Abstract
Generation of induced pluripotent stem cells (iPSCs) with naive pluripotency is important for their applications in regenerative medicine. In female iPSCs, acquisition of naive pluripotency is coupled to X chromosome reactivation (XCR) during somatic cell reprogramming, and live cell monitoring of XCR is potentially useful for analyzing how iPSCs acquire naive pluripotency. Here we generated female mouse embryonic stem cells (ESCs) that carry the enhanced green fluorescent protein (EGFP) and humanized Kusabira-Orange (hKO) genes inserted into an intergenic site near either the Syap1 or Taf1 gene on both X chromosomes. The ESC clones, which initially expressed both EGFP and hKO, inactivated one of the fluorescent protein genes upon differentiation, indicating that the EGFP and hKO genes are subject to X chromosome inactivation (XCI). When the derived somatic cells carrying the EGFP gene on the inactive X chromosome (Xi) were reprogrammed into iPSCs, the EGFP gene on the Xi was reactivated when pluripotency marker genes were induced. Thus, the fluorescent protein genes inserted into an intergenic locus on both X chromosomes enable live cell monitoring of XCI during ESC differentiation and XCR during reprogramming. This is the first study that succeeded live cell imaging of XCR during reprogramming.
Abbreviations: iPSCs, induced pluripotent stem cells; ESCs, embryonic stem cells; XCR, X chromosome reactivation; XCI, X chromosome inactivation; EGFP, enhanced green fluorescent protein; hKO, humanized Kusabira Orange; Xist, X-inactive specific transcript; Xi, inactive X chromosome; Xa, active X chromosome
Keywords: X chromosome reactivation, Reprogramming, Live cell imaging, CRISPR/Cas9
1. Introduction
iPSCs, generated by introduction of defined reprogramming factors into somatic cells [1], hold great promise for regenerative medicine and drug development [2]. However, iPSC generation is beset by inefficiency of reprogramming and heterogeneity of obtained cell populations [3]. It is therefore important to improve the efficiency of somatic cell reprogramming and select for iPSCs with full pluripotency.
iPSCs and ESCs display two distinct phases of pluripotency, the primed and naive states [4]. The ESCs in the naive state show higher ability to differentiate than those in the primed state [4], and generation of iPSCs with the pluripotency equivalent to the naive state is critical for their application in regenerative medicine. The naive state of pluripotency can be distinguished from the primed state by cell morphology, gene expression pattern, dependence on growth factors as well as, in the case of female cells, the presence of two active X chromosomes [4].
In eutherian mammals, female cells possess two X chromosomes, one of which is epigenetically inactivated during the early phase of embryonic development by a dosage compensation mechanism termed XCI [5]. XCI strictly depends on the X-inactive specific transcript (Xist) gene encoding a non-coding RNA, which plays a central role in inactivating the X chromosome in cis. Female somatic cells possess one active X chromosome (Xa) and one Xi, but once reprogrammed into the fully pluripotent state, female somatic cells reactivate Xi by a reverse process termed XCR [6]. Recent studies showed that XCR is closely coupled to acquisition of pluripotency by iPSCs [6], [7]. Thus, monitoring XCR may enable evaluation of pluripotency acquisition by iPSCs during somatic cell reprogramming.
Here we used the CRISPR/Cas9 system [8], [9] to generate female ESCs that carry the EGFP gene on one X chromosome and the hKO gene on the other. The obtained ESC clones expressed both EGFP and hKO, one of which was repressed in a random mode upon differentiation, concurrent with up-regulation of the Xist expression. The derived somatic cells that expressed only hKO were reprogrammed by Sendai virus expressing OCT4, SOX2, KLF4, and c-MYC [10], and found to initiate expression of EGFP when pluripotency marker genes were induced.
2. Materials and methods
2.1. Plasmids and guide RNAs
pX330-U6-Chimeric_BB-CBh-hSpCas9 (#42230) was purchased from Addgene. pPyCAG-EGFP-IP and pPyCAG-EGFP-IZ were generous gifts from Dr. Hitoshi Niwa (RIKEN CDB). Guide RNAs (gRNAs) were designed using CRISPRdirect (https://crispr.dbcls.jp), and the gRNAs that had the minimum potential off-target effects were chosen for the S and T locus (Fig. 1A, B and Supplementary Table 1). The B6N mouse Bac clones B6Ng01-177J10 (for the S locus) and B6Ng01-316J16 (for the T locus) were provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan.
