The molecular conformation is stabilized via intramolecular C—H⋯O and C—H⋯N contacts. The supramolecular structure is mainly governed by C—H⋯N hydrogen-bonded centrosymmetric dimers, C—H⋯O and C—H⋯S hydrogen bonds and S⋯π stacking interactions, which together lead to the formation of a layered crystal packing.
Keywords: crystal structure; 1,2,3-triazole; hydrogen bonding; molecular electrostatic potential; MESP; fingerprint plot
Abstract
The title compound, C15H13N3O2S, crystallizes in the monoclinic space group P21/n and its molecular conformation is stabilized via intramolecular C—H⋯O and C—H⋯N contacts. The supramolecular structure is mainly governed by C—H⋯N hydrogen-bonded centrosymmetric dimers, C—H⋯O and C—H⋯S hydrogen bonds and S⋯π and π–π stacking interactions which, together, lead to the formation of a layered crystal packing. The intermolecular interactions were further evaluated through the molecular electrostatic potential map and Hirshfeld fingerprint analysis.
Chemical context
Compounds containing the 1,2,3-triazole scaffold are considered to be an important class of five-membered N-heterocycles (having two carbon and three nitrogen atoms) because of their unique structural and chemical properties (Kolb & Sharpless, 2003 ▸; Freitas et al., 2014 ▸). In the last few decades, significant attention has been paid to this kind of structural units owing to their versatile applications in the fields of materials science and medicinal chemistry (Zhou & Wang, 2012 ▸; Venugopala et al., 2016 ▸). In addition, 1,2,3-triazoles have also been found to be quite relevant in objective-oriented synthesis (Billing & Nilsson, 2005 ▸), bioconjugation (Speers et al., 2003 ▸) and combinatorial chemistry (Löber et al., 2003 ▸). The geometrical shapes and interaction functions of natural heterocycles and amides can be very similar to those of 1,2,3-triazoles (Thibault et al., 2006 ▸).
In general, the 1,2,3-triazole nucleus is the most fundamental heterocyclic component found in various pharmacologically active agents (Agalave et al., 2011 ▸). In particular, potential pharmaceuticals based on the 1,2,3-triazole ring include anti-HIV (Giffin et al., 2008 ▸), anticancer (Singh et al., 2012 ▸), anti-tubercular (Patpi et al., 2012 ▸), antimicrobial (Demaray et al., 2008 ▸) and antifungal (Lass-Floerl et al., 2011 ▸) agents. This is due to the fact that the 1,2,3-triazole structural unit is stable against metabolic degradation as well as oxidation and reduction in acidic and basic conditions (Ferreira et al., 2010 ▸). Importantly, this special class of structural unit is capable of forming hydrogen-bonding interactions (the N atom acts as an acceptor) as well as π–π stacking and other intermolecular interactions with biological targets to improve their solubility (Lauria et al., 2014 ▸). Hence, it is of extreme importance to explore and understand the supramolecular structure of compounds in which the structural motif is based on 1,2,3-triazole. Keeping in mind the above-mentioned features, we report here the crystal structure and packing analysis of the title compound [5-(4-methoxyphenyl)-2-methyl-2H-1,2,3-triazol-4-yl](thiophen-2-yl)methanone (1).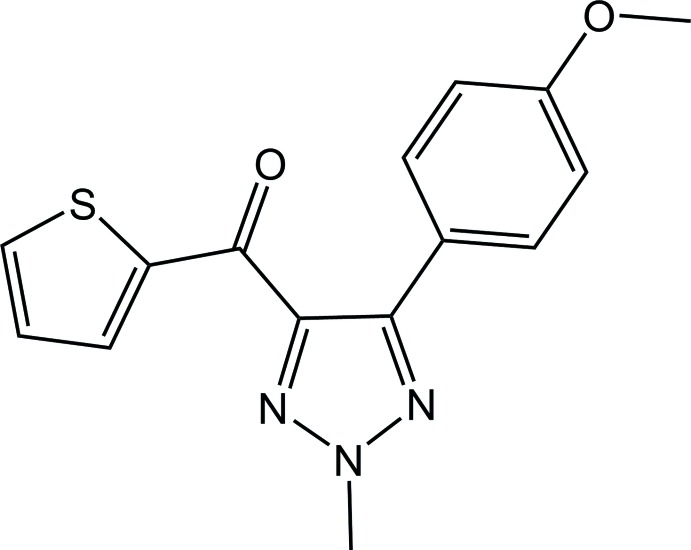
