The title compounds, (CH2)nC3H2O(COOH)2 (n = 4, 5), display intermolecular hydrogen bonding, forming a two-dimensional framework.
Keywords: tartric acid; 1,3-dioxolane; ketals; NMR; hydrogen bonding; crystal structure
Abstract
The title compounds, C9H12O6 and C10H14O6, were formed by careful hydrolysis of the corresponding diethyl esters. Their single crystals were grown from an ethyl acetate/hexane mixture. Crystals of both compounds have monoclinic (P21) symmetry with a single molecule in the asymmetric unit. Both crystal structures are very similar and display four –CO—OH⋯O=C(OH)– hydrogen bonds, forming a two-dimensional double-layered framework.
Chemical context
Transition-metal catalysis has developed as a powerful tool to create a variety of carbon–carbon and carbon–heteroatom bonds. Enantioselective versions of these reactions are especially interesting in the light of the possible pharmaceutical applications. The general route to such processes supposes the use of transition metal complexes with chiral ligands (Yang et al., 2017 ▸). Therefore, easily accessible ligands of this type are of great importance for homogenous catalysis. Chiral phosphine ligands and amino acids are the most popular in this respect (Crassous, 2009 ▸). Examples of chiral carboxylate ligands are also known (Saget et al., 2012 ▸), which can be useful in the synthesis of chiral coordination compounds and materials derived from them (Lam et al., 2011 ▸). Various tartaric acid derivatives, which are also used in organic synthesis as chiral auxiliary agents to create chiral building blocks (Kassai et al., 2000 ▸; Seebach et al., 2001 ▸), might be particularly useful in solving the stated problem. Herein we report the synthesis and structures of two tartaric acid derivatives that may potentially be used as synthetic precursors of chiral transition-metal catalysts.
Condensation of cyclopentanone or cyclohexanone with (2R,3R) diethyl tartrate led to the formation of the corresponding ketals, careful hydrolysis of which allowed us to prepare the title acids (Fig. 1 ▸).
Figure 1.
The synthesis of the title compounds (I) and (II).
Structural commentary
The structures of tartaric acid derivatives (I) and (II) were found as anticipated (Figs. 2 ▸ and 3 ▸, respectively), having a single molecule in the asymmetric unit. The 1,3-dioxolane, cyclopentane [in (I)] and cyclohexane [in (II)] fragments have the usual conformations. The C—C, C—O and C=O bond lengths are within regular distances (Tables 1 ▸ and 2 ▸). A detailed structural and conformational analysis for the crystal structures of some related acetals R′C3H3O2(COR)2 (R = NH2, OAlkyl, OH; substituent R′ is at the 2-position of the 1,3-dioxolane ring) was given by Eissmann et al. (2012 ▸). Although the absolute structures of (I) and (II) cannot be unambiguously determined using the Flack parameter (Flack, 1983 ▸; Parsons, et al., 2013 ▸) with the SHELXL program (Sheldrick, 2015 ▸), the chirality at carbon atoms C2, C3 (2R,3R) is initially known from their synthetic precursor (diethyl l-tartrate), and has been also confirmed for (2R,3R)-diethyl 1,4-dioxaspiro[4.5]decane-2,3-dicarboxylate, and for (II) by optical rotation measurements (see the experimental section). The molecules of (I) and (II) have very similar positions in the unit cells, making the structures nearly isomorphous, but the c axis in (II) is elongated by almost 1.5 Å compared with that in (I) (see Table 5 ▸ below) because of the presence of an additional –(CH2)– unit in the cycloalkane fragment in (II) (see Fig. 4 ▸ for the alignment of the cycloalkane fragments in the unit cell).
Figure 2.
The structure of (2R,3R)-1,4-dioxaspiro[4.4]nonane-2,3-dicarboxylic acid, (I). Displacement ellipsoids are drawn at the 50% probability level.
Figure 3.
The structure of (2R,3R)-1,4-dioxaspiro[4.5]decane-2,3-dicarboxylic acid, (II). Displacement ellipsoids are drawn at the 50% probability level.
Table 1. Selected bond lengths (Å) for (I) .
| O1—C1 | 1.325 (3) | C1—C2 | 1.521 (4) |
| O2—C1 | 1.208 (3) | C2—C3 | 1.541 (4) |
| O3—C4 | 1.222 (3) | C3—C4 | 1.519 (4) |
| O4—C4 | 1.314 (3) | C5—C9 | 1.529 (4) |
| O5—C2 | 1.409 (3) | C5—C6 | 1.539 (4) |
| O5—C5 | 1.443 (3) | C6—C7 | 1.533 (4) |
| O6—C3 | 1.409 (3) | C7—C8 | 1.529 (4) |
| O6—C5 | 1.439 (3) | C8—C9 | 1.522 (4) |
Table 2. Selected bond lengths (Å) for (II) .
| O1—C1 | 1.322 (2) | C2—C3 | 1.541 (2) |
| O2—C1 | 1.208 (2) | C3—C4 | 1.532 (2) |
| O3—C4 | 1.2229 (19) | C5—C6 | 1.519 (2) |
| O4—C4 | 1.3135 (18) | C5—C10 | 1.525 (2) |
| O5—C2 | 1.4107 (18) | C6—C7 | 1.533 (2) |
| O5—C5 | 1.4398 (18) | C7—C8 | 1.526 (3) |
| O6—C3 | 1.4135 (17) | C8—C9 | 1.532 (2) |
| O6—C5 | 1.441 (2) | C9—C10 | 1.538 (2) |
| C1—C2 | 1.529 (2) |
Table 5. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C9H12O6 | C10H14O6 |
| M r | 216.19 | 230.21 |
| Crystal system, space group | Monoclinic, P21 | Monoclinic, P21 |
| Temperature (K) | 100 | 100 |
| a, b, c (Å) | 6.2930 (8), 5.3712 (7), 14.0916 (17) | 6.4272 (8), 5.2976 (6), 15.5678 (19) |
| β (°) | 92.885 (2) | 94.469 (2) |
| V (Å3) | 475.71 (10) | 528.45 (11) |
| Z | 2 | 2 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.13 | 0.12 |
| Crystal size (mm) | 0.21 × 0.07 × 0.03 | 0.39 × 0.15 × 0.05 |
| Data collection | ||
| Diffractometer | Bruker SMART APEXII | Bruker SMART APEXII |
| Absorption correction | Multi-scan (SADABS; Bruker, 2008 ▸) | Multi-scan (SADABS; Bruker, 2008 ▸) |
| T min, T max | 0.827, 0.996 | 0.917, 0.995 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 4142, 2455, 1914 | 4329, 2612, 2503 |
| R int | 0.030 | 0.015 |
| (sin θ/λ)max (Å−1) | 0.682 | 0.682 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.043, 0.083, 1.06 | 0.028, 0.072, 1.05 |
| No. of reflections | 2455 | 2612 |
| No. of parameters | 144 | 153 |
| No. of restraints | 1 | 1 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.29, −0.24 | 0.30, −0.20 |
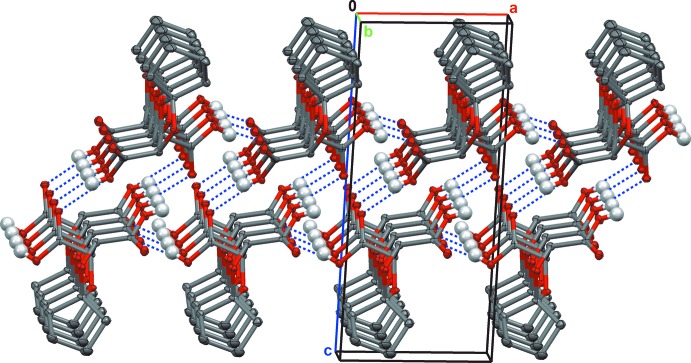
Figure 4.
