Abstract
Dysregulation of chromobox proteins contributes to the progression of human diseases. CBX1 has been implicated in epigenetic control of chromatin structure and gene expression, but its role in human cancers remains largely unknown. Here we show that CBX1 exhibits oncogenic activities in hepatocellular carcinoma (HCC) and indicates poor outcomes. The expression of CBX1 was noticeably increased, at both mRNA and protein levels, in HCC tissues and cell lines, compared with the nontumorous ones. High CBX1 expression was significantly associated with larger tumor size, poor tumor differentiation and tumor vascular invasion. Patients with elevated expression of CBX1 were frequently accompanied with unfavorable overall and disease-free survivals in two independent cohorts consisting of 648 HCC cases. The prognostic value of CBX1 was further confirmed by stratified survival analyses. Multivariate cox regression model suggested CBX1 as an independent factor for overall survival (hazard ratio = 1.735, 95% confident interval: 1.342–2.244, P < .001). In vitro data demonstrated that CBX1 overexpression promoted cell proliferation and migration, whereas the knockdown of CBX1 resulted in the opposite phenotypes. Mechanistically, CBX1 interacted with transcription factor HMGA2 to activate the Wnt/β-Catenin signaling pathway. Suppression of β-Catenin by siRNA or specific inhibitor XAV-939 markedly attenuated CBX1-mediated cell growth. Collectively, our findings indicate that CBX1 functions as an oncogene and may serve as a potential prognostic biomarker in HCC.
Introduction
The clinical outcomes of patients with hepatocellular carcinoma (HCC) have been barely improved, ranking the third most common cause of cancer-related death globally [1], despite new strategies have been applied to the clinic. Options officially available for HCC patients are limited to surgical resection and anti-angiogenic multikinase inhibitor sorafenib [2], which offer litter benefit to the overall survival. The poor 5-year survival rate (about 11%) highlights the need of detailed mechanism of HCC progression. Identification of potential biomarkers that are of clinical implications may provide novel insights for the strategy of HCC management.
Dysregulation of chromobox proteins contributes to the progression of human diseases. Chromobox 1 (CBX1), localized in 17q21.32 of chromosome, belongs to heterochromatin protein 1 (HP1) family that includes three members HP1α (CBX5), HP1β (CBX1), and HP1γ (CBX3) [3]. HP1 proteins were originally described in Drosophila as essential component of heterochromatin and manifest functions of stable epigenetic gene silencing [4]. CBX1 possesses a dimeric, quasi-symmetrical structure and contains a conserved amino terminal chromo-domain and a carboxy terminal chromoshadow-domain [5]. The expression of CBX1 mRNA was much higher in perinatal livers than adult livers [6]. Interestingly, CBX1 was found to play essential role in the maintenance of pluripotency in embryonic stem cells [7]. These data indicate that CBX1 may be involved in the regulation of cell differentiation and proliferation. Several chromatin-remodeling factors and transcriptional regulators, such as Retinoblastoma (Rb1) and HDGFRP2, have been identified as the interacting proteins of CBX1 to exert activities in cellular processes, such as transcriptional activation, chromosome segregation, telomere maintenance, DNA repair, and RNA splicing [8], [9], [10], [11]. It has been demonstrated that the expression of CBX1 was deregulated in human cancers. High expression of CBX1 mRNA in breast cancer was correlated with unfavorable relapse-free survival [12]. However, the role and clinical significance of CBX1 in HCC has not been disclosed.
Using tissue microarray-based immunohistochemistry and in vitro experiments, we showed that the expression of CBX1 was increased in HCC tissues and associated with poor outcomes. Overexpression of CBX1 resulted in the enhancement of HCC cell proliferation and migration via interacting with HMGA2 to trigger the Wnt/β-Catenin signaling pathway. These findings suggest CBX1 as a potential prognostic biomarker for HCC and an oncogene to promote HCC progression.
Materials and Methods
Patients
Archived paraffin-embedded samples of 276 patients with HCC were collected from January 2010 to December 2012 in Dongguan Third People's Hospital and The Affiliated Hexian Memorial Hospital of Southern Medical University, and named as DH cohort. The mean age of patients was 49 years old (ranging from 19 to 79). The median follow-up was 41.5 months. Another 97 paired samples with primary and metastatic tumors were obtained. Thirty-three pairs of fresh tissues were also subjected to the detection of CBX1 mRNA and protein expression. All patients received no chemotherapy or radiotherapy before surgery. The use of human samples was approved by the Research Ethics Committee of Southern Medical University. Other samples and information from The Cancer Genome Atlas (TCGA cohort) and the Oncomine dataset were recruited for the validation of the clinical implication of CBX1 via the following websites: http://www.cbioportal.org and https://www.oncomine.org.
Cell Culture and Transfection
Bel-7404 and Huh7 cells purchased from the Cell Resource Center, Chinese Academy of Science Committee (Shanghai, China), MHCC-97H obtained from the American Type Culture Collection (ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Gaithersburg, MD, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Hyclone, Logan, UT) in a humidified incubator at 37 °C and 5% CO2. The cells were transiently or stably transfected with CBX1 overexpression vector or siRNAs with Lipofectamine 2000, according to the instructions.
