Abstract
Wheat production and sustainability are steadily threatened by pests and pathogens in both wealthy and developing countries. This review is focused on the wheat curl mite (WCM), Aceria tosichella, and its relationship with wheat. WCM is a major pest of wheat and other cereals and a vector of at least four damaging plant viruses (Wheat streak mosaic virus, High plains wheat mosaic virus, Brome streak mosaic virus, and Triticum mosaic virus). The WCM–virus pathosystem causes considerable yield losses worldwide and its severity increases significantly when mixed-virus infections occur. Chemical control strategies are largely ineffective because WCM occupies secluded niches on the plant, e.g., leaf sheaths or curled leaves in the whorl. The challenge of effectively managing this pest–virus complex is exacerbated by the existence of divergent WCM lineages that differ in host-colonization and virus-transmission abilities. We highlight research progress in mite ecology and virus epidemiology that affect management and development of cereal cultivars with WCM- and virus-resistance genes. We also address the challenge of avoiding both agronomically deleterious side effects and selection for field populations of WCM that can overcome these resistance genes. This report integrates the current state of knowledge of WCM–virus-plant interactions and addresses knowledge gaps regarding the mechanisms driving WCM infestation, viral epidemics, and plant responses. We discuss the potential application of molecular methods (e.g., transcriptomics, epigenetics, and whole-genome sequencing) to understand the chemical and cellular interface between the wheat plant and WCM–virus complexes.
Keywords: cereals, eriophyoid mites, pathogen vector, plant viruses, phytophagous mites
Introduction
Wheat, Triticum aestivum L., is the most abundant source of calories and protein in the human diet (Braun et al., 2010; Arzani and Ashraf, 2017). It is grown annually on 215 million acres, an area larger than for any other crop, and remains the most traded on world markets and the most important crop in the 21st Century (Curtis and Halford, 2014).
However, wheat production is affected by a number of pests, including insects, fungi, nematodes, and mites, that can severely reduce yields and lead to crop failures. One of the most important global pests of wheat, occurring in North and South America, Europe, Asia, and Oceania, is the wheat curl mite (WCM), Aceria tosichella Keifer (Figures 1, 2A) which belongs to the superfamily Eriophyoidea (Navia et al., 2013). WCM is minute (about 0.2 mm long) and occupies sheltered niches on the plant, such as leaf sheaths and rolled and curled leaves, making its detection difficult, and limiting its exposure to acaricides (Navia et al., 2013). Moreover, its reproduction by arrhenotokous parthenogenesis (Miller et al., 2012), short developmental time, and high thermal tolerance (Kuczyński et al., 2016) enable great colonization potential.
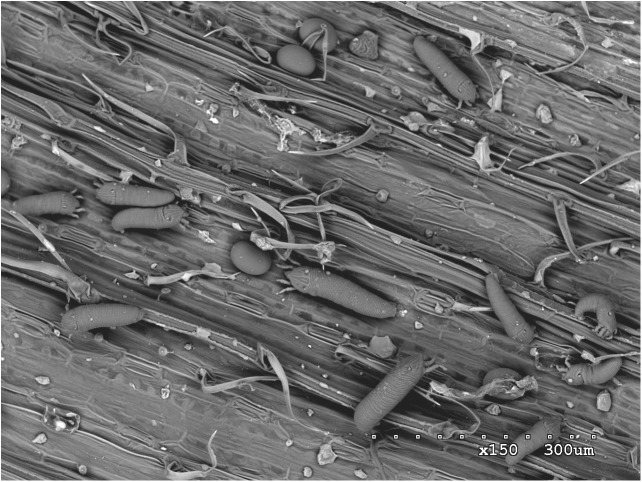
FIGURE 1.
Scanning electron microscopy (SEM) image of wheat curl mite (Aceria tosichella) specimens on a wheat leaf.
FIGURE 2.
Wheat curl mite (WCM) and WSMV symptoms: (A) specimens of WCM on a wheat leaf; (B) leaf curls caused by WCM; and (C) WSMV symptoms on wheat leaf.
The greatest economic impact of WCM results from its ability to transmit at least four damaging plant viruses to several different cereal crops. In this review we integrate the current state of knowledge of WCM–virus-plant interactions and address knowledge gaps regarding the mechanisms driving WCM infestation, viral epidemics, and plant responses.
What Curl Mite Feeding and Virus Transmission
Almost 90 grass species worldwide have been reported as host plants for WCM including cereals such as wheat, oats, barley, pearl millet, corn, and rye, as well as other cultivated (pasture) and uncultivated grasses (Navia et al., 2013). WCM has very short chelicerae (ca. 0.02 mm) and can feed only on leaf epidermal tissues. On wheat they colonize the plant by feeding within the whorl of a developing leaf on thin-walled epidermal tissue known as bulliform cells. Feeding on these cells by mites prevents leaves from unfurling causing leaf curling (Figure 2B) that promotes a humid environment generally preferred by WCM. WCM feeding also reduces photosynthetic capacity (Royalty and Perring, 1996).
The WCM has been shown to be the only transmitter of four distinct viruses to wheat and numerous other grass hosts (Stenger et al., 2016). These viruses occur across two virus families and three virus genera. Slykhuis (1955) first identified WCM as the vector of Wheat streak mosaic virus (family Potyviridae/genus Tritimovirus; acronym WSMV). The mite was also shown to transmit High plains wheat mosaic virus (Fimoviridae/Emaravirus; HPWMoV) (Seifers et al., 1997). Transmission of Brome streak mosaic virus (Potyviridae/Tritimovirus; BrSMV) by WCM was verified by Stephan et al. (2008). Most recently, Seifers et al. (2009) identified the WCM as the vector of Triticum mosaic virus (Potyviridae/Poacevirus; TriMV).
Of these viruses, WSMV is the most widely distributed and studied and it has been identified from every major wheat growing region around the world (Navia et al., 2013). The greatest and most consistent impact from WSMV occurs across the Great Plains of North America with more sporadic impact in other regions. BrSMV has only been found in Europe and no economic impact from the virus has been reported (Stephan et al., 2008).
Wheat streak mosaic virus infection of wheat results in a light and dark green mosaic pattern on the youngest emerged leaves (Figure 2C; Wegulo et al., 2008). As the plant adds new leaves, the newest leaves will first show these subtle mosaic symptoms while older leaves will become progressively more yellow. The tight curling at the leaf edge resulting from mite feeding is often apparent. The severity of symptoms and subsequent yield impact from virus infection in wheat depends on the plant stage at first infection (Hunger et al., 1992; Wosula et al., 2018). Plants infected prior to or during tillering will eventually become severely stunted, discolored, and take on a very prostrate growth pattern. These severe symptoms indicate that extreme yield loss will occur.