Structural commentary
The single-crystal X-ray diffraction study shows that compound 1 crystallizes in the monoclinic space group P21/n with one molecule (Z′ = 1) in the asymmetric unit (Fig. 1 ▸). In the molecular structure, the N-methylated triazol ring is substituted at the two carbon atoms C7 and C8 by a para-methoxy phenyl and a methanone-thienyl ring, respectively, resulting in four conformationally flexible parts in the molecule around the C8—C9, C9—C10, C1—C7 and C4—O1 single bonds (see Fig. 1 ▸). The conformation of the molecule in the crystal is stabilized via intramolecular C2—H2⋯O2 [C2⋯O2 = 2.961 (2) Å] and C11—H11⋯N1 [C11⋯N1 = 2.950 (2) Å] contacts (Fig. 1 ▸; Table 1 ▸). For this reason, the thienyl and triazole rings are nearly coplanar, with an angle of 13.63 (10)° between their mean planes, while the phenyl ring is tilted out from the mean planes of the thienyl and triazole rings by 38.84 (9) and 34.04 (10)°, respectively. It is also important to mention here that the methoxy group attached to C4 is in the same plane as the phenyl ring.
Figure 1.
The asymmetric unit of compound 1 highlighting the intramolecular C—H⋯O and C—H⋯N contacts. Displacement ellipsoids are drawn at the 50% probability level.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C11—H11⋯N1 | 0.95 | 2.41 | 2.950 (2) | 116 |
| C2—H2⋯O2 | 0.95 | 2.42 | 2.961 (2) | 113 |
| C3—H3⋯S1i | 0.95 | 2.96 | 3.810 (2) | 149 |
| C15—H15A⋯O2ii | 0.98 | 2.98 | 3.828 (3) | 146 |
| C15—H15C⋯N3iii | 0.98 | 2.73 | 3.490 (3) | 135 |
| C12—H12⋯N1iv | 0.95 | 2.95 | 3.768 (2) | 145 |
| C13—H13⋯O2v | 0.95 | 2.38 | 3.191 (2) | 143 |
| C14—H14C⋯O1vi | 0.98 | 2.67 | 3.230 (2) | 117 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  ; (vi)
; (vi)  .
.
Supramolecular features
In the crystal, the molecules form two types of centrosymmetric, weak to very weak C—H⋯N hydrogen-bonding dimeric motifs (Table 1 ▸) involving the methyl hydrogen H15C (sp 3) of the methoxy group with the triazol nitrogen N3 [C15⋯N3 = 3.490 (3) Å] and the thiophene hydrogen H12 (sp 2) with the triazol nitrogen N1 [C12⋯N1 = 3.768 (2) Å]. These are extended in an alternate fashion, forming ribbons along the [101] direction (see green and yellow shades in Fig. 2 ▸). Two such adjacent hydrogen-bonded ribbons are connected to each other via Csp 2/sp 3—H⋯O and S⋯C(π) [3.492 (2) Å] interactions along the [010] direction, forming a corrugated sheet perpendicular to the (101) plane (Fig. 2 ▸ and Table 1 ▸). These sheets are further stacked to each other by displaced π–π stacking interactions distances ranging from 3.375 (3) to 3.384 (4) Å through inversion and translational symmetries, and weak C3—H3⋯S1 [C3⋯S1 = 3.810 (2) Å] interactions (Table 1 ▸), leading to the formation of a layered packing arrangement of molecules (Fig. 3 ▸).
Figure 2.
Crystal packing of 1 showing the formation of molecular sheets via two types of centrosymmetric C—H⋯N dimers (shaded in light yellow and green), forming ribbons connected through C—H⋯O and S⋯C(π) interactions.
Figure 3.