Quantitative Real-Time PCR (qRT-PCR)
qRT-PCR was performed to examine the mRNA expression of CBX1. Briefly, complementary DNA was synthesized from the total RNA, using the PrimeScript RT reagent Kit (TAKARA, USA). PCR was performed with SYBR Premix ExTaq (TAKARA, USA). The expression of the endogenous 18S was served as control. The −ΔCt method was applied for the expression of CBX1. Conditions for RT-PCR was set as the following: 95 °C for 10 min, 40 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s and a final extension of 10 min at 72 °C. The primers used in this study were as followings: CBX1 forward: 5′-TTCGAGGCGGAGGTTAGGAG-3′; CBX1 reverse: 5′-GCTCCCATGTGTTGTCCTCA-3′; β-actin, forward: 5′-TGGCACCCAGCACAATGAA-3′ and reverse: 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′. The siRNA for CBX1 was designed as CGGAAGAAGGGUUAUCACAAU.
Western Blot
Proteins extracted from HCC fresh tissue or cells with various treatments were fractionated by SDS-PAGE, transferred to PVDF membrane, and incubated with a primary specific antibody for CBX1 (1:1000, Abcam, ab10478), Phospho-β-Catenin (Ser552) (1:1000, Cell Signaling Technology, #5651), HMGA2 (1:1000, Cell Signaling Technology, #8179) in 5% of non-fat milk, followed by a horse radish peroxidase (HRP)-conjugated anti-rabbit second antibody. ECL detection reagent (Amersham Life Science, Piscataway, NJ, USA) was used to show the results.
Immunohistochemistry (IHC)
IHC staining for CBX1 was performed on a HCC tissue microarray. The expression levels were scored as proportion of immunopositive staining area (0%, 0; 1%-25%, 1; 26%-50%, 2; 51%-75%, 3; 76%-100%, 4) multiplied by intensity of staining (0, negative; 1, weak; 2, moderate; 3, intense). The scores were independently rendered by two pathologists (Dr. Yang YF and Dr. Su SG). The median IHC score (4.5) was chosen as the cut-off value for defining high and low expression.
MTT
Cells were cultured in 96-well plates for 5 days. 20 μl of MTT (5 mg/ml) was added into the wells for 3 h. The formazan crystals were dissolved in DMSO (150 μl/well). The absorbance at 490 nm of each sample was measured. The relative cell growth rate was calculated.
Colony Formation
Five hundred cells were incubated in 6-well plates with DMEM medium plus 500 mg/l G418. After 2 weeks, the colonies were fixed by 4% PFA and stained with 0.05% crystal violet solution for 10 min, washed with PBS, and counted.
Transwell Assay
Cells were cultured with 200 μl serum-free Dulbecco's modified Eagle's medium (DMEM) in the upper compartment of a Transwell chamber (Corning, MA) for 48 h. The migrated cells were stained with 0.1% crystal violet and counted under a microscope. The related fold changes of cell migration were shown by histogram.
Immunofluorescence (IF)
Cells were fixed for 20 min in PBS containing 4% paraformaldehyde, permeabilized in 0.1% Triton X-100 two times, 5 min each, incubated in blocking buffer (3% donkey serum in TBS) for 1 h, and then incubated with antibody for 2 h at room temperature. After washing in PBS three times, 8 min each, cells were incubated with the appropriate fluorochrome-conjugated secondary antibody for 1 h, and observed under a fluorescence microscope.
Coimmunoprecipitation (CoIP)
Proteins were extracted by radioimmunoprecipitation assay buffer supplemented with proteinase inhibitor cocktail. Primary antibodies were added for 2.5 h. Protein A/G beads were added for an additional 2 h. Precipitated proteins were dissolved in SDS loading buffer and fractionated by SDS-PAGE.
Statistical Analyses
Continuous variables were presented as a mean with SEM and analyzed by the Student t test (2-tailed). Kaplan–Meier analysis (log-rank test) was used for survival analysis and univariate analysis. The Cox proportional hazards regression model was used to evaluate the independent prognostic value of CBX1. A P value less than 0.05 was considered statistically significant.
Results
CBX1 Expression is Increased in HCC by qRT-PCR and Western Blot
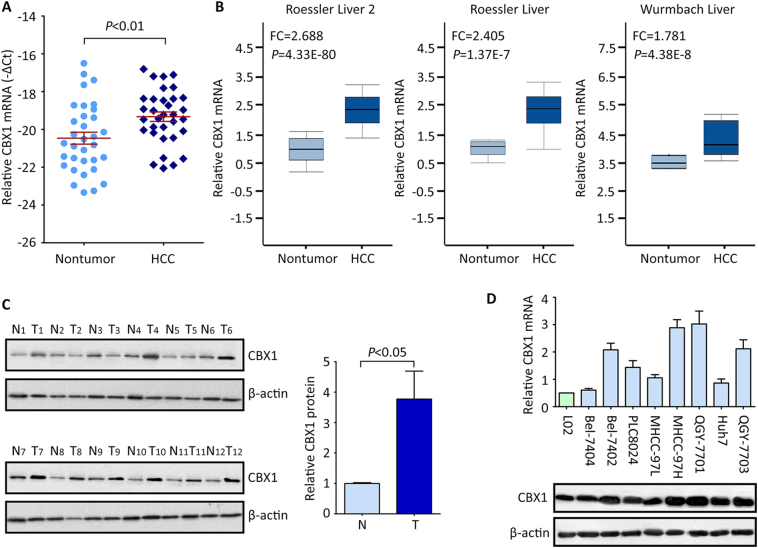
The mRNA expression of CBX1 in HCC tissues was determined by qRT-PCR. Results showed that CBX1 mRNA was up-regulated in 33 HCC tissues, compared to the nontumorous tissues (Figure 1A). This was validated in other studies (Roessler liver, Roessler liver 2 and Wurmbach liver) from Oncomine dataset (Figure 1B). In consistent, the protein level of CBX1 in HCC fresh tissues was on average 3.65-folded higher than that in nontumorous tissues (Figure 1C). 75.0% (9/12) of HCC cases were accompanied with obvious up-regulation of CBX1 protein. Furthermore, the mRNA and protein expressions of CBX1 were elevated in HCC cell lines, compared with those in the immortalized hepatic cell lines L02 (Figure 1D).
