Abstract
BACKGROUND
Vancomycin-resistant enterococci (VRE) are resistant to most classes of antibiotics. Diagnosis of VRE using standard methods takes 2 to 5 days. Development of a rapid PCR-assay that detects and identifies resistant genes in bacteria would provide time-critical information on the presence of VRE in clinical samples allowing early treatment and management of infected patients.
OBJECTIVES
Investigate the use of high resolution melting analysis (HRMA) and 16S-rRNA-PCR approach for rapid and cost-effective identification of VRE.
DESIGN
Descriptive antibiotic susceptibility studies.
SETTING
Manchester Academic Health Sciences Centre and School of Translational Medicine, University of Manchester, UK, and Department of Clinical Laboratory Sciences, Taibah University, Saudi Arabia.
MATERIALS AND METHODS
PCR-HRMA using 16S-rRNA V1-primers was used to detect and identify VRE. DNA from different strains of vancomycin-resistant and -sensitive Enterococcus faecalis (VSE) and Enterococcus faecium were amplified using V1-primer followed by HRMA in a single run. Differentiation of VRE from VSE was based on curve shapes generated against reference organisms (Bacteroides fragilis).
MAIN OUTCOMES MEASURES
Amplification curves and difference plots for VRE and VSE.
RESULTS
Difference plots were generated for all vancomycin-resistant and -sensitive E faecalis and E faecium strains by subtracting their fluorescence melting profile from that of a reference-species B fragilis. A characteristic curve shape was produced by vancomycin-sensitive E faecalis and E faecium. However, vancomycin-resistant strains of these bacteria were associated with a markedly different curve shape facilitating a clear differentiation.
CONCLUSION
The 16S-PCR-HRMA approach has the potential for detecting vancomycin-resistant E faecium and E faecalis. Data with VRE provide the basis for combining VRE identification with pathogens speciation in a rapid, cheap assay able to identify a pathogen as an Enterococcus and whether it is vancomycin-sensitive or -resistant E faecium or E faecalis in a single PCR and HRMA run.
LIMITATIONS
Tested on specific, but not all, reference Enterococcus species and clinical isolates.
Choosing an appropriate antimicrobial therapy for critically ill patients is a major problem for healthcare providers due to the increasing prevalence of drug-resistant microorganisms in the hospital environment and particularly in critical care.1 Treatment and management of patients infected by resistant strains has become more difficult than those infected by susceptible organisms.1–3 Infection caused by resistant organisms increases length of stay and mortality rate, and requires use of more toxic drugs and thus increases overall healthcare costs.3,4 Therefore, the need for clinicians to treat patients with suspected infection as soon as possible means that broad-spectrum antimicrobials are administered until infection is confirmed and the results of an antimicrobial susceptibility tests are available. Early detection and identification of antibiotic-resistant organisms will allow early and optimum treatment and management of the infected patient, and therefore decrease overall healthcare costs.4 The use of broad-spectrum antimicrobials in this way is one possible reason for the increase in antibiotic-resistant bacteria. One of most common antibiotic-resistant bacteria is vancomycin-resistant enterococci (VRE).1,5
The emergence of VRE, particularly Enterococcus faecalis and Enterococcus faecium has become a significant issue in the control of hospital acquired infections worldwide.2,5 A high level vancomycin resistance is usually linked to acquisition of the transmissible genetic elements vanA or vanB, although less common van variants such as vanC have also been identified.2 The vanC gene is usually found in E gallinarum and E casseliflavus/flavescens as these strains naturally have a low-level of resistance to vancomycin.2 VRE are resistant to most major classes of antimicrobial therapy and may transfer the vancomycin resistance gene to other bacteria such as methicillin-resistant Staphylococcus aureus (MRSA).6,7 Inappropriate antibiotic use appears to be the key driver for VRE emergence while longer hospital stay, and increased colonisation are important in increasing the prevalence of VRE infection.5,8 Studies about prevalence and outcomes of infection in ICUs worldwide indicate that about 5% of all confirmed infections in ICU worldwide are caused by VRE.9,10 The high morbidity and mortality and increased costs of care associated with VRE means that early detection is crucial to facilitate optimal treatment and management of the infected patient and ensure they are quickly isolated to reduce the risk of onward transmission.2,5,6 Laboratory diagnosis of VRE infection using standard culture and antibiotic susceptibility testing is a time-consuming process which can take 2 to 5 days to complete.2,5 Therefore, there is an urgent need to develop rapid and cost-effective molecular assays that can provide time-critical information on the presence of VRE in clinical samples. Molecular approaches using real-time PCR have been developed to provide results in less time by detection and identification of mainly the resistance genes in bacteria. The development of molecular methods that could rapidly detect and identify pathogens and determine their likely antimicrobial susceptibility would be helpful in the early treatment and management of patients with infection. PCR-based approaches using multiple primers and labelled probes have been applied to detection of van genes including commercial assays aimed primarily at screening of faecal or rectal samples.11,12 High resolution melting analysis (HRMA), a technique for characterising PCR amplicons based on differences in melting profile (curve shape), identifies a wide spectrum of clinically important pathogens from clinical isolates and broth culture.13,14 The aim of the present study was to investigate the use of HRMA combined with broad range 16S rRNA PCR as an alternative approach for the rapid and cost-effective differentiation of vancomycin-sensitive and -resistant Enterococci strains.
