The crystal structure of [Mn(TMPA)(Ac)(CH3OH)]BPh4 (TMPA = tris(pyridin-2-ylmethyl)amine, Ac = acetate, BPh4 = tetraphenylborate) has been determined. The structure reveals a seven-coordinate MnII center with distorted, pentagonal bipyramidal geometry.
Keywords: crystal structure, manganese(II), tripodal ligand, seven-coordinate
Abstract
Structural analysis of (acetato-κ2 O,O′)(methanol-κO)[tris(pyridin-2-ylmethyl)amine-κ4 N,N′,N′′,N′′′]manganese(II) tetraphenylborate, [Mn(C2H3O2)(C18H18N4)(CH3OH)](C24H20B) or [Mn(TMPA)(Ac)(CH3OH)]BPh4 [TMPA = tris(pyridin-2-ylmethyl)amine, Ac = acetate, BPh4 = tetraphenylborate] by single-crystal X-ray diffraction reveals a complex cation with tetradentate coordination of the tripodal TMPA ligand, bidentate coordination of the Ac ligand and monodentate coordination of the methanol ligand to a single MnII center, balanced in charge by the presence of a tetraphenylborate anion. The MnII complex has a distorted pentagonal–bipyramidal geometry, in which the central amine nitrogen and two pyridyl N atoms of the TMPA ligand, and two oxygen atoms of the acetate ligand occupy positions in the pentagonal plane, while the third pyridyl nitrogen of TMPA and the oxygen from the methanol ligand occupy the axial positions. Within the complex, the acetate O atoms participate in weak C—H⋯O hydrogen-bonding interactions with neighboring pyridyl moieties. In the crystal, complexes form dimers by pairs of O—H⋯O hydrogen bonds between the coordinated methanol of one complex and an acetate oxygen of the other, and weak π-stacking interactions between pyridine rings. Separate dimers then undergo additional π-stacking interactions between the pyridine rings of one moiety and either the pyridine or phenyl rings of another moiety that further stabilize the crystal.
Chemical context
A variety of manganese(II/III) complexes have been studied as structural and functional mimics of superoxide dismutase (SOD) enzymes (Batinić-Haberle et al., 2010 ▸, 2014 ▸; Iranzo, 2011 ▸; Bani & Bencini, 2012 ▸; Miriyala et al., 2012 ▸; Policar, 2016 ▸). The efficacy of these mimics is reliant on their stability in aqueous solution, retention of open or substitutional coordination sites on the manganese ion, and MnIII/MnII redox potential lying in the narrow range of 0.2–0.4 V versus a normal hydrogen electrode (Iranzo, 2011 ▸; Policar, 2016 ▸). These factors are directly related to the nature of the ligands employed, their coordinating atoms, and the geometry of the coordination sphere (Policar, 2016 ▸).
One family of manganese(II) complexes that has been studied incorporates N-centered, tripodal, tetradentate ligands (Policar et al., 2001 ▸; Durot et al., 2005 ▸; Ribeiro et al., 2015 ▸). These ligands can be readily synthesized to provide a variety of N and O donors that give rise to the structural diversity of their metal complexes (Policar et al., 2001 ▸). With that in mind, we have begun to examine manganese(II) complexes with tripodal ligands containing either pyridine or quinoline groups. Herein, we report the synthesis and structural characterization of [Mn(TMPA)(Ac)(CH3OH)]BPh4 [TMPA = tris(pyridin-2-ylmethyl)amine, Ac = acetate, BPh4 = tetraphenylborate]. This compound is prepared by a two-step process (see reaction scheme) in which manganese(II) acetate is reacted with TMPA in a methanol solution, followed by anion exchange with sodium tetraphenylborate. The resulting monomeric complex exhibits notable characteristics including a high coordination number of seven, a distorted pentagonal–bipyramidyl geometry, asymmetric bidentate coordination of the acetate ligand, and coordination by a methanol ligand.
Structural commentary
The title compound (Fig. 1 ▸), which consists of the [Mn(TMPA)(Ac)(CH3OH)]+ monocation and tetraphenylborate counter-anion, crystallizes in the triclinic space group P
 . The manganese(II) ion is heptacoordinate with a geometry that is best described as a distorted pentagonal bipyramid. While this is a high coordination number for a first row transition metal ion, seven-coordinate manganese(II) complexes with N-donor ligands have been described previously (Deroche et al.., 1996 ▸; Policar et al., 2001 ▸; Lessa et al., 2007 ▸; Dees et al., 2007 ▸; Wu et al., 2010 ▸; Lieb et al., 2013 ▸). The TMPA ligand is tetradentate, with its central N2 and two pyridyl nitrogen atoms (N1 and N3) in the pentagonal plane, and the third pyridyl nitrogen (N4) occupying an axial position. The remaining two positions in the pentagonal plane are completed by the bidentate coordination of the acetate ligand (O2 and O3), while the final axial position is occupied by O1 of the methanol ligand. Distortion of the pentagonal–bipyramidal geometry of the coordination sphere is produced by the bite angles of the TMPA and acetate chelate rings. For example, the N2—Mn1—N4 bond angle [75.20 (4)°] of the five-membered metallacycle spanning an equatorial and axial position, is significantly reduced from 90° (Table 1 ▸). This results in a trans O1—Mn1—N4 angle of 166.95 (5)°. Likewise, the O2—Mn1—O3 bond angle [54.74 (4)°] that results from bidentate coordination of the acetate ligand is significantly reduced from the ideal 72° bond angle within the pentagonal plane. The O2—Mn1—O3 plane is also twisted outside of the the pentagonal plane by approximately 10° as a result of weak intramolecular C—H⋯O hydrogen-bonding interactions with neighboring pyridyl rings (Table 2 ▸). What is perhaps most remarkable about the bidentate coordination of the acetate ligand is how asymmetric it is. The Mn1—O2 and Mn1—O3 bond lengths differ from each other by 0.3005 Å. This does not appear to result from steric hindrance, but may be due to an intermolecular hydrogen-bonding interaction between the O2 acetate oxygen of one complex and the hydroxyl hydrogen of the coordinated methanol of another, having the effect of lengthening the Mn1—O2 bond. The bond between the manganese(II) ion and the central TMPA nitrogen, Mn1—N2 is also considerably long at 2.4092 (13) Å. This elongation has been observed in other manganese(II) complexes with tripodal, tetradentate ligands (Deroche et al., 1996 ▸; Wu et al., 2010 ▸). The other Mn—O and Mn—N bonds fall into the range 2.2–2.3 Å, which is typical of manganese(II) complexes (Deroche et al., 1996 ▸; Policar et al., 2001 ▸; Lessa et al., 2007 ▸; Dees et al., 2007 ▸; Wu et al., 2010 ▸; Lieb et al., 2012 ▸).
. The manganese(II) ion is heptacoordinate with a geometry that is best described as a distorted pentagonal bipyramid. While this is a high coordination number for a first row transition metal ion, seven-coordinate manganese(II) complexes with N-donor ligands have been described previously (Deroche et al.., 1996 ▸; Policar et al., 2001 ▸; Lessa et al., 2007 ▸; Dees et al., 2007 ▸; Wu et al., 2010 ▸; Lieb et al., 2013 ▸). The TMPA ligand is tetradentate, with its central N2 and two pyridyl nitrogen atoms (N1 and N3) in the pentagonal plane, and the third pyridyl nitrogen (N4) occupying an axial position. The remaining two positions in the pentagonal plane are completed by the bidentate coordination of the acetate ligand (O2 and O3), while the final axial position is occupied by O1 of the methanol ligand. Distortion of the pentagonal–bipyramidal geometry of the coordination sphere is produced by the bite angles of the TMPA and acetate chelate rings. For example, the N2—Mn1—N4 bond angle [75.20 (4)°] of the five-membered metallacycle spanning an equatorial and axial position, is significantly reduced from 90° (Table 1 ▸). This results in a trans O1—Mn1—N4 angle of 166.95 (5)°. Likewise, the O2—Mn1—O3 bond angle [54.74 (4)°] that results from bidentate coordination of the acetate ligand is significantly reduced from the ideal 72° bond angle within the pentagonal plane. The O2—Mn1—O3 plane is also twisted outside of the the pentagonal plane by approximately 10° as a result of weak intramolecular C—H⋯O hydrogen-bonding interactions with neighboring pyridyl rings (Table 2 ▸). What is perhaps most remarkable about the bidentate coordination of the acetate ligand is how asymmetric it is. The Mn1—O2 and Mn1—O3 bond lengths differ from each other by 0.3005 Å. This does not appear to result from steric hindrance, but may be due to an intermolecular hydrogen-bonding interaction between the O2 acetate oxygen of one complex and the hydroxyl hydrogen of the coordinated methanol of another, having the effect of lengthening the Mn1—O2 bond. The bond between the manganese(II) ion and the central TMPA nitrogen, Mn1—N2 is also considerably long at 2.4092 (13) Å. This elongation has been observed in other manganese(II) complexes with tripodal, tetradentate ligands (Deroche et al., 1996 ▸; Wu et al., 2010 ▸). The other Mn—O and Mn—N bonds fall into the range 2.2–2.3 Å, which is typical of manganese(II) complexes (Deroche et al., 1996 ▸; Policar et al., 2001 ▸; Lessa et al., 2007 ▸; Dees et al., 2007 ▸; Wu et al., 2010 ▸; Lieb et al., 2012 ▸).