Figure 1.
CBX1 expression in fresh HCC tissues and cell lines by qRT-PCR and western blot. A. The mRNA expression of CBX1 in 33 pairs of fresh HCC and nontumor tissues (Wilcoxon matched paired test). B. The elevated expression of CBX1 mRNA was confirmed in other studies in Oncomine dataset. C. The protein level of CBX1 in 12 HCC (T) and nontumor (N) samples was determined by western blot. The statistical analysis was conducted to show the significant alternation of CBX1 in HCC tissues. D. CBX1 mRNA and protein expressions in immobilized liver cell line (L02) and HCC cell lines were examined.
CBX1 Expression is Increased in HCC by IHC
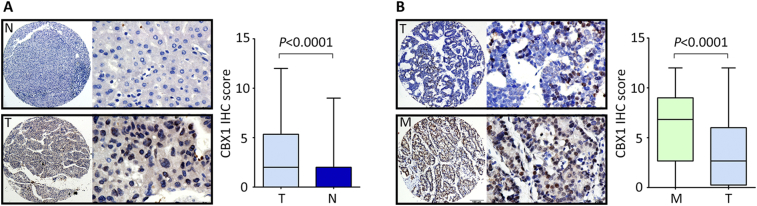
To confirm the expression profile of CBX1 in HCC, a cohort consisting of 276 archived paraffin-embedded HCC cases along with clinical and pathological information were collected to construct tissue microarray (TMA). Results of TMA-based IHC showed that CBX1 was mainly expressed in the nuclear of tumor and nontumor cells (Figure 2A). According to the IHC score, the overall expression of CBX1 was significantly increased in HCC tissues compared to nontumor tissues (Figure 2A). Positive staining of CBX1 in HCC and nontumor tissues were 64.9% (179/276) and 21.4% (59/276), respectively. Increased expression of CBX1 in HCC was observed in 57.6% (159/276) of cases, compared to the corresponding adjacent nontumorous tissues.
Figure 2.
CBX1 expression in paraffin-embedded tissues by IHC. A. The protein expression of CBX1 in 276 HCC samples (T) along with the corresponding adjacent nontumor tissues (N) was examined by IHC. Representative micrographs (left panel) were shown, and reproducibility of the CBX1 IHC score (right panel) in all patients was calculated by the Wilcoxon matched paired test. B. CBX1 expression in 97 HCC cases with tumor metastasis was determined by IHC. Representative photomicrographs (left panel) were shown for the primary tumor (T) and metastatic lesions (M). The IHC scores were presented as mean ± SEM.
To evaluate the expression of CBX1 in metastatic tissues, a cohort containing 97 HCC cases with portal vein embolus was obtained. Results presented that in 88.7% (86/97) of the samples, more immunoreactivity of CBX1 was found in embolus metastases, compared to the primary tumors (Figure 2B). The positive rate of CBX1 staining was 93.8% (91/97) in metastatic tissues. Collectively, these data suggest that the expression of CBX1 was steadily increased in consistent with HCC progression.
