Abstract
Prostate cancer (PCa) is a disease of mutated and misregulated genes. However, primary prostate tumors have relatively few mutations, and only three genes ( ERG, PTEN, and SPOP) are recurrently mutated in more than 10% of primary tumors. On the other hand, metastatic castration-resistant tumors have more mutations, but, with the exception of the androgen receptor gene ( AR), no single gene is altered in more than half of tumors. Structural genomic rearrangements are common, including ERG fusions, copy gains involving the MYC locus, and copy losses containing PTEN. Overall, instead of being associated with a single dominant driver event, prostate tumors display various combinations of modifications in oncogenes and tumor suppressors. This review takes a broad look at the recent advances in PCa research, including understanding the genetic alterations that drive the disease and how specific mutations can sensitize tumors to potential therapies. We begin with an overview of the genomic landscape of primary and metastatic PCa, enabled by recent large-scale sequencing efforts. Advances in three-dimensional cell culture techniques and mouse models for PCa are also discussed, and particular emphasis is placed on the benefits of patient-derived xenograft models. We also review research into understanding how ETS fusions (in particular, TMPRSS2-ERG) and SPOP mutations contribute to tumor initiation. Next, we examine the recent findings on the prevalence of germline DNA repair mutations in about 12% of patients with metastatic disease and their potential benefit from the use of poly(ADP-ribose) polymerase (PARP) inhibitors and immune modulation. Lastly, we discuss the recent increased prevalence of AR-negative tumors (neuroendocrine and double-negative) and the current state of immunotherapy in PCa. AR remains the primary clinical target for PCa therapies; however, it does not act alone, and better understanding of supporting mutations may help guide the development of novel therapeutic strategies.
Keywords: prostate cancer, sequencing, xenograft, immunotherapy, 3D culture, PARP, BRCA
Introduction
Prostate cancer (PCa) is the most commonly diagnosed non-skin cancer in American men and is estimated to account for about 30,000 deaths this year in the USA and at least 10 times as many worldwide 1, 2. The disease is curable when locally confined, but treatment options are limited for metastatic disease. First recognized in the 1940s as an effective therapy for metastatic PCa 3, androgen deprivation remains the primary option for patients with advanced disease; however, tumors invariably relapse into incurable metastatic castration-resistant PCa (mCRPC) 4. Further targeting of the androgen receptor (AR) axis with more effective drugs has extended survival by a few months but leads to resistance, including an increase in once-rare neuroendocrine and non-neuroendocrine/AR-negative tumors 5. Owing to recent large-scale sequencing efforts, there is now a better understanding of the genomic landscape of PCa, including characterization of lower-frequency but nonetheless important mutations in SPOP and DNA repair genes (for example, BRCA2).
This review covers some of the recent advances in understanding PCa, including identification and targeting of key genetic aberrations ( ERG, SPOP, and DNA repair defects), improvements in disease models, the emergence of AR-negative disease, and current immunotherapy strategies. Although AR signaling remains the ultimate driver of most PCa, tumors show an assortment of additional alterations that help promote disease progression and at the same time provide new opportunities for targeting this resilient disease. Although we sought to cover a wide range of topics, many fell beyond the scope of this report. However, several of those important themes can be found in previous reviews, including epigentics 6, 7, diet 8, tumor metabolism 9, 10, biomarkers 11, microRNAs 12, 13, the role of the microenvironment 14, 15, and racial disparities 16.
Genomic analysis of primary tumors
Analysis of PCa at the genome level began around the turn of the century with a wide range of studies using a combination of techniques, such as comparative genomic hybridization, DNA microarray, and targeted sequencing 17, 18. Whole genome sequencing (WGS) efforts began around 2011, including a project that performed WGS of seven primary tumors ( Figure 1) 19. Over the next 2 years, whole exome sequencing (WES) efforts expanded to analyze over a hundred primary tumors 20, 21. A major leap came in 2015 with publication of the data from the PCa branch of The Cancer Genome Atlas (TCGA), a landmark study that published molecular characterization (genomic, epigenomic, and proteomic) of 333 primary prostate tumors 22. Another large-scale study (published in 2017) is the Genomic Hallmarks of Prostate Cancer, which includes WGS for 200 primary tumors and WES for an additional 277 23. Two 2018 studies performed WGS on 92 and 93 additional primary tumors, generating more useful data for analysis 24, 25. One issue that can arise when comparing datasets from different studies is a lack of uniform pipeline analysis (that is, data standardization, normalization, and statistical cutoffs). A 2018 report sought to tackle this problem by re-analyzing 1,013 available WES datasets (680 primary and 333 metastatic) using a common analysis pipeline 26. As a direct result of these transformative studies, researchers finally have an encompassing view of the genetic landscape of primary PCa that provides an important point of reference for understanding this complex disease.
Figure 1. Timeline (not to scale) of key prostate cancer whole genome sequencing and whole exome sequencing studies 19– 26, 27– 30.
One of these studies included 10 hormone treatment-naïve metastatic tumors (metastatic prostate cancer) 24. *A total of 293 primary tumors were analyzed in this study, but 200 were included in a previous study and not included here 23. The study by Armenia et al. 26 is a uniform re-analysis of previous whole exome sequencing studies, including many of those listed here. mCRPC, metastatic castration-resistant prostate cancer; mPC, metastatic prostate cancer; TCGA, The Cancer Genome Atlas; WES, whole exome sequencing; WGS, whole genome sequencing.
At a broad glance, prostate tumors have, on average, fewer mutations (0.7 per Mb) than other common cancers, such as breast (1.2 per Mb), bladder (7.1 per Mb), colorectal (3.1 per Mb), and melanoma (12.1 per Mb) 31. Despite having relatively few point mutations, PCa is characterized by a high rate of genomic instability and chromosomal rearrangments 32. The most frequent genomic aberration in primary tumors is a chromosomal rearrangement fusing strong AR-regulated promoters with ETS family genes (62%), resulting in their prominent overexpression ( Table 1) 22. Although multiple ETS fusions have been identified, the most common is TMPRSS2-ERG, which arises from an approximately 3 Mb deletion on chromosome 21 that brings the androgen-regulated TMPRSS2 promoter upstream of ERG 22, 33. In addition, about 3% of primary tumors show mutation/deletion of ERF, an ETS repressive cofactor, providing another mechanism for increasing ETS activity without their overexpression 36, 37. Other common genomic alterations in primary tumors include loss (usually by deletion) of PTEN (17%), point mutations in SPOP (11%), mutation or deletion of TP53 (8%), and amplification of MYC (7%) ( Table 1) 22. Many of the findings from these sequencing efforts confirmed previously known alterations (for example, ERG, MYC, and PTEN) 38, but they also revealed some less-frequent, novel PCa mutations (for example, SPOP, IDH1, MED12, and FOXA1) 21, 22. The majority of these studies also included mRNA expression or DNA methylation data or both. Information concerning changes in gene expression, mutations, deletions, and amplifications in human PCa can be readily queried via the cBioPortal web tool ( www.cbioportal.org) 34, 35.
Table 1. Common genomic aberrations in primary prostate cancer.
| Gene | Primary
tumors altered, percentage |
Type of
mutation |
|---|---|---|
| ETS family a | 62 | Fusion/Amp |
| ERG | 46 | Fusion |
| PTEN | 17 | Homdel/Mut |
| SPOP | 11 | Mut |
| TP53 | 8 | Homdel/Mut |
| MYC | 7 | Amp |
| AR | 1 | Amp |
| RB1 | 1 | Homdel/Mut |
A selection of common alterations in primary prostate tumors. cBioPortal 34, 35 was used to query the TCGA (The Cancer Genome Atlas) data set, which contains 333 primary tumor samples 22. Data were queried specifically for the type of alterations listed in the third column. Amp, genomic amplification; Homdel, homozygous deletion; Mut, nonsynonymous mutation. a ERG, ETV1/4/5, FLI1.
Genomic analysis of metastatic tumors
Understanding the genomic landscape of primary tumors has many benefits (for example, understanding tumor origin, aiding prognosis, and revealing therapy options), but there is also a more practical need to understand the lethal form of disease, mCRPC. Androgen deprivation and AR signaling inhibitors (ARSis) (for example, abiraterone and enzalutamide) are initially quite effective, but tumors eventually develop resistance via various mechanisms, including (but not limited to) intra-tumoral androgen synthesis, AR amplification, AR ligand-binding domain mutations, or expression of constitutively active AR splice variants 4, 39. Sequencing efforts with metastatic tumors ( Figure 1) identified enrichment of some mutations seen in primary disease, including amplification/mutation of AR (61%), amplification of MYC (20%), and deletion/mutation of TP53 (47%) and PTEN (41%) ( Table 2) 27– 30. mCRPC tumors have about five times as many mutations as primary tumors (2.3~4.4 versus 0.7~1.0 per Mb) 24, 28, 31 and include several new mutations, a selection of which is summarized in Table 2. One key advantage of having sequencing data from hundreds of tumors is the ability to use bioinformatics to recognize and cluster low-frequency, recurrent mutations across multiple genes in a single pathway. At the pathway level, mCRPC tumors have frequent alterations in AR signaling (71%), PI3K/PTEN (49%), WNT (18%), cell cycle (21%), and DNA repair (13%) 28. Furthermore, about 21% of mCRPC tumors have amplified HEY1, which is an important target of the NOTCH pathway ( Table 2) 38.
Table 2. Common genomic aberrations in metastatic prostate cancer.
| Gene | mCRPC
altered, percentage |
Chromosome | Type of
mutation |
|---|---|---|---|
| AR | 61 | Xq | Amp/Mut |
| ETS family a | 49 | - | Fusion/Amp |
| ERG | 35 | 21q | Fusion |
| TP53 | 47 | 17p | Homdel/Mut |
| PTEN | 43 | 10q | Homdel/Mut |
| HEY1 | 21 | 8q | Amp |
| E2F5 | 21 | 8q | Amp |
| MYC | 20 | 8q | Amp |
| RB1 | 17 | 13q | Homdel/Mut |
| FOXA1 | 14 | 14q | Amp/Mut |
| CHD1 | 11 | 5q | Homdel/Mut |
| FOXO1 | 11 | 13q | Homdel/Mut |
| BRCA2 | 11 | 13q | Homdel/Mut |
| MED12 | 9 | Xq | Amp/Mut |
| SPOP | 8 | 17q | Mut |
| ATM | 8 | 11q | Homdel/Mut |
| PIK3CA | 8 | 3q | Amp/Mut |
| CDK12 | 5 | 17q | Mut |
A selection of common alterations in metastatic castration-resistant prostate cancer (mCRPC). cBioPortal 34, 35 was used to query three mCRPC data sets containing 347 tumors from 263 patients 27– 29. The second column shows the percentage of patients with a tumor carrying the alteration. Data were queried specifically for the type of alterations listed in the third column. Amp, genomic amplification; Homdel, homozygous deletion; Mut, nonsynonymous mutation. a ERG, ETV1/4/5, FLI1.
Other recent advances in analyzing mCRPC include circulating tumor cell (CTC) isolation and single-cell sequencing 40– 42. In a 2015 report, the authors used single-cell RNA sequencing on 76 CTCs from 12 patients with mCRPC and found enrichment in expression of stem cell genes, non-canonical WNT signaling, and a range of AR splice variants, sometimes even within the same cell 41. In patients with multiple metastases, tumors usually share common driver mutations and appear to either be clonal or show convergent selection for therapy resistance 29, 43. Moreover, one report analyzed sequencing of multiple metastases within patients and observed that many seeded from an earlier metastasis 43. The authors also found that metastases within a patient are likely to share tumor suppressor loss-of-function mutations (for example, PTEN and TP53) but often show unique AR pathway alterations 43. Although targeting AR by androgen deprivation and ARSi leads to temporary success, there is a clear need to consider other targets, and these recent genomic studies have helped provide some candidates.
Three-dimensional culture models
PCa research has advanced with a relatively small collection of commonly used cell lines (and their derivatives), the vast majority of which were isolated from metastatic tumors (for example, LNCaP, VCaP, PC3, and DU145) 44. The overwhelming majority of cells in human PCas most resemble luminal epithelial cells and have some basal marker expression 38. Unlike the mouse prostate, normal human luminal epithelial cells rarely proliferate and most come from bipotent progenitors in the basal layer 45. It is difficult to establish and maintain human luminal epithelial cells in culture; however, luminal-like cells can be differentiated from basal/intermediate cells, which can be maintained in culture 46– 49. Extracellular matrix conditions can have a significant impact on cell survival and growth. For example, plating PCa cells on laminin can activate integrin α6 (also known as CD49f), which aids invasion and survival 50, 51. It is not clear why primary tumors and normal luminal cells take so poorly to tissue culture conditions. It is likely that something within the in vivo microenvironment has not been properly replicated in culture (paracrine factors, cell–cell interactions, and so on). As a way to better mimic the in vivo microenvironment, research has expanded into three-dimensional (3D) culture systems.