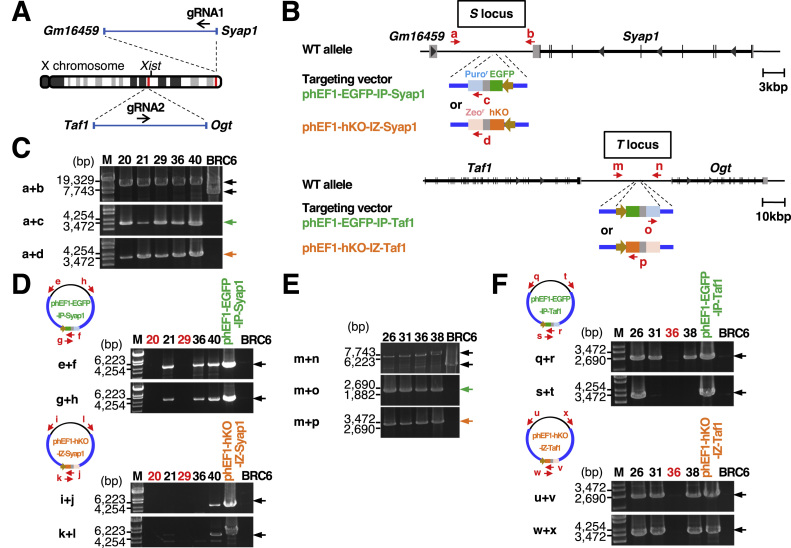
Fig. 1.
Knock-in of the EGFP and hKO genes driven by the human EF-1α promoter into the X chromosomes of mouse female ESCs. (A) Locations of the S and T loci on the mouse X chromosome are indicated by red bars. The location of the Xist gene is also indicated. The black arrows indicate the position and orientation of the guide RNAs (gRNA1 and gRNA2) used for the CRISPR/Cas9 system. (B) The intergenic sites between the Gm16459 and Syap1 genes (“S locus” in this study) and between the Taf1 and Ogt genes (“T locus” in this study) on the mouse X chromosome were chosen for insertion of the EGFP-IRES-Puror or hKO-IRES-Zeor cassette, which is driven by the human EF-1α promoter to express EGFP or hKO, respectively. The positions of the primers for genomic PCR are indicated by red arrows (a-d, m-p). (C) Genomic PCR analyses of the inserted fluorescent protein genes at the S locus in isolated ESC clones. BRC6 indicates the original female mouse ESCs used to insert the fluorescent protein genes. The primer sets used for PCR analyses are shown on the left. (D) Detection of random integration of the targeting vectors in the genome of isolated clones. The positions of primers for PCR analyses are indicated by red arrows (e-h for phEF1-EGFP-IP-Syap1, i-l for phEF1-hKO-IZ-Syap1). (E) Genomic PCR analyses of the inserted fluorescent protein genes at the T locus in isolated ESC clones. (F) Detection of random integration of the targeting vectors in the genome of isolated ESC clones. The positions of primers for PCR are indicated by red arrows (q-t for phEF1-EGFP-IP-Taf1, u-x for phEF1-hKO-IZ-Taf1).
2.2. Construction of plasmids
Complementary pairs of oligonucleotides encoding the gRNAs were annealed and inserted into the BbsI site of pX330-U6-Chimeric_BB-CBh-hSpCas9 to prepare Cas9/gRNA-expression vectors. The targeting vectors to knock-in the fluorescent protein genes into the S or T locus were constructed using pPyCAG-EGFP-IP and pPyCAG-EGFP-IZ. The CAG promoters were replaced by the human elongation factor-1α (EF-1α) promoter, and the EGFP gene in pPyCAG-EGFP-IZ was replaced by the hKO gene. The DNA fragments spanning the target site of gRNA1 (S locus) or gRNA2 (T locus) were isolated from the B6N mouse Bac clones and inserted into the upstream of the fluorescent protein gene and downstream of the drug-resistant gene (Supplementary Table 2).
