Abstract
Background
Renal oncocytomas (ROs) are benign epithelial tumors of the kidney whereas chromophobe renal cell carcinoma (chRCCs) are malignant renal tumors. The latter constitute 5–7% of renal neoplasias. ROs and chRCCs show pronounced molecular and histological similarities, which renders their differentiation demanding. We aimed for the differential proteome profiling of ROs and early-stage chRCCs in order to better understand distinguishing protein patterns.
Methods
We employed formalin-fixed, paraffin-embedded samples (six RO cases, six chRCC cases) together with isotopic triplex dimethylation and a pooled reference standard to enable cohort-wide quantitative comparison. For lysosomal-associated membrane protein 1 (LAMP1) and integrin alpha-V (ITGAV) we performed corroborative immunohistochemistry (IHC) in an extended cohort of 42 RO cases and 31 chRCC cases.
Results
At 1% false discovery rate, we identified > 3900 proteins, of which > 2400 proteins were consistently quantified in at least four RO and four chRCC cases. The proteomic expression profiling discriminated ROs and chRCCs and highlighted established features such as accumulation of mitochondrial proteins in ROs together with emphasizing the accumulation of endo-lysosomal proteins in chRCCs. In line with the proteomic data, IHC showed enrichment of LAMP1 in chRCC and of ITGAV in RO.
Conclusion
We present one of the first differential proteome profiling studies on ROs and chRCCs and highlight differential abundance of LAMP1 and ITGAV in these renal tumors.
Electronic supplementary material
The online version of this article (10.1186/s12014-018-9200-6) contains supplementary material, which is available to authorized users.
Keywords: Renal cell tumors, Formalin-fixation, Paraffin embedment, Proteomics, Immunohistochemistry
Background
Chromophobe renal cell carcinoma (chRCC) constitute 5–7% of renal neoplasias [1]. ChRCC are thought to originate from cell of the distal nephron [2]. Their overall prognosis is more favourable than for renal clear cell carcinomas with a 5-year survival rate of > 75% [1, 3]. However, this is still a malignant tumor entity with the potential for recurrence or metastatic spread. Renal oncocytomas (ROs) are benign epithelial tumors of the kidney. They constitute up to 7% of all adult renal tumors [4]. ROs have been first described as late as 1942 [5] and clinical reports have remained scarce until the 1970s [6]. Differentiation between RO and chRCC in pathological routine practice is often considered challenging [4, 7]. This is because of strong similarities in morphology, growth pattern and localization of benign ROs and malignant chRCCs. For this reason, distinguishing biomarkers are actively being researched. In this context, system-wide, omics-type expression profiling studies are emerging as a strong and unbiased approach. Several of such expression studies aimed at a comprehensive differential profiling of various renal cell tumors, also including clear cell renal cell carcinoma (ccRCCs) or papillary renal cell carcinoma. Often, such studies involved only low numbers of RO and chRCC cases and aimed at their collective distinction from other renal cell tumors rather than molecularly distinguishing ROs and chRCCs.
Yusenko et al. probed genome alterations and DNA copy number variants to specifically differentiate ROs and chRCCs. They identified several genomic alterations that differ between chRCCs and ROs [8]. A second genomic study identified differing chromosomal abnormalities with regard to chromosome 19 in ROs or chRCCs [9]. The resulting gene expression effects affected oxygen sensing. This is an intriguing parallel to ccRCCs which frequently carry somatic, inactivating mutations of the Von Hippel-Lindau gene, ultimately leading to the expression of hypoxia-related genes and promotion of tumorigenesis [10]. On the genomic level, Joshi et al. [11] distinguish two types of ROs and link chromosomal abnormalities involving chromosome 1, X or Y, and/or 14 and 21 with the potential for progression from RO to chRCC. Rohan et al. [12] performed a global transcriptomic profiling of ROs and chRCCs. Major expression differences were found with regard to transcript encoding proteins involved in vesicular transport and cell junction. On the protein level, multiple protein biomarkers have been suggested (reviewed in [4]) but to date there has not been an unbiased, differential proteomic profiling of ROs and chRCCs. However, proteomic profiling is gaining interest for the investigation of malignancies due to the limited correlation between mRNA and protein levels [13, 14]. Formalin-fixed, paraffin-embedded (FFPE) samples are a valuable resource for proteomic profiling [15–17], enabling retrospective profiling of clinic-pathologically annotated specimens [18, 19]. In the present study, we employed “FFPE proteomics” for the differential proteomic profiling of RO and chRCC cases for which we find noticeable differences. In a larger cohort comprising > 70 RO and chRCC cases, we corroborate elevated presence of lysosomal-associated membrane protein 1 in chRCCs and elevated presence of integrin alpha-V in ROs.
Methods
Ethics statement
The study was approved by the Ethics Committee of the University Medical Center Freiburg (311/12). Before study inclusion, all patient data were anonymized.
Histopathological diagnosis for renal oncocytoma or chromophobe renal cell carcinoma
The diagnosis for all RO or chRCC cases used in this study was based on histopathologic parameters (cytoplasm, cell membrane, perinuclear halo, tumor border and septae) and corroborated by immunohistochemistry (IHC) for CD117, cytokeratin-7, and vimentin. These IHC stainings were part of the routine immunohistopathological diagnosis with the corresponding antibodies being supplied by Dako (Hamburg, Germany). We focused on chRCC cases that displayed diffuse-membranous expression of cytokeratin-7 whereas its expression was largely absent in the RO cases [1, 7]. Moreover, the RO and chRCC cases displayed membranous expression of CD117 [7]. Finally, the RO and chRCC cases were vimentin-negative [1].
Tissue collection, sample preparation, liquid chromatography-tandem mass spectrometry (LC–MS/MS), and data analysis
FFPE tissue specimens of six ROs and six chRCCs were used as described previously [15, 19], including microscopically controlled macrodissection to remove areas of necrosis, fibrosis, hemorrhage, and inflammation. For quantitative comparison, triplex isotopic dimethylation of primary amines was employed [20], distinguishing RO, chRCC, and a pooled mix that serves as a standard similar to the Super-SILAC approach [21]. Samples were further fractionated by strong cation exchange chromatography as described [22]. LC–MS/MS was performed using a Q-Exactive plus (Thermo Scientific) mass spectrometer coupled to an Easy nanoLC 1000 (Thermo Scientific) as described previously [18]. MS data were analyzed by MaxQuant version 1.5.28 [23] as described previously [19]. Proteins were only further considered if they were identified and quantified in at least four RO samples and four chRCC samples. Due to this strict requirement, we also included proteins that were identified and quantified by single peptides in individual samples. Files obtained by MaxQuant were further processed using RStudio v.0.99.446 (R Foundation for Statistical Computing, Vienna, Austria) as previously described [24]. Reverse and potential contaminants entries were removed. Ratios were log2 transformed, normalized by centering, and a linear model was fitted using the limma package [25].
Immunohistochemical analysis
IHC analysis was performed with an extended patient cohort, comprising 42 RO cases and 31 chRCC cases. IHC was performed for lysosomal-associated membrane protein 1 (LAMP1) and integrin alpha-V (ITGAV). Slices of 2 µm thickness from FFPE tissue samples were prepared using a Leica RM2255 microtome. Heat induced antigen retrieval was performed at pH 9.0. Primary antibodies were rabbit polyclonal to LAMP1 (Abcam, ab24170, stock concentration 1.0 mg/ml) or rabbit monoclonal to ITGAV (Abcam, EPR16800, stock concentration 1.0 mg/ml). For incubation, primary antibodies were diluted in Zytomed dilution buffer (ZUC025-500; 1:300 for the LAMP1 antibody, 1:7000 for the ITGAV antibody). Incubation time was 60 min. Visualization was performed using DAKO Envision Flex+, Mouse, high pH (Link) Detecting System (K800221-2). Sections were counterstained with hematoxylin for 1 min, dehydrated in an ascending alcohol concentration and covered with xylol and coverslipping film (Tissue-TekR 4770). For evaluation, two experienced pathologists reviewed LAMP1 and ITGAV expression in RO or chRCC tumor cells using a semi-quantitative scoring system covering absence of signal (score 0), weak detection (score 1), medium detection (score 2), and strong detection (score 3). For every sample, the fractional area of the different detection levels (scores 0–3) was determined and reported as percentage of the total tumor area under investigation.
