Abstract
Background: Hepatocellular carcinoma (HCC) is one of the most aggressive cancers, with limited new diagnostic and therapeutic measures. This study aimed to investigate the utility of specific serum and exosome lncRNAs as biomarkers for early diagnosis of HCC.
Methods: The relative expression levels of eight selected lncRNAs in serum were evaluated by quantitative reverse transcription-PCR (qRT-PCR) in the training and validation sets of HCC patients and matched healthy controls. Additionally, the stability, specificity and diagnostic efficiency of these lncRNAs were evaluated to determine their potential as biomarkers. The levels of the final validated lncRNAs in exosome and urine samples of 15 HCC patients and 15 healthy controls were examined for source and path analysis.
Results: LINC00161 was significantly upregulated in serum samples of HCC patients and showed excellent stability and specificity (P< 0.001, fold change=2.85). The area under the receiver operating characteristic (ROC) curve of the validated lncRNA signature was 0.794 (95% CI, 0.712-0.877). LINC00161 expression was detected in serum exosome, exosome-free, and urine samples, and its levels in serum exosome were upregulated in patients with HCC as compared to controls (P= 0.011, fold change=4.27).
Conclusions: Our results indicated that circulating exosomal LINC00161 in serum may be a novel biomarker for HCC. LINC00161 is derived from exosomes into serum and may at least be partly metabolized through urine.
Keywords: serum, exosome, long non coding RNAs, biomarker, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) ranks as the fifth most common cancer globally. HCC mortality rates rose by almost 3% every year between 2010 and 2014, in contrast to declining trends for the four major cancers1, 2. Though surgical treatment for HCC is available in the initial stage of the disease, most patients present with intrahepatic and extrahepatic metastasis upon diagnosis, at which point the efficacy of radiotherapy and chemotherapy is limited3. The most widely used screening tools for HCC are imaging techniques or serum tumor biomarkers, such as analysis of alpha-fetoprotein (AFP). However, imaging techniques are less efficient for detecting early HCC, and AFP is limited by its low diagnostic value in the clinical practice4. Hence, there is urgency in identifying novel diagnostic and prognostic molecular markers and potential new targets for therapies to improve patient survival.
Long noncoding RNAs (lncRNAs) are non-protein coding transcripts with more than 200 nucleotides in length. There is growing evidence that lncRNAs can participate in various biological processes involved in tumorigenesis, proliferation and metastasis 5-8. Previous studies have suggested that lncRNAs can act as diagnostic biomarkers for multiple cancers, such as cancers of the colorectum, gastric, prostate, bladder, and clear cell renal cell 9-12. Recent studies showed that lncRNAs including LINC00462, CCAT1, CCAT2, HOTAIR, MALAT1, LINC00161, and SPRY4-IT1 were significantly upregulated in HCC tumor tissues compared with non-tumor tissues13-19. Our previous study showed that UCA1 was significantly upregulated in HCC tumor tissues compared with non-tumor tissues20. However, the aberrant expression of these lncRNAs in serum of HCC patients is not clear. Furthermore, the origin of these lncRNAs has not been clarified yet.
Circulating exosomes are small membrane vesicles (30-150 nm) that are released into the extracellular environment upon fusion of multivesicular bodies with the serum or plasma membrane 21, 22. These vesicles, loaded with proteins and unique RNA, have a wide range of biological functions, including cell-to-cell communication and signaling. Although various proteins and miRNA have been isolated from exosomes as potential diagnostic, prognostic, or therapeutic biomarkers 23-25, little is known about serum exosomal lncRNAs in HCC and many lncRNAs in exosomes have not been adequately investigated.
