Abstract
Transcriptional and post-transcriptional regulation shapes the transcriptome and proteome changes induced by various cellular signaling cascades. MicroRNAs (miRNAs) are small regulatory RNAs that are approximately 22 nucleotides long, which direct the post-transcriptional regulation of diverse target genes and control cell states. Transforming growth factor (TGF)-β family is a multifunctional cytokine family, which plays many regulatory roles in the development and pathogenesis of diverse diseases, including fibrotic disease, cardiovascular disease and cancer. Previous studies have shown that the TGF-β pathway includes the miRNA pathway as an important component of its downstream signaling cascades. Multiple studies of epithelial–mesenchymal transition (EMT)-related miRNAs have highlighted that miRNAs constitute the intrinsic bistable molecular switches of cell states by forming double negative feedback loops with EMT-inducing transcription factors. This may be important for understanding the reversibility of EMT at the single-cell level, the presence of distinct EMT transition states and the intra- and inter-tumor heterogeneity of cancer cell phenotypes. In the present review, I summarize the connection between TGF-β signaling and the miRNA pathway, placing particular emphasis on the regulation of miRNA expression by TGF-β signaling, the modulation of TGF-β signaling by miRNAs, the miRNA-mediated modulation of EMT and endothelial–mesenchymal transition as well as the crosstalk between miRNA and TGF-β pathways in the tumor microenvironment.
Keywords: miRNA, TGF-β, BMP, EMT, EndMT, tumor microenvironment
1. Introduction
The regulation of transcriptomes is a major consequence of various intracellular signaling cascades that are located downstream of growth factors and cytokines, which often represents their biological activities. These signaling cascades are usually directly linked to their corresponding transcription factors, which mediate the repression or activation of target genes. However, it is now clear that this transcriptional regulation is closely associated with epigenetic modification and post-transcriptional regulation, with the resulting transcriptomes being shaped by complex multilayered regulatory networks.
MicroRNAs (miRNAs) are small regulatory non-coding RNAs, which are approximately 22 nucleotides long. miRNAs form an RNA–protein complex with Argonaute proteins, recognize multiple target mRNAs via sequence complementarity and repress target RNAs, thereby serving as the major players in the post-transcriptional gene regulation of diverse species [1]. In humans, many conserved miRNAs display preferentially conserved interactions with hundreds of target mRNAs and thus, they exhibit diverse effects on transcriptomes [2]. To date, hundreds of distinct miRNAs have been identified in humans using stringent criteria, while thousands of human genes have been shown to be miRNA targets [2,3,4]. Reflecting the diversity of their targets and their involvement in various biological pathways, miRNAs have been shown to regulate multiple cellular pathways and to play important roles in multiple aspects of development, physiology and disease pathogenesis [5]. For example, multiple loss-of-function studies have shown that depletion of miRNA genes leads to developmental, physiological and/or behavioral abnormalities [1,6]. Furthermore, miRNAs have also been implicated in many biological characteristics of cancer (i.e., hallmarks of cancer) [7].
Significant evidence has indicated that miRNAs are involved in the post-transcriptional gene regulation in various conserved cellular signaling cascades [8,9]. These signaling cascades include the transforming growth factor (TGF)-β, Notch, Hedgehog and mitogen-activated protein kinase (MAPK) pathways. The downstream targets of these pathways include multiple downstream miRNAs, which provide positive or negative feedback to downstream signal mediators or transcription factors [10,11].
The TGF-β family is a multifunctional cytokine family that regulates multiple cellular functions, including cell growth, differentiation, adhesion, migration and death [12,13]. TGF-β signaling has many regulatory roles in development, with alterations in this signaling pathway having been associated with the pathogenesis of various diseases, including fibrotic disease, cardiovascular disease and cancer [14,15,16,17]. Previous studies have shown that the TGF-β signaling pathway embraces the miRNA pathway as an important component of its downstream signaling cascades [18,19,20,21,22,23,24]. In the present review, I summarize the connection between TGF-β signaling and miRNAs with a particular focus on: (1) regulation of miRNA expression by TGF-β signaling; (2) modulation of TGF-β signaling by miRNAs; (3) miRNA-mediated regulation of cell state transitions, such as epithelial–mesenchymal transition (EMT) and endothelial–mesenchymal transition (EndMT); and (4) crosstalk between miRNA and TGF-β pathways in cancer.
2. Biogenesis of miRNAs
In mammals, many miRNAs are first generated as a part of a longer primary miRNA transcript (pri-miRNA) transcribed by RNA polymerase II [25]. A hairpin structure of pri-miRNAs is cleaved by the microprocessor complex, which is composed of RNase III Drosha and its cofactor DGCR8 in the nucleus. The microprocessor is a heterotrimeric complex with one Drosha and two DGCR8 molecules that generates stem-loop structured RNAs, which are termed pre-miRNAs, by cleaving the stem region of the hairpin structure of pri-miRNAs [26]. Several proteins have been shown to be associated with the microprocessor complex and regulate its processing activity [27,28,29]. Pre-miRNAs are transported to the cytoplasm by Exportin 5 and RAN-GTP, where they undergo further cleavage by the RNase III enzyme Dicer [30,31]. Dicer processes the pre-miRNA to a double-stranded ~22-nucleotide product that is known as the miRNA duplex, which consists of the miRNA guide strand and passenger strand. In mammals, Dicer associates with the partner proteins HIV transactivation response element (TAR)-binding protein (TRBP) and protein activator of the interferon-induced protein kinase (PACT) [32].
The miRNA duplex is sequentially loaded onto Argonaute proteins (Ago1–Ago4 in mammals) with the help of HSP70/90 chaperone machinery [33]. Finally, one strand, whose 5′-end is captured by the MID domain of Argonaute, is retained in the complex, while the other strand protrudes from Argonaute to form the mature RNA-induced silencing complex. By convention, the more abundant strand is referred to as the miRNA or guide strand, whereas the less abundant strand is known as the miRNA* or the passenger strand. The selection of which strand serves as the guide strand is an asymmetric process. In mammals, this asymmetric selection is directed by the Argonaute proteins [34]. Mammalian Argonaute proteins select strands with 5′-uridine/adenosine and thermodynamically unstable 5′-ends with its two sensor regions, which come into contact with the 5′-nucleobase and 5′-phosphate(s) of prospective guide strands, respectively. Thus, the asymmetry of mammalian miRNAs shows unique patterns that reflect two independent molecular rules for the 5′-end nucleotide identity and thermodynamic stability of each miRNA duplex [34]. Many miRNA duplexes yield a single dominant mature miRNA but often provide both strands as mature miRNAs [3].
Thus, the biogenesis of miRNAs consists of multiple steps, including transcription, processing by Drosha and Dicer and Argonaute loading. This is further influenced by modification by RNA editing, RNA methylation, uridylation, adenylation and RNA decay. The expression of each miRNA species can be regulated at each step [29,35,36]. In fact, the post-transcriptional regulation of miRNA biogenesis has been well studied, while many RNA binding proteins (RBPs) and factors have been reported as post-transcriptional or co-transcriptional modulators of miRNA biogenesis [35,36,37,38,39,40].
3. Function and Expression of miRNAs
In the recent version of miRBase (miRBase v21), 1193 mouse and 1881 human miRNA gene annotations have been registered [41]. Using stringent criteria, the numbers of confidently identified canonical miRNA genes are 475 and 519 in mice and humans, respectively [1,3,4].
In canonical miRNA-based targeting, the recognition of target mRNAs by miRNAs is mediated primarily through sequence complementarity between the miRNA seed (miRNA nucleotides 2–7) and its target site in the 3′-untranslated region (UTR) of target mRNAs [42]. Due to dependence on short seed sequences for target recognition, each miRNA can potentially target hundreds of genes and the emergence of miRNA target sites in 3′-UTRs has influenced 3′-UTR evolution [43,44]. Accordingly, many conserved miRNAs have been shown to exhibit preferentially conserved interactions with hundreds of target mRNAs [2]. The evolution of 3′-UTRs has been associated with the evolutionary conservation of tissue-specific expression patterns of tissue-specific miRNAs [43,44].