Results and discussion
Experimental set-up
We aimed for a differential, quantitative proteome characterization of RO and chRCC using FFPE samples for which we have shown amenability to quantitative proteomic analysis using isotope-coded dimethylation [15]. Our cohort for the proteomic analysis comprised 12 cases; six RO and six chRCC. In order to enable cohort-wide comparison, we also included a pooled sample (comprised of RO and chRCC tissue) as a spike-in reference standard against which every other sample can be compared. This set-up is reminiscent of the Super-SILAC technique, in which metabolically labelled proteomes serve as spike-in reference standard [21]. A triplex labeling scheme was employed (Fig. 1a, actual labeling setup in Additional file 1: Table S1). In a second cohort (42 RO cases; 31 chRCC cases) we employed immunohistochemistry to probe the expression of LAMP1 and ITGAV. Since the benign oncocytomas are very rarely invasive (which constitutes their benign nature), we focused on chRCC cases for which tumor growth was still confined to the kidney (T1 or T2). An overview of the patient characteristics is provided in Table 1.
Fig. 1.
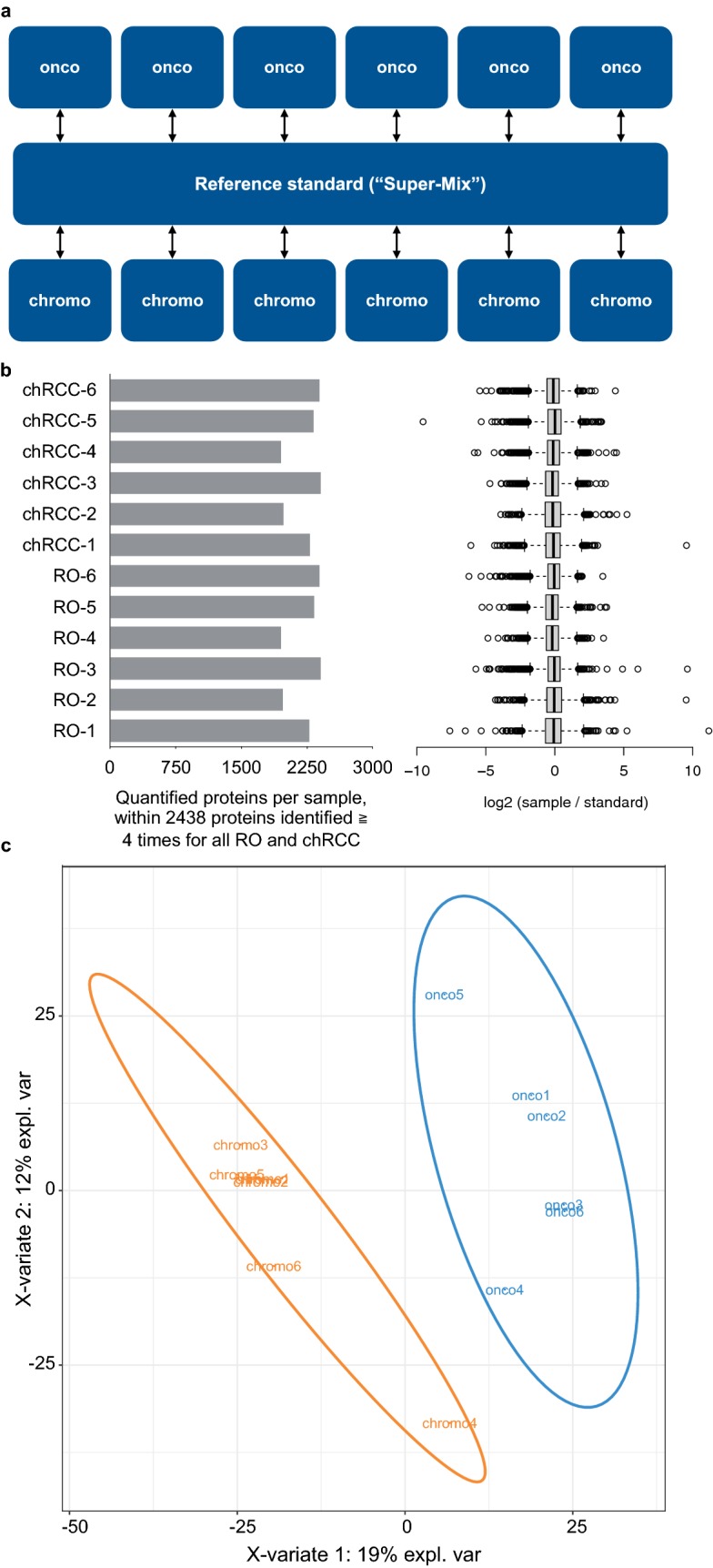
a Samples of chRCC tissue or RO tissue were collected from FFPE specimens and, post-trypsination, differentially labeled by isotopic, formaldehyde-based dimethylation. A differentially labeled, pooled reference standard was also included. b > 2400 proteins were identified (false-discovery rate < 1%) and quantified in at least four RO and four chRCC samples. Within this core proteome, each chRCC and RO case contributed a comparable number of protein identifications and quantitations. Also, the log-transformed sample/standard ratios were comparable for all cases. Whiskers extend to data points that are less than 1.5 × IQR away from 1st/3rd quartile. c Supervised partial least squares discriminant analysis separates the RO and chRCC proteome profiles
Table 1.
Patient characteristics
| Proteomics | Immunohistochemistry (no overlap with cases used for proteomic analysis) | |||
|---|---|---|---|---|
| Renal oncocytomas | Chromophobe renal cell carcinomas | Renal oncocytomas | Chromophobe renal cell carcinomas | |
| N | 6 | 6 | 42 | 31 |
| Tumor size/T classification | ||||
| ≤ 4 cm/1a | 5 | 4 | 25 | 21 |
| ≤ 7 cm/1b | 1 | 2 | 15 | 6 |
| ≤ 10 cm/2a | 0 | 0 | 1 | 3 |
| > 10 cm/2b | 0 | 0 | 1 | 1 |
| Number of simultaneous tumorsa | ||||
| 1 | 6 | 6 | 38 | 31 |
| 2 | 0 | 0 | 3 | 0 |
| 6 | 0 | 0 | 1 | 0 |
| Affected kidney | ||||
| Left | 1 | 2 | 21 | 16 |
| Right | 4 | 3 | 19 | 12 |
| Not documented | 1 | 1 | 2 | 3 |
| Gender | ||||
| Male | 3 | 3 | 24 | 15 |
| Female | 3 | 3 | 18 | 16 |
| Age | ||||
| Average | 66 | 56 | 64 | 60 |
| Range | 57–80 | 42–74 | 42–88 | 34–83 |
aIf simulatenous tumors were present, only one tumor per patient was used for the analyses
Proteome profiling of RO and chRCC
LC–MS/MS analysis enabled the identification (at a false discovery rate < 1%) of > 3900 proteins. Of these, > 2400 proteins were identified and quantified in at least four RO and four chRCC cases. Within this “core proteome”, each case allowed for the identification and quantification of a comparable number of proteins (Fig. 1b). Likewise, the protein quantitation profiles (log-transformed sample/standard ratios) are similar (Fig. 1b). We conclude that proteome coverage and—quantitation allows for the comparison of the RO and chRCC proteome profiles. We employed partial least squares discriminant analysis (PLS-DA) as an initial step to probe whether RO and chRCC display distinguishable proteome profiles. Supervised PLS-DA of the proteomic dataset (Fig. 1c) clearly separates RO and chRCC in an unbiased manner, thus corroborating their initial distinction. Noteworthy, RO and chRCC were primarily distinguished by histopathological parameters and cytokeratin-7 IHC as part of routine diagnosis. Further approaches such as S100A1 IHC or Hale colloidal iron staining [7] have not been routinely employed, which however is not meant to question their discriminatory power.