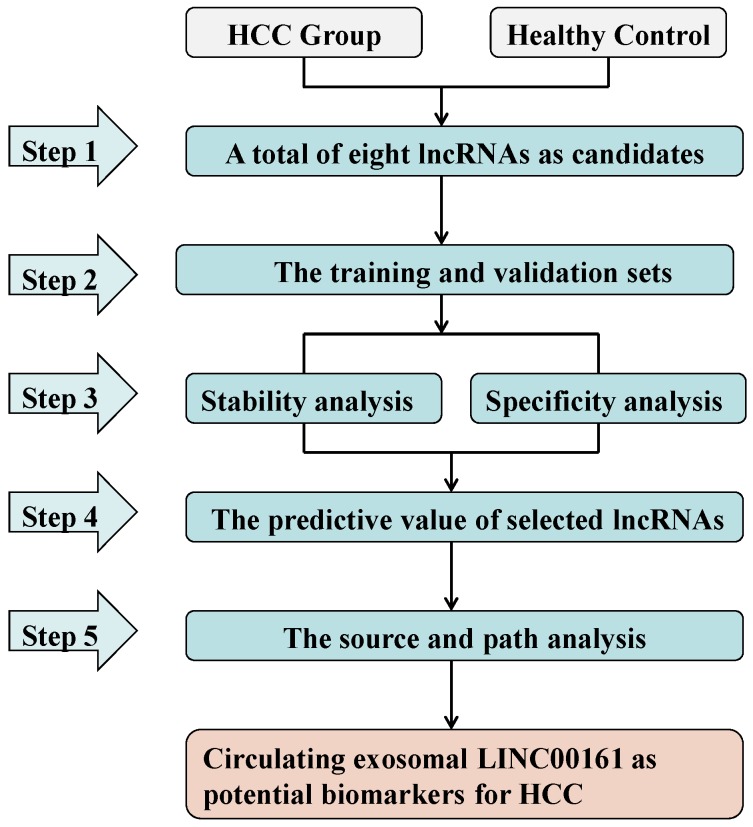
In the present study, we adopted a five-stage strategy for identification of serum and exosome biomarkers corresponding to HCC-related lncRNAs (Fig. 1). First, we selected eight lncRNAs (LINC00462, CCAT1, CCAT2, HOTAIR, LINC00161, SPRY4-IT1, MALAT1 and UCA1) as candidates on the basis of available literature. Subsequently the relative expression levels of these candidates in serum samples were tested in the training and validation sets of HCC patients and matched healthy controls. Consequently, the analysis of stability and specificity were investigated in these aberrantly expressed lncRNAs. Additionally, we investigated the selected lncRNAs for their diagnostic value and their association with clinical, histological, and pathological features in HCC. Finally, we analyzed the source and path of the selected lncRNAs by examining their expression in serum exosomes and urine in a set of 15 HCC and healthy controls.
Figure 1.
Study design of the workflow.
Materials and Methods
The literature analysis to retrieve lncRNAs involved in HCC
We searched the literature for the purpose of identifying serum lncRNAs as potential biomarkers. Some previous studies showed that LINC00462, CCAT1, CCAT2, HOTAIR, LINC00161, and SPRY4-IT1 were upregulated significantly in HCC tumor tissues compared with non-tumor tissues. To the best of our knowledge, the regulatory functions of these six lncRNAs in HCC serum samples have not been clarified yet. Similarly, the regulatory capacity of MALAT1 and UCA1 in HCC serum remain unknown even though their expression is upregulated in HCC tissue 17, 26. However, another report indicated that MALAT1 and UCA1 exhibited similar expression between tumor tissues and non-tumor tissues in HCC patients27. Thus, we selected eight lncRNAs (LINC00462, CCAT1, CCAT2, HOTAIR, LINC00161, SPRY4-IT1, MALAT1, and UCA1) as candidates for this research.
Study subjects and samples preparation
Peripheral blood samples from total 40 participants were recruited for the training set in our study including 20 HCC preoperative and 20 age- and sex-matched healthy controls. Then, differential expression analysis of the selected lncRNAs was tested in 56 HCC preoperative and 56 age- and sex-matched healthy controls. Blood samples were collected in serum separator tubes, followed by centrifugation at 4,000 rpm for 10 min to collect serum. The serum samples were then divided into multiple aliquots of 500 µL each and stored at -80°C until further analysis. Samples were obtained from patients with HCC, who were subjected to clinical therapy at Nanjing First Hospital and the Second Affiliated Hospital of Nanjing Medical University. Controls were from healthy volunteers without any health problems during their routine health check-ups. All subjects included in this study provided written informed consent, and the project protocols were performed in accordance with the guidelines of the Ethics Committee of Nanjing First Hospital and the Second Affiliated Hospital of Nanjing Medical University.
Stability and specificity analysis
The serum samples of 5 healthy controls were divided into 3 aliquots of 500 µL, which were stored at -80°C, 4°C, and room temperature for 24 hours respectively. Afterwards the levels of these samples were investigated to analyze their stability.
To analyze the specificity of screened lncRNAs, another 20 participants with chronic hepatitis were enrolled. The levels of these lncRNAs were tested in healthy controls, patients with chronic hepatitis, and HCC patients.
Exosome isolation
Exosomes were isolated using the Total Exosome Isolation Kit (Invitrogen, USA). The serum samples were thawed in a 25°C water bath and centrifuged at 2000 × g for 30 minutes to remove cells and debris. Next, 100 μL Total Exosome Isolation reagent was added to 500 μL serum, and the mixture was vortexed until it was homogenous. After incubation at 4°C for 30 minutes, the sample was centrifuged at 10000 × g for 10 minutes at room temperature. The exosome pellet was dissolved in 100 µL PBS.