In animals, target repression through partial complementarity involves two modes of target repression, mRNA decay and translational inhibition although they do not target degradation by Argonaute-dependent endonucleolytic cleavage triggered by extensive complementarity [1]. Canonical miRNA-mediated target repression largely depends on the adapter proteins of Argonaute, TNRC6A/B/C. These adapter proteins induce shortening of the poly(A) sequence, subsequent mRNA destabilization and translational repression. It has been reported that mRNA destabilization rather than translation repression dominates target repression by mammalian miRNAs in post-embryonic cells, except for in the early embryo [45,46,47].
Genome-wide expression profiling, including miRNA microarray and small RNA sequencing analyses, has identified hundreds of miRNAs at detectable levels in a single cell type. However, only a small number of miRNAs predominate the total miRNA expression in multiple cell types [48,49]. This is also the case for miRNA function, while a few abundant miRNAs mediate the marked target repression at the population level [50,51]. Importantly, loss-of-function studies have highlighted the importance of such tissue-specific abundant miRNAs in the development and pathogenesis of disease [6]. We have recently shown that these quantitative features of miRNA-mediated gene regulation can be explained by an association between super-enhancers and miRNA genes [52]. Super-enhancers are the major drivers of tissue-specific highly biased miRNA expression and function. Moreover, super-enhancers can also explain alterations of the multiple miRNAs that are involved in tumor pathogenesis [52].
4. Overview of TGF-β Signaling
The TGF-β family consists of three TGF-β isoforms, activins, nodal, bone morphogenetic proteins (BMPs) and growth and differentiation factors [12,13]. These members regulate multiple cellular functions, including cell growth, differentiation, adhesion, migration and death in a context-dependent and cell type-specific manner [13,53,54].
TGF-β family ligands bind to two different types of serine-threonine kinase receptors, which are the type II and type I receptors (TβR-II and TβR-I for TGF-β). Upon ligand binding, specific tetrameric type II/type I receptor complexes are activated to transduce downstream signals. Type I receptors phosphorylate the receptor-regulated Smads (R-Smads). Typically, TGF-β isoforms and activins trigger the phosphorylation of Smad2 and Smad3 (activin/TGF-β-specific R-Smads), while BMPs trigger the phosphorylation of Smad1, Smad5 and Smad8 (BMP-specific R-Smads). Thus, TGF-β family members can be largely divided into two groups according to the downstream R-Smads [12]. Subsequently, R-Smads form complexes with a common-partner Smad (co-Smad). Smad4 is the only co-Smad in mammals that is shared with the TGF-β family signaling pathways. The R-Smad/co-Smad complexes accumulate in the nucleus and regulate the transcription of various target genes together with a range of transcription factors and transcriptional modulators [53,54]. The inhibitory Smads (I-Smads), which are namely Smad6 and Smad7, can inhibit R-Smad activation and overall signal strength [55]. Additionally, TGF-β activates several Smad-independent signal transduction pathways, including the phosphatidylinositol 3 kinase (PI3K)-Akt and MAPK pathways. These are referred to as non-Smad signaling pathways [56,57].
TGF-β signaling has been shown to play multiple important roles in cancer progression [15,16,17,54]. These can be largely categorized into four distinct processes: (1) growth modulation, particularly escape from growth inhibition by TGF-β at the early stage of cancer initiation; (2) enhanced synthesis of the extracellular matrix and fibrosis by TGF-β in the tumor microenvironment; (3) promotion of EMT and/or metastasis by TGF-β; and (4) immune suppression by TGF-β in the tumor microenvironment. TGF-β has been proposed to function both as a tumor suppressor and a tumor promoter, depending on the cancer type and the stage of cancer progression [15]. It is thought that TGF-β serves as a tumor suppressor through the inhibition of cell growth in the early stages of carcinogenesis. On the other hand, it has been suggested that TGF-β promotes tumor progression in the advanced stages of cancer by enhancing cancer cell migration, tissue fibrosis and/or immune suppression [58]. To date, it has been suggested that multiple miRNAs contribute to these processes in relation to TGF-β signaling [18,19,20,21,22,23,24].
5. Regulation of miRNA Expression and Biogenesis by TGF-β Signaling
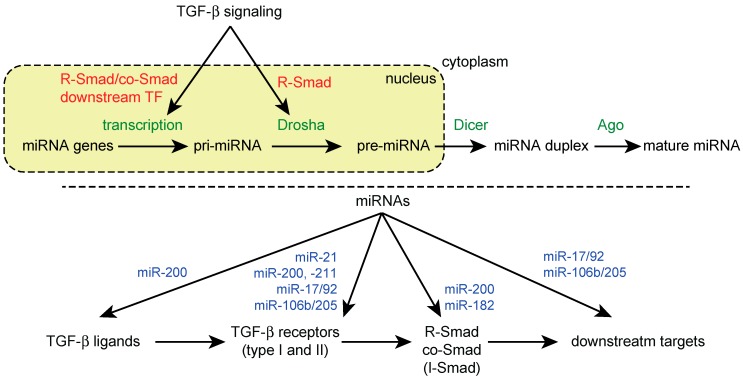
Multiple studies have shown that treatment with TGF-β family ligands induces changes in the expression levels of multiple miRNAs in different cell types [18,19,20,21,22,23,24]. TGF-β signaling has been shown to modulate miRNA expression at both the transcriptional and post-transcriptional levels (Figure 1, top). In particular, early studies have shown that R-Smads directly influence miRNA biogenesis by modulating Drosha-mediated pri-miRNA processing, which demonstrates an unexpected transcription-independent function of Smads [59,60].
Figure 1.
Reciprocal crosstalk between TGF-β signaling and miRNA machinery. (Top) Regulation of miRNA biogenesis by TGF-β signaling; (Bottom) Regulation of TGF-β signaling components by miRNAs. Examples of miRNAs targeting TGF-β signaling components are shown.
TGF-β and BMP signaling have been shown to increase the expression of mature miR-21 in human vascular smooth muscle and breast cancer cells [59]. This increase in miR-21 expression is accompanied by the upregulation of pre-miR-21 but not pri-miR-21. Treatment with TGF-β and BMP4 has been shown to induce an association between R-Smads and the cofactor of the microprocessor p68 on pri-miR-21, which is thought to facilitate pri-miR processing. In contrast to the canonical Smad pathway, the co-Smad Smad4 is not required for this ligand-dependent pri-miRNA processing [59]. The pre-miRNAs of TGF-β/BMP-regulated miRNAs contain a conserved sequence, CAGAC, which is similar to the Smad binding element found in the promoters of TGF-β/BMP-regulated genes [60]. Smads directly interact with this RNA-Smad binding element (R-SBE) through their N-terminal MH1 domain, while TGF-β/BMP-induced pri-miRNA processing depends on R-SBE. R-SBE has been identified in several other TGF-β/BMP-induced miRNAs, including miR-105, miR-199a, miR-215, miR-421 and miR-529 [60]. Additionally, it has been suggested that a Smad4-independent mechanism can mediate TGF-β-induced miR-181a upregulation [61], although both Smad4-independent post-transcriptional and Smad4-dependent transcriptional mechanisms are thought to be important in the TGF-β-dependent induction of the miR-181 family [62,63]. This type of post-transcriptional regulation of miRNA biogenesis has also been demonstrated for other transcriptional regulators, including p53 and Nanog [64,65]. These observations suggest that microprocessor cofactors, such as p68 and p72, as well as other RBPs serve as the molecular interfaces for integrating intracellular signaling pathways into the miRNA pathway [29,37].
TGF-β/BMP-regulated miRNAs can be extended to other miRNAs in different cell types. The miRNAs upregulated by TGF-β signaling include miR-21, the miR-181 family, miR-10b, the miR-17/92 cluster, miR-155, miR-192, the miR-23/24/27 cluster, miR-216/217, miR-494 and miR-182 [18,24,66,67,68,69,70,71,72,73]. The miRNAs downregulated by TGF-β signaling include the miR-200 family, miR-203, let-7, miR-34a and miR-584 [18,24,74,75,76,77]. In some cases, transcriptional mechanisms have been investigated. In normal murine mammary gland (NMuMG) epithelial cells, TGF-β activates the miR-155 promoter and induces miR-155 expression through Smad4 [68]. miR-494 is also induced by TGF-β at the pri-miRNA level in a Smad4-dependent manner, while miR-494 shows reduced expression in pancreatic cancer cells with a loss of Smad4 [72].