The packing of (I) parallel to (010). Two interacting molecular layers are shown. Only the H atoms involved in hydrogen bonding (blue dashed lines) have been included. Displacement ellipsoids are drawn at the 50% probability level.
Supramolecular features
The molecules of both structures are packed in two-dimensional frameworks by four –CO—OH⋯O=C(OH)– hydrogen bonds between neighboring carboxyl groups (Tables 3 ▸ and 4 ▸). The packing diagrams for (I) (Figs. 4 ▸, 5 ▸ a) are nearly identical to those of (II) (not shown). The molecules form double layers parallel to the ab plane and sterically shielded from other layers by the cycloalkane fragments (Fig. 4 ▸). Hydrogen-bonded chains within the same layer are formed via two interactions involving the O1—H1 and O3 atoms of each molecule. These chains are interconnected into a two-dimensional hydrogen-bonded double-layered framework parallel to (001) by the O4—H4 and O2 atoms. The complicated structure of the two-dimensional double-layered framework is shown in Fig. 5 ▸ a, but it can best be visualized in the simplified scheme in Fig. 5 ▸ b. It might be noted that some weak C—H⋯O intermolecular interactions are also present (see the supporting information).
Table 3. Hydrogen-bond geometry (Å, °) for (I) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O3i | 0.78 (4) | 1.87 (4) | 2.620 (3) | 159 (4) |
| O4—H4⋯O2ii | 0.84 (4) | 1.92 (4) | 2.723 (3) | 159 (3) |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Table 4. Hydrogen-bond geometry (Å, °) for (II) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O3i | 0.84 (3) | 1.80 (3) | 2.6230 (16) | 164 (3) |
| O4—H4⋯O2ii | 0.85 (3) | 1.88 (3) | 2.7116 (16) | 164 (2) |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Figure 5.
(a) The packing of (I) parallel to (001). Two interacting molecular layers are shown. Only the H atoms involved in hydrogen bonding (blue dashed lines) have been included. Displacement ellipsoids are drawn at the 50% probability level. (b) The simplified structure of the two-dimensional double-layered framework. Molecules (circles) and hydrogen bonds (solid lines) within the same layers are shown in the same colour (blue or red). Hydrogen bonds between two layers are shown as solid black lines.
Database survey
Twenty crystal structures of tartaric acid ester derivatives possessing the 1,3-dioxolane cycle, R′R′′C3H2O2(COOR)2, are known to date [Cambridge Structural Database (CSD) Version 5.39, latest update Feb 2018; Groom et al., 2016 ▸]. There are 10 crystal structures of esters bearing one substituent R′ (R′′ = H) at the 2-position of the 1,3-dioxolane fragment (acetals): CSD refcodes DAZJET (Lee et al., 1999 ▸), LACREM, LACRUC (Roush et al., 1992 ▸), LEPHAR, LEPHEV (Eissmann et al., 2012 ▸), OLEGAN (Karisalmi et al., 2003 ▸), WEGXOW (Belokon’ et al., 2005 ▸), XEYSEA (Jiang et al., 2007 ▸), YAXHIQ (Lv et al., 2012 ▸) and YIVGUF (Barrett et al., 1995 ▸). The crystal structures of esters with two substituents R′ and R′′ (ketals) are represented by GAGHAY, GUHGUL (Pelphrey et al., 2004 ▸), KEMRID (Wink & Dewan, 1990 ▸), MIWDIF (Ates & Curran, 2001 ▸), NAFWEW (Mikołajczyk et al., 1996 ▸), QOTVUQ (Maezaki et al., 2000 ▸), VICXOU/VICXOU10 (Giordano et al., 1990 ▸; Ianelli et al., 1992 ▸), VIHVAL (Linker et al., 2013 ▸) and VUCHAC, VUCHEG (Ianelli et al., 1992 ▸). The crystal structures of 14 related amide derivatives R′R′′C3H2O2(CONR 2)2 are also known (see the CSD and also Eissmann et al., 2012 ▸ and references therein). However, established crystal structures of related acids, R′R′′C3H2O2(COOH)2, are limited to only one structure with R′ = –C6H4-4-COOH and R′′ = H (LEPHIZ; Eissmann et al., 2012 ▸). This fact can be explained by some subtle problems with the individual isolation of pure acid samples because of the facile hydrolysis of the 1,3-dioxolane fragment during their preparation. Therefore, the synthesis and especially the crystallization of R′R′′C3H2O2(COOH)2 acids is a challenging task.
Synthesis and crystallization
General experimental remarks
(+)-Diethyl l-tartrate [Sigma–Aldrich, >99%, found [α]D 297K = +12° (acetone, 20.5mg ml−1); lit. data [α]D 293K = +10° (ethanol, 53 mg ml−1), see Černý, 1977 ▸] was used as purchased. 1H and 13C{1H} NMR spectra were recorded with Bruker AM-300 and Bruker DRX-500 spectrometers in CDCl3 (Cambridge Isotope Laboratories, Inc., 99.8% 2H) and in acetone-d 6 (Sigma–Aldrich, 99.9 atom % 2H).