2.3. Transfection of female mouse ESCs
Female mouse ESCs, BRC6 (RIKEN BRC, AES0010), were seeded at 5 × 105 cells/well on SNL feeder cells harboring the puromycin-resistant gene in a 6-well plate and cultured at 37 °C under 5% CO2 for 5 h in DMEM supplemented with 1 mM sodium pyruvate (Nacalai tesque, Inc.), 15% KnockOut Serum Replacement (KSR) (Thermo Fisher Scientific Inc.), nonessential amino acids (NEAA) (Wako Pure Chemical Industries, Ltd.), 0.1 mM 2-mercaptoethanol (2-ME) (Thermo Fisher Scientific Inc.) and 1000 U/mL LIF (Wako Pure Chemical Industries, Ltd.). Two micrograms each of pX330-Cas9/gRNA expression vectors and two different targeting vectors (phEF1-EGFP-IP-Syap1 and phEF1-hKO-IZ-Syap1, or phEF1-EGFP-IP-Taf1 and phEF1-hKO-IZ-Taf1) were mixed with 10 μl of Lipofectamine 2000, and the mixture was added to the ESCs. The cells were treated with 1 µg/mL puromycin for 5 days followed by treatment with 50 µg/mL zeocin for 3 days to isolate EGFP+/hKO+ ESC clones.
2.4. Genotype analysis of isolated ESC clones
Genomic DNAs were extracted from the isolated EGFP+/hKO+ ESC clones, which were cultured without feeder cells for 5 days prior to DNA extraction to avoid contamination with feeder cells. The ESCs were lysed in the presence of 0.5 μg/μl proteinase K, and the genome DNA was extracted by phenol/chloroform/isoamyl alcohol and precipitated by isopropanol. The location and sequences of primer sets used for PCR-based genotype analyses are shown in Fig. 1 as well as in Supplementary Table 3.
2.5. Differentiation of the EGFP+/hKO+ ESC clones
The EGFP+/hKO+ ESC clones were grown on SNL feeder cells in a 100 mm dish until ~80% confluency, and then trypsinized and suspended in the DMEM supplemented with 20% Fetal Bovine Serum (FBS), NEAA and 0.1 µM 2-ME. The cell suspension was transferred to a 100 mm cell culture dish and incubated for 20 min to remove feeder cells. Then, the supernatant containing the EGFP+/hKO+ ESCs was collected and plated into a 100 mm non-coated bacterial dish (AGC TECHNO GLASS CO., LTD.) for formation of embryoid bodies (EBs). After 5 days, EBs were trypsinized and filtrated through a 100 µm-cell strainer (BD Falcon). The filtrated cells were cultured on a collagen Type I-coated dish (AGC TECHNO GLASS CO., LTD.) in the presence of 50 µg/mL zeocin to select hKO+ cells.
2.6. Reprogramming of the EGFP+/hKO+ ESC-derived somatic cells
The isolated hKO+ cells were seeded in a 24-well plate at 2.5 × 104 cells/well in DMEM plus 10% FBS and cultured at 37 °C under 5% CO2 for 12 h. The cells were then infected with the Sendai virus which expresses KLF4, OCT4, SOX2 and c-MYC (SeVdp(KOSM) [11]) for 16 h at 32 °C to induce reprograming. The virus-infected cells were trypsinized and cultured on SNL-feeder cells in Knockout DMEM (Thermo Fisher Scientific Inc.) supplemented with 15% KSR, 2 mM GlutaMAX (Thermo Fisher Scientific Inc.), NEAA, 55 μM 2-ME, 100 units/mL penicillin, 100 µg/mL streptomycin (Nacalai tesque, Inc.) and 1000 U/mL LIF for 7 days. The culture medium was replaced by 1:1 mixture of DMEM/F12 (Nacalai tesque, Inc.) and Neurobasal medium (Thermo Fisher Scientific Inc.) supplemented with N2 supplement (Thermo Fisher Scientific Inc.), B27 supplement (Thermo Fisher Scientific Inc.), 2 mM GlutaMax (Thermo Fisher Scientific Inc.), 0.1 mM NEAA, 0.1 mM 2-ME, 0.05% BSA (Thermo Fisher Scientific Inc.), 1000 U/mL LIF, 1 μM MEK inhibitor PD0325901, 3 μM GSK3β inhibitor CHIR99021, 100 units/mL penicillin and 100 μg/mL streptomycin for continuous culture of iPSCs.