Exosome identification
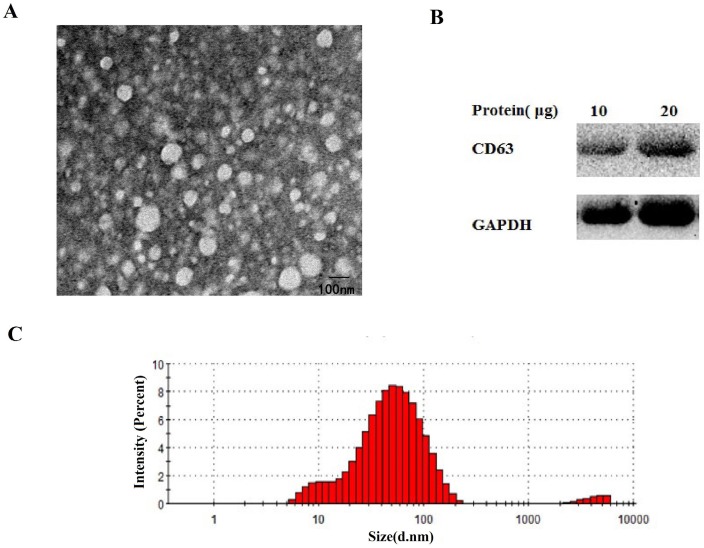
To identify the isolation of the exosomes from the serum, transmission electron microscopy (TEM), nanosize, and Western blotting analyses were performed. Exosomes were fixed in 2.5% glutaraldehyde solution for at least 2 hours. Next, 10 μL of the diluted mixtures were transferred to a cleaned copper net. Images were obtained by TEM (JEM-1010, Jeol, Japan) after dyeing with 2% phosphotungstic acid solution. For nanosize analysis, isolated exosome samples were diluted 2000-fold and resuspended in PBS for size distribution analysis using a ZETASIZER Nano series-Nano-ZS (Malvern Instruments Ltd, UK) according to the manufacturer's instructions.
For Western blot analysis, exosomal pellets from the serum were treated with RIPA lysis buffer (KeyGen Biotech, Nanjing, China). Different amounts of proteins were loaded and resolved using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Antibodies for CD63, GAPDH were obtained respectively from abcam (Cambridge, England) and Bioss (Beijing, China). After incubation with horseradish peroxidase-conjugated secondary antibodies (Biosharp, China), protein bands were visualized using enhanced chemiluminescence (Vazyme Biotech Co, China).
RNA isolation from serum, urine and exosome
Serum, urine and exosome RNA were respectively extracted using TRIzol reagent (Invitrogen, USA) according to the manufacture's instruction. The extracted RNA was eluted with 14 µL RNase-free water. RNA quality and quantity were determined by the spectrophotometer OD-1000+ (OneDrop, USA).
Quantitative Real-time RT-PCR Assay
To confirm the expression of the eight lncRNAs and internal control, reverse transcription of total RNAs were carried out using the reverse transcription kit (Vazyme Biotech Co, China). Samples were measured using the HiScript® II Q RT SuperMix for qPCR (Vazyme Biotech Co, China) on the StepOnePlus™ Real-Time System (Thermo Fisher Scientific, USA). The reactions were initiated with denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. The specificity of the PCR amplification was validated by the presence of a single peak in the melt-curve analysis. β-actin was used as an internal control while the cel-miR-39 was conducted as the external control. The relative expression levels of lncRNAs were analyzed by the 2-ΔΔCT method.
Source and path analysis
A total of 30 participants were enrolled from whom total 90 samples were collected for the lncRNA source and path analysis. Three samples were collected from each participant, including 15 serum, 15 serum exosomes, and 15 urine from the 15 HCC patients. Age- and sex- matched healthy controls were included (3 samples each from 15 participants).
Statistical analysis
SPSS Statistics software 22.0 (IBM, Chicago, USA) was used to perform statistical analyses. All data are presented as mean ± SD. The difference of lncRNA levels were determined by the Mann-Whitney U test. Associations of serum lncRNAs with clinicopathologic parameters were examined by the χ2 test. To evaluate the predictive value of selected lncRNAs for HCC, we applied the ROC curve. A p value < 0.05 was considered statistically significant.