Figure 1.
Molecular structure of [Mn(TMPA)(Ac)(CH3OH)]BPh4 [TMPA = tris(pyridin-2-ylmethyl)amine, Ac = acetate, BPh4 = tetraphenylborate] with atom labels. Displacement ellipsoids are drawn at the 30% probability level.
Table 1. Selected geometric parameters (Å, °).
| Mn1—O1 | 2.1941 (12) | Mn1—N2 | 2.4092 (13) |
| Mn1—O2 | 2.5009 (12) | Mn1—N3 | 2.3022 (13) |
| Mn1—O3 | 2.2004 (13) | Mn1—N4 | 2.2496 (13) |
| Mn1—N1 | 2.2769 (15) | ||
| O1—Mn1—O2 | 81.52 (4) | O2—Mn1—O3 | 54.74 (4) |
| O1—Mn1—N4 | 166.95 (5) | N2—Mn1—N4 | 75.20 (4) |
Table 2. Hydrogen-bond geometry and π–π stacking interactions (Å, °).
Cg1, Cg2, Cg3, Cg4, Cg6, and Cg7 are the centroids of the N1/C4–C8, N3/C1/C9/C10–C12, N4/C14–C18, C22–C27, C34–C39, and C40–C45 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O2i | 0.86 (1) | 1.79 (1) | 2.6480 (17) | 176 (2) |
| C8—H8⋯O2 | 0.95 | 2.45 | 3.056 (2) | 121 |
| C12—H12⋯O3 | 0.95 | 2.35 | 2.987 (2) | 124 |
| C2—H2B⋯Cg6ii | 0.99 | 2.70 | 3.6260 (18) | 156 |
| C38—H38⋯Cg4iii | 0.95 | 2.81 | 3.7135 (19) | 158 |
| C42—H42⋯Cg3iv | 0.95 | 2.96 | 3.659 (2) | 131 |
| Cg1⋯Cg7iv | 4.2073 (11) | |||
| Cg2⋯Cg3 | 4.6125 (10) | |||
| Cg3⋯Cg4v | 4.2267 (12) | |||
| Cg4⋯Cg6iii | 5.0645 (11) |
Symmetry codes: (i)  ; (ii)
; (ii) 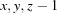 ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Supramolecular features
Within the crystal, dimerization of complexes occurs by the formation of a pair of intermoleclular O—H⋯O hydrogen bonds (Table 2 ▸) between the coordinated methanol of one complex and an acetate oxygen of another (Fig. 2 ▸) forming an  (12) ring-motif interaction. Within a dimer, weak π-stacking interactions between pyridine rings (Cg2⋯Cg3) can be detected. Separate dimers then undergo additional π-stacking between the pyridine rings of one moiety and the phenyl rings of a second (Cg1⋯Cg7 and Cg3⋯Cg4) as well as between the pyridine rings of separate moieties (Cg4⋯Cg6) [where Cg1, Cg2, Cg3, Cg4, Cg6, and Cg7 are the centroids of the N1/C4–C8, N3/C1/C9/C10–C12, N4/C14–C18, C22–C27, C34–C39, and C40–C45 rings, respectively] that further stabilize the crystal packing. In addition, weak slipped parallel C—H⋯π [C2—H2B⋯Cg6, X—H, π = 62°; C38—H38⋯Cg4, X—H, π = 61°; C42—H42⋯Cg3, X—H, π = 38°] (Table 2 ▸) intermolecular interactions are also present and contibute additionally to the crystal packing.
(12) ring-motif interaction. Within a dimer, weak π-stacking interactions between pyridine rings (Cg2⋯Cg3) can be detected. Separate dimers then undergo additional π-stacking between the pyridine rings of one moiety and the phenyl rings of a second (Cg1⋯Cg7 and Cg3⋯Cg4) as well as between the pyridine rings of separate moieties (Cg4⋯Cg6) [where Cg1, Cg2, Cg3, Cg4, Cg6, and Cg7 are the centroids of the N1/C4–C8, N3/C1/C9/C10–C12, N4/C14–C18, C22–C27, C34–C39, and C40–C45 rings, respectively] that further stabilize the crystal packing. In addition, weak slipped parallel C—H⋯π [C2—H2B⋯Cg6, X—H, π = 62°; C38—H38⋯Cg4, X—H, π = 61°; C42—H42⋯Cg3, X—H, π = 38°] (Table 2 ▸) intermolecular interactions are also present and contibute additionally to the crystal packing.
Figure 2.
A view along the b axis of the crystal packing of the title compound. The intramolecular O—H⋯O and intermolecular C—H⋯O hydrogen bonds (Table 2 ▸) are shown as dashed lines.
Database survey
A search of the Cambridge Structural Database (Version 5.39; last update May 2018; Groom et al. 2016 ▸) for manganese(II) complexes containing TMPA revealed 17 structures related to the title compound. Twelve of these are dimeric in nature and contain a variety of bridging ligands (Oshio et al., 1993 ▸; Xiang et al., 1998 ▸; Shin et al., 2010 ▸; Barros et al., 2013 ▸; Khullar & Mandal, 2013 ▸), including one with bridging acetate ligands (Oshio et al., 1993 ▸). The remaining five structures are monomeric and include monodentate ligands in addition to TMPA (Oshio et al., 1993 ▸; Hitomi et al., 2005 ▸; Duboc et al., 2008 ▸; Shin et al., 2010 ▸; Ogo et al., 2014 ▸). Of the 17 structures, 16 are six-coordinate with respect to the manganese(II) centers, while the remaining structure has a five-coordinate manganese(II) center. None of these structures reveal coordination numbers greater than six. However, a separate literature search identified an eight-coordinate complex in which one manganese(II) ion is coordinated to two tetradentate TMPA ligands (Gultneh et al., 1993 ▸).
Synthesis and crystallization
All chemicals were obtained from commercial sources and used without further preparation. The water used was deionized. The 1H NMR spectrum was recorded with a JEOL ECX-300 NMR spectrometer and referenced against the 1H peak of the chloroform solvent. IR spectra were recorded with a Perkin Elmer Spectrum 100 FT–IR.
Tris(pyridin-2-ylmethyl)amine (TMPA). In a 250 mL round-bottom flask, 10 g (61 mmol) picolyl chloride hydrochloride was dissolved in 20 mL H2O and cooled to 273 K in an ice bath. A solution of 5.0 g (120 mmol) NaOH in 20 mL H2O was added dropwise under stirring. Following this, a solution of 2-methylaminopyridine (3.3 g, 31 mmol) in CH2Cl2 (40 mL) was added. The reaction mixture was then removed from the ice bath, capped, and allowed to stir vigorously for five days. The CH2Cl2 layer was then separated, washed twice with brine, and dried over anhydrous sodium sulfate. The solution was filtered and concentrated on a rotary evaporator producing 6.5 g of a red–brown oil that solidified upon cooling. The crude product was chromatographed on alumina (chromatographic grade, 80–200 mesh) eluting with 20:1 ethyl acetate/methanol, producing 4.9 g (55%) of a pure, golden oil that solidified upon standing. 1H NMR (CDCl3, 300 MHz) δ 3.88 (s, 6H), 7.15 (t, 3H), 7.57–7.69 (m, 6H), 8.53 (d, 3H).