MATERIALS AND METHODS
After developing the PCR-HRMA approach with a multi-decision strategy for detection and identification of 21 common bloodstream pathogens as described by Ozbak and co-workers in 2012,15 the PCR-HRMA approach was consequently tested for its ability for detecting and identifying VRE, one of most common antibiotic-resistant organisms in bloodstream infections. We used different primer sets from the primers used by Ozbak and colleagues, so as to produce a package that would be useful in routine molecular microbiology laboratory for rapid identification of the most common bloodstream pathogens and detection of resistance from clinical blood samples. Other experiments were carried out to confirm the presence of standard vancomycin-resistant Enterococcus strains by detecting their van genes and determining whether vanA or vanB.
This study was done at Manchester Academic Health Sciences Centre and School of Translational Medicine (University of Manchester, UK), and at the Department of Clinical Laboratory Sciences (Taibah University, Saudi Arabia) to investigate the use of HRMA and the 16S-rRNA-PCR approach for rapid and cost-effective identification of VRE. PCR-HRMA using 16S-rRNA V1-primers16 ( Table 1) were used to detect and identify VRE. This study was conducted on five reference strains that belong to two groups: vancomycin-resistant E faecalis and E faecium strains (E faecium ATCC 19434 (VRE), E faecalis ATCC 29212 (VRE) and E faecalis ATCC 51299 (VRE)), and vancomycin-sensitive E faecalis and E faecium strains (E faecalis NCTC 77 (VSE), E faecium NCIMB 2699 (VSE)).
Table 1.
Information and sequence data for V1, vanA and vanB primer pairs.
| Primer Pair (PP) | Sequence (5′-3′) | Purification | Melting temperature (°C) | |
|---|---|---|---|---|
|
| ||||
| V1 | F | GYGGCGNACGGGTGAGTAA | HPLC | 58.0 |
| R | TTACCCCACCAACTAGC | HPLC | 60.5 | |
| vanA | F | TATGATGGCCGCTGCAGGTA | HPLC | 55.8 |
| R | CGGTGAAATTATCCCAAGTGGC | HPLC | 58.2 | |
| vanB | F | GCCATGCAAAACCGGGAAAG | HPLC | 59.6 |
| R | CAAGCGATTTCGGGCTGTGA | HPLC | 60.1 | |
Source of bacterial strains
Reference strains for the two groups included in this study; vancomycin-resistant Enterococcus (VRE) strains and vancomycin-sensitive Enterococcus (VSE) strains, and the standard strain of Bacteroides fragilis were purchased from Pro-Lab (Neston, South Wirral, Cheshire, UK), NCIMB Limited or the National Collection of Type Cultures (NCTC; Health Protection Agency, Porton Down, UK). These referenced bacterial strains were as follow: Bacteroides fragilis ATCC 25285, VSE strains (E. faecalis NCTC 77, E faecium NCIMB 2699) and VRE strains (E faecium ATCC 19434, E faecalis ATCC 29212, E faecalis ATCC 51299).
Culture and DNA extraction
All strains used in this study were grown under aerobic conditions except B fragilis ATCC 25285, which was grown under anaerobic condition at 37°C in 10 mL lysogeny broth (LB) (containing 10 g/L bacto-tryptone, 5 g/L yeast extract 10g/L NaCl) and were then subcultured overnight on blood agar (containing based on SIFMA-ALDRICH 10 g/L meat extract, 10 g/L peptone, 10 g/L sodium chloride and 5 g/L agar) under the appropriate conditions as mentioned previously. Cultures were centrifuged at 3000g for 10 minutes and DNA was extracted from the bacterial pellet using High Pure PCR Template Preparation Kit (Roche Diagnostics) according to the manufacturer’s protocol. DNA extracts were aliquoted into sterile microtubes and stored at −20°C (https://lifescience.roche.com/documents/High-Pure-PCR-Template-Preparation-Kit.pdf).
PCR confirmation of vanA/vanB expression
To confirm the expression of van genes (for identification of VRE) associated with VRE standards strains used in this study, extracted DNA was amplified and analysed by real-time PCR on a LightCycler 480 using previously published gene-specific vanA (Forward TATGATGGCCGCTGCAGGTA; Reverse CGGTGAAATTATCCCAAGTGGC) and vanB (Forward GCCATGCAAAACCGGGAAAG; Reverse CAAGCGATTTCGGGCTGTGA) primer sets for detection of van genes17 (Table 1) in a 20 μL reaction volume also containing DNA extract (250 fg-250 ng), 0.3 μM forward and reverse primers, MgCl2 at a final concentration of 3 μM and 10 μL (1X) LightCycler 480 High Resolution Melting Master mix. Negative controls containing PCR grade water instead of bacterial DNA extract were included. vanA and vanB primer sets were optimised to run in the following PCR conditions: activation step at 95°C for 10 minutes followed by 30 cycles of 95°C for 10 seconds, 55°C for 10 seconds and 72°C for 10 seconds. Reactions containing nuclease-free water were also run as negative controls. Each experiment was performed at least three times.