2.7. Reverse transcription and quantitative real-time PCR
Total RNA was extracted from the iPSCs using Sepasol-RNA I Super G (Nacalai tesque, Inc.) according to the manufacture's instruction. To avoid contamination with feeder cells, the hKO+ cells-derived iPSCs were cultured without feeder cells for 5 days prior to RNA extraction. Reverse transcription was performed using Superscript III First-Strand Synthesis System (Thermo Fisher Scientific Inc.), and the mRNA levels of various marker genes were measured by quantitative real-time PCR using GoTaq qPCR Master Mix (Promega Corp.). The mRNA level of γ-tubulin was used to normalize the obtained data. The primers used in this study are listed in Supplementary Table 4.
3. Results
3.1. Knock-in of fluorescent reporter genes into both X chromosomes of female ESCs
To visualize XCR in live cells during somatic cell reprogramming, we first generated female ESCs that express EGFP from one X chromosome and hKO from the other. To insert the EGFP and hKO genes into the genome, we avoided protein-coding genes as an insertion site because of their potential effect as a facilitator or inhibitor on the reprogramming process when iPSCs are generated [12]. Instead, we chose two intergenic sites near the Syap1 or Taf1 gene on the X chromosome (Fig. 1A). These sites were chosen because the insertion sites, which we term S and T loci, are near the genes, Syap1 and Taf1, respectively, that are subject to XCI [13]. In addition, database search of National Center for Biotechnology Information (NCBI) showed that the genes surrounding the S locus (Syap1, Txlng, Rbbp7, and Ctps2) and the T locus (Taf1, Nono, Zmym3, and Med12) do not exhibit strong tissue- or developmental stage-specific expression pattern. Moreover, the GeneProf database (http://www.geneprof.org/) [14] showed that these loci are sandwiched between CTCF binding sites together with at least one of these surrounding genes. Thus, the EGFP and hKO genes that are inserted into the S and T loci were expected to obey XCI and XCR in a similar manner to the surrounding genes.
We inserted the EGFP and hKO gene into each S locus of both X chromosomes in mouse female ESCs, using the CRISPR/Cas9 system [8], [9]. The gRNA1/Cas9 expression vector was introduced into female ESCs together with two different targeting vectors, each of which contained either the EGFP or hKO gene driven by the human EF-1α promoter as well as a drug-resistant gene, puromycin- or zeocin-resistant gene, respectively (Fig. 1B). The single-colored ESCs were removed by sequential selections with puromycin and zeocin to obtain EGFP+/hKO+ ESC colonies. Among the isolated 50 clones, 33 clones that grew well were genotyped, and most of the isolated clones had the inserted gene(s) at the S locus. However, only five ESC clones (No. 20, 21, 29, 36, and 40) had the EGFP and hKO genes at each S locus on both X chromosomes (Fig. 1C) while other clones had only the EGFP or hKO gene on both X chromosomes. PCR analysis using the primer sets shown in Fig. 1D showed that phEF1-EGFP-IP-Syap1 was randomly inserted in the genome in three clones (No. 21, 36, and 40) (e+f and g+h) and phEF1-hKO-IZ-Syap1 in one clone (No. 40) (i + j and k + l) (Fig. 1D). These results show that two ESC clones (No. 20 and 29, hereafter called “S20″ and “S29″) have the EGFP and hKO genes at each S locus on both X chromosomes without any random insertion in the genome.
We also inserted the EGFP and hKO genes into another intergenic site near the Taf1 gene (T locus) (Fig. 1A and B). After co-transfection of mouse female ESCs with the gRNA2/Cas9 expression vector together with the two different targeting vectors harboring the EGFP-IRES-Puror or hKO-IRES-Zeor gene (Fig. 1B), EGFP+/hKO+ colonies were selected sequentially by puromycin and zeocin. Genotyping revealed that four ESC clones (No. 26, 31, 36, and 38) had the EGFP and hKO genes at each T locus on both X chromosomes (Fig. 1E). However, as shown in Fig. 1F, only one ESC clone (No. 36, hereafter called “T36″) was free of a randomly inserted vector. Thus, we obtained three ESC clones that expressed both EGFP and hKO from the S locus (S20 and S29) or the T locus (T36) using the CRISPR/Cas9 system.