Synthesis of (2R,3R)-diethyl 1,4-dioxaspiro[4.5]decane-2,3-dicarboxylate
A 1000 ml round-bottomed flask equipped with a reflux condenser and a Dean–Stark trap was charged with diethyl l-tartrate (85.56 ml, 500 mmol), cyclohexanone (51.82 ml, 500 mmol), toluene (600 ml) and p-toluenesulfonic acid monohydrate (2.80 g, 147 mmol). The mixture was refluxed for 62 h. The resulting dark-brown mixture was washed with a saturated aqueous solution of NaHCO3 (2 × 100ml) and with water (2 × 100ml). The organic layer was dried over anhydrous Na2SO4. The solvent was removed on a rotary evaporator. The obtained dark-brown oil was distilled under reduced pressure (388–391 K, 250 Pa). The yield of the colourless liquid was 84% (120.25 g, 420 mmol). ηD 293K = 1.4625, [α]D 297K = −28.7 (acetone, 20.5 mg ml−1) [Lit. data ηD 293K = 1.4605, [α]D 293K = −35.57 (Tsuzuki, 1937 ▸)]. 1H NMR (CDCl3) δ: 1.12 (t, 6H, CH3—CH2–O), 1.30–1.45 (m, 10H, –C5 H10–), 4.10 (quartet, 4H, CH3—CH2—O), 4.55 (s, 2H, CH).
Synthesis of (2R,3R)-diethyl 1,4-dioxaspiro[4.4]nonane-2,3-dicarboxylate
The synthesis of (2R,3R)-diethyl 1,4-dioxaspiro[4.4]nonane-2,3-dicarboxylate was carried out analogously to that of (2R,3R)-diethyl 1,4-dioxaspiro[4.5]decane-2,3-dicarboxylate, starting from 85.47 ml (500 mmol) of diethyl l-tartrate, 44.23 ml (500 mmol) of cyclopentanone, 600 ml of toluene and 2.80 g (14.7 mmol) of p-toluenesulfonic acid monohydrate. The yield of the colourless liquid after vacuum distillation (383–385 K, 265 Pa) was 78% (106.08 g, 390 mmol). 1H NMR (CDCl3) δ: 1.17 (t, 6H, CH3—CH2—O), 1.51–1.63 (m, 4H, –C4 H8–), 1.64–1.77 (m, 2H, –C4 H8–), 1.77–1.91 (m, 2H, –C4 H8–), 4.12 (quartet, 4H, CH3—CH2—O), 4.57 (s, 2H, CH).
Synthesis and crystallization of (2R,3R)-1,4-dioxaspiro[4.5]decane-2,3-dicarboxylic acid, (II)
A 100 ml round-bottomed flask was charged with 2.130 g (7.52 mmol) of (2R,3R)-diethyl 1,4-dioxaspiro[4.5]decane-2,3-dicarboxylate, 22.5 ml of THF, 22.5 ml of methanol and 22.5 ml of 2 M aqueous solution of LiOH. The reaction mixture was stirred for 6 h. It was then washed with diethyl ether (3 × 20 ml). The aqueous solution was acidified with a 2 M solution of HCl to pH ≃ 1 at 273 K. The formed acid was extracted with ethyl acetate (3 × 20 ml). The organic layer was dried over Na2SO4. The solution was removed on a rotary evaporator. The yield of the resulting white powder was 72% (1.250 g, 5.43 mmol). M.p. = 413K, [α]D 297K = −27.3 (acetone, 20.5 mg ml−1) [Lit. data [α]D 20 = −24.0, ethanol, 304 mg ml−1 (Innis & Lamaty, 1977 ▸)]. 1H NMR (acetone-d 6) δ: 1.36–1.44 (m, 2H, —C5 H10—), 1.53-1.74 (m, 8H, –C5 H10–), 4.82 (s, 2H, CH), 7.0 (br.s, 2H, –COOH). 13C{1H} NMR (acetone-d 6) δ: 24.6, 25.6, 36.8, 77.7, 114.7, 171.7. Crystals of (II) were grown from an ethyl acetate/hexane (1:1 v/v) mixture.
Synthesis and crystallization of (2R,3R)-1,4-dioxaspiro[4.4]nonane-2,3-dicarboxylic acid, (I)
The synthesis of (I) was carried out analogously to that of (II), starting from 2.723 g (10 mmol) of (2R,3R)-diethyl 1,4-dioxaspiro[4.4]nonane-2,3-dicarboxylate, 22.5 ml of THF, 22.5 ml of methanol and 22.5 ml of 2 M aqueous solution of LiOH. The yield of the resulting white powder was 50% (1.081 g, 5 mmol). 1H NMR (acetone-d 6) δ: 1.59–1.72 (m, 4H, –C4 H8–), 1.74–1.87 (m, 2H, –C4 H8–), 1.90–2.02 (m, 2H, –C4 H8–), 4.78 (s, 2H, CH), 7.5 (br.s, 2H, -COOH). 13C{1H} NMR (acetone-d 6) δ: 24.0, 37.4, 77.9, 123.6, 171.4. Crystals of (I) were grown from an ethyl acetate/hexane (1:1 v/v) mixture.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 5 ▸. The positions of all non-H and the hydroxy H atoms were found from the electron difference density maps. These atoms were refined with individual anisotropic (non-H) or isotropic (hydroxy H) displacement parameters. The positions of the other H atoms were also found from the difference map but they were positioned geometrically (C—H distance = 0.99 Å for methylene, 1.00 Å for tertiary hydrogen atoms) and refined as riding atoms with U iso(H) = 1.2U eq(C). Reflection (001) in (II) was affected by the beam stop, and was therefore omitted from the refinement.