Correlation of CBX1 Expression and Prognosis of Patients with HCC
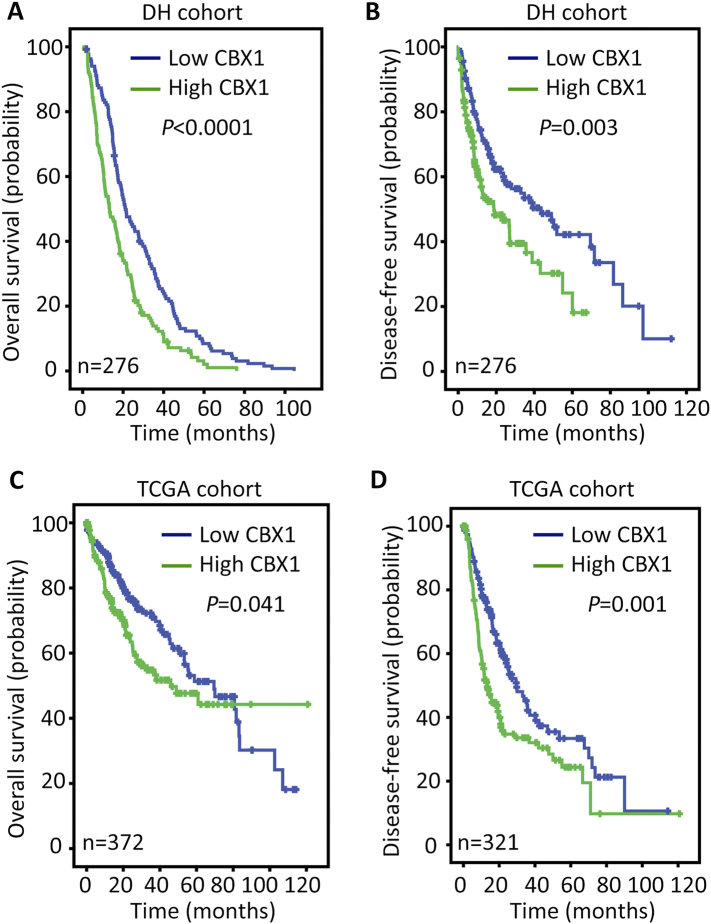
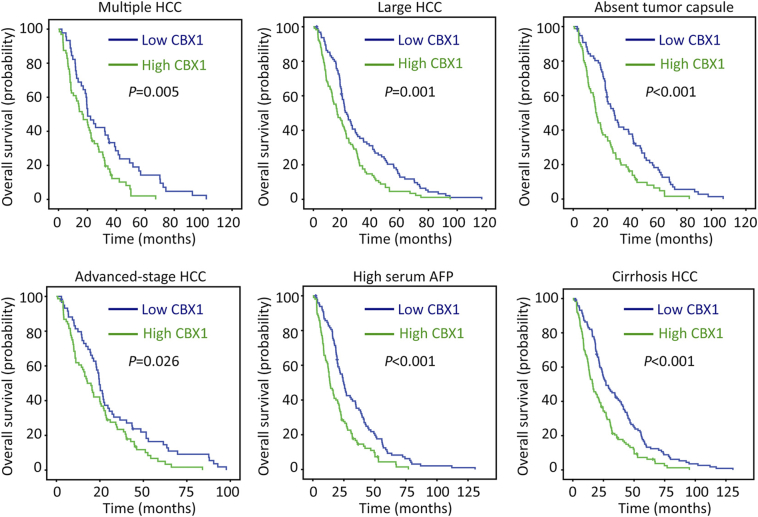
The clinical significance of CBX1 in HCC was next determined. According to the median of CBX1 IHC score (4.5), patients were separated into two groups: high CBX1 expression and low CBX1 expression. Statistical analyses indicated the patients with high CBX1 expression were frequently accompanied with larger tumor, poor differentiated tumor, more tumor lesions and present of tumor vascular invasion (Table 1). Kaplan–Meier analyses were conducted to reveal the prognostic value of CBX1 in HCC. The median survival time was 30.1 months for patients with high CBX1 expression and 18.1 months for the other patients. High expression of CBX1 was associated with shorter overall and disease-free survivals (Figure 3, A and B). This was validated in TCGA cohort, indicating that patients with overexpression of CBX1 mRNA were likely with a shorter living time and a higher tendency of disease progression (Figure 3, C and D). The stratified survival analyses confirmed that CBX1 expression was significantly correlated with overall survival in subgroups of HCC patients defined by pathological features that contributes to poor outcomes (Figure 4). In addition, after adjusting for the prognostic factors established in the univariate analysis, multiple Cox regression analysis suggest CBX1 as an independent factor for unfavorable overall survival (hazard ratio, HR = 1.735, 95% confidence interval, 95% CI: 1.342–2.244, P < .001) (Table 2).
Table 1.
Correlation of clinicopathological parameters and CBX1 expression.
| Variable | CBX1 expression |
|||
|---|---|---|---|---|
| All cases | Low expression | High expression | P valuea | |
| Age (years)b | 0.073 | |||
| < 49 | 132 | 72 (54.5%) | 60 (45.5%) | |
| ≥ 49 | 144 | 63 (43.8%) | 81(56.3%) | |
| Gender | 0.767 | |||
| Male | 250 | 123 (49.2%) | 127 (50.8%) | |
| Female | 26 | 12 (46.2%) | 14 (53.8%) | |
| HBsAg | 0.350 | |||
| Positive | 221 | 105 (47.5%) | 116 (52.5%) | |
| Negative | 55 | 30 (54.5%) | 25 (45.5%) | |
| AFP (ng/ml) | 0.830 | |||
| < 20 | 72 | 36 (50.0%) | 36 (50.0%) | |
| ≥ 20 | 204 | 99 (48.5%) | 105 (51.5%) | |
| Cirrhosis | 0.750 | |||
| Yes | 243 | 118 (48.6%) | 125 (51.4%) | |
| No | 33 | 17 (51.5%) | 16 (48.5%) | |
| Tumor size (cm) | 0.028 | |||
| < 5 | 64 | 39 (60.9%) | 25 (39.1%) | |
| ≥ 5 | 212 | 96 (45.3%) | 116 (54.7%) | |
| Tumor multiplicity | 0.041 | |||
| Single | 167 | 90 (53.9%) | 77 (46.1%) | |
| Multiple | 109 | 45 (41.3%) | 64 (58.7%) | |
| Differentiation | 0.031 | |||
| Well-Moderate | 207 | 109 (52.7%) | 98 (47.3%) | |
| Poor-undifferentiated | 69 | 26 (37.7%) | 43 (62.3%) | |
| TNM | 0.116 | |||
| I-II | 140 | 75 (53.6%) | 65 (46.4%) | |
| III-IV | 136 | 60 (44.1%) | 76 (55.9%) | |
| Vascular invasion | 0.025 | |||
| Yes | 213 | 112 (52.6%) | 101 (47.4%) | |
| No | 63 | 23 (36.5%) | 40 (63.5%) | |
| Tumor Capsule | 0.504 | |||
| Absent | 163 | 77 (47.2%) | 86 (52.8%) | |
| Present | 113 | 58 (51.3%) | 55 (48.7%) | |
Chi-square test;
Median age; AFP, alpha-fetoprotein; HBV, hepatitis B virus.