3.2. The EGFP and hKO genes at the intergenic loci on the X chromosome are subject to XCI inactivation during ESC differentiation
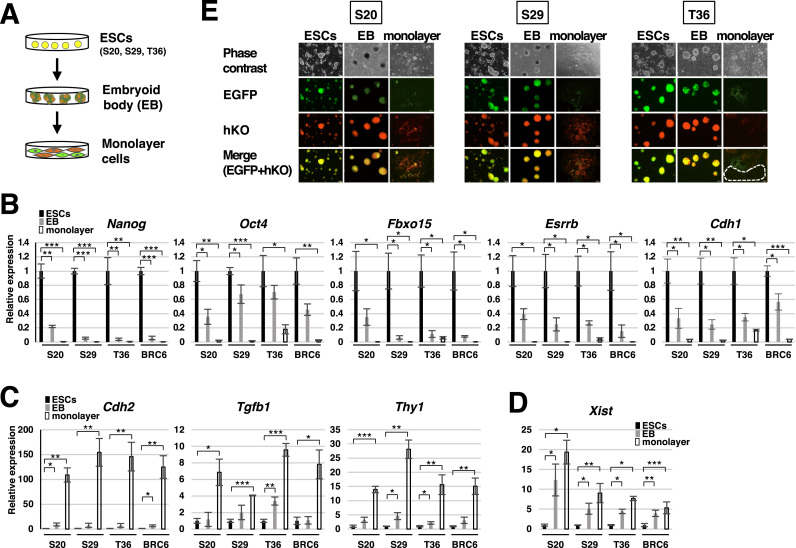
To confirm that the EGFP and hKO genes inserted into the intergenic sites are subject to XCI, we differentiated the three ESC clones (S20, S29, and T36) through embryoid body (EB) formation into monolayer cells (Fig. 2A). Quantitative RT-PCR analyses of the embryoid bodies and monolayer cells showed that expression of pluripotency marker genes (Nanog, Oct4, Fbxo15, Esrrb, and Cdh1) decreased (Fig. 2B) while that of somatic cell marker genes (Cdh2, Tgfb1, and Thy1) increased (Fig. 2C), indicating differentiation of ESCs. Expression of the Xist gene also increased in EBs and monolayer cells derived from ESC clones (S20, S29, and T36), indicating that XCI occurred during differentiation (Fig. 2D). As shown in Fig. 2E, ESC clones (S20, S29, and T36), which initially expressed both EGFP and hKO, gradually lost expression of either EGFP or hKO during EB formation. When the EB-derived cells were allowed to further differentiate as a monolayer, the cells expressed only EGFP or hKO, showing that one of the fluorescent protein genes on the X chromosome was inactivated due to XCI. Some somatic cells differentiated from the T36 ESC clone were found to lose expression of both EGFP and hKO (Fig. 2E, white dotted line), suggesting that the gene inserted into the T locus may be repressed independent of XCI. These results indicate that the fluorescent protein genes driven by the human EF-1α promoter are subject to XCI even when inserted into intergenic sites of the X chromosome, and the fluorescent genes at the S locus may be more suitable than those at the T locus for observing XCI.
Fig. 2.
Observation of XCI in the ESCs carrying the EGFP and hKO genes on the X chromosome. (A) Differentiation of the EGFP+/hKO+ ESCs via embryoid body formation into monolayer cells and the fluorescent patters of the derived somatic cells. ESCs expressed both EGFP and hKO before differentiation, and either EGFP or hKO became inactivated randomly by XCI upon differentiation. (B-D) Expression of pluripotency marker genes (B), somatic cell marker genes (C) and the Xist gene (D) in S20, S29, and T36 clones during differentiation (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001). BRC6 indicates the original female mouse ESCs. (E) Morphology and fluorescent images of the S20, S29, and T36 clones during differentiation. A white-dotted area indicates the cells that expressed neither EGFP nor hKO.