Supplementary Material
Crystal structure: contains datablock(s) I, II, global. DOI: 10.1107/S2056989018009593/eb2009sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018009593/eb2009Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989018009593/eb2009IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989018009593/eb2009Isup4.cml
Supporting information file. DOI: 10.1107/S2056989018009593/eb2009IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
Equipment from the collective exploitation center ‘New petrochemical processes, polymer composites and adhesives’ of TIPS RAS was used.
supplementary crystallographic information
(2R,3R)-1,4-Dioxaspiro[4.4]nonane-2,3-dicarboxylic acid (I) . Crystal data
| C9H12O6 | F(000) = 228 |
| Mr = 216.19 | Dx = 1.509 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.2930 (8) Å | Cell parameters from 621 reflections |
| b = 5.3712 (7) Å | θ = 3–29° |
| c = 14.0916 (17) Å | µ = 0.13 mm−1 |
| β = 92.885 (2)° | T = 100 K |
| V = 475.71 (10) Å3 | Needle, colourless |
| Z = 2 | 0.21 × 0.07 × 0.03 mm |
(2R,3R)-1,4-Dioxaspiro[4.4]nonane-2,3-dicarboxylic acid (I) . Data collection
| Bruker SMART APEXII diffractometer | 2455 independent reflections |
| Radiation source: fine-focus sealed tube | 1914 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.030 |
| ω scans | θmax = 29.0°, θmin = 2.9° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −8→8 |
| Tmin = 0.827, Tmax = 0.996 | k = −7→7 |
| 4142 measured reflections | l = −14→19 |
(2R,3R)-1,4-Dioxaspiro[4.4]nonane-2,3-dicarboxylic acid (I) . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.043 | Hydrogen site location: mixed |
| wR(F2) = 0.083 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.031P)2] where P = (Fo2 + 2Fc2)/3 |
| 2455 reflections | (Δ/σ)max < 0.001 |
| 144 parameters | Δρmax = 0.29 e Å−3 |
| 1 restraint | Δρmin = −0.24 e Å−3 |
(2R,3R)-1,4-Dioxaspiro[4.4]nonane-2,3-dicarboxylic acid (I) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
(2R,3R)-1,4-Dioxaspiro[4.4]nonane-2,3-dicarboxylic acid (I) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.3838 (4) | 0.5521 (4) | 0.45124 (15) | 0.0153 (5) | |
| H1 | 0.271 (6) | 0.587 (8) | 0.468 (3) | 0.039 (13)* | |
| O2 | 0.3808 (3) | 0.8867 (4) | 0.35486 (14) | 0.0156 (5) | |
| O3 | 0.9421 (3) | 0.1722 (4) | 0.44956 (14) | 0.0133 (5) | |
| O4 | 1.0688 (3) | 0.2205 (4) | 0.30548 (14) | 0.0141 (5) | |
| H4 | 1.173 (6) | 0.144 (7) | 0.332 (2) | 0.026 (10)* | |
| O5 | 0.7890 (3) | 0.7805 (3) | 0.30380 (14) | 0.0123 (5) | |
| O6 | 0.6993 (3) | 0.4106 (4) | 0.23156 (13) | 0.0127 (4) | |
| C1 | 0.4679 (4) | 0.7060 (5) | 0.38994 (19) | 0.0108 (6) | |
| C2 | 0.6954 (4) | 0.6301 (5) | 0.3722 (2) | 0.0113 (6) | |
| H2 | 0.782988 | 0.639980 | 0.433289 | 0.014* | |
| C3 | 0.7155 (4) | 0.3667 (5) | 0.33022 (19) | 0.0103 (6) | |
| H3 | 0.591522 | 0.263933 | 0.348493 | 0.012* | |
| C4 | 0.9202 (4) | 0.2424 (5) | 0.3672 (2) | 0.0105 (6) | |
| C5 | 0.7661 (4) | 0.6609 (5) | 0.2121 (2) | 0.0124 (6) | |
| C6 | 0.6025 (5) | 0.7952 (6) | 0.1454 (2) | 0.0174 (7) | |
| H6A | 0.481549 | 0.683619 | 0.127887 | 0.021* | |
| H6B | 0.547171 | 0.945453 | 0.176471 | 0.021* | |
| C7 | 0.7213 (5) | 0.8670 (7) | 0.0571 (2) | 0.0276 (8) | |
| H7A | 0.704227 | 0.737514 | 0.007304 | 0.033* | |
| H7B | 0.669165 | 1.027909 | 0.030732 | 0.033* | |
| C8 | 0.9535 (5) | 0.8873 (7) | 0.0939 (2) | 0.0220 (7) | |
| H8A | 1.051505 | 0.869535 | 0.041494 | 0.026* | |
| H8B | 0.981059 | 1.048463 | 0.126290 | 0.026* | |
| C9 | 0.9776 (4) | 0.6711 (6) | 0.1634 (2) | 0.0161 (6) | |
| H9A | 1.097196 | 0.700938 | 0.210352 | 0.019* | |
| H9B | 1.003536 | 0.513540 | 0.129416 | 0.019* |
(2R,3R)-1,4-Dioxaspiro[4.4]nonane-2,3-dicarboxylic acid (I) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0145 (11) | 0.0154 (11) | 0.0167 (13) | 0.0014 (9) | 0.0066 (9) | 0.0018 (10) |
| O2 | 0.0148 (10) | 0.0150 (10) | 0.0172 (12) | 0.0050 (9) | 0.0011 (8) | 0.0010 (10) |
| O3 | 0.0147 (10) | 0.0140 (10) | 0.0112 (11) | 0.0015 (8) | 0.0007 (8) | 0.0017 (9) |
| O4 | 0.0120 (10) | 0.0154 (11) | 0.0153 (11) | 0.0031 (8) | 0.0034 (8) | 0.0025 (9) |