Stacking of hydrogen-bonded molecular sheets via π–π interactions (dotted lines) in compound 1. Hydrogen atoms are omitted for clarity.
Analysis of molecular electrostatic potential and Hirshfeld fingerprint plots
A deeper insight into intermolecular interactions can be obtained from molecular electrostatic potential (MESP), and two-dimensional fingerprint plots (McKinnon et al., 2007 ▸) mapped on the Hirshfeld surface (Spackman & Jayatilaka, 2009 ▸). All the plots were computed using the programme CrystalExplorer 17.5 (Turner et al., 2017 ▸). The MESP plot of compound 1 (Fig. 4 ▸) shows that the centres of both the triazole and thiophene five-membered rings have nearly neutral ESP values (0.000 and −0.002 a.u., respectively), while the benzene ring is highly electronegative (−0.028 a.u.) compared to the two heterocyclic rings. This electrostatic complementarity among the rings leads to favourable stacking interactions in the crystal packing as a result of a layered supramolecular architecture. Intermolecular hydrogen-bond donors and acceptors appear as blue (positive ESP) and red (negative ESP) regions, respectively, on the surface (Fig. 4 ▸). The two-dimensional fingerprint plots and the contributions of individual interatomic contacts toward the overall crystal packing are shown in Fig. 5 ▸. It is observed that several directional hydrogen-bonding contacts such as N⋯H (7.7%), O⋯H (11.0%), S⋯H (6.3%) along with C⋯H (18.5%), H⋯H (41.6%) and other interatomic contacts stabilize the crystal packing of compound 1.
Figure 4.
MESP of compound 1 mapped over the Hirshfeld surface with a scale of −0.03 a.u. (red) through 0.00 (white) to +0.03 a.u. (blue). The ESP values (in a.u.) for the centre of each ring are given.
Figure 5.
Two-dimensional full fingerprint plots and decomposed fingerprint plots over the Hirshfeld surface for various intermolecular atom–atom contacts in compound 1. The numbers in red indicate the percentage contributions of each contact.
Database survey
A Cambridge Structural Database (Version 5.39, update May 2018; Groom et al., 2016 ▸) search for the (2-methyl-2H-1,2,3-triazol-4-yl)(thiophen-2-yl)methanone subunit resulted in one hit (SONFIM; Girish et al., 2014 ▸). Like compound 1, the molecular conformation of SONFIM is also stabilized by intramolecular C—H⋯O and C—H⋯N hydrogen bonds. The supramolecular structure of SONFIM is primarily determined by intermolecular C—H⋯O and C—H⋯π hydrogen bonds, while C—H⋯N hydrogen bonding plays a secondary role in the overall stabilization of the crystal packing.
Synthesis and crystallization
The title compound was synthesized according to the procedure described elsewhere (Girish et al., 2014 ▸). Single crystals of the pure compound were grown by slow evaporation of a toluene solution at room temperature (297–301 K).
