Abstract
BACKGROUND
Autosomal recessive primary microcephaly (MCPH) is a clinically and genetically heterogeneous disorder. Patients with MCPH exhibit reduced occipito-frontal head circumference and non-progressive intellectual disability. To date, 17 genes have been known as an underlying cause of MCPH in humans. ASPM (abnormal spindle-like, microcephaly associated) is the most commonly mutated MCPH gene.
OBJECTIVE
Identify the genetic defect underlying MCPH in a Saudi family.
DESIGN
A cross-sectional clinical genetic study of a Saudi family.
SETTING
Madinah Maternity and Children Hospital and Centre for Genetics and Inherited Diseases, Taibah University.
PATIENTS AND METHODS
A molecular analysis was carried out on DNA samples from 10 individuals of a Saudi family segregating MCPH. DNA was isolated from the peripheral blood of 10 individuals, including 2 patients, and whole exome sequencing was performed using the Nextera Rapid Capture kit and NextSeq500 instrument. VariantStudio was used to filter and prioritize variants.
MAIN OUTCOME MEASURE(S)
Detection of mutation in the ASPM gene in a family segregating autosomal recessive primary microcephaly.
RESULTS
A novel homozygous splice-site variant (c.3742-1G>C) in the ASPM gene was identified. The variant is predicted to have an effect on splicing. Human Splice Finder, an in silico tool, predicted skipping of exon 16 due to this variant.
CONCLUSION
Skipping of exon 16 may change the order and number of IQ motifs in the ASPM protein leading to typical MCPH phenotype.
LIMITATIONS
Single family study.
Autosomal recessive primary microcephaly (MCPH) is a neurodevelopmental disorder. Mutations in genes that are involved in brain growth and development like proliferation or differentiation of progenitor cells, or cell apoptosis can lead to MCPH.1 MCPH is marked by a decreased brain size associated with a significantly decreased occipito-frontal circumference (OFC), which is greater than two standard deviations (SD) below the mean values for age, sex and origin.2 A head circumference less than 3 SD below the age and sex is generally taken as the clinical demarcation of MCPH.3 The prevalence of MCPH varies worldwide ranging between 1.3 to 150 per 100 000 births depending on level of consanguinity and population type.4 In MCPH, the brain size is reduced substantially though the overall brain architecture is not much affected.5
To date, 17 genes (MCPH1-MCPH17) have been discovered; mutations lead to MCPH.6 Most of the mutations that are reported in these genes are nonsense mutations or frameshift mutations that produce non-functional truncated proteins.7 Mutations in the abnormal spindle-like microcephaly-associated (ASPM) gene cause the most prevalent form of microcephaly (25–50%) known as MCPH5.2,8–23 The genetic basis of brain evolution in primates including humans, involves the ASPM gene.24–27 The ASPM protein is mostly found in the nucleus, but moves to the spindle poles during mitosis, indicating its possible role in keeping the integrity of spindle/centrosome.28
The advent of state of the art techniques like next generation sequencing made it possible to discover the underlying genetic causes of inherited disorders with greater precision and so broaden the existing spectrum of genetic mutations. There are quite a few reports on the syndromic form of genetic microcephaly in Saudi families.29,30 Recently, a splice donor site mutation in a CIT gene was identified in a Saudi family with a nonsyndromic genetic form of microcephaly.31 To the best of our knowledge, this is the only gene known for nonsyndromic form of microcephaly in a Saudi family.
In this study, we report a novel, homozygous, splicesite acceptor mutation affecting exon 16 of the ASPM gene, causing nonsyndromic form of microcephaly in a Saudi family. The mutation has been identified using the approach of whole exome sequencing of the index patient.
PATIENTS AND METHODS
Permission to undertake this study was granted by Ethical Review Committee of Taibah University Almadinah Almunawwarah, Saudi Arabia. Before commencing the study, informed written consent for genetic analysis was obtained from parents. Parents denied consent for publishing clinical photographs. Genetic analysis including DNA extraction, whole exome sequencing and Sanger sequencing was performed in the Centre for Genetics and Inherited Diseases, Taibah University.