Prostate cell culture in 3D (that is, spheroids, prostaspheres, and organoids) has aided research by providing more physiologically relevant conditions and allowing more complex cultures 52– 54. Organoids can be derived from tumors or normal prostate cells, which can recapitulate basal, intermediate, and luminal cells as well as tumor initiation events such as prostatic intra-epithelial neoplasia (PIN) 55– 59. 3D culture can also incorporate different cell types, such as combining epithelial cells plus stroma or cancer cells plus osteoblasts 60– 62. Growing primary human tumors in 3D remains a difficult task, but metastatic tumors have been cultured with some success 52, 58, 63. Innovative studies using organoid cultures have also improved our understanding of prostate tumor initiation and cell of origin 64, 65. A 2016 report provides a protocol for growing prostate organoids using a fairly complex serum-free medium with a variety of growth factors and inhibitors 52, 58, 66. Interestingly, many cell lines behave differently in 2D versus 3D culture; for example, LNCaP cells have higher docetaxel resistance in 3D 67, 68. Though technically challenging, these new culture methods allow better modeling of normal and tumor epithelial structure. However, better understanding of prostate cell biology is needed so we can more efficiently culture prostate tissues, especially primary tumors.
Genetically modified mouse models
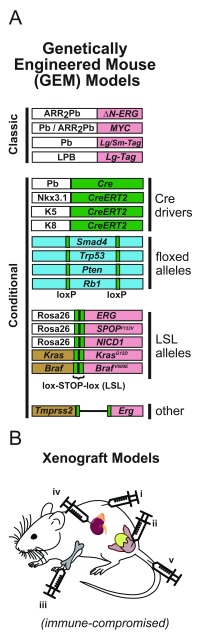
Mouse models have been extremely useful for studying disease initiation and progression in vivo and can broadly be separated into two categories: genetically engineered mouse (GEM) models and xenograft models ( Figure 2) 69. GEM models rely on engineering the mouse genome to knockout or express specific genes, which can be done globally (classic) or in specific tissues (conditional) via tissue-specific promoter-driven expression of Cre recombinase paired with floxed (flanked-loxP) target alleles ( Figure 2A). For prostate-specific expression, the most commonly used promoter is the rat Probasin promoter (Pb), the Large Pb (LPB), or the related ARR 2Pb, which contains Pb plus enhancer elements for higher expression 72, 88. Another common driver for conditional models is a tamoxifen-inducible knock-in Cre (CreERT2) at the Nkx3.1 locus, which is more specific for prostate luminal cells but carries the caveat of losing one functional copy of the gene 74. With CreERT2 models, Cre is still made only in the promoter-specified tissues but must be activated by the addition of tamoxifen (which causes nuclear localization) and thus grants far greater temporal control of recombination. Other valuable CreERT2 drivers include the basal keratins 5 and 14 (K5 and K14) and the luminal keratin 8 (K8) 75, 76, 89, 90. These keratin promoters provide basal/luminal specificity in the prostate but are also expressed in many other epithelial tissues. These models have been especially useful for lineage-tracing experiments, which use a brief pulse of tamoxifen to tag epithelial cells with fluorescent proteins and follow them as they divide and differentiate over time 76.
Figure 2. Overview of mouse models of prostate cancer.
( A) Genetically engineered mouse models. Classic models use the prostate-specific Probasin (Pb) or ARR 2Pb promoter to drive expression of oncogenes, including MYC 70, 71 and N-terminally truncated ERG 71. In the classic TRAMP model, Pb is used to drive expression of large and small SV40 T-antigen (Tag) 72. The LADY models use the Large Pb (LPB) promoter to drive large T-antigen only 73. Conditional models use prostate-specific Cre recombinase expression with loxP tagged alleles. Cre is most frequently driven by Probasin (Pb-Cre4 line) or a knock-in tamoxifen-inducible Cre at the Nkx3.1 locus (Nkx3-1 CreERT2 line) 74. Basal (K5) or luminal (K8) keratin promoters can been used to drive layer-specific expression in the prostate but also are expressed in other epithelial tissues 75, 76. Flanked loxP (floxed) sites can be used to induce loss-of-function deletions in endogenous tumor suppressor genes, including Smad4 77, Trp53 78, Pten 79, and Rb1 80. Lox-STOP-lox (LSL) alleles use Cre to remove an upstream STOP codon and allow expression of an oncogene. For constitutive expression, genes can be knocked-in at the ubiquitously expressed Rosa26 locus (for example, ERG 81, SPOP [ F133V] 82, Notch1 intra-cellular domain [NICD1] 83). Alternately, mutant genes can be knocked-in at the endogenous locus to maintain normal transcriptional regulation (for example, Kras [ G12D] 84 and Braf [ V600E] 85). There is also a model where loxP sites are used to delete the intergenic space between Tmprss2 and Erg, thereby mimicking the fusion observed in tumors 86, 87. Color coding: white = promoter, green = Cre or lox, blue = endogenous tumor suppressor, red = oncogene, brown = other endogenous gene. ( B) Xenograft models. Cells can be injected into immunocompromised mice via multiple methods: (i) subcutaneous, (ii) prostate (orthotopic for primary tumors), (iii) intra-tibial (orthotopic for bone metastatic tumors), (iv) renal capsule, and (v) tail vein.
There are GEM models matching many of the common alterations observed in human prostate tumors, including MYC overexpression 70, 91, Pten loss 79, 92, ERG overexpression 81, 86, 93, and SPOP mutation 82 ( Figure 2A). The first-generation mouse models of PCa (TRAMP and LADY) used Pb-driven SV40 T-antigen (Tag), which promotes massive proliferation and creates tumors displaying partial neuroendocrine differentiation 72, 73. These models have regained some popularity recently, as neuroendocrine tumors are becoming more common in human PCa. Some prostate GEM models give rise to metastasis, but only one, the LPB-Tag/Pb-Hepsin model expressing SV40 and cell-surface protease Hepsin, reliably metastasizes to bone (up to 40% by 23 weeks of age), the most frequent site in human patients 77, 94– 96. Other GEM models used for PCa are included in Figure 2A and have been previously reviewed 97– 99. In summary, recent advancements in GEM models enable the study of autochthonously developing PCa in immune-competent animals, but they lack the complexity of human genetics and human prostate biology and rely on contrived genetic manipulations. To study human tumors in vivo, experiments rely on xenograft models.
Xenograft mouse models
With xenograft models, human samples (tissue or cell line) are implanted into immune-compromised mice. Samples can be engrafted via a variety of routes, and the most common is subcutaneous, orthotopic, renal capsule, or tail vein ( Figure 2B) 100. Subcutaneous grafts allow easy injection and monitoring of tumor growth, while orthotopic injections benefit from a proper microenvironment at the cost of more difficult injection and monitoring. Orthotopic injections can be made into the prostate (for primary tumors) or the metastatic site, including intra-tibial injections for studying bone metastasis ( Figure 2B) 101. Renal capsule implant is somewhat of a compromise between subcutaneous and orthotopic: a moderately difficult grafting site that is favorable for prostate tissue growth 100. Lastly, tail vein injections require single-cell suspensions and enable investigation of extravasation and metastasis establishment 102.
Patient-derived xenografts (PDXs) specifically use human tumor samples for engrafting into mice 63, 103. These models allow propagation of tumors (metastatic and primary) that do not grow well in culture; however, PDX tumors require continual passage in mice, which adds considerable cost 63. The PCa field has had successful PDX models since the late 1990s, but the procedure is laborious and most reports describe only few (<10) established lines 104. The number of available PDX lines was greatly expanded with the LuCaP series, which was first reported in 1996 with two lines and currently consists of 21 ongoing founder lines from a variety of samples, including four primary tumors and 17 metastases 63, 105. Moreover, 10 of the lines have undergone castration in mice to yield castration-resistant variants 63. The overall initial take rate of the LuCaP PDX lines was about 10% and, once established, most lines have a take rate of about 60% to 80% and take 4 to 16 weeks to reach maximum size (~1,000 mg). Genomic analysis of the LuCaP tumors revealed that most maintained their genomic profile from the original patient sample 63. The lines contain a variety of hallmark mutations, including AR amplification (eight lines), PTEN loss (eight heterozygous and four homozygous), RB1 loss (10 heterozygous and six homozygous), TMPRSS2-ERG fusion (10), BRCA2 homozygous loss (one), and neuroendocrine subtype (four) 63. PDX models continue to evolve and enable researchers to test a variety of therapeutic strategies against a range of genomic tumor backgrounds and to better understand tumor resistance mechanisms.
Progress in understanding the role of ETS factors
One of the unique genomic alterations in PCa is the recurrent fusions involving strong AR-regulated promotors to ETS family transcription factors (most frequently, ERG) 27, 33. In normal prostate tissue, ERG is expressed at very low levels, but it is overexpressed in PIN and adenocarcinoma 106, 107. The most frequent fusion (caused by a 3 Mb deletion) joins the TMPRSS2 promoter upstream of ERG ( Figure 3A), although other fusions have been observed with alternate promoters (for example, FOXA1, FOXP1, EST14, and HERVK17) and ETS family members ( ETV1, ETV4, ETV5, and FLI1) 22, 27, 108. Likewise, loss of the ETS family transcriptional repressor ERF (though much less common) can also stimulate oncogenic ETS activity 36. ETS fusions are observed at similar frequencies in primary and mCRPC tumors 28, tend to co-occur with PTEN loss 22, and are mutually exclusive with each other and SPOP mutations 21, 22, 82. Though ETS factors (especially ERG) are frequently altered in PCa, research is ongoing to understand exactly what role they play in disease initiation or progression or both.
Figure 3. Overview of ERG fusion and targeted therapies.
( A) The most common ETS fusion arises from a 3 Mb deletion on chromosome 21, which brings the androgen receptor (AR)-regulated TMPRSS2 promoter (light red) upstream of the ERG gene (dark blue), usually clipping the first three to five exons of ERG in the process. ( B) A schematic showing mechanisms of anti-ERG therapies. Inhibitory peptides block ERG binding to DNA and cause protein destabilization 110. Verteporfin blocks YAP1, a downstream target of ERG 93. YK-4-279 blocks ERG interaction with RNA helicase A (RHA), thereby disrupting transcription of targets 111, 112. Lastly, trabectedin binds minor grooves in ERG binding sites and disrupts its binding to target promoters 113.
GEM models have been used as one way to investigate the role of ERG in PCa development. A 2007 report used mice with Pb-driven overexpression of ETV1 (Pb- ETV1) and observed PIN 109. Other researchers found that high ERG overexpression leads to PIN lesions and disorganization of the basal cell layer in adult animals, that more lesions form as the mice age, and that there is a partially penetrant tumor phenotype in old (16- to 24-month-old) mice 71, 93. The expression level of ERG is critical, as mouse strains with lower levels of ERG overexpression do not develop tumors 71. One study found that expression of ERG in heterozygous Pten mice causes high-grade PIN at 2 months and invasive carcinoma by 6 months and that heterozygous Pten mice without ERG developed PIN at 8 months and no adenocarcinoma 114. Similarly, compared with Pten control mice, mice with conditional prostate-specific deletion of Pten and overexpression of ERG display statistically significant acceleration of prostate tumor development 81. Thus, ERG overexpression is a weak driver of tumor development in mice, but it can accelerate tumor development in the context of Pten downregulation 114, 115.