3.3. Live cell imaging of XCR during reprogramming
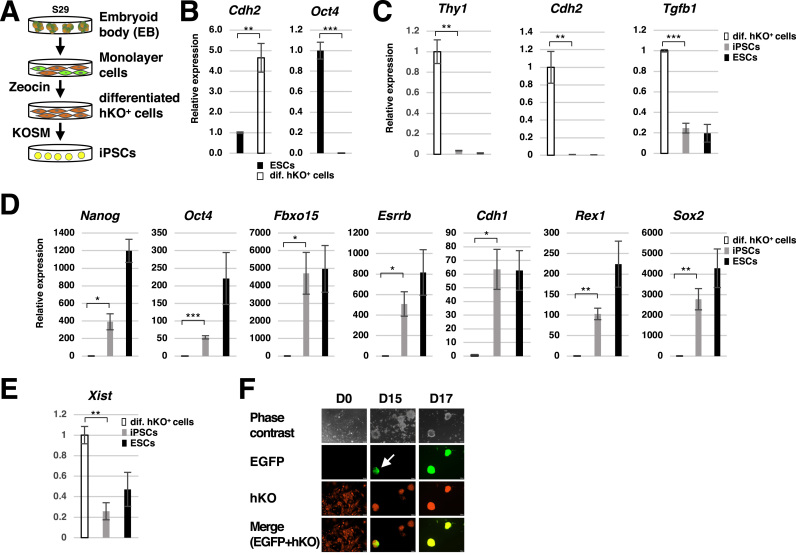
XCR is a reverse process of XCI and occurs in female cells during somatic cell reprogramming [6]. To examine whether the S29 ESCs could be used for detecting XCR in live cells in vitro, we isolated hKO-positive (hKO+) differentiated cells and performed reprogramming. As shown in Fig. 3A, the EB-derived somatic cells were selected by zeocin to isolate only hKO+ cells, which carried the hKO gene on the Xa and the EGFP gene on the Xi. The isolated cells expressed a somatic cell marker gene (Cdh2) but not a pluripotency marker gene (Oct4) (Fig. 3B). The hKO+ cells were then infected with Sendai virus that expresses KLF4, OCT4, SOX2, and c-MYC to induce reprogramming [10]. The virus-infected cells formed colonies, which showed repression of somatic cell marker genes (Thy1, Cdh2, and Tgfb1) (Fig. 3C) and induction of pluripotency marker genes (Nanog, Oct4, Fbxo15, Esrrb, Cdh1, Rex1, and Sox2) (Fig. 3D). Moreover, expression of the Xist gene was down-regulated to a similar level of female ESCs (Fig. 3E). These results indicate that the hKO+ somatic cells were reprogrammed into iPSCs and underwent XCR, which is indicative of the fully pluripotent state [15]. Observation of EGFP and hKO signals in these cells revealed that some colonies started to show the EGFP signal by day 15, which gradually became more homogeneous by day 17, indicating XCR occurred between day 15 and day 17 of reprogramming (Fig. 3F). Thus, the S29 ESCs can be used to monitor the dynamic process of XCR during reprogramming in live cells in a non-invasive manner.
Fig. 3.
Live cell imaging of XCR during reprogramming. (A) Schematic illustration of reprogramming somatic cells derived from S29 ESCs. The hKO+ monolayer cells derived from S29 ESCs were selected by zeocin and then reprogrammed into iPSCs by infection with the Sendai virus expressing KLF4, OCT4, SOX2, and c-MYC (KOSM). (B) Expression of somatic cell marker (Cdh2) and pluripotency marker (Oct4) genes in the differentiated hKO+ cells derived from S29 ESCs. (C-E) Expression of somatic cell marker genes (C), pluripotency marker genes (D) and the Xist gene (E) in iPSCs reprogrammed from hKO+ somatic cells (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001). (F) Morphology and fluorescent images of iPSCs reprogrammed from hKO+ somatic cells at day 0, day 15 and day 17 of reprogramming. The white arrow indicates an EGFP+ colony, which appeared around day 15.