[Mn(TMPA)(Ac)(CH3OH)]BPh4. In a 100 mL round-bottom flask, 0.41 g (1.4 mmol) TMPA was dissolved in 10 mL of methanol. To this solution, 0.35 g (1.4 mmol) of manganese(II) acetate tetrahydrate was added, and the solution was brought to reflux for 20 minutes. A solution of 0.48 g (1.4 mmol) of sodium tetraphenylborate in 10 mL of methanol was then added dropwise to the warm reaction mixture. A precipitate formed during this addition. The reaction mixture was cooled to room temperature and filtered to produce tan microcrystals that were washed twice with cold methanol and air dried to give 0.75 g (74%) of product. The filtrate was then capped and placed in the refrigerator to promote further crystallization. After several days, crystals suitable for X-ray diffraction formed, which gave an IR spectrum identical to the original product. IR (ATR, cm−1) 3000–3053 (aromatic C—H, w), 1589 (C—O, s), 1425 (C—O, s), 731 (BPh4 −, s), 701 (BPh4 −, s).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The hydroxy H atom was located in a difference-Fourier map and refined with the distance restraint O1—H1 = 0.85 ± 0.01 and with U iso(H) = 1.2U eq(O). C-bound H atoms were positioned geometrically and refined as riding: C—H = 0.95–0.99 Å with U iso(H) = 1.2U eq(C) or 1.5U eq(C-methyl).
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | [Mn(C2H3O2)(C18H18N4)(CH4O)](C24H20B) |
| M r | 755.60 |
| Crystal system, space group | Triclinic, P

|
| Temperature (K) | 173 |
| a, b, c (Å) | 11.3885 (8), 11.7598 (7), 15.6703 (10) |
| α, β, γ (°) | 82.041 (5), 70.671 (6), 85.870 (5) |
| V (Å3) | 1960.5 (2) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.38 |
| Crystal size (mm) | 0.44 × 0.38 × 0.26 |
| Data collection | |
| Diffractometer | Rigaku Oxford Diffraction |
| Absorption correction | Multi-scan (CrysAlis PRO; Agilent, 2014 ▸) |
| T min, T max | 0.836, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 24707, 12901, 9324 |
| R int | 0.029 |
| (sin θ/λ)max (Å−1) | 0.763 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.047, 0.122, 1.03 |
| No. of reflections | 12901 |
| No. of parameters | 492 |
| No. of restraints | 3 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.36, −0.30 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989018009611/tx2006sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018009611/tx2006Isup2.hkl
CCDC reference: 1853486
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| [Mn(C2H3O2)(C18H18N4)(CH4O)](C24H20B) | Z = 2 |
| Mr = 755.60 | F(000) = 794 |
| Triclinic, P1 | Dx = 1.280 Mg m−3 |
| a = 11.3885 (8) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 11.7598 (7) Å | Cell parameters from 6236 reflections |
| c = 15.6703 (10) Å | θ = 3.5–32.2° |
| α = 82.041 (5)° | µ = 0.38 mm−1 |
| β = 70.671 (6)° | T = 173 K |
| γ = 85.870 (5)° | Prism, orange |
| V = 1960.5 (2) Å3 | 0.44 × 0.38 × 0.26 mm |
Data collection
| Rigaku Oxford Diffraction diffractometer | 12901 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 9324 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.029 |
| Detector resolution: 16.0416 pixels mm-1 | θmax = 32.8°, θmin = 3.1° |
| ω scans | h = −16→16 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2014) | k = −17→17 |
| Tmin = 0.836, Tmax = 1.000 | l = −23→23 |
| 24707 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.047 | H-atom parameters constrained |
| wR(F2) = 0.122 | w = 1/[σ2(Fo2) + (0.0476P)2 + 0.5222P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.004 |
| 12901 reflections | Δρmax = 0.36 e Å−3 |
| 492 parameters | Δρmin = −0.30 e Å−3 |
| 3 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Mn1 | 0.56866 (2) | 0.16825 (2) | 0.32745 (2) | 0.02659 (7) | |
| O1 | 0.63017 (13) | −0.01175 (10) | 0.34698 (8) | 0.0391 (3) | |
| H1 | 0.613 (2) | −0.0420 (12) | 0.4030 (7) | 0.059* | |
| O2 | 0.41990 (12) | 0.11321 (10) | 0.48365 (8) | 0.0385 (3) | |
| O3 | 0.36694 (11) | 0.14978 (11) | 0.36100 (8) | 0.0388 (3) | |
| N1 | 0.68813 (14) | 0.21241 (12) | 0.40889 (10) | 0.0360 (3) | |
| N2 | 0.77041 (12) | 0.22294 (11) | 0.22297 (9) | 0.0287 (3) | |
| N3 | 0.57535 (12) | 0.13981 (10) | 0.18346 (8) | 0.0267 (3) | |
| N4 | 0.54497 (12) | 0.35519 (11) | 0.27955 (8) | 0.0263 (3) | |
| C1 | 0.68402 (14) | 0.15094 (12) | 0.11539 (10) | 0.0270 (3) | |
| C2 | 0.79743 (15) | 0.16420 (15) | 0.14101 (11) | 0.0344 (3) | |
| H2A | 0.8349 | 0.0873 | 0.1520 | 0.041* | |
| H2B | 0.8593 | 0.2084 | 0.0894 | 0.041* | |
| C3 | 0.85963 (16) | 0.18541 (15) | 0.27130 (12) | 0.0367 (4) | |
| H3A | 0.9397 | 0.2234 | 0.2387 | 0.044* | |
| H3B | 0.8749 | 0.1014 | 0.2717 | 0.044* | |
| C4 | 0.81202 (17) | 0.21393 (14) | 0.36774 (12) | 0.0365 (4) | |
| C5 | 0.8922 (2) | 0.23550 (16) | 0.41321 (15) | 0.0491 (5) | |
| H5 | 0.9795 | 0.2366 | 0.3829 | 0.059* | |
| C6 | 0.8438 (3) | 0.25523 (18) | 0.50269 (16) | 0.0596 (6) | |
| H6 | 0.8973 | 0.2682 | 0.5355 | 0.071* | |
| C7 | 0.7165 (3) | 0.25604 (18) | 0.54439 (15) | 0.0568 (6) | |