Results
The expression of eight lncRNAs in HCC serum
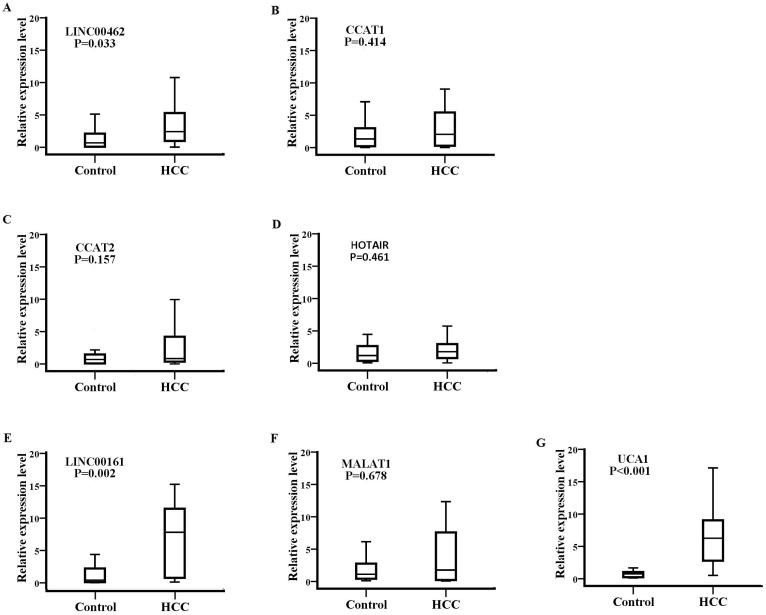
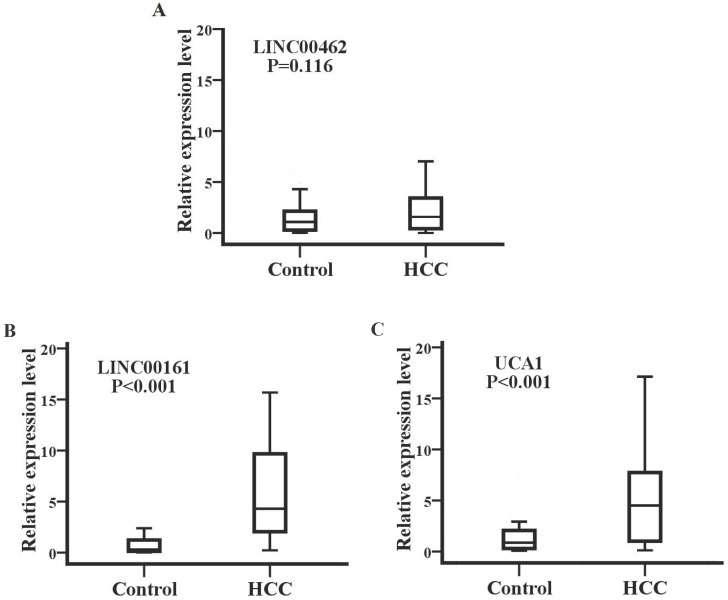
Detection of SPRY4-IT1 significantly varied across technical triplicates possibly due to its high Ct value beyond 36. The other seven lncRNAs (LINC00462, CCAT1, CCAT2, HOTAIR, LINC00161, MALAT1, and UCA1) could be stably amplified. As presented in the training set (Fig. 2), the relative expression levels of LINC00462, LINC00161, and UCA1 were markedly increased in patients with HCC in contrast to controls (all P< 0.05). No statistical significance was demonstrated for the expression of CCAT1, CCAT2, HOTAIR, and MALAT1 between the two groups (P=0.414, 0.157, 0.461 and 0.678 respectively). We further investigated the expression levels of the four significantly upregulated lncRNAs in 56 HCC preoperative and 56 age- and sex-matched healthy controls. As shown in Fig. 3, LINC00161 and UCA1 were significantly upregulated in HCC samples (all P< 0.001).
Figure 2.
The expression of eight lncRNAs between patients with HCC and healthy controls. The relative expression levels of LINC00462(A), CCAT1(B), CCAT2(C), HOTAIR(D), LINC00161(E), MALAT1(F) and UCA1(G) were analyzed in HCC and healthy groups by qRT-PCR. LINC00462, LINC00161 and UCA1(P=0.033, P=0.002, P<0.001 respectively) were confirmed as the candidate lncRNAs in training set analysis.
Figure 3.
LINC00161 and UCA1 were up-regulated in validation set analysis. The relative expression levels of LINC00462(A), LINC00161(B) and UCA1(C) were analyzed in HCC and healthy groups by qRT-PCR. Serum LINC00161 and UCA1 of HCC patients was significantly higher than that of controls (P<0.001).