Another avenue of research involves defining specific targets and mechanisms of ETS factors in prostate tumorigenesis. There is evidence that ERG can act in part by modulating AR transcriptional activity, although the mechanism may depend on PTEN status 81, 116, 117. ERG has also been reported to positively regulate MYC 115, 118 and NOTCH 119, both of which are key for prostate differentiation and tumor development, suggesting that ERG may have a role in disrupting terminal differentiation 38, 120, 121. In addition, ERG overexpression leads to increased endoplasmic reticulum stress in LNCaP cells and the prostates of aged Pb- ERG mice 122. Transcriptional analysis of ERG overexpression in mice revealed upregulation of a YAP1 gene signature, suggesting an interaction with the Hippo tumor suppressor pathway 93. Mechanistically, it was found that YAP1 and TAZ, the transcriptional effectors negatively regulated by the Hippo pathway, are normally expressed at very low levels in human prostate luminal cells but they can be transcriptionally re-activated by overexpressed ERG and ETV1 93, 123– 125. Knockdown and constitutive activation experiments established YAP1 as a key functional target of ERG in prostate cells in vitro 93, 123– 125. Moreover, expression of constitutively active YAP1 in the mouse prostate is sufficient to drive PIN and partially penetrant tumor formation in older mice, similar to the effects of ERG overexpression 93. Knockdown of Erf in mouse prostate organoids upregulates an ERG signature and, when combined with Pten knockout, leads to tumor formation upon subcutaneous engraftment 36. With these recent studies, some of the functions of ERG in PCa are becoming clearer, although much remains to be discovered. Overall, ETS factors appear to be important drivers in PCa development, but their full effects may be seen only in the context of other alterations (for example, AR, PTEN, MYC, and NOTCH), and cross-talk between these pathways is still being investigated.
Targeting of ETS factors
Direct targeting of transcription factors is notoriously difficult but not impossible 126. One route is to target a downstream effector of ERG that is more amenable to inhibitors, such as YAP, which can be inhibited by verteporfin ( Figure 3B). Verteporfin treatment decreases VCaP (ERG +) xenograft tumor growth in mice 93. In efforts to target ERG directly, phage-display library screens have been used to identify 12 ERG inhibitory peptides 110. Two of the peptides were modified (to improve cell permeability and localize to the nucleus) and tested in vitro and in vivo. The peptides disrupted DNA binding, destabilized ERG ( Figure 3B), decreased VCaP and PC3-ERG invasion, and reduced VCaP xenograft growth 110.
Important lessons may be learned from another cancer type with recurrent ETS fusions, Ewing’s sarcoma, in which more than 90% of tumors are driven by a rearrangement fusing the EWSR1 gene to FLI1, a paralog of ERG 127, 128. ETS fusions do occur in some other cancers, but it is not a frequent event 129. YK-4-279, a FLI1 inhibitor initially developed for Ewing’s sarcoma, was reported to shrink PCa tumors in ERG + mouse xenograft models ( Figure 3B) 111, 112. Another potential candidate from the Ewing’s sarcoma field is trabectedin (and its second-generation analogue lurbinectedin), which works in part by binding DNA minor grooves in ETS binding sites and disrupting EWS-FLI1 binding at target promoters ( Figure 3B) 113, 130, 131. Very few studies (and only two phase II clinical trials) have investigated trabectedin in PCa, but the results were disappointing 132, 133. However, patients were not initially stratified by ERG status and the study did not use the newer drug lurbinectedin. These studies demonstrate that there are multiple ways to target ERG, directly or indirectly, and these therapies may be an effective option for patients with ETS + prostate tumors.
DNA repair mutations in prostate cancer
Despite having a low burden of point mutations compared with other cancers, PCa has a high rate of genomic instability (amplifications, deletions, and chromosomal rearrangements) 32. Genomic instability is a result of DNA damage, which can arise from many sources, including (but certainly not limited to) DNA replication stress, alkylating agents, mitotic chromosome segregation errors, and radiation 23, 134, 135. DNA can also be damaged as a result of transcriptional stress; AR has been reported to recruit topoisomerase enzymes to counter DNA torsional stress caused by transcription and enhancer looping 136, 137. An extreme form of genomic instability is chromothripsis, which occurs in about 20% to 30% of primary prostate tumors and involves acute chromosome shattering and reassembly, causing deletions and rearrangements 23, 138. Damage that breaks the phosphate backbone or requires repair via base excision repair, mismatch repair, or nucleotide excision repair will lead to single-strand breaks (SSBs) 134. If SSBs occur close together on opposite strands, double-strand breaks (DSBs) can occur, which are more severe and must be repaired by homologous recombination (HR) or non-homologous end joining (NHEJ) ( Figure 4). HR can occur only if a sister chromatid is present (late S or G 2 phase) and uses the non-damaged DNA as a template for error-free repair of the damaged chromatid, whereas NHEJ can repair DSBs at any cell cycle stage (predominantly G 0/G 1) but has the possibility of introducing deletions or insertions 139, 140. Further information on DNA damage-sensing and repair mechanisms can be found in several recent reviews 134, 141, 142.
Figure 4. Simplified DNA repair pathway diagram.
Single-strand breaks (SSBs) are recognized by a handful of proteins, including poly (ADP-ribose) polymerase (PARP) and RPA. PARP helps recruit DNA repair machinery to repair SSBs (blue shading). ATR is recruited to the site of damage and activates (phosphorylates) a variety of damage-sensing mediators, including CHK1, which in turn can activate p53 and, depending on other signals, pause the cell cycle until damage is repaired or induce apoptosis. Double-strand breaks (DSBs) recruit a variety of factors, including the M/R/N complex (MRE11, RAD50, and NBS1). This complex recruits and activates ATM, which phosphorylates DSB-sensing mediators, including CHK2. 53BP1 binds to M/R/N on loose DNA ends and promotes non-homologous end joining (NHEJ) repair (orange shading). During late S/G 2 phase, BRCA1 can be activated by ATM and compete with 53BP1 for binding at the M/R/N complex and aid resection of DNA ends to promote homologous recombination (HR) repair (green shading).
Germline mutations in DNA repair genes are responsible for a variety of human hereditary diseases, many of which include a predisposition to cancer 143, 144. Two such genes are the key signaling kinases in the DNA damage response—ATM (primarily activated by DSB) and ATR (primarily activated by SSB)—which can activate other signaling proteins, including CHK1 ( CHEK1) and CHK2 ( CHEK2) ( Figure 4) 142. Two other important DNA repair genes are BRCA1 and BRCA2, for which single-copy germline mutations increase the risk of multiple cancers, most significantly breast and ovarian cancer 145– 147. Mechanistically, BRCA1/BRCA2 are crucial for recruiting RAD51, which is required for HR. BRCA1 has multiple roles, including promoting loose-end resection and aiding RAD51 loading onto DNA 148. A recent study suggests that BRCA1 competes with 53BP1 for binding at DSBs and helps determine whether repair is shunted toward NHEJ or HR ( Figure 4) 149.
In 2016, a multi-institutional study sequenced the germline DNA of nearly 700 men with mCRPC and observed that 11.8% of patients carried a germline mutation in a DNA repair gene, most frequently BRCA2 (5.3%), CHEK2 (1.9%), or ATM (1.6%) ( Table 3) 150. Furthermore, somatic metastatic tumor sequencing through the SU2C/PCF landscape project determined that about 20% of metastatic tumors have a DNA repair gene aberration 28. BRCA2 germline mutations occur at about 0.3% in the general population and, though not enriched among all primary tumors, correlate with high-grade disease 22, 150– 152. For example, BRCA2 tumors are more likely to show a pattern of intra-ductal carcinoma, which involves large tumor-filled prostate ducts with intact basal layers and correlates with poor prognosis 153– 155. This knowledge has led to an ongoing discussion about whether all men who present with metastatic PCa should be screened for BRCA status as well as those with localized disease where biopsies demonstrate intra-ductal carcinoma patterning 154– 156.
Table 3. DNA repair mutation rate in tumors and germline.
| Gene | Tumor | Germline | |||
|---|---|---|---|---|---|
| Primary | mCRPC | Overall | Primary | mCRPC | |
| ATM | 6 | 8 | 0.3 | 1 | 1.6 |
| ATR | 0.3 | 0.8 | 0.1 | 0 | 0.3 |
| CHEK2 | 3 | 4 | 0.6 | 0.4 | 1.9 |
| BRCA1 | 1.2 | 0.8 | 0.2 | 0.6 | 0.9 |
| BRCA2 | 3 | 11 | 0.3 | 0.2 | 5.4 |
Mutation frequencies (percentages) for selected DNA repair genes in primary tumors and metastatic castration-resistant prostate cancer (mCRPC) and in the germline of normal, primary, or mCRPC patients. Tumor mutation frequencies were calculated the same way as in Table 1 and Table 2. Germline mutation frequencies include 692 mCRPC patients, 499 primary, and 53,105 overall (exome aggregation consortium) 150. Shading: light blue: 0–0.9%, dark blue: 1–2.9%, light red: 3–4.9%, dark red: ≥5%.
Other studies have confirmed similar rates (about 8–12%) of germline DNA repair defects in patients with mCRPC but had conflicting results as to whether germline mutant patients respond better to anti-androgen therapy 157– 160. Providing evidence against better outcomes are reports that germline mutant patients did not respond any better to initial androgen deprivation 157 and patients with mCRPC saw no additional benefit from abiraterone or enzalutamide 158. However, other studies observed that DNA repair-deficient mCRPC tumors responded better to abiraterone 160 and mCRPC patients with germline BRCA1/BRCA2/ATM mutations showed a better rate of greater than 90% prostate-specific antigen (PSA) reduction (78% versus 28%) and overall survival at 4 years (~75% versus ~25%) on ARSi therapy 159. Although germline DNA repair mutations account for only about 12% of patients with mCRPC, it will be important to better understand how these patients will respond to anti-androgen therapies. Furthermore, the identification of DNA repair mutations (whether germline or somatic) may open a window to new therapeutic options for thousands of the roughly 30,000 men who succumb to metastatic disease every year in the US and also identify family members at increased risk for cancer 1.
Targeting poly(ADP-ribose) polymerase
For patients with tumors deficient in DSB repair, there is strong rationale for targeting poly(ADP-ribose) polymerase (PARP), a family of proteins that are required for sensing and repairing SSBs ( Figure 4) 161. Without PARP, SSBs will cause stalling of replication forks during DNA replication that leads to DSBs, which then require HR or NHEJ for repair. Thus, cells lacking BRCA1 or BRCA2 must rely on error-prone NHEJ for DSB repair and are highly sensitive to loss of PARP 161– 163. In 2014, the US Food and Drug Administration (FDA) approved olaparib, a PARP inhibitor (PARPi), for the treatment of BRCA-mutant ovarian cancer, where it was found to extend average progression-free survival from 4.3 to 11.2 months 164. Since then, two other PARPis have received FDA approval for BRCA-deficient ovarian cancer: rucaparib and niraparib. Currently, those and other PARPis are in various clinical trials and studies are investigating whether the combination of PARPis with DNA-crosslinking drugs (that is, platinum-based chemotherapeutics) will yield better patient outcomes 161, 165. Testing of PARPis in PCa patients with BRCA mutations is ongoing. The TOPARP trial (Trial of PARP Inhibition in Prostate Cancer) tested olaparib in men with mCRPC and reported exciting preliminary results demonstrating response rates of 88% (14/16) in those men with a DNA repair defect and 6% (2/33) in those men without, strongly supporting treatment stratification based on DNA repair deficiency 166. Currently, searches for “PARP inhibitor” in “prostate cancer” on ClinicalTrials.gov yield 13 clinical trials; only two of these trials are completed, and there are no public results yet 167, 168.
Although most studies are ongoing, a 2018 report used retrospective analysis of an earlier study 150 to examine whether PCa patients with germline DNA repair defects had different responses to ARSi, docetaxel, or PARPi 158. Of the previously treated patients, 60 out of 390 had germline mutations and 36% of those were treated with PARPi (some with platinum chemotherapeutic as well). Germline status had no statistically significant correlations with response to docetaxel, ARSi (abiraterone/enzalutamide), or PARPi 158. Being a retrospective analysis, this study comes with multiple caveats, including the lack of initial patient stratification, inconsistent treatment methods, and a relatively small number of patients with germline mutations who received PARPi treatment (total of 22, of which 16 were BRCA2 mutant). In addition, the study did not have information about the tumor mutation landscape, so there could yet be a correlation between DNA repair mutations and PARPi response that was masked by confounding factors (for example, ETS fusion, AR, or PTEN status) 158. Ongoing trials may yet prove to be beneficial for patients with germline DNA repair defects, and further research is needed to better understand how these mutations affect response to androgen deprivation and other PCa therapy resistance.