Figure 3.
Correlation of high CBX1 expression and poor prognosis in HCC. Probabilities overall survival (A,C) and disease-free survival (B,D) of HCC patients were analyzed using Kaplan–Meier survival analysis (log-rank test) in DH and TCGA cohorts.
Figure 4.
Correlation of CBX1 expression with overall survival in pathological HCC subgroups. Stratified survival analyses were performed in subgroups defined by the parameters that contribute to poor prognosis of HCC patients, using Kaplan–Meier survival analysis (log rank test).
Table 2.
Univariate and multivariate analyses of clinicopathological and CBX1 expression for overall survival in overall cohort (n = 276).
| Variables | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Overall survival | ||||
| Age (<49 vs. ≥49 years) | 0.981 (0.771–1.248) | 0.874 | ||
| Gender (female vs. male) | 1.046 (0.692–1.583) | 0.830 | ||
| HBV (positive vs. negative) | 1.156 (0.850–1.570) | 0.356 | ||
| Liver cirrhosis (yes vs. no) | 0.865 (0.600–1.248) | 0.439 | ||
| Tumor size (<5 vs. ≥5 cm) | 1.506 (1.126–2.014) | 0.006 | 1.110 (0.790–1.560) | 0.547 |
| Tumor multiplicity (single vs. multiple) | 1.342 (1.047–1.720) | 0.020 | 0.893(0.658–1.210) | 0.465 |
| Tumor Capsule (absent vs. present) | 0.538 (0.415–0.695) | 0.000 | 0.671(0.505–0.892) | 0.006 |
| AFP (<20 vs. ≥20 ng/mL) | 1.548 (1.174–2.042) | 0.002 | 1.349 (1.005–1.810) | 0.047 |
| Vascular invasion (no vs. yes) | 2.177 (1.622–2.923) | 0.000 | 1.444 (1.032–2.020) | 0.032 |
| Grade | 1.375 (1.038–1.823) | 0.027 | 1.052 (0.780–1.418) | 0.741 |
| TNM (I vs. II-IV) | 1.875 (1.459–2.409) | 0.000 | 1.255 (0.870–1.810) | 0.225 |
| Relapse (yes vs. no) | 0.676 (0.529–0.865) | 0.002 | 0.813 (0.622–1.062) | 0.128 |
| CBX1 expression (low vs. high) | 1.712 (1.337–2.191) | 0.000 | 1.735 (1.342–2.244) | 0.000 |
AFP, a-fetoprotein; HBsAg, hepatitis B surface antigen; HR, hazard ratio; CI, confidence interval.
CBX1-Mediated Oncogenic Activities in HCC
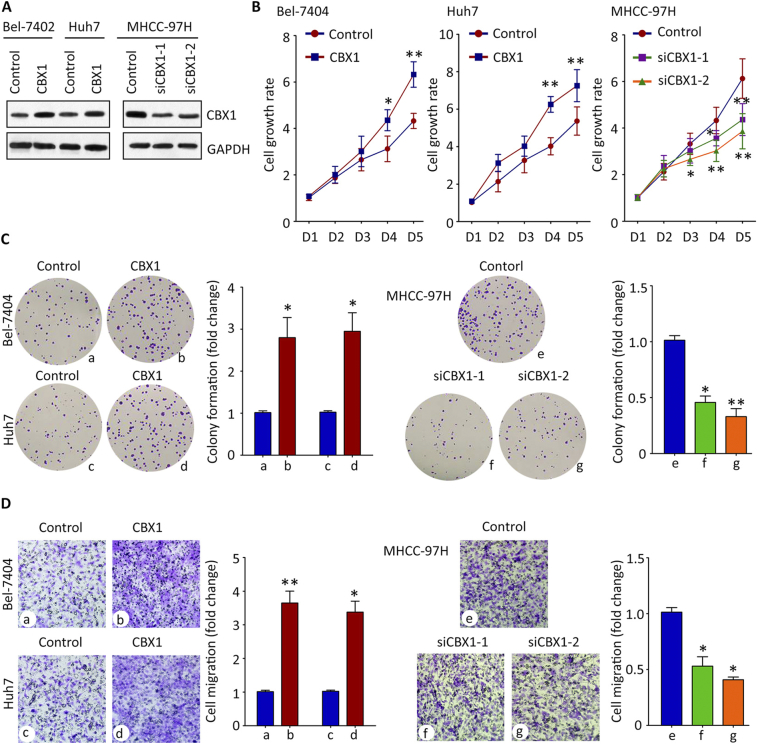
The role of CBX1 in HCC progression was next investigated. Since CBX1 expression was closely associated with tumor size and vascular invasion, the effect of CBX1 on HCC cell proliferation and migration was determined. According to the expression of CBX1 in HCC cell lines (Figure 1D), CBX1 was overexpressed in Bel-7402 and Huh7 cells, and knocked down in MHCC-97H cells (Figure 5A). Results of MTT assays showed that the cell growth rates were accelerated in cells with CBX1 overexpression, but slowed down in cells with CBX1 silencing (Figure 5B). Colony formation assays were used to confirm that CBX1 was capable of promoting HCC cell growth: more foci were induced by exogenous CBX1 (Figure 5C). The impact of CBX1 in cell migration was assessed by Transwell assays. Ectopic CBX1 expression markedly enhanced the ability of cell movement, resulting in more migrated cells, compared to the control (Figure 5D). These data suggest CBX1 exhibits oncogenic activities in HCC.