A Saudi family with two affected individuals having primary microcephaly was recruited for this study. The affected individual was examined clinically as well as radiologically by a paediatric neurologist at Madinah Maternity and Children Hospital, Almadinah. The family pedigree consisting of four generations was drawn after querying the concerned family.
DNA extraction
Blood samples from 10 individuals (III-2, III-3, III-4, III-5, III-6, III-7, IV-1, IV-2, IV-3, IV-5) including two affected individuals (IV-1, IV-2) were collected in EDTA containing vacutainers. Genomic DNA extraction was done using the QIAquick DNA extraction kit. DNA quantification and concentration was determined by Nanodrop spectrophotometer (Green BioResearch Baton Rouge, LA, USA) and Qubit fluorometer (ThermoFisher Scientific Inc, Massachusetts, USA). The DNA integrity was resolved through 1% agarose gel electrophoresis.
Laboratory investigations
Laboratory investigations including measurement of amino acid levels, creatine and creatinine level as well as blood and urine profile were performed to exclude metabolic disorders like phenylketonuria, phosphoglycerate dehydrogenase deficiency and 2-ketoglutaric aciduria. Thyroid function tests were performed to measure TSH, T4, T3 and free T4. Ophthalmic examination was performed to exclude issues related to eye including bilateral macular and perimacular lesions as well as optic nerve abnormalities. Diagnostic measurements of head circumference (HC) of the affected members and magnetic resonance imaging (MRI) of the brain was done at Madinah Maternity and Children Hospital.
Whole exome sequencing
DNA from the affected members (IV-1, IV-2) of the family was subjected to whole exome sequencing. Library preparation and exome enrichment was done by using Nextra Rapid capture Exome Kit that captures 214 405 exons and splice-site with 98.3% RefSeq coverage. DNA sequencing and cluster generation was carried out on NextSeq500 machine (Illumina, San Diego, California, USA).
In brief, 50 ng of genomic DNA was fragmented enzymatically to generate 150–200 bp fragments, followed by tagging with paired end adaptor sequences (tagmentation). The tagmented DNA was then purified and amplified. Resultant library was purified using magnetic beads. The targeted regions were captured using whole exome oligos followed by amplification of the enriched library through polymerase chain reaction. A Qubit fluorometer was used to quantify the enriched library while average library size distribution was calculated through Agilent Bioanalyzer. For cluster generation and sequencing, the quantified DNA library was loaded on the flow cell of the Illumina NextSeq500 instrument. NextSeq500 instrument generates bcl files which were subsequently converted to fastq files using the BCL2FASTQ tool. We used BaseSpace incorporated BWA aligner to align fastq files to the reference genome using BWA-MEM algorithm. Variants calling was done by using genome analysis tool kit (GATK). Genomic variants were filtered and annotated by VariantStudio (Illumina, San Diego, California, USA).
Sanger sequencing
The confirmation of the variants discovered by WES was done by Sanger sequencing. Ensembl genome browser (http://asia.ensembl.org/index.html) was used to download the 10877 base pairs genomic sequence of the ASPM gene (ENST00000367409.8). Primers were designed using primer3 software (http://frodo.wi.mit.edu/primer3/) to amplify the variants along with their flanking sites. Resultant sequence variants were identified using BIOEDIT sequence alignment editor version 6.0.7 (Ibis Biosciences Inc., Carlsbad, CA, USA).