Real-time PCR for high resolution melting analysis
DNA amplification was performed on the LightCycler 480 real-time PCR platform (Roche Applied Science) using pan-bacterial 16S rRNA primers (V1-forward: GYGGCGNACGGGTGAGTAA and V1-reverse: TTACCCCACCAACTAGC primers).16 Primers were HPLC purified (Sigma-Aldrich, Dorset, UK). PCR reactions in a final volume of 20 μL contained the following: DNA extract (250 fg-250 ng), 0.3 μM forward and reverse primers, MgCl2 at a final concentration of 3 μM and 10 μL (1X) LightCycler 480 High Resolution Melting Master mix containing FastStart Taq DNA polymerase, reaction buffer and dNTP mix (Roche Diagnostics; Burgess Hill, UK). Negative controls containing PCR grade water instead of bacterial DNA extract were included. The same conditions were used for all PCR reactions: activation step at 95°C for 10 minutes followed by 30 cycles of 95°C for 10 seconds, 62°C for 10 seconds and 72°C for 10 seconds. Reactions containing nuclease-free water were also run as negative controls (https://pim-eservices.roche.com/LifeScience/Document/7fbff181-4394-e611-6a9e-00215a9b3428).
High resolution melting analysis
HRMA was performed using the the Lightcycler 480 Gene Scanning Software. Amplified DNA was heated at 95°C for 1 minute, followed by cooling at 40°C for 1 minute. HRMA was performed over the range from 65°C to 95°C, rising at 0.02 °C/s with 25 acquisitions/°C to obtain a high resolution melting profile giving an accurate melting temperature of the amplicon. Melting curves were normalised and temperature shifted allowing difference plots to be derived that compare the melting profile given by a particular bacterial species with that of another (reference) species. In this study, Bacteroides fragilis was used as the reference organism for generating difference plots of vancomycin-resistant and -sensitive Enterococcus strains. Reactions containing purified B fragilis DNA (10 pg) were included in each PCR run. Analytical sensitivity of the RT-PCR-HRMA assay was determined over a million-fold range (250 fg to 250 ng) by serial dilution of DNA from vancomycin sensitive and resistant strains.
RESULTS
Confirmation of expression of vanA or vanB gene
A real-time PCR assay was optimised and used to detect the genotype of vancomycin-resistant E faecalis and E faecium strains used in this study (the presence or absence of van genes based on gene-specific primers for confirmation of van genes expression using previously published specific primer sets vanA and vanB).17 The resistance and expression of vanA ( E faecium ATCC 19434 and E faecalis ATCC 29212) and vanB (E faecalis ATCC 51299), the standard vancomycin-resistant strains of Enterococcus species used in this study, were confirmed. However, no amplification was seen with any of the standard vancomycin-sensitive strains of Enterococci (E faecalis NCTC 77, E faecium NCIMB 2699) using vanA and vanB genes primer sets (Table 2).
Table 2.
Results of van genotyping for VSE and VRE standard strains.
| Reference strains | van genotype | |
|---|---|---|
|
| ||
| E faecalis | NCTC 77 | Negative |
| E faecium | NCIMB 2699 | Negative |
| E faecium | ATCC 19434 | vanA |
| E faecalis | ATCC 29212 | vanA |
| E faecalis | ATCC 51299 | vanB |
Differentiation of VRE and VSE Strains by RT-PCR-HRMA approach
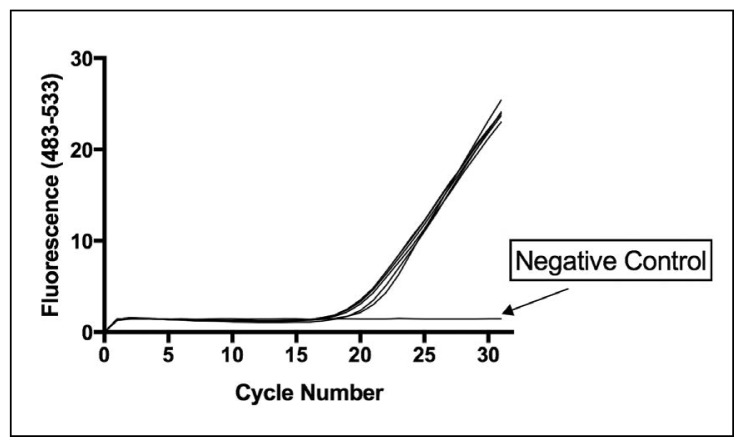
Initial studies used standard strains of vancomycin-resistant and -sensitive E faecalis and E faecium to validate the RT-PCR-HRMA approach. Both vancomycin-sensitive and resistant strains used in this study were amplified with equal efficiency by the V1 16S rRNA primers. Figure 1 shows the PCR-HRMA amplification curves of some vancomycin resistant (VRE) and sensitive (VSE) Enterococcus species used in this study as well as illustrates the amplification curve for B fragilis. However, no amplification was seen for nuclease-free water (Negative control).
Figure 1.