Quantitative proteome differences and distinct proteome motifs of RO and chRCC
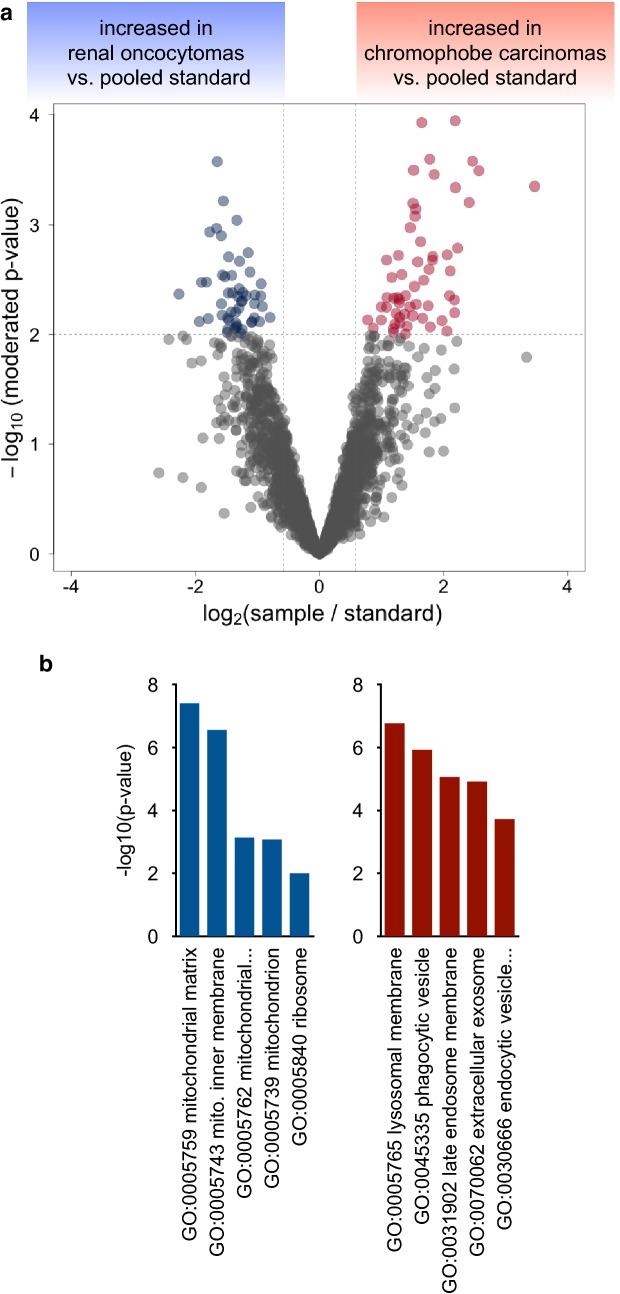
To identify proteins that are significantly enriched in either RO or chRCC (as compared to the pooled standard), we employed a linear model as implemented in the limma statistical package [26], which is particularly powerful with regard to multiple testing correction and prevention of false-positive discoveries in the analysis of omics-style data. We consider proteins with a limma moderated p value < 0.01 and protein identification based on at least three peptide-spectrum-matches to be significantly enriched in either RO or chRCC (as compared to the pooled standard). These criteria resulted in 51 proteins being significantly enriched in RO tissue and 59 proteins being significantly enriched in chRCC tissue (Tables 2 and 3). The corresponding volcano plot is visualized in Fig. 2a; all limma moderated p-values and average log2 ratios are listed in Additional file 2: Table S2. In line with earlier reports [1, 7] and our IHC-assisted confirmation of RO or chRCC diagnosis, there was significant enrichment of cytokeratin-7 in the chRCC cases (p ≪ 0.01).
Table 2.
Proteins found to be significantly enriched in RO
| Average log2 (RO/standard) | Average log2 (chRCC/standard) | p-value (limma moderated) | Uniprot ID | Protein names |
|---|---|---|---|---|
| 1.09 | − 0.57 | 0.00 | P06756 | Integrin alpha-V (Vitronectin receptor) (Vitronectin receptor subunit alpha) (CD antigen CD51) [Cleaved into: Integrin alpha-V heavy chain; Integrin alpha-V light chain] |
| 0.35 | − 1.21 | 0.00 | Q9H9J2 | 39S ribosomal protein L44, mitochondrial (L44mt) (MRP-L44) (EC 3.1.26.-) (Mitochondrial large ribosomal subunit protein mL44) |
| 0.46 | − 0.87 | 0.00 | O15382 | Branched-chain-amino-acid aminotransferase, mitochondrial (BCAT(m)) (EC 2.6.1.42) (Placental protein 18) (PP18) |
| 0.38 | − 1.28 | 0.00 | P51398 | 28S ribosomal protein S29, mitochondrial (MRP-S29) (S29mt) (Death-associated protein 3) (DAP-3) (Ionizing radiation resistance conferring protein) (Mitochondrial small ribosomal subunit protein mS29) |
| 0.40 | − 1.38 | 0.00 | P16219 | Short-chain specific acyl-CoA dehydrogenase, mitochondrial (SCAD) (EC 1.3.8.1) (Butyryl-CoA dehydrogenase) |
| 0.42 | − 1.17 | 0.00 | Q6PI48 | Aspartate–tRNA ligase, mitochondrial (EC 6.1.1.12) (Aspartyl-tRNA synthetase) (AspRS) |
| 0.50 | − 0.66 | 0.00 | Q9BYD6 | 39S ribosomal protein L1, mitochondrial (L1 mt) (MRP-L1) (Mitochondrial large ribosomal subunit protein uL1m) |
| 0.36 | − 1.11 | 0.00 | P30048 | Thioredoxin-dependent peroxide reductase, mitochondrial (EC 1.11.1.15) (Antioxidant protein 1) (AOP-1) (HBC189) (Peroxiredoxin III) (Prx-III) (Peroxiredoxin-3) (Protein MER5 homolog) |
| 0.33 | − 0.96 | 0.00 | P50897 | Palmitoyl-protein thioesterase 1 (PPT-1) (EC 3.1.2.22) (Palmitoyl-protein hydrolase 1) |
| 0.33 | − 0.79 | 0.00 | Q96GK7 | Fumarylacetoacetate hydrolase domain-containing protein 2A (EC 3.-.-.-) |
| 0.04 | − 1.53 | 0.00 | P55084 | Trifunctional enzyme subunit beta, mitochondrial (TP-beta) [Includes: 3-ketoacyl-CoA thiolase (EC 2.3.1.16) (Acetyl-CoA acyltransferase) (Beta-ketothiolase)] |
| 0.40 | − 1.02 | 0.00 | Q96EL3 | 39S ribosomal protein L53, mitochondrial (L53mt) (MRP-L53) (Mitochondrial large ribosomal subunit protein mL53) |
| 0.47 | − 1.06 | 0.00 | Q96PE7 | Methylmalonyl-CoA epimerase, mitochondrial (EC 5.1.99.1) (DL-methylmalonyl-CoA racemase) |
| 0.04 | − 1.79 | 0.00 | Q8TCS8 | Polyribonucleotide nucleotidyltransferase 1, mitochondrial (EC 2.7.7.8) (3′–5′ RNA exonuclease OLD35) (PNPase old-35) (Polynucleotide phosphorylase 1) (PNPase 1) (Polynucleotide phosphorylase-like protein) |
| 0.97 | − 0.93 | 0.00 | Q96P44 | Collagen alpha-1(XXI) chain |
| 0.33 | − 0.61 | 0.00 | P08559 | Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial (EC 1.2.4.1) (PDHE1-A type I) |
| 0.47 | − 0.83 | 0.00 | Q8N5M1 | ATP synthase mitochondrial F1 complex assembly factor 2 (ATP12 homolog) |
| 0.23 | − 1.01 | 0.00 | Q8N0X4 | Citramalyl-CoA lyase, mitochondrial (EC 4.1.3.25) (Beta-methylmalate synthase) (EC 2.3.3.-) (Citrate lyase subunit beta-like protein) (Citrate lyase beta-like) (Malate synthase) (EC 2.3.3.9) |
| 0.41 | − 1.00 | 0.00 | Q9HD33 | 39S ribosomal protein L47, mitochondrial (L47mt) (MRP-L47) (Mitochondrial large ribosomal subunit protein uL29 m) (Nasopharyngeal carcinoma metastasis-related protein 1) |
| 0.47 | − 1.00 | 0.00 | Q16822 | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial (PEPCK-M) (EC 4.1.1.32) |
| 1.01 | − 1.26 | 0.00 | P02792 | Ferritin light chain (Ferritin L subunit) |
| 0.27 | − 0.77 | 0.00 | Q9UIJ7 | GTP:AMP phosphotransferase AK3, mitochondrial (EC 2.7.4.10) (Adenylate kinase 3) (AK 3) (Adenylate kinase 3 alpha-like 1) |
| 0.46 | − 0.73 | 0.00 | Q9NYK5 | 39S ribosomal protein L39, mitochondrial (L39mt) (MRP-L39) (39S ribosomal protein L5, mitochondrial) (L5mt) (MRP-L5) (Mitochondrial large ribosomal subunit protein mL39) |
| 0.37 | − 0.58 | 0.00 | P11177 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial (PDHE1-B) (EC 1.2.4.1) |
