Abstract
Following recent advances in behavioral and psychiatric epigenetics, researchers are increasingly using epigenetic methods to study prenatal exposure to maternal mood disorder and its effects on fetal and newborn neurobehavior. Despite notable progress, various methodological limitations continue to obscure our understanding of the epigenetic mechanisms underpinning prenatal exposure to maternal mood disorder on newborn neurobehavioral development. Here we detail this problem, discussing limitations of the currently dominant analytical approaches (i.e., candidate epigenetic and epigenome-wide association studies), then present a solution that retains many benefits of existing methods while minimizing their shortcomings — epigenetic pathway analysis. We argue that the application of pathway-based epigenetic approaches that target DNA methylation at transcription factor binding sites could substantially deepen our mechanistic understanding of how prenatal exposures influence newborn neurobehavior.
Interest in behavioral epigenetics is currently booming in response to its potential to identify mechanisms through which environmental exposures become biologically embedded to affect infant behavioral and psychological traits (Boyce & Kobor, 2015; Kinsella & Monk, 2009). This opportunity is afforded by recent breakthroughs in our understanding of, and ability to investigate epigenetics—molecular processes occurring on and around the genome that regulate gene activity without changing the underlying DNA sequence (Bird, 2007). Human developmental researchers studying epigenetic processes have already achieved notable successes linking maternal experience to prenatal origins of health, with robust case studies showing epigenetic mechanisms partially mediate the effects of some prenatal maternal exposures (e.g., nutrition) on subsequent child health and behavior (Heijmans et al., 2008; Painter et al., 2008; Tobi et al., 2009). Despite such advances, mechanisms underpinning associations between prenatal maternal mood disorder and newborn behavioral outcomes remain poorly understood. This is unfortunate given evidence that mood disorders are disproportionately prevalent among pregnant women and appear to confer risk for altered fetal development and early childhood behavior (Kinsella & Monk, 2009).
We first provide a general overview of mechanisms by which prenatal exposure to maternal mood disorder may affect fetal and newborn neurobehavior in part via epigenetic processes. We then outline the two dominant approaches for examining epigenetic effects: the candidate epigenetic approach and epigenome-wide association studies (EWAS). We describe advantages and disadvantages of these approaches and briefly outline studies that have used these methods to examine associations between prenatal exposures and fetal and newborn behavioral outcomes. Recently, concerns have been raised about the use of the word “epigenetics” as being “ambiguous, over-encompassing, and uncoupled from its historical roots” (Greally, 2018, p. 207). We respond to this important criticism by advancing a functional epigenetic pathway approach that capitalizes on the strengths, and minimizes the weaknesses, of both candidate epigenetic and EWAS methods.
Prenatal exposure to maternal mood disorder and neurobehavioral outcomes: The role of the HPA axis
There is no question that exposure to maternal mood disorder during early childhood is a risk factor for the development of psychopathology in the child (Goodman et al., 2011). Exposure to maternal mood disorder, such as depression, is associated with altered neuroendocrine stress responses in offspring (Essex, Klein, Cho, & Kalin, 2002) and a more fearful and reactive temperament (Davis et al., 2007). These children are also more likely to exhibit internalizing symptoms in early childhood and anxiety in adolescence (Goodman et al., 2011). Importantly, there is evidence that this risk may be partially transmitted from mother to child in utero (Monk et al., 2016). This route of risk transmission is especially important to understand given that fetal exposure to maternal mood disorder may program fetal and newborn neurobehavior and thus vulnerability to the development of psychological disorders later in life—a third pathway beyond shared genes and postnatal environmental expsures by which the next generation inherits familial vulnerability to psychopathology (Monk, Spicer, & Champagne, 2012).
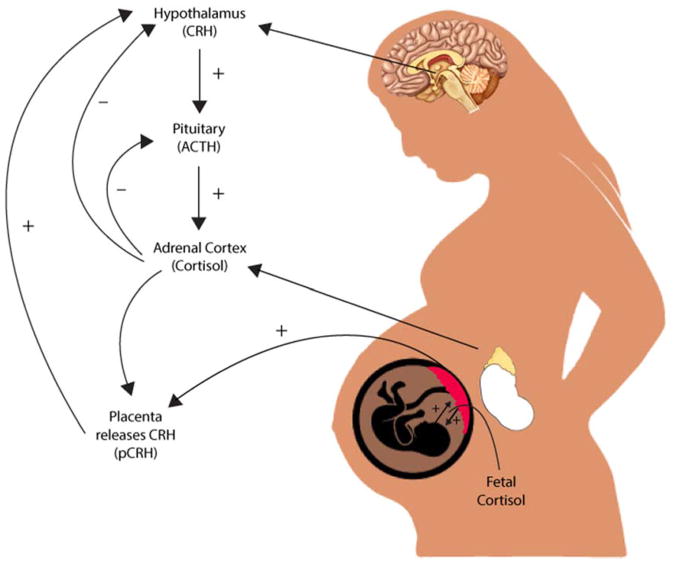
The HPA axis is the most widely studied mechanism linking prenatal exposure to maternal mood and newborn neurobehavior. These associations are far from clear and there are additional mechanisms of interest, such as immune system functioning and the microbiome (Beijers, Buitelaar, & de Weerth, 2014), but few have generated as much empirical evidence as the HPA axis. Pregnant women with mood disorders are more likely to have greater circulating levels of cortisol in the blood compared to their non-depressed peers (Murphy et al., 2015). They also exhibit greater concentrations of corticotropin releasing hormone (CRH) in cerebrospinal fluid, which are reduced following ECT treatment (Kasckow, Baker, & Geracioti, 2001). When the HPA axis is activated, CRH is released from the hypothalamus, which stimulates the synthesis of ACTH by the pituitary and leads to the eventual release of glucocorticoids (cortisol) by the adrenal glands (see Figure 1). However, there is substantial variability in mood disorders. More recent hypotheses suggest that pregnant women exhibiting “atypical” depression—characterized by rejection sensitivity, agitation, irritability, and mood reactivity—exhibit a blunted diurnal cortisol response and lower cortisol levels upon awakening (Glynn, Davis, & Sandman, 2013).
Figure 1.