Additionally, the downstream transcription factors of TGF-β signaling have been shown to regulate multiple miRNAs at the transcriptional level. This is easily recognized with EMT-related miRNAs. As discussed later, several miRNAs, including the miR-200 family, miR-203 and miR-216/217, are suppressed by EMT-related transcriptional regulators, such as ZEB1 (also known as δEF1), ZEB2 (also known as SIP1), SNAIL, SLUG and E-box-dependent mechanisms, which are located downstream of TGF-β signaling [71,74,78,79,80,81].
6. Regulation of TGF-β Signaling by miRNAs
TGF-β signaling and miRNA pathways have been shown to exhibit reciprocal crosstalk. Consistent with the diversity of miRNA target genes, the computational prediction of miRNA targets suggests that multiple components of the TGF-β signaling pathway are targeted by multiple miRNAs. In fact, several miRNAs have been experimentally validated to be modulators of TGF-β signaling at multiple levels by targeting ligands, receptors, R-Smad, co-Smad, I-Smad and non-Smad pathway components as well as downstream targets of TGF-β signaling (Figure 1, bottom) [18,20,24].
Many miRNAs targeting TGF-β receptors, especially the type II receptor, have been identified in various cancer types. These miRNAs, which include miR-21, the miR-17/92 cluster, miR-106b, miR-211 and miR-590 [82,83,84,85,86,87], are frequently oncogenic [20,24]. These miRNAs promote proliferation, invasion or resistance to chemotherapeutic drugs in different cancer cell types.
Intriguingly, some TGF-β-regulated miRNAs target TGF-β signaling components, thus comprising the feedback regulation of TGF-β signaling. The miR-200 family, which consists of TGF-β-downregulated miRNAs, targets TGF-β, TβR-I and Smad2 [88,89]. Thus, the downregulation of the miR-200 family by TGF-β enhances TGF-β signaling and induction of EMT. miR-182, a TGF-β-inducible miRNA, targets Smad7 and thus, modulates a negative feedback loop of TGF-β signaling as Smad7 is also induced by TGF-β [90]. As a result, miR-182 potentiates the TGF-β-induced EMT and metastasis of cancer cells.
The miRNAs also regulate TGF-β signaling by convergently suppressing a range of downstream TGF-β target genes. The miR-106b/205 cluster that is composed of E2F1-regulated miRNAs targets the TGF-β downstream effectors, p21Waf1/Cip1 and Bim, and disrupt TGF-β-dependent cell cycle arrest and apoptosis in gastric cancer [91]. Additionally, the miR-17/92 cluster, which targets multiple TGF-β signaling components, such as TβR-I, Smad2 and Smad4, suppresses the TGF-β-responsive genes p21Waf1/Cip1 and Bim. This ultimately inhibits the effects of TGF-β signaling [84,92,93].
7. Control of Epithelial Identity and Epithelial–Mesenchymal Transition (EMT) by the miR-200 Family
The dynamic and reversible regulation of cell identity is important for the organization of complex multicellular systems and for adaptation under diverse physiological and pathological conditions. EMT, which converts the epithelial cells into mesenchymal cells, is a well-known example of such cellular plasticity, with the regulation of EMT being involved in the development and pathogenesis of cancer [94]. TGF-β is known to induce EMT. Previous studies have identified multiple miRNAs that are involved in EMT and have shown that both downstream transcription factors and miRNAs orchestrate the conserved TGF-β-inducible EMT programs. In this section, I focus on the miR-200 family as its role in EMT has been extensively studied [78,79,80,81].
The miR-200 family consists of five miRNAs with similar seed sequences: miR-200b, miR-200c and miR-429 with the seed sequence AAUACU and miR-200a and miR-141 with the seed sequence AACACU, which differ by a single nucleotide [95,96,97]. These miRNAs are encoded in two transcriptional clusters, which are namely the miR-200b/200a/429 and miR-200c/141 clusters. Consistent with their roles in epithelial identity, these miRNA clusters are closely associated with super-enhancers found in digestive organs [52].
Members of the miR-200 family are markedly downregulated following TGF-β treatment in multiple EMT models using various cell lines, including Madin Darby canine kidney (MDCK) epithelial cells, NMuMG epithelial cells and MCF10A mammary epithelial cells [78,80,98]. Forced expression of the miR-200 family is sufficient for preventing the TGF-β-induced EMT in MDCK cells and inducing mesenchymal–epithelial transition (MET) in mesenchymal cells. This suggests that the miR-200 family acts as an enforcer of epithelial identity [78,79,80,81]. The promoter regions of the miR-200b/200a/429 and miR-200c/141 clusters are repressed by the TGF-β-induced transcriptional repressors ZEB1 and ZEB2 during EMT through ZEB-type E-box elements [81,99]. Furthermore, prolonged exposure to TGF-β can induce reversible DNA methylation of miR-200 family promoters [88]. Conversely, the miR-200 family targets the 3′-UTRs of ZEB1/2 [78,79,80,81]. Thus, ZEB1/2 and the miR-200 family form a double negative feedback loop, which controls epithelial identity (Figure 2a). Additionally, the maintenance of a stable mesenchymal phenotype requires TGF-β autocrine signaling, which is supported by the release of miR-200-mediated repression of TGF-β ligand production in MDCK cells. This suggests a more complex feedback network involving TGF-β, ZEB1/2 and the miR-200 family [88]. The clinical importance of this feedback loop is supported by a positive correlation between ZEB1/2 and TGF-β; by negative correlations between miR-200 and TGF-β and between miR-200 and ZEB1/2 in breast cancer [78,88]; and by an association between high miR-200 expression and better clinical outcomes in multiple cancer types [100].
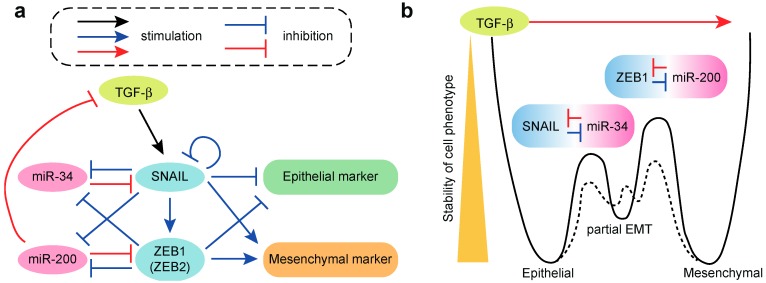
Figure 2.
Roles of miRNAs in TGF-β-induced epithelial–mesenchymal transition (EMT). (a) EMT circuits containing a SNAIL-miR-34 loop and ZEB1-miR-200 loop; (b) A relationship between two miRNA-transcription factor (TF) modules and three EMT states. Solid line indicates the cell state transitions among epithelial, partial EMT and mesenchymal phenotypes, which are controlled by two miRNA-TF molecular switches. Dotted line suggests the alternative transition states generated by additional molecular switches.
An unbiased investigation of miR-200 targets using Ago2 HITS-CLIP (high-throughput sequencing after cross-linked immunoprecipitation) has identified hundreds of miR-200a and miR-200b targets. This study has revealed an important effect of miR-200 on actin cytoskeleton dynamics [101]. Thus, the miR-200 family is important for maintaining epithelial identity and prevents cell migration by regulating a core EMT transcriptional program and multiple regulators of Rho-ROCK signaling, focal adhesion, matrix metalloproteinase activity and invadopodia formation. In addition to their regulation of cell motility and metastasis, the miR-200 family also plays important roles in regulating drug resistance in cancer therapy [95,96,97]. The crosstalk between miR-200 and ZEB1/2 has also been demonstrated in stem cell biology. The induction of the miR-200 family and MET occurs in the initial phase of the reprogramming of somatic cells to induced pluripotent stem (iPS) cells, facilitating their generation [102,103].