PCR-HRMA amplification curves of vancomycin-resistant (VRE) and -sensitive (VSE) E faecalis and E faecium. Amplification curves for VRE (E faecium ATCC 19434, E faecalis ATCC 51299 and E faecalis ATCC 29212) and the VSE standard strains (E faecalis NCTC 77 and E faecium NCIMB 2699). Each assay contained 250 pg of bacterial DNA. B fragilis was also included as the reference organism for generating VRE/VSE difference curves. B fragilis amplifies with the same efficiency as the Enterococcus strains. Nuclease-free water was used as a negative control (NC).
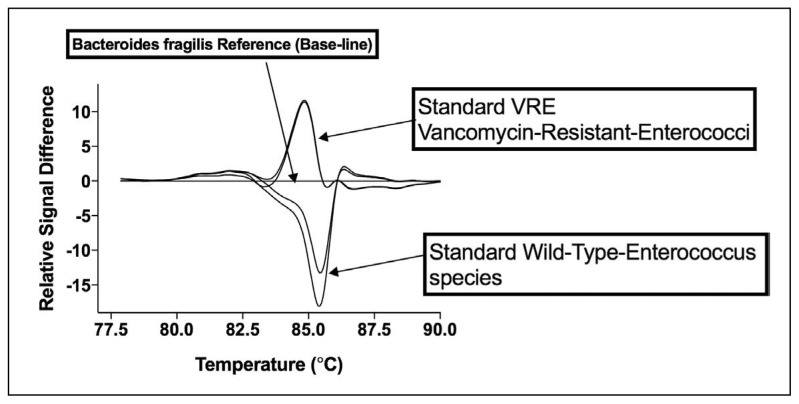
Figure 2 shows results following PCR analysis by HRMA. Difference plots were generated for each organism (vancomycin-resistant and -sensitive E faecalis and E faecium) by subtracting their fluorescence melting profile from that of a reference species B fragilis. A characteristic difference curve shape was produced by vancomycin-sensitive strains of E faecalis and E faecium. Vancomycin-resistant strains of these bacteria were associated with a markedly different curve shape facilitating a simple differentiation of resistant and sensitive strains. These data provide evidence that amplification of a region of the 16S followed by HRMA analysis can be used to differentiate vancomycin-resistant E faecalis and E faecium with vanA and vanB genotypes from vancomycin sensitive organisms.
Figure 2.
PCR-HRMA difference plots of VRE and VSE E faecalis and E faecium generated against B fragilis using V1 primer set. Difference curve plots for (A) VSE (NCTC 77, NCIMB 2699) and (B) vanA/B VRE (ATCC 19434, ATCC 51299, ATCC 29212) standard strains of E faecalis and E faecium. Each assay contained 250 pg of bacterial DNA. B fragilis was also included as the reference organism for generating VRE/VSE difference curves (plots). B fragilis amplifies with the same efficiency as the E faecalis and E faecium strains.
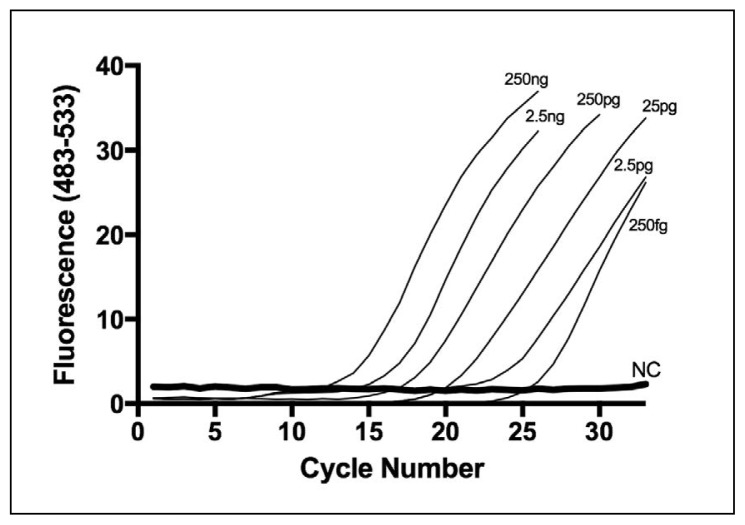
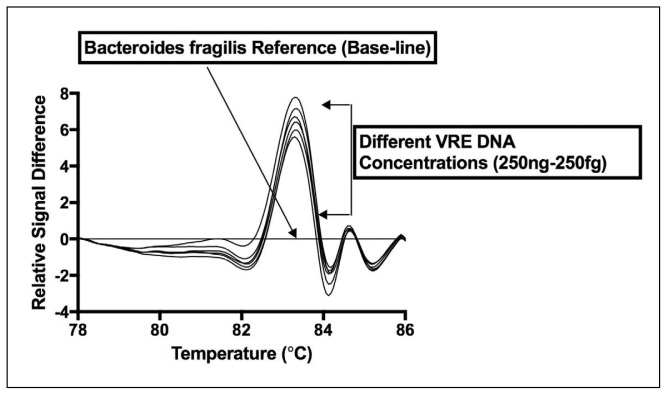
Figures 3 and 4 demonstrate the effect of DNA concentration on amplification curve and difference curve shape, consecutively, produced by the vancomycin-resistant Enterococcus strain (e.g. E faecalis ATCC 29212). All DNA concentrations for vancomycin-resistant E faecalis (ATCC 29212) ranging from 250 ng down to 250 fg were amplified using PCR-HRMA and V1 primer set (Figure 3). The difference curve shape was highly reproducible and no significant variation in shape was observed over a million-fold range of DNA concentration (250 fg to 250 ng per assay) (Figure 4). Similar data were obtained with the other reference VRE and wild-type Enterococcus strains used in this study (data not shown).
Figure 3.
Concentration dependence of RT-PCR-HRMA using the V1 primer set: Amplification curves for different quantities of DNA from VRE E faecalis ATCC 29212 (250 fg to 250 ng) using the V1 primer pair.
Figure 4.
Analytical sensitivity of VRE PCR-HRMA assay using the V1 primer set. Data show that the characteristic difference curve shape for VRE (using DNA from VRE E faecalis strain ATCC 29212) is maintained over a 6-log concentration range (250 fg to 250 ng DNA).
DISCUSSION
As described previously, Ozbak and co-authors15 provided evidence that the PCR-HRMA approach has potential as a low-cost alternative to other molecular approaches for identification of the 21 bacterial pathogens responsible for bloodstream infection (including E faecalis and E faecium). When combined with a suitable pathogen pre-amplification step for blood culture bottles, PCR-HRMA can detect and speciate low-level infection (e.g. 10 CFU/mL) and significantly faster than would be possible by the traditional clinical microbiological approach. However, a deficiency of most current molecular approaches to pathogen detection is that they often provide very limited information on antibiotic susceptibility.17,18 Currently, antibiotic susceptibility determined by standard culture techniques is routinely provided to clinicians to help direct treatment before more specific information becomes available. As with detection and speciation of pathogens, molecular determination of antibiotic resistance has the potential to decrease significantly the time required for determination of antibiotic resistance. We studied the ability of the 16S rRNA based PCR-HRMA approach in the detection of VRE, which is one of the most important antibiotic resistances encountered in the setting of bloodstream infection. Data presented in this study suggests that the 16S PCR-HRMA assay has the potential for rapid identification of vancomycin-resistant E faecalis and E faecium. This speculation needs to be confirmed in a representative clinical sample and supported by appropriate statistical analysis.
The use of PCR-based DNA detection of antibiotic resistance is possible if the genetic differences in the genes that confer resistance are known. Molecular approaches have been established for detection of antibiotic resistant genes using mainly PCR techniques,17,18 but also microarray.19,20 For instance, direct detection of the mecA gene, the primary determinant of methicillin resistance, has been described in several studies, but most require confirmation of positive culture before detection.21,22 These assays reliably detect mecA expression. The gene mecA is located on the chromosome but not on plasmids.23 Furthermore, in most cases, PCR assays for MRSA are limited by an inability to differentiate MRSA from methicillin-resistant coagulase negative Staphylococci (CoNS) requiring other assays to differentiate S aureus from CoNS (usually culture) which adds to the assay time. Similar limitations apply to current PCR-based approaches to detection of ESBLs and VRE. Several assays have been described, but these require confirmation and identification of the presence of gram-negative organisms (for ESBLs) or gram-positive Enterococcus species (for VRE) in the sample by additional techniques prior to analysis of resistance genes.24–26 Moreover, PCR-based approaches have been also developed for detection of van genes in VRE including commercial assays aimed primarily at screening of fecal samples and rectal swabs.11,12 However, these assays lack sensitivity and have been applied only to rectal or faecal specimens. Traditional and automated antibiotic sensitivity testing methods are not able to detect low-level vancomycin resistant Enterococci (mainly vanC genotype) and certain strains of vanB genotype, which indicates the need for developing rapid molecular approaches to detect all strains of VRE in clinical samples.27,28
The advantages offered by the PCR-HRMA approach, including speed, relative technical simplicity compared to probe-based techniques and low cost,13–15 makes it an attractive potential approach for detection of antibiotic resistance. HRMA has been used successfully to identify resistant genes in several bacteria. Recently, Hemarajata and his team developed in 2015 a HRMA-RT-PCR for detection and differentiation of OXA-48-Like-β-lactamases in carbapenem-resistant Enterobacteriaceae.29 Other applications of HRMA in this area include detection of rifaximin-resistance in Clostridium difficile,30 identification of nosocomial ESBL-producing E coli including ST131,31 detection of antimicrobial resistance in Neisseria gonorrhoeae,32 and detection of sequence type 131 E coli.33 These PCR-HRMA approaches have the potential to be used as tests to determine efficiently and rapidly some antibiotic resistance genes in pathogens and therefore could facilitate the timely administration of appropriate antimicrobial therapy for the infected patient.
The decision to investigate the potential application of 16S PCR-HRMA for the detection of common antibiotic resistance mechanisms relevant to bloodstream pathogens is based on several factors. Firstly, there is emerging evidence that signals for resistance to several antibiotics can be found on the 16S ribosomal gene. Markers of antibiotic resistance can be identified from sequence analysis of this region or some resistant genes may be located on the 16S ribosomal gene.34–36 Richardson reported that identification of antibiotic resistance is possible for some species using 16S rRNA sequential and thermodynamic properties.34 Several plasmid-encoded 16S rRNA methylases have emerged in clinical isolates of gram-negative bacilli.35,37 The emergence of pan-aminoglycoside-resistant, 16S rRNA methylase-producing, gram-negative bacteria has been increasingly reported in recent years.35,37 Secondly, if signals for antibiotic resistance were found using 16S-based PCR-HRMA it would be possible to develop a combined HRMA assay that was able to identify the pathogen species (as detailed by Ozbak)15 and at the same time provide evidence for antibiotic resistance without separate or additional steps. This is an important goal for molecular diagnostic assays of infection. For instance, SeptiFast (Roche Diagnostics), a multiplex real-time PCR platform that is designed to detect and identify a range of sepsis-related pathogens in blood, includes MRSA identification based on mecA38 but this requires a second PCR assay and no other antibiotic resistances are covered. Similarly, VYOO from SIRS-Lab is designed to detect a wide range of sepsis pathogens and includes several resistance genes including mecA and van genes but there are currently no published studies of the clinical performance of this test. I was fortunate in being able to test the usefulness of HRMA to detect resistance signals because of the availability of several different 16S primer sets that had already been optimised for HRMA and shown to work well for pathogen speciation in reports by Ozbak and colleagues in 2012.15
The data presented in this report suggest that the 16S PCR-HRMA approach developed for pathogen speciation has the potential to reliably differentiate vancomycin-resistant E faecalis and E faecium strains carrying the most common van genes (vanA and vanB), from vancomycin-sensitive strains using the V1 16S primers based on differences in melting curve shapes that are generated against B fragilis.
The finding of a “signature” for VRE in the 16S region is interesting and unexpected. The signature does not likely represent a sequence difference due to the presence of van genes. These genes are associated with plasmid uptake and are not located in the 16S region, but they are located on plasmids or on the chromosome.39,40 The implication is that HRMA is picking up a secondary change in the 16S region perhaps due to a phenotypic difference in Enterococci in association with the acquisition of van genes. An alternative explanation is that the gene encoding on different parts of the bacterial genome account for differences in detection, which could be due to their mobility.
Interestingly, strains with different van genotypes produce similar curve shapes. This appears to be a novel finding with no other studies showing a “VRE-signal” in the 16S region. Further studies are required to confirm the importance of this finding. The differences in curve shape in HRMA should reflect sequence differences in the amplified DNA. It will be important therefore to sequence the 16S product from vancomycin-sensitive and -resistant organisms including both reference standards and clinical isolates, to try to determine whether a sequence variation exists and whether the location is within the 16S gene. If a sequence difference is present in resistant organisms, further work is necessary to identify the phenotypic consequence of this variation and how it relates to vancomycin resistance. This could provide a better understanding of the secondary effects of acquiring resistance genes such as van. It would also be advantageous to apply this assay to a broader range of clinical isolates. Further testing of the developed VRE-HRMA assay in this study on clinical isolates of Enterococcus species is important for validation of this assay. It will also be advantageous to show that the same differences in melting curve shape are observed when resistant and sensitive isolates from different clinical sites are analysed using the 16S PCR-HRMA assay. Moreover, it is important to investigate whether this approach would also be useful in identifying vancomycin resistant E casseliflavus and E gallinarum expressing the vanC gene. It would be interesting if both E casseliflavus and E gallinarum showed the same curve shape as vancomycin-resistant E faecium and E faecalis.
In conclusion, this work has shown the potential application of the 16S PCR-HRMA approach for rapidly detecting and identifying resistant pathogens, notably vancomycin-resistant E faecium and E faecalis. The data on VRE provide a basis for combining VRE identification with pathogen speciation (as described by Ozbak15) in a rapid, low cost assay that is able to identify an organism as an Enterococcus and determine whether E faecium or E faecalis is resistant to vancomycin. This is not possible in current microbiological testing where identification of pathogens and antibiotic sensitivity require quite different and separate approaches that take a considerable time. The cost per sample in a single PCR-HRMA run (including DNA extraction, HRMA master mix and primer pairs) is estimated as about $10 (USD)15 which is quite cheap especially when compared to other advanced molecular approaches such as sequencing. The results presented in this study suggest that 16S-PCR-HRMA assay could be useful for epidemiological studies on Enterococcus genotypic distribution and therefore alert us to the dissemination of resistance. This study has also raised interesting questions about whether the acquisition of van genes brings about secondary changes in the bacterial genome.
Footnotes
Funding: None.
CONFLICT OF INTEREST: None.
REFERENCES
- 1.Weiner L, Webb A, Limbago B, Dudeck M, Patel J, Kallen A, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol. 2016;37:1288–1301. doi: 10.1017/ice.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat rev Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadak M, Cramp E, Ashiru-Oredope D. Antimicrobial Resistance and Stewardship in National Action Plans. Curr Treat Options Infect Dis. 2016;8:57–71. [Google Scholar]
- 4.Dellinger R, LeRTvy M, Rhodes A, Annane D, Gerlach H, Jaeschke R, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Intensive Care Med. 2013;39:165–228. doi: 10.1007/s00134-012-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Driscoll T, Crank CW. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist. 2015;8:217–230. doi: 10.2147/IDR.S54125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arias CA, Contreras GA, Murray BE. Management of multidrug-resistant enterococcal infections. Clin Microb Infect. 2010;16:555–562. doi: 10.1111/j.1469-0691.2010.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Niederhäusern S, Bondi M, Messi P, Iseppi R, Sabia C, Manicardi G, et al. Vancomycin-resistance transferability from vanA enterococci to Staphylococcus aureus. Curr Microbiol. 2011;62:1363–1367. doi: 10.1007/s00284-011-9868-6. [DOI] [PubMed] [Google Scholar]
- 8.Shnoy ES, Paras ML, Noubary F, Walensky RP, Hooper DC. Natural history of colonization with methicillin-resistant S. aureus (MRSA) and vancomycin-resistant Enterococci (VRE): a systematic review. BMC Infect Dis. 2014;14:177. doi: 10.1186/1471-2334-14-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanberger H, Antonelli M, Holmbom M, Lipman J, Pickkers P, Leone M, et al. Infections, antibiotic treatment and mortality in patients admitted to ICUs in countries considered to have high levels of antibiotic resistance compared to those with low levels. BMC Infect Dis. 2014;14:513. doi: 10.1186/1471-2334-14-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincent J, Rello J, Marshall J, Silva E, Anzueto A, Martin C, et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 11.Gazin M, Lammens C, Goossens H, Malhotra-Kumar S. Evaluation of GeneOhm VanR and Xpert vanA/vanB molecular assays for the rapid detection of vancomycin-resistant Enterococci. Eur J Clin Microbiol Infect Dis. 2012;31:273–276. doi: 10.1007/s10096-011-1306-y. [DOI] [PubMed] [Google Scholar]
- 12.Bourdon N, Bérenger R, Lepoultier R, Mouet A, Lesteven C, Borgey F, et al. Rapid detection of vancomycin-resistant Enterococci from rectal swabs by the Cepheid Xpert vanA/vanB. Diagn Microbiol Infect Dis. 2010;67:291–293. doi: 10.1016/j.diagmicrobio.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Cheng J, Huang C, Lin C, Chen C, Chang Y, Chang S, et al. Rapid detection and identification of clinically important bacteria by high resolution melting analysis after broad range ribosomal RNA real time PCR. Clin Chem. 2006;52:1997–2004. doi: 10.1373/clinchem.2006.069286. [DOI] [PubMed] [Google Scholar]
- 14.Yang S, Ramachandran P, Rothman R, Hsieh Y, Hardick A, Won H, et al. Rapid identification of biothreat and other clinically relevant bacterial species by use of universal PCR coupled with high resolution melting analysis. J Clin Microbiol. 2009;47:2252–2255. doi: 10.1128/JCM.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozbak H, Dark P, Maddi S, Chadwich P, Warhurst G. Combined molecular gram typing and high-resolution melting analysis for rapid identification of a syndromic panel of bacteria responsible for sepsis-associated bloodstream infection. J Mol Diagn. 2012;14:176–184. doi: 10.1016/j.jmoldx.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Hindiyeh MY, Smollan G, Gefen-Halevi S, Mendelson E, Keller N. Molecular Detection of Antibiotic Resistance Genes from Positive Blood Cultures. Methods Mol Biol. 2015;1237:97–108. doi: 10.1007/978-1-4939-1776-1_10. [DOI] [PubMed] [Google Scholar]
- 17.Arbour N, Weirich A, Cornejo-Palma D, Prevost S, Ramotar K, Harder CJ. Real-time PCR detection of VRE. Spartan Bioscience Inc; 2008. pp. 1–3. AN0019, version 1.1. [Google Scholar]
- 18.Abbott A, Fang F. Molecular Detection of Antibacterial Drug Resistance. In: Jorgensen James H, Pfaller Michael A, Carroll Karen C, Funke Guido, Landry Marie Louise, Richter Sandra S, Warnock David W., editors. Manual Clin Microbiol. Eleventh Edition. 2015. pp. 1379–1389. [Google Scholar]
- 19.Frye J, Lindsey R, Rondeau G, Porwollik S, Long F, McClelland M, et al. Development of a DNA Microarray to Detect Antimicrobial Resistance Genes Identified in the National Center for Biotechnology Information Database. Microb Drug Resist. 2010;16:9–19. doi: 10.1089/mdr.2009.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Card R, Zhang J, Das P, Cook C, Woodford N, Anjum MF. Evaluation of al. Expanded Microarray for Detecting Antibiotic Resistance Genes in a Broad Range of Gram-Negative Bacterial Pathogens. Antimicrob Agents Chemother. 2013;57:458–465. doi: 10.1128/AAC.01223-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fosheim G, Nicholson A, Albrecht V, Limbago B. Multiplex real-time PCR assay for detection of methicillin-resistant Staphylococcus aureus and associated toxingenes. J Clin Microbiol. 2011;49:3071–3073. doi: 10.1128/JCM.00795-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirvonen J, Nevalainen M, Tissari P, Salmenlinna S, Rantakokko-Jalava K, Kaukoranta S. Rapid confirmation of suspected methicillin-resistant Staphylococcus aureus colonies on chromogenic agars by a new commercial PCR assay, the Geno-mEra MRSA/SA Diagnose. Eur J Clin Microbiol Infect Dis. 2012;3:1961–1968. doi: 10.1007/s10096-011-1527-0. [DOI] [PubMed] [Google Scholar]
- 23.Berger-Bächi B. Expression of resistance to methicillin. Trends Microbiol. 1994;2:389–393. doi: 10.1016/0966-842x(94)90617-3. [DOI] [PubMed] [Google Scholar]
- 24.Jyoti S, Meera S, Pallab R. Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res. 2010;132:332–336. [PubMed] [Google Scholar]
- 25.Ellem J, Partridge S, Iredell J. Efficient direct extended-spectrum β-lactamase detection by multiplex real-time PCR: accurate assignment of phenotype by use of a limited set of genetic markers. J Clin Microbiol. 2011;49:3074–3077. doi: 10.1128/JCM.02647-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaschke AJ, Heyrend C, Byington CL, Fisher MA, Barker E, Garrone NF, et al. Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the Film Array system. Diagn Microbiol Infect Dis. 2012;74:349–355. doi: 10.1016/j.diagmicrobio.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park Y. Evaluation of VITEK 2, MicroScan, and Phoenix for identification of clinical isolates and reference strains. Diagn Microbiol Infect Dis. 2011;70:442–447. doi: 10.1016/j.diagmicrobio.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 28.Jenkins SG, Schuetz AN. Current concepts in laboratory testing to guide antimicrobial therapy. Mayo Clin Proc. 2012;87:290–308. doi: 10.1016/j.mayocp.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemarajata P, Yang S, Hindler JA, Humphries RM. Development of a Novel Real-Time PCR Assay with High-Resolution Melt Analysis To Detect and Differentiate OXA-48-Like β-Lactamases in Carbapenem-Resistant Enterobacteriaceae. Antimicrob Agents Chemother. 2015;59:5574–5580. doi: 10.1128/AAC.00425-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pecavar V, Blaschitz M, Hufnagl P, Zeinzinger J, Fiedler A, Allerberger F, et al. High-resolution melting analysis of the single nucleotide polymorphism hot-spot region in the rpoB gene as an indicator of reduced susceptibility to rifaximin in Clostridium difficile. Med Microbiol. 2012;61:780–785. doi: 10.1099/jmm.0.041087-0. [DOI] [PubMed] [Google Scholar]
- 31.Woksepp H, Jernberg C, Tärnberg M, Ryberg A, Brolund A, Nordvall M, et al. High-resolution melting-curve analysis of ligation-mediated real-time PCR for rapid evaluation of an epidemiological outbreak of extended-spectrum-beta-lactamase-producing Escherichia coli. J Clin Microbiol. 2011;49:4032–4039. doi: 10.1128/JCM.01042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dona V, Kasraian S, Lupo A, Guilarte YN, Hauser C, Furrer H, et al. Multiplex real-time PCR assay with high-resolution melting analysis for characterization of antimicrobial resistance in Neisseria gonorrhoeae. J Clin Microbiol. 2016;54:2074–2081. doi: 10.1128/JCM.03354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrison L, Hanson N. High-Resolution Melting Analysis for Rapid Detection of Sequence Type 131 Escherichia coli. Antimicrob Agents Chemother. 2017;61:e00265–17. doi: 10.1128/AAC.00265-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson C. Antibiotic resistant characteristics from 16S rRNA. 2012. [Accessed: 12 September 2017]. Available from Cornell University Library. <arXiv:1209.5801>. [Google Scholar]
- 35.Wachino J, Arakawa Y. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updat. 2012;15:133–148. doi: 10.1016/j.drup.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Tada T, Shimada K, Satou K, Hirano T, Pokhrel BM, Sherchand JB, et al. Pseudomonas aeruginosa clinical isolates in Nepal coproducing metallo-β-lactamases and 16S rRNA methyltransferases. Antimicrob Agents Chemother. 2017;61:e00694–17. doi: 10.1128/AAC.00694-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamane K, Wachino J, Suzuki S, Shibata N, Kato H, Shibayama K, et al. 16S rRNA Methylase-producing, Gram-negative pathogens, Japan[10] Emerg Infecti [11]Dis. 2007;13:642–646. doi: 10.3201/eid1304.060501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehmann L, Hunfeld K, Emrich T, Haberhausen G, Wissing H, Hoeft A, et al. Multiplex real time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med Microbiol & Immunol. 2008;197:313–324. doi: 10.1007/s00430-007-0063-0. [DOI] [PubMed] [Google Scholar]
- 39.Abadia Patiño L, Courvalin P, Perichon B. vanE gene cluster of vancomycin-resistant Enterococcus faecalis BM4405. J Bacteriol. 2002;184:6457–6464. doi: 10.1128/JB.184.23.6457-6464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sujatha S, Praharaj I. Glycopeptide Resistance in Gram-Positive Cocci: A Review. Interdiscip Perspect Infect Dis. 2012:10. doi: 10.1155/2012/781679. Article ID 781679. [DOI] [PMC free article] [PubMed] [Google Scholar]