| 0.18 | − 1.13 | 0.00 | Q9UFN0 | Protein NipSnap homolog 3A (NipSnap3A) (Protein NipSnap homolog 4) (NipSnap4) (Target for Salmonella secreted protein C) (TassC) |
| 0.36 | − 0.89 | 0.00 | P12694 | 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial (EC 1.2.4.4) (Branched-chain alpha-keto acid dehydrogenase E1 component alpha chain) (BCKDE1A) (BCKDH E1-alpha) |
| 0.52 | − 0.75 | 0.01 | Q9Y3B7 | 39S ribosomal protein L11, mitochondrial (L11 mt) (MRP-L11) (Mitochondrial large ribosomal subunit protein uL11m) |
| 0.33 | − 0.72 | 0.01 | Q8N490 | Probable hydrolase PNKD (EC 3.-.-.-) (Myofibrillogenesis regulator 1) (MR-1) (Paroxysmal nonkinesiogenic dyskinesia protein) (Trans-activated by hepatitis C virus core protein 2) |
| 0.54 | − 1.05 | 0.01 | P52815 | 39S ribosomal protein L12, mitochondrial (L12 mt) (MRP-L12) (5c5-2) (Mitochondrial large ribosomal subunit protein bL12m) |
| 0.40 | − 0.53 | 0.01 | P26038 | Moesin (Membrane-organizing extension spike protein) |
| 0.67 | − 0.64 | 0.01 | Q9Y619 | Mitochondrial ornithine transporter 1 (Solute carrier family 25 member 15) |
| 0.22 | − 1.21 | 0.01 | P22033 | Methylmalonyl-CoA mutase, mitochondrial (MCM) (EC 5.4.99.2) (Methylmalonyl-CoA isomerase) |
| 0.60 | − 0.76 | 0.01 | P20674 | Cytochrome c oxidase subunit 5A, mitochondrial (Cytochrome c oxidase polypeptide Va) |
| 0.80 | − 0.77 | 0.01 | Q15111 | Inactive phospholipase C-like protein 1 (PLC-L1) (Phospholipase C-deleted in lung carcinoma) (Phospholipase C-related but catalytically inactive protein) (PRIP) |
| 0.40 | − 0.40 | 0.01 | P27348 | 14-3-3 protein theta (14-3-3 protein T-cell) (14-3-3 protein tau) (Protein HS1) |
| 0.29 | − 0.76 | 0.01 | Q5TEU4 | Arginine-hydroxylase NDUFAF5, mitochondrial (EC 1.-.-.-) (NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 5) (Putative methyltransferase NDUFAF5) (EC 2.1.1.-) |
| 0.01 | − 1.47 | 0.01 | P10809 | 60 kDa heat shock protein, mitochondrial (EC 3.6.4.9) (60 kDa chaperonin) (Chaperonin 60) (CPN60) (Heat shock protein 60) (HSP-60) (Hsp60) (HuCHA60) (Mitochondrial matrix protein P1) (P60 lymphocyte protein) |
| 0.62 | − 1.18 | 0.01 | Q96CU9 | FAD-dependent oxidoreductase domain-containing protein 1 (EC 1.-.-.-) |
| 0.46 | − 1.01 | 0.01 | P10606 | Cytochrome c oxidase subunit 5B, mitochondrial (Cytochrome c oxidase polypeptide Vb) |
| 0.33 | − 0.77 | 0.01 | P04179 | Superoxide dismutase [Mn], mitochondrial (EC 1.15.1.1) |
| 1.09 | − 0.85 | 0.01 | O14773 | Tripeptidyl-peptidase 1 (TPP-1) (EC 3.4.14.9) (Cell growth-inhibiting gene 1 protein) (Lysosomal pepstatin-insensitive protease) (LPIC) (Tripeptidyl aminopeptidase) (Tripeptidyl-peptidase I) (TPP-I) |
| 0.34 | − 0.63 | 0.01 | P21953 | 2-oxoisovalerate dehydrogenase subunit beta, mitochondrial (EC 1.2.4.4) (Branched-chain alpha-keto acid dehydrogenase E1 component beta chain) (BCKDE1B) (BCKDH E1-beta) |
| 0.25 | − 0.85 | 0.01 | P03928 | ATP synthase protein 8 (A6L) (F-ATPase subunit 8) |
| 0.18 | − 1.17 | 0.01 | P30042 | ES1 protein homolog, mitochondrial (Protein GT335) (Protein KNP-I) |
| 0.44 | − 0.90 | 0.01 | P09001 | 39S ribosomal protein L3, mitochondrial (L3 mt) (MRP-L3) (Mitochondrial large ribosomal subunit protein uL3m) |
| 0.80 | − 0.55 | 0.01 | Q15067 | Peroxisomal acyl-coenzyme A oxidase 1 (AOX) (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (Straight-chain acyl-CoA oxidase) (SCOX) |
| 0.37 | − 1.01 | 0.01 | O75947 | ATP synthase subunit d, mitochondrial (ATPase subunit d) |
| 0.29 | − 0.97 | 0.01 | P82933 | 28S ribosomal protein S9, mitochondrial (MRP-S9) (S9 mt) (Mitochondrial small ribosomal subunit protein uS9m) |
| 0.38 | − 1.11 | 0.01 | P09669 | Cytochrome c oxidase subunit 6C (Cytochrome c oxidase polypeptide VIc) |
| 0.38 | − 1.11 | 0.01 | Q3ZCW2 | Galectin-related protein (Lectin galactoside-binding-like protein) |
| 0.37 | − 0.90 | 0.01 | Q9Y3D9 | 28S ribosomal protein S23, mitochondrial (MRP-S23) (S23mt) (Mitochondrial small ribosomal subunit protein mS23) |
| 1.09 | − 0.57 | 0.00 | P06756 | Integrin alpha-V (Vitronectin receptor) (Vitronectin receptor subunit alpha) (CD antigen CD51) [Cleaved into: Integrin alpha-V heavy chain; Integrin alpha-V light chain] |
| 0.35 | − 1.21 | 0.00 | Q9H9J2 | 39S ribosomal protein L44, mitochondrial (L44mt) (MRP-L44) (EC 3.1.26.-) (Mitochondrial large ribosomal subunit protein mL44) |
| 0.46 | − 0.87 | 0.00 | O15382 | Branched-chain-amino-acid aminotransferase, mitochondrial (BCAT(m)) (EC 2.6.1.42) (Placental protein 18) (PP18) |
| 0.38 | − 1.28 | 0.00 | P51398 | 28S ribosomal protein S29, mitochondrial (MRP-S29) (S29mt) (Death-associated protein 3) (DAP-3) (Ionizing radiation resistance conferring protein) (Mitochondrial small ribosomal subunit protein mS29) |
| 0.40 | − 1.38 | 0.00 | P16219 | Short-chain specific acyl-CoA dehydrogenase, mitochondrial (SCAD) (EC 1.3.8.1) (Butyryl-CoA dehydrogenase) |
| 0.42 | − 1.17 | 0.00 | Q6PI48 | Aspartate–tRNA ligase, mitochondrial (EC 6.1.1.12) (Aspartyl-tRNA synthetase) (AspRS) |
| 0.50 | − 0.66 | 0.00 | Q9BYD6 | 39S ribosomal protein L1, mitochondrial (L1 mt) (MRP-L1) (Mitochondrial large ribosomal subunit protein uL1m) |
Table 3.