SPOP mutation
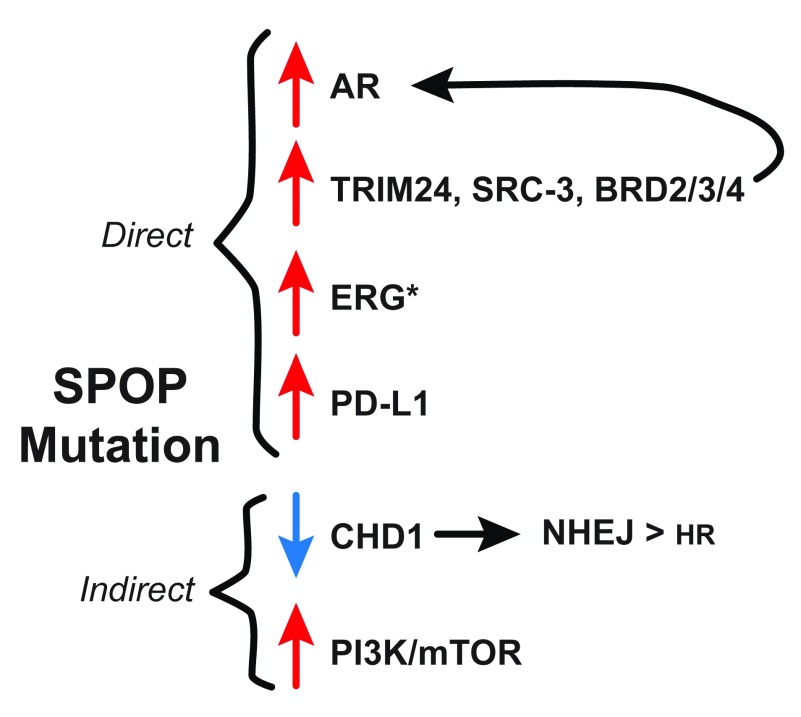
One of the novel PCa alterations elucidated by genomic sequencing efforts is mutation of SPOP 21. Heterozygous point mutations in SPOP occur in about 10% of primary and metastatic tumors ( Table 1 and Table 2) and are mutually exclusive to PTEN loss and ETS rearrangements. SPOP is an adapter component of the CUL3 E3-ligase complex, which has multiple degradation targets, including AR and its co-activators SRC-3 ( NCOA3) and TRIM24 ( Figure 5) 169– 172. Other recently identified direct targets of SPOP are the BET family proteins: BRD2/3/4 173– 175. BET proteins are transcriptional co-activators that upregulate a variety of oncogenes, including AR, MYC, and ERG 173. BET inhibitors are being investigated for use in PCa 181, but SPOP mutant tumors with elevated BET expression may be more resistant to such therapies 173, 181. Earlier this year, PD-L1 was also identified as an SPOP target, which has implications for immunotherapy and will be discussed later 179.
Figure 5. SPOP mutations and prostate cancer.
SPOP adapts the CUL3 E3-ligase complex to directly target and degrade AR and the AR co-activators TRIM24, SRC-3, and BRD2/3/4 170– 176. Thus, loss of SPOP increases both AR and its cofactors, leading to increased AR signaling. (*) ERG has also been reported as a direct SPOP target, although this is a debated topic 176– 178. SPOP has been shown to target PD-L1, a key target of checkpoint inhibitor immunotherapies 179. For indirect mechanisms, SPOP mutation strongly correlates with deletion of CHD1, which leads to 53BP1 stabilization and preference for error-prone NHEJ DSB repair 58, 180. SPOP mutation also upregulates PI3K/mTOR signaling via an unknown mechanism, which aids tumor growth and survival 83. AR, androgen receptor; DSB, double-strand break; NHEJ, non-homologous end joining.
ERG has also been reported as an SPOP target, although there are conflicting data in the literature for this relationship 176– 178. In support of the connection, two reports from 2015 identified ERG as a direct SPOP degradation target. One report found that knockdown of SPOP or CUL3 in PC3 and DU145 cells increased ERG (but not ETV1 or ERF) protein expression by increasing protein half-life and that this could be blocked by inhibition of casein kinase I 177. The authors also used a tissue microarray with immunohistochemistry (IHC) (for ERG expression) and in situ hybridization (for TMPRSS2-ERG fusions) to identify 14 ERG-positive/fusion-negative primary tumors, of which five (36%) had SPOP mutations, suggesting that SPOP mutation may upregulate ERG without fusion events 177. The second study found that SPOP knockdown increased ERG in LNCaP, C4-2, PC3, and 22RV1 lines 176. Both studies reported that SPOP targets a recognition site coded by the fourth exon of ERG that is usually disrupted in TMPRSS2-ERG fusions ( Figure 3), thus suggesting that fusions not only cause transcriptional upregulation but also increase protein stability 176, 177.
In 2017, a study using a GEM model of SPOP mutation (F133V) observed high-grade PIN at 6 months in Pten −/+ mice and prostate tumors in Pten −/− mice by 12 months 82. Additionally, the authors found that SPOP mutation alone increases PI3K/mTOR signaling. Furthermore, while PI3K signaling has negative feedback on AR, Pten −/−/SPOP F133V tumors maintain Ar and express high levels of its transcriptional targets ( Fkbp5, Psca, and Nkx3.1) 82. Thus, SPOP appears to have key intersections between AR and PI3K pathways which may help explain its importance in PCa ( Figure 5). These findings, however, do not provide a reasonable explanation for the mutual exclusivity between PTEN loss and SPOP mutation in human PCa. In a 2018 report from the same group, the authors investigated potential effects on ERG with their GEM model. Unlike the investigators in the 2015 studies 176, 177, who observed that SPOP mutation or knockdown in PCa lines increased ERG protein, this group did not observe upregulated Erg or an Erg transcriptional signature in their SPOP F133V mice 178. In addition, they detected ERG (by IHC) in only one out of 22 SPOP-mutant human tumor samples, leading them to conclude that mutant SPOP was not regulating ERG 178. One important lesson from these conflicting studies is that loss of SPOP protein does not appear to have the same functional consequence as overexpression of a mutant. Importantly, SPOP is rarely deleted in PCa tumors and the vast majority of mutations are in-frame missense point mutations. Clearly, additional research is needed to fully characterize SPOP mutants and understand how they differ in their interactions with wild-type SPOP targets.
SPOP has also been linked to DNA repair. SPOP mutant tumors have especially high rates of chromosomal rearrangements and share a transcriptional signature with BRCA1 loss, implicating SPOP in genomic instability 182. Furthermore, one of the common associations with SPOP mutations is deletion of CHD1, which is involved in DSB repair and whose loss correlates with poor survival 24, 180, 183. A query on cBioPortal using all available PCa datasets shows that CHD1 loss and SPOP mutation are significantly correlated ( p <0.01). Specifically, 57% of tumors with CHD1 deletion (68/119) have an SPOP mutation and 29% of tumors with SPOP mutations (68/233) have CHD1 deletion. A 2017 report demonstrated that CHD1-null cells (mouse stem cells and 22RV1) are more sensitive to olaparib (PARPi) and carboplatin and have increased 53BP1 protein stability, which promotes error-prone NHEJ repair ( Figure 4) 180. Furthermore, PTEN deletion is mutually exclusive to CHD1 deletion, and PTEN-null tumors require CHD1 for proliferation and survival 184. Thus, CHD1 loss may partly explain why SPOP mutations are exclusive of PTEN loss. In summary, SPOP has been implicated in many key PCa pathways (AR, MYC, ERG, PI3K, and DNA repair) and work has only recently begun to uncover specific targets and oncogenic mechanisms. Better understanding of SPOP, including its connections with ERG and CHD1, will be needed to help choose successful targeting strategies.
Androgen receptor-negative prostate cancer
The vast majority of prostate tumors depend on the AR pathway for survival 28. However, there are small subsets of PCa whose frequency appears to be increasing, including neuroendocrine PCa (NEPC) 185 and double-negative PCa (DNPC) 186, which lack AR expression and therefore are not sensitive to androgen deprivation or ARSi.
Primary prostate tumors often show regions of neuroendocrine foci, but predominantly NEPC tumors (also referred to as ‘small cell’) are rare at initial diagnosis (<2%) 5, 187, 188. However, since the advent of new ARSi therapies (abiraterone in 2011 and enzalutamide in 2012) 189, 190, there has been an increase in NEPC, which now accounts for about 15% of mCRPC and has become a mechanism of ARSi resistance 185, 186, 191. These tumors are typically more aggressive and are characterized by their lack of AR and expression of neuroendocrine-associated genes, such as chromogranin A ( CHGA) and synaptophysin ( SYP) 5, 185. NEPC tumors often show upregulation of stem-associated genes (for example, SOX2 and MYCN) 192– 194 and upregulation of genes associated with epithelial–mesenchymal transition (for example, SNAI1 and VIM) 5 and frequently lose expression of TP53 and RB1 195. Moreover, these molecular changes are also directly implicated in resistance to AR-targeting therapies 192, 195. Interestingly, NEPC tumors have TMPRSS2-ERG fusions at about the same rate as adenocarcinomas; however, owing to the lack of AR signaling, the expression levels of ERG are low in these tumors. These findings suggest that NEPC represents trans-differentiation from an androgen-responsive, epithelial-derived precursor, as opposed to the possibility that they originate from normal neuroendocrine cells, which make up less than 1% of cells in the normal prostate 38, 185.
In addition to NEPC, there is a recently identified subtype called DNPC that lacks AR and NEPC markers 186, 196. For example, recent RNA sequencing and pathway analysis with mCRPC samples from 96 patients treated before or after 2012 (the advent of abiraterone/enzalutamide) identified an increase in patients with NEPC (6.3% to 13.3%) as well as an even larger increase in DNPC (5.4% to 23.3%) 186. It is also possible that DNPC is not an entirely distinct subset of PCa but rather an intermediate step on the way from adenocarcinoma to NEPC. An AR-negative LNCaP line (LNCaP APIPC), which lacks CHGA and SYP, was used to investigate the mechanisms driving DNPC. This line has diminished AKT signaling and relies on an upregulated autocrine FGF8 → FGFR → ERK signaling pathway for survival 186. Analysis of human tumor data and PDX lines with DNPC confirmed a pattern of upregulated FGFs (FGF1/8/9), FGFRs (FGFR1/2/3/4), and an ERK signature. Furthermore, LNCaP APIPC xenografts are sensitive to FGFR inhibitors (CH-5183284, PD173074) 186. Thus, targeting the FGFR/ERK signaling axis may be beneficial for patients with DNPC, although it has yet to be tested in clinical trials. Thus, while AR-negative PCa accounts for a minority of prostate tumors, they are becoming more common and will require a different therapeutic strategy than classic AR-positive PCa.
Immunotherapy
Recently, cancer immunotherapy has received significant attention and has demonstrated great potential across different types of cancer. Immunotherapies can broadly be grouped into three strategies: cancer vaccines, immune checkpoint inhibitors, and engineered live immune cell components. Cancer vaccines use tumor-specific proteins to generate a targeted immune response or tag tumors with a lethal, targetable protein. There are a handful of vaccine-based trials for PCa, although the only currently FDA-approved therapy is Sipuleucel-T (also known as Provenge ®) 197. Sipuleucel-T uses prostatic acid phosphatase as a tumor antigen and has shown about 4- to 5-month extended survival for patients with mCRPC, while patients with lower baseline PSA level had even greater (about 13-month) survival 198, 199. Several additional PCa vaccines are being tested, and detailed information about those studies can be obtained in the cited reviews 197, 200.
A second branch of immunotherapy is checkpoint inhibition, which attempts to re-activate cancer-targeting T cells that have been disarmed by tumors. The primary targets for checkpoint inhibition are CTLA-4, PD1, and the PD1 ligands PD-L1/PD-L2. An early checkpoint inhibitor is ipilimumab, which targets CTLA-4 and was approved by the FDA in 2011 for the treatment of melanoma. Ipilimumab has been tested in PCa trials with mixed results; it was able to delay progression but did not extend overall survival 197, 201. However, this study had a small subset of patients (two out of 400) who had a complete response (>4 years) 202. Other checkpoint inhibitors (for example, nivolumab, pembrolizumab, and atezolizumab) target PD1 or PD-L1/L2 200. In general, tumors with high mutational burden generate more novel proteins (neoantigens) and respond more favorably to immunotherapy 203, 204. A small subset of mCRPC tumors have mismatch repair defects (5% rate of deletion/mutation of MSH2 or MSH6 or both) 27– 29 and exhibit a hypermutated phenotype 205. Interestingly, mismatch repair defects appear to be enriched in the most aggressive primary tumors. In one study, 40% of samples with intra-ductal carcinoma (four out of 10) showed loss of MSH2, MSH6, or MLH1 206. Another investigation analyzed 1,133 primary and NEPC tumors by IHC via tissue microarray and observed that 8% of Gleason pattern five tumors (seven out of 91) had loss of MSH2 protein (due to technical issues, MSH6 and MHL1 were not included) 207. In 2017, the FDA approved pembrolizumab for the treatment of solid metastatic tumors with mismatch repair-defects, providing a new option for some patients with mCRPC 208.