3.4. Visualizing XCR to monitor acquisition of pluripotency during reprogramming
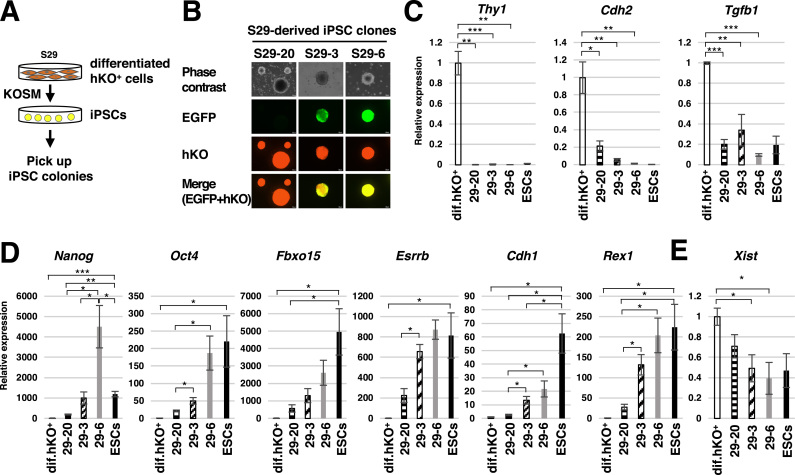
When we isolated iPSC colonies reprogrammed from hKO+ somatic cells that were derived from S29 ESCs, we obtained colonies that were morphologically indistinguishable but nonetheless showed different expression patterns of EGFP (Fig. 4A and B). Given that XCR is a late event in reprogramming when iPSCs acquire pluripotency [15], we further examined the relationship between expression of both fluorescent protein genes and the acquisition of pluripotency. We isolated three iPSC clones generated from hKO+ somatic cells (Fig. 4A and B); S29-20 colonies did not express EGFP at all whereas S29-3 and S29-6 colonies expressed EGFP in a mosaic or homogeneous pattern, respectively. All three clones showed low expression of somatic cell marker genes (Thy1, Cdh2, and Tgfb1) (Fig. 4C), indicating that they progressed past the early stage of reprogramming [16]. By contrast, the three clones showed increased expression of pluripotency marker genes (Fig. 4D), which are induced during reprogramming [16]. Except for Nanog, there was no significant difference in the expression levels of the pluripotency marker genes between S29-3 and S29-6 (Fig. 4D), which show mosaic and homogeneous EGFP fluorescence, respectively (Fig. 4B). However, S29-20 iPSCs, which failed to express EGFP, had significantly lower expression levels of Nanog, Oct4, Esrrb, Cdh1, and Rex1 (Fig. 4D). Expression of the Xist gene was significantly decreased in S29-3 and S29-6 but not in S29-20, indicating that XCR occurred in S29-3 and S29-6 but not in S29-20 during reprogramming (Fig. 4E). These results indicate that EGFP expression from the Xi, even if it is mosaic, reflects XCR and acquisition of pluripotency at a late stage of reprogramming.
Fig. 4.
XCR and acquisition of pluripotency in female iPSCs. (A) Schematic illustration of isolation of iPSC clones reprogrammed from S29-derived hKO+ somatic cells. (B) Morphology and fluorescent images of isolated iPSC clones that exhibit no expression (S29-20), heterogeneous expression (S29-3), or homogeneous expression (S29-6) of EGFP. The colonies were isolated at day 19 of reprogramming. (C-E) Expression of somatic cell marker genes (C), pluripotency marker genes (D) and the Xist gene (E) in the iPSC clones (n = 3, *P < 0.05, **P < 0.01, ***P < 0.001).
4. Discussion
Here we describe female ESCs that carry the EGFP and hKO genes at intergenic sites on both X chromosomes. The EGFP and hKO genes inserted into an intergenic site near the Syap1 gene are subject to both XCI during ESC differentiation and XCR during somatic cell reprogramming and thus allow live cell monitoring of XCI and XCR.
Previous studies reported insertion of fluorescent genes into the coding region of the Pgk1 and Hprt genes, which obey XCI during mouse development [17], [18]. In these studies, inserting the fluorescent genes into the Pgk1 and Hprt genes does not appear to compromise normal mouse development. However, many genes have been shown to potentially influence the reprogramming efficiency [19]. Therefore, to avoid inadvertent effects on iPSC generation by fluorescent protein genes inserted within a gene, we tested the feasibility of inserting the fluorescent protein genes into intergenic sites where neighboring genes are subject to XCI. Although the EGFP and hKO genes inserted near the Taf1 gene showed unstable expression upon differentiation (Fig. 2E), the fluorescent protein genes inserted near the Syap1 gene underwent XCI and XCR in a predicted manner. Thus, if properly chosen and experimentally tested, an intergenic site on the X chromosome allows a foreign gene to obey XCI and XCR.
During reprogramming of S29 ESC-derived somatic cells, reactivation of the EGFP gene accompanied down-regulation of the Xist gene and induction of pluripotency marker genes, demonstrating that acquisition of pluripotency is visualized in live cells by fluorescence imaging of EGFP and hKO (Fig. 3, Fig. 4). Moreover, iPSCs that fail to activate the EGFP gene from Xi showed low expression of pluripotency marker genes, consistent with the close coupling between pluripotency and XCR [15]. Thus, the ESCs with the EGFP and hKO genes on both X chromosomes, especially the S29 ESCs described here, may become an important tool for high-throughput screenings for factors and culture conditions that promote the acquisition of pluripotency by iPSCs.