Proteins found to be significantly enriched in chRCC
| Average log2 (RO/standard) | Average log2 (chRCC/standard) | p-value (limma moderated) | Uniprot ID | Protein names |
|---|---|---|---|---|
| − 0.83 | 0.59 | 0.01 | P11117 | Lysosomal acid phosphatase (LAP) (EC 3.1.3.2) |
| − 0.66 | 0.21 | 0.01 | P04075 | Fructose-bisphosphate aldolase A (EC 4.1.2.13) (Lung cancer antigen NY-LU-1) (Muscle-type aldolase) |
| − 0.81 | 0.27 | 0.01 | P09972 | Fructose-bisphosphate aldolase C (EC 4.1.2.13) (Brain-type aldolase) |
| − 0.82 | 0.38 | 0.00 | P07355 | Annexin A2 (Annexin II) (Annexin-2) (Calpactin I heavy chain) (Calpactin-1 heavy chain) (Chromobindin-8) (Lipocortin II) (Placental anticoagulant protein IV) (PAP-IV) (Protein I) (p36) |
| − 1.06 | 0.52 | 0.00 | Q96BM9 | ADP-ribosylation factor-like protein 8A (ADP-ribosylation factor-like protein 10B) (Novel small G protein indispensable for equal chromosome segregation 2) |
| − 0.73 | 0.83 | 0.01 | P15289 | Arylsulfatase A (ASA) (EC 3.1.6.8) (Cerebroside-sulfatase) [Cleaved into: Arylsulfatase A component B; Arylsulfatase A component C] |
| − 0.99 | 0.24 | 0.01 | P61421 | V-type proton ATPase subunit d 1 (V-ATPase subunit d 1) (32 kDa accessory protein) (V-ATPase 40 kDa accessory protein) (V-ATPase AC39 subunit) (p39) (Vacuolar proton pump subunit d 1) |
| − 0.56 | 0.70 | 0.00 | P16070 | CD44 antigen (CDw44) (Epican) (Extracellular matrix receptor III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (HUTCH-I) (Heparan sulfate proteoglycan) (Hermes antigen) (Hyaluronate receptor) (Phagocytic glycoprotein 1) (PGP-1) (Phagocytic glycoprotein I) (PGP-I) (CD antigen CD44) |
| − 1.21 | 0.63 | 0.00 | P08962 | CD63 antigen (Granulophysin) (Lysosomal-associated membrane protein 3) (LAMP-3) (Melanoma-associated antigen ME491) (OMA81H) (Ocular melanoma-associated antigen) (Tetraspanin-30) (Tspan-30) (CD antigen CD63) |
| − 0.87 | 0.42 | 0.01 | P12532 | Creatine kinase U-type, mitochondrial (EC 2.7.3.2) (Acidic-type mitochondrial creatine kinase) (Mia-CK) (Ubiquitous mitochondrial creatine kinase) (U-MtCK) |
| − 0.95 | 0.81 | 0.00 | P53634 | Dipeptidyl peptidase 1 (EC 3.4.14.1) (Cathepsin C) (Cathepsin J) (Dipeptidyl peptidase I) (DPP-I) (DPPI) (Dipeptidyl transferase) [Cleaved into: Dipeptidyl peptidase 1 exclusion domain chain (Dipeptidyl peptidase I exclusion domain chain); Dipeptidyl peptidase 1 heavy chain (Dipeptidyl peptidase I heavy chain); Dipeptidyl peptidase 1 light chain (Dipeptidyl peptidase I light chain)] |
| − 1.00 | 0.50 | 0.01 | P00167 | Cytochrome b5 (Microsomal cytochrome b5 type A) (MCB5) |
| − 1.20 | 0.55 | 0.01 | O75911 | Short-chain dehydrogenase/reductase 3 (EC 1.1.1.300) (DD83.1) (Retinal short-chain dehydrogenase/reductase 1) (retSDR1) (Retinol dehydrogenase 17) (Short chain dehydrogenase/reductase family 16C member 1) |
| − 0.64 | 0.55 | 0.01 | O60884 | DnaJ homolog subfamily A member 2 (Cell cycle progression restoration gene 3 protein) (Dnj3) (Dj3) (HIRA-interacting protein 4) (Renal carcinoma antigen NY-REN-14) |
| − 1.04 | 0.74 | 0.00 | Q9UK22 | F-box only protein 2 |
| − 1.57 | 0.84 | 0.00 | P04066 | Tissue alpha-L-fucosidase (EC 3.2.1.51) (Alpha-L-fucosidase I) (Alpha-L-fucoside fucohydrolase 1) (Alpha-L-fucosidase 1) |
| − 0.88 | 0.52 | 0.00 | Q96C23 | Aldose 1-epimerase (EC 5.1.3.3) (Galactose mutarotase) |
| − 0.84 | 0.67 | 0.00 | Q9P2T1 | GMP reductase 2 (GMPR 2) (EC 1.7.1.7) (Guanosine 5′-monophosphate oxidoreductase 2) (Guanosine monophosphate reductase 2) |
| − 0.82 | 0.86 | 0.00 | P63096 | Guanine nucleotide-binding protein G(i) subunit alpha-1 (Adenylate cyclase-inhibiting G alpha protein) |
| − 1.37 | 0.81 | 0.00 | P15586 | N-acetylglucosamine-6-sulfatase (EC 3.1.6.14) (Glucosamine-6-sulfatase) (G6S) |
| − 1.00 | 0.64 | 0.00 | Q9NRV9 | Heme-binding protein 1 (p22HBP) |
| − 2.02 | 0.21 | 0.00 | P16401 | Histone H1.5 (Histone H1a) (Histone H1b) (Histone H1 s-3) |
| − 1.70 | 0.35 | 0.01 | P80365 | Corticosteroid 11-beta-dehydrogenase isozyme 2 (EC 1.1.1.-) (11-beta-hydroxysteroid dehydrogenase type 2) (11-DH2) (11-beta-HSD2) (11-beta-hydroxysteroid dehydrogenase type II) (11-HSD type II) (11-beta-HSD type II) (NAD-dependent 11-beta-hydroxysteroid dehydrogenase) (11-beta-HSD) (Short chain dehydrogenase/reductase family 9C member 3) |
| − 3.15 | 0.32 | 0.00 | P08729 | Keratin, type II cytoskeletal 7 (Cytokeratin-7) (CK-7) (Keratin-7) (K7) (Sarcolectin) (Type-II keratin Kb7) |
| − 1.64 | 0.41 | 0.00 | P11279 | Lysosome-associated membrane glycoprotein 1 (LAMP-1) (Lysosome-associated membrane protein 1) (CD107 antigen-like family member A) (CD antigen CD107a) |
| − 1.56 | 0.61 | 0.00 | P13473 | Lysosome-associated membrane glycoprotein 2 (LAMP-2) (Lysosome-associated membrane protein 2) (CD107 antigen-like family member B) (LGP-96) (CD antigen CD107b) |
| − 1.43 | 1.04 | 0.00 | O00462 | Beta-mannosidase (EC 3.2.1.25) (Lysosomal beta A mannosidase) (Mannanase) (Mannase) |
| − 0.99 | 0.53 | 0.00 | Q9H8H3 | Methyltransferase-like protein 7A (EC 2.1.1.-) (Protein AAM-B) |
| − 1.45 | 0.34 | 0.01 | Q92597 | Protein NDRG1 (Differentiation-related gene 1 protein) (DRG-1) (N-myc downstream-regulated gene 1 protein) (Nickel-specific induction protein Cap43) (Reducing agents and tunicamycin-responsive protein) (RTP) (Rit42) |
| − 1.66 | 0.44 | 0.00 | Q5TFE4 | 5′-nucleotidase domain-containing protein 1 (EC 3.1.3.-) |
| − 0.91 | 0.63 | 0.00 | Q92882 | Osteoclast-stimulating factor 1 |
| − 1.21 | 1.36 | 0.00 | Q15124 | Phosphoglucomutase-like protein 5 (Aciculin) (Phosphoglucomutase-related protein) (PGM-RP) |
| − 0.29 | 0.49 | 0.01 | A6NDG6 | Glycerol-3-phosphate phosphatase (G3PP) (EC 3.1.3.21) (Aspartate-based ubiquitous Mg(2 +)-dependent phosphatase) (AUM) (EC 3.1.3.48) (Phosphoglycolate phosphatase) (PGP) |
| − 1.15 | 1.04 | 0.00 | Q86T03 | Type 1 phosphatidylinositol 4,5-bisphosphate 4-phosphatase (Type 1 PtdIns-4,5-P2 4-Ptase) (EC 3.1.3.78) (PtdIns-4,5-P2 4-Ptase I) (Transmembrane protein 55B) |
| − 0.95 | 0.44 | 0.01 | Q8NHP8 | Putative phospholipase B-like 2 (EC 3.1.1.-) (76 kDa protein) (p76) (LAMA-like protein 2) (Lamina ancestor homolog 2) (Phospholipase B domain-containing protein 2) [Cleaved into: Putative phospholipase B-like 2 32 kDa form; Putative phospholipase B-like 2 45 kDa form] |