| O5 | 0.0164 (10) | 0.0109 (10) | 0.0100 (11) | −0.0014 (8) | 0.0030 (8) | −0.0012 (8) |
| O6 | 0.0178 (10) | 0.0102 (9) | 0.0099 (11) | −0.0003 (8) | −0.0003 (8) | −0.0005 (9) |
| C1 | 0.0146 (13) | 0.0093 (12) | 0.0085 (14) | −0.0004 (11) | −0.0002 (11) | −0.0040 (12) |
| C2 | 0.0136 (14) | 0.0087 (13) | 0.0117 (16) | 0.0012 (10) | 0.0015 (11) | −0.0005 (11) |
| C3 | 0.0127 (13) | 0.0083 (12) | 0.0103 (15) | −0.0005 (10) | 0.0021 (11) | 0.0001 (12) |
| C4 | 0.0100 (13) | 0.0063 (13) | 0.0151 (16) | −0.0005 (10) | 0.0001 (11) | −0.0031 (12) |
| C5 | 0.0171 (14) | 0.0110 (13) | 0.0091 (15) | 0.0011 (11) | 0.0019 (11) | −0.0020 (12) |
| C6 | 0.0183 (15) | 0.0182 (14) | 0.0155 (17) | 0.0050 (12) | −0.0003 (13) | −0.0006 (13) |
| C7 | 0.0310 (18) | 0.035 (2) | 0.0167 (18) | 0.0068 (16) | 0.0021 (14) | 0.0092 (16) |
| C8 | 0.0258 (16) | 0.0225 (16) | 0.0185 (18) | 0.0003 (14) | 0.0089 (13) | 0.0071 (15) |
| C9 | 0.0164 (14) | 0.0163 (15) | 0.0160 (17) | 0.0014 (12) | 0.0047 (12) | 0.0012 (14) |
(2R,3R)-1,4-Dioxaspiro[4.4]nonane-2,3-dicarboxylic acid (I) . Geometric parameters (Å, º)
| O1—C1 | 1.325 (3) | C3—H3 | 1.0000 |
| O1—H1 | 0.78 (4) | C5—C9 | 1.529 (4) |
| O2—C1 | 1.208 (3) | C5—C6 | 1.539 (4) |
| O3—C4 | 1.222 (3) | C6—C7 | 1.533 (4) |
| O4—C4 | 1.314 (3) | C6—H6A | 0.9900 |
| O4—H4 | 0.84 (4) | C6—H6B | 0.9900 |
| O5—C2 | 1.409 (3) | C7—C8 | 1.529 (4) |
| O5—C5 | 1.443 (3) | C7—H7A | 0.9900 |
| O6—C3 | 1.409 (3) | C7—H7B | 0.9900 |
| O6—C5 | 1.439 (3) | C8—C9 | 1.522 (4) |
| C1—C2 | 1.521 (4) | C8—H8A | 0.9900 |
| C2—C3 | 1.541 (4) | C8—H8B | 0.9900 |
| C2—H2 | 1.0000 | C9—H9A | 0.9900 |
| C3—C4 | 1.519 (4) | C9—H9B | 0.9900 |
| C1—O1—H1 | 117 (3) | O5—C5—C6 | 111.9 (2) |
| C4—O4—H4 | 108 (2) | C9—C5—C6 | 106.2 (2) |
| C2—O5—C5 | 109.3 (2) | C7—C6—C5 | 106.0 (2) |
| C3—O6—C5 | 109.7 (2) | C7—C6—H6A | 110.5 |
| O2—C1—O1 | 125.4 (2) | C5—C6—H6A | 110.5 |
| O2—C1—C2 | 124.0 (2) | C7—C6—H6B | 110.5 |
| O1—C1—C2 | 110.5 (2) | C5—C6—H6B | 110.5 |
| O5—C2—C1 | 112.8 (2) | H6A—C6—H6B | 108.7 |
| O5—C2—C3 | 102.6 (2) | C8—C7—C6 | 103.9 (3) |
| C1—C2—C3 | 113.9 (2) | C8—C7—H7A | 111.0 |
| O5—C2—H2 | 109.1 | C6—C7—H7A | 111.0 |
| C1—C2—H2 | 109.1 | C8—C7—H7B | 111.0 |
| C3—C2—H2 | 109.1 | C6—C7—H7B | 111.0 |
| O6—C3—C4 | 115.5 (2) | H7A—C7—H7B | 109.0 |
| O6—C3—C2 | 102.8 (2) | C9—C8—C7 | 103.1 (3) |
| C4—C3—C2 | 110.9 (2) | C9—C8—H8A | 111.1 |
| O6—C3—H3 | 109.1 | C7—C8—H8A | 111.1 |
| C4—C3—H3 | 109.1 | C9—C8—H8B | 111.1 |
| C2—C3—H3 | 109.1 | C7—C8—H8B | 111.1 |
| O3—C4—O4 | 123.4 (2) | H8A—C8—H8B | 109.1 |
| O3—C4—C3 | 121.0 (2) | C8—C9—C5 | 104.8 (2) |
| O4—C4—C3 | 115.6 (2) | C8—C9—H9A | 110.8 |
| O6—C5—O5 | 105.2 (2) | C5—C9—H9A | 110.8 |
| O6—C5—C9 | 112.9 (2) | C8—C9—H9B | 110.8 |
| O5—C5—C9 | 109.6 (2) | C5—C9—H9B | 110.8 |
| O6—C5—C6 | 111.2 (2) | H9A—C9—H9B | 108.9 |
| C5—O5—C2—C1 | −95.7 (2) | C3—O6—C5—O5 | −9.5 (3) |
| C5—O5—C2—C3 | 27.4 (3) | C3—O6—C5—C9 | 110.0 (2) |
| O2—C1—C2—O5 | −5.2 (4) | C3—O6—C5—C6 | −130.8 (2) |
| O1—C1—C2—O5 | 177.0 (2) | C2—O5—C5—O6 | −12.6 (3) |
| O2—C1—C2—C3 | −121.7 (3) | C2—O5—C5—C9 | −134.3 (2) |
| O1—C1—C2—C3 | 60.5 (3) | C2—O5—C5—C6 | 108.2 (2) |
| C5—O6—C3—C4 | −95.4 (3) | O6—C5—C6—C7 | −120.1 (3) |
| C5—O6—C3—C2 | 25.5 (3) | O5—C5—C6—C7 | 122.6 (3) |
| O5—C2—C3—O6 | −32.0 (2) | C9—C5—C6—C7 | 3.0 (3) |
| C1—C2—C3—O6 | 90.3 (3) | C5—C6—C7—C8 | −26.1 (3) |
| O5—C2—C3—C4 | 92.0 (3) | C6—C7—C8—C9 | 39.3 (3) |
| C1—C2—C3—C4 | −145.7 (2) | C7—C8—C9—C5 | −37.6 (3) |
| O6—C3—C4—O3 | −172.3 (2) | O6—C5—C9—C8 | 143.4 (3) |
| C2—C3—C4—O3 | 71.3 (3) | O5—C5—C9—C8 | −99.7 (3) |
| O6—C3—C4—O4 | 8.6 (3) | C6—C5—C9—C8 | 21.4 (3) |
| C2—C3—C4—O4 | −107.8 (3) |
(2R,3R)-1,4-Dioxaspiro[4.4]nonane-2,3-dicarboxylic acid (I) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O3i | 0.78 (4) | 1.87 (4) | 2.620 (3) | 159 (4) |
| O4—H4···O2ii | 0.84 (4) | 1.92 (4) | 2.723 (3) | 159 (3) |
| C2—H2···O3iii | 1.00 | 2.34 | 3.315 (4) | 166 |
| C3—H3···O2iv | 1.00 | 2.43 | 3.358 (3) | 155 |
Symmetry codes: (i) −x+1, y+1/2, −z+1; (ii) x+1, y−1, z; (iii) −x+2, y+1/2, −z+1; (iv) x, y−1, z.