RESULTS
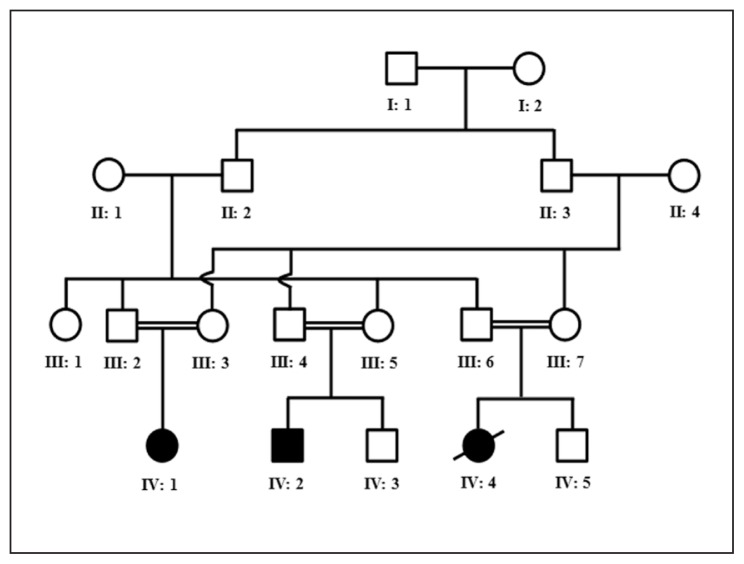
A consanguineous Saudi family, inheriting autosomal recessive primary microcephaly with two living affected members born to phenotypically normal parents, was recruited for this study (Figure 1). The age of the affected individuals IV-1 and IV-2 were 18 and 15 years, respectively. Affected individual IV-4 exhibited similar clinical features to individuals IV-1 and IV-2. She died at the age of 9 after developing high grade fever. Laboratory investigations including measurement of amino acid levels, creatine and creatinine level as well as blood and urine profile excluded any metabolic disorder. Thyroid function tests revealed a normal functioning thyroid. Ophthalmic examination excluded issues related to eye/vision. Clinical examination showed a small head with bi-temporal narrowing. Occipitofrontal circumference (OFC) for affected individuals IV-1 and IV-2 were 41cM and 45 cM, respectively. Both parents showed normal intelligence scores and normal HC. Sloping forehead in both cases was evident. Both affected children showed intellectual disability and speech delay, although no hearing impairment was observed. Patient IV-2 developed epilepsy at the age of 6 years and is currently taking carbamazepine and valproic acid. Magnetic resonance imaging (MRI) of the brain revealed a parieto-occipital and left temporo-occipital gyration defects in patient IV-2. Both patients showed short stature (height is less than 2.5 SD).
Figure 1.
Pedigree drawing of the family with primary microcephaly. Open squares and circles represent unaffected males and females, respectively. Filled squares and circles represent affected individuals. Double lines between symbols are representative of consanguineous unions.
Exome data analysis
The resulting VCF (variant call format) file contains approximately 85 000 variants. These variants were filtered based on quality, frequency, genomic position, protein effect, pathogenicity and previous associations with the phenotype. Candidate variants were expected to follow an autosomal recessive inheritance pattern. Rare, potentially harmful variants present in the homozygous state were of primary interest due to the positive consanguinity for this family. Looking for rare homo/hemizygous variants within the protein coding regions of all genes that have previously been associated with MCPH phenotype including MCPH1, WDR62, CDK5RAP2, CASC5, ASPM, CENPJ, STIL, CEP135, CEP152, ZNF335, PHC1, CDK6 CENPE, SASS6, MFSD2A, ANKLE2 and CIT yielded one plausible candidate (c.3742-1G>C) in the ASPM gene. The genomic position of this variant is chr1:197091174 according to the Ensembl transcript number ENST00000294732. Mutations in ASPM are associated with autosomal recessive primary microcephaly, type 5. Thereafter, rare, potentially harmful variants present in those genes in (compound) heterozygous state were further considered. However, this approach did not yield additional plausible candidates. The analysis was then expanded to all genes, whereby we first focused on potentially harmful homozygous variants and then moved on to potential candidates in the (compound) heterozygous state. However, none of these approaches yielded good candidate variants.