A recent study suggests that even tumors with low PD-L1 may be targetable using combination therapy. CDK4 was reported to negatively regulate PD-L1 via SPOP, and CDK4/6 inhibitors can cause upregulation of PD-L1 in mouse tissue and breast cancer xenografts 179. The authors went on to show that 80% of SPOP-mutant tumors (12 out of 15) had high PD-L1 by IHC staining versus 10% of the non-mutant SPOP tumors. Thus, SPOP-mutant tumors are likely to express PD-L1 and benefit from checkpoint inhibitory therapy, while other tumors may be driven to express PD-L1 by CDK4/6 inhibition.
Another recent report focused on targeting myeloid-derived suppressor cells (MDSCs), which can shield tumors from T cells 209. A GEM model with Pb-driven knockout of Pten, Trp53, and Smad4 was developed, and combinations of checkpoint inhibitors (anti-CTLA4 and anti-PD1 antibodies) plus inhibitors against multiple tyrosine kinases (dasatinib and cabozantinib) and PI3K (dactolisib/BEZ235) were evaluated. The authors found that combination therapy (checkpoint + tyrosine kinase + PI3K inhibition) had a major effect on decreasing tumor burden. Furthermore, they went on to discover that a key mechanism of this therapy was decreased cytokine production and MDSC tumor infiltration caused by the inhibition of tyrosine kinases and PI3K, which in turn sensitized the tumors to the checkpoint inhibition 209.
The third branch of immunotherapy uses engineered immune cells, including chimeric antigen receptor T-cell (CAR-T) therapy. CAR-T involves isolating patient immune cells and genetically engineering them to express a chimeric protein fusing a tumor-recognizing antibody region with a T-cell activation domain 210. The engineered cells are then grafted back into the patient. CAR-T cells can directly recognize tumors and trigger activation. This therapy has been extremely successful for treating B-cell acute lymphoblastic leukemia and B-cell lymphoma 210, 211. Trials with CAR-T therapies for PCa are under way and primarily involve using PSMA and PSCA as targeting antigens 212, 213. The multiple immunotherapy strategies of cancer vaccines, checkpoint inhibitors, and CAR-T continue to improve. Meanwhile, patient stratification based on tumor mutational burden, PD1/PD-L expression, and tumor-enriched antigens such as PSMA and PSCA will help direct these therapies to the patients most likely to benefit.
Conclusions
Owing to large-scale sequencing efforts ( Figure 1), the PCa field now has a near-comprehensive view of the mutational landscape of human PCa. These studies revealed that ETS fusions are the most frequent mutation in primary tumors, occurring in a little over half of cases ( Figure 3). Likewise, the second most common alteration, PTEN loss, occurs in about a quarter of primary tumors. PCa shows great resilience in evading androgen deprivation therapy and finding ways to maintain the AR pathway, but there is a growing number of tumors that do not express AR and rely on alternate survival mechanisms 4.
Perhaps the key takeaway is that human prostate tumors are driven by a combination of alterations in a handful of signaling pathways. Understanding the role of these pathways in tumor initiation, progression, and therapeutic resistance will be critical in the future. In order to functionally test the role and mechanisms of these signaling pathways, there is a need to continually improve existing cell culture and animal models. Recently developed GEM models and organoid culture conditions can provide great opportunities for studying disease initiation, metastasis, and testing therapies against patient-derived tumors. As has become clear with other cancers, there will almost certainly be no single effective therapy for PCa. While most tumors will likely still benefit from improved AR-targeting therapies, it will be important to recognize subsets of tumors that may benefit from targeting other supporting mutations.
Abbreviations
AR, androgen receptor; ARSi, androgen receptor signaling inhibitor; CAR-T, chimeric antigen receptor T-cell; CTC, circulating tumor cell; DNPC, double-negative prostate cancer; DSB, double-strand break; FDA, US Food and Drug Administration; GEM, genetically engineered mouse; HR, homologous recombination; IHC, immunohistochemistry; LPB, large probasin promoter; LSL, Lox-STOP-Lox; mCRPC, metastatic castration-resistant prostate cancer; MDSC, myeloid-derived suppressor cell; NEPC, neuroendocrine prostate cancer; NHEJ, non-homologous end joining; PARP, poly(ADP-ribose) polymerase; PARPi, poly(ADP-ribose) polymerase inhibitor; Pb, Probasin; PCa, prostate cancer; PDX, patient-derived xenograft; PIN, prostatic intra-epithelial neoplasia; SSB, single-strand break; TCGA, The Cancer Genome Atlas; WES, whole exome sequencing; WGS, whole genome sequencing.
Acknowledgments
We would like to thank Payel Chatterjee for helpful discussion and notes on DNA damage repair pathways. We apologize to colleagues where space constraints precluded citing their work.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Christopher E Barbieri, The Caryl and Israel Englander Institute for Precision Medicine of Weill Cornell Medicine, and New York-Presbyterian Hospital, New York, USA
Emmanuel S Antonarakis, Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, MD, USA
Funding Statement
This work was funded by the National Institutes of Health: P30CA015704, P50CA097186, and R01CA176844.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
References
- 1. Siegel RL, Miller KD, Jemal A: Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 2. Wong MC, Goggins WB, Wang HH, et al. : Global Incidence and Mortality for Prostate Cancer: Analysis of Temporal Patterns and Trends in 36 Countries. Eur Urol. 2016;70(5):862–74. 10.1016/j.eururo.2016.05.043 [DOI] [PubMed] [Google Scholar]
- 3. Huggins C, Hodges CV: Studies on prostatic cancer: I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. 1941. J Urol. 2002;168(1):9–12. 10.1016/S0022-5347(05)64820-3 [DOI] [PubMed] [Google Scholar]
- 4. Coutinho I, Day TK, Tilley WD, et al. : Androgen receptor signaling in castration-resistant prostate cancer: a lesson in persistence. Endocr Relat Cancer. 2016;23(12):T179–T197. 10.1530/ERC-16-0422 [DOI] [PubMed] [Google Scholar]
- 5. Davies AH, Beltran H, Zoubeidi A: Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat Rev Urol. 2018;15(5):271–86. 10.1038/nrurol.2018.22 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Sheahan AV, Ellis L: Epigenetic reprogramming: A key mechanism driving therapeutic resistance. Urol Oncol. 2018; pii: S1078-1439(17)30650-6. 10.1016/j.urolonc.2017.12.021 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 7. Kgatle MM, Kalla AA, Islam MM, et al. : Prostate Cancer: Epigenetic Alterations, Risk Factors, and Therapy. Prostate Cancer. 2016;2016:5653862. 10.1155/2016/5653862 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 8. Labbé DP, Zadra G, Ebot EM, et al. : Role of diet in prostate cancer: the epigenetic link. Oncogene. 2015;34(36):4683–91. 10.1038/onc.2014.422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lindqvist LM, Tandoc K, Topisirovic I, et al. : Cross-talk between protein synthesis, energy metabolism and autophagy in cancer. Curr Opin Genet Dev. 2018;48:104–11. 10.1016/j.gde.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 10. Zabala-Letona A, Arruabarrena-Aristorena A, Martín-Martín N, et al. : mTORC1-dependent AMD1 regulation sustains polyamine metabolism in prostate cancer. Nature. 2017;547(7661):109–13. 10.1038/nature22964 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 11. Alford AV, Brito JM, Yadav KK, et al. : The Use of Biomarkers in Prostate Cancer Screening and Treatment. Rev Urol. 2017;19(4):221–34. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 12. Bonci D, Coppola V, Patrizii M, et al. : A microRNA code for prostate cancer metastasis. Oncogene. 2016;35(9):1180–92. 10.1038/onc.2015.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fabris L, Ceder Y, Chinnaiyan AM, et al. : The Potential of MicroRNAs as Prostate Cancer Biomarkers. Eur Urol. 2016;70(2):312–22. 10.1016/j.eururo.2015.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ganguly SS, Li X, Miranti CK: The host microenvironment influences prostate cancer invasion, systemic spread, bone colonization, and osteoblastic metastasis. Front Oncol. 2014;4:364. 10.3389/fonc.2014.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sfanos KS, Yegnasubramanian S, Nelson WG, et al. : The inflammatory microenvironment and microbiome in prostate cancer development. Nat Rev Urol. 2018;15(1):11–24. 10.1038/nrurol.2017.167 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 16. McGinley KF, Tay KJ, Moul JW: Prostate cancer in men of African origin. Nat Rev Urol. 2016;13(2):99–107. 10.1038/nrurol.2015.298 [DOI] [PubMed] [Google Scholar]
- 17. Rhodes DR, Barrette TR, Rubin MA, et al. : Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002;62(15):4427–33. [PubMed] [Google Scholar]
- 18. Taylor BS, Schultz N, Hieronymus H, et al. : Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. 10.1016/j.ccr.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Berger MF, Lawrence MS, Demichelis F, et al. : The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–20. 10.1038/nature09744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baca SC, Prandi D, Lawrence MS, et al. : Punctuated evolution of prostate cancer genomes. Cell. 2013;153(3):666–77. 10.1016/j.cell.2013.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Barbieri CE, Baca SC, Lawrence MS, et al. : Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–9. 10.1038/ng.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cancer Genome Atlas Research Network: The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–25. 10.1016/j.cell.2015.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fraser M, Sabelnykova VY, Yamaguchi TN, et al. : Genomic hallmarks of localized, non-indolent prostate cancer. Nature. 2017;541(7637):359–64. 10.1038/nature20788 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. Wedge DC, Gundem G, Mitchell T, et al. : Sequencing of prostate cancers identifies new cancer genes, routes of progression and drug targets. Nat Genet. 2018;50(5):682–92. 10.1038/s41588-018-0086-z [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Espiritu SMG, Liu LY, Rubanova Y, et al. : The Evolutionary Landscape of Localized Prostate Cancers Drives Clinical Aggression. Cell. 2018;173(4):1003–1013.e15. 10.1016/j.cell.2018.03.029 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Armenia J, Wankowicz SAM, Liu D, et al. : The long tail of oncogenic drivers in prostate cancer. Nat Genet. 2018;50(5):645–51. 10.1038/s41588-018-0078-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grasso CS, Wu Y, Robinson DR, et al. : The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–43. 10.1038/nature11125 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Robinson D, Van Allen EM, Wu YM, et al. : Integrative Clinical Genomics of Advanced Prostate Cancer. Cell. 2015;162(2):454. 10.1016/j.cell.2015.06.053 [DOI] [PubMed] [Google Scholar]
- 29. Kumar A, Coleman I, Morrissey C, et al. : Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22(4):369–78. 10.1038/nm.4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beltran H, Prandi D, Mosquera JM, et al. : Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22(3):298–305. 10.1038/nm.4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lawrence MS, Stojanov P, Mermel CH, et al. : Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. 10.1038/nature12912 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Barbieri CE, Rubin MA: Genomic rearrangements in prostate cancer. Curr Opin Urol. 2015;25(1):71–6. 10.1097/MOU.0000000000000129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tomlins SA, Rhodes DR, Perner S, et al. : Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8. 10.1126/science.1117679 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Cerami E, Gao J, Dogrusoz U, et al. : The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao J, Aksoy BA, Dogrusoz U, et al. : Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bose R, Karthaus WR, Armenia J, et al. : ERF mutations reveal a balance of ETS factors controlling prostate oncogenesis. Nature. 2017;546(7660):671–5. 10.1038/nature22820 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Huang FW, Mosquera JM, Garofalo A, et al. : Exome Sequencing of African-American Prostate Cancer Reveals Loss-of-Function ERF Mutations. Cancer Discov. 2017;7(9):973–83. 10.1158/2159-8290.CD-16-0960 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Frank SB, Miranti CK: Disruption of prostate epithelial differentiation pathways and prostate cancer development. Front Oncol. 2013;3:273. 10.3389/fonc.2013.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mahajan K, Malla P, Lawrence HR, et al. : ACK1/TNK2 Regulates Histone H4 Tyr88-phosphorylation and AR Gene Expression in Castration-Resistant Prostate Cancer. Cancer Cell. 2017;31(6):790–803.e8. 10.1016/j.ccell.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 40. Miyamoto DT, Lee RJ, Stott SL, et al. : Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2(11):995–1003. 10.1158/2159-8290.CD-12-0222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miyamoto DT, Zheng Y, Wittner BS, et al. : RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349(6254):1351–6. 10.1126/science.aab0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang L, Ma F, Chapman A, et al. : Single-Cell Whole-Genome Amplification and Sequencing: Methodology and Applications. Annu Rev Genomics Hum Genet. 2015;16:79–102. 10.1146/annurev-genom-090413-025352 [DOI] [PubMed] [Google Scholar]