8. miRNA Regulation and miRNA-Mediated Bistable Switches of the TGF-β-Mediated EMT
Similar to the miR-200 family, additional miRNAs have also been shown to be associated with EMT. miR-205 is downregulated after TGF-β treatment and suppresses ZEB1/2 like the miR-200 family in MDCK cells [78]. ZEB1 has also been shown to repress several other miRNAs, such as miR-203 and miR-183, which cooperatively repress stem cell factors and inhibit the stemness property in cancer cells and mouse embryonic stem cells [104]. Additionally, several TGF-β-upregulated miRNAs, such as miR-181, miR-155 and miR-10b, promote EMT [61,66,68]. Other EMT-related miRNAs include miR-30, the miR-34 family and miR-223 [89,105,106,107].
The members of the miR-34 family, which are namely miR-34a and miR-34b/c, are suppressed by the EMT-inducing transcription factor SNAIL and conversely, the miR-34 family targets the 3′-UTR of SNAIL [105,106]. Thus, SNAIL and the miR-34 family comprise another double negative feedback loop controlling EMT, which is similar to the ZEB1/2-miR-200 feedback loop (Figure 2a). Detailed quantitative measurement of the dynamics of the TGF-β-induced EMT in MCF10A cells at the population and single-cell levels suggests that the two transcription factor-miRNA double negative feedback loops (i.e., the SNAIL-miR-34 and ZEB1-miR-200 loops) form bistable cell fate switches in the epithelial phenotype, partial EMT phenotype and mesenchymal phenotype (Figure 2a) [98,108]. These three phenotypes are characterized by high and low; medium and medium; and low and high abundance of E-cadherin and vimentin, respectively. The time course measurements of the TGF-β-induced EMT have suggested a two-step process among the three cell phenotypes (Figure 2b). In the first step, the epithelial cells transit to the partial EMT state and the SNAIL-miR-34 bistable switch is activated, which leads to reduced expression of E-cadherin and increased expression of vimentin. In the second step, the cells transit from the partial EMT state to the mesenchymal stat, and the ZEB1-miR-200 bistable switch is activated, which leads to reduced expression of E-cadherin and robust induction of vimentin expression [98]. In MCF-10A cells, it appears that the transition from epithelial to partial EMT state is reversible, whereas the transition from partial EMT to the mesenchymal state is irreversible, especially at high concentrations of TGF-β. In summary, in MCF10A cells, the TGF-β-induced EMT proceeds through the stepwise activation of the SNAIL-miR-34 and ZEB1-miR-200 modules in the initial and later phases, respectively [98,108]. This model will be important in furthering our understanding of the reversibility of EMT at the single-cell level and in interpreting the heterogeneity in EMT phenotypes, including partial EMT, in vivo and in clinical samples.
9. Regulation of TGF-β-Induced EndMT by miRNAs
TGF-β also plays important roles in the plasticity of other cell types. TGF-β is a major inducer of EndMT, which is a process that is similar to EMT. This process converts differentiated endothelial cells into mesenchymal cells [109]. EndMT is observed in cardiac development and diverse pathological conditions, including tissue fibrosis, cavernous malformations, vein stenosis and graft remodeling and cancer [110,111,112,113,114]. Several reports have recently described changes in the expression levels of multiple miRNAs during EndMT and the regulatory roles of several miRNAs in EndMT in different endothelial cells [115,116].
In the embryonic heart, miR-23 is necessary for restricting endocardial cushion formation by inhibiting TGF-β-induced EndMT and extracellular hyaluronic acid production [117]. miR-126, which is specifically expressed in endothelial cells and closely associated with super-enhancers found in human umbilical endothelial cells [52], has been reported to inhibit the TGF-β-induced EndMT through modulation of the PIK3R2-PI3K/Akt signaling pathway [118]. This suggests an analogous role of miRNAs in maintaining endothelial cell identity in addition to an essential role of the miR-200 family in epithelial cells. In a recent report, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) long non-coding RNA (lncRNA) was reported to be induced by TGF-β, which modulates the TGF-β-induced EndMT by suppressing miR-145. This suggests a complex network between lncRNAs and small RNAs [119]. Other reports have described the regulatory roles of several miRNAs, such as miR-21, miR-302c, miR-18a and miR-20a, in EndMT [120,121,122,123].
We have also reported that several miRNAs modulate EndMT in MS-1 mouse pancreatic microvascular endothelial cells [124,125]. Constitutively active miR-31 positively regulates the induction of mesenchymal markers during the TGF-β-induced EndMT in MS-1 endothelial cells without concomitant effects on the induction of TGF-β target genes and downregulation of endothelial markers. Additionally, miR-31 potentiates the induction of various chemokines and cytokines, including CCL17, CX3CL1, CXCL16, interleukin (IL)-6 and Angptl2, which has been designated as the EndMT-associated secretory phenotype (EndMT-SP). TGF-β induces alternative polyadenylation-mediated exclusion of the internal poly(A) sequence in the 3′-UTR of Stk40, which is a negative regulator of the NF-κB pathway. This enhances miR-31-dependent Stk40 suppression without concomitant miR-31 induction [124]. We also identified miR-27b, a member of the miR-23/24/27 cluster, as a TGF-β-inducible miRNA in MS-1 cells and observed that inhibition of miR-27 suppressed the induction of mesenchymal markers by TGF-β treatment [125].
10. TGF-β-Related miRNAs and the Tumor Microenvironment
miRNAs have diverse functions in cancer progression and are linked to wide aspects of cancer hallmarks. We have previously shown that the alterations of super-enhancers are observed for various cancer-related miRNAs associated with cancer hallmark traits [52]. Previous studies have demonstrated multifaceted contributions of multiple miRNAs to the roles of TGF-β signaling in cancer progression, including modulation of growth, EMT and metastasis, which have been discussed earlier (Figure 3) [18,19,20,21,22,23,24]. Additionally, recent advances in miRNA research have revealed that miRNAs regulate not only the properties of cancer cells but also the tumor microenvironment to facilitate tumor metastasis, resistance to cancer therapy and immune suppression [126]. This is also the case for TGF-β-related miRNAs. In this section, I summarize the non-cell-autonomous functions of TGF-β-related miRNAs and their roles in the cancer microenvironment (Figure 3).
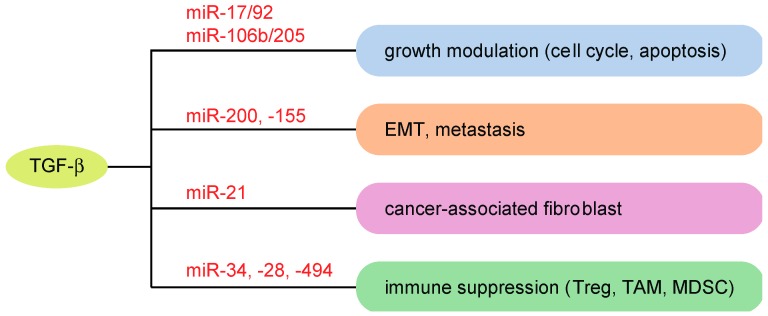
Figure 3.
Roles of TGF-β-related miRNAs in cancer progression. Examples of miRNAs modulating TGF-β-associated cancer properties are shown. Treg: regulatory T cells; TAM: tumor-associated macrophages; MDSC: myeloid-derived suppressor cells.
Alterations of miRNAs in cancer cells can foster a favorable microenvironment for cancer cells through the altered production of factors secreted from cancer cells. In hepatocellular carcinoma (HCC), increased TGF-β signaling, which is associated with persistent hepatitis B virus (HBV) infection, results in suppression of miR-34a and subsequently enhances the production of the chemokine CCL22, which promotes the recruitment of regulatory T cells [127]. In HBV-positive HCC and portal vein tumor thrombus, miR-34a expression levels are inversely correlated with the expression levels of CCL22 and FoxP3, which strengthens the importance of the TGF-β-miR-34a-CCL22 axis [127]. Additionally, in HCC, TGF-β secreted from tumor-associated macrophages (TAMs) suppresses miR-28 in cancer cells [128]. Decreased expression of miR-28 results in increased secretion of its target IL-34 and enhances the infiltration of TAMs. This miRNA-mediated feedback loop between cancer cells and TAMs modulates HCC metastasis.