| H7 | 0.6811 | 0.2711 | 0.6059 | 0.068* | |
| C8 | 0.6414 (2) | 0.23465 (16) | 0.49564 (12) | 0.0444 (4) | |
| H8 | 0.5537 | 0.2357 | 0.5243 | 0.053* | |
| C9 | 0.69333 (16) | 0.14408 (13) | 0.02599 (10) | 0.0325 (3) | |
| H9 | 0.7714 | 0.1523 | −0.0211 | 0.039* | |
| C10 | 0.58756 (18) | 0.12519 (14) | 0.00635 (11) | 0.0370 (4) | |
| H10 | 0.5917 | 0.1203 | −0.0546 | 0.044* | |
| C11 | 0.47552 (17) | 0.11350 (14) | 0.07615 (12) | 0.0356 (4) | |
| H11 | 0.4014 | 0.0999 | 0.0644 | 0.043* | |
| C12 | 0.47377 (15) | 0.12198 (13) | 0.16317 (11) | 0.0304 (3) | |
| H12 | 0.3965 | 0.1148 | 0.2112 | 0.037* | |
| C13 | 0.76954 (15) | 0.34957 (14) | 0.20102 (12) | 0.0343 (3) | |
| H13A | 0.8208 | 0.3691 | 0.1365 | 0.041* | |
| H13B | 0.8098 | 0.3824 | 0.2389 | 0.041* | |
| C14 | 0.64285 (14) | 0.40629 (13) | 0.21531 (10) | 0.0265 (3) | |
| C15 | 0.63108 (16) | 0.51242 (14) | 0.16752 (11) | 0.0350 (4) | |
| H15 | 0.7008 | 0.5458 | 0.1207 | 0.042* | |
| C16 | 0.51625 (18) | 0.56882 (15) | 0.18919 (14) | 0.0424 (4) | |
| H16 | 0.5064 | 0.6423 | 0.1580 | 0.051* | |
| C17 | 0.41608 (17) | 0.51760 (15) | 0.25640 (13) | 0.0389 (4) | |
| H17 | 0.3367 | 0.5555 | 0.2731 | 0.047* | |
| C18 | 0.43378 (15) | 0.41047 (13) | 0.29869 (11) | 0.0304 (3) | |
| H18 | 0.3642 | 0.3739 | 0.3435 | 0.036* | |
| C19 | 0.33901 (15) | 0.11975 (13) | 0.44512 (10) | 0.0298 (3) | |
| C20 | 0.20607 (17) | 0.09291 (19) | 0.49920 (13) | 0.0473 (5) | |
| H20A | 0.1625 | 0.1621 | 0.5240 | 0.071* | |
| H20B | 0.2037 | 0.0322 | 0.5493 | 0.071* | |
| H20C | 0.1652 | 0.0669 | 0.4596 | 0.071* | |
| C21 | 0.6314 (2) | −0.10245 (16) | 0.29597 (14) | 0.0516 (5) | |
| H21A | 0.6816 | −0.0813 | 0.2322 | 0.077* | |
| H21B | 0.5461 | −0.1167 | 0.2996 | 0.077* | |
| H21C | 0.6675 | −0.1721 | 0.3208 | 0.077* | |
| C22 | 0.87749 (14) | 0.44057 (12) | 0.82076 (11) | 0.0275 (3) | |
| C23 | 0.88441 (18) | 0.54783 (14) | 0.76863 (13) | 0.0385 (4) | |
| H23 | 0.9513 | 0.5613 | 0.7131 | 0.046* | |
| C24 | 0.7964 (2) | 0.63588 (16) | 0.79538 (17) | 0.0520 (5) | |
| H24 | 0.8044 | 0.7079 | 0.7584 | 0.062* | |
| C25 | 0.6982 (2) | 0.61887 (17) | 0.87495 (16) | 0.0518 (5) | |
| H25 | 0.6373 | 0.6783 | 0.8924 | 0.062* | |
| C26 | 0.68858 (18) | 0.51532 (17) | 0.92930 (14) | 0.0433 (4) | |
| H26 | 0.6213 | 0.5030 | 0.9847 | 0.052* | |
| C27 | 0.77791 (15) | 0.42884 (14) | 0.90271 (11) | 0.0328 (3) | |
| H27 | 0.7712 | 0.3586 | 0.9419 | 0.039* | |
| C28 | 0.90673 (13) | 0.22992 (12) | 0.76412 (9) | 0.0239 (3) | |
| C29 | 0.77827 (14) | 0.22214 (13) | 0.78503 (10) | 0.0260 (3) | |
| H29 | 0.7251 | 0.2787 | 0.8177 | 0.031* | |
| C30 | 0.72484 (15) | 0.13489 (15) | 0.75994 (11) | 0.0323 (3) | |
| H30 | 0.6369 | 0.1328 | 0.7760 | 0.039* | |
| C31 | 0.79897 (16) | 0.05108 (15) | 0.71164 (11) | 0.0343 (4) | |
| H31 | 0.7629 | −0.0084 | 0.6942 | 0.041* | |
| C32 | 0.92625 (16) | 0.05606 (14) | 0.68956 (11) | 0.0334 (3) | |
| H32 | 0.9787 | −0.0005 | 0.6564 | 0.040* | |
| C33 | 0.97842 (15) | 0.14322 (13) | 0.71539 (11) | 0.0299 (3) | |
| H33 | 1.0664 | 0.1442 | 0.6994 | 0.036* | |
| C34 | 1.02371 (13) | 0.28206 (12) | 0.87796 (10) | 0.0238 (3) | |
| C35 | 1.05266 (15) | 0.16588 (13) | 0.89976 (11) | 0.0305 (3) | |
| H35 | 1.0374 | 0.1108 | 0.8660 | 0.037* | |
| C36 | 1.10278 (16) | 0.12836 (15) | 0.96888 (12) | 0.0370 (4) | |
| H36 | 1.1217 | 0.0490 | 0.9809 | 0.044* | |
| C37 | 1.12527 (15) | 0.20534 (16) | 1.02014 (12) | 0.0361 (4) | |
| H37 | 1.1606 | 0.1798 | 1.0668 | 0.043* | |
| C38 | 1.09545 (15) | 0.32066 (15) | 1.00249 (11) | 0.0327 (3) | |
| H38 | 1.1088 | 0.3747 | 1.0379 | 0.039* | |
| C39 | 1.04616 (14) | 0.35686 (13) | 0.93316 (11) | 0.0286 (3) | |
| H39 | 1.0265 | 0.4363 | 0.9223 | 0.034* | |
| C40 | 1.09510 (15) | 0.37221 (12) | 0.70190 (10) | 0.0277 (3) | |
| C41 | 1.21400 (15) | 0.38527 (13) | 0.70467 (11) | 0.0314 (3) | |
| H41 | 1.2281 | 0.3688 | 0.7616 | 0.038* | |
| C42 | 1.31390 (17) | 0.42153 (16) | 0.62752 (13) | 0.0414 (4) | |
| H42 | 1.3934 | 0.4299 | 0.6329 | 0.050* | |
| C43 | 1.2972 (2) | 0.44500 (17) | 0.54396 (13) | 0.0484 (5) | |
| H43 | 1.3649 | 0.4689 | 0.4912 | 0.058* | |
| C44 | 1.1809 (2) | 0.43339 (18) | 0.53774 (13) | 0.0509 (5) | |
| H44 | 1.1680 | 0.4498 | 0.4804 | 0.061* | |
| C45 | 1.08230 (18) | 0.39766 (16) | 0.61519 (12) | 0.0403 (4) | |
| H45 | 1.0030 | 0.3902 | 0.6091 | 0.048* | |
| B1 | 0.97542 (15) | 0.33157 (13) | 0.79157 (11) | 0.0245 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Mn1 | 0.02860 (12) | 0.02566 (12) | 0.02341 (11) | −0.00437 (8) | −0.00563 (9) | −0.00115 (8) |
| O1 | 0.0554 (8) | 0.0279 (6) | 0.0311 (6) | 0.0023 (5) | −0.0115 (6) | −0.0019 (5) |
| O2 | 0.0386 (7) | 0.0386 (7) | 0.0363 (6) | −0.0014 (5) | −0.0102 (5) | −0.0024 (5) |
| O3 | 0.0376 (7) | 0.0460 (7) | 0.0263 (6) | −0.0104 (5) | −0.0018 (5) | 0.0002 (5) |
| N1 | 0.0463 (9) | 0.0316 (7) | 0.0326 (7) | −0.0079 (6) | −0.0160 (6) | −0.0003 (5) |