Validation through Sanger sequencing and segregation analysis
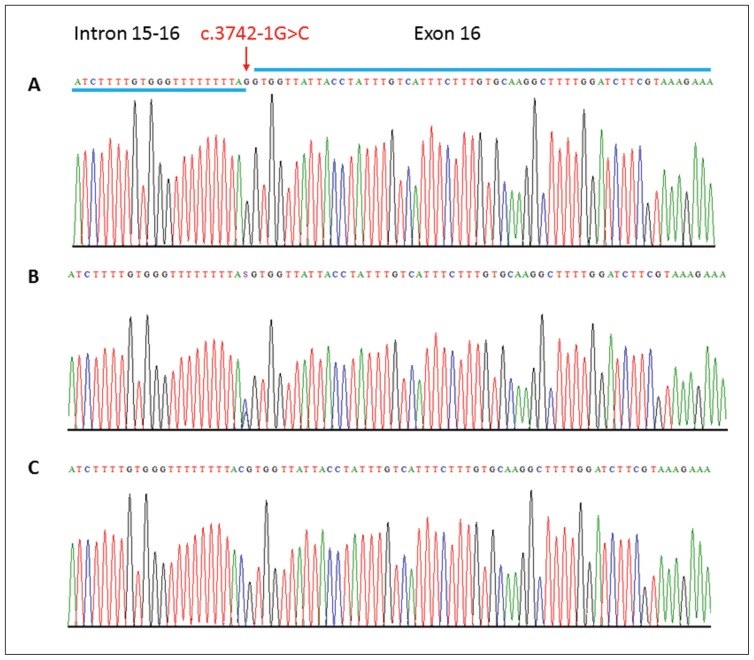
Intron 15–16 and exon 16 of the ASPM gene was sequenced by the Sanger approach in all 10 available DNA samples of a family members to validate the identified mutation. Sanger sequencing validates the exome discovered splice-site mutation in both affected individuals and confirmed the segregation of same mutation in the whole family. Unaffected parents were found heterozygous for this mutation (Figure 2).
Figure 2.
Sequence analysis of the ASPM gene variant. Partial DNA sequence of the ASPM gene identified a homozygous splice-site variant (c.3742-1G>C) in the affected individuals (Panel C) and heterozygous in the carriers (Panel B). Arrows indicate position of the mutation.
In silico analysis of splice-site variant
The variant identified in this study in the ASPM gene occurs at a splice acceptor site. In silico analysis with different splice-site effect prediction tools including human splicing finder (http://www.umd.be/HSF3/), SpliceView (http://bioinfo.itb.cnr.it/oriel/splice-view.html), Sroogle (http://sroogle.tau.ac.il/, SplicePort (http://spliceport.cbcb.umd.edu/), Spliceman,32 GeneID,33 MaxEnt,34 ASSP,35 and NNSPLICE36 predicted this variant to abolish the splice acceptor site.
DISCUSSION
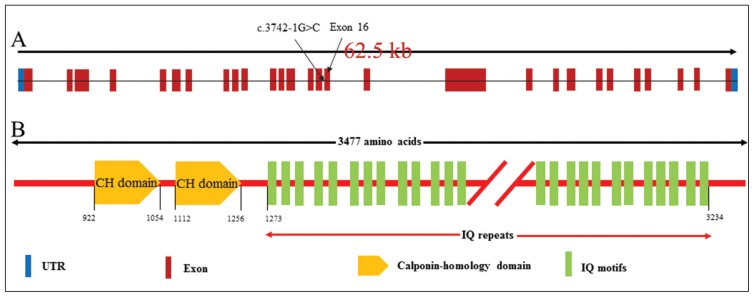
A novel homozygous variant altering the splice-site acceptor sequence (c.3742-1G>C) preceding exon 16 of the ASPM gene was detected in two patients with MCPH. The variant perfectly segregates with the disease phenotype in a family. This variant is predicted to cause skipping of exons 16. Skipping of exon 16 is predicted to change the order and number of IQ motifs in the resulting protein leading to typical MCPH phenotype (Figure 3).
Figure 3.
Genomic organization of ASPM exons. Arrow head indicates the position of mutation identified in this study (A). Schematic representation of the organization of ASPM protein domains (B). CH: calponin-homology domains, IQ: calmodulin binding domains.