TGF-β also modulates the functions of various stromal cells in the tumor microenvironment through miRNA-mediated gene regulation. TGF-β induces miR-494 in myeloid-derived suppressor cells (MDSCs), with this miRNA required for the accumulation and function of tumor-expanded MDSCs through PTEN suppression [129]. Inhibition of miR-494 reversed the ability of MDSCs and suppressed tumor growth and metastasis. TGF-β also promotes the induction of MDSCs by upregulating miR-21 and miR-155 [130]. Additionally, TGF-β reduces the tumor cytolysis activity of natural killer (NK) cells and abrogates its perforin polarization [131]. This inhibition of NK cell function is mediated by the induction of miR-183 by TGF-β and suppression of its target gene DAP12, which is important for surface NK receptor stabilization and downstream signals. Furthermore, it has been suggested that TGF-β-related miRNAs modulate the TGF-β-induced formation of cancer-associated fibroblasts. TGF-β-inducible miR-21 targets Smad7 and potentiates TGF-β-induced cancer formation [132].
Taken together, these findings suggest that miRNAs contribute to the modulation of the tumor microenvironment and immune suppression by TGF-β signaling through multiple mechanisms, including non-cell-autonomous functions in cancer cells and proximal functions in tumor stromal cells.
11. Concluding Remarks
In this review, I summarized the relationships between TGF-β signaling and the miRNA pathway. Previous studies have produced considerable conceptual advances regarding: (1) the regulation of miRNA expression by TGF-β signaling; (2) the modulation of TGF-β signaling by miRNAs; (3) the miRNA-mediated regulation of cell state transitions, which include EMT and EndMT; and (4) the crosstalk between miRNA and TGF-β pathways in cancer.
Crosstalk between TGF-β signaling and the miRNA biogenesis machinery has identified the unique transcription-independent roles of R-Smads, which suggests intimate crosstalk between transcriptional regulation and RNA processing. In fact, a recent study demonstrated that Smad2/3 facilitate the conversion of adenosine to N6-methyladenosine (m6A) on target RNA through the recruitment of the m6A methyltransferase complex, expanding the transcription-independent roles of R-Smads [133].
Studies regarding EMT-related miRNAs have highlighted that miRNAs constitute intrinsic bistable molecular switches in conjunction with EMT-inducing transcription factors [98,108]. This may be critical for modulating the reversibility of cell plasticity and the variability in cell differentiation. Further investigation of these feedback modules may increase our understanding of the reversibility of EMT at the single-cell level; of the intra- and inter-tumor heterogeneity of cancer phenotypes [134]; and of recently reported distinct EMT transition states [135]. Moreover, this may also provide further insight into several open questions regarding the EMT in vivo and in clinical samples [136].
Finally, TGF-β-related miRNAs may serve as potential therapeutic targets in various diseases, while clinically suitable methods of delivering small RNAs to various tissues should be improved [137,138]. Further studies of transcriptional and post-transcriptional regulation will yield a more comprehensive view of the alterations of the gene regulatory network modulated by TGF-β signaling and provide a basis for the development of new diagnostic and therapeutic approaches, which can be combined with improvements in the analytical analyses of miRNA networks [139,140,141].
Abbreviations
| Ago | Argonaute |
| BMP | bone morphogenetic protein |
| co-Smad | common-partner Smad |
| ECM | extracellular matrix |
| EMT | epithelial–mesenchymal transition |
| EndMT | endothelial–mesenchymal transition |
| EndMT-SP | EndMT-associated secretory phenotype |
| GDF | growth and differentiation factor |
| HBV | hepatitis B virus |
| HCC | hepatocellular carcinoma |
| HITS-CLIP | high-throughput sequencing after cross-linked immunoprecipitation |
| I-Smad | inhibitory Smad |
| lncRNA | long non-coding RNA |
| MALAT1 | metastasis-associated lung adenocarcinoma transcript 1 |
| MAPK | mitogen-activated protein kinase |
| MDCK | Madin Darby canine kidney |
| MDSC | myeloid-derived suppressor cell |
| MET | mesenchymal–epithelial transition |
| MMP | matrix metalloproteinase |
| NK | natural killer |
| NMuMG | normal murine mammary gland |
| PACT | protein activator of the interferon-induced protein kinase |
| PI3K | phosphatidylinositol 3 kinase |
| pri-miRNA | primary miRNA |
| RBP | RNA binding protein |
| R-Smad | receptor-regulated Smad |
| SBE | Smad binding element |
| TAM | tumor-associated macrophages |
| TβR | TGF-β receptor |
| TGF-β | transforming growth factor-β |
| TRBP | HIV transactivation response element (TAR)-binding protein |
| 3′-UTR | 3′ untranslated region |
Funding
This work was supported by the Uehara Memorial Foundation Research Fellowship and the Osamu Hayaishi Memorial Scholarship for Study Abroad (H.I.S.) and Takeda Science Foundation (courtesy of Akira Saito, Department of Respiratory Medicine, Graduate School of Medicine, The University of Tokyo).
Conflicts of Interest
The author declares no conflict of interest.
References
- 1.Bartel D.P. Metazoan microRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiang H.R., Schoenfeld L.W., Ruby J.G., Auyeung V.C., Spies N., Baek D., Johnston W.K., Russ C., Luo S., Babiarz J.E., et al. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fromm B., Billipp T., Peck L.E., Johansen M., Tarver J.E., King B.L., Newcomb J.M., Sempere L.F., Flatmark K., Hovig E., et al. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu. Rev. Genet. 2015;49:213–242. doi: 10.1146/annurev-genet-120213-092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Park C.Y., Choi Y.S., McManus M.T. Analysis of microRNA knockouts in mice. Hum. Mol. Genet. 2010;19:R169–R175. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lujambio A., Lowe S.W. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichimura A., Ruike Y., Terasawa K., Tsujimoto G. miRNAs and regulation of cell signaling. FEBS J. 2011;278:1610–1618. doi: 10.1111/j.1742-4658.2011.08087.x. [DOI] [PubMed] [Google Scholar]
- 9.Inui M., Martello G., Piccolo S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell. Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 10.Hornstein E., Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 11.Ebert M.S., Sharp P.A. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyazawa K., Shinozaki M., Hara T., Furuya T., Miyazono K. Two major Smad pathways in TGF-β superfamily signalling. Genes Cells. 2002;7:1191–1204. doi: 10.1046/j.1365-2443.2002.00599.x. [DOI] [PubMed] [Google Scholar]
- 13.Morikawa M., Derynck R., Miyazono K. TGF-β and the TGF-β family: Context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng X.M., Nikolic-Paterson D.J., Lan H.Y. TGF-β: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016;12:325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 15.Bierie B., Moses H.L. Tumour microenvironment: TGFβ: The molecular Jekyll and Hyde of cancer. Nat. Rev. Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 16.Pickup M., Novitskiy S., Moses H.L. The roles of TGFβ in the tumour microenvironment. Nat. Rev. Cancer. 2013;13:788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W., Ten Dijke P. Immunoregulation by members of the TGFβ superfamily. Nat. Rev. Immunol. 2016;16:723–740. doi: 10.1038/nri.2016.112. [DOI] [PubMed] [Google Scholar]
- 18.Butz H., Racz K., Hunyady L., Patocs A. Crosstalk between TGF-β signaling and the microRNA machinery. Trends Pharmacol. Sci. 2012;33:382–393. doi: 10.1016/j.tips.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Bowen T., Jenkins R.H., Fraser D.J. MicroRNAs, transforming growth factor β-1, and tissue fibrosis. J. Pathol. 2013;229:274–285. doi: 10.1002/path.4119. [DOI] [PubMed] [Google Scholar]