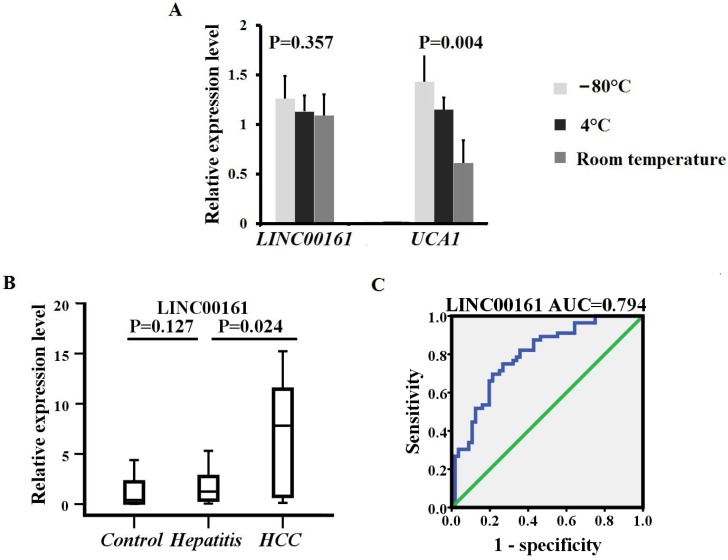
Stability and specificity detection of two lncRNAs in human serum
We examined the stability of exosomes derived from serum obtained from five healthy controls, which were divided into 3 aliquots at -80°C, 4°C, and room temperature for 12 hours, respectively. The expression levels of UCA1 were significantly downregulated at 4°C and room temperature after 12 hours when compared with samples at -80°C. However, all temperature incubation conditions had minimal effects on the concentration of LINC00161, demonstrating that the LINC00161 was sufficiently stable in human serum (Fig. 4A). The expression of the validated lncRNA was further analyzed in 20 chronic hepatitis patients. The relative expression levels of LINC00161 were downregulated in patients with chronic hepatitis in contrast to HCC controls. No statistical significance was demonstrated for the expression of LINC00161 in chronic hepatitis patients compared with healthy controls demonstrating that LINC00161 was sufficiently specific in HCC serum (Fig. 4B).
Figure 4.
The stability, specificity and diagnostic value analysis of selected lncRNA. (A)The relative expression levels of LINC00161 were stable after storing at -80°C, 4°C and room temperature for 12 hours respectively. (B) The relative expression levels of LINC00161 in control, hepatitis and HCC. (C) ROC analysis of LINC00161 in differentiating HCC from healthy controls. Kruskal-Wallis test was used to determine statistical significance. *P < 0.05.
Accuracy of serum parameters for predicting HCC by ROC analysis
Next, the diagnostic value of the selected lncRNA (LINC00161) was evaluated by ROC curve analyses by analyzing the results of 112 participants (56 HCC and 56 controls) in validation sets. As shown in Fig.4C, the area under the curve (AUC) of the validated LINC00161 was 0.794 (95% CI, 0.712-0.877). The sensitivities and specificities were 75.0% and 73.2% respectively.
Correlation between the serum levels of LINC00161 and clinical characteristics
As presented in Table 1, we assessed the correlation between the validated LINC00161 expression and the clinical features of HCC patients. There was no statistically significant correlation between the LINC00161 and clinical pathological information such as age, gender, chronic hepatitis, cirrhosis, tumor size and tumor number (P > 0.05). However, LINC00161 expression was significantly associated with serum AFP concentration and TNM stage (P < 0.05).
Table 1.
Association between LINC00161 relative expression and clinical features of HCC patients.
| Features | n | LINC00161 | ||
|---|---|---|---|---|
| High | Low | P value | ||
| Age(years) | ||||
| <50 | 22 | 6 | 16 | 0.203 |
| ≥50 | 34 | 15 | 19 | |
| Gender | ||||
| Male | 50 | 21 | 29 | 0.708 |
| Female | 6 | 3 | 3 | |
| Underlying disease | ||||
| Chronic hepatitis | 32 | 12 | 20 | 0.747 |
| Others | 24 | 8 | 16 | |
| Cirrhosis | ||||
| Negative | 32 | 12 | 20 | 0.350 |
| Positive | 24 | 12 | 12 | |
| Serum AFP(ng/ml) | ||||
| <200 | 23 | 16 | 7 | 0.002 |
| ≥200 | 33 | 9 | 24 | |
| Tumor size | ||||
| <5 cm | 19 | 10 | 9 | 0.389 |
| ≥5 cm | 37 | 15 | 22 | |
| Tumor number | ||||
| Solitary | 18 | 6 | 12 | 0.789 |
| Multiple | 38 | 14 | 24 | |
| TNM stage | ||||
| I and II | 21 | 14 | 7 | 0.019 |
| III and IV | 35 | 14 | 21 | |
Size and protein expression analysis to confirm the isolation of exosomes from serum
As shown in Fig. 5A-C, the round shaped microvesicles were about 30-150 nm in diameter in the exosome enriched fraction. Additionally, CD63 was used to identify serum exosomes (Fig. 5B).