| N2 | 0.0265 (6) | 0.0273 (6) | 0.0319 (7) | −0.0024 (5) | −0.0086 (5) | −0.0034 (5) |
| N3 | 0.0292 (6) | 0.0236 (6) | 0.0241 (6) | −0.0041 (5) | −0.0042 (5) | −0.0019 (5) |
| N4 | 0.0280 (6) | 0.0262 (6) | 0.0251 (6) | −0.0031 (5) | −0.0085 (5) | −0.0036 (5) |
| C1 | 0.0289 (7) | 0.0203 (6) | 0.0270 (7) | −0.0010 (5) | −0.0030 (6) | −0.0016 (5) |
| C2 | 0.0253 (8) | 0.0398 (9) | 0.0348 (8) | −0.0005 (6) | −0.0030 (6) | −0.0107 (7) |
| C3 | 0.0305 (8) | 0.0372 (9) | 0.0446 (10) | −0.0005 (7) | −0.0156 (7) | −0.0037 (7) |
| C4 | 0.0451 (10) | 0.0272 (8) | 0.0427 (9) | −0.0071 (7) | −0.0231 (8) | 0.0026 (6) |
| C5 | 0.0594 (13) | 0.0370 (10) | 0.0644 (13) | −0.0122 (8) | −0.0396 (11) | 0.0031 (9) |
| C6 | 0.0905 (19) | 0.0443 (11) | 0.0657 (15) | −0.0172 (11) | −0.0548 (14) | 0.0016 (10) |
| C7 | 0.0952 (19) | 0.0447 (11) | 0.0406 (11) | −0.0183 (11) | −0.0340 (12) | −0.0010 (8) |
| C8 | 0.0651 (13) | 0.0388 (9) | 0.0310 (9) | −0.0133 (8) | −0.0169 (9) | 0.0000 (7) |
| C9 | 0.0385 (9) | 0.0268 (7) | 0.0245 (7) | −0.0002 (6) | −0.0002 (6) | −0.0030 (6) |
| C10 | 0.0500 (11) | 0.0342 (8) | 0.0276 (8) | −0.0006 (7) | −0.0125 (7) | −0.0070 (6) |
| C11 | 0.0402 (9) | 0.0337 (8) | 0.0373 (9) | −0.0029 (7) | −0.0160 (7) | −0.0092 (7) |
| C12 | 0.0299 (8) | 0.0296 (7) | 0.0296 (7) | −0.0064 (6) | −0.0054 (6) | −0.0042 (6) |
| C13 | 0.0279 (8) | 0.0296 (8) | 0.0400 (9) | −0.0061 (6) | −0.0049 (7) | 0.0011 (6) |
| C14 | 0.0288 (7) | 0.0273 (7) | 0.0247 (7) | −0.0056 (6) | −0.0092 (6) | −0.0034 (5) |
| C15 | 0.0370 (9) | 0.0350 (8) | 0.0344 (8) | −0.0118 (7) | −0.0154 (7) | 0.0057 (6) |
| C16 | 0.0436 (10) | 0.0337 (9) | 0.0539 (11) | −0.0046 (7) | −0.0266 (9) | 0.0104 (8) |
| C17 | 0.0343 (9) | 0.0360 (9) | 0.0493 (10) | 0.0015 (7) | −0.0194 (8) | −0.0017 (7) |
| C18 | 0.0288 (8) | 0.0315 (8) | 0.0306 (8) | −0.0032 (6) | −0.0086 (6) | −0.0043 (6) |
| C19 | 0.0329 (8) | 0.0229 (7) | 0.0279 (7) | −0.0030 (6) | −0.0017 (6) | −0.0038 (5) |
| C20 | 0.0340 (9) | 0.0643 (13) | 0.0331 (9) | −0.0098 (8) | 0.0030 (7) | −0.0010 (8) |
| C21 | 0.0766 (15) | 0.0312 (9) | 0.0400 (10) | −0.0003 (9) | −0.0088 (10) | −0.0067 (7) |
| C22 | 0.0298 (8) | 0.0228 (7) | 0.0353 (8) | −0.0003 (5) | −0.0167 (6) | −0.0068 (6) |
| C23 | 0.0437 (10) | 0.0255 (8) | 0.0508 (10) | −0.0020 (7) | −0.0224 (8) | −0.0015 (7) |
| C24 | 0.0655 (14) | 0.0262 (9) | 0.0791 (15) | 0.0092 (8) | −0.0450 (13) | −0.0071 (9) |
| C25 | 0.0553 (13) | 0.0427 (11) | 0.0744 (15) | 0.0252 (9) | −0.0403 (12) | −0.0301 (10) |
| C26 | 0.0363 (9) | 0.0539 (11) | 0.0498 (11) | 0.0155 (8) | −0.0217 (8) | −0.0294 (9) |
| C27 | 0.0329 (8) | 0.0333 (8) | 0.0356 (8) | 0.0059 (6) | −0.0139 (7) | −0.0121 (6) |
| C28 | 0.0258 (7) | 0.0229 (6) | 0.0228 (6) | −0.0036 (5) | −0.0074 (5) | −0.0022 (5) |
| C29 | 0.0254 (7) | 0.0287 (7) | 0.0241 (7) | −0.0019 (5) | −0.0082 (5) | −0.0032 (5) |
| C30 | 0.0270 (8) | 0.0430 (9) | 0.0277 (7) | −0.0105 (6) | −0.0078 (6) | −0.0048 (6) |
| C31 | 0.0387 (9) | 0.0379 (9) | 0.0272 (7) | −0.0170 (7) | −0.0077 (6) | −0.0061 (6) |
| C32 | 0.0371 (9) | 0.0296 (8) | 0.0327 (8) | −0.0054 (6) | −0.0066 (7) | −0.0112 (6) |
| C33 | 0.0261 (7) | 0.0310 (8) | 0.0336 (8) | −0.0037 (6) | −0.0081 (6) | −0.0095 (6) |
| C34 | 0.0187 (6) | 0.0241 (6) | 0.0263 (7) | −0.0019 (5) | −0.0035 (5) | −0.0043 (5) |
| C35 | 0.0314 (8) | 0.0282 (7) | 0.0323 (8) | 0.0028 (6) | −0.0098 (6) | −0.0084 (6) |
| C36 | 0.0355 (9) | 0.0330 (8) | 0.0415 (9) | 0.0100 (7) | −0.0134 (7) | −0.0047 (7) |
| C37 | 0.0270 (8) | 0.0482 (10) | 0.0347 (8) | 0.0034 (7) | −0.0130 (7) | −0.0046 (7) |
| C38 | 0.0282 (8) | 0.0393 (9) | 0.0331 (8) | −0.0051 (6) | −0.0109 (6) | −0.0080 (7) |
| C39 | 0.0268 (7) | 0.0258 (7) | 0.0334 (8) | −0.0046 (5) | −0.0087 (6) | −0.0053 (6) |
| C40 | 0.0312 (8) | 0.0214 (7) | 0.0298 (7) | −0.0058 (5) | −0.0072 (6) | −0.0047 (5) |
| C41 | 0.0291 (8) | 0.0293 (7) | 0.0326 (8) | −0.0060 (6) | −0.0035 (6) | −0.0072 (6) |
| C42 | 0.0326 (9) | 0.0410 (9) | 0.0434 (10) | −0.0115 (7) | 0.0009 (7) | −0.0089 (8) |
| C43 | 0.0513 (12) | 0.0440 (10) | 0.0369 (10) | −0.0175 (8) | 0.0060 (8) | −0.0043 (8) |
| C44 | 0.0677 (14) | 0.0531 (12) | 0.0283 (9) | −0.0187 (10) | −0.0109 (9) | 0.0040 (8) |
| C45 | 0.0447 (10) | 0.0445 (10) | 0.0324 (9) | −0.0129 (8) | −0.0128 (7) | −0.0008 (7) |
| B1 | 0.0241 (8) | 0.0207 (7) | 0.0282 (8) | −0.0029 (6) | −0.0069 (6) | −0.0041 (6) |
Geometric parameters (Å, º)
| Mn1—O1 | 2.1941 (12) | C20—H20A | 0.9800 |
| Mn1—O2 | 2.5009 (12) | C20—H20B | 0.9800 |
| Mn1—O3 | 2.2004 (13) | C20—H20C | 0.9800 |
| Mn1—N1 | 2.2769 (15) | C21—H21A | 0.9800 |
| Mn1—N2 | 2.4092 (13) | C21—H21B | 0.9800 |
| Mn1—N3 | 2.3022 (13) | C21—H21C | 0.9800 |
| Mn1—N4 | 2.2496 (13) | C22—C23 | 1.397 (2) |
| O1—H1 | 0.863 (9) | C22—C27 | 1.401 (2) |
| O1—C21 | 1.415 (2) | C22—B1 | 1.648 (2) |
| O2—C19 | 1.251 (2) | C23—H23 | 0.9500 |
| O3—C19 | 1.2541 (19) | C23—C24 | 1.395 (3) |
| N1—C4 | 1.343 (2) | C24—H24 | 0.9500 |