- 20.Guo L., Zhang Y., Zhang L., Huang F., Li J., Wang S. MicroRNAs, TGF-β signaling, and the inflammatory microenvironment in cancer. Tumor Biol. 2016;37:115–125. doi: 10.1007/s13277-015-4374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W., Zhou S., Mao L., Zhang H., Sun D., Zhang J., Li J., Tang J.H. Crosstalk between TGF-β signaling and miRNAs in breast cancer metastasis. Tumor Biol. 2016;37:10011–10019. doi: 10.1007/s13277-016-5060-8. [DOI] [PubMed] [Google Scholar]
- 22.Kang H. Role of microRNAs in TGF-β signaling pathway-mediated pulmonary fibrosis. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18122527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulei D., Mehterov N., Ling H., Stanta G., Braicu C., Berindan-Neagoe I. The “good-cop bad-cop” TGF-β role in breast cancer modulated by non-coding RNAs. Biochim. Biophys. Acta. 2017;1861:1661–1675. doi: 10.1016/j.bbagen.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Janakiraman H., House R.P., Gangaraju V.K., Diehl J.A., Howe P.H., Palanisamy V. The long (lncRNA) and short (miRNA) of It: TGFβ-mediated control of RNA-binding proteins and noncoding RNAs. Mol. Cancer Res. 2018;16:567–579. doi: 10.1158/1541-7786.MCR-17-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen T.A., Jo M.H., Choi Y.G., Park J., Kwon S.C., Hohng S., Kim V.N., Woo J.S. Functional anatomy of the human microprocessor. Cell. 2015;161:1374–1387. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 28.Shiohama A., Sasaki T., Noda S., Minoshima S., Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp. Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki H.I., Miyazono K. Emerging complexity of microRNA generation cascades. J. Biochem. 2011;149:15–25. doi: 10.1093/jb/mvq113. [DOI] [PubMed] [Google Scholar]
- 30.Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/S0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 31.Hutvagner G., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y., Yeo J., Lee J.H., Cho J., Seo D., Kim J.S., Kim V.N. Deletion of human tarbp2 reveals cellular microRNA targets and cell-cycle function of TRBP. Cell Rep. 2014;9:1061–1074. doi: 10.1016/j.celrep.2014.09.039. [DOI] [PubMed] [Google Scholar]
- 33.Iwasaki S., Kobayashi M., Yoda M., Sakaguchi Y., Katsuma S., Suzuki T., Tomari Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell. 2010;39:292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki H.I., Katsura A., Yasuda T., Ueno T., Mano H., Sugimoto K., Miyazono K. Small-RNA asymmetry is directly driven by mammalian Argonautes. Nat. Struct. Mol. Biol. 2015;22:512–521. doi: 10.1038/nsmb.3050. [DOI] [PubMed] [Google Scholar]
- 35.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell. Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 36.Lin S., Gregory R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki H.I., Miyazono K. Dynamics of microRNA biogenesis: Crosstalk between p53 network and microRNA processing pathway. J. Mol. Med. 2010;88:1085–1094. doi: 10.1007/s00109-010-0650-1. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki H.I., Miyazono K. Control of microRNA maturation by p53 tumor suppressor and MCPIP1 ribonuclease. Enzymes. 2012;32:163–183. doi: 10.1016/B978-0-12-404741-9.00008-8. [DOI] [Google Scholar]
- 39.Suzuki H.I., Arase M., Matsuyama H., Choi Y.L., Ueno T., Mano H., Sugimoto K., Miyazono K. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol. Cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki H.I., Katsura A., Miyazono K. A role of uridylation pathway for blockade of let-7 microRNA biogenesis by Lin28B. Cancer Sci. 2015;106:1174–1181. doi: 10.1111/cas.12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozomara A., Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartel D.P. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 44.Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 45.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bazzini A.A., Lee M.T., Giraldez A.J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., Guenther M.G., Johnston W.K., Wernig M., Newman J., et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullokandov G., Baccarini A., Ruzo A., Jayaprakash A.D., Tung N., Israelow B., Evans M.J., Sachidanandam R., Brown B.D. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bosson A.D., Zamudio J.R., Sharp P.A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki H.I., Young R.A., Sharp P.A. Super-enhancer-mediated RNA processing revealed by integrative microRNA network analysis. Cell. 2017;168:1000–1014 e1015. doi: 10.1016/j.cell.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Massague J. TGFβ signalling in context. Nat. Rev. Mol. Cell. Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.David C.J., Massague J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell. Biol. 2018 doi: 10.1038/s41580-018-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyazawa K., Miyazono K. Regulation of TGF-β family signaling by inhibitory Smads. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moustakas A., Heldin C.H. Non-Smad TGF-β signals. J. Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y.E. Non-Smad Signaling Pathways of the TGF-β Family. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyazono K., Ehata S., Koinuma D. Tumor-promoting functions of transforming growth factor-β in progression of cancer. Upsal. J. Med. Sci. 2012;117:143–152. doi: 10.3109/03009734.2011.638729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis B.N., Hilyard A.C., Lagna G., Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davis B.N., Hilyard A.C., Nguyen P.H., Lagna G., Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol. Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor M.A., Sossey-Alaoui K., Thompson C.L., Danielpour D., Schiemann W.P. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. J. Clin. Investig. 2013;123:150–163. doi: 10.1172/JCI64946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang B., Hsu S.H., Majumder S., Kutay H., Huang W., Jacob S.T., Ghoshal K. TGFβ-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29:1787–1797. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y., Yu Y., Tsuyada A., Ren X., Wu X., Stubblefield K., Rankin-Gee E.K., Wang S.E. Transforming growth factor-β regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene. 2011;30:1470–1480. doi: 10.1038/onc.2010.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki H.I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 65.Bourguignon L.Y., Spevak C.C., Wong G., Xia W., Gilad E. Hyaluronan-CD44 interaction with protein kinase C(epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the Production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J. Biol. Chem. 2009;284:26533–26546. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han X., Yan S., Weijie Z., Feng W., Liuxing W., Mengquan L., Qingxia F. Critical role of miR-10b in transforming growth factor-β1-induced epithelial-mesenchymal transition in breast cancer. Cancer Gene Ther. 2014;21:60–67. doi: 10.1038/cgt.2013.82. [DOI] [PubMed] [Google Scholar]
- 67.Tili E., Michaille J.J., Liu C.G., Alder H., Taccioli C., Volinia S., Calin G.A., Croce C.M. GAM/ZFp/ZNF512B is central to a gene sensor circuitry involving cell-cycle regulators, TGFβ effectors, Drosha and microRNAs with opposite oncogenic potentials. Nucleic Acids Res. 2010;38:7673–7688. doi: 10.1093/nar/gkq637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kong W., Yang H., He L., Zhao J.J., Coppola D., Dalton W.S., Cheng J.Q. MicroRNA-155 is regulated by the transforming growth factor β/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chung A.C., Huang X.R., Meng X., Lan H.Y. miR-192 mediates TGF-β/Smad3-driven renal fibrosis. J. Am. Soc. Nephrol. 2010;21:1317–1325. doi: 10.1681/ASN.2010020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang S., He X., Ding J., Liang L., Zhao Y., Zhang Z., Yao X., Pan Z., Zhang P., Li J., et al. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-β-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int. J. Cancer. 2008;123:972–978. doi: 10.1002/ijc.23580. [DOI] [PubMed] [Google Scholar]