The HPA axis regulation in the maternal-placental-fetal unit. During pregnancy, the placenta releases corticotropin-releasing hormone (CRH) into the fetal and maternal bloodstream. Both maternal and fetal cortisol increases production of placental CRH, which rises across the course of gestation, and leads to increases in maternal and fetal cortisol.
In typical human pregnancies, maternal cortisol levels are five to 10 times higher than cortisol levels of the fetus (Ishimoto & Jaffe, 2011). Higher-than-normal levels of circulating cortisol in the maternal bloodstream could cross the placenta (which typically filters out 80–90% of maternal cortisol), thus exposing the fetus to excess cortisol and potentially reprogramming the fetal HPA axis (Sandman & Davis, 2012). When a pregnant woman experiences stress, glucocorticoids also activate CRH expression in the placenta, leading to greater synthesis and release of cortisol in the fetal and maternal bloodstream. The release of placental CRH does not inhibit the maternal HPA axis but instead leads to greater increases in cortisol until parturition (Figure 1; Sandman, Glynn, & Davis, 2016)). There is a great deal of evidence suggesting that placental CRH production may be accelerated by an adverse intrauterine environment characterized by high levels of prenatal stress exposure (Sandman et al., 2016). Greater CRH release, for instance, occurs when placental cells are exposed to cortisol, catecholamines, and proinflammatory cytokines (Sandman & Davis, 2012), many of which are also associated with maternal mood disorder. These elevations could alter fetal brain structures such as the amygdala, as is the case in animal models (Salm et al., 2004) and human studies (Buss et al., 2012).
CRH exposure also affects fetal HPA axis development, especially near term, by stimulating the release of fetal cortisol from the fetal adrenal axis (Sirianni, Rehman, Carr, Parker, & Rainey, 2005). An increase in CRH, which accompanies typical human pregnancy, also impacts fetal responses to novelty and greater fetal heart rate reactivity, suggesting that the fetal nervous system is adversely affected by greater than typical increases in CRH (Sandman et al., 2016). There are also effects of maternal stress on newborn stress regulation, fearful temperament, and brain development (Sandman et al., 2016). Notably, children exposed to high levels of maternal cortisol in utero were twice as likely to exhibit borderline or clinically significant levels of anxiety in childhood (Sandman & Davis, 2012) and had a right amygdala that was 6.4% larger than those with lower exposure (Buss et al., 2012). Greater exposure to high levels of maternal cortisol in the second half of gestation was also related to a larger, prolonged cortisol response to stress in neonates (Buss et al., 2012).
There are a number of important epigenetic mechanisms that may underlie the association of prenatal exposure to maternal mood disorder on fetal and newborn neurobehavioral outcomes, including histone modification, genomic imprinting, and DNA methylation. Here we focus on DNA methylation given recent breakthroughs using this platform in behavioral epigenetics (Weaver et al., 2004), and the enhanced feasibility of integrating methylation assays into behavioral and biomedical studies (Adkins, Rasmussen, & Docherty, 2017). DNA methylation refers to the attachment of a small molecule (i.e., methyl group) to a specific DNA position (i.e., genomic locus). While the full range of DNA methylation’s functional effects are unknown, one of its primary functions is to silence the expression of the associated gene by blocking the interaction of regulatory molecules to the methylated DNA (Bird, 2007). That is, when a methyl group is attached to a specific locus in a given gene’s promoter region, specifically transcription factor binding sites, it can block the process of gene expression, therefore silencing the anticipated activity of that gene (Bird, 2007). Conversely, methylation in other regions of the genome, gene bodies, for instance, can instead increase gene expression (Jones, 2012).
Recent calls have been made to expand focus beyond the HPA axis (Beijers et al., 2014), with which we concur. We therefore describe the functional epigenetic pathway targeting the HPA axis as a proof of principle, but describe how this pathway approach can generalize to any known pathway, or even examine the full range of known biological pathways (Docherty, Moscati, Adkins, et al., 2017; Docherty, Moscati, Dick, et al., 2017). We argue that a bioinformatics approach to analyzing epigenetic data that focuses on a specific, theory-driven biological pathway of interest will advance the field of behavioral epigenetics and provide much needed mechanistic insights into how epigenetic processes could be implicated in the development of neurobehavioral disorders. We further highlight the importance of targeting CpG sites associated with known transcription factor binding sites, given that it is in these regions that DNA methylation is likely to lead to differences in gene expression (Bird, 2007).
Below we review advantages and disadvantages of candidate epigenetic approaches, some of which have found associations between DNA methylation at transcription factor binding sites and newborn neurobehavior. We assert that this epigenetic pathway approach targeting transcription factor binding sites is likely to significantly advance our mechanistic understanding of biological embedding, and add much needed specificity to programming theories related to prenatal exposures and neurobehavioral development (Greally, 2018; Lappalainen & Greally, 2017).
Candidate epigenetic studies: Advantages and disadvantages
Candidate epigenetic studies have, until recently, dominated the nascent field of behavioral epigenetics (Conradt, 2017; Lester, Conradt, & Marsit, 2016). Indeed, much of the empirical work to date has focused on a single candidate—the glucocorticoid receptor gene, NR3c1, which is involved in reuptake of cortisol throughout the body, but most notably in the hippocampus (Herman & Spencer, 1998). Research into the topic began with the observation of maternal effects on offspring differences in behavior and HPA stress responses in rodent models (Meaney, 2010). In these models, maternal behavior was shown to influence HPA responses to stress in offspring through tissue-specific effects on gene expression (Weaver et al., 2004), including glucocorticoid receptor expression in hippocampal brain areas. This region is of interest given that glucocorticoid activation inhibits HPA activity via negative feedback inhibition (Herman & Spencer, 1998). These findings were further supported by evidence that familial dysfunction and childhood adversity are also linked to altered HPA stress response in humans, which, in turn, is associated with increased risk for various types of psychopathology (De Bellis et al., 1994; Heim & Nemeroff, 2001). Building on this body of research, Meaney and colleagues combined rodent models with emerging epigenetic technologies to show that adult offspring who were born to mothers who engaged in low rates of maternal care show increased methylation at a glucocorticoid receptor gene (NR3c1) promoter in hippocampus, which upregulates HPA stress response in adulthood (Meaney, 2010). Recently, increased methylation at the NR3c1 promoter has been observed in human suicide victims, and studied in relation to psychosocial stress in humans, with mixed results (McGowan et al., 2009; Turecki & Meaney, 2016).