MCPH is a heterogenetic disorder with numerous genes known to cause the MCPH phenotype when mutated. ASPM gene (MCPH5) occupies the most prominent position in disease aetiology of MCPH owing to its mutations rate, which is as high as 40%.19 Different ethnic populations show different prevalences for the MCPH5 locus. For instance, in Pakistan this toll falls in the range of 43–86%,15 in Indian populations it is 33.5%,12 while in Iranian population it is reported to be 13.3%.21
The ASPM protein is essential for normal mitotic spindle function in embryonic neuroblasts. By promoting the division of neural progenitor cells during early brain development, ASPM helps determine the total number of neurons and the overall size of the brain.10 Defects in the ASPM protein are associated with autosomal recessive microcephaly, and mild to severe mental retardation. Delayed motor and speech development are frequent symptoms of this condition.9,19 Mutations in ASPM are associated with autosomal recessive primary microcephaly, type 5 (MCPH5), which is characterized by decreased occipitofrontal circumference, usually less than 3 SD of the mean, present at birth and associated with mental retardation and speech delay. Other features may include short stature or mild seizures. MCPH5 is associated with a simplification of the cerebral cortical gyral pattern in some cases, which is considered within the phenotypic spectrum of primary microcephaly. 5,9,20 Pathogenic ASPM mutations are typically truncating (nonsense, frameshift or splice affecting variants). Hence, the probable disease mechanism leading to primary microcephaly was nonsense mediated decay of the ASPM mRNA leading to a pronounced reduction of the protein in neuroepithelial cells.19
The detected novel variant is located at a splice-site acceptor of the ASPM gene (between exons 15–16). The variant is predicted to have an effect on splicing according to the scores of Jian et al (SC Ada, SC RF).37 In addition, an alteration of splicing due to this variant is also predicted by the in silico tool Human Splice Finder (see http://www.umd.be/HSF3/) (the in silico analysis indicates that this variant could lead to a broken WT acceptor site, most probably affecting splicing). Functional experiments are however needed to assess this hypothesis. Interestingly, a few mutations in the close proximity of this variant are described as pathogenic (p.Glu1266Term, rs199422161; splice variant, c.3741+1G>A, rs199422160; R1271*; rs140602858), providing further support for this variant as a plausible candidate to explain the patient’s phenotype in this family.11,19 Nicholas et al19 have studied cohorts of microcephalic children to extend both the phenotype and the mutation spectrum and identified a Caucasian patient carrying a homozygous splice-site mutation (c.3741+1G>A) only one residue away from the variant detected here, hence affecting the same splice-site. In conclusion, this variant is likely to contribute to the primary microcephaly reported for this patient.
Acknowledgments
Authors are thankful for the cooperation of family members.
Footnotes
Authors’ contribution
KMA recruited the family and performed phenotyping; AMA extracted DNA; JAH, KR and AMA performed exome sequencing; AM, MIS and SB analysed the exome data; JAH, AMA performed Sanger sequencing; KR analysed Sanger reads.
Conflict of Interest
None declared.
REFERENCES
- 1.Barkovich AJ, Kuzniecky RI, Jackson GD, Guerrini R, Dobyns WB. A developmental and genetic classification for malformations of cortical development. Neurology. 2005;65:1873–87. doi: 10.1212/01.wnl.0000183747.05269.2d. [DOI] [PubMed] [Google Scholar]
- 2.Kaindl AM, Passemard S, Kumar P, Kraemer N, Issa L, Zwirner A, et al. Many roads lead to primary autosomal recessive microcephaly. Prog Neurobiol. 2010;90(3):363–83. doi: 10.1016/j.pneurobio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Passemard S, Kaindl AM, Verloes A. Microcephaly. Handbook of Clinical neurology, Vol. III. Pediatric Neurology Part I. 2013:129–141. doi: 10.1016/B978-0-444-52891-9.00013-0. [DOI] [PubMed] [Google Scholar]