(2R,3R)-1,4-Dioxaspiro[4.5]decane-2,3-dicarboxylic acid (II) . Crystal data
| C10H14O6 | Dx = 1.447 Mg m−3 |
| Mr = 230.21 | Melting point: 413(1) K |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.4272 (8) Å | Cell parameters from 361 reflections |
| b = 5.2976 (6) Å | θ = 3–29° |
| c = 15.5678 (19) Å | µ = 0.12 mm−1 |
| β = 94.469 (2)° | T = 100 K |
| V = 528.45 (11) Å3 | Needle, colourless |
| Z = 2 | 0.39 × 0.15 × 0.05 mm |
| F(000) = 244 |
(2R,3R)-1,4-Dioxaspiro[4.5]decane-2,3-dicarboxylic acid (II) . Data collection
| Bruker SMART APEXII diffractometer | 2612 independent reflections |
| Radiation source: fine-focus sealed tube | 2503 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.015 |
| ω scans | θmax = 29.0°, θmin = 2.6° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −8→8 |
| Tmin = 0.917, Tmax = 0.995 | k = −7→5 |
| 4329 measured reflections | l = −21→20 |
(2R,3R)-1,4-Dioxaspiro[4.5]decane-2,3-dicarboxylic acid (II) . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.028 | Hydrogen site location: mixed |
| wR(F2) = 0.072 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0411P)2 + 0.0783P] where P = (Fo2 + 2Fc2)/3 |
| 2612 reflections | (Δ/σ)max < 0.001 |
| 153 parameters | Δρmax = 0.30 e Å−3 |
| 1 restraint | Δρmin = −0.20 e Å−3 |
(2R,3R)-1,4-Dioxaspiro[4.5]decane-2,3-dicarboxylic acid (II) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
(2R,3R)-1,4-Dioxaspiro[4.5]decane-2,3-dicarboxylic acid (II) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.38484 (18) | 0.5840 (2) | 0.45476 (7) | 0.0142 (2) | |
| H1 | 0.275 (4) | 0.638 (6) | 0.4745 (17) | 0.046 (8)* | |
| O2 | 0.37180 (17) | 0.9308 (2) | 0.37124 (7) | 0.0150 (2) | |
| O3 | 0.92950 (17) | 0.2135 (2) | 0.45684 (7) | 0.0124 (2) | |
| O4 | 1.05961 (17) | 0.2630 (2) | 0.32878 (7) | 0.0134 (2) | |
| H4 | 1.158 (4) | 0.171 (5) | 0.3519 (15) | 0.027 (6)* | |
| O5 | 0.76550 (18) | 0.8366 (2) | 0.32166 (7) | 0.0118 (2) | |
| O6 | 0.70176 (17) | 0.4547 (2) | 0.25475 (7) | 0.0116 (2) | |
| C1 | 0.4607 (2) | 0.7451 (3) | 0.40041 (9) | 0.0104 (3) | |
| C2 | 0.6846 (2) | 0.6758 (3) | 0.38313 (9) | 0.0099 (3) | |
| H2 | 0.774689 | 0.688258 | 0.438295 | 0.012* | |
| C3 | 0.7100 (2) | 0.4108 (3) | 0.34451 (9) | 0.0099 (3) | |
| H3 | 0.588324 | 0.303877 | 0.357764 | 0.012* | |
| C4 | 0.9117 (2) | 0.2852 (3) | 0.38196 (10) | 0.0103 (3) | |
| C5 | 0.7505 (2) | 0.7152 (3) | 0.23867 (9) | 0.0109 (3) | |
| C6 | 0.9603 (2) | 0.7413 (3) | 0.20088 (9) | 0.0145 (3) | |
| H6A | 1.069512 | 0.658928 | 0.239569 | 0.017* | |
| H6B | 0.996255 | 0.922393 | 0.196647 | 0.017* | |
| C7 | 0.9553 (3) | 0.6204 (4) | 0.11126 (11) | 0.0208 (4) | |
| H7A | 1.090749 | 0.648573 | 0.086579 | 0.025* | |
| H7B | 0.934204 | 0.435992 | 0.116324 | 0.025* | |
| C8 | 0.7800 (3) | 0.7324 (4) | 0.05127 (10) | 0.0233 (4) | |
| H8A | 0.775479 | 0.646187 | −0.005267 | 0.028* | |
| H8B | 0.807902 | 0.913725 | 0.041928 | 0.028* | |
| C9 | 0.5691 (3) | 0.7025 (4) | 0.08968 (10) | 0.0191 (3) | |
| H9A | 0.535628 | 0.520808 | 0.094378 | 0.023* | |
| H9B | 0.458492 | 0.782178 | 0.050960 | 0.023* | |
| C10 | 0.5734 (2) | 0.8254 (3) | 0.17929 (10) | 0.0151 (3) | |
| H10A | 0.593000 | 1.009889 | 0.173941 | 0.018* | |
| H10B | 0.438602 | 0.795642 | 0.204323 | 0.018* |
(2R,3R)-1,4-Dioxaspiro[4.5]decane-2,3-dicarboxylic acid (II) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0135 (5) | 0.0139 (6) | 0.0160 (5) | 0.0025 (5) | 0.0065 (4) | 0.0026 (5) |
| O2 | 0.0139 (5) | 0.0142 (6) | 0.0172 (5) | 0.0044 (4) | 0.0029 (4) | 0.0027 (5) |
| O3 | 0.0127 (5) | 0.0125 (6) | 0.0122 (5) | 0.0009 (4) | 0.0014 (4) | 0.0004 (4) |
| O4 | 0.0115 (5) | 0.0133 (6) | 0.0157 (5) | 0.0032 (4) | 0.0036 (4) | 0.0017 (5) |
| O5 | 0.0156 (5) | 0.0092 (5) | 0.0111 (5) | −0.0026 (4) | 0.0036 (4) | −0.0012 (4) |
| O6 | 0.0156 (5) | 0.0087 (5) | 0.0104 (5) | −0.0011 (4) | 0.0005 (4) | 0.0002 (4) |
