Abstract
Scrub typhus threatens one billion people in the Asia-Pacific area and cases have emerged outside this region. It is caused by infection with any of the multitude of strains of the bacterium Orientia tsutsugamushi. A vaccine that affords heterologous protection and a commercially-available molecular diagnostic assay are lacking. Herein, we determined that the nucleotide and translated amino acid sequences of outer membrane protein A (OmpA) are highly conserved among 51 O. tsutsugamushi isolates. Molecular modeling revealed the predicted tertiary structure of O. tsutsugamushi OmpA to be very similar to that of the phylogenetically-related pathogen, Anaplasma phagocytophilum, including the location of a helix that contains residues functionally essential for A. phagocytophilum infection. PCR primers were developed that amplified ompA DNA from all O. tsutsugamushi strains, but not from negative control bacteria. Using these primers in quantitative PCR enabled sensitive detection and quantitation of O. tsutsugamushi ompA DNA from organs and blood of mice that had been experimentally infected with the Karp or Gilliam strains. The high degree of OmpA conservation among O. tsutsugamushi strains evidences its potential to serve as a molecular diagnostic target and justifies its consideration as a candidate for developing a broadly-protective scrub typhus vaccine.
Keywords: scrub typhus, Orientia tsutsugamushi, Rickettsia, Rickettsiales, outer membrane protein A, Anaplasma
1. Introduction
Scrub typhus is an acute, febrile, and potentially deadly disease caused by infection with the larval Leptotrombidium mite-vectored bacterium, Orientia tsutsugamushi. Long known to be endemic to the Asia-Pacific, a densely-populated region where more than one million cases are estimated to occur annually, scrub typhus affects all organs and the central nervous system. Clinical manifestations can include eschar, fever, myalgia, maculopapular rash, lymphadenopathy, pneumonitis, meningitis, and coagulopathies that can result in circulatory system collapse (reviewed in [1,2]). The disease can account for up to 20% of all acute undifferentiated febrile episodes and up to 27% of blood culture-negative fever patients in endemic areas [3,4,5]. Its non-specific clinical presentation makes clinical diagnosis very difficult [6]. Mortality rates range from less than 1.4% to approximately 70%, depending on prior patient immune status, whether proper antibiotic treatment is initiated in a timely manner, and the bacterial strain causing the infection [1,7,8,9]. Recent outbreaks in the Asia-Pacific [10,11,12,13,14,15,16,17,18,19,20,21,22,23] as well as evidence for scrub typhus or scrub typhus-like infections in Cameroon, Kenya, Congo, Djibouti, Tanzania, Chile, and Peru signify these illnesses as both emerging and reemerging diseases of global importance [24,25,26,27,28,29,30].
The genus Orientia is a member of the order Rickettsiales, which contains other arthropod vector-transmitted pathogens, including Anaplasma, Ehrlichia, and Rickettsia. Until recently, the genus consisted of a single species, Orientia tsutsugamushi, which contained multiple antigenically-distinct strains [31]. In 2010, a second agent, Candidatus Orientia chuto, was discovered in a febrile patient presenting with a scrub typhus-like illness that had been acquired in the United Arab Emirates [32]. The extensive immunogenic diversity among O. tsutsugamushi strains has contributed to the inability to develop a scrub typhus vaccine that achieves heterologous protection despite more than seven decades’ worth of efforts [6]. No commercially-available molecular diagnostic assay for the disease exists. Serology-based tests suffer from a high seroprevalence baseline among populations living in scrub typhus-endemic areas. While polymerase chain reaction (PCR)-based tests can overcome limitations of serologic assays, only a limited number of Orientia spp. nucleic acid sequences have been explored for their potential as molecular diagnostic targets [33,34,35,36,37].
Outer membrane protein A (OmpA; also referred to as peptidoglycan-associated lipoprotein) is conserved among most Gram-negative bacteria and contributes to the virulence of Gram-negative pathogens, especially their abilities to adhere to and invade host cells [38,39,40,41,42,43,44,45]. Antisera raised against entire OmpA proteins or specific binding domains thereof for Anaplasma spp., Ehrlichia chaffeensis, and R. conorii inhibit bacterial invasion of host cells in vitro [38,41,42,44,45]. These Rickettsiales members express OmpA during infection of human patients and/or experimentally infected animals [38,44,46]. Several Rickettsiales species and strains have stretches of ompA DNA sequences that exhibit high degrees of identity [44,45,47,48], which suggests their potential as effective nucleic acid-based diagnostic targets. Limited evidence suggests that OmpA antibodies offer at least some protection from rickettsial infections in vivo [49]. While O. tsutsugamushi Ikeda expresses OmpA during infection of mammalian host cells in vitro [50], ompA conservation among Orientia spp., and whether these bacteria express ompA during in vivo infection, have yet to be examined.
In this study, we determined that ompA DNA and translated amino acid sequences are highly conserved among 51 geographically-diverse O. tsutsugamushi isolates. Molecular modeling revealed the predicted tertiary structure of O. tsutsugamushi OmpA to be very similar to that of Anaplasma phagocytophilum OmpA, including the location of a helix and residues thereof that are essential for Anaplasma spp. OmpA function. A PCR primer pair was developed that amplified ompA DNA from all O. tsutsugamushi strains examined and enabled sensitive detection and quantitation of O. tsutsugamushi ompA DNA from organs and blood of experimentally-infected mice. The high degree of conservation of OmpA among O. tsutsugamushi isolates suggests that it be considered both as a diagnostic target and potential antigen for developing a broadly-protective scrub typhus vaccine.
2. Materials and Methods
2.1. O. tsutsugamushi DNA Samples Examined in This Study
Nearly all of the O. tsutsugamushi strains examined in this study have been previously described [32,51,52,53,54,55,56,57,58,59,60,61,62,63,64]. The isolates, their countries of origin, publication in which they were originally reported, and their ompA GenBank accession numbers and locus tags are listed in Table 1.
Table 1.
O. tsutsugamushi isolates used in this study.
| Isolate | Geographic Origin | Reference | ompA GenBank Accession Number or Locus Tag |
|---|---|---|---|
| 18-032460 | Malaysia | [63] | MH167971 |
| AFC3 | Thailand | [56] | MH167972 |
| AFC30 | Thailand | NR | MH167973 |
| AFPL12 | Thailand | NR | MH167974 |
| Boryong | South Korea | [54] | OTBS_RS08365 |
| Brown | Australia | [61] | MH167975 |
| Calcutta | India | NR | MH167976 |
| Citrano | Australia | [61] | MH167977 |
| CRF09 | Northern Thailand | [52] | MH167978 |
| CRF58 | Northern Thailand | [52] | MH167979 |
| CRF136 | Northern Thailand | [52] | MH167980 |
| CRF158 | Northern Thailand | [52] | MH167981 |
| FPW1038 | Western Thailand | [53] | MH167982 |
| FPW2016 | Western Thailand | [53] | MH167983 |
| FPW2049 | Western Thailand | [53] | MH167984 |
| Gilliam | Burma | [57] | MH167985 |
| Ikeda | Japan | [58] | OTT_RS06375 |
| Karp | New Guinea | [55] | OTSKARP_0358 |
| Kato | Japan | [64] | OTSKATO_0610 |
| LNT1153 | Northwestern Laos | [51] | MH167986 |
| LNT1189 | Northwestern Laos | [51] | MH167987 |
| LNT1301 | Northwestern Laos | [51] | MH167988 |
| LNT1310 | Northwestern Laos | [51] | MH167989 |
| MAK110 | Taiwan | [62] | MH167990 |
| MAK119 | Taiwan | [62] | MH167991 |
| MAK243 | Taiwan | [62] | MH167992 |
| SV400 | Southern Laos | [51] | MH167993 |
| SV445 | Southern Laos | [51] | MH167994 |
| SV484 | Southern Laos | [51] | MH167995 |
| TA763 | Thailand | [59] | OTSTA763_0977 |
| TH1812 | Thailand | [59] | MH167996 |
| TH1817 | Thailand | [59] | MH167997 |
| TM2328 | Central Laos | [51] | MH167998 |
| TM2395 | Central Laos | [51] | MH167999 |
| TM2494 | Central Laos | [51] | MH168000 |
| TM2532 | Central Laos | [51] | MH168001 |
| TM2766 | Central Laos | [51] | MH168002 |
| TM2950 | Central Laos | [51] | MH168003 |
| TM3115 | Central Laos | [51] | MH168004 |
| UT76 | Northeastern Thailand | [53] | MH168005 |
| UT125 | Northeastern Thailand | [53] | MH168006 |
| UT169 | Northeastern Thailand | [53] | MH168007 |
| UT177 | Northeastern Thailand | [53] | MH168008 |
| UT219 | Northeastern Thailand | [53] | MH168009 |
| UT336 | Northeastern Thailand | [53] | MH168010 |
| UT340 | Northeastern Thailand | [53] | MH168011 |
| UT418 | Northeastern Thailand | [53] | MH168012 |
| UT559 | Northeastern Thailand | [53] | MH168013 |
| UT652 | Northeastern Thailand | [53] | MH168014 |
| Volner | New Guinea | [60] | MH168015 |
| Wood | Australia | [61] | MH168016 |
NR: no previous published report.
2.2. PCR, Cloning, and DNA Sequence Analyses
PCR was performed using isolated O. tsutsugamushi strain DNA and MyTaq polymerase Red (Bioline, Taunton, MA, USA) following the manufacturer’s instructions. Following an initial denaturing step at 95 °C for 1 min, thermal cycling conditions were 35 cycles of 95 °C for 15 s, 55 °C for 15 s, and 72 °C for 10 s, followed by a final extension at 72 °C for 20 s. Amplicons were analyzed in 2.0% agarose gels in 40 mM tris-acetate-2 mM EDTA (pH 8.5). Primer sequences targeting ompA were designed according to ompA (OTT_RS06375) of the annotated O. tsutsugamushi Ikeda genome [65] and are listed in Table 2. DNA samples that yielded amplicons of the expected sizes were again subjected to PCR using the appropriate primer sets and Platinum HiFi Taq polymerase (Thermo Fisher, Waltham, MA, USA) according to the manufacturer’s instructions. Platinum HiFi Taq polymerase thermal cycling conditions consisted of an initial denaturation step of 94 °C for 2 min, followed by 34 cycles of 94 °C for 15 s, 55 °C for 30 s, and a final extension step at 68 °C for 1 min. The resulting amplicons were subjected to agarose gel electrophoresis, after which they were visualized using a Blue View Transilluminator (Vernier Biotechnology, Beaverton, OR, USA), excised, and purified using the QIAquickGel Extraction Kit (Qiagen, Valencia, CA, USA). The purified PCR products were TA-cloned into pCR2.1-TOPO using the TOPO TA Cloning kit (Thermo Fisher). Clones were transformed into chemically competent TOPO Escherichia coli and incubated for 1 h in SOC medium (Thermo Fisher) at 37 °C with agitation at 250 RPM. Aliquots of each culture were plated onto Luria-Bertani (LB) agar plates containing 0.1 mg/mL ampicillin and incubated at 37°C overnight. Colony PCR using vector-derived M13F and M13R primers was performed to identify colonies that harbored plasmids containing inserts of the expected size. Plasmids isolated from PCR-positive colonies using the PerfectPrep Spin Mini Kit (5 Prime, Gaithersburg, MD, USA) were sequenced using M13F and M13R primers by Genewiz (South Plainfield, NJ, USA) and the provided sequences were analyzed using the Lasergene 7.1 software package (DNASTAR, Madison, WI, USA).
Table 2.
Oligonucleotide primers used in this study.
| Primer Designation a | Sequence (5′ to 3′) |
|---|---|
| OTT_RS06375-1F | ATGATTAAAAAGTCAATTATTAGTATATGTGTATTAGTGC |
| OTT_RS06375-615R | CTATGCTATATTACTTTTAATAATTGTGACAGACC |
| OTT_RS06375-64F | TGTTTATGGCAAAGATCTAAACATAGTAAC |
| ompA-57F | GTGGAAATGTTTATGGCAAAGATCTAAAC |
| ompA-260R | GCTTGTAAAAACTGTTCATGCTTTACATC |
| Eubacterial 16S-F | GTTCGGAATTACTGGGCGTA |
| Eubacterial 16S-R | AATTAAACCGCATGCTCCAC |
| R17K-135 | ATGAATAAACAACGKbCANGGHACAC |
| R17K-249 | RAAGTAATGCRCCTACACCTACTC |
a F and R refer to primers that bind to the sense and antisense strand, respectively. The number immediately preceding the F or R denotes the first nucleotide position where the primer binds. b Degenerate positions contained equal molar base concentrations of the following nucleotides: (K), guanine and thymine, (N), adenine, guanine, thymine, and cytosine; (H), adenine, guanine, and thymine; (R) adenine and guanine.
2.3. In Silico Analyses and GenBank Accession Numbers of O. tsutsugamushi Strain ompA Sequences
O. tsutsugamushi strain ompA sequences were aligned using MegAlign and translated using EditSeq, both of which are part of the Lasergene 7.1 software package (DNASTAR). New O. tsutsugamushi strain ompA sequences identified herein have been deposited in GenBank with accession numbers listed in Table 1. Sequence distances and percent similarity of O. tsutsugamushi OmpA proteins were calculated using the ClustalW option in MegAlign. Heat maps indicating similarity/diversity of the O. tsutsugamushi strain OmpA nucleotide and translated protein sequences were generated using HEATMAP hierarchical clustering web tool (www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html/). To predict a putative tertiary structure for O. tsutsugamushi Ikeda OmpA, the mature (minus the signal sequence) Ikeda sequence (residues 21 to 204) was threaded onto solved crystal structures of proteins with similar sequences using the PHYRE2 (Protein Homology/analogy Recognition Engine, version 2.0) server (www.sbg.bio.ic.ac.uk/phyre2) [66]. Six templates (c4zhvB (bacterial signaling protein Yfib), c5jirB (oop family OmpA-OmpF porin; Treponema pallidum protein tp0624), c3s0yA (periplasmic domain of motility protein B (MotB) residues 64-256), c2l26A (uncharacterized Mycobacterium tuberculosis membrane protein rv0899/mt0922), c2k1sA (folded C-terminal fragment of YiaD from E. coli), and c5wtlB (periplasmic portion of outer membrane protein 2a (OmpA) from Capnocytophaga gingivalis)) were selected to model OmpA based on heuristics to maximize confidence, percentage amino acid identity, and alignment coverage. For the final model, 92% of the protein was modeled at greater than 90% confidence. Residues that vary among the OmpA translated sequences identified in this study were denoted on the O. tsutsugamushi Ikeda OmpA PHYRE2 model using the PyMOL algorithm (pymol.org/educational).
2.4. Confirmation of ompA PCR Primer Specificity
Based on an alignment of all ompA sequences determined herein, primers ompA-57F and ompA-260R (Table 2) were designed to amplify nucleotides 57 to 260 based on O. tsutsugamushi Ikeda ompA (OTT_RS06375) [65]. To confirm that the primers were specific for O. tsutsugamushi ompA, they were utilized in PCR reactions that included genomic DNA from Rickettsia species (R. africae, R. akari, R. australis, R. conorii, R. montanensis, R. parkeri, R. rhipicephali, R. rickettsii, R. sibirica, R. prowazekii, R. typhi, and R. amblyommii), as well as other bacterial species (Proteus mirabilis, E. coli, Legionella pneumophila, Bartonella vinsonii, Neorickettsia risticii, N. sennetsu, Francisella persica, A. phagocytophilum, E. chaffeensis, Coxiella burnetii, Chlamydia trachomatis, and Chlamydia pneumoniae) using thermal cycling conditions and agarose gel electrophoresis described above. To verify that the control templates and thermal cycling conditions were amenable for PCR amplification, reactions were simultaneously performed on the Rickettsia species using degenerate primers that targeted the Rickettsia 17-kDa gene, R17K-135F and R17K-249R [36], and on the other bacterial species using primers that targeted a conserved eubacterial 16S rRNA sequence (Table 2) [36].
2.5. Mice
Female six- to eight-week old CD-1 Swiss outbred mice (Charles River Laboratories, Wilmington, MA, USA) were housed in animal biosafety level (ABSL)-2 laboratories prior to inoculation. Two days prior to inoculation, they were relocated to an ABSL-3 laboratory to adapt to their new surroundings. The mice were intradermally inoculated with 103 MuID50 of O. tsutsugamushi Karp or Gilliam strains produced from liver-spleen homogenate of infected CD-1 mice into the dorsum of the right ear as previously described [67]. Sterile PBS was used as mock inoculum to inject negative control animals [67]. In some cases, Swiss CD-1 mice were intraperitoneally inoculated with 103 MuID50 of O. tsutsugamushi Karp. At various days post-infection, the mice were euthanized, and organs or blood were harvested for DNA isolation [68]. All animal research was performed under the approval of the Institutional Animal Care and Use Committee at the Naval Medical Research Center (Protocol Number: 11-IDD-26).
2.6. Quantitative Real-Time PCR (qPCR)
To generate an ompA DNA standard, ompA nucleotides 1 to 615 were amplified using the OTT_RS06375-1F/615R primer set and Platinum HiFi Taq polymerase. The amplicon was gel-purified and cloned into pCR2.1-TOPO as described above. Concentration (in ng/µL) of the resulting plasmid, pCR2.1-ompA, was determined by UV spectrophotometry. The concentration was converted to copies/µL using the EndMemo DNA/RNA Copy Number Calculator (http://endmemo.com/bio/dnacopynum.php). To evaluate the sensitivity of the ompA-57F/260R primer set, triplicate 20 µL reactions containing either dilutions of pCR2.1-ompA ranging from 1 × 106 to 1 × 10−2 copies/µL or no template were subjected to qPCR using SsoFastEvaGreenSupermix (Bio-Rad, Hercules, CA, USA) in a CFX96 Real-Time System thermocycler (Bio-Rad). Thermal cycling conditions consisted of an initial denaturation step of 95 °C for 3 min, followed by 40 cycles of 95 °C for 10 s and 55 °C for 30 s. The ability of the ompA-57F/260R primer set to detect ompA in DNA isolated from mouse tissues recovered from O. tsutsugamushi infected mice was also assessed. Using the DNeasy Blood and Tissue kit (Qiagen), DNA was isolated from the heart, kidney, liver, lung, spleen, and blood harvested on various days post-infection from Swiss CD-1 mice that had been infected with either O. tsutsugamushi Gilliam or Karp or uninfected control mice [68]. Fifty ng of mouse tissue DNA template per sample was subjected to qPCR exactly as described for the ompA DNA standards. Infrequently, an individual ompA-57F/260R reaction using uninfected mouse tissue DNA as template or no-template control would generate a Cq value at cycle 36.5 or later. In such experiments, only earlier Cq values generated for infected samples were considered as specifically amplifying ompA.
3. Results
3.1. Analyses of O. tsutsugamushi ompA DNA and Translated Amino Acid Sequences
DNA samples recovered from 51 geographically-diverse O. tsutsugamushi isolates (Table 1) were subjected to PCR using primer set OTT_RS06375-1F/615R (Table 2), which targets the full-length ompA gene (OTT_RS06375) of annotated O. tsutsugamushi strain Ikeda. [65]. Amplicons of the expected size were generated for all O. tsutsugamushi isolates except for SV400 and UT125. A second primer set specific for OTT_RS06375 nucleotides 64 to 615 successfully amplified the targeted ompA region from SV400 and UT125. Amplicons generated using the OTT_RS06375-1F/615R and OTT_RS06375-64F/615R primer sets were cloned and sequenced. The ompA nucleotide and translated amino acid sequences were deposited in GenBank. The nucleotide and amino acid identities ranged from 93.6% to 100.0% and 90.6% to 100.0%, respectively (Table S1, Figure 1 and Figure 2). SV400 and UT125 were excluded from heat map analyses because only partial sequences had been obtained for them. Aligning all OmpA amino acid sequences revealed that several segments thereof were 100% conserved among the isolates (Figure 3).
Figure 1.
Heat map of percent nucleotide identity of the ompA DNA sequences among 49 O. tsutsugamushi isolates. The heat maps were generated using the pairwise identity matrix tables with hierarchical clustering method. The names of the isolates are provided on the right side and bottom of the heat map. Dendrograms are on the left side and on top of the heat map.
Figure 2.
Heat map of percent identity of the OmpA translated amino acid sequences among 49 O. tsutsugamushi isolates. The heat maps were generated using the pairwise identity matrix tables with hierarchical clustering method. The names of the isolates are provided on the right side and bottom of the heat map. Dendrograms are on the left side and on top of the heat map.
Figure 3.
Alignment of translated O. tsutsugamushi isolate OmpA sequences. Presented is a ClustalW alignment of the translated amino acid sequences of all 51 O. tsutsugamushi isolates in this study. Amino acid differences relative to the majority (consensus) sequence are denoted by black-shaded white text. Sequence gaps relative to the majority are indicated by dashes. Residues 1–21 for SV400 and UT125 could not be predicted because ompA could be amplified by OTT_RS06375-64F/615R, but not OTT_RS06375-1F/615R.
3.2. Molecular Modeling of OmpA
OmpA proteins of A. phagocytophilum and A. marginale, which are in the order Rickettsiales with O. tsutsugamushi, contribute to their abilities to bind and invade mammalian host cells [41,44]. We previously demonstrated that these two OmpA proteins’ tertiary structures are highly similar and residues that are critical for adhesin function are conserved between them and are presented as part of surface-exposed alpha helices [41,44,45]. To predict the tertiary structure of O. tsutsugamushi OmpA, molecular modeling of Ikeda OmpA residues 21 to 204 (minus the signal sequence) as a representative naturally-occurring OmpA sequence was performed using the PHYRE2 recognition server (www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi) [69], which generates three-dimensional models for protein sequences and threads them on known crystal structures. The resulting model is presented in Figure 4A. The O. tsutsugamushi OmpA tertiary structure was evaluated for similarity to Anaplasma spp. OmpA. Because the A. phagocytophilum and A. marginale OmpA models are nearly identical [41], the O. tsutsugamushi OmpA predicted structure was compared only to that of A. phagocytophilum. Threading the two models onto each other using PyMOL revealed their folded portions to be very similar structurally with the exception of O. tsutsugamushi residues 21 through 50, which are disordered (Figure 4B). Notably, O. tsutsugamushi OmpA bears a surface-exposed alpha helix that overlays with the functionally-essential surface-exposed alpha helix of A. phagocytophilum OmpA. Moreover, aligning the A. phagocytophilum (L59KGPGKKVILELVEQL74) and A. marginale OmpA binding domains (I53KGSGKKVLLGLVERM68) with the analogous region of O. tsutsugamushi OmpA (L103SEESKRVLRAQSAWL118) indicated conservation of several residues, including a lysine that is critical for A. phagocytophilum OmpA and A. marginale OmpA adhesin function [41,45] (Figure 4C). This region is identical among all O. tsutsugamushi translated OmpA sequences in this study with the exception of residue 116 (Figure 3 and Figure 4). Thus, O. tsutsugamushi OmpA is predicted to exhibit high structural similarity to OmpA proteins of other Rickettsiales members that are important for infection, its surface-exposed alpha helix is identical among all O. tsutsugamushi isolates examined herein, and residues of the alpha helix are identical to those of Anaplasma spp. OmpA proteins that are key for pathogenicity.
Figure 4.
Molecular modeling of O. tsutsugamushi OmpA reveals structural similarity with A. phagocytophilum OmpA and conservation of residues that are critical for Anaplasma spp. OmpA function. (A) Predicted tertiary structure for O. tsutsugamushi OmpA. The OmpA mature protein sequence was predicted using the PHYRE2 algorithm. The cyan portion corresponds to residues 103 to 118, which are analogous to residues 59 to 74 and 53 to 68 of A. phagocytophilum and A. marginale OmpA predicted tertiary structures, respectively, that form receptor-binding domains. Magenta residues are those that vary among the 51 O. tsutsugamushi OmpA sequences studied herein. (B) Overlay of O. tsutsugamushi and A. phagocytophilum predicted OmpA tertiary structures. A. phagocytophilum OmpA residues are labeled dark blue except for residues 59 to 74 (labeled orange) that lie within a surface-exposed alpha helix and constitute the receptor binding domain. O. tsutsugamushi OmpA residues are labeled gray except for residues 103 to 118 (labeled cyan) that are analogous to A. phagocytophilum OmpA residues 59 to 74. (C) Alignment of O. tsutsugamushi (Ot) OmpA residues 103 to 118, A. phagocytophilum (Ap) amino acids 59 to 74, and A. marginale (Am) residues 53 to 68. Identical residues among the three sequences are shaded gray. Similar amino acids are underlined. Functionally essential residues in the Anaplasma spp. OmpA proteins are in red text.
3.3. Development of a Primer Set That Specifically Amplifies an ompA Sequence Unique to O. tsutsugamushi
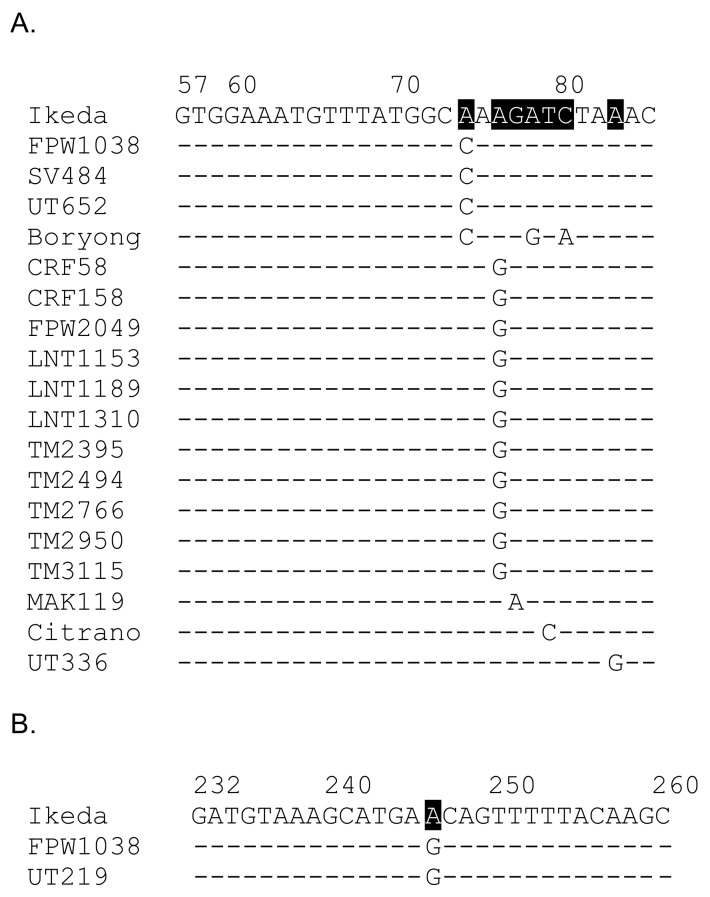
Next, whether a primer set could be devised that would amplify a segment of O. tsutsugamushi ompA from all O. tsutsugamushi isolates in this study was examined. A BLASTN search against GenBank using the O. tsutsugamushi Ikeda OTT_RS06375 sequence as query determined that nucleotides 64 to 279 were unique to O. tsutsugamushi isolates. Examination of nucleotides flanking and within this region for sequences that would have annealing temperatures compatible with thermal cycling conditions denoted nucleotides 57 to 85 (primer ompA-57F; Table 2) and 232 to 260 (primer ompA-260R; Table 2) as potential primers. For nucleotides 57 to 85, 32 of the 51 isolates had a sequence that was identical to the consensus, 17 had a single nucleotide mismatch, and one (Boryong) had three nucleotide mismatches (Figure 5A). For isolates SV400 and UT125, which ompA sequences were identified using primer set OTT_RS06375-64F/615R and therefore began with nucleotide 64, identity of nucleotides 57 to 63 could not be determined. However, nucleotides 64 to 85 for these two isolates exactly matched the consensus. All isolates exhibited 100% identity among nucleotides 232 to 260 except for two (FPW1038 and UT219), each of which had a single nucleotide mismatch (Figure 5B). For all isolate target sequences of the ompA-57F/260R primer set that had nucleotide mismatches, only one (UT336) exhibited a nucleotide mismatch near the 3′ end of either primer. Importantly, the UT336 nucleotide mismatch for ompA-57F did not occur within the final two nucleotides. Thus, it was expected that the primer set would amplify ompA from all 51 isolates.
Figure 5.
Nucleotide mismatches in the binding sites for ompA-57F and ompA-260R among O. tsutsugamushi isolates examined in this study. The ompA-57F (A) and ompA-260R (B) sequences, both of which correspond to O. tsutsugamushi Ikeda nucleotides, are listed. Below each are the corresponding nucleotides of where each primer would bind for any isolate in this study that has at least one mismatch. Numbers above the sequences refer to the nucleotide positions in Ikeda ompA. Dashes indicate identical nucleotides to those of ompA-57F and ompA-260R. Mismatches in the primer sequences are denoted by black-shaded white text with the specific nucleotide difference indicated per sequence below.
To confirm the efficacy of the ompA-57F/260R primer set, it was evaluated for the ability to amplify its target from six representative isolates that had one or more nucleotide mismatches at the primer binding sites (Boryong, LNT1153, SV484, UT336, MAK119, Citrano) plus SV400 and UT125, for which it was unknown whether ompA-57F would bind. A band of the expected size was generated for all eight samples, but not for negative control reaction that lacked DNA template (Figure 6). Next, whether the primer set would non-specifically generate products from a variety of eubacterial human pathogens, including A. phagocytophilum, C. burnetii, C. trachomatis, C. pneumoniae, and several Rickettsia spp. was examined. The primer set failed to yield PCR products from the 23 samples examined but produced an amplicon of the expected size from O. tsutsugamushi Ikeda (Figure 7A), which has a sequence that is identical to the consensus binding sites for both primers. Because PCR products could be generated from the eubacterial spp. using primers targeting eubacterial 16S rRNA and the rickettsial gene encoding 17-kDa protein [36] (Figure 7B,C), it could be concluded that the integrity of these samples was sufficient to allow for DNA amplification and that the only reason the ompA-57F/260R primer set did not generate PCR products for them was specifically due to disparity in the primer binding sites. Overall, these data demonstrate that the ompA-57F/260R specifically amplifies its DNA target sequence and does so even if it contains limited nucleotide mismatches.
Figure 6.
Primer set ompA-57F/260R amplifies ompA sequences having one to three nucleotide mismatches in the primer binding sites. DNA isolated from tissue culture cells infected with each of the indicated O. tsutsugamushi isolates or minus template control ((−) ctrl) were subjected to PCR analysis using ompA-57F/260R primers. The numbers to the left of the panel correspond to DNA ladder sizes. Data are representative of three experiments with similar results.
Figure 7.
Confirmation of ompA-57F/260R specificity. DNA samples from eubacterial and Rickettsia spp. were subjected to PCR using (A) ompA-57F/260R. The eubacterial and Rickettsia spp. DNA samples were also subjected to PCR using primer sets targeting eubacterial 16S rRNA (B) and the rickettsial gene encoding the 17-kDa protein (C) to verify sample integrity. For the experiment in (A), O. tsutsugamushi Ikeda DNA was included as a positive control. A minus template control ((−) ctrl) was included in each experiment. Vertical lines between lanes indicate that irrelevant lanes from the gel images were removed. Data are representative of two experiments with similar results.
3.4. Evaluation of the O. tsutsugamushi ompA-Specific Primer Set in qPCR
To determine the detection limit of ompA-57F/260R, the primer set was evaluated by qPCR using plasmid pCR2.1-ompA as template, which has ompA nucleotides 1 to 615 inserted, serially-diluted from 1 × 106 to 1 × 10−2 copies per reaction. The primers detected ompA as low as 101 copies (R2 = 0.995) (Figure 8A). The experiment was repeated with reactions containing pCR2.1-ompA diluted from 100 to 3.5 copies. The primers detected ompA DNA at a concentration as low as 3.5 copies (R2 = 0.950) (Figure 8B). Thus, ompA-57F/260R has an approximate detection limit of 3.5 copies.
Figure 8.
Primer set specific for O. tsutsugamushi OmpA is capable of amplifying low copy number ompA DNA standards. Standard curves generated by ompA-57F/260R amplification of ompA DNA standards diluted ten-fold from 1 × 106 to 1 × 10−2 (A) and from 100 to 3.5 copies (B). Data are representative of two experiments with similar results.
Next, the ompA-57F/260R primer set was evaluated using qPCR for the ability to detect and quantify ompA copies in DNA samples isolated from organs harvested from Swiss CD-1 mice infected with O. tsutsugamushi Gilliam. The CD-1 mouse intradermal inoculation model was recently demonstrated to exhibit features of early scrub typhus infection in humans, including distant organ dissemination. Gilliam was one such strain that was evaluated using this model [68]. Reactions performed on the same plate with pCR2.1-ompA diluted ten-fold from 1 × 106 to 1 × 100 copies allowed for copy number quantitation. Negative control reactions consisted of those containing DNA isolated from organs of mock-inoculated mice and those lacking DNA template. The primers detected ompA at copy numbers of 71.0 ± 5.2, 39.3 ± 22.2, 184.0 ± 22.2, and 181.0 ± 49.8 in kidney, liver, lung, and spleen DNA samples, respectively, recovered on Day 6 from a mouse that had been intradermally injected with O. tsutsugamushi Gilliam, but failed to amplify ompA from DNA isolated from heart of the infected mouse or from a mock-inoculated mouse (Figure 9A). When qPCR was performed on DNA samples isolated on Days 10, 14, and 21 from organs of mice that had been intradermally inoculated with O. tsutsugamushi Karp, ompA DNA was detected in all samples and at the highest levels for each organ in samples isolated on day 14 (Figure 9B). Copy numbers of ompA for these samples ranged from 22.4 ± 5.0 (heart 10 days post-infection) to 1890 ± 219.0 (lung 14 days post-infection). Lungs having the highest level of ompA DNA is consistent with the bacterial burden being the greatest in the lungs in this mouse model [68]. The primer sets also detected ompA DNA in blood drawn from mice that had been intraperitoneally inoculated with the Karp strain, again with Day 14 samples having the highest ompA levels (Figure 9C). Intraperitoneal inoculation resulted in a high bacteremia. Only one of three mice survived until Day 21 following the intraperitoneal inoculation route. These data demonstrate the ability of ompA-57F/260R to amplify and quantify ompA copies in DNA samples recovered from organs and blood of O. tsutsugamushi-infected mice.
Figure 9.
The ompA-57F/260R primer set amplifies ompA in qPCR from DNA recovered from the organs or blood of O. tsutsugamushi-infected mice. Fifty ng of DNA isolated on the indicated day post-infection (dpi) from the noted organs (A,B) or blood (C) from Swiss CD-1 mice that had been mock inoculated or that had been infected with O. tsutsugamushi Gilliam (A) or Karp (B,C). Each bar represents the mean + SD ompA copy number per 50 ng DNA recovered from an individual mouse analyzed in triplicate. Data are representative of at least three experiments with similar results.
4. Discussion
Scrub typhus is a global health concern for which neither a vaccine that provides heterologous protection nor a reliable diagnostic assay exists. To effectively protect against or detect the diversity of O. tsutsugamushi strains, the bacterial target must be highly conserved. OmpA satisfies this criterion, as it displays 93.6% to 100.0% and 90.6% to 100.0% conservation at the nucleotide and protein levels, respectively, among the isolates in this study that originated from multiple Asia-Pacific locations.
While the role of OmpA in O. tsutsugamushi pathobiology is unclear, studies of other Rickettsiales members’ OmpA proteins offer precedents that O. tsutsugamushi OmpA likely contributes to and could be immunologically targeted to inhibit infection. OmpA proteins of A. phagocytophilum, A. marginale, E. chaffeensis, and R. conorii are each on the bacterial surface, participate in host cell entry, and can be targeted by antibodies to inhibit infection in vitro [38,41,42,44,45]. Patients naturally infected with A. phagocytophilum or R. conorii and animals experimentally infected with A. phagocytophilum or E. chaffeensis develop antibodies that recognize recombinant forms of the respective OmpA proteins in serological assays [38,44,46], indicating that Rickettsiales bacteria express OmpA during in vivo infection. The abilities of A. phagocytophilum and A. marginale OmpA to mediate bacterial adhesion to, and invasion of, host cells, rely on receptor-binding domains that consist of specific lysine and glycine residues within the proteins’ structurally-conserved, surface-exposed alpha helices. Antisera specific for these binding domains inhibits Anaplasma spp. infection of host cells [41,45]. Strikingly, the predicted O. tsutsugamushi OmpA tertiary structure is very similar to that of A. phagocytophilum OmpA, so much so that their aforementioned alpha-helices and a lysine residue thereof overlay when the proteins are superimposed on each other. It will be important to confirm whether O. tsutsugamushi OmpA is surface-exposed and if antiserum raised against the full-length protein or its alpha helix amino acids 103–118 inhibits infection of host cells.
A R. prowazekii ompA deletion mutant retains the ability to productively infect mice [70], likely due to OmpA being one of many OMPs that cooperatively function to mediate invasion of host cells [71,72,73]. Nonetheless, it is worth investigating whether immunization against OmpA offers protection from O. tsutsugamushi challenge. A humoral immune response against OmpA or a key portion thereof together with other conserved OMPs could inhibit bacterial entry into host cells, which would essentially lead to its demise due to its obligatory intracellular lifestyle. This very concept has been demonstrated for blocking A. phagocytophilum infection in vitro: whereas OmpA binding domain antibody alone reduces infection by approximately 25%, an antibody cocktail targeting the binding domains of OmpA together with two additional OMPs nearly eliminates infection of host cells [44,45]. Anti-OmpA antibodies could also eradicate O. tsutsugamushi load in vivo via complement-mediated killing or opsonophagocytosis. Indeed, although the exact mechanism is unclear, guinea pigs immunized with a recombinant form of truncated R. heilongjiangensis OmpA exhibited reduced bacterial load, organ pathology, and interstitial pneumonia following challenge with R. heilongjiangensis or R. rickettsii, compared to sham-immunized animals [49].
Exploiting the high degree of ompA nucleotide conservation facilitated development of primer set ompA-57F/260R, which specifically amplified its target from all O. tsutsugamushi isolates examined herein, including ompA sequences having up to three nucleotide mismatches in the primer binding sites. In qPCR, ompA-57F/260R detected ompA at a copy number as low as 3.5. This sensitivity level rivaled or exceeded that reported for other qPCR assays [34,74,75]. Moreover, these primers detected ompA in DNA isolated from organs or blood of O. tsutsugamushi-infected mice. The ability of ompA-57F/260R to detect ompA in the presence of excess host tissue-derived DNA evidences its potential for sensitively detecting O. tsutsugamushi ompA in DNA isolated from scrub typhus patient-derived samples such as eschar swabs, blood, or buffy coats, as has been demonstrated for other qPCR assays [34,74,76,77,78,79,80,81].
5. Conclusions
The high degree of nucleotide and amino acid conservation of OmpA among diverse O. tsutsugamushi isolates and its structural similarity to other Rickettsiales OmpA proteins that have been successfully targeted to inhibit bacterial invasion of host cells, argue for its consideration as a vaccine candidate that could provide heterologous protection and as a molecular target that could be useful in diagnosing scrub typhus infections.
Acknowledgments
We are grateful to Jere W. McBride and Madison Rogan (University of Texas Medical Branch, Galveston, TX, USA), Stacey D. Gilk (Indiana University School of Medicine, Indianapolis, IN, USA), and Elizabeth A. Rucks (University of Nebraska Medical Center, Omaha, NE, USA) for providing DNA isolated from host cells infected with E. chaffeensis, C. burnetii, or Chlamydia spp., respectively. This study was supported by the National Institutes of Health (grants AI123346 and AI128152 to Jason A. Carlyon), American Heart Association (grant 13GRNT16810009 to Jason A. Carlyon and grant 13PRE16840032 to Lauren VieBrock), the U.S. Military Infectious Diseases Research Program (grant A1230 to Allen L. Richards), and Virginia Commonwealth University (Presidential Research Quest Fund grant to Jason A. Carlyon). The views expressed in this presentation are those of the authors and do not necessarily represent the official policy or position of the Department of the Navy, Department of Defense, U.S. Government.
Supplementary Materials
The following are available online at http://www.mdpi.com/2414-6366/3/2/63/s1, Table S1: Nucleotide and amino acid identity values for O. tsutsugamushi OmpA sequences.
Author Contributions
J.A.C., S.M.E., L.V., A.L.R., and R.T.M. conceived and designed the experiments and analyses; S.M.E., H.E.A., L.V., A.L.-F., and S.C. performed the experiments; J.A.C., S.M.E., L.V., H.E.A., R.S.G., and J.A.C. analyzed the data; D.P., A.L.R., and J.J. contributed reagents and materials; J.A.C. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- 1.Bonell A., Lubell Y., Newton P.N., Crump J.A., Paris D.H. Estimating the burden of scrub typhus: A systematic review. PLoS Negl. Trop. Dis. 2017;11:e0005838. doi: 10.1371/journal.pntd.0005838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu G., Walker D.H., Jupiter D., Melby P.C., Arcari C.M. A review of the global epidemiology of scrub typhus. PLoS Negl. Trop. Dis. 2017;11:e0006062. doi: 10.1371/journal.pntd.0006062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasper M.R., Blair P.J., Touch S., Sokhal B., Yasuda C.Y., Williams M., Richards A.L., Burgess T.H., Wierzba T.F., Putnam S.D. Infectious etiologies of acute febrile illness among patients seeking health care in south-central Cambodia. Am. J. Trop. Med. Hyg. 2012;86:246–253. doi: 10.4269/ajtmh.2012.11-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suttinont C., Losuwanaluk K., Niwatayakul K., Hoontrakul S., Intaranongpai W., Silpasakorn S., Suwancharoen D., Panlar P., Saisongkorh W., Rolain J.M., et al. Causes of acute, undifferentiated, febrile illness in rural Thailand: Results of a prospective observational study. Ann. Trop. Med. Parasitol. 2006;100:363–370. doi: 10.1179/136485906X112158. [DOI] [PubMed] [Google Scholar]
- 5.Phongmany S., Rolain J.M., Phetsouvanh R., Blacksell S.D., Soukkhaseum V., Rasachack B., Phiasakha K., Soukkhaseum S., Frichithavong K., Chu V., et al. Rickettsial infections and fever, Vientiane, Laos. Emerg. Infect. Dis. 2006;12:256–262. doi: 10.3201/eid1202.050900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valbuena G., Walker D.H. Approaches to vaccines against Orientia tsutsugamushi. Front. Cell. Infect. Microbiol. 2012;2:170. doi: 10.3389/fcimb.2012.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly D.J., Richards A.L., Temenak J., Strickman D., Dasch G.A. The past and present threat of rickettsial diseases to military medicine and international public health. Clin. Infect. Dis. 2002;34:S145–S169. doi: 10.1086/339908. [DOI] [PubMed] [Google Scholar]
- 8.Taylor A.J., Paris D.H., Newton P.N. A systematic review of mortality from untreated scrub typhus (Orientia tsutsugamushi) PLoS Negl. Trop. Dis. 2015;9:e0003971. doi: 10.1371/journal.pntd.0003971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varghese G.M., Abraham O.C., Mathai D., Thomas K., Aaron R., Kavitha M.L., Mathai E. Scrub typhus among hospitalised patients with febrile illness in South India: Magnitude and clinical predictors. J. Infect. 2006;52:56–60. doi: 10.1016/j.jinf.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Rodkvamtook W., Kuttasingkee N., Linsuwanon P., Sudsawat Y., Richards A.L., Somsri M., Sangjun N., Chao C.C., Davidson S., Wanja E., et al. Scrub typhus outbreak in Chonburi Province, Central Thailand, 2013. Emerg. Infect. Dis. 2018;24:361–365. doi: 10.3201/eid2402.171172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sedhain A., Bhattarai G.R. Renal manifestation in scrub typhus during a major outbreak in central Nepal. Indian J. Nephrol. 2017;27:440–445. doi: 10.4103/ijn.IJN_133_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vivian Thangaraj J.W., Mittal M., Verghese V.P., Kumar C.P.G., Rose W., Sabarinathan R., Pandey A.K., Gupta N., Murhekar M. Scrub typhus as an etiology of acute febrile illness in Gorakhpur, Uttar Pradesh, India, 2016. Am. J. Trop. Med. Hyg. 2017;97:1313–1315. doi: 10.4269/ajtmh.17-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takhar R.P., Bunkar M.L., Arya S., Mirdha N., Mohd A. Scrub typhus: A prospective, observational study during an outbreak in Rajasthan, India. Natl. Med. J. India. 2017;30:69–72. [PubMed] [Google Scholar]
- 14.Tshokey T., Graves S., Tshering D., Phuntsho K., Tshering K., Stenos J. Scrub typhus outbreak in a remote primary school, Bhutan, 2014. Emerg. Infect. Dis. 2017;23:1412–1414. doi: 10.3201/eid2308.162021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Usha K., Kumar E., Kalawat U., Kumar B.S., Chaudhury A., Gopal D.V. Molecular characterization of Orientia tsutsugamushi serotypes causing scrub typhus outbreak in southern region of Andhra Pradesh, India. Indian J. Med. Res. 2016;144:597–603. doi: 10.4103/0971-5916.200886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marks M., Joshua C., Longbottom J., Longbottom K., Sio A., Puiahi E., Jilini G., Stenos J., Dalipanda T., Musto J. An outbreak investigation of scrub typhus in Western Province, Solomon Islands, 2014. West. Pac. Surveill. Response J. 2016;7:6–9. doi: 10.5365/wpsar.2015.6.3.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jakharia A., Borkakoty B., Biswas D., Yadav K., Mahanta J. Seroprevalence of scrub typhus infection in Arunachal Pradesh, India. Vector Borne Zoonotic Dis. 2016;16:659–663. doi: 10.1089/vbz.2016.1970. [DOI] [PubMed] [Google Scholar]
- 18.Harris P.N.A., Oltvolgyi C., Islam A., Hussain-Yusuf H., Loewenthal M.R., Vincent G., Stenos J., Graves S. An outbreak of scrub typhus in military personnel despite protocols for antibiotic prophylaxis: Doxycycline resistance excluded by a quantitative PCR-based susceptibility assay. Microbes Infect. 2016;18:406–411. doi: 10.1016/j.micinf.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Park J.H. Recent outbreak of scrub typhus in North Western part of India. Indian J. Med. Microbiol. 2016;34:114. doi: 10.4103/0255-0857.167682. [DOI] [PubMed] [Google Scholar]
- 20.Sharma R., Krishna V.P., Manjunath, Singh H., Shrivastava S., Singh V., Dariya S.S., Soni M., Sharma S. Analysis of two outbreaks of scrub typhus in Rajasthan: A clinico-epidemiological study. J. Assoc. Physicians India. 2014;62:24–29. [PubMed] [Google Scholar]
- 21.Hu J., Tan Z., Ren D., Zhang X., He Y., Bao C., Liu D., Yi Q., Qian W., Yin J., et al. Clinical characteristics and risk factors of an outbreak with scrub typhus in previously unrecognized areas, Jiangsu province, China 2013. PLoS ONE. 2015;10:e0125999. doi: 10.1371/journal.pone.0125999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L., Bi Z., Kou Z., Yang H., Zhang A., Zhang S., Meng X., Zheng L., Zhang M., Yang H., et al. Scrub typhus caused by Orientia tsutsugamushi Kawasaki-related genotypes in Shandong Province, northern China. Infect. Genet. Evol. 2015;30:238–243. doi: 10.1016/j.meegid.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 23.Wei Y., Luo L., Jing Q., Li X., Huang Y., Xiao X., Liu L., Wu X., Yang Z. A city park as a potential epidemic site of scrub typhus: A case-control study of an outbreak in Guangzhou, China. Parasites Vectors. 2014;7:513. doi: 10.1186/s13071-014-0513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weitzel T., Dittrich S., Lopez J., Phuklia W., Martinez-Valdebenito C., Velasquez K., Blacksell S.D., Paris D.H., Abarca K. Endemic scrub typhus in South America. N. Engl. J. Med. 2016;375:954–961. doi: 10.1056/NEJMoa1603657. [DOI] [PubMed] [Google Scholar]
- 25.Horton K.C., Jiang J., Maina A., Dueger E., Zayed A., Ahmed A.A., Pimentel G., Richards A.L. Evidence of Rickettsia and Orientia infections among abattoir workers in Djibouti. Am. J. Trop. Med. Hyg. 2016;95:462–465. doi: 10.4269/ajtmh.15-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thiga J.W., Mutai B.K., Eyako W.K. High seroprevalence of antibodies against spotted fever and scrub typhus bacteria in patients with febrile Illness, Kenya. Emerg. Infect. Dis. 2015;21:688–691. doi: 10.3201/eid2104.141387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balcells M.E., Rabagliati R., Garcia P., Poggi H., Oddo D., Concha M., Abarca K., Jiang J., Kelly D.J., Richards A.L., et al. Endemic scrub typhus-like illness, Chile. Emerg. Infect. Dis. 2011;17:1659–1663. doi: 10.3201/eid1709.100960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groen J., Nur Y.A., Dolmans W., Ligthelm R.J., Osterhaus A.D. Scrub and murine typhus among Dutch travellers. Infection. 1999;27:291–292. [PubMed] [Google Scholar]
- 29.Osuga K., Kimura M., Goto H., Shimada K., Suto T. A case of tsutsugamushi disease probably contracted in Africa. Eur. J. Clin. Microbiol. Infect. Dis. 1991;10:95–96. doi: 10.1007/BF01964418. [DOI] [PubMed] [Google Scholar]
- 30.Kocher C., Jiang J., Morrison A.C., Castillo R., Leguia M., Loyola S., Ampuero J.S., Cespedes M., Halsey E.S., Bausch D.G., et al. Serologic evidence of scrub typhus in the Peruvian Amazon. Emerg. Infect. Dis. 2017;23:1389–1391. doi: 10.3201/eid2308.170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly D.J., Fuerst P.A., Ching W.M., Richards A.L. Scrub typhus: The geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin. Infect. Dis. 2009;48(Suppl. 3):S203–S230. doi: 10.1086/596576. [DOI] [PubMed] [Google Scholar]
- 32.Izzard L., Fuller A., Blacksell S.D., Paris D.H., Richards A.L., Aukkanit N., Nguyen C., Jiang J., Fenwick S., Day N.P., et al. Isolation of a novel Orientia species (O. chuto sp. nov.) from a patient infected in Dubai. J. Clin. Microbiol. 2010;48:4404–4409. doi: 10.1128/JCM.01526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janardhanan J., Trowbridge P., Varghese G.M. Diagnosis of scrub typhus. Expert Rev. Anti-Infect. Ther. 2014;12:1533–1540. doi: 10.1586/14787210.2014.974559. [DOI] [PubMed] [Google Scholar]
- 34.Paris D.H., Aukkanit N., Jenjaroen K., Blacksell S.D., Day N.P. A highly sensitive quantitative real-time PCR assay based on the groEL gene of contemporary Thai strains of Orientia tsutsugamushi. Clin. Microbiol. Infect. 2009;15:488–495. doi: 10.1111/j.1469-0691.2008.02671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D.M., Kim H.L., Park C.Y., Yang T.Y., Lee J.H., Yang J.T., Shim S.K., Lee S.H. Clinical usefulness of eschar polymerase chain reaction for the diagnosis of scrub typhus: A prospective study. Clin. Infect. Dis. 2006;43:1296–1300. doi: 10.1086/508464. [DOI] [PubMed] [Google Scholar]
- 36.Jiang J., Chan T.C., Temenak J.J., Dasch G.A., Ching W.M., Richards A.L. Development of a quantitative real-time polymerase chain reaction assay specific for Orientia tsutsugamushi. Am. J. Trop. Med. Hyg. 2004;70:351–356. [PubMed] [Google Scholar]
- 37.Furuya Y., Yoshida Y., Katayama T., Yamamoto S., Kawamura A., Jr. Serotype-specific amplification of Rickettsia tsutsugamushi DNA by nested polymerase chain reaction. J. Clin. Microbiol. 1993;31:1637–1640. doi: 10.1128/jcm.31.6.1637-1640.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Z., Miura K., Popov V.L., Kumagai Y., Rikihisa Y. Insights into the CtrA regulon in development of stress resistance in obligatory intracellular pathogen Ehrlichia chaffeensis. Mol. Microbiol. 2011;82:1217–1234. doi: 10.1111/j.1365-2958.2011.07885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Confer A.W., Ayalew S. The OmpA family of proteins: Roles in bacterial pathogenesis and immunity. Vet. Microbiol. 2013;163:207–222. doi: 10.1016/j.vetmic.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 40.Godlewska R., Wisniewska K., Pietras Z., Jagusztyn-Krynicka E.K. Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: Function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol. Lett. 2009;298:1–11. doi: 10.1111/j.1574-6968.2009.01659.x. [DOI] [PubMed] [Google Scholar]
- 41.Hebert K.S., Seidman D., Oki A.T., Izac J., Emani S., Oliver L.D., Jr., Miller D.P., Tegels B.K., Kannagi R., Marconi R.T., et al. Anaplasma marginale outer membrane protein A is an adhesin that recognizes sialylated and fucosylated glycans and functionally depends on an essential binding domain. Infect. Immun. 2017;85:e00968-16. doi: 10.1128/IAI.00968-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hillman R.D., Jr., Baktash Y.M., Martinez J.J. OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with α2β1 integrin. Cell. Microbiol. 2013;15:727–741. doi: 10.1111/cmi.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnan S., Prasadarao N.V. Outer membrane protein A and OprF: Versatile roles in Gram-negative bacterial infections. FEBS J. 2012;279:919–931. doi: 10.1111/j.1742-4658.2012.08482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ojogun N., Kahlon A., Ragland S.A., Troese M.J., Mastronunzio J.E., Walker N.J., Viebrock L., Thomas R.J., Borjesson D.L., Fikrig E., et al. Anaplasma phagocytophilum outer membrane protein A interacts with sialylated glycoproteins to promote infection of mammalian host cells. Infect. Immun. 2012;80:3748–3760. doi: 10.1128/IAI.00654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seidman D., Hebert K.S., Truchan H.K., Miller D.P., Tegels B.K., Marconi R.T., Carlyon J.A. Essential domains of Anaplasma phagocytophilum invasins utilized to infect mammalian host cells. PLoS Pathog. 2015;11:e1004669. doi: 10.1371/journal.ppat.1004669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Do E.J., Kim J.E., Park J.M., Lee K.M., Jung M.Y., Lee H.J., Cho H.W., Choi Y.J., Lee S.H., Park K.H., et al. Development of recombinant OmpA and OmpB proteins as diagnostic antigens for rickettsial disease. Microbiol. Immunol. 2009;53:368–374. doi: 10.1111/j.1348-0421.2009.00142.x. [DOI] [PubMed] [Google Scholar]
- 47.Fournier P.E., Roux V., Raoult D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Pt 3Int. J. Syst. Bacteriol. 1998;48:839–849. doi: 10.1099/00207713-48-3-839. [DOI] [PubMed] [Google Scholar]
- 48.Gracner M., Avsic-Zupanc T., Punda-Polic V., Dolinsek J., Bouyer D., Walker D.H., Zavala-Castro J.E., Bradaric N., Crocquet-Valdes P.A., Duh D. Comparative ompA gene sequence analysis of Rickettsia felis-like bacteria detected in Haemaphysalis sulcata ticks and isolated in the mosquito C6/36 cell line. Clin. Microbiol. Infect. 2009;15(Suppl. 2):265–266. doi: 10.1111/j.1469-0691.2008.02228.x. [DOI] [PubMed] [Google Scholar]
- 49.Jiao Y., Wen B., Chen M., Niu D., Zhang J., Qiu L. Analysis of immunoprotectivity of the recombinant OmpA of Rickettsia heilongjiangensis. Ann. N. Y. Acad. Sci. 2005;1063:261–265. doi: 10.1196/annals.1355.042. [DOI] [PubMed] [Google Scholar]
- 50.Beyer A.R., Rodino K.G., VieBrock L., Green R.S., Tegels B.K., Oliver L.D., Jr., Marconi R.T., Carlyon J.A. Orientia tsutsugamushi Ank9 is a multifunctional effector that utilizes a novel GRIP-like Golgi localization domain for Golgi-to-endoplasmic reticulum trafficking and interacts with host COPB2. Cell. Microbiol. 2017;19:e12727. doi: 10.1111/cmi.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayxay M., Castonguay-Vanier J., Chansamouth V., Dubot-Peres A., Paris D.H., Phetsouvanh R., Tangkhabuanbutra J., Douangdala P., Inthalath S., Souvannasing P., et al. Causes of non-malarial fever in Laos: A prospective study. Lancet Glob. Health. 2013;1:e46–e54. doi: 10.1016/S2214-109X(13)70008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paris D.H., Blacksell S.D., Nawtaisong P., Jenjaroen K., Teeraratkul A., Chierakul W., Wuthiekanun V., Kantipong P., Day N.P. Diagnostic accuracy of a loop-mediated isothermal PCR assay for detection of Orientia tsutsugamushi during acute scrub typhus infection. PLoS Negl. Trop. Dis. 2011;5:e1307. doi: 10.1371/journal.pntd.0001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blacksell S.D., Luksameetanasan R., Kalambaheti T., Aukkanit N., Paris D.H., McGready R., Nosten F., Peacock S.J., Day N.P. Genetic typing of the 56-kDa type-specific antigen gene of contemporary Orientia tsutsugamushi isolates causing human scrub typhus at two sites in north-eastern and western Thailand. FEMS Immunol. Med. Microbiol. 2008;52:335–342. doi: 10.1111/j.1574-695X.2007.00375.x. [DOI] [PubMed] [Google Scholar]
- 54.Chang W.H., Kang J.S., Lee W.K., Choi M.S., Lee J.H. Serological classification by monoclonal antibodies of Rickettsia tsutsugamushi isolated in Korea. J. Clin. Microbiol. 1990;28:685–688. doi: 10.1128/jcm.28.4.685-688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rights F.L., Smadel H.I., Jackson E.B. Studies on scrub typhus (tsutsugamushi disease). III. Heterogeneity of strains of R. tsutsugamushi, as demonstrated by cross-vaccination studies. J. Exp. Med. 1948;87:339–351. doi: 10.1084/jem.87.4.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watt G., Kantipong P., Jongsakul K., Watcharapichat P., Phulsuksombati D. Azithromycin activities against Orientia tsutsugamushi strains isolated in cases of scrub typhus in Northern Thailand. Antimicrob. Agents Chemother. 1999;43:2817–2818. doi: 10.1128/aac.43.11.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bengtson I.A. Apparent serological heterogeneity among strains of tsutsugamushi disease (scrub typhus) Public Health Rep. 1945;60:1483–1488. doi: 10.2307/4585496. [DOI] [PubMed] [Google Scholar]
- 58.Tamura A., Takahashi K., Tsuruhara T., Urakami H., Miyamura S., Sekikawa H., Kenmotsu M., Shibata M., Abe S., Nezu H. Isolation of Rickettsia tsutsugamushi antigenically different from Kato, Karp, and Gilliam strains from patients. Microbiol. Immunol. 1984;28:873–882. doi: 10.1111/j.1348-0421.1984.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 59.Elisberg B.L., Campbell J.M., Bozeman F.M. Antigenic diversity of Rickettsia tsutsugamushi: Epidemiologic and ecologic significance. J. Hyg. Epidemiol. Microbiol. Immunol. 1968;12:18–25. [PubMed] [Google Scholar]
- 60.Philip C.B., Woodward T.E., Sullivan R.R. Tsutsugamushi disease (scrub or mite-borne typhus) in the Philippine Islands during American re-occupation in 1944–45. Am. J. Trop. Med. Hyg. 1946;26:229–242. doi: 10.4269/ajtmh.1946.s1-26.229. [DOI] [PubMed] [Google Scholar]
- 61.Carley J.G., Doherty R.L., Derrick E.H., Pope J.H., Emanuel M.L., Ross C.J. The investigation of fevers in North Queensland by mouse inoculation, with particular reference to scrub typhus. Australas. Ann. Med. 1955;4:91–99. doi: 10.1111/imj.1955.4.2.91. [DOI] [PubMed] [Google Scholar]
- 62.Olson J.G., Bourgeois A.L. Rickettsia tsutsugamushi infection and scrub typhus incidence among Chinese military personnel in the Pescadores Islands. Am. J. Epidemiol. 1977;106:172–175. doi: 10.1093/oxfordjournals.aje.a112448. [DOI] [PubMed] [Google Scholar]
- 63.Taylor A., Sivarajah A., Kelly D.J., Lewis G.E., Jr. An analysis of febrile illnesses among members of the Malaysian Police Field Force. Mil. Med. 1986;151:442–445. doi: 10.1093/milmed/151.8.442. [DOI] [PubMed] [Google Scholar]
- 64.Shishido A., Ohtawara M., Tateno S., Mizuno S., Ogura M., Kitaoka M. The nature of immunity against scrub typhus in mice. I. The resistance of mice, surviving subcutaneous infection of scrub typhus rickettsia, to intraperitoneal reinfection of the same agent. Jpn. J. Med. Sci. Biol. 1958;11:383–399. doi: 10.7883/yoken1952.11.383. [DOI] [Google Scholar]
- 65.Nakayama K., Yamashita A., Kurokawa K., Morimoto T., Ogawa M., Fukuhara M., Urakami H., Ohnishi M., Uchiyama I., Ogura Y., et al. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 2008;15:185–199. doi: 10.1093/dnares/dsn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kelley L.A., Sternberg M.J. Partial protein domains: Evolutionary insights and bioinformatics challenges. Genome Biol. 2015;16:100. doi: 10.1186/s13059-015-0663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan T.C., Jiang J., Temenak J.J., Richards A.L. Development of a rapid method for determining the infectious dose (ID)50 of Orientia tsutsugamushi in a scrub typhus mouse model for the evaluation of vaccine candidates. Vaccine. 2003;21:4550–4554. doi: 10.1016/S0264-410X(03)00505-X. [DOI] [PubMed] [Google Scholar]
- 68.Sunyakumthorn P., Paris D.H., Chan T.C., Jones M., Luce-Fedrow A., Chattopadhyay S., Jiang J., Anantatat T., Turner G.D., Day N.P., et al. An intradermal inoculation model of scrub typhus in Swiss CD-1 mice demonstrates more rapid dissemination of virulent strains of Orientia tsutsugamushi. PLoS ONE. 2013;8:e54570. doi: 10.1371/journal.pone.0054570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noriea N.F., Clark T.R., Hackstadt T. Targeted knockout of the Rickettsia rickettsii OmpA surface antigen does not diminish virulence in a mammalian model system. MBio. 2015;6:e00323-15. doi: 10.1128/mBio.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riley S.P., Goh K.C., Hermanas T.M., Cardwell M.M., Chan Y.G., Martinez J.J. The Rickettsia conorii autotransporter protein Sca1 promotes adherence to nonphagocytic mammalian cells. Infect. Immun. 2010;78:1895–1904. doi: 10.1128/IAI.01165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cardwell M.M., Martinez J.J. The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect. Immun. 2009;77:5272–5280. doi: 10.1128/IAI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan Y.G., Cardwell M.M., Hermanas T.M., Uchiyama T., Martinez J.J. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell. Microbiol. 2009;11:629–644. doi: 10.1111/j.1462-5822.2008.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim D.M., Park G., Kim H.S., Lee J.Y., Neupane G.P., Graves S., Stenos J. Comparison of conventional, nested, and real-time quantitative PCR for diagnosis of scrub typhus. J. Clin. Microbiol. 2011;49:607–612. doi: 10.1128/JCM.01216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tantibhedhyangkul W., Wongsawat E., Silpasakorn S., Waywa D., Saenyasiri N., Suesuay J., Thipmontree W., Suputtamongkol Y. Use of multiplex real-time PCR to diagnose scrub typhus. J. Clin. Microbiol. 2017;55:1377–1387. doi: 10.1128/JCM.02181-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le Viet N., Laroche M., Thi Pham H.L., Viet N.L., Mediannikov O., Raoult D., Parola P. Use of eschar swabbing for the molecular diagnosis and genotyping of Orientia tsutsugamushi causing scrub typhus in Quang Nam province, Vietnam. PLoS Negl. Trop. Dis. 2017;11:e0005397. doi: 10.1371/journal.pntd.0005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Premaratna R., Blanton L.S., Samaraweera D.N., de Silva G.N., Chandrasena N.T., Walker D.H., de Silva H.J. Genotypic characterization of Orientia tsutsugamushi from patients in two geographical locations in Sri Lanka. BMC Infect. Dis. 2017;17:67. doi: 10.1186/s12879-016-2165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chao C.C., Belinskaya T., Zhang Z., Ching W.M. Development of recombinase polymerase amplification assays for detection of Orientia tsutsugamushi or Rickettsia typhi. PLoS Negl. Trop. Dis. 2015;9:e0003884. doi: 10.1371/journal.pntd.0003884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watthanaworawit W., Turner P., Turner C., Tanganuchitcharnchai A., Richards A.L., Bourzac K.M., Blacksell S.D., Nosten F. A prospective evaluation of real-time PCR assays for the detection of Orientia tsutsugamushi and Rickettsia spp. for early diagnosis of rickettsial infections during the acute phase of undifferentiated febrile illness. Am. J. Trop. Med. Hyg. 2013;89:308–310. doi: 10.4269/ajtmh.12-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Prakash J.A., Reller M.E., Barat N., Dumler J.S. Assessment of a quantitative multiplex 5′ nuclease real-time PCR for spotted fever and typhus group rickettsioses and Orientia tsutsugamushi. Clin. Microbiol. Infect. 2009;15(Suppl. 2):292–293. doi: 10.1111/j.1469-0691.2008.02242.x. [DOI] [PubMed] [Google Scholar]
- 81.Singhsilarak T., Leowattana W., Looareesuwan S., Wongchotigul V., Jiang J., Richards A.L., Watt G. Short report: Detection of Orientia tsutsugamushi in clinical samples by quantitative real-time polymerase chain reaction. Am. J. Trop. Med. Hyg. 2005;72:640–641. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.