| N1—C8 | 1.341 (2) | C24—C25 | 1.373 (3) |
| N2—C2 | 1.475 (2) | C25—H25 | 0.9500 |
| N2—C3 | 1.467 (2) | C25—C26 | 1.375 (3) |
| N2—C13 | 1.481 (2) | C26—H26 | 0.9500 |
| N3—C1 | 1.3406 (19) | C26—C27 | 1.389 (2) |
| N3—C12 | 1.334 (2) | C27—H27 | 0.9500 |
| N4—C14 | 1.3428 (19) | C28—C29 | 1.396 (2) |
| N4—C18 | 1.342 (2) | C28—C33 | 1.404 (2) |
| C1—C2 | 1.498 (2) | C28—B1 | 1.651 (2) |
| C1—C9 | 1.383 (2) | C29—H29 | 0.9500 |
| C2—H2A | 0.9900 | C29—C30 | 1.392 (2) |
| C2—H2B | 0.9900 | C30—H30 | 0.9500 |
| C3—H3A | 0.9900 | C30—C31 | 1.386 (2) |
| C3—H3B | 0.9900 | C31—H31 | 0.9500 |
| C3—C4 | 1.504 (3) | C31—C32 | 1.378 (2) |
| C4—C5 | 1.386 (3) | C32—H32 | 0.9500 |
| C5—H5 | 0.9500 | C32—C33 | 1.389 (2) |
| C5—C6 | 1.372 (3) | C33—H33 | 0.9500 |
| C6—H6 | 0.9500 | C34—C35 | 1.404 (2) |
| C6—C7 | 1.379 (4) | C34—C39 | 1.406 (2) |
| C7—H7 | 0.9500 | C34—B1 | 1.644 (2) |
| C7—C8 | 1.377 (3) | C35—H35 | 0.9500 |
| C8—H8 | 0.9500 | C35—C36 | 1.391 (2) |
| C9—H9 | 0.9500 | C36—H36 | 0.9500 |
| C9—C10 | 1.378 (3) | C36—C37 | 1.379 (3) |
| C10—H10 | 0.9500 | C37—H37 | 0.9500 |
| C10—C11 | 1.379 (2) | C37—C38 | 1.387 (2) |
| C11—H11 | 0.9500 | C38—H38 | 0.9500 |
| C11—C12 | 1.375 (2) | C38—C39 | 1.385 (2) |
| C12—H12 | 0.9500 | C39—H39 | 0.9500 |
| C13—H13A | 0.9900 | C40—C41 | 1.389 (2) |
| C13—H13B | 0.9900 | C40—C45 | 1.403 (2) |
| C13—C14 | 1.505 (2) | C40—B1 | 1.643 (2) |
| C14—C15 | 1.386 (2) | C41—H41 | 0.9500 |
| C15—H15 | 0.9500 | C41—C42 | 1.398 (2) |
| C15—C16 | 1.382 (3) | C42—H42 | 0.9500 |
| C16—H16 | 0.9500 | C42—C43 | 1.372 (3) |
| C16—C17 | 1.379 (3) | C43—H43 | 0.9500 |
| C17—H17 | 0.9500 | C43—C44 | 1.378 (3) |
| C17—C18 | 1.374 (2) | C44—H44 | 0.9500 |
| C18—H18 | 0.9500 | C44—C45 | 1.391 (3) |
| C19—C20 | 1.502 (2) | C45—H45 | 0.9500 |
| O1—Mn1—O2 | 81.52 (4) | C18—C17—C16 | 118.46 (16) |
| O1—Mn1—O3 | 101.03 (5) | C18—C17—H17 | 120.8 |
| O1—Mn1—N1 | 88.33 (5) | N4—C18—C17 | 122.91 (15) |
| O1—Mn1—N2 | 92.28 (5) | N4—C18—H18 | 118.5 |
| O1—Mn1—N3 | 88.03 (4) | C17—C18—H18 | 118.5 |
| O1—Mn1—N4 | 166.95 (5) | O2—C19—O3 | 120.79 (15) |
| O2—Mn1—O3 | 54.74 (4) | O2—C19—C20 | 120.41 (15) |
| O3—Mn1—N1 | 132.87 (5) | O3—C19—C20 | 118.79 (16) |
| O3—Mn1—N2 | 152.00 (5) | C19—C20—H20A | 109.5 |
| O3—Mn1—N3 | 83.91 (5) | C19—C20—H20B | 109.5 |
| O3—Mn1—N4 | 88.91 (5) | C19—C20—H20C | 109.5 |
| N1—Mn1—O2 | 81.88 (5) | H20A—C20—H20B | 109.5 |
| N1—Mn1—N2 | 71.41 (5) | H20A—C20—H20C | 109.5 |
| N1—Mn1—N3 | 142.97 (5) | H20B—C20—H20C | 109.5 |
| N2—Mn1—O2 | 152.77 (5) | O1—C21—H21A | 109.5 |
| N3—Mn1—O2 | 133.76 (4) | O1—C21—H21B | 109.5 |
| N3—Mn1—N2 | 71.94 (5) | O1—C21—H21C | 109.5 |
| N4—Mn1—O2 | 111.30 (4) | H21A—C21—H21B | 109.5 |
| N4—Mn1—N1 | 91.07 (5) | H21A—C21—H21C | 109.5 |
| N2—Mn1—N4 | 75.20 (4) | H21B—C21—H21C | 109.5 |
| N4—Mn1—N3 | 84.61 (4) | C23—C22—C27 | 115.29 (15) |
| Mn1—O1—H1 | 115.2 (11) | C23—C22—B1 | 124.68 (15) |
| C21—O1—Mn1 | 129.43 (12) | C27—C22—B1 | 120.02 (13) |
| C21—O1—H1 | 106.3 (11) | C22—C23—H23 | 118.9 |
| C19—O2—Mn1 | 85.21 (9) | C24—C23—C22 | 122.17 (19) |
| C19—O3—Mn1 | 99.20 (11) | C24—C23—H23 | 118.9 |
| C4—N1—Mn1 | 117.72 (11) | C23—C24—H24 | 119.9 |
| C8—N1—Mn1 | 123.44 (13) | C25—C24—C23 | 120.28 (19) |
| C8—N1—C4 | 118.83 (16) | C25—C24—H24 | 119.9 |
| C2—N2—Mn1 | 109.13 (9) | C24—C25—H25 | 120.2 |
| C2—N2—C13 | 112.02 (13) | C24—C25—C26 | 119.65 (17) |
| C3—N2—Mn1 | 106.06 (10) | C26—C25—H25 | 120.2 |
| C3—N2—C2 | 110.69 (13) | C25—C26—H26 | 120.2 |
| C3—N2—C13 | 110.45 (13) | C25—C26—C27 | 119.6 (2) |
| C13—N2—Mn1 | 108.28 (9) | C27—C26—H26 | 120.2 |
| C1—N3—Mn1 | 118.77 (10) | C22—C27—H27 | 118.5 |
| C12—N3—Mn1 | 122.76 (10) | C26—C27—C22 | 122.97 (17) |
| C12—N3—C1 | 118.23 (13) | C26—C27—H27 | 118.5 |
| C14—N4—Mn1 | 117.29 (10) | C29—C28—C33 | 114.96 (13) |
| C18—N4—Mn1 | 123.01 (10) | C29—C28—B1 | 124.89 (13) |
| C18—N4—C14 | 118.46 (13) | C33—C28—B1 | 120.14 (13) |
| N3—C1—C2 | 116.93 (14) | C28—C29—H29 | 118.6 |
| N3—C1—C9 | 122.07 (15) | C30—C29—C28 | 122.70 (14) |
| C9—C1—C2 | 120.91 (14) | C30—C29—H29 | 118.6 |
| N2—C2—C1 | 112.94 (13) | C29—C30—H30 | 119.7 |
| N2—C2—H2A | 109.0 | C31—C30—C29 | 120.52 (15) |
| N2—C2—H2B | 109.0 | C31—C30—H30 | 119.7 |
| C1—C2—H2A | 109.0 | C30—C31—H31 | 120.8 |
| C1—C2—H2B | 109.0 | C32—C31—C30 | 118.45 (14) |
| H2A—C2—H2B | 107.8 | C32—C31—H31 | 120.8 |
| N2—C3—H3A | 109.4 | C31—C32—H32 | 119.8 |
| N2—C3—H3B | 109.4 | C31—C32—C33 | 120.48 (15) |
| N2—C3—C4 | 111.33 (14) | C33—C32—H32 | 119.8 |
| H3A—C3—H3B | 108.0 | C28—C33—H33 | 118.6 |
| C4—C3—H3A | 109.4 | C32—C33—C28 | 122.87 (15) |
| C4—C3—H3B | 109.4 | C32—C33—H33 | 118.6 |
| N1—C4—C3 | 116.60 (15) | C35—C34—C39 | 114.69 (14) |
| N1—C4—C5 | 121.68 (18) | C35—C34—B1 | 124.20 (13) |
| C5—C4—C3 | 121.66 (18) | C39—C34—B1 | 121.01 (13) |
| C4—C5—H5 | 120.4 | C34—C35—H35 | 118.7 |
| C6—C5—C4 | 119.1 (2) | C36—C35—C34 | 122.57 (15) |
| C6—C5—H5 | 120.4 | C36—C35—H35 | 118.7 |