Figure 5.
The identify analysis of serum exosomes. Transmission electron microscopy(A), western blotting (B) and nanosight(C) analysis were performed to confirm the isolation of exosomes from serum.
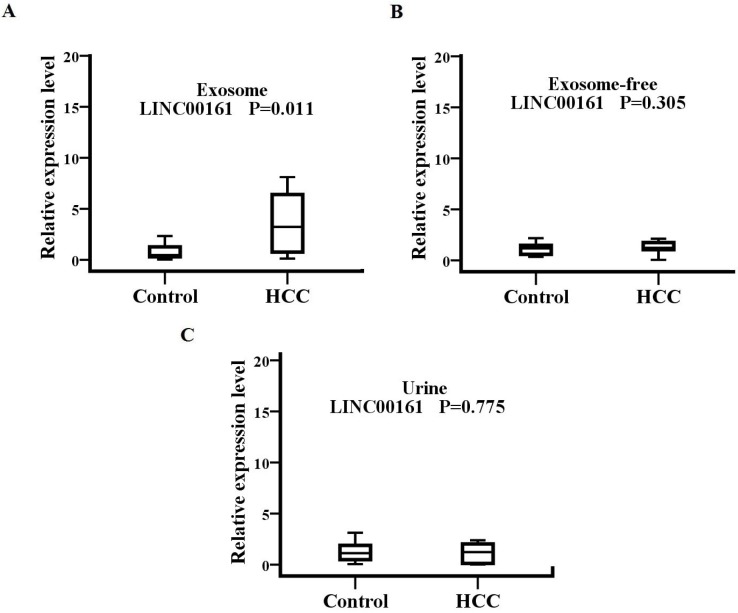
The source and path analysis of LINC00161
To analyze the source and path of LINC00161, samples from 15 HCC patients and 15 healthy controls were examined, including serum exosome, exosome-free, and urine (Fig.6). The serum exosomal expression levels of LINC00161 were upregulated in patients with HCC in contrast to controls. No statistical significance was detected for the expression of LINC00161 in HCC urine and serum exosome-free samples compared with controls, respectively. However, a statistical significance was demonstrated in serum exosomal expression levels of LINC00161 in HCC as compared to controls (P< 0.05).
Figure 6.
The souce and path analysis of LINC00161. The relative expression levels of LINC00161 in serum exosome(A), exosome-free(B) and urine(C)were analyzed in healthy and HCC groups by qRT-PCR. The serum exosomal expression levels of LINC00161 were up-regulated in patients with HCC in contrast to controls(P=0.011).
Discussion
HCC is one of the most common malignancies worldwide, with high mortality rates. Lack of early diagnosis results in limited effective therapeutic options and is associated with extremely poor outcomes for HCC patients28. Thus, new diagnosis measures for HCC are urgently needed. Along with an extensive characterization of the protein-coding genome of HCC, recent research has focused on the potential cancer-related lncRNAs and their role in the modulation of tumor development29. Alterations of many lncRNAs in cancer tissues have been shown in patients with HCC13-15, 17-19, 26. However, the regulation of these lncRNAs in serum and exosome of HCC patients remains largely unknown. Furthermore, the origin of these lncRNAs has not been clarified yet.
In light of these observations, we used an integrative approach based on clinical validation together with computational methods to seek insights into the diagnosis biomarkers of HCC. Firstly, eight lncRNAs (LINC00462, CCAT1, CCAT2, HOTAIR, LINC00161, SPRY4-IT1, MALAT1 and UCA1) were selected as candidates retrieved from literature.
Then, we analyzed the relative expression levels of the candidate lncRNAs in the training and validation sets of HCC patients and matched healthy controls by qRT-PCR and 2-ΔΔCT method algorithms. The serum levels of LINC00161 and UCA1 were significantly increased in HCC patients, which were consistent with results from recent reports and our previous study in HCC tissue18, 20. However, the increased serum level of UCA1 was not the same as this reported in some literature. Yang Z et al. reported that UCA1 was similarly expressed between tumor tissues and non-tumor tissues in HCC patients (P=0.375)27. A few reasons could explain the contrasting results, including small sample size, heterogeneity of tumors, samples with different clinical features, easily degraded samples, and unstable expression.