- 43. Gundem G, Van Loo P, Kremeyer B, et al. : The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520(7547):353–7. 10.1038/nature14347 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Sobel RE, Sadar MD: Cell lines used in prostate cancer research: a compendium of old and new lines--part 1. J Urol. 2005;173(2):342–59. 10.1097/01.ju.0000141580.30910.57 [DOI] [PubMed] [Google Scholar]
- 45. Moad M, Hannezo E, Buczacki SJ, et al. : Multipotent Basal Stem Cells, Maintained in Localized Proximal Niches, Support Directed Long-Ranging Epithelial Flows in Human Prostates. Cell Rep. 2017;20(7):1609–22. 10.1016/j.celrep.2017.07.061 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Collins AT, Habib FK, Maitland NJ, et al. : Identification and isolation of human prostate epithelial stem cells based on alpha(2)beta(1)-integrin expression. J Cell Sci. 2001;114(Pt 21):3865–72. [DOI] [PubMed] [Google Scholar]
- 47. Litvinov IV, Vander Griend DJ, Xu Y, et al. : Low-calcium serum-free defined medium selects for growth of normal prostatic epithelial stem cells. Cancer Res. 2006;66(17):8598–607. 10.1158/0008-5472.CAN-06-1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lamb LE, Knudsen BS, Miranti CK: E-cadherin-mediated survival of androgen-receptor-expressing secretory prostate epithelial cells derived from a stratified in vitro differentiation model. J Cell Sci. 2010;123(Pt 2):266–76. 10.1242/jcs.054502 [DOI] [PubMed] [Google Scholar]
- 49. Niranjan B, Lawrence MG, Papargiris MM, et al. : Primary culture and propagation of human prostate epithelial cells. Methods Mol Biol. 2013;945:365–82. 10.1007/978-1-62703-125-7_22 [DOI] [PubMed] [Google Scholar]
- 50. Sroka IC, Anderson TA, McDaniel KM, et al. : The laminin binding integrin alpha6beta1 in prostate cancer perineural invasion. J Cell Physiol. 2010;224(2):283–8. 10.1002/jcp.22149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lamb LE, Zarif JC, Miranti CK: The androgen receptor induces integrin α6β1 to promote prostate tumor cell survival via NF-κB and Bcl-xL Independently of PI3K signaling. Cancer Res. 2011;71(7):2739–49. 10.1158/0008-5472.CAN-10-2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Drost J, Karthaus WR, Gao D, et al. : Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc. 2016;11(2):347–58. 10.1038/nprot.2016.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang S, Gao D, Chen Y: The potential of organoids in urological cancer research. Nat Rev Urol. 2017;14(7):401–14. 10.1038/nrurol.2017.65 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Chu JH, Yu S, Hayward SW, et al. : Development of a three-dimensional culture model of prostatic epithelial cells and its use for the study of epithelial-mesenchymal transition and inhibition of PI3K pathway in prostate cancer. Prostate. 2009;69(4):428–42. 10.1002/pros.20897 [DOI] [PubMed] [Google Scholar]
- 55. Bello-DeOcampo D, Kleinman HK, Deocampo ND, et al. : Laminin-1 and alpha6beta1 integrin regulate acinar morphogenesis of normal and malignant human prostate epithelial cells. Prostate. 2001;46(2):142–53. [DOI] [PubMed] [Google Scholar]
- 56. Garraway IP, Sun W, Tran CP, et al. : Human prostate sphere-forming cells represent a subset of basal epithelial cells capable of glandular regeneration in vivo. Prostate. 2010;70(5):491–501. 10.1002/pros.21083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen S, Principessa L, Isaacs JT: Human prostate cancer initiating cells isolated directly from localized cancer do not form prostaspheres in primary culture. Prostate. 2012;72(13):1478–89. 10.1002/pros.22503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gao D, Vela I, Sboner A, et al. : Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159(1):176–87. 10.1016/j.cell.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 59. Wang M, Nagle RB, Knudsen BS, et al. : A basal cell defect promotes budding of prostatic intraepithelial neoplasia. J Cell Sci. 2017;130(1):104–10. 10.1242/jcs.188177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stock K, Estrada MF, Vidic S, et al. : Capturing tumor complexity in vitro: Comparative analysis of 2D and 3D tumor models for drug discovery. Sci Rep. 2016;6: 28951. 10.1038/srep28951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sieh S, Taubenberger AV, Lehman ML, et al. : Paracrine interactions between LNCaP prostate cancer cells and bioengineered bone in 3D in vitro culture reflect molecular changes during bone metastasis. Bone. 2014;63:121–31. 10.1016/j.bone.2014.02.001 [DOI] [PubMed] [Google Scholar]
- 62. Wang R, Xu J, Juliette L, et al. : Three-dimensional co-culture models to study prostate cancer growth, progression, and metastasis to bone. Semin Cancer Biol. 2005;15(5):353–64. 10.1016/j.semcancer.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 63. Nguyen HM, Vessella RL, Morrissey C, et al. : LuCaP Prostate Cancer Patient-Derived Xenografts Reflect the Molecular Heterogeneity of Advanced Disease an--d Serve as Models for Evaluating Cancer Therapeutics. Prostate. 2017;77(6):654–71. 10.1002/pros.23313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Park JW, Lee JK, Phillips JW, et al. : Prostate epithelial cell of origin determines cancer differentiation state in an organoid transformation assay. Proc Natl Acad Sci U S A. 2016;113(16):4482–7. 10.1073/pnas.1603645113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chua CW, Shibata M, Lei M, et al. : Single luminal epithelial progenitors can generate prostate organoids in culture. Nat Cell Biol. 2014;16(10):951–61, 1–4. 10.1038/ncb3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Karthaus WR, Iaquinta PJ, Drost J, et al. : Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159(1):163–75. 10.1016/j.cell.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 67. Chambers KF, Mosaad EM, Russell PJ, et al. : Correction: 3D Cultures of Prostate Cancer Cells Cultured in a Novel High-Throughput Culture Platform Are More Resistant to Chemotherapeutics Compared to Cells Cultured in Monolayer. PLoS One. 2015;10(4):e0125641. 10.1371/journal.pone.0125641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Edmondson R, Adcock AF, Yang L: Influence of Matrices on 3D-Cultured Prostate Cancer Cells' Drug Response and Expression of Drug-Action Associated Proteins. PLoS One. 2016;11(16):e0158116. 10.1371/journal.pone.0158116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Frese KK, Tuveson DA: Maximizing mouse cancer models. Nat Rev Cancer. 2007;7(9):645–58. 10.1038/nrc2192 [DOI] [PubMed] [Google Scholar]
- 70. Ellwood-Yen K, Graeber TG, Wongvipat J, et al. : Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4(3):223–38. 10.1016/S1535-6108(03)00197-1 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 71. Klezovitch O, Risk M, Coleman I, et al. : A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105(6):2105–10. 10.1073/pnas.0711711105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Greenberg NM, DeMayo F, Finegold MJ, et al. : Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92(8):3439–43. 10.1073/pnas.92.8.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kasper S, Sheppard PC, Yan Y, et al. : Development, progression, and androgen-dependence of prostate tumors in probasin-large T antigen transgenic mice: a model for prostate cancer. Lab Invest. 1998;78(3):319–33. [PubMed] [Google Scholar]
- 74. Wang X, Kruithof-de Julio M, Economides KD, et al. : A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461(7263):495–500. 10.1038/nature08361 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Van Keymeulen A, Rocha AS, Ousset M, et al. : Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479(7372):189–93. 10.1038/nature10573 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. Ousset M, Van Keymeulen A, Bouvencourt G, et al. : Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat Cell Biol. 2012;14(11):1131–8. 10.1038/ncb2600 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 77. Ding Z, Wu C, Chu GC, et al. : SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470(7333):269–73. 10.1038/nature09677 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. Ding Z, Wu CJ, Jaskelioff M, et al. : Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell. 2012;148(5):896–907. 10.1016/j.cell.2012.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Lesche R, Groszer M, Gao J, et al. : Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32(2):148–9. 10.1002/gene.10036 [DOI] [PubMed] [Google Scholar]
- 80. Zhou Z, Flesken-Nikitin A, Corney DC, et al. : Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res. 2006;66(16):7889–98. 10.1158/0008-5472.CAN-06-0486 [DOI] [PubMed] [Google Scholar]
- 81. Chen Y, Chi P, Rockowitz S, et al. : ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013;19(8):1023–9. 10.1038/nm.3216 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 82. Blattner M, Liu D, Robinson BD, et al. : SPOP Mutation Drives Prostate Tumorigenesis In Vivo through Coordinate Regulation of PI3K/mTOR and AR Signaling. Cancer Cell. 2017;31(3):436–51. 10.1016/j.ccell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 83. Kwon OJ, Zhang L, Wang J, et al. : Notch promotes tumor metastasis in a prostate-specific Pten-null mouse model. J Clin Invest. 2016;126(7):2626–41. 10.1172/JCI84637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mulholland DJ, Kobayashi N, Ruscetti M, et al. : Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res. 2012;72(7):1878–89. 10.1158/0008-5472.CAN-11-3132 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 85. Wang J, Kobayashi T, Floc'h N, et al. : B-Raf activation cooperates with PTEN loss to drive c-Myc expression in advanced prostate cancer. Cancer Res. 2012;72(18):4765–76. 10.1158/0008-5472.CAN-12-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Baena E, Shao Z, Linn DE, et al. : ETV1 directs androgen metabolism and confers aggressive prostate cancer in targeted mice and patients. Genes Dev. 2013;27(6):683–98. 10.1101/gad.211011.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Linn DE, Penney KL, Bronson RT, et al. : Deletion of Interstitial Genes between TMPRSS2 and ERG Promotes Prostate Cancer Progression. Cancer Res. 2016;76(7):1869–81. 10.1158/0008-5472.CAN-15-1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yan Y, Sheppard PC, Kasper S, et al. : Large fragment of the probasin promoter targets high levels of transgene expression to the prostate of transgenic mice. Prostate. 1997;32(2):129–39. [DOI] [PubMed] [Google Scholar]
- 89. Vasioukhin V, Degenstein L, Wise B, et al. : The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci U S A. 1999;96(15):8551–6. 10.1073/pnas.96.15.8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kwon OJ, Zhang L, Ittmann MM, et al. : Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin. Proc Natl Acad Sci U S A. 2014;111(5):E592–600. 10.1073/pnas.1318157111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang X, Lee C, Ng PY, et al. : Prostatic neoplasia in transgenic mice with prostate-directed overexpression of the c-myc oncoprotein. Prostate. 2000;43(4):278–85. 10.1002/1097-0045(20000601)43:4%3c278::AID-PROS7%3e3.0.CO;2-4 [DOI] [PubMed] [Google Scholar]
- 92. Di Cristofano A, Pesce B, Cordon-Cardo C, et al. : Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19(4):348–55. 10.1038/1235 [DOI] [PubMed] [Google Scholar]
- 93. Nguyen LT, Tretiakova MS, Silvis MR, et al. : ERG Activates the YAP1 Transcriptional Program and Induces the Development of Age-Related Prostate Tumors. Cancer Cell. 2015;27(6):797–808. 10.1016/j.ccell.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Klezovitch O, Chevillet J, Mirosevich J, et al. : Hepsin promotes prostate cancer progression and metastasis. Cancer Cell. 2004;6(2):185–95. 10.1016/j.ccr.2004.07.008 [DOI] [PubMed] [Google Scholar]
- 95. Abate-Shen C, Banach-Petrosky WA, Sun X, et al. : Nkx3.1; Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res. 2003;63(14):3886–90. [PubMed] [Google Scholar]