- 4.Woods CG, Parker A. Investigating microcephaly. Arch Dis Child. 2013;98:707–13. doi: 10.1136/archdischild-2012-302882. [DOI] [PubMed] [Google Scholar]
- 5.Woods CG, Bond J, Enard W. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am J Hum Genet. 2005;76:717–28. doi: 10.1086/429930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdel-Hamid MS, Ismail MF, Darwish HA, Effat LK, Zaki MS, Abdel-Salam GMH. Molecular and Phenotypic Spectrum of ASPM-Related Primary Microcephaly: Identification of Eight Novel Mutations. Am J Med Genet Part A. 2016;9999A:1–8. doi: 10.1002/ajmg.a.37724. [DOI] [PubMed] [Google Scholar]
- 7.Barbelanne M, Tsang WY. Molecular and Cellular Basis of Autosomal Recessive Primary Microcephaly. BioMed Res Int. 2014:1–13. doi: 10.1155/2014/547986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts E, Hampshire DJ, Pattison L, Springell K, Jafri H, Corry P, et al. Autosomal recessive primary microcephaly: an analysis of locus heterogeneity and phenotypic variation. Journal of Medical Genet. 2002;39:718–21. doi: 10.1136/jmg.39.10.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Passemard S, Titomanlio L, Elmaleh M, Afenjar A, Alessandri JL, Andria G, et al. Expanding the clinical and neuroradiologic phenotype of primary microcephaly due to ASPM mutations. Neurology. 2009;73(12):962–9. doi: 10.1212/WNL.0b013e3181b8799a. [DOI] [PubMed] [Google Scholar]
- 10.Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32(2):316–20. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 11.Bond J, Scott S, Hampshire DJ, Springel K, Corry P, Abramowicz MJ, et al. Protein-truncating mutations in ASPM cause variable reduction in brain size. Am J Hum Genet. 2003;73(5):1170–77. doi: 10.1086/379085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar A, Blanton SH, Babu M, Markandaya M, Girimaji SC. Genetic analysis of primary microcephaly in Indian families: novel ASPM mutations. Clinical Genet. 2004;66(4):341–48. doi: 10.1111/j.1399-0004.2004.00304.x. [DOI] [PubMed] [Google Scholar]
- 13.Pichon B, Vankerckhove S, Bourrouillou G, Duprez L, Abramowicz MJ. A translocation breakpoint disrupts the ASPM gene in a patient with primary microcephaly. Eur J Hum Genet. 2004;12(5):419–21. doi: 10.1038/sj.ejhg.5201169. [DOI] [PubMed] [Google Scholar]
- 14.Shen J, Eyaid W, Mochida GH, Al-Moayyad F, Bodell A, Woods CG, et al. ASPM mutations identified in patients with primary microcephaly and seizures. J Med Genet. 2005;42(9):725–29. doi: 10.1136/jmg.2004.027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gul A, Hassan MJ, Mahmood S, Chen W, Rahmani S, Naseer MI, et al. Genetic studies of autosomal recessive primary microcephaly in 33 Pakistani families: novel sequence variants in ASPM gene. Neurogenetics. 2006;7(2):105–10. doi: 10.1007/s10048-006-0042-4. [DOI] [PubMed] [Google Scholar]
- 16.Gul A, Tariq M, Khan MN, Hassan MJ, Ali G, Ahmad W. Novel protein-truncating mutations in the ASPM gene in families with autosomal recessive primary microcephaly. J Neurogenetics. 2007;21(3):153–63. doi: 10.1080/01677060701508594. [DOI] [PubMed] [Google Scholar]
- 17.Desir J, Cassart M, David P, van Bogaert P, Abramowicz M. Primary microcephaly with ASPM mutation shows simplified cortical gyration with antero-posterior gradient pre-and post-natally. Am J Med Genet Part A. 2008;146(11):1439–1443. doi: 10.1002/ajmg.a.32312. [DOI] [PubMed] [Google Scholar]
- 18.Muhammad F, Baig SM, Hansen L, Hussain MS, Inayat A, Aslam M, et al. Compound heterozygous aspm mutations in Pakistani MCPH families. Am J Med Genet Part A. 2009;149(5):926–30. doi: 10.1002/ajmg.a.32749. [DOI] [PubMed] [Google Scholar]
- 19.Nicholas AK, Swanson EA, Cox JJ, Karbani G, Malik S, Springell K, et al. The molecular landscape of ASPM mutations in primary microcephaly. J Med Genet. 2009;46(4):249–53. doi: 10.1136/jmg.2008.062380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saadi A, Borck G, Boddaert N, Chekkour MC, Imessaoudene B, Munnich A, et al. Compound heterozygous ASPM mutations associated with microcephaly and simplified cortical gyration in a consanguineous Algerian family. Eur J Med Genet. 2009;52(4):180–84. doi: 10.1016/j.ejmg.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Darvish H, Esmaeeli-Nieh S, Monajemi GB, Mohseni M, Ghasemi-Firouzabadi S, Abedini SS, et al. A clinical and molecular genetic study of 112 Iranian families with primary microcephaly. J Med Genet. 2010;47(12):823–28. doi: 10.1136/jmg.2009.076398. [DOI] [PubMed] [Google Scholar]
- 22.Kousar R, Nawaz H, Khurshid M, Ali G, Khan SU, Mir H, et al. Mutation analysis of the ASPM gene in 18 Pakistani families with autosomal recessive primary microcephaly. J Child Neur. 2010;25(6):715–20. doi: 10.1177/0883073809346850. [DOI] [PubMed] [Google Scholar]
- 23.Jamieson CR, Fryns JP, Jacobs J, Matthijs G, Abramowicz MJ. Primary autosomal recessive microcephaly: MCPH5 maps to 1q25-q32. Am J Hum Genet. 2000;67(6):1575–77. doi: 10.1086/316909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans PD, Anderson JR, Vallender EJ, Gilbert SL, Malcom CM, Dorus S, et al. Adaptive evolution of aspm, a major determinant of cerebral cortical size in humans. Hum Mol Genet. 2004;13:489–94. doi: 10.1093/hmg/ddh055. [DOI] [PubMed] [Google Scholar]
- 25.Kouprina N, Pavlicek A, Mochida GH, Solomon G, Gersch W, Yoon YH, et al. Accelerated evolution of the aspm gene controlling brain size begins prior to human brain expansion. PLoS Biol. 2004;2(5):E126. doi: 10.1371/journal.pbio.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mekel-Bobrov N, Gilbert SL, Evans PD, Vallender EJ, Anderson JR, Hudson RR, et al. Ongoing adaptive evolution of aspm, a brain size determinant in homo sapiens. Science. 2005;309:1720–22. doi: 10.1126/science.1116815. [DOI] [PubMed] [Google Scholar]
- 27.Montgomery SH, Mundy NI. Evolution of aspm is associated with both increases and decreases in brain size in primates. Evolution. 2012;66:927–32. doi: 10.1111/j.1558-5646.2011.01487.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhong X, Liu L, Zhao A, Pfeifer GP, Xu X. The abnormal spindle-like, microcephaly-associated (ASPM) gene encodes a centrosomal protein. Cell Cycle. 2005;4(9):1227–29. doi: 10.4161/cc.4.9.2029. [DOI] [PubMed] [Google Scholar]
- 29.Alrayes N, Mohamoud HSA, Ahmed S, Almramhi MM, Shuaib TM, Wang J, et al. The alkylglycerol monooxygenase (AGMO) gene previously involved in autism also causes a novel syndromic form of primary microcephaly in a consanguineous Saudi family. J Neurol Sci. 2016;363:240–44. doi: 10.1016/j.jns.2016.02.063. [DOI] [PubMed] [Google Scholar]
- 30.Alkuraya FS, Cai X, Emery C, Mochida GH, Al-Dosari MS, Felie JM, et al. Human Mutations in NDE1 Cause Extreme Microcephaly with Lissencephaly. Am J Hum Genet. 2011;88:536–47. doi: 10.1016/j.ajhg.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basit S, Al-Harbi KM, Alhijji SA, Albalawi AM, Alharby E, Eldardear A, Samman MI. CIT, a gene involved in neurogenic cytokinesis, is mutated in human primary microcephaly. Hum Genet. 2016;135(10):1199–207. doi: 10.1007/s00439-016-1724-0. [DOI] [PubMed] [Google Scholar]
- 32.Lim KH, Fairbrother WG. Spliceman - a computational web server that predicts sequence variations in pre-mRNA splicing. Bioinformatics. 2012;28:1031–32. doi: 10.1093/bioinformatics/bts074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parra G, Blanco E, Guigó R. Geneid in Drosophila. Genome Res. 2000;10:511–15. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–94. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 35.Wang M, Marín A. Characterization and prediction of alternative splice-sites. Gene. 2006;366:219–27. doi: 10.1016/j.gene.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 36.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice-site detection in genie. J Comp Biol. 1997;4:311–23. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 37.Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42(22):13534–544. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]