It is noteworthy that the trauma-mood disorder link appears to be mediated partially by epigenetic modifications to NR3c1 in the hippocampus. This finding has served as an important proof of principle for behavioral epigenetic research, and consequently, attracted tremendous resources to epigenetic studies of candidate genes with known biological function. Thus, while this approach has yielded profound insights into the biological embedding of social adversity (Essex et al., 2013; Monk et al., 2012) its success has also led to an overly narrow focus on candidate epigenetic studies, particularly those related to glucocorticoid function. This emphasis on candidate epigenetic studies was due, in part, to the once prohibitive cost of epigenome-wide association studies (EWAS); however, recent technological advances have rendered these studies cost-effective and easily implemented (Adkins et al., 2017).
Nonetheless, a major advantage of the candidate epigenetic approach is that these studies are theoretically-driven with clear hypothesized relations between gene function and outcome. They also provide more precise mechanistic insights into the nature of how environmental exposures or insults may affect later child behavior (Keating, 2016).
The principal disadvantage of candidate epigenetic studies is that they likely over-simplify what is known to be an extremely complex process. While once necessitated by technological limits, it has long been recognized that focus a single gene, locus, or epigenetic mark suffers a profound research limitation in its disregard for the well-established, highly multifactorial, polygenic etiology of complex traits (Lester et al., 2016). In addition, maternal mood disorder is complex and can be associated with multiple comorbid diagnoses (e.g., personality disorders), health practices (e.g., prenatal care), and risk factors (e.g., second-hand smoke exposure). To date there has been a seemingly arbitrary focus on select CpG sites (or a range of CpG sites) within a candidate gene, with relatively little theoretical justification for the selection of those sites, which clearly can introduce Type I errors. Emphasizing the effect on one gene or range of CpG sites obscures the fact that a network of genes are most likely turned on or off by the exposure. Candidate epigenetic approaches stemming from the animal literature are attractive because they demonstrate the importance of how a single gene may be causally implicated in a behavioral outcome. However, we should not be seduced by these elegant studies when trying to translate the animal research to humans since it is impossible to gain comparable experimental control.
Furthermore, we risk being short-sighted with respect to the epigenetic influences on behavior, due to an over-emphasis in the published literature of the effects of a handful of genes on a child phenotype of interest (e.g., NR3c1, 11β-HSD2, and FKBP5). This is concerning given recent evidence that effect sizes are small and often fail correction for multiple comparisons (Breton et al., 2017; Mansell et al., 2016). Another disadvantage is that we typically select a candidate gene because it is involved in a known physiological or biochemical process (Zhu & Zhao, 2007). Yet we have a limited understanding of the biological etiology of complex phenotypes, such as mood disorders, and because of this we may be ignoring a network of genes that play a role in the prenatal origins of depression and other psychopathology. Finally, many candidate epigenetic studies fail to account for cellular heterogeneity. Within any given tissue, including brain and blood, there are many different types of cells, and high levels of DNA methylation found in a particular tissue may be due to the large numbers of particular kinds of cells, such as T-cells, found in the tissue of one individual over another. For instance, candidate DNA methylation studies published using blood samples may actually be confounded by blood heterogeneity between individuals (de Goede et al., 2015). Although there are statistical methods to control for cell type in blood, buccal, and placental samples (Houseman et al., 2012), these controls are rarely used in developmental behavioral epigenetic research (for an exception, see Paquette, Houseman, Green, Lesseur, Armstrong, Lester, & Marsit, 2016).
Of 13 studies identified through a PubMed search on associations between an epigenetic mechanism and fetal or newborn neurobehavior, 11 utilized a candidate gene approach (Table 1; 7/13 focused on NR3c1; 6/13 on HSD11β2). Seven examined how DNA methylation of NR3c1 was related to individual differences in newborn neurobehavior. Greater DNA methylation of NR3c1 likely impacts altered glucocorticoid receptor gene expression in the hippocampus, resulting in greater circulating cortisol in the blood (Meaney, 2010). DNA methylation of NR3c1 has been associated with infant movement and attention (Bromer, Marsit, Armstrong, Padbury, & Lester, 2013), and infant attention, self-regulation, lethargy, and soothability in a group of newborns exposed to maternal smoking in utero (Stroud, Papandonatos, Salisbury, Phipps, Huestis, Miaura, Padbury, Marsit, & Lester, 2016). Interactions between NR3c1 and maternal depression are related to newborn self-regulation, lethargy, and hypotonia (Conradt, Lester, Appleton, Armstrong, & Marsit, 2013).
Table 1.