Refinement
Crystal data, data collection and structure refinement details are given in Table 2 ▸. Hydrogen atoms were positioned geometrically and refined as riding: C—H = 0.98 Å with Ui so(H) =1.5U eq(C) for the methyl group and C—H = 0.95Å with U iso(H) = 1.2U eq(C) for the aromatic C atoms.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C15H13N3O2S |
| M r | 299.34 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 100 |
| a, b, c (Å) | 8.5851 (10), 16.8986 (19), 9.3455 (11) |
| β (°) | 92.465 (4) |
| V (Å3) | 1354.6 (3) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.25 |
| Crystal size (mm) | 0.30 × 0.10 × 0.06 |
| Data collection | |
| Diffractometer | Bruker APEXII D8 Venture CMOS |
| Absorption correction | Multi-scan (SADABS; Bruker, 2012 ▸) |
| T min, T max | 0.619, 0.746 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 17149, 3962, 2914 |
| R int | 0.065 |
| (sin θ/λ)max (Å−1) | 0.705 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.053, 0.114, 1.03 |
| No. of reflections | 3962 |
| No. of parameters | 192 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.46, −0.53 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989018010654/xi2009sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018010654/xi2009Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989018010654/xi2009Isup3.cml
CCDC reference: 1850683
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C15H13N3O2S | F(000) = 624 |
| Mr = 299.34 | Dx = 1.468 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.5851 (10) Å | Cell parameters from 6642 reflections |
| b = 16.8986 (19) Å | θ = 2.4–30.0° |
| c = 9.3455 (11) Å | µ = 0.25 mm−1 |
| β = 92.465 (4)° | T = 100 K |
| V = 1354.6 (3) Å3 | Plate, yellow |
| Z = 4 | 0.30 × 0.10 × 0.06 mm |
Data collection
| Bruker APEXII D8 Venture CMOS diffractometer | 2914 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.065 |
| Absorption correction: multi-scan (SADABS; Bruker, 2012) | θmax = 30.1°, θmin = 2.4° |
| Tmin = 0.619, Tmax = 0.746 | h = −12→11 |
| 17149 measured reflections | k = −23→23 |
| 3962 independent reflections | l = −10→13 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.053 | H-atom parameters constrained |
| wR(F2) = 0.114 | w = 1/[σ2(Fo2) + (0.037P)2 + 1.3716P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max < 0.001 |
| 3962 reflections | Δρmax = 0.46 e Å−3 |
| 192 parameters | Δρmin = −0.53 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.22992 (6) | 0.20914 (3) | 0.39316 (5) | 0.01627 (13) | |
| O1 | 1.18053 (15) | 0.18243 (8) | 0.81322 (14) | 0.0166 (3) | |
| O2 | 0.48083 (16) | 0.18611 (8) | 0.60466 (16) | 0.0200 (3) | |
