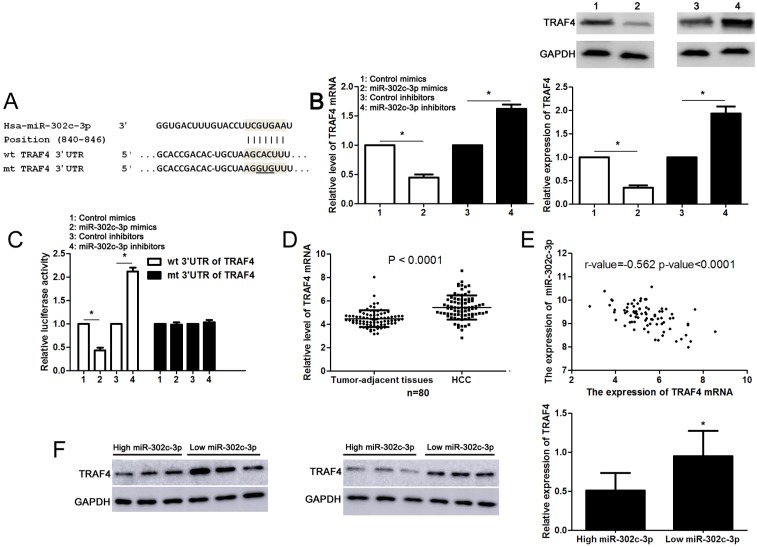
Next, the candidate target genes of miR-302c-3p were predicted using TargetScan (http://www.targetscan.org) and miRanda (microRNA.org). We found that the 3'-UTR of TRAF4 mRNA contained the complementary sequence of miR-302c-3p according to public available database analysis (Figure 4A). Thus, TRAF4 was selected for further experimental validation. Subsequently, we found that miR-302c-3p restoration significantly reduced while miR-302c-3p silencing increased the levels of TRAF4 mRNA and protein in HepG2 cells (P<0.05, respectively, Figure 4B). Then, plasmids with wt 3'-UTR of TRAF4 or mt 3'-UTR of TRAF4 were transfected into HepG2 cells. Notably, miR-302c-3p restoration prominently reduced while miR-302c-3p silencing increased the fluorescence intensity of cells transfected with wt 3'-UTR of TRAF4 (P<0.05, respectively, Figure 4C). While, the fluorescence intensity had no significant variation after being transfected with miR-302c-3p mimics or inhibitors in the mutant group (Figure 4C). Our data further confirmed the overexpression of TRAF4 mRNA in HCC tissues compared to tumor-adjacent tissues (P<0.0001, Figure 4D). Notably, an inverse correlation between TRAF4 mRNA and miR-302c-3p expression was detected in HCC tissues (r=-0.562, P<0.0001, Figure 4E). Furthermore, six HCC tissues with different expression levels of miR-302c-3p were subjected to immunoblotting for TRAF4 expression. Quantitative data revealed that TRAF4 protein expression in HCC tissues with low miR-302c-3p level was prominently higher than that in HCC tissues with high miR-302c-3p level (P<0.05, Figure 4F). Taken together, we firstly disclose miR-302c-3p regulation of TRAF4 in HCC tissues.
Figure 4.
miR-302c-3p regulates TRAF4 expression by directly binding to its 3'-UTR in HCC cells. (A) The complementary sequences of miR-302c-3p were discovered in 3'-UTR of TRAF4 mRNA using TargetScan (http://www.targetscan.org) and miRanda (microRNA.org). The mutagenesis was performed in the complementary sites for the seed region of miR-302c-3p. (B) miR-302c-3p overexpression reduced while miR-302c-3p knockdown increased the expression of TRAF4 in HepG2 cells. n= three repeats with similar results, *P<0.05 by Student's t-test. (C) miR-302c-3p inversely modulated the luciferase activity of plasmids that carried wt rather than mt 3'-UTR of TRAF4 (wt, wild type; mt, mutant type). n= three repeats with similar results, *P<0.05 by Student's t-test. (D) The expression of TRAF4 mRNA in HCC tissues was significantly higher than that in matched tumor-adjacent tissues. n= 80, *P<0.05 by Student's t-test. (E) A negative correlation between TRAF4 mRNA expression and miR-302c-3p level was found in HCC tissues. n= 80, *P<0.05 by Pearson correlation test. (F) Western blotting analysis indicated that the expression of TRAF protein in HCC tissues with high miR-302c level was remarkably lower than that in cases with low miR-302c level. n= 6, *P<0.05 by Student's t-test.