Studies examining associations between epigenetic processes and fetal or newborn neurobehavior
| Authors | Sample size | Tissue type | Genes targeted | Transcription factor binding sites targeted? | Findings |
|---|---|---|---|---|---|
| Appleton, Lester, Armstrong, Lesseur, & Marsit, 2015 | 372 | Placenta | NR3c1 exon 1F (average of 13 CpG sites), HSD11β2, promoter (average of 3 CpG sites) | No | Interactions between NR3c1, 11β-HSD2 predicted habituation, excitability, and asymmetrical reflexes |
| Bromer, Marsit, Armstrong, Padbury, & Lester, 2013 | 186 | Placenta | NR3c1 exon 1F (average of 13 CpG sites) | No | Main effects of DNA methylation of NR3c1 on infant movement and infant attention |
| Conradt, Lester, Appleton, Armstrong, & Marsit, 2013 | 521 | Placenta | NR3c1 CpG2 on exon 1F; HSD11β2, CpG4 on promoter | No | Interactions between DNA methylation of NR3c1 CpG2 and maternal depression predicted self-regulation, lethargy, and hypotonia; Interactions between 11β-HSD2 CpG4 and maternal prenatal anxiety predicted hypotonia |
| Lesseur, Armstrong, Murphy, Appleton, Koestler, Paquette, Lester, & Marsit, 2014 | 444 | Placenta | LEP promoter | No | DNA methylation of LEP related o lethargy and hypotonicity in males |
| Lester, Marsit, Giarraputo, Hawes, LaGasse, & Padbury, 2015 | 67 | Buccal cells | NR3c1 CpG3 on exon 1F; HSD11β2, CpG3 on promoter | Yes (transcription factor NGF1-A and E2F1) | “high risk” neurobehavioral profile associated with differential DNA methylation of NR3c1 CpG3 on exon 1F and 11β-HSD2 CpG3 in promoter |
| Maccani, Koestler, Lester, Houseman, Armstrong, Kelsey, & Marsit, 2015 | 192 | Placenta | EWAS | No | “high risk” neurobehavioral profile associated with DNA methylation differences in 10 loci, 6 of which resided in the EMID2 gene |
| Marsit, Lambertini, Maccani, Koestler, Houseman, Padbury, Lester, & Chen, 2012 | 106 | Placenta | 22 imprinted candidate genes | No (assessed for gene expression only) | Imprinted genes were associated with quality of movement and handling |
| Marsit, Maccani, Padbury, & Lester, 2012 | 185 | Placenta | HSD11β2, promoter (average of 4 CpG sites) | No | DNA methylation of 11β-HSD2 promoter related to quality of movement |
| Monk, Feng, Lee, Krupska, Champagne, & Tycko, 2016 | 61 | Placenta | HSD11β2, NR3C1, and FKBP5 | No | Elevated DNA methylation of HSD11β2 and FKBP5 related to lower coupling of fetal HR and movement |
| Paquette, Houseman, Green, Lesseur, Armstrong, Lester, & Marsit, 2016 | 335 | Placenta | EWAS | No | Infant attention related to DNA methylation of FHIT, ANKRD11 |
| Paquette, Lesseur, Armstrong, Koestler, Appleton, Lester, & Marsit, 2013 | 444 | Placenta | HTR2A | No | HTR2A methylation related to quality of movement and attention |
| Paquette, Lester, Lesseur, Armstrong, Guerin, Appleton, & Marsit, 2015 | 537 | Placenta | NR3c1, HSD11β2, FKBP5, and ADCYAP1R1 | Yes | Increased methylation of NR3c1 and HSD11β2 related to reactive, poorly regulated NNNS profile |
| Stroud, Papandonatos, Salisbury, Phipps, Huestis, Miaura, Padbury, Marsit, & Lester, 2016 | 45 | Placenta | NR3c1, | No | DNA methylation of NR3c1 related to infant attention, self-regulation, lethargy, and soothability |
HSD11β2 may regulate the extent to which the fetus is exposed to maternal cortisol given that this gene functions to convert maternal cortisol to inert cortisone. Greater methylation of this gene should therefore result in increased fetal cortisol exposure. Monk and colleagues (Monk et al., 2016) found that higher levels of maternal stress in the second trimester was related to greater methylation of placental HSD11β2 methylation and, in turn, decreased fetal coupling in the 3rd trimester, thought to reflect impaired central nervous system development. DNA methylation of HSD11β2 has was also associated with quality of movement (Marsit, Maccani, Padbury, & Lester, 2012) and to a reactive, poorly regulated newborn neurobehavioral profile (Paquette, Lester, Lesseur, Armstrong, Guerin, Appleton, & Marsit, 2015). Interactions between HSD11β2 and NR3c1 methylation were associated with habituation, excitability, and poor reflexes (Appleton, Lester, Armstrong, Lesseur, & Marsit, 2015). These findings converge on the premise that exposure to maternal mood disorder may result in increased methylation of placental genes that expose the fetus to greater levels of cortisol in utero, which may then impact the infant’s neuroendocrine system and newborn-child neurobehavior.
Epigenome-wide association studies: Advantages and disadvantages
Epigenome-wide association studies (EWAS) is the emerging dominant analytical paradigm in behavioral epigenetics, and is apt to continue its rapid expansion given significant advantages over candidate epigenetic approaches. Its principle advantage lies in the ability to investigate the entirety of the epigenome in an unbiased fashion, the vast majority of which is poorly characterized and of unknown function (Swedish Schizophrenia Consortium et al., 2015). Such comprehensive epigenomic approaches are critical given our limited understanding of both the neurobiological mechanisms underlying human psychopathology, and the epigenetic mechanisms regulating gene expression (Szyf, 2012). As the effects of prenatal exposures likely affect multiple biological systems and various functionally relevant genomic elements, an optimal approach must be comprehensive (Maccari, Krugers, Morley-Fletcher, Szyf, & Brunton, 2014). Many current EWAS methylation assays satisfy these criteria (Adkins et al., 2017).
A growing body of research has applied EWAS methylation approaches to study psychopathology, typically using array-based methods, which are standardized chips assaying a limited number of specific methylation loci (typically ≲ 850K CpG sites; (Bock, 2012). Fundamentally, such EWAS approaches are analytically agnostic, as they aim to unbiasedly assay the entire methylome. Thus, EWAS is liberated from the narrow restriction of looking only under the lampposts of existing biological knowledge, a fundamental limitation of candidate gene methods (Adkins et al., 2017). EWAS is also critical to hypothesis generation as an inductive discovery tool, the findings of which can then be further refined in deductive, targeted epigenetic analyses. And while most epigenetic studies of mood disorders have used candidate gene approaches, several EWAS, using a variety of technologies and examining hundreds of thousands of methylation markers across a range of tissues, have also been reported (Labonté et al., 2012, 2013; Nagy et al., 2015; Weder et al., 2014). For example, epigenome-wide association studies have implicated several putative risk methylation marks for mood disorders, including loci at CPSF3, LASS2, PRIMA1, ZNF263, ID3, TPPP, GRIN1, STK32C, and ZBTB20 (Davies et al., 2014; Dempster et al., 2014; Weder et al., 2014).
EWAS is, however, not without its disadvantages. EWAS is frequently severely underpowered due to a combination of small sample sizes, small effect sizes, and punitive multiple test corrections (Tsai & Bell, 2015). And while the cost of EWAS has dropped significantly, assays are still relatively expensive and large samples remain rare (Breton et al., 2017). Moreover, the analytical problem of multiple testing in massively dimensional context has only increased as assay sizes have increased from 450K arrays, to 850K arrays, to next-generation sequencing methods assaying tens of millions of sites (Adkins et al., 2017). Relatedly, EWAS is further limited by its failure to take advantage of the massive amounts of high quality, prior information on genomic elements and biological pathways that is freely available through open access bioinformatics databases (Huber et al., 2015).