| C5—C6—H6 | 120.4 | C35—C36—H36 | 119.7 |
| C5—C6—C7 | 119.2 (2) | C37—C36—C35 | 120.60 (16) |
| C7—C6—H6 | 120.4 | C37—C36—H36 | 119.7 |
| C6—C7—H7 | 120.5 | C36—C37—H37 | 120.6 |
| C8—C7—C6 | 119.1 (2) | C36—C37—C38 | 118.89 (15) |
| C8—C7—H7 | 120.5 | C38—C37—H37 | 120.6 |
| N1—C8—C7 | 122.1 (2) | C37—C38—H38 | 120.1 |
| N1—C8—H8 | 119.0 | C39—C38—C37 | 119.87 (15) |
| C7—C8—H8 | 119.0 | C39—C38—H38 | 120.1 |
| C1—C9—H9 | 120.5 | C34—C39—H39 | 118.3 |
| C10—C9—C1 | 118.90 (15) | C38—C39—C34 | 123.35 (14) |
| C10—C9—H9 | 120.5 | C38—C39—H39 | 118.3 |
| C9—C10—H10 | 120.4 | C41—C40—C45 | 114.91 (15) |
| C9—C10—C11 | 119.28 (15) | C41—C40—B1 | 124.17 (14) |
| C11—C10—H10 | 120.4 | C45—C40—B1 | 120.91 (14) |
| C10—C11—H11 | 120.8 | C40—C41—H41 | 118.5 |
| C12—C11—C10 | 118.34 (16) | C40—C41—C42 | 123.04 (16) |
| C12—C11—H11 | 120.8 | C42—C41—H41 | 118.5 |
| N3—C12—C11 | 123.17 (15) | C41—C42—H42 | 120.0 |
| N3—C12—H12 | 118.4 | C43—C42—C41 | 120.06 (18) |
| C11—C12—H12 | 118.4 | C43—C42—H42 | 120.0 |
| N2—C13—H13A | 108.4 | C42—C43—H43 | 120.5 |
| N2—C13—H13B | 108.4 | C42—C43—C44 | 119.05 (17) |
| N2—C13—C14 | 115.31 (13) | C44—C43—H43 | 120.5 |
| H13A—C13—H13B | 107.5 | C43—C44—H44 | 119.9 |
| C14—C13—H13A | 108.4 | C43—C44—C45 | 120.23 (18) |
| C14—C13—H13B | 108.4 | C45—C44—H44 | 119.9 |
| N4—C14—C13 | 118.13 (13) | C40—C45—H45 | 118.6 |
| N4—C14—C15 | 121.82 (15) | C44—C45—C40 | 122.71 (18) |
| C15—C14—C13 | 119.89 (14) | C44—C45—H45 | 118.6 |
| C14—C15—H15 | 120.6 | C22—B1—C28 | 110.31 (12) |
| C16—C15—C14 | 118.85 (16) | C34—B1—C22 | 108.42 (12) |
| C16—C15—H15 | 120.6 | C34—B1—C28 | 110.47 (11) |
| C15—C16—H16 | 120.3 | C40—B1—C22 | 110.69 (12) |
| C17—C16—C15 | 119.44 (16) | C40—B1—C28 | 107.22 (12) |
| C17—C16—H16 | 120.3 | C40—B1—C34 | 109.72 (12) |
| C16—C17—H17 | 120.8 | ||
| Mn1—O2—C19—O3 | −2.16 (14) | C23—C22—B1—C28 | −109.03 (17) |
| Mn1—O2—C19—C20 | 178.52 (15) | C23—C22—B1—C34 | 129.87 (15) |
| Mn1—O3—C19—O2 | 2.48 (16) | C23—C22—B1—C40 | 9.5 (2) |
| Mn1—O3—C19—C20 | −178.19 (13) | C23—C24—C25—C26 | 1.4 (3) |
| Mn1—N1—C4—C3 | −0.32 (19) | C24—C25—C26—C27 | −0.4 (3) |
| Mn1—N1—C4—C5 | −177.45 (13) | C25—C26—C27—C22 | −1.7 (3) |
| Mn1—N1—C8—C7 | 177.02 (14) | C27—C22—C23—C24 | −1.6 (2) |
| Mn1—N2—C2—C1 | 35.27 (16) | C27—C22—B1—C28 | 70.36 (17) |
| Mn1—N2—C3—C4 | −43.91 (15) | C27—C22—B1—C34 | −50.73 (17) |
| Mn1—N2—C13—C14 | −22.55 (17) | C27—C22—B1—C40 | −171.13 (14) |
| Mn1—N3—C1—C2 | 8.77 (17) | C28—C29—C30—C31 | −0.4 (2) |
| Mn1—N3—C1—C9 | −174.82 (11) | C29—C28—C33—C32 | 0.1 (2) |
| Mn1—N3—C12—C11 | 174.96 (12) | C29—C28—B1—C22 | −15.7 (2) |
| Mn1—N4—C14—C13 | −18.61 (18) | C29—C28—B1—C34 | 104.11 (16) |
| Mn1—N4—C14—C15 | 166.15 (12) | C29—C28—B1—C40 | −136.36 (14) |
| Mn1—N4—C18—C17 | −167.80 (13) | C29—C30—C31—C32 | 0.3 (2) |
| N1—C4—C5—C6 | 0.3 (3) | C30—C31—C32—C33 | 0.1 (3) |
| N2—C3—C4—N1 | 32.0 (2) | C31—C32—C33—C28 | −0.2 (3) |
| N2—C3—C4—C5 | −150.90 (16) | C33—C28—C29—C30 | 0.3 (2) |
| N2—C13—C14—N4 | 28.7 (2) | C33—C28—B1—C22 | 163.65 (13) |
| N2—C13—C14—C15 | −155.92 (15) | C33—C28—B1—C34 | −76.48 (17) |
| N3—C1—C2—N2 | −30.8 (2) | C33—C28—B1—C40 | 43.04 (18) |
| N3—C1—C9—C10 | 0.0 (2) | C34—C35—C36—C37 | 0.6 (3) |
| N4—C14—C15—C16 | 2.5 (2) | C35—C34—C39—C38 | 1.4 (2) |
| C1—N3—C12—C11 | 0.6 (2) | C35—C34—B1—C22 | 146.46 (14) |
| C1—C9—C10—C11 | −0.1 (2) | C35—C34—B1—C28 | 25.47 (19) |
| C2—N2—C3—C4 | −162.15 (14) | C35—C34—B1—C40 | −92.54 (16) |
| C2—N2—C13—C14 | 97.84 (16) | C35—C36—C37—C38 | 0.9 (3) |
| C2—C1—C9—C10 | 176.25 (15) | C36—C37—C38—C39 | −1.2 (2) |
| C3—N2—C2—C1 | 151.62 (14) | C37—C38—C39—C34 | 0.0 (2) |
| C3—N2—C13—C14 | −138.27 (15) | C39—C34—C35—C36 | −1.7 (2) |
| C3—C4—C5—C6 | −176.65 (17) | C39—C34—B1—C22 | −37.39 (18) |
| C4—N1—C8—C7 | −1.7 (3) | C39—C34—B1—C28 | −158.39 (13) |
| C4—C5—C6—C7 | −1.7 (3) | C39—C34—B1—C40 | 83.61 (16) |
| C5—C6—C7—C8 | 1.3 (3) | C40—C41—C42—C43 | −0.6 (3) |
| C6—C7—C8—N1 | 0.4 (3) | C41—C40—C45—C44 | −0.1 (3) |
| C8—N1—C4—C3 | 178.47 (15) | C41—C40—B1—C22 | 107.07 (16) |
| C8—N1—C4—C5 | 1.3 (2) | C41—C40—B1—C28 | −132.55 (14) |
| C9—C1—C2—N2 | 152.78 (14) | C41—C40—B1—C34 | −12.55 (19) |
| C9—C10—C11—C12 | 0.4 (2) | C41—C42—C43—C44 | 0.6 (3) |
| C10—C11—C12—N3 | −0.7 (2) | C42—C43—C44—C45 | −0.4 (3) |
| C12—N3—C1—C2 | −176.64 (13) | C43—C44—C45—C40 | 0.1 (3) |
| C12—N3—C1—C9 | −0.2 (2) | C45—C40—C41—C42 | 0.3 (2) |
| C13—N2—C2—C1 | −84.62 (16) | C45—C40—B1—C22 | −72.30 (18) |
| C13—N2—C3—C4 | 73.20 (17) | C45—C40—B1—C28 | 48.07 (18) |
| C13—C14—C15—C16 | −172.67 (16) | C45—C40—B1—C34 | 168.08 (14) |
| C14—N4—C18—C17 | −0.9 (2) | B1—C22—C23—C24 | 177.84 (16) |
| C14—C15—C16—C17 | −1.1 (3) | B1—C22—C27—C26 | −176.78 (15) |
| C15—C16—C17—C18 | −1.1 (3) | B1—C28—C29—C30 | 179.69 (14) |
| C16—C17—C18—N4 | 2.2 (3) | B1—C28—C33—C32 | −179.38 (14) |
| C18—N4—C14—C13 | 173.78 (14) | B1—C34—C35—C36 | 174.68 (15) |
| C18—N4—C14—C15 | −1.5 (2) | B1—C34—C39—C38 | −175.12 (14) |