Figure 5.
CBX1-promoted HCC cell proliferation and migration in vitro. A. The overexpression or knockdown of CBX1 protein in HCC cell lines was confirmed by western blot analysis. B. Cells were transfected with CBX1 overexpression vector or siRNAs on day 1 and day3. The related cell growth rates assessed by MTT assay were indicated. *P < .05, **P < .01. C. Colony formation assays were performed to demonstrate the effect of CBX1 on HCC cell growth. D. Transwell assays were used to show the cell migration in cells with CBX1 overexpression or silence. *P < .05, **P < .01.
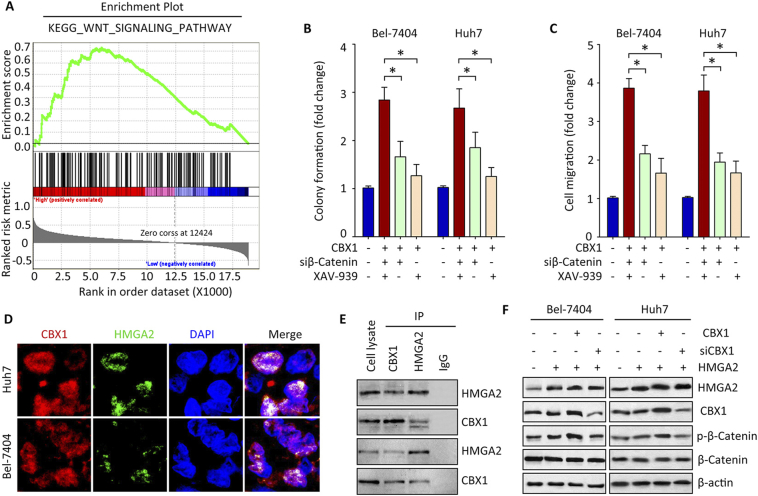
CBX1 Interacts with HMGA2 and Activates Wnt/β-Catenin Signaling Pathway
The underlying mechanism of CBX1-promoted cell growth was next determined. Gene Set Enrichment Analysis (GSEA) was conducted, using The Cancer Genome Atlas (TCGA) data. Results showed that WNT signaling pathway was activated in patients with high expression of CBX1 (Figure 6A), suggesting a role of β-Catenin in CBX1-mediated phenotypes. Suppression of β-Catenin activation by siRNA or specific inhibitor XAV-939 led to the attenuation of CBX1-promoted cell proliferation and migration (Figure 6, B and C). According to the analyses of bioinformatic algorithms, transcription factor HMGA2 was predicted as an interacting partner of CBX1 (data not shown). In HCC cell lines, CBX1 was colocalized with HMGA2 in the nucleus (Figure 6D). CoIP results showed that both CBX1 and HMGA2 was detectable in the precipitate mediated by CBX1 or HMGA2 antibody (Figure 6E). Overexpression of CBX1 enhanced, whereas knockdown of CBX1 weakened the phosphorylation of β-Catenin at Ser552 caused by ectopic expression of HMGA2 (Figure 6F). These findings indicate that CBX1 exerts oncogenic functions via HMGA2-mediated Wnt/β-Catenin signaling pathway.
Figure 6.
Activation of Wnt/β-Catenin pathway by the interaction of CBX1 and HMGA2. A. Gene set enrichment analysis (GSEA) based on the TCGA data indicated that WNT signaling pathway was activated in HCC cases with CBX1 overexpression. B. Cells with CBX1 overexpression were treated with β-Catenin siRNA or inhibitor XAV-939. Colony formation was performed to determine the effect of β-Catenin inhibition on CBX1-mediated cell proliferation. C. Transwell assays were used to examine the cell migration. D. CoIP was performed using specific antibodies for CBX1 and HMGA2 in HCC cells. The colocalization of the two proteins was observed by confocal assays. E. Cells were transfected with HGMA2 overexpression vector and CBX1 overexpression vector or siRNA for 48 h. Western blot was used to examine the expression of β-Catenin phosphorylation at Ser552, CBX1 and HMGA2.
Discussion
Despite of new treatment strategies applied to the clinic management, the outcomes of patients with HCC have been barely improved last decades [13]. The high mortality of HCC due to the extremely high probability of tumor recurrence and metastasis makes HCC a lethal threat globally. To date, interests on searching for biomarkers useful for diagnosis and personalized therapy of HCC have been accumulating. Here, we that CBX1 was of clinical implications for prognostic prediction and HCC intervention. Overexpression of CBX1 was found in HCC tissues, correlated with poor overall and disease-free survivals, and promoted the HCC cell proliferation and migration.