While accounting for correlated tests using false discovery rates can substantially increase power compared to Bonferroni correction (Benjamini, Y. & Hochberg, Y., 1995; Swedish Schizophrenia Consortium et al., 2015), it remains the case that effect sizes must be large in order to be detected in methylome-wide context (i.e., p ≲ 5×10−6). Unfortunately, effect sizes in EWAS, similar to genome-wide association studies (GWAS), tend to be small (Breton et al., 2017), necessitating large samples to robustly detect individual effects. For these reasons, to date, EWAS has suffered replication problems, particularly in psychiatry where few findings have replicated in large independent samples (McCarthy et al., 2008). In addition to limitations in statistical power, the agnosticism of the EWAS approach also limits the approach by ignoring the immense body of high-quality prior information that is publicly available in bioinformatics databases and published summary statistics. This has prevented EWAS approaches from boosting power through aggregating individual small effects into functional pathways and polygenic risk scores (de Moor et al., 2015; Docherty, Moscati, Dick, et al., 2017), while also limiting our ability to extract mechanistic biological inferences from EWAS findings.
Two out of the 13 studies testing epigenetic-neurobehavioral associations used EWAS (Maccani et al., 2015; Paquette et al., 2016). Maccani and colleagues found that DNA methylation differences between newborns who were characterized as “high risk” due to their poor regulation; 6 out of 19 of these loci came from a gene EMID2, which is implicated in asthma and neural tube patterning. Paquette and colleagues found that DNA methylation of FHIT and ANKRD11, associated with brain and neurobehavioral development and placental physiology, were related to infant attention. These EWAS studies are likely to yield novel genes and genetic regions associated with newborn neurobehavior. However, it is not clear why some of these genes may be involved in neurobehavioral development. We turn now to a description of the functional epigenetic pathway that leverages the strengths of the candidate epigenetic and EWAS methods to interrogate epigenetic alterations in a known biological pathway of interest.
Functional epigenetic pathway analyses
We argue here for a hybrid candidate epigenetic/EWAS approach that we term functional epigenetic pathway. This approach capitalizes on the advantages of the candidate epigenetic and EWAS techniques while minimizing their disadvantages. It is theory-driven and utilizes state-of-the art bioinformatic techniques to provide mechanistic insights into how an exposure may be related to an epigenetic phenomenon and phenotypic outcome. We should emphasize that while most of the literature to date has examined associations between maternal mood disorders, genes that regulate HPA axis functioning and fetal and newborn neurobehavior, the physiological data linking maternal mood and HPA axis functioning have been mixed.
A functional epigenetic pathway may advance the science of prenatal programming research in two ways: first, by targeting a known biological pathway of interest, and second by examining transcription factor binding sites. It is important to target transcription factor binding sites because of their role in gene expression. DNA methylation can lead to the silencing of the anticipated activity of a gene but only if DNA methylation occurs close to transcription factor binding sites. Therefore this approach responds to recent concerns raised about the definition of the term epigenetic since this functional approach assumes that the phenotype of interest is mediated by DNA methylation at transcription factor binding sites (Greally, 2018). Given their importance for regulating gene expression, it is surprising that these regions are typically not targeted in behavioral epigenetic research. Of the 13 studies reviewed investigating the possible epigenetic basis for effects of prenatal exposure to maternal mood disorder on fetal and newborn neurobehavior, two focus on these transcription factor binding sites (see Table 1). This may be because the field of behavioral epigenetics is in its infancy, and initial investigations focused on finding any possible associations between a prenatal exposure, DNA methylation, and a behavioral outcome. We argue that to advance the field of behavioral epigenetics, it is important to target DNA methylation of transcription factor binding sites, as opposed to other regions on the gene that may not have any functional significance, with the important caveat that, in the absence of tests of this mediation at the cellular level, we cannot infer direction of effect (e.g., that DNA methylation → transcription factor binding or vice-versa; Lappalainen & Greally, 2017).
Our goal is to redirect the field of prenatal programing and behavioral epigenetics to focus on a functional epigenetic pathway approach targeting the HPA axis. This would be accomplished first through a comprehensive literature review identifying published associations between genes and the biological pathway of interest. This theory-driven approach adds specificity to current prenatal programming theories (Nigg, 2016) by targeting epigenetics marks along a hypothesized biological pathway of interest (in this case, the HPA axis). There are also public pathway databases available (for example the Broad Institute’s Molecular Signatures Database v6.0 (MSigDB; Subramanian et al., 2005) that allow researchers to identify co-regulated gene networks in, for example, genes associated with the HPA axis. Following the identification of relevant genes, EWAS can be conducted that could generate additional novel sites associated with the biological pathway of interest. This information can be pooled together to generate a long list of specific genes and gene regions that, after controlling for multiple comparison, are associated with the biological pathway, termed the functional epigenetic pathway. The functional epigenetic pathway should then be replicated in an independent sample. If successful, this pathway could generate novel therapeutic targets for mitigating the risk of prenatal exposure to maternal mood on neurobehavioral development.
It is also possible to take a polygenic risk score approach to identifying which CpG sites may have strong associations with either prenatal exposure to maternal mood disorder or newborn neurobehavior. While the polygenic risk score approach has recently become enormously popular and productive in the analysis of GWAS genotype data (Gandal, Leppa, Won, Parikshak, & Geschwind, 2016), there has been very little application of the approach to genome-wide methylation data. Thus, polygenic risk score holds tremendous promise for epigenetic research, and is currently limited only by the size of existing training data (e.g., prior EWAS results for maternal mood). This approach leverages techniques used in bioinformatics that are already being applied in cancer research and molecular biology, but have yet to be applied to the study of human behavior.
Limitations and Conclusions
In this paper we have introduced a novel approach to studying epigenetic processes. The functional epigenetic pathway method uses bioinformatics to identify epigenetic markers empirically rather than using traditional candidate epigenetic or EWAS methods. This technique capitalizes on the strengths of both the candidate epigenetic and EWAS methods by being theory-driven while also opening up the exploration process to discover larger epigenetic networks. Ideally, such networks will be functionally related and associated with known biological mechanisms underlying problematic newborn neurobehavior. However, it is also possible that novel pathways or interconnections will be uncovered with this technique, enriching our understanding of biological underpinnings of complex phenotypes. The functional epigenetic pathway approach proposed here builds upon similar work in the genetics literature, which has also increasingly applied bioinformatics to uncover gene networks.