| − 0.74 | 0.55 | 0.00 | O15162 | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2 +)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b) |
| − 0.72 | 0.56 | 0.00 | O15305 | Phosphomannomutase 2 (PMM 2) (EC 5.4.2.8) |
| − 0.53 | 0.47 | 0.01 | P35813 | Protein phosphatase 1A (EC 3.1.3.16) (Protein phosphatase 2C isoform alpha) (PP2C-alpha) (Protein phosphatase IA) |
| − 1.20 | 0.63 | 0.00 | P42785 | Lysosomal Pro-X carboxypeptidase (EC 3.4.16.2) (Angiotensinase C) (Lysosomal carboxypeptidase C) (Proline carboxypeptidase) (Prolylcarboxypeptidase) (PRCP) |
| − 1.03 | 0.26 | 0.01 | P11216 | Glycogen phosphorylase, brain form (EC 2.4.1.1) |
| − 0.69 | 0.48 | 0.00 | P61106 | Ras-related protein Rab-14 |
| − 0.68 | 0.78 | 0.00 | Q9H0U4 | Ras-related protein Rab-1B |
| − 0.58 | 0.51 | 0.00 | Q9UL25 | Ras-related protein Rab-21 |
| − 0.80 | 0.44 | 0.01 | P51149 | Ras-related protein Rab-7a |
| − 1.64 | 0.18 | 0.00 | P51151 | Ras-related protein Rab-9A |
| − 0.69 | 0.31 | 0.01 | P61224 | Ras-related protein Rap-1b (GTP-binding protein smg p21B) |
| − 1.03 | 0.63 | 0.01 | P61225 | Ras-related protein Rap-2b |
| − 1.04 | 0.30 | 0.00 | Q8TC12 | Retinol dehydrogenase 11 (EC 1.1.1.300) (Androgen-regulated short-chain dehydrogenase/reductase 1) (HCV core-binding protein HCBP12) (Prostate short-chain dehydrogenase/reductase 1) (Retinal reductase 1) (RalR1) (Short chain dehydrogenase/reductase family 7C member 1) |
| − 0.69 | 0.61 | 0.01 | Q14108 | Lysosome membrane protein 2 (85 kDa lysosomal membrane sialoglycoprotein) (LGP85) (CD36 antigen-like 2) (Lysosome membrane protein II) (LIMP II) (Scavenger receptor class B member 2) (CD antigen CD36) |
| − 1.01 | 0.36 | 0.01 | P01011 | Alpha-1-antichymotrypsin (ACT) (Cell growth-inhibiting gene 24/25 protein) (Serpin A3) [Cleaved into: Alpha-1-antichymotrypsin His-Pro-less] |
| − 0.82 | 0.80 | 0.00 | Q9HAT2 | Sialate O-acetylesterase (EC 3.1.1.53) (H-Lse) (Sialic acid-specific 9-O-acetylesterase) |
| − 1.69 | 0.49 | 0.01 | Q6IA17 | Single Ig IL-1-related receptor (Single Ig IL-1R-related molecule) (Single immunoglobulin domain-containing IL1R-related protein) (Toll/interleukin-1 receptor 8) (TIR8) |
| − 0.78 | 0.77 | 0.00 | Q00796 | Sorbitol dehydrogenase (EC 1.1.1.14) (L-iditol 2-dehydrogenase) |
| − 1.60 | 0.50 | 0.00 | Q13488 | V-type proton ATPase 116 kDa subunit a isoform 3 (V-ATPase 116 kDa isoform a3) (Osteoclastic proton pump 116 kDa subunit) (OC-116 kDa) (OC116) (T-cell immune regulator 1) (T-cell immune response cDNA7 protein) (TIRC7) (Vacuolar proton translocating ATPase 116 kDa subunit a isoform 3) |
| − 0.29 | 1.17 | 0.01 | Q9UGI8 | Testin (TESS) |
| − 0.58 | 0.62 | 0.01 | O76062 | Delta(14)-sterol reductase (Delta-14-SR) (EC 1.3.1.70) (Another new gene 1 protein) (C-14 sterol reductase) (Putative sterol reductase SR-1) (Sterol C14-reductase) (Transmembrane 7 superfamily member 2) |
| − 1.92 | 0.05 | 0.01 | Q9NUM4 | Transmembrane protein 106B |
| − 0.71 | 0.38 | 0.00 | Q12792 | Twinfilin-1 (Protein A6) (Protein tyrosine kinase 9) |
| − 0.40 | 1.13 | 0.00 | O15498 | Synaptobrevin homolog YKT6 (EC 2.3.1.-) |
Fig. 2.
a Statistical analysis using linear models as implemented in Limma distinguishes significantly (p < 0.01) affected proteins. b Gene ontology (confined to “cellular compartment”) analysis of proteins that were found to be significantly increased in either RO or chRCC
In order to classify the differentially regulated proteins in RO or chRCC tissue, we performed a gene ontology (GO) enrichment analysis, with a focus on the “cellular compartment” annotation [27, 28]. We chose the TopGO algorithm to minimize GO term redundancy [29–31]. For RO tissue, this analysis points towards an enrichment of mitochondrial proteins whereas lysosomal proteins appear to be enriched in chRCC tissue (Fig. 2b). High abundance of—potentially respiration-defective—mitochondria has been previously named as a distinguishing feature of renal oncocytomas [11, 32, 33]. Likewise lysosomal defects have been identified as a hallmark of ROs [11]. Likewise, an under-representation of endocytotic proteins in ROs has been reported [12] and the presence of vesicular proteins of the tetraspanin family has been proposed as a chRCC marker [8]. We conclude that our proteome profiling reflects hallmark features of ROs and chRCCs.
Differential expression of LAMP1 in renal oncocytomas and chromophobe renal cell carcinoma
Lysosomal defects have been reported for ROs [34] but lysosomal marker proteins have not yet been used to distinguish ROs and chRCCs by IHC. In our proteomic dataset, we noticed significant enrichment of lysosome-associated membrane proteins (LAMPs) 1– 3 in chRCC (Table 3). Of these, we chose LAMP1 for IHC analysis in an extended cohort comprising 42 RO cases and 31 chRCC cases. Although LAMP-2 and -3 were even stronger enriched in chRCCs, we opted for LAMP-1 since we had previously probed LAMP-1 as a prototypical marker of lysosomal biology [35, 36]. Exemplary stainings are shown in Fig. 3a. In the RO cases, we noticed elevated levels of tumor cells with a heterogenous distribution of LAMP1, comprising single or multiple LAMP-1 clusters with focal or apical localization. In contrast, LAMP1 within chRCC tumor cells was often present in a diffuse cytosolic manner (Fig. 3a, b). Similarly different staining patterns (diffuse in chRCC; apical/polar in RO) have been previously reported for the related protein LAMP-3 (referred to as CD63 in the study) [4, 37]. In addition to the different localization, we used a semi-quantitative scoring system to evaluate the LAMP1 presence, covering absence of signal (score 0), weak detection (score 1), medium detection (score 2), and strong detection (score 3). For cells displaying heterogenous LAMP1 staining, the predominant staining intensity was considered; this typically being the weaker signal. Our analysis highlighted that ROs are characterized by weaker LAMP1 presence in contrast to the elevated LAMP1 presence in chRCCs (Fig. 3c). The IHC analysis thus corroborates the proteomic result.