Identification of proteins with prognostic value benefits the tumor classification. The dysregulation of HP1 proteins and their clinical significances have been documents in human cancers. Elevated expression of CBX3 was found in prostate cancer [14], breast cancer [15], lung cancer and colorectal cancer [16]. Overexpression of CBX3 was correlated with poor prognosis in tongue squamous cell carcinoma [17], non-small cell lung cancer [18] and prostate cancer [14]. Increased CBX5 expression was reported in prostate cancer [19], [20] and breast cancer [15]. Furthermore, CBX1 was upregulated in prostate cancer [21] and breast cancer [12]. High expression of CBX1 transcript was associated with poor recurrence-free survival of patients with breast cancer [12]. On the other hand, CBX1 was reduced in thyroid carcinomas [22]. In the present study, CBX1 was markedly induced in HCC tissues. Patients with high CBX1 mRNA or protein expression survived shorter and experienced a shorter interval of tumor relapse or metastasis in two cohorts containing 648 patients. This might be attributed to high CBX1 expression was closely associated with larger tumor size, poor tumor differentiation and tumor vascular invasion. In addition, after adjusting for the prognostic factors established in the univariate analysis, a significant correlation between CBX1 expression and overall survival was confirmed. Collectively, these data indicate HP1 proteins as promising biomarkers for prediction of post-surgical prognosis of patients with malignances.
It has been well known that dysregulation of proteins involved in chromatin structure is responsible for the initiation and progression of human cancers. Our in vitro studies demonstrated that CBX1 exerted oncogenic activities in HCC. Ectopic expression of CBX1 in HCC cells enhanced the cell viability, colony formation and cell migration, whereas the silence of CBX1 led to the opposite phenotypes. Mechanism studies demonstrated that CBX1 physically bound to the transcription factor HMGA2 to trigger Wnt/β-Catenin signaling. The role of HMGA2 in the modulation of β-Catenin activation has been reported in human cancers [23], [24]. However, the mechanism via which CBX1 enhanced the effect of HMGA2 on Wnt/β-Catenin pathway to promote HCC cell proliferation and migration was still unclear. Knockdown of CBX1 conferred breast cancer cells sensitivity to PARP inhibitor treatment [25]. CBX1 was functionally associated with Suv4–20 h2 and H4K20me3, two histone methyltransferases contributing to tumor cell growth [26], [27]. ChIP-seq study revealed that CBX1 potentially suppressed the expression of PLK3 and SLCO1B3 that contributing to hepatocarcinogenesis [28], [29]. However, Yi et al. reported that depletion of CBX1 facilitated the cell movement in colon cancer via inducing the expression of MMP2 [30]. Therefore, the effect of CBX1 in cancer progression seems to be contradictory and requires further investigations. Taken together, our data indicate that CBX1 functions as an oncogene and may serve as a potential prognostic biomarker in HCC.
Footnotes
Conflict of interest: All authors declare no conflict of interest.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics. CA Cancer J Clin. 2012;65(2015):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Zhu AX, Park JO, Ryoo BY, Yen CJ, Poon R, Pastorelli D, Blanc JF, Chung HC, Baron AD, Pfiffer TE. Ramucirumab versus placebo as second-line treatment in patients with advanced hepatocellular carcinoma following first-line therapy with sorafenib (REACH): a randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol. 2015;16:859–870. doi: 10.1016/S1470-2045(15)00050-9. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman PD. New partners for HP1 in transcriptional gene silencing. Mol Cell. 2011;41:1–2. doi: 10.1016/j.molcel.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eissenberg JC, Elgin SC. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 6.Lu H, Cui JY, Gunewardena S, Yoo B, Zhong XB, Klaassen CD. Hepatic ontogeny and tissue distribution of mRNAs of epigenetic modifiers in mice using RNA-sequencing. Epigenetics. 2012;7:914–929. doi: 10.4161/epi.21113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattout A, Aaronson Y, Sailaja BS, Raghu Ram EV, Harikumar A, Mallm JP, Sim KH, Nissim-Rafinia M, Supper E, Singh PB. Heterochromatin Protein 1beta (HP1beta) has distinct functions and distinct nuclear distribution in pluripotent versus differentiated cells. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yearim A, Gelfman S, Shayevitch R, Melcer S, Glaich O, Mallm JP, Nissim-Rafinia M, Cohen AH, Rippe K, Meshorer E. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 2015;10:1122–1134. doi: 10.1016/j.celrep.2015.01.038. [DOI] [PubMed] [Google Scholar]