Our novel approach to investigating how epigenetic processes may be implicated in prenatal exposure to maternal mood disorder is not without its limitations. First, it is extremely difficult to ascertain direction of effect in human behavioral epigenetic studies and any associations between DNA methylation and a phenotype of interest should be tested using cellular models that test for cellular reprogramming as a consequence of the exposure of interest (Lappalainen & Greally, 2017). Second, we begin by proposing only one biological pathway, the HPA axis, which has received a disproportionate amount of attention in the early life stress literature. There are mechanisms other than the HPA axis that should also be interrogated using a functional epigenetic pathway approach. A full review of additional stress response systems and pathways is beyond the scope of this paper, but the immune system and inflammation, serotonin system, imprinted genes, and mitochondrial biology will likely yield additional insights as to how prenatal maternal mood disorder exposure may alter the fetal epigenome and affect future child health.
Third, we have documented here that in the prenatal stress literature, there is an overreliance on the HPA axis as a mechanism by which prenatal exposure to maternal mood disorder may impact newborn neurobehavioral health that has yielded inconsistent results. Nevertheless, we are proposing to interrogate a network of genes implicated in HPA axis functioning to determine if methylation of these genes could explain the emergence of newborn neurobehavioral and downstream psychiatric problems. We argue that the reason for the mixed findings may be because we have not used the integrated functional epigenetic assay approach, which is hypothesis-driven and leverages a key biological pathway that is implicated in some manner in research on maternal mood disorders. By testing the validity of this assay, we may ultimately be able to determine whether the HPA axis is a key mediator linking prenatal stress with impaired newborn neurobehavioral and gestational health (e.g., preterm birth) outcomes.
Fourth, we propose to examine epigenetic effects in tissues other than the brain, such as the placenta, buccal cells, or blood. There is no established protocol for placenta collection, which is limited by the fact that placental RNA quality is affected by processing time post-delivery. In addition, there are disagreements as to whether cord blood should be used for DNA methylation analyses since the presence of nucleated red blood cells could contaminate cell populations and skew DNA methylation profiles. However, the presence of these disparate cell types can be accounted for in post-hoc statistical analyses. Single-cell sequencing can also be used to characterize different cell types (e.g., scRNA-seq; Lappalainen & Greally, 2017). Relatedly, placenta and blood samples include a mixture of different cell types, though this concern can also be raised when studying brain tissue. Nevertheless, if we hope to incorporate epigenetic processes as biomarkers for pathophysiological processes related to HPA axis development in the fetus, peripheral tissues must be used.
Fifth, we described this approach in the context of maternal mood disorder, which has also received a disproportionate amount of attention in the epigenetic literature (Conradt, 2017). It is important to note that depression is a highly heterogeneous diagnosis. In addition to symptom-level heterogeneity (i.e., the fact that people can meet diagnostic threshold via unique combinations of symptoms), there are many distinct etiological pathways leading to depression and different comorbidities. For example, it is possible that depression in the context of antisocial personality disorder would have a different effect on child outcomes than depression in the context of anxiety. Depressive symptoms can also arise secondary to life stress, health problems, and any number of other life circumstances. Future epigenetic research should focus less on specific psychiatric diagnoses and instead emphasize broad trait-level vulnerabilities, such as emotion dysregulation or impulsivity. Such traits may be more closely linked to genetic and epigenetic processes relative to complex distal phenotypes such as depression.
We believe that testing this novel functional epigenetic pathway has potential to benefit public health and the field of behavioral epigenetics. Particularly exciting to us is the possibility that other researchers to apply this pathway approach to their own research questions that implicate the HPA axis in the expression of disorder. There are a number of additional stress and immune-related pathways that could be also be interrogated using the methods described above. Furthermore, the use of tools, such as epigenetic aging studies, could bring specificity to how prenatal experiences become biologically embedded. Ultimately, we expect that the application of this functional epigenetic network approach will improve prevention efforts aimed at reducing the effects of maternal mood exposure prenatally and could result in novel therapeutic targets to buffer the fetus against maternal mood dysregulation, stress, and other risk processes.
Footnotes
Conflict of Interest: The authors report no conflict of interest.
Financial disclosure: This manuscript was supported by the National Institute of Mental Health under Award Number R21MH109777 (to S.C., E.C.) and a Career Development Award from the National Institute on Drug Abuse 7K08DA038959-02 (to E.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health, National Institute on Drug Abuse, or the National Institutes of Health.
References
- Adkins DE, Rasmussen KM, Docherty AR. Social epigenetics and human behavior. In: Hopcroft R, editor. Oxford Handbook of Evolution, Biology, and Society. London: Oxford University Press; 2017. Oxford Handbook of Evolution, Biology, and Society. London: OUP. [Google Scholar]
- Beijers R, Buitelaar JK, de Weerth C. Mechanisms underlying the effects of prenatal psychosocial stress on child outcomes: beyond the HPA axis. European Child & Adolescent Psychiatry. 2014;23(10):943–956. doi: 10.1007/s00787-014-0566-3. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B - Methodological. 1995;57(1):289–300. [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bock C. Analysing and interpreting DNA methylation data. Nature Reviews Genetics. 2012;13(10):705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- Boyce WT, Kobor MS. Development and the epigenome: the “synapse” of gene-environment interplay. Developmental Science. 2015;18(1):1–23. doi: 10.1111/desc.12282. [DOI] [PubMed] [Google Scholar]