| N1 | 0.35646 (18) | −0.00987 (9) | 0.68224 (18) | 0.0160 (3) | |
| N3 | 0.57606 (18) | −0.03357 (10) | 0.81538 (18) | 0.0158 (3) | |
| N2 | 0.43569 (18) | −0.05846 (9) | 0.76814 (18) | 0.0162 (3) | |
| C1 | 0.7388 (2) | 0.08137 (10) | 0.7775 (2) | 0.0130 (4) | |
| C11 | 0.1653 (2) | 0.06385 (11) | 0.4486 (2) | 0.0137 (4) | |
| H11 | 0.167530 | 0.012226 | 0.488904 | 0.016* | |
| C4 | 1.0365 (2) | 0.14778 (10) | 0.8101 (2) | 0.0130 (4) | |
| C3 | 0.9476 (2) | 0.15862 (11) | 0.6832 (2) | 0.0132 (4) | |
| H3 | 0.988464 | 0.188552 | 0.607394 | 0.016* | |
| C7 | 0.5901 (2) | 0.03848 (11) | 0.7557 (2) | 0.0132 (4) | |
| C5 | 0.9775 (2) | 0.10385 (11) | 0.9210 (2) | 0.0150 (4) | |
| H5 | 1.037167 | 0.096550 | 1.007982 | 0.018* | |
| C10 | 0.2700 (2) | 0.12300 (11) | 0.4876 (2) | 0.0128 (4) | |
| C6 | 0.8294 (2) | 0.07061 (11) | 0.9028 (2) | 0.0147 (4) | |
| H6 | 0.789370 | 0.039832 | 0.977967 | 0.018* | |
| C2 | 0.8005 (2) | 0.12602 (11) | 0.6675 (2) | 0.0131 (4) | |
| H2 | 0.740456 | 0.134022 | 0.580881 | 0.016* | |
| C8 | 0.4519 (2) | 0.05345 (11) | 0.6724 (2) | 0.0138 (4) | |
| C9 | 0.4044 (2) | 0.12489 (11) | 0.5897 (2) | 0.0141 (4) | |
| C15 | 1.2758 (2) | 0.17414 (14) | 0.9413 (2) | 0.0245 (5) | |
| H15A | 1.224395 | 0.199444 | 1.020945 | 0.037* | |
| H15B | 1.376982 | 0.199448 | 0.928427 | 0.037* | |
| H15C | 1.291563 | 0.117828 | 0.962413 | 0.037* | |
| C12 | 0.0541 (2) | 0.08913 (12) | 0.3416 (2) | 0.0160 (4) | |
| H12 | −0.026154 | 0.056184 | 0.301494 | 0.019* | |
| C13 | 0.0753 (2) | 0.16613 (12) | 0.3025 (2) | 0.0173 (4) | |
| H13 | 0.011053 | 0.192791 | 0.232674 | 0.021* | |
| C14 | 0.3821 (2) | −0.13873 (11) | 0.7969 (2) | 0.0213 (4) | |
| H14A | 0.273221 | −0.144368 | 0.762375 | 0.032* | |
| H14B | 0.390158 | −0.148830 | 0.900188 | 0.032* | |
| H14C | 0.446872 | −0.176823 | 0.747341 | 0.032* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0164 (2) | 0.0123 (2) | 0.0201 (3) | −0.00008 (18) | 0.00098 (18) | 0.00335 (19) |
| O1 | 0.0124 (6) | 0.0188 (7) | 0.0183 (7) | −0.0030 (5) | −0.0029 (5) | 0.0017 (6) |
| O2 | 0.0164 (7) | 0.0107 (6) | 0.0325 (8) | −0.0021 (5) | −0.0028 (6) | 0.0009 (6) |
| N1 | 0.0151 (8) | 0.0118 (7) | 0.0209 (9) | 0.0003 (6) | −0.0009 (7) | 0.0019 (7) |
| N3 | 0.0124 (8) | 0.0154 (8) | 0.0195 (8) | −0.0002 (6) | −0.0003 (6) | 0.0005 (7) |
| N2 | 0.0135 (8) | 0.0125 (8) | 0.0223 (9) | −0.0006 (6) | −0.0013 (7) | 0.0036 (7) |
| C1 | 0.0136 (9) | 0.0092 (8) | 0.0162 (10) | 0.0008 (7) | 0.0014 (7) | −0.0018 (7) |
| C11 | 0.0150 (9) | 0.0120 (8) | 0.0143 (9) | 0.0017 (7) | 0.0010 (7) | −0.0010 (7) |
| C4 | 0.0123 (8) | 0.0094 (8) | 0.0173 (9) | 0.0006 (7) | 0.0001 (7) | −0.0019 (7) |