miR-302c-3p inhibits AKT-mediated EMT in HCC cells
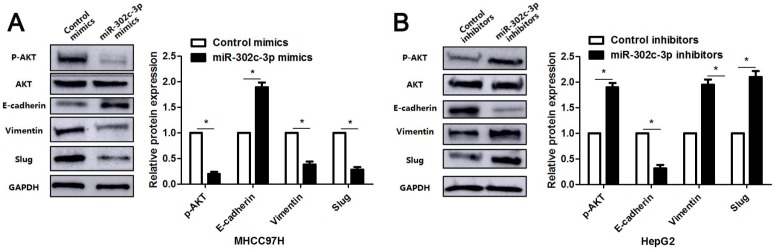
Since previous study reports that TRAF4 contributes to HCC cell metastasis via PI3K/AKT-mediated epithelial-mesenchymal transition (EMT) 20. As an upstream regulator of TRAF4, the regulatory effect of miR-302c-3p on AKT-mediated EMT in HCC cells was subsequently determined. As expected, miR-302c-3p overexpression reduced the expression of p-AKT and mesenchymal markers (Vimentin and Slug), and increased the level of epithelial marker (E-cadherin) in MHCC97H cells (P<0.05, respectively, Figure 5). Furthermore, miR-302c-3p knockdown promoted AKT signaling activation and EMT progression in HepG2 cells (P<0.05, respectively, Figure 5). Thus, miR-302c-3p is a regulator of EMT process in HCC cells.
Figure 5.
miR-302c-3p suppresses AKT-mediated EMT in HCC cells. MHCC97H cells were transfected with miR-302c-3p mimics and control mimics, respectively. miR-302c-3p overexpression reduced the levels of p-AKT, Vimentin and Slug, and increased E-cadherin expression. HepG2 cells were transfected with miR-302c-3p inhibitors and control inhibitors, respectively. miR-302c-3p knockdown promoted the activation of AKT signaling pathway and EMT progression. n= three repeats with similar results, *P<0.05 by Student's t-test.
TRAF4 mediates the role of miR-302c-3p in HCC cells
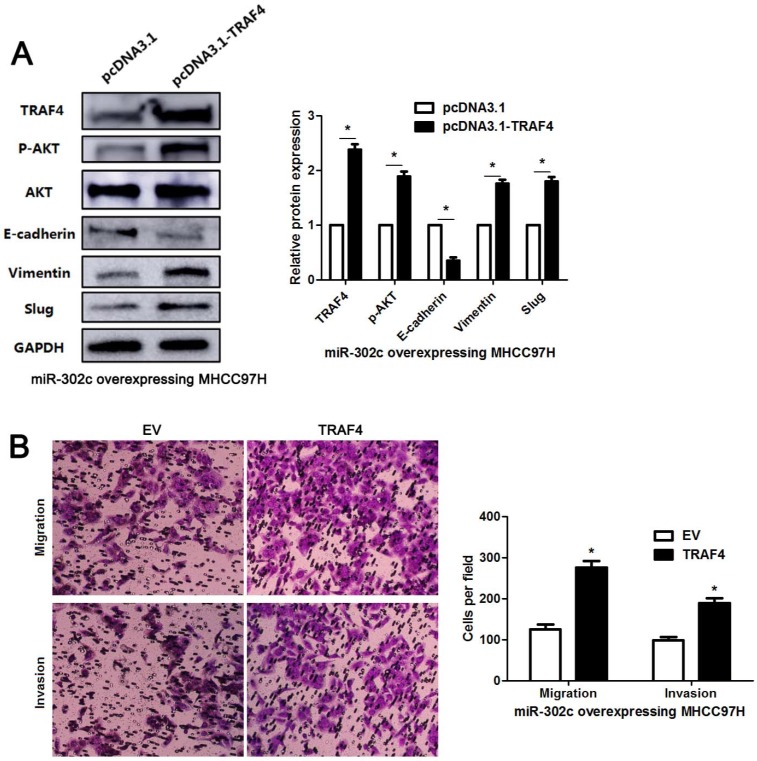
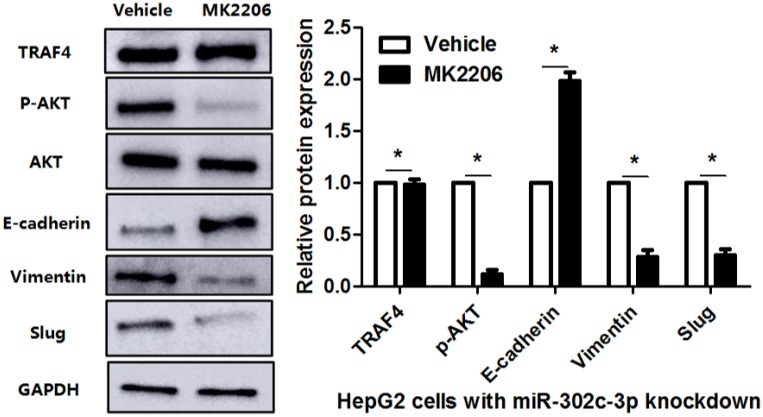
To clarify whether TRAF4 was a downstream effector of miR-302c-3p, the rescue experiments were performed in HCC cells. TRAF4 expression was restored in miR-302c-3p overexressing MHCC97H cells (P<0.05, Figure 6A). TRAF4 restoration increased the levels of p-AKT, Vimentin and Slug, and reduced E-cadherin expression in MHCC97H cells with miR-302c-3p overexpression (P<0.05. respectively, Figure 6A). Functional experiments revealed that TRAF4 restoration reversed the inhibitory effect of miR-302c-3p on migration and invasion of MHCC97H cells (P<0.05, respectively, Figure 6B). An AKT inhibitor, MK2206, was used to block AKT activation in HepG2 cells with miR-302c-3p knockdown (P<0.05, Figure 7). Furthermore, inactivation of AKT increased E-cadherin expression and reduced the levels of Vimentin and Slug in miR-302c-3p silencing HepG2 cells (P<0.05, respectively, Figure 7). These results suggest that TRAF4 is a downstream functional effector of miR-302c-3p in HCC.