Conflicts of interest
The authors declare no conflict of interest.
Funding
This work was supported by JSPS KAKENHI Grant Numbers JP16K07244, JP17H05063, and JP16K08610 (to K.N., Y.H., an A.F., respectively), and JP17H04036 and JP17K19339 (to K.H.) as well as by Program to Disseminate Tenure Tracking System by MEXT (to K.N.).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.07.007.
Contributor Information
Aya Fukuda, Email: fukudaa@md.tsukuba.ac.jp.
Koji Hisatake, Email: kojihisa@md.tsukuba.ac.jp.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Wu S.M., Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat. Cell Biol. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cahan P., Daley G.Q. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat. Rev. Mol. Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nichols J., Smith A. Naive and primed pluripotent states. Cell. Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 5.Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 6.Pasque V., Plath K. X chromosome reactivation in reprogramming and in development. Curr. Opin. Cell Biol. 2015;37:75–83. doi: 10.1016/j.ceb.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payer B., Lee J.T. Coupling of X-chromosome reactivation with the pluripotent stem cell state. RNA Biol. 2014;11:798–807. doi: 10.4161/rna.29779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA – guided dna endonuclease in adaptice bacterial immunity. Science. 2012;337:816–822. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H., La Russa M., Qi L.S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura K., Sano M., Ohtaka M., Furuta B., Umemura Y., Nakajima Y., Ikehara Y., Kobayashi T., Segawa H., Takayasu S., Sato H., Motomura K., Uchida E., Kanayasu-Toyoda T., Asashima M., Nakauchi H., Yamaguchi T., Nakanishia M. Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming. J. Biol. Chem. 2011;286:4760–4771. doi: 10.1074/jbc.M110.183780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishimura K., Kato T., Chen C., Oinam L., Shiomitsu E., Ayakawa D., Ohtaka M., Fukuda A., Nakanishi M., Hisatake K. Manipulation of KLF4 expression generates iPSCs paused at successive stages of reprogramming. Stem Cell Rep. 2014;3:915–929. doi: 10.1016/j.stemcr.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebrahimi B. Reprogramming barriers and enhancers: strategies to enhance the efficiency and kinetics of induced pluripotency. Cell Regen. 2015;4 doi: 10.1186/s13619-015-0024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang F., Babak T., Shendure J., Disteche C.M. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbritter F., Vaidya H.J., Tomlinson S.R. GeneProf: analysis of high-throughput sequencing experiments. Nat. Methods. 2012;9:7–8. doi: 10.1038/nmeth.1809. [DOI] [PubMed] [Google Scholar]
- 15.Pasque V., Tchieu J., Karnik R., Uyeda M., Sadhu Dimashkie A., Case D., Papp B., Bonora G., Patel S., Ho R., Schmidt R., McKee R., Sado T., Tada T., Meissner A., Plath K. X chromosome reactivation dynamics reveal stages of reprogramming to pluripotency. Cell. 2014;159:1681–1697. doi: 10.1016/j.cell.2014.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J., Bar-Nur O., Cheloufi S., Stadtfeld M., Figueroa M.E., Robinton D., Natesan S., Melnick A., Zhu J., Ramaswamy S., Hochedlinger K. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi S., Hosoi Y., Shiura H., Yamagata K., Takahashi S., Fujihara Y., Kohda T., Okabe M., Ishino F. Live imaging of X chromosome reactivation dynamics in early mouse development can discriminate naïve from primed pluripotent stem cells. Development. 2016;143:2958–2964. doi: 10.1242/dev.136739. [DOI] [PubMed] [Google Scholar]
- 18.Wu H., Luo J., Yu H., Rattner A., Mo A., Wang Y., Smallwood P.M., Erlanger B., Wheelan S.J., Nathans J. Cellular resolution maps of X chromosome inactivation: implications for neural development, function, and disease. Neuron. 2014;81:103–119. doi: 10.1016/j.neuron.2013.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin H., Diaz A., Blouin L., Lebbink R.J., Patena W., Tanbun P., Leproust E.M., McManus M.T., Song J.S., Ramalho-Santos M. Systematic identification of barriers to human Ipsc generation. Cell. 2014;158:449–461. doi: 10.1016/j.cell.2014.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material