Herein, we demonstrated that the concentration of LINC00161 was sufficiently stable in different temperatures, while the levels of UCA1 were downregulated at high temperature. Thus the instability of UCA1 in serum maybe one of the main reasons that explains the discrepancy in different reports. Further analysis in chronic hepatitis patients compared with healthy and HCC controls demonstrated that LINC00161 was sufficiently specific in HCC serum.
Moreover, ROC curve analyses demonstrated that LINC00161 could differentiate HCC from healthy controls, indicating that LINC00161might be a novel potential diagnostic candidate for HCC. The expression of LINC00161 in serum was markedly correlated with the concentration of serum AFP and TNM stage. LINC00161 expression has been evaluated in HCC tissue. Xu L et al. showed that the expression level of LINC00161 was significantly higher in HCC tissues, and LINC00161 expression was an independent prognostic factor for overall survival. Moreover, those authors indicated that LINC00161 might serve as an oncogenic gene and play a pivotal role in promoting tumor migration and invasion in HCC18. That data and those from the present study indicate that detection of the expression levels of LINC00161, both in serum and tissue, can contribute to assessing the tumor stage and progression for HCC. Nonetheless, we should note the relative ease and non-invasive nature in collecting serum samples compared to tumor tissues.
Finally, we examined the levels of LINC00161 in serum exosomes, exosome-free, and urine from 15 HCC patients and 15 healthy controls for the source and path analysis. In recent years, increasing studies have focused on exosomes, which are a subclass of extracellular vesicles involved in intercellular communication released by most cell types and various body fluids30. Cells can trigger cancer-related disorders after the recognition and uptake of circulating exosomal lncRNAs, providing indications for early tumor biopsy, diagnosis, and treatment31. The validated lncRNA was all detected in serum exosomes, exosome-free, and urine, and we found that the levels of LINC00161 in serum exosomes were upregulated in patients with HCC in contrast to controls. No statistical significance was detected for the expression of the LINC00161 in exosome-free and urine samples in HCC to controls. These results demonstrated that the upregulated LINC00161 in HCC serum was derived from exosomes into serum. The LINC00161 may at least be partly metabolized through urine.
To the best of our knowledge, the present study is the first to report that the expression of LINC00161 was upregulated in serum and exosome of HCC patients and was associated with TNM stage in serum. The origin of the validated lncRNA was clarified, at least partly. As a result, circulating exosomal LINC00161 can act as a novel biomarker in the diagnosis of HCC.
Conclusion
In summary, as a member of lncRNA family, LINC00161 was significantly expressed in serum and exosome of HCC patients. The levels of LINC00161 suggest a significant prediction of tumor growth and metastasis in HCC. The upregulated LINC00161 expression in HCC serum was derived from exosomes into serum, and may at least be partly metabolized through urine. Our findings indicate that serum and exosome LINC00161 can serve as a potential non-invasive biomarker for predicting the diagnosis of HCC. Besides, our data provides new insight into the role of lncRNAs from serum exosome. However, this study is limited by the small sample size. Further exosomal high throughput sequencing and large-scale studies are needed to confirm these findings and elucidate the potential role of LINC00161 in HCC.
Acknowledgments
This study was supported by grants from the National Nature Science Foundation of China (No.81472027,81501820) to SKW and YQP, Key Project of Science and Technology Development of Nanjing Medicine (No.ZDX16005), the International Exchange and Cooperation Foundation of Nanjing Medical University(No.C063), the Natural Science Foundation for colleges of Jiangsu Province (No.17KJB310007), the Research and Innovation Program for graduate students of Jiangsu Province(No.KYCX17_1234), the Science and Technology Support Project of Jiangsu Province (No.BE2017763), the Science and Technology Development Foundation of Nanjing Medical University of China (No.2015NJMUZD037), Innovation team of Jiangsu provincial health-strengthening engineering by science and education to SKW.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Mortality GBD, Causes of Death C. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385(9963):117–71. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar N, Choudhary NS, Saraf N. et al. Hepatocellular Carcinoma Presenting as Dyspnea at Rest due to Innumerable Lung Metastasis. J Clin Exp Hepatol. 2016;6(4):319–320. doi: 10.1016/j.jceh.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou L, Liu J, Luo F. Serum tumor markers for detection of hepatocellular carcinoma. World J Gastroenterol. 2006;12(8):1175–81. doi: 10.3748/wjg.v12.i8.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin J, Luo W, Zeng X. et al. UXT-AS1-induced alternative splicing of UXT is associated with tumor progression in colorectal cancer. Am J Cancer Res. 2017;7(3):462–472. [PMC free article] [PubMed] [Google Scholar]