- 71.Kato M., Putta S., Wang M., Yuan H., Lanting L., Nair I., Gunn A., Nakagawa Y., Shimano H., Todorov I., et al. TGF-β activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat. Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L., Li Z., Kong X., Xie D., Jia Z., Jiang W., Cui J., Du Y., Wei D., Huang S., et al. Down-regulation of microRNA-494 via loss of SMAD4 increases FOXM1 and β-catenin signaling in pancreatic ductal adenocarcinoma cells. Gastroenterology. 2014;147:485–497 e418. doi: 10.1053/j.gastro.2014.04.048. [DOI] [PubMed] [Google Scholar]
- 73.Song L., Liu L., Wu Z., Li Y., Ying Z., Lin C., Wu J., Hu B., Cheng S.Y., Li M., et al. TGF-β induces miR-182 to sustain NF-kappaB activation in glioma subsets. J. Clin. Investig. 2012;122:3563–3578. doi: 10.1172/JCI62339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding X., Park S.I., McCauley L.K., Wang C.Y. Signaling between transforming growth factor β (TGF-β) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J. Biol. Chem. 2013;288:10241–10253. doi: 10.1074/jbc.M112.443655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pandit K.V., Corcoran D., Yousef H., Yarlagadda M., Tzouvelekis A., Gibson K.F., Konishi K., Yousem S.A., Singh M., Handley D., et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dogar A.M., Towbin H., Hall J. Suppression of latent transforming growth factor (TGF)-β1 restores growth inhibitory TGF-β signaling through microRNAs. J. Biol. Chem. 2011;286:16447–16458. doi: 10.1074/jbc.M110.208652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fils-Aime N., Dai M., Guo J., El-Mousawi M., Kahramangil B., Neel J.C., Lebrun J.J. MicroRNA-584 and the protein phosphatase and actin regulator 1 (PHACTR1), a new signaling route through which transforming growth factor-β Mediates the migration and actin dynamics of breast cancer cells. J. Biol. Chem. 2013;288:11807–11823. doi: 10.1074/jbc.M112.430934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 79.Park S.M., Gaur A.B., Lengyel E., Peter M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Korpal M., Lee E.S., Hu G., Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu Y., Kanwar S.S., Patel B.B., Oh P.S., Nautiyal J., Sarkar F.H., Majumdar A.P. MicroRNA-21 induces stemness by downregulating transforming growth factor β receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis. 2012;33:68–76. doi: 10.1093/carcin/bgr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mishra S., Deng J.J., Gowda P.S., Rao M.K., Lin C.L., Chen C.L., Huang T., Sun L.Z. Androgen receptor and microRNA-21 axis downregulates transforming growth factor β receptor II (TGFBR2) expression in prostate cancer. Oncogene. 2014;33:4097–4106. doi: 10.1038/onc.2013.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mestdagh P., Bostrom A.K., Impens F., Fredlund E., Van Peer G., De Antonellis P., von Stedingk K., Ghesquiere B., Schulte S., Dews M., et al. The miR-17-92 microRNA cluster regulates multiple components of the TGF-β pathway in neuroblastoma. Mol. Cell. 2010;40:762–773. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang H., Liu J., Zong Y., Xu Y., Deng W., Zhu H., Liu Y., Ma C., Huang L., Zhang L., et al. miR-106b aberrantly expressed in a double transgenic mouse model for Alzheimer’s disease targets TGF-β type II receptor. Brain Res. 2010;1357:166–174. doi: 10.1016/j.brainres.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 86.Chu T.H., Yang C.C., Liu C.J., Lui M.T., Lin S.C., Chang K.W. miR-211 promotes the progression of head and neck carcinomas by targeting TGFβRII. Cancer Lett. 2013;337:115–124. doi: 10.1016/j.canlet.2013.05.032. [DOI] [PubMed] [Google Scholar]
- 87.Jiang X., Xiang G., Wang Y., Zhang L., Yang X., Cao L., Peng H., Xue P., Chen D. MicroRNA-590-5p regulates proliferation and invasion in human hepatocellular carcinoma cells by targeting TGF-β RII. Mol. Cells. 2012;33:545–551. doi: 10.1007/s10059-012-2267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gregory P.A., Bracken C.P., Smith E., Bert A.G., Wright J.A., Roslan S., Morris M., Wyatt L., Farshid G., Lim Y.Y., et al. An autocrine TGF-β/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol. Biol. Cell. 2011;22:1686–1698. doi: 10.1091/mbc.e11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Braun J., Hoang-Vu C., Dralle H., Huttelmaier S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. 2010;29:4237–4244. doi: 10.1038/onc.2010.169. [DOI] [PubMed] [Google Scholar]
- 90.Yu J., Lei R., Zhuang X., Li X., Li G., Lev S., Segura M.F., Zhang X., Hu G. MicroRNA-182 targets SMAD7 to potentiate TGFβ-induced epithelial-mesenchymal transition and metastasis of cancer cells. Nat. Commun. 2016;7:13884. doi: 10.1038/ncomms13884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Petrocca F., Visone R., Onelli M.R., Shah M.H., Nicoloso M.S., de Martino I., Iliopoulos D., Pilozzi E., Liu C.G., Negrini M., et al. E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 92.Xiao C., Srinivasan L., Calado D.P., Patterson H.C., Zhang B., Wang J., Henderson J.M., Kutok J.L., Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Inomata M., Tagawa H., Guo Y.M., Kameoka Y., Takahashi N., Sawada K. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113:396–402. doi: 10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- 94.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell. Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Humphries B., Yang C. The microRNA-200 family: Small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6:6472–6498. doi: 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Senfter D., Madlener S., Krupitza G., Mader R.M. The microRNA-200 family: Still much to discover. Biomol. Concepts. 2016;7:311–319. doi: 10.1515/bmc-2016-0020. [DOI] [PubMed] [Google Scholar]
- 97.Mutlu M., Raza U., Saatci O., Eyupoglu E., Yurdusev E., Sahin O. miR-200c: A versatile watchdog in cancer progression, EMT, and drug resistance. J. Mol. Med. 2016;94:629–644. doi: 10.1007/s00109-016-1420-5. [DOI] [PubMed] [Google Scholar]
- 98.Zhang J., Tian X.J., Zhang H., Teng Y., Li R., Bai F., Elankumaran S., Xing J. TGF-β-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci. Signal. 2014;7:ra91. doi: 10.1126/scisignal.2005304. [DOI] [PubMed] [Google Scholar]
- 99.Bracken C.P., Gregory P.A., Kolesnikoff N., Bert A.G., Wang J., Shannon M.F., Goodall G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 100.Lee J.S., Ahn Y.H., Won H.S., Sun S., Kim Y.H., Ko Y.H. Prognostic Role of the microRNA-200 family in various carcinomas: A systematic review and meta-analysis. BioMed Res. Int. 2017;2017:1928021. doi: 10.1155/2017/1928021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bracken C.P., Li X., Wright J.A., Lawrence D.M., Pillman K.A., Salmanidis M., Anderson M.A., Dredge B.K., Gregory P.A., Tsykin A., et al. Genome-wide identification of miR-200 targets reveals a regulatory network controlling cell invasion. EMBO J. 2014;33:2040–2056. doi: 10.15252/embj.201488641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 103.Wang G., Guo X., Hong W., Liu Q., Wei T., Lu C., Gao L., Ye D., Zhou Y., Chen J., et al. Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc. Natl. Acad. Sci. USA. 2013;110:2858–2863. doi: 10.1073/pnas.1212769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wellner U., Schubert J., Burk U.C., Schmalhofer O., Zhu F., Sonntag A., Waldvogel B., Vannier C., Darling D., zur Hausen A., et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 105.Kim N.H., Kim H.S., Li X.Y., Lee I., Choi H.S., Kang S.E., Cha S.Y., Ryu J.K., Yoon D., Fearon E.R., et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J. Cell Biol. 2011;195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Siemens H., Jackstadt R., Hunten S., Kaller M., Menssen A., Gotz U., Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 107.Ma J., Fang B., Zeng F., Ma C., Pang H., Cheng L., Shi Y., Wang H., Yin B., Xia J., et al. Down-regulation of miR-223 reverses epithelial-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Oncotarget. 2015;6:1740–1749. doi: 10.18632/oncotarget.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tian X.J., Zhang H., Xing J. Coupled reversible and irreversible bistable switches underlying TGFβ-induced epithelial to mesenchymal transition. Biophys. J. 2013;105:1079–1089. doi: 10.1016/j.bpj.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pardali E., Sanchez-Duffhues G., Gomez-Puerto M.C., Ten Dijke P. TGF-β-induced endothelial-mesenchymal transition in fibrotic diseases. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18102157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Markwald R.R., Fitzharris T.P., Smith W.N. Sturctural analysis of endocardial cytodifferentiation. Dev. Biol. 1975;42:160–180. doi: 10.1016/0012-1606(75)90321-8. [DOI] [PubMed] [Google Scholar]