In the North American Great Plains co-infection of the viruses is common (Burrows et al., 2009; Byamukama et al., 2013, 2016) and may result in more spotted appearance on leaves but distinguishing symptoms of co-infections is not possible. Co-infection of WSMV and TriMV have been shown to increase the severity of symptoms and yield impacts (Tatineni et al., 2010; Byamukama et al., 2012, 2014). HPWMoV is not manually transmissible and this has limited study of this virus both independently and in combination with other viruses (Tatineni et al., 2014; Stenger et al., 2016).
WCM Diversity and its Implications
Understanding the relationships between WCM, viruses, and their hosts is challenging since WCM is a cryptic species complex. It includes multiple lineages that are distinguishable using mitochondrial (mtDNA COI, 16S) and nuclear (28S rDNA D2, ITS1–ITS2, and ANT) DNA sequences, differing also in host preference (Skoracka et al., 2012, 2013; Miller et al., 2013; Szydło et al., 2015). Some lineages are highly host-specific and locally distributed, whereas others are generalists with wider geographic ranges (Skoracka et al., 2014). Two WCM genotypes associated with wheat are the most polyphagous and widespread, having been found in the Middle East, Europe, Australia, and North and South America (Skoracka et al., 2014; Wosula et al., 2016). They are known as type 1 and type 2 in Australia (Carew et al., 2009) with corresponding genotypes occurring in North America (Hein et al., 2012), as well as in Europe and South America where they are known as MT-8 and MT-1, respectively (Skoracka et al., 2014). Hereafter this latter nomenclature will be used.
In North America these two lineages have been shown to transmit WSMV (Wosula et al., 2016). However, MT-1 had a higher reproductive capacity in the presence of WSMV and vectored it more efficiently than MT-8 (Seifers et al., 2002; Siriwetwiwat, 2006; Oliveira-Hofman et al., 2015). In Australia, among these two lineages only MT-1 has been observed to transmit WSMV (Schiffer et al., 2009). MT-1 is also the most effective vector of HPWMoV and TriMV (Seifers et al., 2002; McMechan et al., 2014; Wosula et al., 2016). Mixed-virus infections further confound virus–mite studies, e.g., transmission by MT-1 was more frequent from WSMV infected source plants than from those co-infected with TriMV (Oliveira-Hofman et al., 2015).
MT-8 and MT-1 have been found coexisting in mixed populations in wheat-producing areas in North America, Australia, and Europe, where plants from a single wheat field contained both MT-1 and MT-8 (Siriwetwiwat, 2006; Schiffer et al., 2009; Hein et al., 2012; Skoracka et al., 2017), further complicating management of viruses vectored by WCM. This sympatry combined with differential virus-transmission accentuates the need for efficient identification methods.
WCM Management
To date, research to manage this mite–virus complex has focused mainly on the development of classical host plant resistance (HPR) to both the mite and viruses by introgressing favorable traits from resistant germplasm into advanced breeding lines (Whelan and Hart, 1988; Chen et al., 1998; Harvey et al., 2003; Malik et al., 2003a; Hakizimana et al., 2004; Carrera et al., 2012; Carver et al., 2016), in addition to cultural practices such as planting date and summer control of volunteer wheat plants (McMechan and Hein, 2016). The search for genes conferring WSMV resistance to wheat began shortly after the virus was identified in the 1950s (McKinney and Sando, 1951). With few sources of resistance available in wheat, the search eventually targeted close relatives culminating with the chromosome translocation of the Wsm1 gene from Thinopyrum intermedium (Host) Barkworth & D.R. Dewey to the short arm of chromosome 4D in wheat (Friebe et al., 1991).
Continued efforts resulted in release of the first germplasm: KS96HW10-3 (Seifers et al., 1995) and first commercial cultivar ‘Mace’ (Graybosch et al., 2009) with the Wsm1 gene. This gene has demonstrated resistance to both WSMV and TriMV (Friebe et al., 2009), however, its value has been limited due to linkage drag that reduces yields (Sharp et al., 2002). Similar issues have impacted a second gene, Wsm3, transferred into wheat from T. intermedium but efforts continue to improve its effectiveness and identify genetic markers (Friebe et al., 2009; Danilova et al., 2017).
A germplasm line, CO96093-2, was identified by Haley et al. (2002) as resistant to WSMV, but the gene’s origin was uncertain. Lu et al. (2011) found this gene to be a new gene (Wsm2) of wheat origin. Four varieties have thus far been released with the Wsm2 gene: ‘RonL’ (Seifers et al., 2007), ‘Snowmass’ (Haley et al., 2011), ‘Clara CL’ (Martin et al., 2014), and ‘Oakley CL’ (Zhang et al., 2015). Studies with both Wsm1 and Wsm2 have demonstrated that both genes are temperature-sensitive with high levels of resistance below 20°C but breaking down as temperatures approach 25°C (Seifers et al., 1995, 2007).
Additional sources of WSMV resistance in wheat have recently been identified and hold promise for incorporation into commercial wheats (Seifers et al., 2007, 2013), including increased temperature stable resistance (Fahim et al., 2012a; Kumssa et al., 2017). Lu et al. (2011) has hypothesized the presence of a minor gene in wheat that confers partial resistance or tolerance in some commercial cultivars.
Early efforts to identify resistance to the WCM in wheat were not successful (Harvey and Livers, 1975), and this led to efforts to target close wheat relatives for resistance. Thus far, four WCM-resistance genes have been identified. The earliest of these genes (Cmc3) was translocated to wheat from rye (Secale cereale L.) (Martin et al., 1983; Malik et al., 2003a). It was present in ‘TAM 107,’ a commercial release that became widely used in the 1980s and 1990s across the Great Plains (Porter et al., 1987). However, the extensive use of TAM 107 led to loss of effectiveness of the gene (Harvey et al., 1995, 1997). A mite-resistance gene (Cmc1) translocated from Aegilops tauschii (Coss.) Schmal. to wheat (Thomas and Conner, 1986; Whelan and Thomas, 1989) has been used to develop breeding material (Cox et al., 1999) and the recent release of ‘Radiant’ in Canada (Thomas et al., 2012). A third source of resistance, Thinopyrum ponticum (Podp.) Barkworth & D.R. Dewey, contributed with gene Cmc2 (Whelan and Hart, 1988). A second gene originating from A. tauschii (Cmc4) was found to be independent of Cmc1 (Cox et al., 1999; Malik et al., 2003a) and has been used in the breeding release OK05312 (Carver et al., 2016). Additional resistance genes have been proposed but not yet isolated from common wheat (Harvey and Martin, 1992), rye (Cainong et al., 2010), and A. tauschii (Malik et al., 2003b; Dhakal et al., 2017).