- 6.Yan X, Zhang D, Wu W, Mesenchymal Stem Cells Promote Hepatocarcinogenesis via lncRNA-MUF Interaction with ANXA2 and miR-34a. Cancer Res; 2017. [DOI] [PubMed] [Google Scholar]
- 7.Yang L, Lin C, Jin C. et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 9.Shi J, Li X, Zhang F. et al. Circulating lncRNAs associated with occurrence of colorectal cancer progression. Am J Cancer Res. 2015;5(7):2258–65. [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K, Shi H, Xi H. et al. Genome-Wide lncRNA Microarray Profiling Identifies Novel Circulating lncRNAs for Detection of Gastric Cancer. Theranostics. 2017;7(1):213–227. doi: 10.7150/thno.16044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martens-Uzunova ES, Bottcher R, Croce CM. et al. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65(6):1140–51. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Ellinger J, Alam J, Rothenburg J. et al. The long non-coding RNA lnc-ZNF180-2 is a prognostic biomarker in patients with clear cell renal cell carcinoma. Am J Cancer Res. 2015;5(9):2799–807. [PMC free article] [PubMed] [Google Scholar]
- 13.Gong J, Qi X, Zhang Y. et al. Long noncoding RNA linc00462 promotes hepatocellular carcinoma progression. Biomed Pharmacother. 2017;93:40–47. doi: 10.1016/j.biopha.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Deng L, Yang SB, Xu FF. et al. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F, Bai G, Li Y. et al. A positive feedback loop of long noncoding RNA CCAT2 and FOXM1 promotes hepatocellular carcinoma growth. Am J Cancer Res. 2017;7(7):1423–1434. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Xing Z, Mani SK. et al. RNA helicase DEAD box protein 5 regulates Polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology. 2016;64(4):1033–48. doi: 10.1002/hep.28698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou Z, Xu X, Fu X. et al. HBx-related long non-coding RNA MALAT1 promotes cell metastasis via up-regulating LTBP3 in hepatocellular carcinoma. Am J Cancer Res. 2017;7(4):845–856. [PMC free article] [PubMed] [Google Scholar]
- 18.Xu LC, Chen QN, Liu XQ. et al. Up-regulation of LINC00161 correlates with tumor migration and invasion and poor prognosis of patients with hepatocellular carcinoma. Oncotarget. 2017;8(34):56168–56173. doi: 10.18632/oncotarget.17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jing W, Gao S, Zhu M. et al. Potential diagnostic value of lncRNA SPRY4-IT1 in hepatocellular carcinoma. Oncol Rep. 2016;36(2):1085–92. doi: 10.3892/or.2016.4859. [DOI] [PubMed] [Google Scholar]
- 20.Wang F, Ying HQ, He BS. et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6(10):7899–917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Niel G, Porto-Carreiro I, Simoes S. et al. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140(1):13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Zeringer E, Barta T, Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos Trans R Soc Lond B Biol Sci; 2014. p. 369. (1652) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugimachi K, Matsumura T, Hirata H. et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112(3):532–8. doi: 10.1038/bjc.2014.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moris D, Beal EW, Chakedis J. et al. Role of exosomes in treatment of hepatocellular carcinoma. Surg Oncol. 2017;26(3):219–228. doi: 10.1016/j.suronc.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Li X, Sun W. et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017;397:33–42. doi: 10.1016/j.canlet.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Xiao JN, Yan TH, Yu RM. et al. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol. 2017;143(6):981–990. doi: 10.1007/s00432-017-2370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z, Lu Y, Xu Q. et al. HULC and H19 Played Different Roles in Overall and Disease-Free Survival from Hepatocellular Carcinoma after Curative Hepatectomy: A Preliminary Analysis from Gene Expression Omnibus. Dis Markers. 2015;2015:191029. doi: 10.1155/2015/191029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fong ZV, Tanabe KK. The clinical management of hepatocellular carcinoma in the United States, Europe, and Asia: a comprehensive and evidence-based comparison and review. Cancer. 2014;120(18):2824–38. doi: 10.1002/cncr.28730. [DOI] [PubMed] [Google Scholar]
- 29.Klingenberg M, Matsuda A, Diederichs S. et al. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67(3):603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Ruivo CF, Adem B, Silva M. et al. The Biology of Cancer Exosomes: Insights and New Perspectives. Cancer Res. 2017;77(23):6480–6488. doi: 10.1158/0008-5472.CAN-17-0994. [DOI] [PubMed] [Google Scholar]
- 31.Fan Q, Yang L, Zhang X. et al. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2017;414:107–115. doi: 10.1016/j.canlet.2017.10.040. [DOI] [PubMed] [Google Scholar]