Fig. 3.
a Exemplary immunohistochemistry (IHC) of LAMP1 in RO and chRCC. b Fraction of RO or chRCC cells displaying a heterogenous staining pattern for LAMP1 (statistical significance determined using the two-way Student t-test; values are mean ± standard deviation). b Overview of LAMP1 IHC in the extended cohort using a semi-quantitative scoring system covering absence of intensity (score 0), weak detection (score 1), medium detection (score 2), and strong detection (score 3). Statistical significance determined using the two-way Student t-test; values are mean ± standard deviation; scale bar is 100 μm
To assess a possible correlation of LAMP1 staining with clinical-pathological parameters of chRCC such as overall survival and T1–T4 staging, we are referring to data of the The Human Protein Atlas/Pathology Atlas [38, 39] since our cohort focuses on T1 and T2 stages. LAMP1 showed a tendency for shortened overall survival upon elevated expression but no correlation with tumor stage.
Differential expression of ITGAV in renal oncocytomas and chromophobe renal cell carcinomas
Integrin biology has been rarely reported as a differentiating feature that discriminates RO and chRCC. In our proteomic data, we noticed significant enrichment of ITGAV in the RO cases, which we further investigated by IHC in the extended IHC cohort (see above). In good agreement with the proteomic data, there was comparably strong presence of ITGAV in the RO cases while ITGAV presence in chRCC was sparse (Fig. 4a). This was further corroborated by a semi-quantitative analysis (see above for details) of different staining ITGAV staining intensities and their fraction of the tumor area under investigation (Fig. 4b).
Fig. 4.
a Exemplary immunohistochemistry (IHC) of ITGAV in RO and chRCC. b Overview of ITGAV IHC in the extended cohort using a semi-quantitative scoring system covering absence of intensity (score 0), weak detection (score 1), medium detection (score 2), and strong detection (score 3). Statistical significance determined using the two-way Student t-test; values are mean ± standard deviation, scale bar is 100 μm
Several transcriptome studies found comparable transcript levels of ITGAV, both when comparing RO and chRCC as well as when extending the comparison to other renal neoplasms [8, 12, 40, 41]. However, as previously outlined, there is increasing evidence that mRNA abundance and protein levels only display a limited correlation [13, 14], e.g. due to differential synthesis and degradation rates, thus emphasizing the importance of direct protein analysis by means of mass spectrometry or immuno-detection.
To assess a possible correlation of ITGAV staining with clinical-pathological parameters of chRCC such as overall survival and T1–T4 staging, we are referring to data of the The Human Protein Atlas/Pathology Atlas [38, 39] since our cohort focuses on T1 and T2 stages. ITGAV showed a tendency for shortened overall survival upon elevated expression but no correlation with tumor stage.
Conclusion
We present one of the first proteomic profiling studies to differentiate renal oncocytomas and chromophobe renal cell carcinomas. We found distinguishable proteome profiles, which reflect previously annotated, discriminating features of ROs and chRCCs. Moreover, we identified novel protein candidates for which differential expression between ROs and chRCCs has not yet been described. Using an extended cohort of > 70 RO and chRCC cases, we corroborate strong presence of ITGAV in RO and of LAMP1 in chRCC. Methodologically, our work further validates the robustness of using FFPE material for retrospective quantitative proteomics as a first step for differential marker identification. Extension to further variants of renal cell neoplasms [32] is an intriguing outlook.
Additional files
Additional file 1: Table S1. Labeling scheme setup.
Additional file 2: Table S2. Overview of the > 2400 proteins were identified (false-discovery rate < 1%) and quantified in at least four RO and four chRCC samples.
Authors’ contributions
VD, MW, CAJ, OS conceived the study; BH, VD performed experiments; VD, BH, CS, LK, MW, CAJ, OS analysed data. All authors read and approved the final manuscript.
Acknowledgements
Tissue sampling, archiving, annotation, and accessibility for the presented research was supported by the tumor biobank of the Comprehensive Cancer Center Freiburg (CCCF) and the German Cancer Consortium (DKTK), partner site Freiburg.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [42] partner repository with the dataset identifier PXD009300. Reviewer account details: Username: reviewer06581@ebi.ac.uk, Password: 0PRnXlya.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the University Medical Center Freiburg (311/12). Before study inclusion, all patient data were anonymized.
Funding
OS acknowledges support by Deutsche Forschungsgemeinschaft (SCHI 871/5, SCHI 871/8, SCHI 871/9, SCHI 871/11, INST 39/900-1, and SFB850-Project Z1), the Excellence Initiative of the German Federal and State Governments (EXC 294, BIOSS), the European Research Council (PoC 780730, ProteaseNter), and the German-Israeli Foundation (Grant No. 1444).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ccRCC
cell renal cell carcinoma
- chRCC
chromophobe renal cell carcinoma
- FFPE
formalin-fixed, paraffin-embedded
- IHC
immunohistochemistry
- ITGAV
integrin alpha-V
- LAMP1
lysosomal-associated membrane protein 1
- LC–MS/MS
liquid chromatography–tandem mass spectrometry
- RO
renal oncocytomas
Contributor Information
Vanessa Drendel, Email: vanessadrendel@web.de.
Bianca Heckelmann, Email: bianca.rueb@gmx.de.
Christoph Schell, Email: christoph.schell@uniklinik-freiburg.de.
Lucas Kook, Email: kook.lucas@web.de.
Martin L. Biniossek, Email: martin.biniossek@mol-med.uni-freiburg.de
Martin Werner, Email: martin.werner@uniklinik-freiburg.de.
Cordula A. Jilg, Phone: +49 761 270 28930, Email: Cordula.Jilg@uniklinik-freiburg.de
Oliver Schilling, Phone: +49 761 203 9615, Email: oliver.schilling@mol-med.uni-freiburg.de.