- Breton CV, Marsit CJ, Faustman E, Nadeau K, Goodrich JM, Dolinoy DC, … Murphy SK. Small-Magnitude Effect Sizes in Epigenetic End Points are Important in Children’s Environmental Health Studies: The Children’s Environmental Health and Disease Prevention Research Center’s Epigenetics Working Group. Environmental Health Perspectives. 2017;125(4) doi: 10.1289/EHP595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss C, Davis EP, Shahbaba B, Pruessner JC, Head K, Sandman CA. Maternal cortisol over the course of pregnancy and subsequent child amygdala and hippocampus volumes and affective problems. Proceedings of the National Academy of Sciences. 2012;109(20):E1312–E1319. doi: 10.1073/pnas.1201295109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt E. Using Principles of Behavioral Epigenetics to Advance Research on Early-Life Stress. Child Development Perspectives. 2017;11(2):107–112. doi: 10.1111/cdep.12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Krause L, Bell JT, Gao F, Ward KJ, Wu H, … Wang J. Hypermethylation in the ZBTB20 gene is associated with major depressive disorder. Genome Biology. 2014;15(4):R56. doi: 10.1186/gb-2014-15-4-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EP, Glynn LM, Schetter CD, Hobel C, Chicz-Demet A, Sandman CA. Prenatal Exposure to Maternal Depression and Cortisol Influences Infant Temperament. Journal of the American Academy of Child & Adolescent Psychiatry. 2007;46(6):737–746. doi: 10.1097/chi.0b013e318047b775. [DOI] [PubMed] [Google Scholar]
- De Bellis MD, Chrousos GP, Dorn LD, Burke L, Helmers K, Kling MA, … Putnam FW. Hypothalamic-pituitary-adrenal axis dysregulation in sexually abused girls. The Journal of Clinical Endocrinology & Metabolism. 1994;78(2):249–255. doi: 10.1210/jcem.78.2.8106608. [DOI] [PubMed] [Google Scholar]
- de Goede OM, Razzaghian HR, Price EM, Jones MJ, Kobor MS, Robinson WP, Lavoie PM. Nucleated red blood cells impact DNA methylation and expression analyses of cord blood hematopoietic cells. Clinical Epigenetics. 2015;7(1) doi: 10.1186/s13148-015-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor MHM, van den Berg SM, Verweij KJH, Krueger RF, Luciano M, Arias Vasquez A, … Boomsma DI. Meta-analysis of Genome-wide Association Studies for Neuroticism, and the Polygenic Association With Major Depressive Disorder. JAMA Psychiatry. 2015;72(7):642. doi: 10.1001/jamapsychiatry.2015.0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster EL, Wong CCY, Lester KJ, Burrage J, Gregory AM, Mill J, Eley TC. Genome-wide Methylomic Analysis of Monozygotic Twins Discordant for Adolescent Depression. Biological Psychiatry. 2014;76(12):977–983. doi: 10.1016/j.biopsych.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty AR, Moscati A, Adkins DE, Wallace GT, Kumar G, Riley BP, … Bacanu S-A. Proof of concept: Molecular prediction of schizophrenia risk. 2017 doi: 10.1101/129213. [DOI]
- Docherty AR, Moscati A, Dick D, Savage JE, Salvatore JE, Cooke M, … Kendler KS. Polygenic prediction of the phenome, across ancestry, in emerging adulthood. 2017 doi: 10.1101/124651. [DOI] [PMC free article] [PubMed]
- Essex MJ, Thomas Boyce W, Hertzman C, Lam LL, Armstrong JM, Neumann SMA, Kobor MS. Epigenetic Vestiges of Early Developmental Adversity: Childhood Stress Exposure and DNA Methylation in Adolescence: Epigenetic Vestiges of Early Adversity. Child Development. 2013;84(1):58–75. doi: 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex MJ, Klein MH, Cho E, Kalin NH. Maternal stress beginning in infancy may sensitize children to later stress exposure: effects on cortisol and behavior. Biological Psychiatry. 2002;52(8):776–784. doi: 10.1016/s0006-3223(02)01553-6. [DOI] [PubMed] [Google Scholar]
- Gandal MJ, Leppa V, Won H, Parikshak NN, Geschwind DH. The road to precision psychiatry: translating genetics into disease mechanisms. Nature Neuroscience. 2016;19(11):1397–1407. doi: 10.1038/nn.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn LM, Davis EP, Sandman CA. New insights into the role of perinatal HPA-axis dysregulation in postpartum depression. Neuropeptides. 2013;47(6):363–370. doi: 10.1016/j.npep.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Goodman SH, Rouse MH, Connell AM, Broth MR, Hall CM, Heyward D. Maternal Depression and Child Psychopathology: A Meta-Analytic Review. Clinical Child and Family Psychology Review. 2011;14(1):1–27. doi: 10.1007/s10567-010-0080-1. [DOI] [PubMed] [Google Scholar]
- Greally JM. A user’s guide to the ambiguous word “epigenetics. Nature Reviews Molecular Cell Biology. 2018;19(4):207–208. doi: 10.1038/nrm.2017.135. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, … Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim C, Nemeroff CB. The role of childhood trauma in the neurobiology of mood and anxiety disorders: preclinical and clinical studies. Biological Psychiatry. 2001;49(12):1023–1039. doi: 10.1016/S0006-3223(01)01157-X. [DOI] [PubMed] [Google Scholar]
- Herman JP, Spencer R. Regulation of hippocampal glucocorticoid receptor gene transcription and protein expression in vivo. Journal of Neuroscience. 1998;18(18):7462–7473. doi: 10.1523/JNEUROSCI.18-18-07462.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, … Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13(1):86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, … Morgan M. Orchestrating high-throughput genomic analysis with Bioconductor. Nature Methods. 2015;12(2):115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto H, Jaffe RB. Development and Function of the Human Fetal Adrenal Cortex: A Key Component in the Feto-Placental Unit. Endocrine Reviews. 2011;32(3):317–355. doi: 10.1210/er.2010-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Reviews Genetics. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kasckow JW, Baker D, Geracioti TD. Corticotropin-releasing hormone in depression and post-traumatic stress disorder. Peptides. 2001;22(5):845–851. doi: 10.1016/S0196-9781(01)00399-0. [DOI] [PubMed] [Google Scholar]
- Keating DP. Transformative Role of Epigenetics in Child Development Research: Commentary on the Special Section. Child Development. 2016;87(1):135–142. doi: 10.1111/cdev.12488. [DOI] [PubMed] [Google Scholar]
- Kinsella MT, Monk C. Impact of Maternal Stress, Depression and Anxiety on Fetal Neurobehavioral Development. Clinical Obstetrics and Gynecology. 2009;52(3):425–440. doi: 10.1097/GRF.0b013e3181b52df1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté B, Suderman M, Maussion G, Lopez JP, Navarro-Sánchez L, Yerko V, … Turecki G. Genome-Wide Methylation Changes in the Brains of Suicide Completers. American Journal of Psychiatry. 2013;170(5):511–520. doi: 10.1176/appi.ajp.2012.12050627. [DOI] [PubMed] [Google Scholar]