| C1 | 0.0114 (6) | 0.0102 (7) | 0.0097 (6) | −0.0006 (5) | 0.0010 (5) | −0.0031 (6) |
| C2 | 0.0101 (6) | 0.0087 (7) | 0.0110 (6) | 0.0007 (5) | 0.0020 (5) | 0.0006 (5) |
| C3 | 0.0097 (6) | 0.0090 (7) | 0.0111 (6) | 0.0015 (5) | 0.0011 (5) | −0.0007 (5) |
| C4 | 0.0099 (6) | 0.0066 (7) | 0.0145 (7) | −0.0003 (5) | 0.0008 (5) | −0.0011 (5) |
| C5 | 0.0125 (6) | 0.0095 (7) | 0.0108 (6) | 0.0003 (5) | 0.0020 (5) | −0.0007 (6) |
| C6 | 0.0141 (7) | 0.0158 (8) | 0.0142 (6) | −0.0018 (6) | 0.0048 (5) | −0.0016 (6) |
| C7 | 0.0203 (8) | 0.0265 (10) | 0.0165 (7) | −0.0028 (7) | 0.0079 (6) | −0.0053 (7) |
| C8 | 0.0288 (9) | 0.0291 (10) | 0.0126 (7) | −0.0068 (8) | 0.0054 (6) | −0.0003 (7) |
| C9 | 0.0208 (8) | 0.0239 (10) | 0.0122 (7) | −0.0016 (7) | −0.0008 (6) | 0.0028 (7) |
| C10 | 0.0149 (7) | 0.0158 (8) | 0.0145 (7) | 0.0015 (6) | 0.0000 (5) | 0.0029 (6) |
(2R,3R)-1,4-Dioxaspiro[4.5]decane-2,3-dicarboxylic acid (II) . Geometric parameters (Å, º)
| O1—C1 | 1.322 (2) | C5—C10 | 1.525 (2) |
| O1—H1 | 0.84 (3) | C6—C7 | 1.533 (2) |
| O2—C1 | 1.208 (2) | C6—H6A | 0.9900 |
| O3—C4 | 1.2229 (19) | C6—H6B | 0.9900 |
| O4—C4 | 1.3135 (18) | C7—C8 | 1.526 (3) |
| O4—H4 | 0.85 (3) | C7—H7A | 0.9900 |
| O5—C2 | 1.4107 (18) | C7—H7B | 0.9900 |
| O5—C5 | 1.4398 (18) | C8—C9 | 1.532 (2) |
| O6—C3 | 1.4135 (17) | C8—H8A | 0.9900 |
| O6—C5 | 1.441 (2) | C8—H8B | 0.9900 |
| C1—C2 | 1.529 (2) | C9—C10 | 1.538 (2) |
| C2—C3 | 1.541 (2) | C9—H9A | 0.9900 |
| C2—H2 | 1.0000 | C9—H9B | 0.9900 |
| C3—C4 | 1.532 (2) | C10—H10A | 0.9900 |
| C3—H3 | 1.0000 | C10—H10B | 0.9900 |
| C5—C6 | 1.519 (2) | ||
| C1—O1—H1 | 112 (2) | C5—C6—H6A | 109.4 |
| C4—O4—H4 | 109.4 (15) | C7—C6—H6A | 109.4 |
| C2—O5—C5 | 109.65 (12) | C5—C6—H6B | 109.4 |
| C3—O6—C5 | 109.70 (12) | C7—C6—H6B | 109.4 |
| O2—C1—O1 | 125.41 (14) | H6A—C6—H6B | 108.0 |
| O2—C1—C2 | 123.59 (14) | C8—C7—C6 | 110.88 (15) |
| O1—C1—C2 | 110.89 (13) | C8—C7—H7A | 109.5 |
| O5—C2—C1 | 112.10 (12) | C6—C7—H7A | 109.5 |
| O5—C2—C3 | 103.28 (11) | C8—C7—H7B | 109.5 |
| C1—C2—C3 | 114.66 (12) | C6—C7—H7B | 109.5 |
| O5—C2—H2 | 108.9 | H7A—C7—H7B | 108.1 |
| C1—C2—H2 | 108.9 | C7—C8—C9 | 110.75 (14) |
| C3—C2—H2 | 108.9 | C7—C8—H8A | 109.5 |
| O6—C3—C4 | 114.38 (12) | C9—C8—H8A | 109.5 |
| O6—C3—C2 | 103.84 (12) | C7—C8—H8B | 109.5 |
| C4—C3—C2 | 111.08 (12) | C9—C8—H8B | 109.5 |
| O6—C3—H3 | 109.1 | H8A—C8—H8B | 108.1 |
| C4—C3—H3 | 109.1 | C8—C9—C10 | 110.87 (14) |
| C2—C3—H3 | 109.1 | C8—C9—H9A | 109.5 |
| O3—C4—O4 | 123.68 (14) | C10—C9—H9A | 109.5 |
| O3—C4—C3 | 120.74 (14) | C8—C9—H9B | 109.5 |
| O4—C4—C3 | 115.58 (13) | C10—C9—H9B | 109.5 |
| O5—C5—O6 | 105.80 (12) | H9A—C9—H9B | 108.1 |
| O5—C5—C6 | 107.89 (12) | C5—C10—C9 | 110.35 (14) |
| O6—C5—C6 | 111.46 (13) | C5—C10—H10A | 109.6 |
| O5—C5—C10 | 111.57 (13) | C9—C10—H10A | 109.6 |
| O6—C5—C10 | 108.05 (13) | C5—C10—H10B | 109.6 |
| C6—C5—C10 | 111.92 (13) | C9—C10—H10B | 109.6 |
| C5—C6—C7 | 110.97 (13) | H10A—C10—H10B | 108.1 |
| C5—O5—C2—C1 | −99.25 (14) | C2—O5—C5—O6 | −12.62 (15) |
| C5—O5—C2—C3 | 24.70 (14) | C2—O5—C5—C6 | −132.02 (13) |
| O2—C1—C2—O5 | −6.9 (2) | C2—O5—C5—C10 | 104.65 (14) |
| O1—C1—C2—O5 | 176.76 (12) | C3—O6—C5—O5 | −6.39 (15) |
| O2—C1—C2—C3 | −124.22 (16) | C3—O6—C5—C6 | 110.62 (13) |
| O1—C1—C2—C3 | 59.43 (16) | C3—O6—C5—C10 | −126.00 (13) |
| C5—O6—C3—C4 | −100.36 (14) | O5—C5—C6—C7 | −178.91 (14) |
| C5—O6—C3—C2 | 20.87 (14) | O6—C5—C6—C7 | 65.34 (17) |
| O5—C2—C3—O6 | −27.60 (14) | C10—C5—C6—C7 | −55.80 (19) |
| C1—C2—C3—O6 | 94.65 (14) | C5—C6—C7—C8 | 55.80 (19) |
| O5—C2—C3—C4 | 95.81 (13) | C6—C7—C8—C9 | −56.5 (2) |
| C1—C2—C3—C4 | −141.94 (13) | C7—C8—C9—C10 | 56.9 (2) |
| O6—C3—C4—O3 | −172.39 (14) | O5—C5—C10—C9 | 176.83 (13) |
| C2—C3—C4—O3 | 70.47 (18) | O6—C5—C10—C9 | −67.27 (16) |
| O6—C3—C4—O4 | 7.99 (19) | C6—C5—C10—C9 | 55.82 (18) |