The value of mite-resistance lies in the potential for reduced virus transmission and spread through the field, as well as in the reduction of mite buildup in the volunteer wheat that serves as a bridge host to the following wheat crop (Martin et al., 1984; Conner et al., 1991; Harvey et al., 2005). However, mite response to resistance genes has often been variable (Harvey et al., 1999) and the stability of resistance genes is a concern due to the apparent adaptation to Cmc3 by mite populations (Harvey et al., 1995, 1997, 1999). Greater understanding of the variability in mite genotype responses to resistance genes is needed to evaluate potential stability of resistance genes. Genetic characterization of the mites used in resistance studies has become critical to understanding mite-gene response (Richardson et al., 2014; Aguirre-Rojas et al., 2017; Dhakal et al., 2017). Future efforts to pyramid Wsm and Cmc genes may enhance the utility and stability of these management options.
Molecular tools, such as in situ hybridization and genetic marker maps have improved the efficiency and precision of HPR introgression efforts. In addition, RNAi techniques have been used to produce transgenic wheat lines with resistance to WSMV (Fahim et al., 2010, 2012b; Cruz et al., 2014) and TriMV (Shoup Rupp et al., 2016) although no commercial wheat cultivars with this resistance have been released. With current advances in DNA sequencing technology, the whole genome sequences (WGSs) of wheat, WCM, WSMV, HPWMoV, TriMV, and BrSMV (Gustafson et al., 1987; Seifers et al., 2008; Stewart, 2016; Tatineni et al., 2016; Zimin et al., 2017) are all now available, presenting the opportunity to study these tripartite host–vector–virus relationships at the level of genome sequence and gene expression.
Future Directions
Wheat–WCM Interactions
Like many eriophyoid mites that attack grasses, WCM is vagrant, i.e., inhabiting the leaf surface rather than inducing galls, and there is very little published information regarding its direct molecular or physiological interactions with its hosts. Given the availability of its genome sequence and those of several of its hosts, such as wheat (Zimin et al., 2017), maize (Schnable et al., 2009), and barley (Mascher et al., 2017), WCM is a good candidate to be a model for such studies in grass-infesting Eriophyoidea. For example, using available genomic and transcriptomic (Ozsolak and Milos, 2011; Jänes et al., 2015) resources, it will be possible to determine whether the ability of polyphagous genotypes (e.g., MT-1, MT-8) to change from one host to another is genetically or epigenetically (Laird, 2011) controlled. Similarly, the factors that determine which plant species are accepted by a host-specific WCM genotype can be dissected (Gompert et al., 2010; Narum et al., 2013). Moreover, novel genomic technologies and high-throughput phenotyping of wheat varieties can accelerate germplasm improvement (see Mondal et al., 2016 for examples).
Proteomic analyses of rice leaves from control plants and those infested with Schizotetranychus oryzae (Acari: Tetranychidae) revealed a wide range of intracellular physiological changes induced by this mite although the specific source(s) of induction (e.g., salivary components) are not known (Buffon et al., 2016). Similar analyses of WCM on one or more of its hosts could take advantage of the mite’s and host plants’ genomic resources, as well as recent techniques developed to characterize the salivary proteins of a tetranychid mite (Jonckheere et al., 2016), to assess mite–host interactions from both sides. Effects of individual proteins could be assessed through knockout genotypes created by the CRISPR-Cas9 mutagenesis (Ran et al., 2013). Complementary studies of other eriophyoids and mite species from other families that attack cereal crops would identify similarities and differences in these interactions that could shed light on prospective control strategies against multiple mite species, e.g., via RNAi in the host plant to block production of essential mite proteins.
WCM–Virus Interactions
Regarding the ability of mites to transmit WSMV, a genotyping-by-sequencing study (e.g., Narum et al., 2013) incorporating all known WCM genotypes with variable WSMV transmission ability and anchored to an annotated WGS of WCM would identify candidate genomic regions associated with WSMV transmission variability. This could also be used to explore the differential transmission of TriMV and HPWMoV by WCM genotypes. Complementary transcriptomic and epigenetic studies could further identify the candidate gene(s) involved in this variability and tease apart genetic and epigenetic factors.
Different strains of WSMV have also been detected that are differentially transmitted by individual WCM genotypes (Wosula et al., 2016). Mutations to the helper component proteinase (HC-Pro) gene of WSMV have been shown to alter transmission from mite to plant or prevent it altogether (Stenger et al., 2006; Young et al., 2007) although the precise physiological mechanism of transmission is unknown. Given that WSMV is a circulative virus that is transmitted via the salivary glands of WCM (Paliwal, 1980), the development of salivary protein characterization techniques (Jonckheere et al., 2016) may enable association of specific WCM salivary proteins with successful or unsuccessful WSMV transmission. If other WCM-transmitted viruses have a persistent circulative type of relationship with the vector, understanding the mechanisms (receptors) by which these viruses cross the midgut epithelium and salivary gland barriers to reach the stylet channel may yield basic information regarding the traffic of these viruses within the mite body.
WCM Colonization Potential
The spread of WCM and its associated plant viruses to cereal-producing regions worldwide is of increasing scientific and economic importance (Navia et al., 2013). Colonization and invasive potential of any organism is inevitably associated with its dispersal ability and its degree of ecological specialization (Ehrlich, 1986). WCM disperses passively by air currents (Sabelis and Bruin, 1996) and wheat-associated lineages are characterized by low host-specificity (Skoracka et al., 2013). Generalists with high dispersal ability are typically successful invaders (Wilson et al., 2009). But relationships between WCM dispersal potential, degree of host specialization, and colonization success have never been tested. To do so, it will be necessary to understand the mechanisms of successful WCM wheat colonization, including long-established and recent invasions. Research on the relationship between WCM host specialization and dispersal ability revealed trade-offs in plant performance between different host plant species after mite dispersal (Laska et al., 2017). Also it has been shown that a small number of WCM specimens landing on wheat plants after aerial dispersal (about 2% of an initial source population) were able to establish dense colonies very quickly, indicating great colonization potential (Kiedrowicz et al., 2017). Understanding how interactions between dispersal and local adaptation shape WCM distribution is crucial because predicting spread of potentially invasive organisms, particularly under current anthropogenic environmental changes, is a key to managing pest outbreaks and minimizing ecosystem degradation.