Figure 6.
TRAF4 restoration reverses the tumor suppressive role of miR-302c-3p in MHCC97H cells. (A) miR-302c-3p overexpressing MHCC97H cells were transfected with TRAF4 expression plasmid and control plasmid, respectively. TRAF4 restoration promoted the activation of AKT signaling pathway and EMT progression. n= three repeats with similar results, *P<0.05 by Student's t-test. (B) The number of migrated and invaded MHCC97H cells was reduced by miR-302c-3p overexpression and subsequently reversed by TRAF4 restoration. n= three repeats with similar results, *P<0.05 by Student's t-test.
Figure 7.
AKT inhibition suppresses miR-302c-3p silencing-induced EMT of HepG2 cells. HepG2 cells with miR-302c-3p knockdown were treated with MK2206, an AKT inhibitor, and DMSO, respectively. Western blot analysis indicated that MK2206 treatment significantly reduced the levels of p-AKT, Vimenting and Slug, and increased E-cadherin expression. n= three repeats with similar results, *P<0.05 by Student's t-test.
Discussion
Deregulated expression of miR-302c-3p has been found in various human cancers including breast cancer 13, glioma 15, 21 and gastric cancer 22. Meanwhile, aberrant expression of miR-302c-3p is associated with prognostic features of human cancer. For instance, low miR-302c-3p expression is positively correlated with more invasive depth, advanced TNM tumor stage and absent of lymph node metastasis in gastric cancer 22. Glioma patients with decreased miR-302c-3p expression had high Karnofsky performance status (KPS) score and WHO grade 21. In the present study, under-expression of miR-302c-3p was confirmed by qRT-PCR in our cohort of HCC cases. Low miR-302c-3p expression was frequently associated with multiple tumor nodes, venous infiltration and advanced TNM tumor stage. Notably, decreased expression of miR-302c-3p in HCC tissues indicated a remarkable reduced overall survival of patients based on our follow up data and TCGA data. The prognostic significance of miR-302c-3p is also reported in glioma, gastric cancer and adenocarcinomas of the gastroesophageal junction 21-23. Thus, these data suggest miR-302c-3p as a promising prognostic biomarker in HCC.