| C3 | 0.0143 (9) | 0.0117 (8) | 0.0136 (9) | 0.0019 (7) | 0.0019 (7) | −0.0002 (7) |
| C7 | 0.0139 (9) | 0.0110 (8) | 0.0148 (9) | 0.0012 (7) | 0.0020 (7) | −0.0002 (7) |
| C5 | 0.0143 (9) | 0.0150 (9) | 0.0154 (9) | 0.0009 (7) | −0.0012 (7) | 0.0002 (7) |
| C10 | 0.0121 (9) | 0.0105 (8) | 0.0160 (9) | 0.0017 (7) | 0.0028 (7) | 0.0008 (7) |
| C6 | 0.0173 (9) | 0.0123 (9) | 0.0145 (9) | −0.0005 (7) | 0.0022 (7) | 0.0008 (7) |
| C2 | 0.0132 (9) | 0.0124 (8) | 0.0136 (9) | 0.0032 (7) | −0.0004 (7) | −0.0010 (7) |
| C8 | 0.0138 (9) | 0.0099 (8) | 0.0178 (10) | −0.0005 (7) | 0.0009 (7) | −0.0010 (7) |
| C9 | 0.0115 (9) | 0.0116 (8) | 0.0193 (10) | 0.0012 (7) | 0.0028 (7) | −0.0015 (7) |
| C15 | 0.0189 (10) | 0.0338 (12) | 0.0203 (11) | −0.0080 (9) | −0.0066 (8) | 0.0034 (9) |
| C12 | 0.0144 (9) | 0.0170 (9) | 0.0164 (10) | −0.0014 (7) | −0.0015 (7) | −0.0028 (8) |
| C13 | 0.0160 (9) | 0.0204 (10) | 0.0156 (10) | 0.0025 (7) | 0.0000 (7) | −0.0005 (8) |
| C14 | 0.0199 (10) | 0.0116 (9) | 0.0322 (12) | −0.0030 (8) | −0.0010 (9) | 0.0070 (8) |
Geometric parameters (Å, º)
| S1—C13 | 1.706 (2) | C3—C2 | 1.380 (3) |
| S1—C10 | 1.7292 (19) | C3—H3 | 0.9500 |
| O1—C4 | 1.368 (2) | C7—C8 | 1.413 (3) |
| O1—C15 | 1.427 (2) | C5—C6 | 1.394 (3) |
| O2—C9 | 1.230 (2) | C5—H5 | 0.9500 |
| N1—N2 | 1.317 (2) | C10—C9 | 1.466 (3) |
| N1—C8 | 1.353 (2) | C6—H6 | 0.9500 |
| N3—N2 | 1.334 (2) | C2—H2 | 0.9500 |
| N3—C7 | 1.347 (2) | C8—C9 | 1.481 (3) |
| N2—C14 | 1.461 (2) | C15—H15A | 0.9800 |
| C1—C6 | 1.390 (3) | C15—H15B | 0.9800 |
| C1—C2 | 1.398 (3) | C15—H15C | 0.9800 |
| C1—C7 | 1.475 (3) | C12—C13 | 1.365 (3) |
| C11—C10 | 1.383 (3) | C12—H12 | 0.9500 |
| C11—C12 | 1.419 (3) | C13—H13 | 0.9500 |
| C11—H11 | 0.9500 | C14—H14A | 0.9800 |
| C4—C5 | 1.388 (3) | C14—H14B | 0.9800 |
| C4—C3 | 1.394 (3) | C14—H14C | 0.9800 |
| C13—S1—C10 | 91.62 (9) | C1—C6—H6 | 119.2 |
| C4—O1—C15 | 117.45 (15) | C5—C6—H6 | 119.2 |
| N2—N1—C8 | 103.66 (15) | C3—C2—C1 | 120.72 (17) |
| N2—N3—C7 | 104.09 (15) | C3—C2—H2 | 119.6 |
| N1—N2—N3 | 116.16 (15) | C1—C2—H2 | 119.6 |
| N1—N2—C14 | 122.16 (16) | N1—C8—C7 | 108.49 (16) |
| N3—N2—C14 | 121.29 (16) | N1—C8—C9 | 121.73 (16) |
| C6—C1—C2 | 118.34 (17) | C7—C8—C9 | 129.73 (17) |
| C6—C1—C7 | 120.11 (17) | O2—C9—C10 | 119.57 (17) |
| C2—C1—C7 | 121.13 (17) | O2—C9—C8 | 119.53 (17) |
| C10—C11—C12 | 112.23 (17) | C10—C9—C8 | 120.88 (16) |
| C10—C11—H11 | 123.9 | O1—C15—H15A | 109.5 |
| C12—C11—H11 | 123.9 | O1—C15—H15B | 109.5 |
| O1—C4—C5 | 124.87 (17) | H15A—C15—H15B | 109.5 |
| O1—C4—C3 | 115.02 (17) | O1—C15—H15C | 109.5 |
| C5—C4—C3 | 120.10 (17) | H15A—C15—H15C | 109.5 |
| C2—C3—C4 | 120.21 (18) | H15B—C15—H15C | 109.5 |