Author Contributions
AS, BR, and GH designed the conception, wrote the manuscript, and read and approved the submitted version with equal contribution.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Alicja Laska (Population Ecology Lab, Adam Mickiewicz University) for invaluable help in preparation of graphics, and reviewers for their valuable remarks that improved the manuscript.
Footnotes
Funding. AS was supported by National Science Centre in Poland (Grant No. UMO-2016/21/B/NZ8/00786).
References
- Aguirre-Rojas L. M., Khalaf L. K., Garces-Carrera S., Sinha D. K., Chuang W., Smith C. M. (2017). Resistance to wheat curl mite in arthropod-resistant rye-wheat translocation lines. Agronomy 7:74 10.3390/agronomy7040074 [DOI] [Google Scholar]
- Arzani A., Ashraf M. (2017). Cultivated ancient wheats (Triticum spp.): a potential source of health-beneficial food products. Compr. Rev. Food Sci. Food Saf. 16 477–488. 10.1111/1541-4337.12262 [DOI] [PubMed] [Google Scholar]
- Braun H. J., Atlin G., Payne T. (2010). “Multi-location testing as a tool to identify plant response to global climate change,” in Climate Change and Crop Production, ed. Reynolds M. P. (London: CAB International; ), 115–138. 10.1079/9781845936334.0115 [DOI] [Google Scholar]
- Buffon G., Blasi R., Adamski J. M., Ferla N. J., Berger M., Santi L., et al. (2016). Physiological and molecular alterations promoted by Schizotetranychus oryzae mite infestation in rice leaves. J. Proteome Res. 15 431–446. 10.1021/acs.jproteome.5b00729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows M., Franc G., Rush C., Blunt T., Ito D., Kinzer K., et al. (2009). Occurrence of viruses in wheat in the Great Plains region, 2008. Plant Health Prog. 1–7. 10.1094/PHP-2009-0706-01-RS [DOI] [Google Scholar]
- Byamukama E., Seifers D. L., Hein G. L., De Wolf E., Tisserat N. A., Langham M. C., et al. (2013). Occurrence and distribution of Triticum mosaic virus in the central great plains. Plant Dis. 97 21–29. 10.1094/PDIS-06-12-0535-RE [DOI] [PubMed] [Google Scholar]
- Byamukama E., Tatineni S., Hein G. L., Graybosch R. A., Baenziger P. S., French R., et al. (2012). Effects of single and double infections of winter wheat by Triticum mosaic virus and Wheat streak mosaic virus on yield determinants. Plant Dis. 96 859–864. 10.1094/PDIS-11-11-0957-RE [DOI] [PubMed] [Google Scholar]
- Byamukama E., Tatineni S., Hein G. L., McMechan A. J., Wegulo S. N. (2016). Incidence of Wheat streak mosaic virus, Triticum mosaic virus, and Wheat mosaic virus in wheat curl mites recovered from maturing winter wheat spikes. Plant Dis. 100 318–323. 10.1094/PDIS-06-15-0692-RE [DOI] [PubMed] [Google Scholar]
- Byamukama E., Wegulo S. N., Tatineni S., Hein G. L., Graybosch R. A., Baenziger P. S., et al. (2014). Quantification of yield loss caused by Triticum mosaic virus and Wheat streak mosaic virus in winter wheat under field conditions. Plant Dis. 98 127–133. 10.1094/PDIS-04-13-0419-RE [DOI] [PubMed] [Google Scholar]
- Cainong J. C., Zavatsky L. E., Chen M. S., Johnson J., Friebe B., Gill B. S., et al. (2010). Wheat-rye T2BS 2BL-2RL recombinants with resistance to Hessian Fly (H21). Crop Sci. 50 920–925. 10.2135/cropsci2009.06.0310 [DOI] [Google Scholar]
- Carew M., Schiffer M., Umina P., Weeks A., Hoffmann A. (2009). Molecular markers indicate that the wheat curl mite, Aceria tosichella Keifer, may represent a species complex in Australia. Bull. Entomol. Res. 99 479–486. 10.1017/S0007485308006512 [DOI] [PubMed] [Google Scholar]
- Carrera S. G., Davis H., Aguirre-Rojas L., Murugan M., Smith C. M. (2012). Multiple categories of resistance to wheat curl mite (Acari: Eriophyidae) expressed in accessions of Aegilops tauschii. J. Econ. Entomol. 105 2180–2186. 10.1603/EC12252 [DOI] [PubMed] [Google Scholar]
- Carver B. F., Smith C. M., Chang W., Hunger R. M., Edwards J. T., Yan L., et al. (2016). Registration of OK 05312, a high-yielding hard winter wheat donor of Cmc4 for wheat curl mite resistance. J. Plant Regist. 10 75–79. 10.3198/jpr2015.04.0026crg [DOI] [Google Scholar]
- Chen Q., Conner R. L., Ahmad F., Laroche A., Fedak G., Thomas J. B. (1998). Molecular characterization of the genome composition of partial amphiploids derived from Triticum aestivum x Thinopyrum ponticum and T. aestivum x Th. intermedium as sources of resistance to Wheat streak mosaic virus and its vector, Aceria tosichella. Theor. Appl. Genet. 97 1–8. 10.1007/s001220050860 [DOI] [Google Scholar]
- Conner R. L., Thomas J. B., Whelan E. D. P. (1991). Comparison of mite resistance for control of wheat streak mosaic. Crop Sci. 31 315–318. 10.2135/cropsci1991.0011183X003100020018x [DOI] [Google Scholar]
- Cox T. S., Bockus W. W., Gill B. S., Sears R. G., Harvey T. L., Leath S., et al. (1999). Registration of KS96WGR40 hard red winter wheat germplasm resistant to wheat curl mite, stagonospora leaf blotch, and septoria leaf blotch. Crop Sci. 39:597 10.2135/cropsci1999.0011183X003900020070x [DOI] [Google Scholar]
- Cruz L. F., Shoup Rupp J. L., Trick H. N., Fellers J. P. (2014). Stable resistance to Wheat streak mosaic virus in wheat mediated by RNAi. In Vitro Cell. Dev. Biol. Plant 50 665–672. 10.1007/s11627-014-9634-0 [DOI] [Google Scholar]