- 111.Zeisberg E.M., Tarnavski O., Zeisberg M., Dorfman A.L., McMullen J.R., Gustafsson E., Chandraker A., Yuan X., Pu W.T., Roberts A.B., et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 2007;13:952–961. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 112.Maddaluno L., Rudini N., Cuttano R., Bravi L., Giampietro C., Corada M., Ferrarini L., Orsenigo F., Papa E., Boulday G., et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 2013;498:492–496. doi: 10.1038/nature12207. [DOI] [PubMed] [Google Scholar]
- 113.Cooley B.C., Nevado J., Mellad J., Yang D., Hilaire C.S., Negro A., Fang F., Chen G., San H., Walts A.D., et al. TGF-β signaling mediates endothelial-to-mesenchymal transition (EndMT) during vein graft remodeling. Sci. Transl. Med. 2014;6:227ra234. doi: 10.1126/scitranslmed.3006927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zeisberg E.M., Potenta S., Xie L., Zeisberg M., Kalluri R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007;67:10123–10128. doi: 10.1158/0008-5472.CAN-07-3127. [DOI] [PubMed] [Google Scholar]
- 115.Ghosh A.K., Nagpal V., Covington J.W., Michaels M.A., Vaughan D.E. Molecular basis of cardiac endothelial-to-mesenchymal transition (EndMT): Differential expression of microRNAs during EndMT. Cell Signal. 2012;24:1031–1036. doi: 10.1016/j.cellsig.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Srivastava S.P., Koya D., Kanasaki K. MicroRNAs in kidney fibrosis and diabetic nephropathy: Roles on EMT and EndMT. BioMed Res. Int. 2013;2013:125469. doi: 10.1155/2013/125469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lagendijk A.K., Goumans M.J., Burkhard S.B., Bakkers J. MicroRNA-23 restricts cardiac valve formation by inhibiting Has2 and extracellular hyaluronic acid production. Circ. Res. 2011;109:649–657. doi: 10.1161/CIRCRESAHA.111.247635. [DOI] [PubMed] [Google Scholar]
- 118.Zhang J., Zhang Z., Zhang D.Y., Zhu J., Zhang T., Wang C. microRNA 126 inhibits the transition of endothelial progenitor cells to mesenchymal cells via the PIK3R2-PI3K/Akt signalling pathway. PLoS ONE. 2013;8:e83294. doi: 10.1371/journal.pone.0083294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xiang Y., Zhang Y., Tang Y., Li Q. MALAT1 Modulates TGF-β1-induced endothelial-to-mesenchymal transition through downregulation of miR-145. Cell. Physiol. Biochem. 2017;42:357–372. doi: 10.1159/000477479. [DOI] [PubMed] [Google Scholar]
- 120.Kumarswamy R., Volkmann I., Jazbutyte V., Dangwal S., Park D.H., Thum T. Transforming growth factor-β-induced endothelial-to-mesenchymal transition is partly mediated by microRNA-21. Arterioscler. Thromb. Vasc. Biol. 2012;32:361–369. doi: 10.1161/ATVBAHA.111.234286. [DOI] [PubMed] [Google Scholar]
- 121.Zhu K., Pan Q., Jia L.Q., Dai Z., Ke A.W., Zeng H.Y., Tang Z.Y., Fan J., Zhou J. MiR-302c inhibits tumor growth of hepatocellular carcinoma by suppressing the endothelial-mesenchymal transition of endothelial cells. Sci. Rep. 2014;4:5524. doi: 10.1038/srep05524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Geng H., Guan J. MiR-18a-5p inhibits endothelial-mesenchymal transition and cardiac fibrosis through the Notch2 pathway. Biochem. Biophys. Res. Commun. 2017;491:329–336. doi: 10.1016/j.bbrc.2017.07.101. [DOI] [PubMed] [Google Scholar]
- 123.Correia A.C., Moonen J.R., Brinker M.G., Krenning G. FGF2 inhibits endothelial-mesenchymal transition through microRNA-20a-mediated repression of canonical TGF-β signaling. J. Cell Sci. 2016;129:569–579. doi: 10.1242/jcs.176248. [DOI] [PubMed] [Google Scholar]
- 124.Katsura A., Suzuki H.I., Ueno T., Mihira H., Yamazaki T., Yasuda T., Watabe T., Mano H., Yamada Y., Miyazono K. MicroRNA-31 is a positive modulator of endothelial-mesenchymal transition and associated secretory phenotype induced by TGF-β. Genes Cells. 2016;21:99–116. doi: 10.1111/gtc.12323. [DOI] [PubMed] [Google Scholar]
- 125.Suzuki H.I., Katsura A., Mihira H., Horie M., Saito A., Miyazono K. Regulation of TGF-β-mediated endothelial-mesenchymal transition by microRNA-27. J. Biochem. 2017;161:417–420. doi: 10.1093/jb/mvx017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Suzuki H.I., Katsura A., Matsuyama H., Miyazono K. MicroRNA regulons in tumor microenvironment. Oncogene. 2015;34:3085–3094. doi: 10.1038/onc.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang P., Li Q.J., Feng Y., Zhang Y., Markowitz G.J., Ning S., Deng Y., Zhao J., Jiang S., Yuan Y., et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhou S.L., Hu Z.Q., Zhou Z.J., Dai Z., Wang Z., Cao Y., Fan J., Huang X.W., Zhou J. miR-28-5p-IL-34-macrophage feedback loop modulates hepatocellular carcinoma metastasis. Hepatology. 2016;63:1560–1575. doi: 10.1002/hep.28445. [DOI] [PubMed] [Google Scholar]
- 129.Liu Y., Lai L., Chen Q., Song Y., Xu S., Ma F., Wang X., Wang J., Yu H., Cao X., et al. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. J. Immunol. 2012;188:5500–5510. doi: 10.4049/jimmunol.1103505. [DOI] [PubMed] [Google Scholar]
- 130.Li L., Zhang J., Diao W., Wang D., Wei Y., Zhang C.Y., Zen K. MicroRNA-155 and MicroRNA-21 promote the expansion of functional myeloid-derived suppressor cells. J. Immunol. 2014;192:1034–1043. doi: 10.4049/jimmunol.1301309. [DOI] [PubMed] [Google Scholar]
- 131.Donatelli S.S., Zhou J.M., Gilvary D.L., Eksioglu E.A., Chen X., Cress W.D., Haura E.B., Schabath M.B., Coppola D., Wei S., et al. TGF-β-inducible microRNA-183 silences tumor-associated natural killer cells. Proc. Natl. Acad. Sci. USA. 2014;111:4203–4208. doi: 10.1073/pnas.1319269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li Q., Zhang D., Wang Y., Sun P., Hou X., Larner J., Xiong W., Mi J. MiR-21/Smad 7 signaling determines TGF-β1-induced CAF formation. Sci. Rep. 2013;3:2038. doi: 10.1038/srep02038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bertero A., Brown S., Madrigal P., Osnato A., Ortmann D., Yiangou L., Kadiwala J., Hubner N.C., de Los Mozos I.R., Sadee C., et al. The SMAD2/3 interactome reveals that TGFβ controls m(6)A mRNA methylation in pluripotency. Nature. 2018;555:256–259. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Matak A., Lahiri P., Ford E., Pabst D., Kashofer K., Stellas D., Thanos D., Zatloukal K. Stochastic phenotype switching leads to intratumor heterogeneity in human liver cancer. Hepatology. 2017 doi: 10.1002/hep.29679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S., et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 136.Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 137.Sotillo E., Thomas-Tikhonenko A. Shielding the messenger (RNA): MicroRNA-based anticancer therapies. Pharmacol. Ther. 2011;131:18–32. doi: 10.1016/j.pharmthera.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lieberman J., Sharp P.A. Harnessing RNA interference for therapy: The silent treatment. JAMA. 2015;313:1207–1208. doi: 10.1001/jama.2015.1241. [DOI] [PubMed] [Google Scholar]
- 139.Suzuki H.I., Mihira H., Watabe T., Sugimoto K., Miyazono K. Widespread inference of weighted microRNA-mediated gene regulation in cancer transcriptome analysis. Nucleic Acids Res. 2013;41:e62. doi: 10.1093/nar/gks1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Suzuki H.I., Matsuyama H., Noguchi M., Yao T., Komatsu N., Mano H., Sugimoto K., Miyazono K. Computational dissection of distinct microRNA activity signatures associated with peripheral T cell lymphoma subtypes. Leukemia. 2013;27:2107–2111. doi: 10.1038/leu.2013.121. [DOI] [PubMed] [Google Scholar]
- 141.Suzuki H.I., Spengler R.M., Grigelioniene G., Kobayashi T., Sharp P.A. Deconvolution of seed and RNA-binding protein crosstalk in RNAi-based functional genomics. Nat. Genet. 2018;50:657–661. doi: 10.1038/s41588-018-0104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]