| C2—C3—H3 | 119.9 | C13—C12—C11 | 112.45 (17) |
| C4—C3—H3 | 119.9 | C13—C12—H12 | 123.8 |
| N3—C7—C8 | 107.60 (16) | C11—C12—H12 | 123.8 |
| N3—C7—C1 | 118.64 (16) | C12—C13—S1 | 112.53 (15) |
| C8—C7—C1 | 133.56 (17) | C12—C13—H13 | 123.7 |
| C4—C5—C6 | 118.99 (17) | S1—C13—H13 | 123.7 |
| C4—C5—H5 | 120.5 | N2—C14—H14A | 109.5 |
| C6—C5—H5 | 120.5 | N2—C14—H14B | 109.5 |
| C11—C10—C9 | 132.22 (17) | H14A—C14—H14B | 109.5 |
| C11—C10—S1 | 111.18 (14) | N2—C14—H14C | 109.5 |
| C9—C10—S1 | 116.60 (13) | H14A—C14—H14C | 109.5 |
| C1—C6—C5 | 121.63 (18) | H14B—C14—H14C | 109.5 |
| C8—N1—N2—N3 | −0.6 (2) | C4—C5—C6—C1 | −1.0 (3) |
| C8—N1—N2—C14 | −173.55 (18) | C4—C3—C2—C1 | −0.4 (3) |
| C7—N3—N2—N1 | 0.5 (2) | C6—C1—C2—C3 | −0.1 (3) |
| C7—N3—N2—C14 | 173.46 (18) | C7—C1—C2—C3 | −172.69 (17) |
| C15—O1—C4—C5 | −1.4 (3) | N2—N1—C8—C7 | 0.5 (2) |
| C15—O1—C4—C3 | 179.19 (17) | N2—N1—C8—C9 | −177.17 (17) |
| O1—C4—C3—C2 | 179.69 (16) | N3—C7—C8—N1 | −0.3 (2) |
| C5—C4—C3—C2 | 0.2 (3) | C1—C7—C8—N1 | 174.3 (2) |
| N2—N3—C7—C8 | −0.1 (2) | N3—C7—C8—C9 | 177.16 (19) |
| N2—N3—C7—C1 | −175.62 (16) | C1—C7—C8—C9 | −8.3 (4) |
| C6—C1—C7—N3 | −30.9 (3) | C11—C10—C9—O2 | 179.7 (2) |
| C2—C1—C7—N3 | 141.55 (18) | S1—C10—C9—O2 | 0.5 (2) |
| C6—C1—C7—C8 | 155.0 (2) | C11—C10—C9—C8 | 1.3 (3) |
| C2—C1—C7—C8 | −32.5 (3) | S1—C10—C9—C8 | −177.88 (14) |
| O1—C4—C5—C6 | −178.95 (17) | N1—C8—C9—O2 | 166.57 (18) |
| C3—C4—C5—C6 | 0.4 (3) | C7—C8—C9—O2 | −10.5 (3) |
| C12—C11—C10—C9 | −178.84 (19) | N1—C8—C9—C10 | −15.0 (3) |
| C12—C11—C10—S1 | 0.4 (2) | C7—C8—C9—C10 | 167.85 (19) |
| C13—S1—C10—C11 | −0.13 (15) | C10—C11—C12—C13 | −0.5 (2) |
| C13—S1—C10—C9 | 179.21 (15) | C11—C12—C13—S1 | 0.4 (2) |
| C2—C1—C6—C5 | 0.8 (3) | C10—S1—C13—C12 | −0.14 (16) |
| C7—C1—C6—C5 | 173.47 (17) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C11—H11···N1 | 0.95 | 2.41 | 2.950 (2) | 116 |
| C2—H2···O2 | 0.95 | 2.42 | 2.961 (2) | 113 |
| C3—H3···S1i | 0.95 | 2.96 | 3.810 (2) | 149 |
| C15—H15A···O2ii | 0.98 | 2.98 | 3.828 (3) | 146 |
| C15—H15C···N3iii | 0.98 | 2.73 | 3.490 (3) | 135 |
| C12—H12···N1iv | 0.95 | 2.95 | 3.768 (2) | 145 |
| C13—H13···O2v | 0.95 | 2.38 | 3.191 (2) | 143 |
| C14—H14C···O1vi | 0.98 | 2.67 | 3.230 (2) | 117 |
Symmetry codes: (i) x+1, y, z; (ii) x+1/2, −y+1/2, z+1/2; (iii) −x+2, −y, −z+2; (iv) −x, −y, −z+1; (v) x−1/2, −y+1/2, z−1/2; (vi) −x+3/2, y−1/2, −z+3/2.
Funding Statement
This work was funded by Indian Institute of Science Education and Research Bhopal grant to S. Bhandary. National Research Foundation grants 96807 and 98884. Durban University of Technology grant to K. N. Venugopalac.