- Curtis T., Halford N. G. (2014). Food security: the challenge of increasing wheat yield and the importance of not compromising food safety. Ann. Appl. Biol. 164 354–372. 10.1111/aab.12108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova T. V., Zhang G., Liu W., Friebe B., Gill B. S. (2017). Homoeologous recombination-based transfer and molecular cytogenetic mapping of a wheat streak mosaic virus and Triticum mosaic virus resistance gene Wsm3 from Thinopyrum intermedium to wheat. Theor. Appl. Genet. 130 549–556. 10.1007/s00122-016-2834-8 [DOI] [PubMed] [Google Scholar]
- Dhakal S., Tan C., Paezold L., Fuentealba M. P., Rudd J. C., Blaser B. C., et al. (2017). Wheat curl mite resistance in hard winter wheat in the US great plains. Crop Sci. 57 53–61. 10.2135/cropsci2016.02.0121 [DOI] [Google Scholar]
- Ehrlich P. R. (1986). “Which animal will invade?,” in Ecology of Biological Invasions of North American and Hawaii, eds Mooney H. A., Drake J. A. (New York, NY: Springer-Verlag; ), 79–95. 10.1007/978-1-4612-4988-7_5 [DOI] [Google Scholar]
- Fahim M., Ayala-Navarrete L., Millar A. A., Larkin P. J. (2010). Hairpin RNA derived from viral NIa gene confers immunity to Wheat streak mosaic virus infection in transgenic wheat plants. Plant Biotechnol. J. 8 821–834. 10.1111/j.1467-7652.2010.00513.x [DOI] [PubMed] [Google Scholar]
- Fahim M., Mechanicos A., Ayala-Navarrete L., Haber S., Larkin P. J. (2012a). Resistance to wheat streak mosaic virus – a survey of resources and development of molecular markers. Plant Pathol. 61 425–440. 10.1111/j.1365-3059.2011.02542.x [DOI] [Google Scholar]
- Fahim M., Millar A. A., Wood C. C., Larkin P. J. (2012b). Resistance to Wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol. J. 10 150–163. 10.1111/j.1467-7652.2011.00647.x [DOI] [PubMed] [Google Scholar]
- Friebe B., Mukai Y., Dhaliwal H. S., Martin T. J., Gill B. S. (1991). Identification of alien chromatin specifying resistance to wheat streak mosaic and greenbug in wheat germplasm by C-banding and in situ hybridization. Theor. Appl. Genet. 81 381–389. 10.1007/BF00228680 [DOI] [PubMed] [Google Scholar]
- Friebe B., Qi L. L., Wilson D. L., Chang Z. J., Seifers D. L., Martin T. J., et al. (2009). Wheat-Thinopyrum intermedium recombinants resistant to Wheat streak mosaic virus and Triticum mosaic virus. Crop Sci. 49 1221–1226. 10.2135/cropsci2008.09.0513 [DOI] [Google Scholar]
- Gompert Z., Forister M. L., Fordyce J. A., Nice C. C., Williamson R. J., Buerkle C. A. (2010). Bayesian analysis of molecular variance in pyrosequences quantifies population genetic structure across the genome of Lycaeides butterflies. Mol. Ecol. 19 2455–2473. 10.1111/j.1365-294X.2010.04666.x [DOI] [PubMed] [Google Scholar]
- Graybosch R. A., Peterson C. J., Baenziger P. S., Baltensperger D. D., Nelson L. A., Jin Y., et al. (2009). Registration of “Mace” hard red winter wheat. J. Plant Regist. 3 51–56. 10.3198/jpr2008.06.0345crc [DOI] [Google Scholar]
- Gustafson G., Hunter B., Hanau R., Armour S. L., Jackson A. O. (1987). Nucleotide sequence and genetic organization of Barley stripe mosaic virus RNA-γ. 1987. Virology 158 394–406. 10.1016/0042-6822(87)90211-X [DOI] [PubMed] [Google Scholar]
- Hakizimana F., Ibrahim A. M. H., Langham M. A. C., Haley S. D., Rudd J. C. (2004). Diallel analysis of Wheat streak mosaic virus resistance in winter wheat. Crop Sci. 44 89–92. 10.2135/cropsci2004.8900 [DOI] [Google Scholar]
- Haley S. D., Johnson J. J., Peairs F. B., Stromberger J. A., Heaton E. E., Seifert S. A., et al. (2011). Registration of ‘Snowmass’ wheat. J. Plant Regist. 5 87–90. 10.3198/jpr2010.03.0175crc [DOI] [Google Scholar]
- Haley S. D., Martin T. J., Quick J. S., Seifers D. L., Stromberger J. A., Clayshulte S. R., et al. (2002). Registration of CO960293-2 wheat germplasm resistant to Wheat streak mosaic virus and Russian wheat aphid. Crop Sci. 42 1381–1382. 10.2135/cropsci2002.1381 [DOI] [Google Scholar]
- Harvey T. L., Livers R. W. (1975). Resistance to wheat curl mite, Aceria tulipae Keifer, in rye and wheat-rye addition lines. Environ. Entomol. 4 523–526. 10.1093/ee/4.3.523 [DOI] [Google Scholar]
- Harvey T. L., Martin T. J. (1992). Resistance to the wheat curl mite (Acari: Eriophyidae) in common wheat. Cereal Res. Commun. 20 63–66. [Google Scholar]
- Harvey T. L., Martin T. J., Seifers D. L. (2003). Resistance to the wheat curl mite (Acari: Eriophyidae) prevents loss in wheat yield. J. Agric. Urban Entomol. 20 7–10. [Google Scholar]
- Harvey T. L., Martin T. J., Seifers D. L., Sloderbeck P. E. (1995). Adaptation of wheat curl mite (Acari: Eriophyidae) to resistant wheat in Kansas. J. Agric. Entomol. 12 119–125. [Google Scholar]
- Harvey T. L., Martin T. J., Seifers D. L., Sloderbeck P. E. (1997). Change in virulence of wheat curl mite detected on TAM 107 wheat. Crop Sci. 37 624–625. 10.2135/cropsci1997.0011183X003700020052x [DOI] [Google Scholar]
- Harvey T. L., Seifers D. L., Martin T. J., Brown-Guedira G., Gill B. S. (1999). Survival of wheat curl mites on different sources of resistance in wheat. Crop Sci. 39 1887–1889. 10.2135/cropsci1999.3961887x [DOI] [Google Scholar]
- Harvey T. L., Seifers D. L., Martin T. J., Michaud J. P. (2005). Effect of resistance to Wheat streak mosaic virus on transmission efficiency of wheat curl mites. J. Agric. Urban Entomol. 22 1–6. [Google Scholar]
- Hein G. L., French R., Siriwetwiwat B., Amrine J. W. (2012). Genetic characterization of North American populations of wheat curl mite and dry bulb mite. J. Econ. Entomol. 105 1801–1808. 10.1603/EC11428 [DOI] [PubMed] [Google Scholar]
- Hunger R. M., Sherwood J. L., Evans C. K., Montana J. R. (1992). Effects of planting date and inoculation date on severity of wheat streak mosaic in hard red winter wheat cultivars. Plant Dis. 76 1056–1060. 10.1094/PD-76-1056 [DOI] [Google Scholar]