| C22—C23—C24—C25 | −0.4 (3) | B1—C40—C41—C42 | −179.08 (14) |
| C23—C22—C27—C26 | 2.7 (2) | B1—C40—C45—C44 | 179.36 (17) |
Hydrogen-bond geometry (Å, º)
Cg1, Cg2, Cg3, Cg4, Cg6, and Cg7 are the centroids of the N1/C4–C8, N3/C1/C9/C10–C12, N4/C14–C18, C22–C27, C34–C39, and C40–C45 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O2i | 0.86 (1) | 1.79 (1) | 2.6480 (17) | 176 (2) |
| C8—H8···O2 | 0.95 | 2.45 | 3.056 (2) | 121 |
| C12—H12···O3 | 0.95 | 2.35 | 2.987 (2) | 124 |
| C2—H2B···Cg6ii | 0.99 | 2.70 | 3.6260 (18) | 156 |
| C38—H38···Cg4iii | 0.95 | 2.81 | 3.7135 (19) | 158 |
| C42—H42···Cg3iv | 0.95 | 2.96 | 3.659 (2) | 131 |
| Cg1···Cg7iv | 4.2073 (11) | |||
| Cg2···Cg3 | 4.6125 (10) | |||
| Cg3···Cg4v | 4.2267 (12) | |||
| Cg4···Cg6iii | 5.0645 (11) |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) x, y, z−1; (iii) −x+2, −y+1, −z+2; (iv) −x+2, −y+1, −z+1; (v) −x+1, −y+1, −z+1.
Selected bond lengths (Å) and angles (°) of the title compound
| Mn1—O1 | 2.1941 (12) | Mn1—N4 | 2.2496 (13) |
| Mn1—O2 | 2.5009 (12) | O1—Mn1—N4 | 166.95 (5) |
| Mn1—O3 | 2.2004 (13) | O2—Mn1—O3 | 54.74 (4) |
| Mn1—N1 | 2.2769 (15) | N2—Mn1—N4 | 75.20 (4) |
| Mn1—N2 | 2.4092 (13) | O1—Mn1—O2 | 81.52 (4) |
| Mn1—N3 | 2.3022 (13) |
Funding Statement
This work was funded by NSF-MRI grant CHE-1039027 to Jerry P. Jasinski.
References
- Agilent (2014). CrysAlis PRO. Agilent Technologies Ltd, Yarnton, England.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Bani, D. & Bencini, A. (2012). Curr. Med. Chem. 19, 4431–4444. [DOI] [PubMed]
- Barros, W. P., Inglis, R., Nichol, G. S., Rajeshkumar, T., Rajaraman, G., Piligkos, S., Stumpf, H. O. & Brechin, E. K. (2013). Dalton Trans. 42, 16510–16517. [DOI] [PubMed]
- Batinić-Haberle, I., Rebouças, J. S. & Spasojević, I. (2010). Antioxid. Redox Signal. 13, 877–918. [DOI] [PMC free article] [PubMed]
- Batinić-Haberle, I., Tovmasyan, A., Roberts, E. R. H., Vujasković, Z., Leong, K. W. & Spasojevic, I. (2014). Antioxid. Redox Signal. 20, 2372–2415. [DOI] [PMC free article] [PubMed]
- Dees, A., Zahl, A., Puchta, R., van Eikema Hommes, N. J. R., Heinemann, F. W. & Ivanović-Burmazović, I. (2007). Inorg. Chem. 46, 2459–2470. [DOI] [PubMed]
- Deroche, A., Morgenstern-Badarau, I., Cesario, M., Guilhem, J., Keita, B., Nadjo, L. & Houée-Levin, C. (1996). J. Am. Chem. Soc. 118, 4567–4573.
- Duboc, C., Collomb, M.-N., Pécaut, J., Deronzier, A. & Neese, F. (2008). Chem. Eur. J. 14, 6498–6509. [DOI] [PubMed]
- Durot, S., Policar, C., Cisnetti, F., Lambert, F., Renault, J.-P., Pelosi, G., Blain, G., Korri-Youssoufi, H. & Mahy, J.-P. (2005). Eur. J. Inorg. Chem. pp. 3513–3523.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Gultneh, Y., Farooq, A., Karlin, K. D., Liu, S. & Zubiet, J. (1993). Inorg. Chim. Acta, 211, 171–175.
- Hitomi, Y., Ando, A., Matsui, H., Ito, T., Tanaka, T., Ogo, S. & Funabiki, T. (2005). Inorg. Chem. 44, 3473–3478. [DOI] [PubMed]
- Iranzo, O. (2011). Bioorg. Chem. 39, 73–87. [DOI] [PubMed]
- Khullar, S. & Mandal, S. K. (2013). CrystEngComm, 15, 6652–6662.
- Lessa, J. A., Horn, A. Jr, Pinheiro, C. B., Farah, L. L., Eberlin, M. N., Benassi, M., Catharino, R. R. & Fernandes, S. (2007). Inorg. Chem. Commun. 10, 863–866.
- Lieb, D., Friedel, F. C., Yawer, M., Zahl, A., Khusniyarov, M. M., Heinemann, F. W. & Ivanovíc-Burmazović, I. (2013). Inorg. Chem. 52, 222–236. [DOI] [PubMed]
- Miriyala, S., Spasojević, I., Tovmasyan, A., Salvemini, D., Vujasković, Z., St. Clair, D. & Batinić-Haberle, I. (2012). Biochim. Biophys. Acta, 1822, 794–814. [DOI] [PMC free article] [PubMed]
- Ogo, S., Wantanabe, Y. & Funabiki, T. (2014). Private Communication (Refcode 117555). CCDC, Cambridge, England.
- Oshio, H., Ino, E., Mogi, I. & Ito, T. (1993). Inorg. Chem. 32, 5697–5703.
- Policar, C. (2016). Redox-Active Therapeutics, edited by I. Batinić-Haberle, J. Robouças & I. Spasojević, pp. 125–164. Switzerland: Springer International Publishing.
- Policar, C., Durot, S., Lambert, F., Cesario, M., Ramiandrasoa, F. & Morgenstern-Badarau, I. (2001). Eur. J. Inorg. Chem. pp. 1807–1818.
- Ribeiro, T., Fernandes, C., Melo, K. V., Ferreira, S. S., Lessa, J. A., Franco, R. W. A., Schenk, G., Pereira, M. D. & Horn, A. Jr (2015). Free Radical Biol. Med. 80, 67–76. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst A71, 3–8.
- Sheldrick, G. M. (2015). Acta Cryst C71, 3–8.
- Shin, B. K., Kim, M. & Han, J. (2010). Polyhedron, 29, 2560–2568.
- Wu, H., Yuan, J., Qi, B., Kong, J., Kou, F., Jiaa, F., Fan, X. & Wang, Y. (2010). Z. Naturforsch. Teil B, 65, 1097–1100.
- Xiang, D. F., Duan, C. Y., Tan, X. S., Liu, Y. J. & Tang, W. X. (1998). Polyhedron, 17, 2647–2653.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989018009611/tx2006sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018009611/tx2006Isup2.hkl
CCDC reference: 1853486
Additional supporting information: crystallographic information; 3D view; checkCIF report