References
- Agalave, S. G., Maujan, S. R. & Pore, V. S. (2011). Chem. Asian J. 6, 2696–2718. [DOI] [PubMed]
- Billing, J. F. & Nilsson, U. J. (2005). J. Org. Chem. 70, 4847–4850. [DOI] [PubMed]
- Bruker (2012). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Burla, M. C., Caliandro, R., Carrozzini, B., Cascarano, G. L., Cuocci, C., Giacovazzo, C., Mallamo, M., Mazzone, A. & Polidori, G. (2015). J. Appl. Cryst. 48, 306–309.
- Demaray, J. A., Thuener, J. E., Dawson, M. N. & Sucheck, S. J. (2008). Bioorg. Med. Chem. Lett. 18, 4868–4871. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Ferreira, S. B., Sodero, A. C. R., Cardoso, M. F. C., Lima, E. S., Kaiser, C. R., Silva, F. P. & Ferreira, V. F. (2010). J. Med. Chem. 53, 2364–2375. [DOI] [PubMed]
- Freitas, L. B. de O., Borgati, T. F., de Freitas, R. P., Ruiz, A. L. T. G., Marchetti, G. M., de Carvalho, J. E., da Cunha, E. F. F., Ramalho, T. C. & Alves, R. B. (2014). Eur. J. Med. Chem. 84, 595–604. [DOI] [PubMed]
- Giffin, M. J., Heaslet, H., Brik, A., Lin, Y.-C., Cauvi, G., Wong, C.-H., McRee, D. E., Elder, J. H., Stout, C. D. & Torbett, B. E. (2008). J. Med. Chem. 51, 6263–6270. [DOI] [PMC free article] [PubMed]
- Girish, Y. R., Sharath Kumar, K. S., Muddegowda, U., Lokanath, N. K., Rangappa, K. S. & Shashikanth, S. (2014). RSC Adv. 4, 55800–55806.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Kolb, H. C. & Sharpless, K. B. (2003). Drug Discovery Today, 8, 1128–1137. [DOI] [PubMed]
- Lass-Floerl, C. (2011). Drugs, 71, 2405–2419. [DOI] [PubMed]
- Lauria, A., Delisi, R., Mingoia, F., Terenzi, A., Martorana, A., Barone, G. & Almerico, A. M. (2014). Eur. J. Org. Chem. 2014, 3289–3306.
- Löber, S., Rodriguez-Loaiza, P. & Gmeiner, P. (2003). Org. Lett. 5, 1753–1755. [DOI] [PubMed]
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. 3814–3816. [DOI] [PubMed]
- Patpi, S. R., Pulipati, L., Yogeeswari, P., Sriram, D., Jain, N., Sridhar, B., Murthy, R., Anjana Devi, T., Kalivendi, S. V. & Kantevari, S. (2012). J. Med. Chem. 55, 3911–3922. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Singh, P., Raj, R., Kumar, V., Mahajan, M. P., Bedi, P. M. S., Kaur, T. & Saxena, A. K. (2012). Eur. J. Med. Chem. 47, 594–600. [DOI] [PubMed]
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Speers, A. E., Adam, G. C. & Cravatt, B. F. (2003). J. Am. Chem. Soc. 125, 4686–4687. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Thibault, R. J., Takizawa, K., Lowenheilm, P., Helms, B., Mynar, J. L., Fréchet, J. M. J. & Hawker, C. J. (2006). J. Am. Chem. Soc. 128, 12084–12085. [DOI] [PubMed]
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer 17.5. University of Western Australia.
- Venugopala, K. N., Rao, D., Bhandary, S., Pillay, M., Chopra, D., Aldhubiab, B. E., Attimarad, M., Alwassil, O. I., Harsha, S. & Mlisana, K. (2016). Drug. Des. Dev. Ther. 10, 2681–2690. [DOI] [PMC free article] [PubMed]
- Zhou, C.-H. & Wang, Y. (2012). Curr. Med. Chem. 19, 239–280. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989018010654/xi2009sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018010654/xi2009Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989018010654/xi2009Isup3.cml
CCDC reference: 1850683
Additional supporting information: crystallographic information; 3D view; checkCIF report