- Jänes J., Hu F., Lewin A., Turro E. (2015). A comparative study of RNA-seq analysis strategies. Brief. Bioinform. 16 932–940. 10.1093/bib/bbv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckheere W., Dermauw W., Zhurov V., Wybouw N., Van den Bulcke J., Villarroel C. A., et al. (2016). The salivary protein repertoire of the polyphagous spider mite Tetranychus urticae: a quest for effectors. Mol. Cell. Proteomics 15 3594–3613. 10.1074/mcp.M116.058081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowicz A., Kuczyński L., Laska A., Lewandowski M., Proctor H., Skoracka A. (2017). “Dispersal strategies in passively spreading phytophagous mites,”in Proceedings of the ASAB Easter Conference 2017, Liverpool, 875. [Google Scholar]
- Kuczyński L., Rector B. G., Kiedrowicz A., Lewandowski M., Szydło W., Skoracka A. (2016). Thermal niches of two invasive genotypes of the wheat curl mite Aceria tosichella: congruence between physiological and geographical distribution data. PLoS One 11:e0154600. 10.1371/journal.pone.0154600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumssa T. T., Zhao D., Bai G., Zhang G. (2017). Resistance to Wheat streak mosaic virus and Triticum mosaic virus in wheat lines carrying Wsm1 and Wsm3. Eur. J. Plant Pathol. 147 709–712. 10.1007/s10658-016-1021-8 [DOI] [Google Scholar]
- Laird P. W. (2011). Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet. 11 191–202. 10.1038/nrg2732 [DOI] [PubMed] [Google Scholar]
- Laska A., Kuczyński L., Radwan J., Lewandowski M., Bharati K., Karpicka-Ignatowska K., et al. (2017). “How to test the evolution of specialization and dispersal: the case study of the invasive wheat curl mite. Aceria tosichella,”in Proceedings of the Polish Evolutionary Conference, Toruń, 42. [Google Scholar]
- Lu H., Price J., Devkota R., Rush C., Rudd J. (2011). A dominant gene for resistance to Wheat streak mosaic virus in winter wheat line CO960293-2. Crop Sci. 51 5–12. 10.2135/cropsci2010.01.0038 [DOI] [Google Scholar]
- Malik R., Brown-Guedira G. L., Smith C. M., Harvey T. L., Gill B. S. (2003a). Genetic mapping of wheat curl mite resistance genes Cmc3 and Cmc4 in common wheat. Crop Sci. 43 644–650. 10.2135/cropsci2003.0644 15942758 [DOI] [Google Scholar]
- Malik R., Smith C. M., Brown-Guedira G. L., Harvey T. L., Gill B. S. (2003b). Assessment of Aegilops tauschii for resistance to biotypes of wheat curl mite (Acari: Eriophyidae). J. Econ. Entomol. 96 1329–1333. 10.1603/0022-0493-96.4.1329 [DOI] [PubMed] [Google Scholar]
- Martin T. J., Harvey T. L., Bender C. G., Seifers D. L. (1984). Control of Wheat streak mosaic virus with vector resistance in wheat. Phytopathology 74 963–964. 10.1094/Phyto-74-963 [DOI] [Google Scholar]
- Martin T. J., Harvey T. L., Bender C. G., Seifers D. L., Hatchett J. H. (1983). Wheat curl mite resistance wheat germplasm. Crop Sci. 23:89 10.2135/cropsci1983.0011183X002300040075x [DOI] [Google Scholar]
- Martin T. J., Zhang G., Fritz A. K., Miller R., Chen M. (2014). Registration of ‘Clara CL’ wheat. J. Plant Regist. 8 38–42. 10.3198/jpr2013.07.0040crc [DOI] [Google Scholar]
- Mascher M., Gundlach H., Himmelback A., Beier S., Twardziok S. O., Wicker T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544 427–433. 10.1038/nature22043 [DOI] [PubMed] [Google Scholar]
- McMechan A. J., Hein G. L. (2016). Planting date and variety selection for management of viruses transmitted by the wheat curl mite (Acari: Eriophyidae). J. Econ. Entomol. 109 70–77. 10.1093/jee/tov311 [DOI] [PubMed] [Google Scholar]
- McMechan A. J., Tatineni S., French R., Hein G. L. (2014). Differential transmission of Triticum mosaic virus by wheat curl mite populations collected in the Great Plains. Plant Dis. 98 806–810. 10.1094/PDIS-06-13-0582-RE [DOI] [PubMed] [Google Scholar]
- Miller A. D., Skoracka A., Navia D., de Mendonca R., Szydło W., Schultz M., et al. (2013). Phylogenetic analyses reveal extensive cryptic speciation and host specialization in an economically important mite taxon. Mol. Phylogenet. Evol. 66 928–940. 10.1016/j.ympev.2012.11.021 [DOI] [PubMed] [Google Scholar]
- Miller A. D., Umina P. A., Weeks A. R., Hoffmann A. A. (2012). Population genetics of the wheat curl mite (Aceria tosichella keifer) in Australia: implications for the management of wheat pathogens. Bull. Entomol. Res. 102 199–212. 10.1017/S0007485311000526 [DOI] [PubMed] [Google Scholar]
- Mondal S., Rutkoski J. E., Velu G., Singh P. K., Crespo-Herrera L. A., Guzmán C., et al. (2016). Harnessing diversity in wheat to enhance grain yield, climate resilience, disease and insect pest resistance and nutrition through conventional and modern breeding approaches. Front. Plant Sci. 7:991. 10.3389/fpls.2016.00991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narum S. R., Buerkle C. A., Davey J. W., Miller M. R., Hohenlohe P. A. (2013). Genotyping-by-sequencing in ecological and conservation genomics. Mol. Ecol. 22 2841–2847. 10.1111/mec.12350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navia D., de Mendonca R. S., Skoracka A., Szydło W., Knihinicki D., Hein G. L., et al. (2013). Wheat curl mite, Aceria tosichella, and transmitted viruses: an expanding pest complex affecting cereal crops. Exp. Appl. Acarol. 59 95–143. 10.1007/s10493-012-9633-y [DOI] [PubMed] [Google Scholar]