References
- 1.Moch H, Humphrey PA, Ulbright TM, Reuter VE, editors. WHO Classification of tumours of the urinary system and male genital organs. 4. Lyon: International Agency for Research on Cancer, Lyon Cedex, Frankreich; 2016. [Google Scholar]
- 2.Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, Buhay C, Kang H, Kim SC, Fahey CC, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amin MB, Paner GP, Alvarado-Cabrero I, Young AN, Stricker HJ, Lyles RH, Moch H. Chromophobe renal cell carcinoma: histomorphologic characteristics and evaluation of conventional pathologic prognostic parameters in 145 cases. Am J Surg Pathol. 2008;32:1822–1834. doi: 10.1097/PAS.0b013e3181831e68. [DOI] [PubMed] [Google Scholar]
- 4.Ng KL, Morais C, Bernard A, Saunders N, Samaratunga H, Gobe G, Wood S. A systematic review and meta-analysis of immunohistochemical biomarkers that differentiate chromophobe renal cell carcinoma from renal oncocytoma. J Clin Pathol. 2016;69:661–671. doi: 10.1136/jclinpath-2015-203585. [DOI] [PubMed] [Google Scholar]
- 5.Zippel L. Zur Kenntnis der Oncocytome. Virchows Arch A Pathol Anat. 1942;308:360–382. doi: 10.1007/BF02593372. [DOI] [Google Scholar]
- 6.Klein MJ, Valensi QJ. Proximal tubular adenomas of kidney with so-called oncocytic features. A clinicopathologic study of 13 cases of a rarely reported neoplasm. Cancer. 1976;38:906–914. doi: 10.1002/1097-0142(197608)38:2<906::AID-CNCR2820380238>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 7.Reuter VE, Argani P, Zhou M, Delahunt B. Best practices recommendations in the application of immunohistochemistry in the kidney tumors: report from the International Society of Urologic Pathology consensus conference. Am J Surg Pathol. 2014;38:e35–e49. doi: 10.1097/PAS.0000000000000258. [DOI] [PubMed] [Google Scholar]
- 8.Yusenko MV, Kuiper RP, Boethe T, Ljungberg B, van Kessel AG, Kovacs G. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer. 2009;9:152. doi: 10.1186/1471-2407-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koeman JM, Russell RC, Tan MH, Petillo D, Westphal M, Koelzer K, Metcalf JL, Zhang Z, Matsuda D, Dykema KJ, et al. Somatic pairing of chromosome 19 in renal oncocytoma is associated with deregulated EGLN2-mediated [corrected] oxygen-sensing response. PLoS Genet. 2008;4:e1000176. doi: 10.1371/journal.pgen.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frew IJ, Moch H. A clearer view of the molecular complexity of clear cell renal cell carcinoma. Annu Rev Pathol. 2015;10:263–289. doi: 10.1146/annurev-pathol-012414-040306. [DOI] [PubMed] [Google Scholar]
- 11.Joshi S, Tolkunov D, Aviv H, Hakimi AA, Yao M, Hsieh JJ, Ganesan S, Chan CS, White E. The genomic landscape of renal oncocytoma identifies a metabolic barrier to tumorigenesis. Cell Rep. 2015;13:1895–1908. doi: 10.1016/j.celrep.2015.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohan S, Tu JJ, Kao J, Mukherjee P, Campagne F, Zhou XK, Hyjek E, Alonso MA, Chen YT. Gene expression profiling separates chromophobe renal cell carcinoma from oncocytoma and identifies vesicular transport and cell junction proteins as differentially expressed genes. Clin Cancer Res. 2006;12:6937–6945. doi: 10.1158/1078-0432.CCR-06-1268. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Fortelny N, Overall CM, Pavlidis P, Freue GVC. Can we predict protein from mRNA levels? Nature. 2017;547:E19–E20. doi: 10.1038/nature22293. [DOI] [PubMed] [Google Scholar]
- 15.Weisser J, Lai ZW, Bronsert P, Kuehs M, Drendel V, Timme S, Kuesters S, Jilg CA, Wellner UF, Lassmann S, et al. Quantitative proteomic analysis of formalin-fixed, paraffin-embedded clear cell renal cell carcinoma tissue using stable isotopic dimethylation of primary amines. BMC Genom. 2015;16:559. doi: 10.1186/s12864-015-1768-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bronsert P, Weisser J, Biniossek ML, Kuehs M, Mayer B, Drendel V, Timme S, Shahinian H, Kusters S, Wellner UF, et al. Impact of routinely employed procedures for tissue processing on the proteomic analysis of formalin-fixed paraffin-embedded tissue. Proteomics Clin Appl. 2014;8:796–804. doi: 10.1002/prca.201300082. [DOI] [PubMed] [Google Scholar]
- 17.Gustafsson OJ, Arentz G, Hoffmann P. Proteomic developments in the analysis of formalin-fixed tissue. Biochim Biophys Acta. 2015;1854:559–580. doi: 10.1016/j.bbapap.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Muller AK, Foll M, Heckelmann B, Kiefer S, Werner M, Schilling O, Biniossek ML, Jilg CA, Drendel V. Proteomic characterization of prostate cancer to distinguish nonmetastasizing and metastasizing primary tumors and lymph node metastases. Neoplasia. 2018;20:140–151. doi: 10.1016/j.neo.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drendel V, Heckelmann B, Chen CY, Weisser J, Espadas G, Schell C, Sabido E, Werner M, Jilg CA, Schilling O. Proteome profiling of clear cell renal cell carcinoma in von Hippel-Lindau patients highlights upregulation of Xaa-Pro aminopeptidase-1, an anti-proliferative and anti-migratory exoprotease. Oncotarget. 2017;8:100066–100078. doi: 10.18632/oncotarget.21929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boersema PJ, Aye TT, van Veen TA, Heck AJ, Mohammed S. Triplex protein quantification based on stable isotope labeling by peptide dimethylation applied to cell and tissue lysates. Proteomics. 2008;8:4624–4632. doi: 10.1002/pmic.200800297. [DOI] [PubMed] [Google Scholar]
- 21.Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods. 2010;7:383–385. doi: 10.1038/nmeth.1446. [DOI] [PubMed] [Google Scholar]
- 22.Shahinian H, Loessner D, Biniossek ML, Kizhakkedathu JN, Clements JA, Magdolen V, Schilling O. Secretome and degradome profiling shows that Kallikrein-related peptidases 4, 5, 6, and 7 induce TGFbeta-1 signaling in ovarian cancer cells. Mol Oncol. 2014;8:68–82. doi: 10.1016/j.molonc.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 24.Gomez-Auli A, Hillebrand LE, Biniossek ML, Peters C, Reinheckel T, Schilling O. Impact of cathepsin B on the interstitial fluid proteome of murine breast cancers. Biochimie. 2016;122:88–98. doi: 10.1016/j.biochi.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004; 3:Article 3. [DOI] [PubMed]
- 27.Gene Ontology C Gene ontology consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexa A, Rahnenfuhrer J. topGO: enrichment analysis for gene ontology. R package version 2240 2016.
- 30.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 31.Gentleman R, Carey V, Dudoit S, Ellis B, Gautier L, Gentry J, Huber W, Irizarry R, Rossini A, Smyth G, Zhang J. The bioconductor project. Technical report Dana Farber Cancer Institute, Boston, MA, USA; 2003.
- 32.Yusenko MV. Molecular pathology of renal oncocytoma: a review. Int J Urol. 2010;17:602–612. doi: 10.1111/j.1442-2042.2010.02574.x. [DOI] [PubMed] [Google Scholar]
- 33.Tickoo SK, Lee MW, Eble JN, Amin M, Christopherson T, Zarbo RJ, Amin MB. Ultrastructural observations on mitochondria and microvesicles in renal oncocytoma, chromophobe renal cell carcinoma, and eosinophilic variant of conventional (clear cell) renal cell carcinoma. Am J Surg Pathol. 2000;24:1247–1256. doi: 10.1097/00000478-200009000-00008. [DOI] [PubMed] [Google Scholar]
- 34.Chen F, Zhang Y, Senbabaoglu Y, Ciriello G, Yang L, Reznik E, Shuch B, Micevic G, De Velasco G, Shinbrot E, et al. Multilevel genomics-based taxonomy of renal cell carcinoma. Cell Rep. 2016;14:2476–2489. doi: 10.1016/j.celrep.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tholen S, Biniossek ML, Gansz M, Ahrens TD, Schlimpert M, Kizhakkedathu JN, Reinheckel T, Schilling O. Double deficiency of cathepsins B and L results in massive secretome alterations and suggests a degradative cathepsin-MMP axis. Cell Mol Life Sci. 2014;71:899–916. doi: 10.1007/s00018-013-1406-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kern U, Wischnewski V, Biniossek ML, Schilling O, Reinheckel T. Lysosomal protein turnover contributes to the acquisition of TGFbeta-1 induced invasive properties of mammary cancer cells. Mol Cancer. 2015;14:39. doi: 10.1186/s12943-015-0313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mete O, Kilicaslan I, Gulluoglu MG, Uysal V. Can renal oncocytoma be differentiated from its renal mimics? The utility of anti-mitochondrial, caveolin 1, CD63 and cytokeratin 14 antibodies in the differential diagnosis. Virchows Arch. 2005;447:938–946. doi: 10.1007/s00428-005-0048-6. [DOI] [PubMed] [Google Scholar]
- 38.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Tissue-based map of the human proteome. Science. 2015;347:126419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 39.Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:1–11. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 40.Higgins JP, Shinghal R, Gill H, Reese JH, Terris M, Cohen RJ, Fero M, Pollack JR, van de Rijn M, Brooks JD. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol. 2003;162:925–932. doi: 10.1016/S0002-9440(10)63887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 42.Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Labeling scheme setup.
Additional file 2: Table S2. Overview of the > 2400 proteins were identified (false-discovery rate < 1%) and quantified in at least four RO and four chRCC samples.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [42] partner repository with the dataset identifier PXD009300. Reviewer account details: Username: reviewer06581@ebi.ac.uk, Password: 0PRnXlya.