- Labonté B, Suderman M, Maussion G, Navaro L, Yerko V, Mahar I, … Turecki G. Genome-wide Epigenetic Regulation by Early-Life Trauma. Archives of General Psychiatry. 2012;69(7) doi: 10.1001/archgenpsychiatry.2011.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Greally JM. Associating cellular epigenetic models with human phenotypes. Nature Reviews Genetics. 2017;18(7):441–451. doi: 10.1038/nrg.2017.32. [DOI] [PubMed] [Google Scholar]
- Lester BM, Conradt E, Marsit C. Introduction to the Special Section on Epigenetics. Child Development. 2016;87(1):29–37. doi: 10.1111/cdev.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccani JZJ, Koestler DC, Lester B, Houseman EA, Armstrong DA, Kelsey KT, Marsit CJ. Placental DNA Methylation Related to Both Infant Toenail Mercury and Adverse Neurobehavioral Outcomes. Environmental Health Perspectives. 2015 doi: 10.1289/ehp.1408561. [DOI] [PMC free article] [PubMed]
- Maccari S, Krugers HJ, Morley-Fletcher S, Szyf M, Brunton PJ. The Consequences of Early-Life Adversity: Neurobiological, Behavioural and Epigenetic Adaptations. Journal of Neuroendocrinology. 2014;26(10):707–723. doi: 10.1111/jne.12175. [DOI] [PubMed] [Google Scholar]
- Mansell T, Vuillermin P, Ponsonby A-L, Collier F, Saffery R, Ryan J Barwon Infant Study Investigator Team. Maternal mental well-being during pregnancy and glucocorticoid receptor gene promoter methylation in the neonate. Development and Psychopathology. 2016;28(4pt2):1421–1430. doi: 10.1017/S0954579416000183. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JPA, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature Reviews Genetics. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, … Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nature Neuroscience. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ. Epigenetics and the biological definition of gene× environment interactions. Child development. 2010;81(1):41–79. doi: 10.1111/j.1467-8624.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- Monk C, Feng T, Lee S, Krupska I, Champagne FA, Tycko B. Distress During Pregnancy: Epigenetic Regulation of Placenta Glucocorticoid-Related Genes and Fetal Neurobehavior. American Journal of Psychiatry. 2016;173(7):705–713. doi: 10.1176/appi.ajp.2015.15091171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk C, Spicer J, Champagne FA. Linking prenatal maternal adversity to developmental outcomes in infants: The role of epigenetic pathways. Development and Psychopathology. 2012;24(4):1361–1376. doi: 10.1017/S0954579412000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SE, Braithwaite EC, Hubbard I, Williams KV, Tindall E, Holmes EA, Ramchandani PG. Salivary cortisol response to infant distress in pregnant women with depressive symptoms. Archives of Women’s Mental Health. 2015;18(2):247–253. doi: 10.1007/s00737-014-0473-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy C, Suderman M, Yang J, Szyf M, Mechawar N, Ernst C, Turecki G. Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Molecular Psychiatry. 2015;20(3):320–328. doi: 10.1038/mp.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg JT. Where Do Epigenetics and Developmental Origins Take the Field of Developmental Psychopathology? Journal of Abnormal Child Psychology. 2016;44(3):405–419. doi: 10.1007/s10802-015-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter RC, Westendorp RGJ, de Rooij SR, Osmond C, Barker DJP, Roseboom TJ. Increased reproductive success of women after prenatal undernutrition. Human Reproduction (Oxford, England) 2008;23(11):2591–2595. doi: 10.1093/humrep/den274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette AG, Houseman EA, Green BB, Lesseur C, Armstrong DA, Lester B, Marsit CJ. Regions of variable DNA methylation in human placenta associated with newborn neurobehavior. Epigenetics. 2016;11(8):603–613. doi: 10.1080/15592294.2016.1195534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salm AK, Pavelko M, Krouse EM, Webster W, Kraszpulski M, Birkle DL. Lateral amygdaloid nucleus expansion in adult rats is associated with exposure to prenatal stress. Developmental Brain Research. 2004;148(2):159–167. doi: 10.1016/j.devbrainres.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Sandman CA, Davis EP. Neurobehavioral risk is associated with gestational exposure to stress hormones. Expert Review of Endocrinology & Metabolism. 2012;7(4):445–459. doi: 10.1586/eem.12.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandman CA, Glynn LM, Davis EP. Fetal Development. Springer International Publishing; 2016. Neurobehavioral consequences of fetal exposure to gestational stress; –229.pp. 265 [Google Scholar]
- Sirianni R, Rehman KS, Carr BR, Parker CR, Rainey WE. Corticotropin-Releasing Hormone Directly Stimulates Cortisol and the Cortisol Biosynthetic Pathway in Human Fetal Adrenal Cells. The Journal of Clinical Endocrinology & Metabolism. 2005;90(1):279–285. doi: 10.1210/jc.2004-0865. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, … Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedish Schizophrenia Consortium. McClay JL, Shabalin AA, Dozmorov MG, Adkins DE, Kumar G, … van den Oord EJCG. High density methylation QTL analysis in human blood via next-generation sequencing of the methylated genomic DNA fraction. Genome Biology. 2015;16(1) doi: 10.1186/s13059-015-0842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. How do environments talk to genes? Nature Neuroscience. 2012;16(1):2–4. doi: 10.1038/nn.3286. [DOI] [PubMed] [Google Scholar]
- Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, … Heijmans BT. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Human Molecular Genetics. 2009;18(21):4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P-C, Bell JT. Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. International Journal of Epidemiology. 2015;44(4):1429–1441. doi: 10.1093/ije/dyv041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G, Meaney MJ. Effects of the Social Environment and Stress on Glucocorticoid Receptor Gene Methylation: A Systematic Review. Biological Psychiatry. 2016;79(2):87–96. doi: 10.1016/j.biopsych.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ICG, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, … Meaney MJ. Epigenetic programming by maternal behavior. Nature Neuroscience. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weder N, Zhang H, Jensen K, Yang BZ, Simen A, Jackowski A, … Kaufman J. Child Abuse, Depression, and Methylation in Genes Involved With Stress, Neural Plasticity, and Brain Circuitry. Journal of the American Academy of Child & Adolescent Psychiatry. 2014;53(4):417–424e5. doi: 10.1016/j.jaac.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Zhao S. Candidate gene identification approach: progress and challenges. Int J Biol Sci. 2007;3(7):420–427. doi: 10.7150/ijbs.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]