- Oliveira-Hofman C., Wegulo S. N., Tatineni S., Hein G. L. (2015). Impact of Wheat streak mosaic virus and Triticum mosaic virus coinfection of wheat on transmission rates by wheat curl mites. Plant Dis. 99 1170–1174. 10.1094/PDIS-08-14-0868-RE [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Milos P. M. (2011). RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12 87–98. 10.1038/nrg2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliwal Y. C. (1980). Relationship of wheat streak mosaic and barley stripe mosaic viruses to vector and nonvector eriophyid mites. Arch. Virol. 63 123–132. 10.1007/BF01320769 [DOI] [PubMed] [Google Scholar]
- Porter K. B., Worrall W. D., Gardenhire J. H., Gilmore E. C., McDaniel M. E., Tuleen N. A. (1987). Registration of “TAM 107” wheat. Crop Sci. 27 818–819. 10.2135/cropsci1987.0011183X002700040050x [DOI] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson K., Miller A. D., Hoffmann A. A., Larkin P. (2014). Potential new sources of wheat curl mite resistance in wheat to prevent the spread of yield-reducing pathogens. Exp. Appl. Acarol. 64 1–19. 10.1007/s10493-014-9808-9 [DOI] [PubMed] [Google Scholar]
- Royalty R. N., Perring T. M. (1996). “Nature of damage and its assessment,” in Eriophyoid Mites: Their Biology, Natural Enemies and Control, eds Lindquist E. E., Sabelis M. W., Bruin J. (Amsterdam: Elsevier Science; ), 493–512. 10.1016/S1572-4379(96)80031-5 [DOI] [Google Scholar]
- Sabelis M. W., Bruin J. (1996). “Evolutionary ecology: life history patterns, food plant choice and dispersal,” in Eriophyoid Mites: Their Biology, Natural Enemies and Control, eds Lindquist E. E., Sabelis M. W., Bruin J. (Amsterdam: Elsevier Science; ), 329–366. 10.1016/S1572-4379(96)80020-0 [DOI] [Google Scholar]
- McKinney H. H., Sando W. J. (1951). Susceptibility and resistance to the Wheat streak-mosaic virus in the genera Triticum, Agropyron, Secale, and certain hybrids. Plant Dis. Rep. 35 476–479. 10.1111/j.1744-7348.2009.00349.x [DOI] [Google Scholar]
- Schiffer M., Umina P., Carew M., Hoffmann A., Rodoni B., Miller A. (2009). The distribution of wheat curl mite (Aceria tosichella) lineages in Australia and their potential to transmit Wheat streak mosaic virus. Ann. Appl. Biol. 155 371–379. 10.1111/j.1744-7348.2009.00349.x [DOI] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., Pasternak S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326 1112–1115. 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Seifers D. L., Harvey T. L., Louie R., Gordon D. T., Martin T. J. (2002). Differential transmission of isolates of the High Plains virus by different sources of wheat curl mites. Plant Dis. 86 138–142. 10.1094/PDIS.2002.86.2.138 [DOI] [PubMed] [Google Scholar]
- Seifers D. L., Harvey T. L., Martin T. J., Jensen S. G. (1997). Identification of the wheat curl mite as the vector of the High Plains virus of corn and wheat. Plant Dis. 81 1161–1166. 10.1094/PDIS.1997.81.10.1161 [DOI] [Google Scholar]
- Seifers D. L., Martin T. J., Haber S. (2013). Temperature sensitive resistance to Wheat streak mosaic virus in CO960333 and KS06HW79 wheat. Plant Dis. 97 983–987. 10.1094/PDIS-10-12-0971-RE [DOI] [PubMed] [Google Scholar]
- Seifers D. L., Martin T. J., Harvey T. L., Fellers J. P., Michaud J. P. (2009). Identification of the wheat curl mite as the vector of Triticum mosaic virus. Plant Dis. 93 25–29. 10.1094/PDIS-93-1-0025 [DOI] [PubMed] [Google Scholar]
- Seifers D. L., Martin T. J., Harvey T. L., Fellers J. P., Stack J. P., Ryba-White M., et al. (2008). Triticum mosaic virus: a new virus isolated from wheat in Kansas. Plant Dis. 92 808–817. 10.1094/PDIS-92-5-0808 [DOI] [PubMed] [Google Scholar]
- Seifers D. L., Martin T. J., Harvey T. L., Gill B. S. (1995). Temperature sensitivity and efficacy of Wheat streak mosaic virus resistance derived from Agropyron intermedium. Plant Dis. 79 1104–1106. 10.1094/PD-90-0623 [DOI] [PubMed] [Google Scholar]
- Seifers D. L., Martin T. J., Harvey T. L., Haber S. (2007). Temperature sensitive Wheat streak mosaic virus resistance identified in KS03HW12 wheat. Plant Dis 91 1029–1033. 10.1094/PDIS-91-8-1029 [DOI] [PubMed] [Google Scholar]
- Sharp G. L., Martin J. M., Lanning S. P., Blake N. K., Brey C. W., Sivamani E., et al. (2002). Field evaluation of transgenic and classical sources of Wheat streak mosaic virus resistance. Crop Sci. 43 105–110. 10.2135/cropsci2002.1050 [DOI] [PubMed] [Google Scholar]
- Shoup Rupp J. L., Cruz L. F., Trick H. N., Fellers J. P. (2016). RNAi-mediated, stable resistance to Triticum mosaic virus in wheat. Crop Sci. 56 1602–1610. 10.2135/cropsci2015.09.0577 [DOI] [Google Scholar]
- Siriwetwiwat B. (2006). Interactions Between the Wheat Curl Mite Aceria Tosichella Keifer (Eriophyidae), and Wheat Streak Mosaic Virus and Distribution of Wheat Curl Mite Biotypes in the Field. Ph.D. thesis, University of Nebraska-Lincoln, Lincoln, NE. 10.1071/IS11037 [DOI] [Google Scholar]
- Skoracka A., Kuczyński L., de Mendonca R., Dabert M., Szydło W., Knihinicki D., et al. (2012). Cryptic species within the wheat curl mite Aceria tosichella (Keifer) (Acari, Eriophyoidea) revealed by mitochondrial, nuclear and morphometric data. Invertebr. Syst. 26 417–433. 10.1071/IS11037 [DOI] [Google Scholar]
- Skoracka A., Kuczyński L., Szydło W., Rector B. (2013). The wheat curl mite Aceria tosichella (Acari: Eriophyoidea) is a complex of cryptic lineages with divergent host ranges: evidence from molecular and plant bioassay data. Biol. J. Linn. Soc. 109 165–180. 10.1111/bij.12024 [DOI] [Google Scholar]