miRNAs are implicated in a variety of cellular processes, including cell cycle progression, proliferation, apoptosis, migration and invasion 24. Increasing studies recognize miR-302c-3p acts as a tumor suppressor in breast cancer, glioma and gastric cancer 15, 16, 25, and inhibits HCC growth by suppressing endothelial cell-mediated angiogenesis 17. Here, we demonstrated that miR-302c-3p played a tumor suppressive role via inhibiting migration and invasion of HCC cells. Interestingly, we found that miR-302c-3p also suppressed proliferation of HCC cells (Supplementary Figure 1). Thus, it is worth to disclose the mechanism underlying the regulatory effect of miR-302c-3p in HCC growth. miRNAs exhibit its functional role by inhibiting various downstream target genes in human cancer. Subsequently, TRAF4 was identified as a candidate target gene of miR-302c-3p. Public available database and luciferase reporter assay further confirmed that TRAF4 was a direct target of miR-302c-3p in HCC cells. TRAF4, a member of TRAFs family, belongs to signal adaptor proteins in the cytoplasm 26. Some studies reveal the vital role of TRAF4 in embryogenesis and central nervous system myelin homeostasis 27, 28. TRAF4 is firstly identified in breast cancer and its overexpression has been verified in numerous cancers including breast cancer, lung cancer, colon cancer and HCC 20, 29-31. Recently, several studies have reported that TRAT4 plays a crucial role in tumorigenesis and progression via regulating multiple signaling pathways, such as PI3K/AKT pathway, transforming growth factor-β (TGF-β) pathway and Wnt/β-catenin20, 32-34. As a direct negative regulator of TRAF4, miR-302c-3p showed an opposite role in regulating AKT signaling pathway and EMT progression in HCC cells. miR-302c-3p reduced the levels of p-AKT, Vimentin and Slug, and increased E-cadherin expression. Notably, TRAF4 restoration reversed the tumor suppressive role of miR-302c-3p in HCC cells. Inactivation of AKT by MK2206 repressed miR-302c-3p knockdown-induced EMT of HCC cells. These data indicate that miR-302c-3p inhibits the metastasis of HCC cells by suppressing TRAF4.
In conclusion, we demonstrate that miR-302c-3p acts as a potential prognostic indicator and plays a tumor suppressive role in HCC. MiR-302c-3p inhibits the phosphorylation of AKT, EMT progression, migration and invasion in HCC cells (Supplementary Figure 2). TRAF4 restoration abrogates the tumor suppressive effect of miR-302c-3p in HCC cells. Thus, these findings suggest a potential therapeutic value of miR-302c-3p/TRAF4 axis for HCC.
Supplementary Material
Supplementary figures.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81772575, 81602179, 81502092); the Zhejiang Provincial Natural Science Foundation of China (LY16H160043, LY13H160039); the Key Program Funds from the Health Department of Zhejiang Province(2015C03030); the General Project Funds from the Health Department of Zhejiang Province (2016KYA022, 2015KYB033).