- 96. Tang X, Mahajan SS, Nguyen LT, et al. : Targeted inhibition of cell-surface serine protease Hepsin blocks prostate cancer bone metastasis. Oncotarget. 2014;5(5):1352–62. 10.18632/oncotarget.1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Valkenburg KC, Williams BO: Mouse models of prostate cancer. Prostate Cancer. 2011;2011: 895238. 10.1155/2011/895238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Grabowska MM, DeGraff DJ, Yu X, et al. : Mouse models of prostate cancer: picking the best model for the question. Cancer Metastasis Rev. 2014;33(2–3):377–97. 10.1007/s10555-013-9487-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rea D, Del Vecchio V, Palma G, et al. : Mouse Models in Prostate Cancer Translational Research: From Xenograft to PDX. Biomed Res Int. 2016;2016: 9750795. 10.1155/2016/9750795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wang Y, Revelo MP, Sudilovsky D, et al. : Development and characterization of efficient xenograft models for benign and malignant human prostate tissue. Prostate. 2005;64(2):149–59. 10.1002/pros.20225 [DOI] [PubMed] [Google Scholar]
- 101. Corey E, Quinn JE, Bladou F, et al. : Establishment and characterization of osseous prostate cancer models: intra-tibial injection of human prostate cancer cells. Prostate. 2002;52(1):20–33. 10.1002/pros.10091 [DOI] [PubMed] [Google Scholar]
- 102. Elkin M, Vlodavsky I: Tail vein assay of cancer metastasis. Curr Protoc Cell Biol. 2001;Chapter 19(1):Unit 19.2. 10.1002/0471143030.cb1902s12 [DOI] [PubMed] [Google Scholar]
- 103. Gao H, Korn JM, Ferretti S, et al. : High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21(11):1318–25. 10.1038/nm.3954 [DOI] [PubMed] [Google Scholar]
- 104. Toivanen R, Taylor RA, Pook DW, et al. : Breaking through a roadblock in prostate cancer research: an update on human model systems. J Steroid Biochem Mol Biol. 2012;131(3–5):122–31. 10.1016/j.jsbmb.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 105. Ellis WJ, Vessella RL, Buhler KR, et al. : Characterization of a novel androgen-sensitive, prostate-specific antigen-producing prostatic carcinoma xenograft: LuCaP 23. Clin Cancer Res. 1996;2(6):1039–48. [PubMed] [Google Scholar]
- 106. Mohamed AA, Tan SH, Mikhalkevich N, et al. : Ets family protein, erg expression in developing and adult mouse tissues by a highly specific monoclonal antibody. J Cancer. 2010;1:197–208. 10.7150/jca.1.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Liu H, Shi J, Wilkerson M, et al. : Immunohistochemical evaluation of ERG expression in various benign and malignant tissues. Ann Clin Lab Sci. 2013;43(1):3–9. [PubMed] [Google Scholar]
- 108. Hermans KG, van der Korput HA, et al. : Truncated ETV1, fused to novel tissue-specific genes, and full-length ETV1 in prostate cancer. Cancer Res. 2008;68(18):7541–9. 10.1158/0008-5472.CAN-07-5930 [DOI] [PubMed] [Google Scholar]
- 109. Tomlins SA, Laxman B, Dhanasekaran SM, et al. : Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448(7153):595–9. 10.1038/nature06024 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 110. Wang X, Qiao Y, Asangani IA, et al. : Development of Peptidomimetic Inhibitors of the ERG Gene Fusion Product in Prostate Cancer. Cancer Cell. 2017;31(4):532–548.e7. 10.1016/j.ccell.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 111. Winters B, Brown L, Coleman I, et al. : Inhibition of ERG Activity in Patient-derived Prostate Cancer Xenografts by YK-4-279. Anticancer Res. 2017;37(7):3385–96. 10.21873/anticanres.11705 [DOI] [PubMed] [Google Scholar]
- 112. Rahim S, Minas T, Hong SH, et al. : A small molecule inhibitor of ETV1, YK-4-279, prevents prostate cancer growth and metastasis in a mouse xenograft model. PLoS One. 2014;9(12):e114260. 10.1371/journal.pone.0114260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Harlow ML, Maloney N, Roland J, et al. : Lurbinectedin Inactivates the Ewing Sarcoma Oncoprotein EWS-FLI1 by Redistributing It within the Nucleus. Cancer Res. 2016;76(22):6657–68. 10.1158/0008-5472.CAN-16-0568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Carver BS, Tran J, Gopalan A, et al. : Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41(5):619–24. 10.1038/ng.370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zong Y, Xin L, Goldstein AS, et al. : ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci U S A. 2009;106(30):12465–70. 10.1073/pnas.0905931106 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 116. Yu J, Yu J, Mani RS, et al. : An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17(5):443–54. 10.1016/j.ccr.2010.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Chng KR, Chang CW, Tan SK, et al. : A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J. 2012;31(12):2810–23. 10.1038/emboj.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sun C, Dobi A, Mohamed A, et al. : TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27(40):5348–53. 10.1038/onc.2008.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mohamed AA, Tan SH, Xavier CP, et al. : Synergistic Activity with NOTCH Inhibition and Androgen Ablation in ERG-Positive Prostate Cancer Cells. Mol Cancer Res. 2017;15(10):1308–17. 10.1158/1541-7786.MCR-17-0058 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 120. Berger PL, Frank SB, Schulz VV, et al. : Transient induction of ING4 by Myc drives prostate epithelial cell differentiation and its disruption drives prostate tumorigenesis. Cancer Res. 2014;74(12):3357–68. 10.1158/0008-5472.CAN-13-3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Frank SB, Berger PL, Ljungman M, et al. : Human prostate luminal cell differentiation requires NOTCH3 induction by p38-MAPK and MYC. J Cell Sci. 2017;130(11):1952–64. 10.1242/jcs.197152 [DOI] [PubMed] [Google Scholar]
- 122. Sreenath TL, Macalindong SS, Mikhalkevich N, et al. : ETS Related Gene mediated Androgen Receptor Aggregation and Endoplasmic Reticulum Stress in Prostate Cancer Development. Sci Rep. 2017;7(1): 1109. 10.1038/s41598-017-01187-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kim TD, Jin F, Shin S, et al. : Histone demethylase JMJD2A drives prostate tumorigenesis through transcription factor ETV1. J Clin Invest. 2016;126(2):706–20. 10.1172/JCI78132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kim TD, Shin S, Janknecht R: ETS transcription factor ERG cooperates with histone demethylase KDM4A. Oncol Rep. 2016;35(6):3679–88. 10.3892/or.2016.4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Liu CY, Yu T, Huang Y, et al. : ETS (E26 transformation-specific) up-regulation of the transcriptional co-activator TAZ promotes cell migration and metastasis in prostate cancer. J Biol Chem. 2017;292(22):9420–30. 10.1074/jbc.M117.783787 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 126. Hagenbuchner J, Ausserlechner MJ: Targeting transcription factors by small compounds--Current strategies and future implications. Biochem Pharmacol. 2016;107:1–13. 10.1016/j.bcp.2015.12.006 [DOI] [PubMed] [Google Scholar]
- 127. Delattre O, Zucman J, Melot T, et al. : The Ewing family of tumors--a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N Engl J Med. 1994;331(5):294–9. 10.1056/NEJM199408043310503 [DOI] [PubMed] [Google Scholar]
- 128. Kovar H, Amatruda J, Brunet E, et al. : The second European interdisciplinary Ewing sarcoma research summit--A joint effort to deconstructing the multiple layers of a complex disease. Oncotarget. 2016;7(8):8613–24. 10.18632/oncotarget.6937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Feng FY, Brenner JC, Hussain M, Chinnaiyan AM: Molecular pathways: targeting ETS gene fusions in cancer. Clin Cancer Res. 2014;20(17):4442–8. 10.1158/1078-0432.CCR-13-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Leal JF, Martínez-Díez M, García-Hernández V, et al. : PM01183, a new DNA minor groove covalent binder with potent in vitro and in vivo anti-tumour activity. Br J Pharmacol. 2010;161(5):1099–110. 10.1111/j.1476-5381.2010.00945.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Larsen AK, Galmarini CM, D'Incalci M: Unique features of trabectedin mechanism of action. Cancer Chemother Pharmacol. 2016;77(4):663–71. 10.1007/s00280-015-2918-1 [DOI] [PubMed] [Google Scholar]
- 132. Acikgoz E, Guven U, Duzagac F, et al. : Enhanced G2/M Arrest, Caspase Related Apoptosis and Reduced E-Cadherin Dependent Intercellular Adhesion by Trabectedin in Prostate Cancer Stem Cells. PLoS One. 2015;10(10):e0141090. 10.1371/journal.pone.0141090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Michaelson MD, Bellmunt J, Hudes GR, et al. : Multicenter phase II study of trabectedin in patients with metastatic castration-resistant prostate cancer. Ann Oncol. 2012;23(5):1234–40. 10.1093/annonc/mdr399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Sirbu BM, Cortez D: DNA damage response: three levels of DNA repair regulation. Cold Spring Harb Perspect Biol. 2013;5(8):a012724. 10.1101/cshperspect.a012724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Mehta A, Haber JE: Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6(9):a016428. 10.1101/cshperspect.a016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Haffner MC, de Marzo AM, Meeker AK, et al. : Transcription-induced DNA double strand breaks: both oncogenic force and potential therapeutic target? Clin Cancer Res. 2011;17(12):3858–64. 10.1158/1078-0432.CCR-10-2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Puc J, Kozbial P, Li W, et al. : Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell. 2015;160(3):367–80. 10.1016/j.cell.2014.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Kovtun IV, Murphy SJ, Johnson SH, et al. : Chromosomal catastrophe is a frequent event in clinically insignificant prostate cancer. Oncotarget. 2015;6(30):29087–96. 10.18632/oncotarget.4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Waters CA, Strande NT, Pryor JM, et al. : The fidelity of the ligation step determines how ends are resolved during nonhomologous end joining. Nat Commun. 2014;5: 4286. 10.1038/ncomms5286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zaboikin M, Zaboikina T, Freter C, et al. : Non-Homologous End Joining and Homology Directed DNA Repair Frequency of Double-Stranded Breaks Introduced by Genome Editing Reagents. PLoS One. 2017;12(1):e0169931. 10.1371/journal.pone.0169931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Jeggo PA, Pearl LH, Carr AM: DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer. 2016;16(1):35–42. 10.1038/nrc.2015.4 [DOI] [PubMed] [Google Scholar]
- 142. Maréchal A, Zou L: DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5(9): pii: a012716. 10.1101/cshperspect.a012716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Nalepa G, Clapp DW: Fanconi anaemia and cancer: an intricate relationship. Nat Rev Cancer. 2018;18(3):168–85. 10.1038/nrc.2017.116 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 144. Fassihi H, Sethi M, Fawcett H, et al. : Deep phenotyping of 89 xeroderma pigmentosum patients reveals unexpected heterogeneity dependent on the precise molecular defect. Proc Natl Acad Sci U S A. 2016;113(9):E1236–45. 10.1073/pnas.1519444113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Bayraktar S, Arun B: BRCA mutation genetic testing implications in the United States. Breast. 2017;31:224–32. 10.1016/j.breast.2016.11.021 [DOI] [PubMed] [Google Scholar]
- 146. Aparicio T, Baer R, Gautier J: DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst). 2014;19:169–75. 10.1016/j.dnarep.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Leão RRN, Price AJ, James Hamilton R: Germline BRCA mutation in male carriers-ripe for precision oncology? Prostate Cancer Prostatic Dis. 2018;21(1):48–56. 10.1038/s41391-017-0018-5 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 148. Prakash R, Zhang Y, Feng W, et al. : Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7(4):a016600. 10.1101/cshperspect.a016600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Liu Y, Cussiol JR, Dibitetto D, et al. : TOPBP1 Dpb11 plays a conserved role in homologous recombination DNA repair through the coordinated recruitment of 53BP1 Rad9. J Cell Biol. 2017;216(3):623–39. 10.1083/jcb.201607031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Pritchard CC, Mateo J, Walsh MF, et al. : Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med. 2016;375(5):443–53. 10.1056/NEJMoa1603144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Gallagher DJ, Gaudet MM, Pal P, et al. : Germline BRCA mutations denote a clinicopathologic subset of prostate cancer. Clin Cancer Res. 2010;16(7):2115–21. 10.1158/1078-0432.CCR-09-2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Mitra A, Fisher C, Foster CS, et al. : Prostate cancer in male BRCA1 and BRCA2 mutation carriers has a more aggressive phenotype. Br J Cancer. 2008;98(2):502–7. 10.1038/sj.bjc.6604132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Tsuzuki T: Intraductal carcinoma of the prostate: a comprehensive and updated review. Int J Urol. 2015;22(2):140–5. 10.1111/iju.12657 [DOI] [PubMed] [Google Scholar]
- 154. Isaacsson Velho P, Silberstein JL, Markowski MC, et al. : Intraductal/ductal histology and lymphovascular invasion are associated with germline DNA-repair gene mutations in prostate cancer. Prostate. 2018;78(5):401–7. 10.1002/pros.23484 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 155. Taylor RA, Fraser M, Livingstone J, et al. : Germline BRCA2 mutations drive prostate cancers with distinct evolutionary trajectories. Nat Commun. 2017;8: 13671. 10.1038/ncomms13671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Mateo J, Boysen G, Barbieri CE, et al. : DNA Repair in Prostate Cancer: Biology and Clinical Implications. Eur Urol. 2017;71(3):417–25. 10.1016/j.eururo.2016.08.037 [DOI] [PubMed] [Google Scholar]