- 9.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 10.Canzio D, Liao M, Naber N, Pate E, Larson A, Wu S, Marina DB, Garcia JF, Madhani HD, Cooke R. A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature. 2013;496:377–381. doi: 10.1038/nature12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baude A, Aaes TL, Zhai B, Al-Nakouzi N, Oo HZ, Daugaard M, Rohde M, Jaattela M. Hepatoma-derived growth factor-related protein 2 promotes DNA repair by homologous recombination. Nucleic Acids Res. 2016;44:2214–2226. doi: 10.1093/nar/gkv1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang YK, Lin HY, Chen CF, Zeng Prognostic values of distinct CBX family members in breast cancer. Oncotarget. 2017;8:92375–92387. doi: 10.18632/oncotarget.21325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China. CA Cancer J Clin. 2015;66(2016):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 14.Slezak J, Truong M, Huang W, Jarrard D. HP1gamma expression is elevated in prostate cancer and is superior to Gleason score as a predictor of biochemical recurrence after radical prostatectomy. BMC Cancer. 2013;13:148. doi: 10.1186/1471-2407-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomsen R, Christensen DB, Rosborg S, Linnet TE, Blechingberg J, Nielsen AL. Analysis of HP1alpha regulation in human breast cancer cells. Mol Carcinog. 2011;50:601–613. doi: 10.1002/mc.20755. [DOI] [PubMed] [Google Scholar]
- 16.Liu M, Huang F, Zhang D, Ju J, Wu XB, Wang Y, Wang Y, Wu Y, Nie M, Li Z. Heterochromatin protein HP1gamma promotes colorectal cancer progression and is regulated by miR-30a. Cancer Res. 2015;75:4593–4604. doi: 10.1158/0008-5472.CAN-14-3735. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Chen W, Fu X, Su X, Yang A. CBX3 promotes tumor proliferation by regulating G1/S phase via p21 downregulation and associates with poor prognosis in tongue squamous cell carcinoma. Gene. 2018;654:49–56. doi: 10.1016/j.gene.2018.02.043. [DOI] [PubMed] [Google Scholar]
- 18.Chang SC, Lai YC, Chen YC, Wang NK, Wang WS, Lai JI. CBX3/heterochromatin protein 1 gamma is significantly upregulated in patients with non-small cell lung cancer. Asia Pac J Clin Oncol. 2017 doi: 10.1111/ajco.12820. [DOI] [PubMed] [Google Scholar]
- 19.Shapiro E, Huang H, Ruoff R, Lee P, Tanese N, Logan SK. The heterochromatin protein 1 family is regulated in prostate development and cancer. J Urol. 2008;179:2435–2439. doi: 10.1016/j.juro.2008.01.091. [DOI] [PubMed] [Google Scholar]
- 20.Ci X, Hao J, Dong X, Choi SYC, Xue H, Wu R, Qu S, Gout PW, Zhang F, Haegert AM. Heterochromatin protein 1alpha mediates development and aggressiveness of neuroendocrine prostate cancer. Cancer Res. 2018;78(10):2691–2704. doi: 10.1158/0008-5472.CAN-17-3677. [DOI] [PubMed] [Google Scholar]
- 21.Ngollo M, Lebert A, Daures M, Judes G, Rifai K, Dubois L, Kemeny JL, Penault-Llorca F, Bignon YJ, Guy L. Global analysis of H3K27me3 as an epigenetic marker in prostate cancer progression. BMC Cancer. 2017;17:261. doi: 10.1186/s12885-017-3256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tretiakova MS, Bond SD, Wheeler D, Contreras A, Kocherginsky M, Kroll TG, Hale TK. Heterochromatin protein 1 expression is reduced in human thyroid malignancy. Lab Investig. 2014;94:788–795. doi: 10.1038/labinvest.2014.68. [DOI] [PubMed] [Google Scholar]
- 23.Ou W, Lv J, Zou X, Yao Y, Wu J, Yang J, Wang Z, Ma Y. Propofol inhibits hepatocellular carcinoma growth and invasion through the HMGA2-mediated Wnt/beta-catenin pathway. Exp Ther Med. 2017;13:2501–2506. doi: 10.3892/etm.2017.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wend P, Runke S, Wend K, Anchondo B, Yesayan M, Jardon M, Hardie N, Loddenkemper C, Ulasov I, Lesniak MS. WNT10B/beta-catenin signalling induces HMGA2 and proliferation in metastatic triple-negative breast cancer. EMBO Mol Med. 2013;5:264–279. doi: 10.1002/emmm.201201320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YH, Liu X, Qiu F, O'Connor TR, Yen Y, Ann DK. HP1beta is a biomarker for breast cancer prognosis and PARP inhibitor therapy. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosch-Presegue L, Raurell-Vila H, Thackray JK, Gonzalez J, Casal C, Kane-Goldsmith N, Vizoso M, Brown JP, Gomez A, Ausio J. Mammalian HP1 Isoforms Have Specific Roles in Heterochromatin Structure and Organization. Cell Rep. 2017;21:2048–2057. doi: 10.1016/j.celrep.2017.10.092. [DOI] [PubMed] [Google Scholar]
- 27.Bromberg KD, Mitchell TR, Upadhyay AK, Jakob CG, Jhala MA, Comess KM, Lasko LM, Li C, Tuzon CT, Dai Y. The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity. Nat Chem Biol. 2017;13:317–324. doi: 10.1038/nchembio.2282. [DOI] [PubMed] [Google Scholar]
- 28.Tell R, Wang QT, Blunier A, Benya RV. Identification of ChIP-seq mapped targets of HP1beta due to bombesin/GRP receptor activation. Clin Epigenetics. 2011;2:331–338. doi: 10.1007/s13148-011-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vavricka SR, Jung D, Fried M, Grutzner U, Meier PJ, Kullak-Ublick GA. The human organic anion transporting polypeptide 8 (SLCO1B3) gene is transcriptionally repressed by hepatocyte nuclear factor 3beta in hepatocellular carcinoma. J Hepatol. 2004;40:212–218. doi: 10.1016/j.jhep.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 30.Yi SA, Ryu HW, Lee DH, Han JW, Kwon SH. HP1beta suppresses metastasis of human cancer cells by decreasing the expression and activation of MMP2. Int J Oncol. 2014;45:2541–2548. doi: 10.3892/ijo.2014.2646. [DOI] [PubMed] [Google Scholar]