Abbreviations
- HCC
hepatocellular carcinoma
- miRNAs
microRNAs
- ncRNAs
non-coding RNAs
- 3'-UTRs
3'-untranslated regions
- ERα
estrogen receptor-alpha
- NK
natural killer
- ULBP2
UL-16 binding protein 2
- MICA/B
MHC class I chain-related A and B
- IL8
interleukin-8
- RACK1
receptor for activated C-kinase 1
- HCV
hepatitis C virus
- HBV
hepatitis B virus
- TRAF4
tumor necrosis factor receptor associated factor 4
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- TCGA
The Cancer Genome Atlas
- EMT
epithelial-mesenchymal transition
- KPS
Karnofsky performance status
- TGF-β
transforming growth factor-β
- AFP
alpha-fetoprotein
- TNM
tumor-node-metastasis.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Diaz-Gonzalez A, Reig M, Bruix J. Treatment of Hepatocellular Carcinoma. Dig Dis. 2016;34:597–602. doi: 10.1159/000445275. [DOI] [PubMed] [Google Scholar]
- 4.Drusco A, Croce CM. MicroRNAs and Cancer: A Long Story for Short RNAs. Advances in cancer research. 2017;135:1–24. doi: 10.1016/bs.acr.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Dou C, Liu Z, Xu M, Jia Y, Wang Y, Li Q. et al. miR-187-3p inhibits the metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting S100A4. Cancer Lett. 2016;381:380–90. doi: 10.1016/j.canlet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Tao J, Liu Z, Wang Y, Wang L, Yao B, Li Q. et al. MiR-542-3p inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting UBE3C. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2017;93:420–8. doi: 10.1016/j.biopha.2017.06.070. [DOI] [PubMed] [Google Scholar]
- 7.Xu Q, Liu X, Liu Z, Zhou Z, Wang Y, Tu J. et al. MicroRNA-1296 inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting SRPK1-mediated PI3K/AKT pathway. Molecular cancer. 2017;16:103. doi: 10.1186/s12943-017-0675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuo H, Wang Y, Wang L, Yao B, Li Q, Wang C. et al. MiR-324-3p promotes tumor growth through targeting DACT1 and activation of Wnt/beta-catenin pathway in hepatocellular carcinoma. Oncotarget. 2017;8:65687–98. doi: 10.18632/oncotarget.20058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu K, Liu Z, Yao B, Han S, Yang W. MicroRNA-519a promotes tumor growth by targeting PTEN/PI3K/AKT signaling in hepatocellular carcinoma. Int J Oncol. 2016;48:965–74. doi: 10.3892/ijo.2015.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Wang Y, Dou C, Sun L, Li Q, Wang L. et al. MicroRNA-1468 promotes tumor progression by activating PPAR-gamma-mediated AKT signaling in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:49. doi: 10.1186/s13046-018-0717-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang C. et al. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Molecular cancer. 2017;16:123. doi: 10.1186/s12943-017-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leivonen SK, Makela R, Ostling P, Kohonen P, Haapa-Paananen S, Kleivi K. et al. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene. 2009;28:3926–36. doi: 10.1038/onc.2009.241. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimoto N, Toyama T, Takahashi S, Sugiura H, Endo Y, Iwasa M. et al. Distinct expressions of microRNAs that directly target estrogen receptor alpha in human breast cancer. Breast Cancer Res Treat. 2011;130:331–9. doi: 10.1007/s10549-011-1672-2. [DOI] [PubMed] [Google Scholar]
- 14.Min D, Lv XB, Wang X, Zhang B, Meng W, Yu F. et al. Downregulation of miR-302c and miR-520c by 1,25(OH)2D3 treatment enhances the susceptibility of tumour cells to natural killer cell-mediated cytotoxicity. Br J Cancer. 2013;109:723–30. doi: 10.1038/bjc.2013.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Wei Y, Tong H, Chen L, Fan Y, Ji Y. et al. MiR-302c-3p suppresses invasion and proliferation of glioma cells via down-regulating metadherin (MTDH) expression. Cancer Biol Ther. 2015;16:1308–15. doi: 10.1080/15384047.2015.1056413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Min L, Wang X, Zhao J, Chen H, Qin J. et al. Loss of RACK1 Promotes Metastasis of Gastric Cancer by Inducing a miR-302c/IL8 Signaling Loop. Cancer Res. 2015;75:3832–41. doi: 10.1158/0008-5472.CAN-14-3690. [DOI] [PubMed] [Google Scholar]