- 157. Annala M, Struss WJ, Warner EW, et al. : Treatment Outcomes and Tumor Loss of Heterozygosity in Germline DNA Repair-deficient Prostate Cancer. Eur Urol. 2017;72(1):34–42. 10.1016/j.eururo.2017.02.023 [DOI] [PubMed] [Google Scholar]
- 158. Mateo J, Cheng HH, Beltran H, et al. : Clinical Outcome of Prostate Cancer Patients with Germline DNA Repair Mutations: Retrospective Analysis from an International Study. Eur Urol. 2018;73(5):687–93. 10.1016/j.eururo.2018.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Antonarakis ES, Lu C, Luber B, et al. : Germline DNA-repair Gene Mutations and Outcomes in Men with Metastatic Castration-resistant Prostate Cancer Receiving First-line Abiraterone and Enzalutamide. Eur Urol. 2018;74(2):218–225. 10.1016/j.eururo.2018.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 160. Hussain M, Daignault-Newton S, Twardowski PW, et al. : Targeting Androgen Receptor and DNA Repair in Metastatic Castration-Resistant Prostate Cancer: Results From NCI 9012. J Clin Oncol. 2018;36(10):991–9. 10.1200/JCO.2017.75.7310 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 161. Ohmoto A, Yachida S: Current status of poly(ADP-ribose) polymerase inhibitors and future directions. Onco Targets Ther. 2017;10:5195–208. 10.2147/OTT.S139336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Bryant HE, Schultz N, Thomas HD, et al. : Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–7. 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- 163. Farmer H, McCabe N, Lord CJ, et al. : Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–21. 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 164. Ledermann J, Harter P, Gourley C, et al. : Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15(8):852–61. 10.1016/S1470-2045(14)70228-1 [DOI] [PubMed] [Google Scholar]
- 165. George A, Kaye S, Banerjee S: Delivering widespread BRCA testing and PARP inhibition to patients with ovarian cancer. Nat Rev Clin Oncol. 2017;14(5):284–96. 10.1038/nrclinonc.2016.191 [DOI] [PubMed] [Google Scholar]
- 166. Mateo J, Carreira S, Sandhu S, et al. : DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373(18):1697–708. 10.1056/NEJMoa1506859 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 167. Fong PC, Boss DS, Yap TA, et al. : Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361(2):123–34. 10.1056/NEJMoa0900212 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 168. Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. : Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33(3):244–50. 10.1200/JCO.2014.56.2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. An J, Wang C, Deng Y, et al. : Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6(4):657–69. 10.1016/j.celrep.2014.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Li C, Ao J, Fu J, et al. : Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30(42):4350–64. 10.1038/onc.2011.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Geng C, He B, Xu L, et al. : Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci U S A. 2013;110(17):6997–7002. 10.1073/pnas.1304502110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Groner AC, Cato L, de Tribolet-Hardy J, et al. : TRIM24 Is an Oncogenic Transcriptional Activator in Prostate Cancer. Cancer Cell. 2016;29(6):846–58. 10.1016/j.ccell.2016.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Dai X, Gan W, Li X, et al. : Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med. 2017;23(9):1063–71. 10.1038/nm.4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Janouskova H, El Tekle G, Bellini E, et al. : Opposing effects of cancer-type-specific SPOP mutants on BET protein degradation and sensitivity to BET inhibitors. Nat Med. 2017;23(9):1046–54. 10.1038/nm.4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Zhang P, Wang D, Zhao Y, et al. : Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat Med. 2017;23(9):1055–62. 10.1038/nm.4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. An J, Ren S, Murphy SJ, et al. : Truncated ERG Oncoproteins from TMPRSS2-ERG Fusions Are Resistant to SPOP-Mediated Proteasome Degradation. Mol Cell. 2015;59(6):904–16. 10.1016/j.molcel.2015.07.025 [DOI] [PubMed] [Google Scholar]
- 177. Gan W, Dai X, Lunardi A, et al. : SPOP Promotes Ubiquitination and Degradation of the ERG Oncoprotein to Suppress Prostate Cancer Progression. Mol Cell. 2015;59(6):917–30. 10.1016/j.molcel.2015.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Shoag J, Liu D, Blattner M, et al. : SPOP mutation drives prostate neoplasia without stabilizing oncogenic transcription factor ERG. J Clin Invest. 2018;128(1):381–6. 10.1172/JCI96551 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 179. Zhang J, Bu X, Wang H, et al. : Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553(7686):91–5. 10.1038/nature25015 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 180. Shenoy TR, Boysen G, Wang MY, et al. : CHD1 loss sensitizes prostate cancer to DNA damaging therapy by promoting error-prone double-strand break repair. Ann Oncol. 2017;28(7):1495–507. 10.1093/annonc/mdx165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Markowski MC, De Marzo AM, Antonarakis ES: BET inhibitors in metastatic prostate cancer: therapeutic implications and rational drug combinations. Expert Opin Investig Drugs. 2017;26(12):1391–7. 10.1080/13543784.2017.1393518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Boysen G, Barbieri CE, Prandi D, et al. : SPOP mutation leads to genomic instability in prostate cancer. eLife. 2015;4: pii: e09207. 10.7554/eLife.09207 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 183. Kari V, Mansour WY, Raul SK, et al. : Loss of CHD1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness. EMBO Rep. 2016;17(11):1609–23. 10.15252/embr.201642352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Zhao D, Lu X, Wang G, et al. : Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature. 2017;542(7642):484–8. 10.1038/nature21357 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 185. Beltran H, Rickman DS, Park K, et al. : Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1(6):487–95. 10.1158/2159-8290.CD-11-0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Bluemn EG, Coleman IM, Lucas JM, et al. : Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell. 2017;32(4):474–489.e6. 10.1016/j.ccell.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 187. Abbas F, Civantos F, Benedetto P, et al. : Small cell carcinoma of the bladder and prostate. Urology. 1995;46(5):617–30. 10.1016/S0090-4295(99)80290-8 [DOI] [PubMed] [Google Scholar]
- 188. Grigore AD, Ben-Jacob E, Farach-Carson MC: Prostate cancer and neuroendocrine differentiation: more neuronal, less endocrine? Front Oncol. 2015;5:37. 10.3389/fonc.2015.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Kluetz PG, Ning Y, Maher VE, et al. : Abiraterone acetate in combination with prednisone for the treatment of patients with metastatic castration-resistant prostate cancer: U.S. Food and Drug Administration drug approval summary. Clin Cancer Res. 2013;19(24):6650–6. 10.1158/1078-0432.CCR-13-2134 [DOI] [PubMed] [Google Scholar]
- 190. Ning YM, Pierce W, Maher VE, et al. : Enzalutamide for treatment of patients with metastatic castration-resistant prostate cancer who have previously received docetaxel: U.S. Food and Drug Administration drug approval summary. Clin Cancer Res. 2013;19(22):6067–73. 10.1158/1078-0432.CCR-13-1763 [DOI] [PubMed] [Google Scholar]
- 191. Watson PA, Arora VK, Sawyers CL: Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15(12):701–11. 10.1038/nrc4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Mu P, Zhang Z, Benelli M, et al. : SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355(6320):84–8. 10.1126/science.aah4307 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 193. Dardenne E, Beltran H, Benelli M, et al. : N-Myc Induces an EZH2-Mediated Transcriptional Program Driving Neuroendocrine Prostate Cancer. Cancer Cell. 2016;30(4):563–77. 10.1016/j.ccell.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Lee JK, Phillips JW, Smith BA, et al. : N-Myc Drives Neuroendocrine Prostate Cancer Initiated from Human Prostate Epithelial Cells. Cancer Cell. 2016;29(4):536–47. 10.1016/j.ccell.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Ku SY, Rosario S, Wang Y, et al. : Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355(6320):78–83. 10.1126/science.aah4199 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 196. Wang W, Epstein JI: Small cell carcinoma of the prostate. A morphologic and immunohistochemical study of 95 cases. Am J Surg Pathol. 2008;32(1):65–71. 10.1097/PAS.0b013e318058a96b [DOI] [PubMed] [Google Scholar]
- 197. Schepisi G, Farolfi A, Conteduca V, et al. : Immunotherapy for Prostate Cancer: Where We Are Headed. Int J Mol Sci. 2017;18(12): pii: E2627. 10.3390/ijms18122627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Kantoff PW, Higano CS, Shore ND, et al. : Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363(5):411–22. 10.1056/NEJMoa1001294 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 199. Schellhammer PF, Chodak G, Whitmore JB, et al. : Lower baseline prostate-specific antigen is associated with a greater overall survival benefit from sipuleucel-T in the Immunotherapy for Prostate Adenocarcinoma Treatment (IMPACT) trial. Urology. 2013;81(6):1297–302. 10.1016/j.urology.2013.01.061 [DOI] [PubMed] [Google Scholar]
- 200. Redman JM, Gulley JL, Madan RA: Combining immunotherapies for the treatment of prostate cancer. Urol Oncol. 2017;35(12):694–700. 10.1016/j.urolonc.2017.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Beer TM, Kwon ED, Drake CG, et al. : Randomized, Double-Blind, Phase III Trial of Ipilimumab Versus Placebo in Asymptomatic or Minimally Symptomatic Patients With Metastatic Chemotherapy-Naive Castration-Resistant Prostate Cancer. J Clin Oncol. 2017;35(1):40–7. 10.1200/JCO.2016.69.1584 [DOI] [PubMed] [Google Scholar]
- 202. Cabel L, Loir E, Gravis G, et al. : Long-term complete remission with Ipilimumab in metastatic castrate-resistant prostate cancer: case report of two patients. J Immunother Cancer. 2017;5:31. 10.1186/s40425-017-0232-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Le DT, Uram JN, Wang H, et al. : PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372(26):2509–20. 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 204. Gubin MM, Artyomov MN, Mardis ER, et al. : Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest. 2015;125(9):3413–21. 10.1172/JCI80008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Pritchard CC, Morrissey C, Kumar A, et al. : Complex MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nat Commun. 2014;5: 4988. 10.1038/ncomms5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Schweizer MT, Cheng HH, Tretiakova MS, et al. : Mismatch repair deficiency may be common in ductal adenocarcinoma of the prostate. Oncotarget. 2016;7(50):82504–10. 10.18632/oncotarget.12697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Guedes LB, Antonarakis ES, Schweizer MT, et al. : MSH2 Loss in Primary Prostate Cancer. Clin Cancer Res. 2017;23(22):6863–74. 10.1158/1078-0432.CCR-17-0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Barroso-Sousa R, Ott PA: PD-1 inhibitors in endometrial cancer. Oncotarget. 2017;8(63):106169–70. 10.18632/oncotarget.22583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Lu X, Horner JW, Paul E, et al. : Effective combinatorial immunotherapy for castration-resistant prostate cancer. Nature. 2017;543(7647):728–32. 10.1038/nature21676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Brudno JN, Kochenderfer JN: Chimeric antigen receptor T-cell therapies for lymphoma. Nat Rev Clin Oncol. 2018;15(1):31–46. 10.1038/nrclinonc.2017.128 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 211. Aldoss I, Bargou RC, Nagorsen D, et al. : Redirecting T cells to eradicate B-cell acute lymphoblastic leukemia: bispecific T-cell engagers and chimeric antigen receptors. Leukemia. 2017;31(4):777–87. 10.1038/leu.2016.391 [DOI] [PubMed] [Google Scholar]
- 212. Junghans RP, Ma Q, Rathore R, et al. : Phase I Trial of Anti-PSMA Designer CAR-T Cells in Prostate Cancer: Possible Role for Interacting Interleukin 2-T Cell Pharmacodynamics as a Determinant of Clinical Response. Prostate. 2016;76(14):1257–70. 10.1002/pros.23214 [DOI] [PubMed] [Google Scholar]
- 213. Priceman SJ, Gerdts EA, Tilakawardane D, et al. : Co-stimulatory signaling determines tumor antigen sensitivity and persistence of CAR T cells targeting PSCA+ metastatic prostate cancer. Oncoimmunology. 2018;7(2):e1380764. 10.1080/2162402X.2017.1380764 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation