Abstract
Targeting of the programmed cell-death 1 ligand 1 (PD-L1) signal pathway is a promising treatment strategy in several cancers. The purpose of this study was to evaluate the clinical significance of PD-L1 in patients with colon adenocarcinoma (COAD). A total of 240 patients who were diagnosed with COAD from The Cancer Genome Atlas (TCGA) RNA-sequencing data and another cohort for pair-matched COAD samples (n = 40) in tissue microarray (TMA) were enrolled in this study. The correlation of PD-L1 or miR-191-5p expression with clinicopathological features and prognosis in patients with COAD was further analyzed using TCGA data and TMA. The Cox proportional hazard regression model was used to evaluate the association of PD-L1 or miR-191-5p expression with overall survival (OS) and tumor recurrence in patients with COAD. The microRNAs (miRNAs) that target PD-L1 gene were identified by bioinformatics and Spearman correlation analysis. We found that PD-L1 expression was increased in COAD tissues and was correlated with poor survival and tumor recurrence in patients with COAD. The increased expression of PD-L1 was attributed to the dysregulation of miR-191-5p expression rather than its genetic or epigenetic alterations. Moreover, the expression of miR-191-5p presented the negative correlation with PD-L1 expression and acted as an independent prognostic factor of OS in patients with COAD. Therefore, PD-L1 may predict poor prognosis and is negatively associated with miR-191-5p expression in patients with COAD.
Keywords: colon adenocarcinoma, miR-191-5p, PD-L1, survival
Introduction
Despite the fact that steady reduction of colorectal cancer (CRC) incidence and mortality has been reported over the past few decades worldwide, CRC still ranks the first place in gastrointestinal malignant tumors and is the second cause of cancer-related deaths.1 The key factors in some signaling pathway are associated with the prognosis of CRC,2 such as programmed cell-death 1 ligand 1 (PD-L1),3 which has been reported as the therapeutic target in multiple malignancies.4 However, the independent role of PD-L1 in colon adenocarcinoma (COAD) tumorigenesis remains unclear. Therefore, more extensive studies are required to identify the prognostic predictor or therapeutic target of COAD, providing effective anti-cancer breakthroughs.
Programmed cell-death protein 1 (PD-1) and its cognate ligand PD-L1 regulate CD8+ cytotoxic T lymphocytes activation upon T-cell receptor and induce this target-cell lysis.5 In this way, PD-L1/PD-1 signaling functions as a co-receptor pathway6 or immune checkpoint, inhibiting the immune system in cancer.5 It is worth noting that PD-L1 is not only related to T-cell responses and tumor microenvironment in vitro and in vivo6–8 but also leveraged by B-cell lymphomas so as to escape from immune recognition.5 Furthermore, PD-L1 is considered as one of the most important cancer therapeutics in multiple clinical experiments, which evidence that its inhibitors, including pembrolizumab, atezolizumab, and nivolumab,9–13 harbor a remarkable therapeutic efficacy in malignancies such as melanoma,9,10 non-small cell lung cancer (NSCLC),11,12 and advanced urothelial carcinoma.13 Therefore, PD-L1 acts a critical role in cancer progression and represents a novel therapeutic target.
MicroRNAs (miRNAs) as single-stranded small non-coding RNAs of ~22 nucleotides regulate gene expression through mediating translational inhibition or promoting degradation of target mRNAs in a wide range of biologic processes14 including gastrointestinal tumors.15 Accumulating evidence shows that dysregulation of miRNAs is involved in multiple cellular processes of CRC, such as tumorigenesis,16 cancer cell viability, apoptosis, and invasion.17 Moreover, miRNAs act as the key regulators of PD-L1 in multiple malignancies.18 MiR-138 represses tumor metastasis in CRC by targeting PD-L1,17 and miR-200 targets PD-L1 gene to suppress lung cancer metastasis by causing CD8+ T-cell immunosuppression.19 These data uncover the regulation link between PD-L1 and miRNAs in cancer.
Although some studies demonstrate that PD-L1 expression is observed in 433 mismatch repair-proficient CRC with strong positivity,20 the clinical significance of PD-L1 and its regulation by miRNAs remain elusive in COAD. In this study, The Cancer Genome Atlas (TCGA) data and tissue microarray (TMA) analysis indicated that increased expression of PD-L1 was correlated with overall survival (OS) in patients with COAD. Taking into account no significant alterations of PD-L1 in genetic and epigenetic modulation, we revealed that the dysregulation of miRNAs as post-transcriptional regulation might lead to the upregulation of PD-L1 in COAD. Furthermore, miR-191-5p was identified to have negative correlation with PD-L1 expression and acted as an independent prognostic factor of OS in patients with COAD.
Materials and methods
Clinical data
The clinicopathological and prognostic data for 240 cases of COAD patients as well as the relative expression levels of PD-L1 and miRNAs (has-miR-191-5p, has-miR-181a-5p, has-miR-181b-5p, has-miR-181c-5p, has-miR-20b-5p, has-miR-23b-3p, has-miR-497-5p, has-miR-363-3p, has-miR-15b-5p, has-miR-145-5p, has-miR-129-5p,has-miR-542-3p, has-miR-20a-5p, has-miR-424-5p, has-miR-106b-5p, has-miR-17-5p, has-miR-15a-5p, has-miR-33a-5p, has-miR-26b-5p, has-miR-136-5p, has-miR-27a-3p, has-miR-194-5p, has-miR-181d-5p, has-miR-224-5p, has-miR-106a-5p, has-miR-103a-3p, has-miR-128-3p, has-miR-27b-3p, has-miR-431-5p, and has-miR-93-5p) were downloaded from TCGA 2015 RNA-sequencing database (https://genome-cancer.ucsc.edu). The protocols used in our study were approved by the Ethics Committee of Shanghai Sixth People’s Hospital. The clinicopathological characteristics of COAD patients for PD-L1 expression were summarized in Supplementary Table S1.
TMA and immunohistochemistry analysis
The TMA of COAD was provided by our gastroenterology lab of Shanghai Sixth People’s Hospital, including 40 COAD samples and corresponding adjacent normal tissues. The protocols used in our study were approved by the Ethics Committee of Shanghai Sixth People’s Hospital. The expression and cellular localization of PD-L1 in COAD tissue cells were determined by immunohistochemistry (IHC) analysis. The COAD tissues were stained for anti-PD-L1 (ab205921, Rabbit monoclonal antibody, Abcam, Cambridge, MA, USA). The procedure of IHC analysis and the quantification of PD-L1 expression were conducted as previously described.21
Identification of miRNAs in cancer tissues
The miRNAs that target 3′UTR of PD-L1 gene were identified in cancer tissues using the StarBase v2.0 (http://starbase.sysu.edu.cn) under the strictly incorporated conditions with cancer type (⩾2) and forecasting software (TargetScan 7.1 and microRNA.org).
Cell culture
Human colon cancer cell lines LoVo, RKO (lowly differentiated colon cancer cell line), HCT116, Caco-2, and normal human colon epithelial cells NCM460 were stored at Digestive Disease Laboratory of Shanghai Sixth People’s Hospital and cultured in Dulbecco’s Modified Eagle medium (DMEM) medium added with 10% heat-inactivated fetal bovine serum (FBS), 100 U/mL of penicillin, and 100 μg/mL of streptomycin (HyClone) in a humidified atmosphere containing 5% CO2 at 37°C.
Quantitative real-time polymerase chain reaction
Total RNA of cell lines was extracted using TRIzol and used to synthesize complementary DNA (cDNA) by reverse transcription through M-MLV (an RNA-dependent DNA polymerase that can be used in cDNA synthesis with long messenger RNA templates) and cDNA amplification using the Power SYBR Green Master Mix kit (Takara, Otsu, Japan). In addition, total RNA for miRNAs was collected using a High Pure miRNA isolation kit (Roche) and real-time polymerase chain reaction (RT-PCR) using a TaqMan MicroRNA Reverse Transcription kit (Life Technologies, Carlsbad, CA, USA). A miScript Primer Assay (QIAGEN) was used for the miR-191-5p and U6. Data were analyzed using the comparative Ct method (2–ΔΔCt). The procedure of quantification of PD-L1 expression in RT-PCR was conducted as previously described.21 The primers are listed in Supplementary Table S2.
MiR-191-5p mimic and inhibitor
miR-191-5p mimic and inhibitor were purchased from Genepharma (Shanghai, China), and the empty vector was used as a control. LoVo and RKO cells were planted in 6-well plates 24 h prior to miR-191-5p mimic or inhibitor transfection with 50%–70% confluence and then were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacture instructions.
Statistical analysis
Statistical analyses were conducted by SPSS 20.0 (SPSS, IBM, Chicago, IL, USA) and GraphPad Prism 7.0. Student’s t-test or chi-square test was used to evaluate the statistical significance for the comparisons of two groups. Pearson’s correlation coefficient analysis was used to analyze the correlations of PD-L1 with miRNAs. OS was defined as the interval between the dates of surgery and death, and OS as well as recurrence curves was analyzed with the Kaplan–Meier method and log-rank test. Univariate analysis and multivariate models were performed using a Cox proportional hazards regression model. P < 0.05 was considered statistically significant.
Results
Upregulation of PD-L1 expression is correlated with poor prognosis in patients with COAD
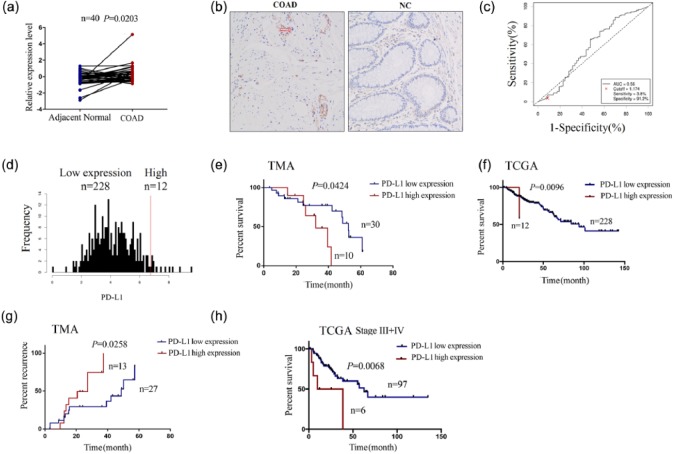
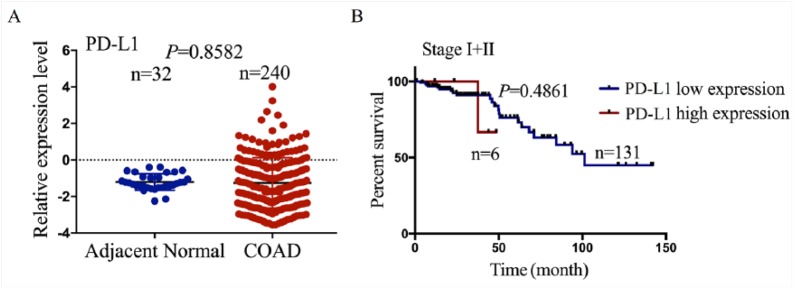
In consistence with previous studies indicating the upregulation of PD-L1 expression in human breast cancer,22 our results illustrated that the expression level of PD-L1 was increased in COAD samples (n = 40) as compared with pair-matched adjacent normal tissues using TMA analysis (Figure 1(a)), while it showing no difference between the patients of unpair-matched COAD samples and adjacent normal tissues (Supplementary Figure S1A). IHC analysis showed that PD-L1 was mainly localized in the cytoplasm, and its positive rate was much higher in COAD tissues than the adjacent normal tissues (P < 0.05, Figure 1(b)). These findings suggested that elevation of PD-L1 expression was a frequent event in COAD.
Figure 1.
Increasing expression of PD-L1 predicts poor survival and recurrence in patients with COAD. (a) TMA analysis of the differentially expressed level of PD-L1 in pair-matched tumor tissues. (b) IHC analysis of the expression level and cellular localization of PD-L1 in COAD tissue cells (200×). (c) The cut-off value and AUC of PD-L1 were assessed in COAD samples (n = 240). (d) The patients with COAD were divided into PD-L1 high expression group and low expression group. (e) Kaplan–Meier analysis of the link of PD-L1 high or low expression with the OS in patients with COAD from TMA data. (f) Kaplan–Meier analysis of the link of PD-L1 high or low expression with the OS in patients with COAD from TCGA data. (g) Kaplan–Meier analysis of the link of PD-L1 high or low expression with the tumor recurrence in patients with COAD from TMA data. (h) Kaplan–Meier plotter analysis of the correlation of PD-L1 high or low expression with the OS of COAD patients with stage III + IV.
Moreover, according to the OS time, OS status, and PD-L1 expression level, we acquired the cut-off value (1.174) of PD-L1 (Figure 1(c)) in COAD samples (n = 240) in TCGA sequencing data and divided the patients into two groups: PD-L1 high expression and PD-L1 low expression (Figure 1(d)). Receiver operating characteristic (ROC) curve and AUC were used to calculate the potential marker of PD-L1 expression, which clarified that the AUC of PD-L1 was 0.56 (Figure 1(c)), indicating that PD-L1 expression might be a potential marker for COAD patients. As shown in Table 1, we further analyzed the correlation of PD-L1 expression with the clinicopathological characteristics and prognosis in COAD patients and found that PD-L1 high expression was associated with the lymphatic invasion (P = 0.0310), but had no correlation with the other factors such as age, gender, pathological stage, and histological type (each P > 0.05). Kaplan–Meier analysis revealed that the patients with PD-L1 high expression were accompanied with shorter OS time (Figure 1(e) and (f)) and higher tumor recurrence (Figure 1(g)) as compared with those with PD-L1 low expression both in TMA analysis and TCGA data. Besides, the patients of late stage (stage III + IV) with PD-L1 high expression possessed poor OS compared with those with PD-L1 low expression (Figure 1(h)), while the patients of early stage (stage I + II) showed no difference between them (Supplementary Figure S1B). Univariate Cox regression analysis displayed that PD-L1 expression, pathologic stage, and lymphatic invasion were associated with OS of COAD patients. Multivariate analyses were further conducted by a Cox proportional hazard model, which showed that pathologic stage rather than PD-L1 expression was an independent prognostic factor for OS of COAD patients (Supplementary Table S3).
Table 1.
Correlation of PD-L1 expression with clinicopathological parameters of COAD patients.
| Variables | Cases (n) | PD-L1 expression | P-value | |
|---|---|---|---|---|
| Low (n = 228) | High (n = 12) | |||
| Age | ||||
| ⩽60 | 86 | 84 (97.7%) | 2 (2.3%) | 0.1563 |
| >60 | 154 | 144 (93.5%) | 10 (6.5%) | |
| Gender | ||||
| Female | 112 | 105 (93.8%) | 7 (6.2%) | 0.4727 |
| Male | 128 | 123 (96.1%) | 5 (3.9%) | |
| Pathologic stage | ||||
| I–II | 137 | 131 (95.6%) | 6 (4.4%) | 0.6110 |
| III–IV | 103 | 97 (94.2%) | 6 (5.8%) | |
| Lymphatic invasion | ||||
| No | 167 | 162 (97.0%) | 5 (3.0%) | 0.0310 |
| Yes | 73 | 66 (90.4%) | 7 (9.6%) | |
| Histological type | ||||
| CMA | 33 | 30 (90.9%) | 3 (9.1%) | 0.2456 |
| CA | 207 | 198 (95.7%) | 9 (4.4%) | |
PD-L1: programmed cell-death 1 ligand 1; COAD: colon adenocarcinoma; CMA: colon mucinous adenocarcinoma; CA: colon adenocarcinoma.
PD-L1 exhibits a negative correlation with miR-191-5p expression in CRC samples
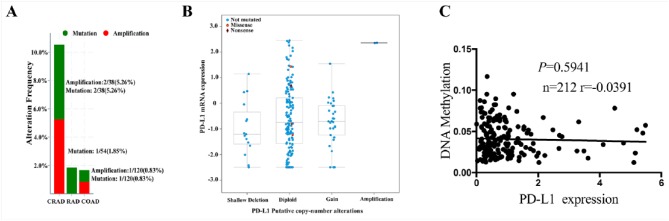
To uncover the reason for PD-L1 upregulation in CRC, we assessed the genetic or epigenetic dysregulation of PD-L1 expression in CRC (n = 212) through cBioPortal for Cancer Genomics,23,24 and it was indicated that PD-L1 had no significant alterations at the genetic (Supplementary Figure S2A and B) and methylation levels (Supplementary Figure S2C), and gene amplification, mutation, copy number, and methylation modification could not account for the upregulation of PD-L1 in CRC.
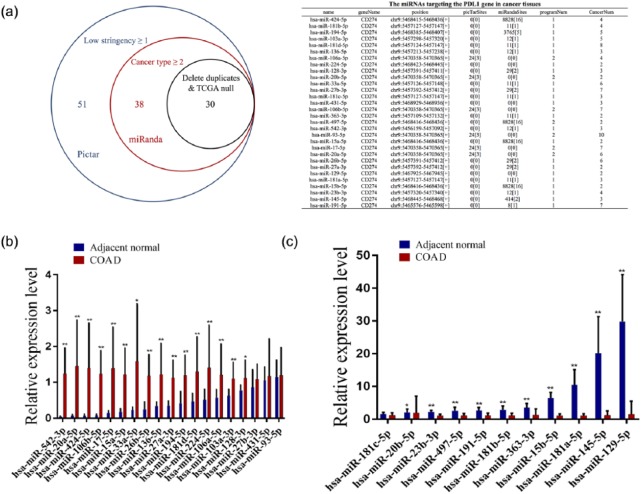
In addition, it is known that miRNAs act by negatively regulating their target genes.25 To illuminate whether PD-L1 expression is regulated by miRNAs, the prediction tools microRNA.org and TargetScan 7.1 were applied to identify the potential miRNAs that target 3′UTR of PD-L1 gene, of which 30 miRNAs have the potential to bind to the 3′UTR of PD-L1 (Figure 2(a)). Then, we assessed the expression levels of these 30 miRNAs in COAD samples, of which the expression levels of 20 miRNAs were increased (Figure 2(b)), but those of 10 miRNAs were substantially decreased in total colon samples as compared with the normal tissues (Figure 2(c)).
Figure 2.
The expression levels of miRNAs that targeting PD-L1 gene in COAD samples: (a) bioinformatic software identification of the 30 miRNAs that target 3′UTR of PD-L1 in cancer tissues. (b, c) TCGA cohort analysis of the differentially expressed levels of these 30 miRNAs in the total tumor tissues. Blue: adjacent normal, Red: COAD, **P < 0.001, and *P < 0.05.
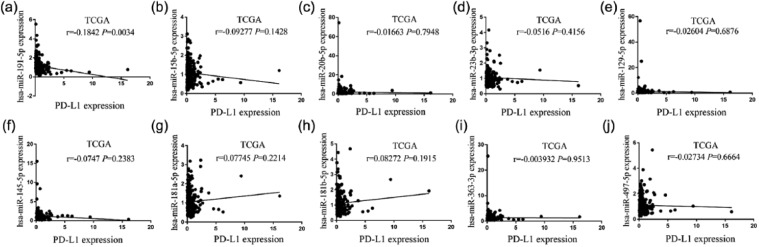
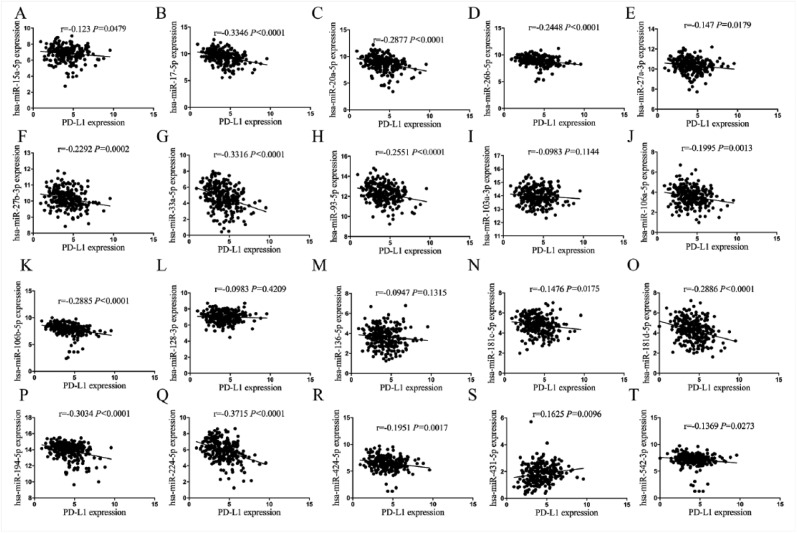
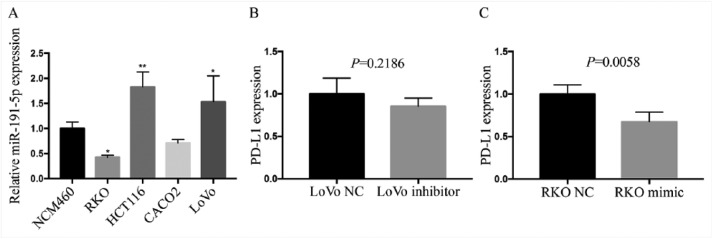
Furthermore, our results showed that among these miRNAs, spearman correlation analysis revealed the association between PD-L1 expression and these 10 downregulated miRNAs (Figures 3(a)–(j)) or other 20 upregulated miRNAs (Supplementary Figure S3) in COAD. These findings suggested that the expression status of PD-L1 varied and was influenced by the different miRNAs in COAD. Only miR-191-5p indicated the significant negative correlation with upregulated PD-L1 expression in COAD samples (r = −0.1842, P = 0.0034, Figure 3(a)). Besides, we first detected the expression levels of miR-191-5p in colon cancer cell lines by quantitative real-time PCR (qRT-PCR), indicating that miR-191-5p possessed higher expression level in LoVo cells and lower expression level in RKO cells compared with the NCM460 (Supplementary Figure S4A). Then, miR-191-5p mimic (100 μM) or inhibitor (100 μM) was respectively transfected into RKO cells with miR-191-5p low expression and LoVo cells with miR-191-5p high expression. After transfection for 48 h, it was revealed by qRT-PCR demonstrated that miR-191-5p overexpression substantially reduced the expression level of PD-L1 in RKO cells, while miR-191 knockdown did not increase its expression in LoVo cells in vitro (Supplementary Figure S4B and C). Although some studies indicated that miR-191-5p expression was reported increased in CRC tissues (n=24),26 our analysis was performed based on a larger size TMA data (n=40), indicating that miR-191-5p displayed a negative correlation with PD-L1 expression in COAD samples and was selected for further investigation.
Figure 3.
Pearson correlation analysis of the correlation of PD-L1 expression with these 10 downregulated miRNAs in COAD samples: (a) Pearson correlation analysis of the correlation of PD-L1 expression with miR-191-5p in COAD samples. (b–j) Pearson correlation analysis of the correlation of PD-L1 expression with other nine downregulated miRNAs in COAD samples.
Decreased expression of miR-191-5p was associated with OS and tumor recurrence in patients with COAD
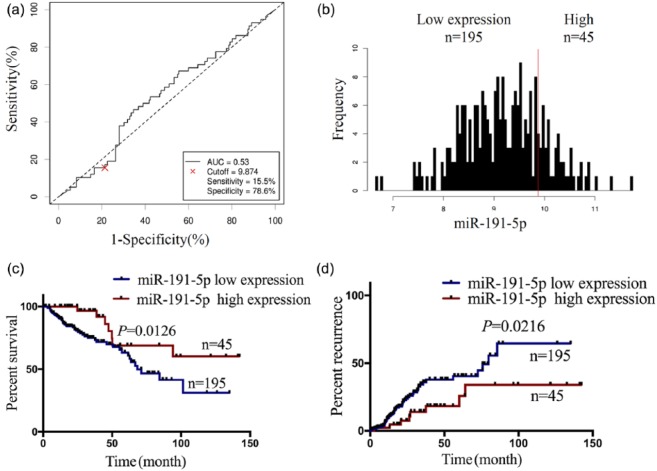
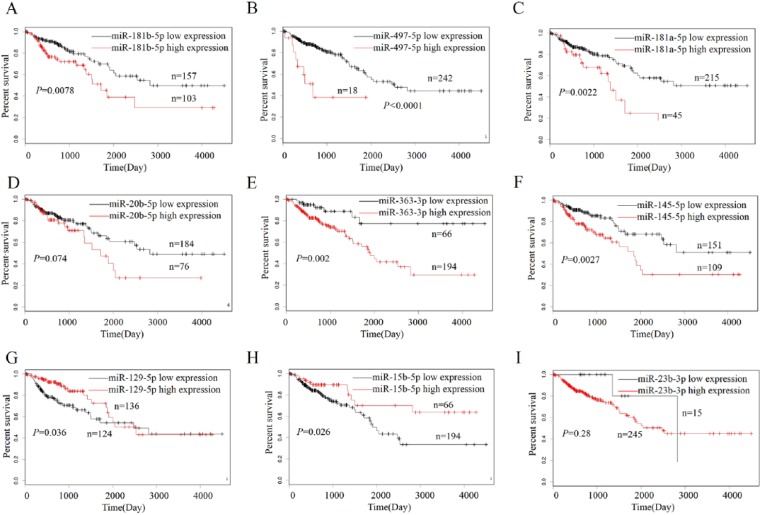
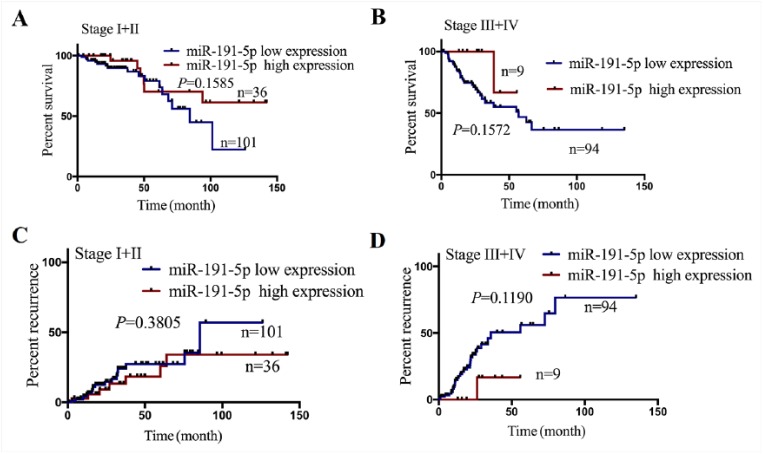
Having defined the decreased expression of miR-191-5p (Figure 2(c)) and its negative correlation with PD-L1 expression (Figure 3(a)) in COAD samples, we further analyzed the relationship between its expression and clinicopathological features or prognosis in patients with COAD. According to the OS time, OS status, and miR-191-5p expression level, the cut-off value (9.874) of miR-191-5p (Figure 4(a)) in colon samples (n = 240) was determined, by which the patients were classified into two groups: miR-191-5p high expression and miR-191-5p low expression (Figure 4(b)). ROC curve was drawn to assess the sensitivity and specificity of miR-191-5p in COAD, which indicated that the AUC, sensitivity, and specificity of miR-191-5p were, respectively, 0.53, 15.5%, and 78.6% (Figure 4(a)), suggesting that miR-191-5p might be a potential marker for COAD patients. Furthermore, we found that miR-191-5p low expression was associated with pathologic stage and (P = 0.001) and lymphatic invasion (P = 0.048), but had no correlation with the age, gender, and histological type (each P > 0.05, Table 2). Although Kaplan–Meier analysis referred that the patients with miR-191-5p, miR-129-5p, and miR-15b-5p low expression presented shorter survival (Figures 4(c), Supplementary Figure S5G and H), only the patients with miR-191-5p low expression indicated the significant negative correlation with PD-L1 expression in COAD samples and presented shorter survival (Figure 4(c)) and higher tumor recurrence (Figure 4(d)) as compared with those with miR-191-5p high expression. Nevertheless, the patients of early stage (stage I + II) or late stage (stage III + IV) with miR-191-5p low expression exhibited no difference in OS (Supplementary Figure S6A and B) and tumor recurrence (Supplementary Figure S6C and D) as compared with those with miR-191-5p high expression. Multivariate analyses illuminated that miR-191-5p expression as well as pathologic stage was an independent prognostic factor of OS in COAD patients (Table 3), but lymphatic invasion rather than miR-191-5p was an independent prognostic factor of tumor recurrence in COAD patients (Supplementary Table S4).
Figure 4.
Decreased expression of miR-191-5p was positively correlated with poor prognosis in patients with COAD. (a) The cut-off value, sensitivity, and specificity of miR-191-5p were determined in COAD samples (n = 240). (b) The patients with COAD were classified into miR-191-5p high expression (n = 45) or low expression group (n = 195). (c) Kaplan–Meier analysis of the association of miR-191-5p high or low expression with the OS in patients with COAD. (d) Kaplan–Meier analysis of the association of miR-191-5p high or low expression with the tumor recurrence in patients with COAD.
Table 2.
Correlation of miR-191-5p expression with clinicopathological parameters of COAD patients.
| Variables | Cases (n) | miR-191-5p expression | P-value | |
|---|---|---|---|---|
| Low (n = 195) | High (n = 45) | |||
| Age | ||||
| ⩽60 | 86 | 73 (84.9%) | 13 (15.1%) | 0.306 |
| >60 | 154 | 122 (79.2%) | 32 (20.8%) | |
| Gender | ||||
| Female | 112 | 85 (75.9%) | 27 (24.1%) | 0.067 |
| Male | 128 | 110 (85.9%) | 18 (14.1%) | |
| Pathologic stage | ||||
| I–II | 137 | 101 (73.7%) | 36 (26.3%) | 0.001 |
| III–IV | 103 | 94 (91.3%) | 9 (8.7%) | |
| Lymphatic invasion | ||||
| No | 167 | 130 (77.8%) | 37 (22.2%) | 0.048 |
| Yes | 73 | 65 (89.0%) | 8 (11.0%) | |
| Histological type | ||||
| CMA | 33 | 30 (90.9%) | 3 (9.1%) | 0.153 |
| CA | 207 | 165 (79.7%) | 42 (20.3%) | |
COAD: colon adenocarcinoma; CMA: colon mucinous adenocarcinoma; CA: colon adenocarcinoma.
Table 3.
Cox regression analysis of miR-191-5p as potential overall survival predictor.
| Variables | Univariate Cox
regression analysis |
Multivariate Cox
regression |
P-value | |
|---|---|---|---|---|
| RR (95% CI) | P-value | RR (95% CI) | ||
| Age (years) | ||||
| ⩽60 versus >60 | 0.807 (0.604–1.077) | 0.146 | ||
| Gender | ||||
| Male versus Female | 0.600 (0.350–1.028) | 0.060 | ||
| Pathologic stage | ||||
| I + II versus III + IV | 0.363 (0.212–0.619) | <0.001 | 0.403 (0.222–0.732) | 0.003 |
| Lymphatic invasion | ||||
| No versus Yes | 0.478 (0.281–0.814) | 0.007 | 0.765 (0.418–1.400) | 0.384 |
| Histological type | ||||
| CMA versus CA | 0.826 (0.405–1.687) | 0.600 | ||
| miR-191-5p expression | ||||
| Low versus High | 0.558 (0.350–0.890) | 0.014 | 0.571 (0.355–0.918) | 0.021 |
RR: relative risk; CI: confidence interval; CMA: colon mucinous adenocarcinoma; CA: colon adenocarcinoma.
Discussion
PD-L1 high expression has been demonstrated in multiple malignancies including NSCLC,27 and breast cancer,22 implicated in tumorigenesis,16 cell viability, apoptosis, and invasion,17 and was correlated with tumor unfavorable features. Consistent with these results in breast cancer22 and NSCLC,27 our study showed that PD-L1 expression was upregulated and positively correlated with lymphatic invasion, poor survival, and tumor recurrence in patients with COAD. However, multivariate analysis did not elucidate PD-L1 as an independent prognostic factor of OS in COAD patients. Although other studies claimed that high expressions of PD-1 and PD-L1 correlates with better prognosis of CRC patients,28 our results were based on analyzing the patients with COAD and sustained by another report, indicating that PD-L1 high expression is related to high-risk prognostic indicator and poor survival in breast cancer.29 These findings suggested that elevation of PD-L1 expression might be involved in COAD tumorigenesis.
We further elucidated the reasons for PD-L1 upregulation in COAD, of which several signaling pathways medate PD-L1 to participate in tumor progression. For example, PI3K/AKT pathway increases PD-L1 expression and inhibiton of this pathway enhances the anti-proliferative effect of interferon (IFN)-β in PD-L1+ lung adenocarcinoma.30 Besides, STAT3 signaling leads to the immunosuppression and tumor progression by upregulating PD-1/PD-L1 axis in head and neck squamous cell carcinoma31 and colitis-associated cancer.32 PD-L1 is a putative biomarker of miR-197/CKS1B/STAT3 axis, and miR-197 sensitizes PD-L1high drug-resistant NSCLC cells to chemotherapy.33 PD-L1 promoter methylation is also reported associated with survival in CRC,34 but we found that PD-L1 had no significant alterations in genetic and epigenetic dysregulation through cBioPortal for Cancer Genomics. Thus, we speculated that PD-L1 might be regulated by miRNAs from the post-transcriptional regulation. We attempted to predict 30 miRNAs that could target PD-L1 gene, of which miR-191-5p had the strong negative relevance with PD-L1 expression in COAD samples. Other studies show that PD-L1 is negatively modulated by miR-138, miR-200c and miR-34a in CRC,17 and acute myeloid leukemia (AML),18 respectively. These results implied that aberrant expression of PD-L1 might be caused by dysregulation of miRNAs in COAD. However, further investigation was needed to confirm the binding of miR-191-5p with PD-L1 and how miR-191-5p affects its expression in COAD cells.
Recently, a variety of studies have demonstrated that miR-191-5p can act as an oncogene or a tumor suppressor in cancer. However, some reports showed that the increased expression of miR-191-5p as the diagnostic marker predicted poor survival in patients with osteosarcoma,35 CRC,36 and AML.37 Overexpression of miR-191-5p facilitates proliferation, invasion, and migration and represses cell apoptosis in hepatocellular carcinoma38 and intrahepatic cholangiocarcinoma by targeting TET1-p53 pathway39 and promotes the epithelial–mesenchymal transition and cancer stem cell-like properties involved in metastatic properties of transformed bronchial epithelial cells.40 However, a few studies show that miR-191-5p suppresses cell proliferation, migration, and invasion and induces cell apoptosis in primary human fibroblasts41 and renal cell carcinoma.42 Moreover, it impairs tumorigenesis and metastasis in aggressive breast cancer43 and inhibits radiation-resistance in lung cancer.44 In our study, multivariate analysis revealed decreased expression of miR-191-5p was associated with poor survival and tumor recurrence in patients with COAD and acted as an independent prognostic factor for OS in COAD patients. Our findings regarding the prognosis and diagnostic potentials of miR-191-5p in COAD were supported by these studies in renal cell carcinoma, breast cancer, and lung cancer,41–44 but opposed by other studies in AML,37 hepatic cell carcinoma,38 and intrahepatic cholangiocarcinoma.39 Therefore, our and other studies indicate that miR-191-5p is a potential biomarker involved in the progression of COAD.
Taken together, PD-L1 expression was upregulated, but miR-191-5p expression was downregulated in COAD samples, and PD-L1 high expression and miR-191-5p low expression were correlated with the poor prognosis in patients with COAD. miR-191-5p was further identified to exhibit a negative correlation with PD-L1 expression and acted as an independent prognostic factor for OS of COAD patients.
Figure S1.
The expression of PD-L1 in patients with COAD and its correlation with overall survival (A) Differentially expressed level of PD-L1 in unpair-matched COAD tumor tissues and adjacent normal tissue. (B) Kaplan-Meier plotter analysis of the correlation of PD-L1 high or low expression with the overall survival in COAD patients with stage I+II.
Figure S2.
The genetic alteration frequency and the methylation levels of PD-L1 in CRC patients. (A) The genetic alteration frequency of PD-L1 amplification, and mutation in different pathological subtypes of CRC patients. CRAD: colorectal adenocarcinoma RAD: rectal adenocarcinoma COAD: colon adenocarcinoma (B) The correlation of PD-L1 gene expression with its putative copy number alterations in CRC samples. (C) The correlation of PD-L1 gene expression with its methylation level in CRC samples.
Figure S3. (A~T).
Pearson correlation analysis of the correlation of PD-L1 expression with these 20 upregulated miRNAs in COAD samples.
Figure S4.
RT-PCR analysis of effect of miR-191-5p on PD-L1 in colon cancer lines. (A) RT-PCR analysis of the expression level of miR-191-5p in colon cell lines and NCM460 cells. (B and C). RT- PCR analysis of the expression levels of miR-191-5p and PD-L1 after the transfection with miR-191-5p inhibitor in LoVo cells or miR-191-5p mimic in RKO cells. ** P < 0.001, * P < 0.05.
Figure S5. (A~I).
Kaplan-Meier analysis of the association of other downregulated miRNAs with the OS in patients with COAD.
Figure S6.
The association of miR-191-5p expression with the OS and tumor recurrence in COAD patients. (A and B) Kaplan-Meier plotter analysis of the correlation of miR-191-5p high or low expression with the tumor overall survival of COAD patients with stage I+II or stage III+IV. (C and D) Kaplan-Meier plotter analysis of the correlation of miR-191-5p high or low expression with the tumor recurrence of COAD patients with stage I+II or stage III+IV.
Table S1.
The clinicopathological data of COAD patients in TCGA.
| Variables | Case (n=240, 100%) |
|---|---|
| Age (years) | |
| ≥ 60 | 86 (35.83%) |
| < 60 | 154 (64.17%) |
| Gender | |
| Male | 112 (46.67%) |
| Female | 128 (53.33%) |
| Pathological stage | |
| I/II | 137 (57.08%) |
| III/IV | 103 (42.92%) |
| Lymphatic invasion | |
| No | 167 (69.58%) |
| Yes | 73 (30.42%) |
| Histological type | |
| CMA | 33 (13.75%) |
| CA | 207 (86.25%) |
CMA: Colon Mucinous Adenocarcinoma, CA: Colon Adenocarcinoma.
Table S2.
List of primers of the genes.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| PD-L1 | 5’-CTGTAGCACTGACATTCATCTTCCG-3’ | 5’-TGCCAATGCTGGATTACGTCTC-3’ |
| β-Actin | 5’-GTGGCCGAGGACTTTGATTG-3’ | 5’-CCTGTAACAACGCATCTCATATT-3’ |
| has-miR-191-5p | 5’- CAACGGAATCCCAAAAGCAGCTG-3’ | 5’-CAACGGAATCCCAAAAGCA -3’ |
| U6 | 5’-GCTTCGGCAGCACATATACTAAAAT-3’ | 5’-CGCTTCACGAATTTGCGTGTCAT-3’ |
Table S3.
Cox regression analysis of PD-L1 as potential overall survival predictor.
| Variables | Univariate Cox regression
analysis |
Multivariate Cox regression
|
||
|---|---|---|---|---|
| RR (95% CI) | P value | RR(95% CI) | P value | |
| Age(years) | ||||
| ≤60 vs. >60 | 1.414 (0.838 to 2.388) | 0.195 | NA | NA |
| Gender | ||||
| Male vs Female | 0.601 (0.350 to 1.031) | 0.065 | NA | NA |
| pathologic stage | ||||
| I+ II vs III+ IV | 0.398 (0.234 to 0.678) | 0.001 | 0.628(0.340 to 1.160) | 0.137 |
| lymphatic invasion | ||||
| No vs Yes | 0.355 (0.209 to 0.602) | < 0.001 | 0.465(0.256 to 0.844) | 0.012 |
| Histological type | ||||
| CMA vs CA | 0.966 (0.437 to 2.132) | 0.932 | NA | NA |
| PD-L1 expression | ||||
| Low vs High | 0.950(0.231 to 3.910) | 0.943 | NA | NA |
| miR-191-5p expression | ||||
| Low vs High | 2.367 (1.112 to 5.037) | 0.025 | 1.896(0.873 to 4.116) | 0.106 |
RR: relative risk, CMA: Colon Mucinous Adenocarcinoma, CA: Colon Adenocarcinoma.
Table S4.
Cox regression analysis of miR-191-5p as potential tumor recurrence predictor.
| Variables | Univariate Cox regression
analysis |
Multivariate Cox regression
|
||
|---|---|---|---|---|
| RR (95% CI) | P value | RR(95% CI) | P value | |
| Age(years) | ||||
| ≤60 vs. >60 | 0.807 (0.604 to 1.077) | 0.146 | NA | NA |
| Gender | ||||
| Male vs Female | 0.600 (0.350 to 1.028) | 0.060 | NA | NA |
| pathologic stage | ||||
| I+ II vs III+ IV | 0.363 (0.212 to 0.619) | < 0.001 | 0.480 (0.263 to0.876) | 0.017 |
| lymphatic invasion | ||||
| No vs Yes | 0.478 (0.281 to 0.814) | 0.007 | 0.715 (0.398 to 1.284) | 0.262 |
| Histological type | ||||
| CMA vs CA | 0.826 (0.405 to 1.687) | 0.600 | NA | NA |
| PDL1 expression | ||||
| Low vs High | 1.637 (1.095 to 2.446) | 0.016 | 1.397 (0.923 to 2.113) | 0.114 |
RR: relative risk, CMA: Colon Mucinous Adenocarcinoma, CA: Colon Adenocarcinoma.
Supplemental Material
Supplemental material, IJI790318__Supplementary_REV1 for Upregulation of PD-L1 predicts poor prognosis and is associated with miR-191-5p dysregulation in colon adenocarcinoma by Xiao-Yu Chen, Jing Zhang, Li-Dan Hou, Rui Zhang, Wei Chen, Hui-Ning Fan, Yan-Xia Huang, Hui Liu and Jin-Shui Zhu in International Journal of Immunopathology and Pharmacology
Acknowledgments
X-Y.C. and J.Z. contributed equally to this work.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval and consent to participate: This study was approved by the Hospital’s Protection of Human Subjects Committee.
Funding: This study was supported by grants from the National Natural Science Foundation of China (No. 81573747), Shanghai Science and Technology Commission Western Medicine Guide project (No. 17411966500), Shanghai Jiao Tong University School of Medicine Transformation Medicine and Innovation Center Research Project (No. TM201723), and Shanghai Jiao Tong University School of Medicine Doctoral Innovation Fund (No. BXJ201737).
References
- 1. Siegel RL, Miller KD, Jemal A. (2018) Cancer statistics, 2018. CA: A Cancer Journal for Clinicians 68(1): 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Frouws MA, van Herk-Sukel MPP, Maas HA, et al. (2017) The mortality reducing effect of aspirin in colorectal cancer patients: Interpreting the evidence. Cancer Treatment Reviews 55: 120–127. [DOI] [PubMed] [Google Scholar]
- 3. Hamada T, Cao Y, Qian ZR, et al. (2017) Aspirin use and colorectal cancer survival according to tumor CD274 (programmed cell death 1 Ligand 1) expression status. Journal of Clinical Oncology 35(16): 1836–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cancer Genome Atlas Research Network, Electronic address:wheeler@bcm.edu; Cancer Genome Atlas Research Network (2017) Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169(7): 1327.e23–1341.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goodman A, Patel SP, Kurzrock R. (2017) PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nature Reviews Clinical Oncology 14(4): 203–220. [DOI] [PubMed] [Google Scholar]
- 6. Freeman GJ, Long AJ, Iwai Y, et al. (2000) Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. Journal of Experimental Medicine 192(7): 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L. (2004) Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nature Reviews Immunology 4(5): 336–347. [DOI] [PubMed] [Google Scholar]
- 8. Zou W, Chen L. (2008) Inhibitory B7-family molecules in the tumour microenvironment. Nature Reviews Immunology 8(6): 467–477. [DOI] [PubMed] [Google Scholar]
- 9. Robert C, Schachter J, Long GV, et al. (2015) Pembrolizumab versus ipilimumab in advanced melanoma. New England Journal of Medicine 372(26): 2521–2532. [DOI] [PubMed] [Google Scholar]
- 10. Robert C, Long GV, Brady B, et al. (2015) Nivolumab in previously untreated melanoma without BRAF mutation. New England Journal of Medicine 372(4): 320–330. [DOI] [PubMed] [Google Scholar]
- 11. Rittmeyer A, Barlesi F, Waterkamp D, et al. (2017) Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet 389(10066): 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hellmann MD, Rizvi NA, Goldman JW, et al. (2017) Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): Results of an open-label, phase 1, multicohort study. The Lancet Oncology 18(1): 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bellmunt J, de Wit R, Vaughn DJ, et al. (2017) Pembrolizumab as second-line therapy for advanced urothelial carcinoma. New England Journal of Medicine 376(11): 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fabris L, Ceder Y, Chinnaiyan AM, et al. (2016) The potential of microRNAs as prostate cancer biomarkers. European Urology 70(2): 312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang J, Jin M, Chen X, et al. (2018) Loss of PPM1F expression predicts tumour recurrence and is negatively regulated by miR-590-3p in gastric cancer. Cell Proliferation. Epub ahead of print 22 February. DOI: 10.1111/cpr.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsuji S, Kawasaki Y, Furukawa S, et al. (2014) The miR-363-GATA6-Lgr5 pathway is critical for colorectal tumourigenesis. Nature Communication 5: 3150. [DOI] [PubMed] [Google Scholar]
- 17. Zhang X-L, Xu L-L, Wang F. (2017) Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biology International 41(9): 1056–1064. [DOI] [PubMed] [Google Scholar]
- 18. Pyzer AR, Stroopinsky D, Rosenblatt J, et al. (2017) MUC1 inhibition leads to decrease in PD-L1 levels via upregulation of miRNAs. Leukemia 31(12): 2780–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen L, Gibbons DL, Goswami S, et al. (2014) Metastasis is regulated via microRNA-200/ZEB1 axis control of tumor cell PD-L1 expression and intratumoral immunosuppression. Nature Communication 5: 5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Droeser RA, Hirt C, Viehl CT, et al. (2013) Clinical impact of programmed cell death ligand 1 expression in colorectal cancer. European Journal of Cancer 49(9): 2233–2242. [DOI] [PubMed] [Google Scholar]
- 21. Zhang J, Wang G, Chu S-J, et al. (2016) Loss of large tumor suppressor 1 promotes growth and metastasis of gastric cancer cells through upregulation of the YAP signaling. Oncotarget 7(13): 16180–16193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noman MZ, Janji B, Abdou A, et al. (2017) The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology 6(1): e1263412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cerami E, Gao J, Dogrusoz U, et al. (2012) The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discovery 2(5): 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gao J, Aksoy BA, Dogrusoz U, et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling 6(269): pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Treiber T, Treiber N, Meister G. (2012) Regulation of microRNA biogenesis and function. Thrombosis and Haemostasis 107(4): 605–610. [DOI] [PubMed] [Google Scholar]
- 26. Xi Y, Formentini A, Chien M, et al. (2006) Prognostic values of microRNAs in colorectal cancer. Biomarker Insights 2: 113–121. [PMC free article] [PubMed] [Google Scholar]
- 27. Hong S, Chen N, Fang W, et al. (2016) Upregulation of PD-L1 by EML4-ALK fusion protein mediates the immune escape in ALK positive NSCLC: Implication for optional anti-PD-1/PD-L1 immune therapy for ALK-TKIs sensitive and resistant NSCLC patients. Oncoimmunology 5(3): e1094598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Y, Liang L, Dai W, et al. (2016) Prognostic impact of programmed cell death-1 (PD-1) and PD-ligand 1 (PD-L1) expression in cancer cells and tumor infiltrating lymphocytes in colorectal cancer. Molecular Cancer 15(1): 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang C, Zhu H, Zhou Y, et al. (2017) Prognostic value of PD-L1 in breast cancer: A meta-analysis. Breast Journal 23(4): 436–443. [DOI] [PubMed] [Google Scholar]
- 30. Gao Y, Yang J, Cai Y, et al. (2018) IFN-γ-mediated inhibition of lung cancer correlates with PD-L1 expression and is regulated by PI3K-AKT signaling. International Journal of Cancer. Epub ahead of print 8 March. DOI: 10.1002/ijc.31357. [DOI] [PubMed] [Google Scholar]
- 31. Bu LL, Yu GT, Wu L, et al. (2017) STAT3 induces immunosuppression by upregulating PD-1/PD-L1 in HNSCC. Journal of Dental Research 96(9): 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grivennikov S, Karin E, Terzic J, et al. (2009) IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15(2): 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fujita Y, Yagishita S, Hagiwara K, et al. (2015) The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Molecular Therapy 23(4): 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goltz D, Gevensleben H, Dietrich J, et al. (2016) PD-L1 (CD274) promoter methylation predicts survival in colorectal cancer patients. Oncoimmunology 6(1): e1257454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang T, Ji F, Dai Z, et al. (2015) Increased expression of microRNA-191 as a potential serum biomarker for diagnosis and prognosis in human osteosarcoma. Cancer biomarkers: Section A of Disease Markers 15(5): 543–550. [DOI] [PubMed] [Google Scholar]
- 36. Qin S, Zhu Y, Ai F, et al. (2014) MicroRNA-191 correlates with poor prognosis of colorectal carcinoma and plays multiple roles by targeting tissue inhibitor of metalloprotease 3. Neoplasma 61(1): 27–34. [PubMed] [Google Scholar]
- 37. Garzon R, Volinia S, Liu C-G, et al. (2008) MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 111(6): 3183–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elyakim E, Sitbon E, Faerman A, et al. (2010) hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Research 70(20): 8077–8087. [DOI] [PubMed] [Google Scholar]
- 39. Li H, Zhou Z-Q, Yang Z-R, et al. (2017) MicroRNA-191 acts as a tumor promoter by modulating the TET1-p53 pathway in intrahepatic cholangiocarcinoma. Hepatology 66: 136–151. [DOI] [PubMed] [Google Scholar]
- 40. Xu W, Ji J, Xu Y, et al. (2015) MicroRNA-191, by promoting the EMT and increasing CSC-like properties, is involved in neoplastic and metastatic properties of transformed human bronchial epithelial cells. Molecular Carcinogenesis 54(S1): E148–E161. [DOI] [PubMed] [Google Scholar]
- 41. Polioudakis D, Abell NS, Iyer VR. (2015) MiR-191 regulates primary human fibroblast proliferation and directly targets multiple oncogenes. PLoS ONE 10(5): e0126535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen P, Pan X, Zhao L, et al. (2018) MicroRNA-191-5p exerts a tumor suppressive role in renal cell carcinoma. Experimental and Therapeutic Medicine 15(2): 1686–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Di Leva G, Piovan C, Gasparini P, et al. (2013) Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genetics 9(3): e1003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu Z, Huang S. (2015) Inhibition of miR-191 contributes to radiation-resistance of two lung cancer cell lines by altering autophagy activity. Cancer Cell International 15(1): 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, IJI790318__Supplementary_REV1 for Upregulation of PD-L1 predicts poor prognosis and is associated with miR-191-5p dysregulation in colon adenocarcinoma by Xiao-Yu Chen, Jing Zhang, Li-Dan Hou, Rui Zhang, Wei Chen, Hui-Ning Fan, Yan-Xia Huang, Hui Liu and Jin-Shui Zhu in International Journal of Immunopathology and Pharmacology