- 17.Zhu K, Pan Q, Jia LQ, Dai Z, Ke AW, Zeng HY. et al. MiR-302c inhibits tumor growth of hepatocellular carcinoma by suppressing the endothelial-mesenchymal transition of endothelial cells. Sci Rep. 2014;4:5524. doi: 10.1038/srep05524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oksuz Z, Serin MS, Kaplan E, Dogen A, Tezcan S, Aslan G. et al. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Mol Biol Rep. 2015;42:713–20. doi: 10.1007/s11033-014-3819-9. [DOI] [PubMed] [Google Scholar]
- 19.Xu Q, Tu J, Dou C, Zhang J, Yang L, Liu X. et al. HSP90 promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance via Thr-328 phosphorylation in hepatocellular carcinoma. Mol Cancer. 2017;16:178. doi: 10.1186/s12943-017-0748-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu K, Wu X, Zang X, Huang Z, Lin Z, Tan W. et al. TRAF4 Regulates Migration, Invasion, and Epithelial-Mesenchymal Transition via PI3K/AKT Signaling in Hepatocellular Carcinoma. Oncol Res. 2017;25:1329–40. doi: 10.3727/096504017X14876227286564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng GD, Wang YU, Zhu XD. Evaluation of miR-302c-3p as prognostic biomarkers in glioma patients. Eur Rev Med Pharmacol Sci. 2016;20:1521–5. [PubMed] [Google Scholar]
- 22.Ma G, Li Q, Dai W, Yang X, Sang A. Prognostic Implications of miR-302a/b/c/d in Human Gastric Cancer. Pathol Oncol Res. 2017;23:899–905. doi: 10.1007/s12253-017-0282-7. [DOI] [PubMed] [Google Scholar]
- 23.Odenthal M, Hee J, Gockel I, Sisic L, Schmitz J, Stoecklein NH. et al. Serum microRNA profiles as prognostic/predictive markers in the multimodality therapy of locally advanced adenocarcinomas of the gastroesophageal junction. Int J Cancer. 2015;137:230–7. doi: 10.1002/ijc.29363. [DOI] [PubMed] [Google Scholar]
- 24.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–33. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C. et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11:R27. doi: 10.1186/bcr2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung JY, Park YC, Ye H, Wu H. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J Cell Sci. 2002;115:679–88. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- 27.Blaise S, Kneib M, Rousseau A, Gambino F, Chenard MP, Messadeq N. et al. In vivo evidence that TRAF4 is required for central nervous system myelin homeostasis. PLoS One. 2012;7:e30917. doi: 10.1371/journal.pone.0030917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masson R, Regnier CH, Chenard MP, Wendling C, Mattei MG, Tomasetto C. et al. Tumor necrosis factor receptor associated factor 4 (TRAF4) expression pattern during mouse development. Mech Dev. 1998;71:187–91. doi: 10.1016/s0925-4773(97)00192-5. [DOI] [PubMed] [Google Scholar]
- 29.Tomasetto C, Regnier C, Moog-Lutz C, Mattei MG, Chenard MP, Lidereau R. et al. Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11-q21.3 region of chromosome 17. Genomics. 1995;28:367–76. doi: 10.1006/geno.1995.1163. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Peng C, Lee MH, Lim D, Zhu F, Fu Y. et al. TRAF4 is a critical molecule for Akt activation in lung cancer. Cancer Res. 2013;73:6938–50. doi: 10.1158/0008-5472.CAN-13-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang K, Wang F, Han JJ. TRAF4 promotes the growth and invasion of colon cancer through the Wnt/beta-catenin pathway. Int J Clin Exp Pathol. 2015;8:1419–26. [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Zhou F, Garcia de Vinuesa A, de Kruijf EM, Mesker WE, Hui L. et al. TRAF4 promotes TGF-beta receptor signaling and drives breast cancer metastasis. Mol Cell. 2013;51:559–72. doi: 10.1016/j.molcel.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Wei D, Wang W, Shen B, Xu S, Cao Y. TRAF4 enhances oral squamous cell carcinoma cell growth, invasion and migration by Wnt-beta-catenin signaling pathway. Int J Clin Exp Pathol. 2015;8:11837–46. [PMC free article] [PubMed] [Google Scholar]
- 34.Yao W, Wang X, Cai Q, Gao S, Wang J, Zhang P. TRAF4 enhances osteosarcoma cell proliferation and invasion by Akt signaling pathway. Oncol Res. 2014;22:21–8. doi: 10.3727/096504014X14077751730351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.