- Skoracka A., Lewandowski M., Rector B. G., Szydło W., Kuczyński L. (2017). Spatial and host-related variation in prevalence and population density of wheat curl mite (Aceria tosichella) cryptic genotypes in agricultural landscapes. PLoS One 12:e0169874. 10.1371/journal.pone.0169874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoracka A., Rector B., Kuczyński L., Szydło W., Hein G., French R. (2014). Global spread of wheat curl mite by its most polyphagous and pestiferous lineages. Ann. Appl. Biol. 165 222–235. 10.1111/aab.12130 [DOI] [Google Scholar]
- Slykhuis J. T. (1955). Aceria tulipae Keifer (Acarina: Eriophyidae) in relation to the spread of wheat streak mosaic. Phytopathology. 45 116–128. 10.1016/j.virol.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Stenger D. C., Hein G. L., French R. (2006). Nested deletion analysis of Wheat streak mosaic virus HC-Pro: mapping of domains affecting polyprotein processing and eriophyid mite transmission. Virology 350 465–474. 10.1016/j.virol.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Stenger D. C., Hein G. L., Tatineni S., French R. (2016). “Eriophyid mite vectors of plant viruses,” in Vector-Mediated Transmission of Plant Pathogens, ed. Brown J. K. (Saint Paul, MN: The American Phytopathological Society; ), 263–274. 10.1007/s00705-007-1065-3 [DOI] [Google Scholar]
- Stephan D., Moeller I., Skoracka A., Ehrig F., Maiss E. (2008). Eriophyid mite transmission and host range of a Brome streak mosaic virus isolate derived from a full-length cDNA clone. Arch. Virol. 153 181–185. 10.1007/s00705-007-1065-3 [DOI] [PubMed] [Google Scholar]
- Stewart L. R. (2016). Sequence diversity of wheat mosaic virus isolates. Virus Res. 213 299–303. 10.1016/j.virusres.2015.11.013 [DOI] [PubMed] [Google Scholar]
- Szydło W., Hein G., Denizhan E., Skoracka A. (2015). Exceptionally high levels of genetic diversity in wheat curl mite (Acari: Eriophyidae) populations from Turkey. J. Econ. Entomol. 108 2030–2039. 10.1093/jee/tov180 [DOI] [PubMed] [Google Scholar]
- Tatineni S., Graybosch R. A., Hein G. L., Wegulo S. N., French R. (2010). Wheat cultivar-specific disease synergism and alteration of virus accumulation during co-infection with Wheat streak mosaic virus and Triticum mosaic virus. Phytopathology. 100 230–238. 10.1094/PHYTO-100-3-0230 [DOI] [PubMed] [Google Scholar]
- Tatineni S., McMechan A. J., Wosula E. N., Wegulo S. N., Graybosch R. A., French R., et al. (2014). An eriophyid mite-transmitted plant virus contains eight genomic RNA segments with unusual heterogeneity in the nucleocapsid protein. J. Virol. 88 11834–11845. 10.1128/JVI.01901-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatineni S., McMechan A. J., Wosula E. N., Wegulo S. N., Graybosch R. A., French R., et al. (2016). An eriophyid mite-transmitted plant virus contains eight genomic RNA segments with unusual heterogeneity in the nucleocapsid protein. J. Virol. 88 11834–11845. 10.1128/JVI.01901-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. B., Conner R. L. (1986). Resistance to colonization by the wheat curl mite in Aegilops squarrosa and its inheritance after transfer to common wheat. Crop Sci. 26 527–530. 10.2135/cropsci1986.0011183X002600030019x [DOI] [Google Scholar]
- Thomas J. B., Conner R. L., Graf R. J. (2012). Radiant hard red winter wheat. Can. J. Plant Sci. 92 169–175. 10.4141/CJPS2011-082 [DOI] [Google Scholar]
- Wegulo S. N., Hein G. L., Klein R. N., French R. C. (2008). Managing Wheat Streak Mosaic.EC1871. Lincoln, NE: University of Nebraska-Lincoln; 10.1139/g88-050 [DOI] [Google Scholar]
- Whelan E. D. P., Hart G. E. (1988). A spontaneous translocation that transfers wheat curl mite resistance from decaploid Agropyron elongatum to common wheat. Genome 30 289–292. 10.1139/g88-050 [DOI] [Google Scholar]
- Whelan E. D. P., Thomas J. B. (1989). Chromosomal location in common wheat of a gene (Cmc1) from Aegilops squarrosa that conditions resistance to colonization by the wheat curl mite. Genome 32 1033–1036. 10.1139/g89-548 [DOI] [Google Scholar]
- Wilson J. R. U., Dormontt E. E., Prentis P. J., Lowe A. J., Richardson D. M. (2009). Something in the way you move: dispersal pathways affect invasion success. Trends Ecol. Evol. 24 136–144. 10.1016/j.tree.2008.10.007 [DOI] [PubMed] [Google Scholar]
- Wosula E. N., McMechan A. J., Knoell E., Tatineni S., Wegulo S. N., Hein G. L. (2018). Impact of timing and method of virus inoculation on the severity of wheat streak mosaic disease. Plant Dis. 102 645–650. 10.1094/PDIS-08-17-1227-RE [DOI] [PubMed] [Google Scholar]
- Wosula E. N., McMechan A. J., Oliveira-Hofman C., Wegulo S. N., Hein G. L. (2016). Differential transmission of two isolates of Wheat streak mosaic virus by five wheat curl mite populations. Plant Dis. 100 154–158. 10.1094/PDIS-03-15-0342-RE [DOI] [PubMed] [Google Scholar]
- Young B. A., Hein G. L., French R., Stenger D. C. (2007). Substitution of conserved cysteine residues in Wheat streak mosaic virus HC-Pro abolishes virus transmission by the wheat curl mite. Arch. Virol. 152 2107–2111. 10.1007/s00705-007-1034-x [DOI] [PubMed] [Google Scholar]
- Zhang G., Martin T. J., Fritz A. K., Miller R., Chen M., Bowden R. L., et al. (2015). Registration of ‘Oakley’ CL wheat. J. Plant Regist. 9 190–195. 10.3198/jpr2014.04.0023crc [DOI] [Google Scholar]
- Zimin A. V., Puiu D., Hall R., Kingan S., Clavijo B. J., Salzberg S. L. (2017). The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. GigaScience 6 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]