| C2—C3—C4—O4 | −109.15 (15) | C8—C9—C10—C5 | −56.08 (19) |
(2R,3R)-1,4-Dioxaspiro[4.5]decane-2,3-dicarboxylic acid (II) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O3i | 0.84 (3) | 1.80 (3) | 2.6230 (16) | 164 (3) |
| O4—H4···O2ii | 0.85 (3) | 1.88 (3) | 2.7116 (16) | 164 (2) |
| C2—H2···O3iii | 1.00 | 2.41 | 3.3818 (19) | 164 |
| C3—H3···O2iv | 1.00 | 2.44 | 3.392 (2) | 160 |
| C6—H6A···O4 | 0.99 | 2.52 | 3.255 (2) | 131 |
Symmetry codes: (i) −x+1, y+1/2, −z+1; (ii) x+1, y−1, z; (iii) −x+2, y+1/2, −z+1; (iv) x, y−1, z.
Funding Statement
This work was funded by Russian Academy of Sciences grant .
References
- Ates, A. & Curran, D. P. (2001). J. Am. Chem. Soc. 123, 5130–5131. [DOI] [PubMed]
- Barrett, A. G. M., Doubleday, W. W., Kasdorf, K., Tustin, G. J., White, A. J. P. & Williams, D. J. (1995). J. Chem. Soc. Chem. Commun. pp. 407–408.
- Belokon’, Yu. N., Gagieva, S. Ch., Sukhova, T. A., Dmitriev, A. V., Lyssenko, K. A., Bravaya, N. M., Bulychev, B. M. & Seebach, D. (2005). Russ. Chem. Bull. 54, 2348–2353.
- Bruker (2008). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Černý, M. (1977). Collect. Czech. Chem. Commun. 42, 3069–3078.
- Crassous, J. (2009). Chem. Soc. Rev. 38, 830–845. [DOI] [PubMed]
- Eissmann, D., Katzsch, F. & Weber, E. (2012). Struct. Chem. 23, 1131–1142.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Giordano, C., Coppi, L. & Restelli, A. (1990). J. Org. Chem. 55, 5400–5402.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Ianelli, S., Nardelli, M., Giordano, C., Coppi, L. & Restelli, A. (1992). Acta Cryst. C48, 1722–1727.
- Innis, C. & Lamaty, G. (1977). Nouv. J. Chim. 1, 503–509.
- Jiang, J., Pan, Y., Wang, D.-C. & Ou-yang, P.-K. (2007). Acta Cryst. E63, o1093–o1094.
- Karisalmi, K., Rissanen, K. & Koskinen, A. M. P. (2003). Org. Biomol. Chem. 1, 3193–3196. [DOI] [PubMed]
- Kassai, C., Juvancz, Z., Bálint, J., Fogassy, E. & Kozma, D. (2000). Tetrahedron, 56, 8355–8359.
- Lam, F. L., Kwong, F. Y. & Chan, A. S. C. (2011). Top. Organomet. Chem. 36, 29–66.
- Lee, D., Sello, J. K. & Schreiber, S. L. (1999). J. Am. Chem. Soc. 121, 10648–10649.
- Linker, T., Fudickar, W., Kelling, A. & Schilde, U. (2013). Z. Kristallogr. New Cryst. Struct. 228, 241–242.
- Lv, C.-L., Chen, J.-H., Zhang, Y.-Z., Lu, D.-Q. & OuYang, P.-K. (2012). Acta Cryst. E68, o1128. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Maezaki, N., Sakamoto, A., Nagahashi, N., Soejima, M., Li, Y.-X., Imamura, T., Kojima, N., Ohishi, H., Sakaguchi, K., Iwata, C. & Tanaka, T. (2000). J. Org. Chem. 65, 3284–3291. [DOI] [PubMed]
- Mikołajczyk, M., Mikina, M., Wieczorek, M. W. & Błaszczyk, J. (1996). Angew. Chem. Int. Ed. Engl. 35, 1560–1562.
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Pelphrey, P. M., Abboud, K. A. & Wright, D. L. (2004). J. Org. Chem. 69, 6931–6933. [DOI] [PubMed]
- Roush, W. R., Ratz, A. M. & Jablonowski, J. A. (1992). J. Org. Chem. 57, 2047–2052.
- Saget, T., Lemouzy, S. J. & Cramer, N. (2012). Angew. Chem. Int. Ed. 51, 2238–2242. [DOI] [PubMed]
- Seebach, D., Beck, A. K. & Heckel, A. (2001). Angew. Chem. Int. Ed. 40, 92–138. [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Tsuzuki, Y. (1937). Bull. Chem. Soc. Jpn, 12, 487–492.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Wink, D. J. & Dewan, J. C. (1990). Acta Cryst. C46, 1058–1061.
- Yang, L., Melot, R., Neuburger, M. & Baudoin, O. (2017). Chem. Sci. 8, 1344–1349. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, II, global. DOI: 10.1107/S2056989018009593/eb2009sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018009593/eb2009Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989018009593/eb2009IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989018009593/eb2009Isup4.cml
Supporting information file. DOI: 10.1107/S2056989018009593/eb2009IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report