Abstract
BACKGROUND
In Saudi Arabia, the epidemiology and clinical significnance of Torque Teno virus (TTV) infection alone and in patients with hepatitis virus infections have not been determined in a single study. In this paper, we molecularly investigated the rate and genotypes of TTV infection among Saudi Arabian blood donors and patients with viral hepatitis. The effect of TTV coinfection on viral hepatitis was also examined.
SUBJECTS AND METHODS
DNA was extracted from the sera of 200 healthy blood volunteers, 45 hepatitis B virus patients, 100 hepatitis C virus patients, 19 hepatitis G virus patients, and 56 non-A-G hepatitis patients. TTV DNA was amplified using primers derived from the ORF1 and 5′UTR regions. The alanine aminotransferase (ALT) level was determined for each specimen. Sequencing of ORF1 amplicons was carried out to investigate TTV genotypes.
RESULTS
Using primers derived from ORF1 and 5′UTR, TTV DNA was detected in 5.5% and 50.5%, respectively, of healthy blood donors, in 2.2% and 88.8% in hepatitis B patients, in 2.0% and 70% of hepatitis C patients, in 15.8% and 100% of hepatitis G patients, in 5.4% and 12.5% of non-A-G hepatitis patients and in 4.8% and 56.4% overall. No detrimental effect of TTV coinfection in viral hepatitis patients was noted. An overall prevalence of 4.8% and 56.4% was established. Phylogenetic analysis indicated that the most common genotype of TTV among Saudis is 2c.
CONCLUSION
The rate of TTV infection among Saudi Arabians seems to be lower than that stated in previous reports on Saudi Arabia and in some other countries. The virus does not seem to worsen the status of those who are suffering from viral hepatitis infection.
A DNA virus recently isolated from the serum of a patient with post-transfusion non-A-E hepatitis was named TT virus (TTV) after the initials of the patient.1,2 Later, it was established that the virus is a non-enveloped virus with a single-stranded, circular genomic DNA of approximately 3.9 kb, coated with proteins, and classified as the first member of the Circovirus genus in the Circinoviridae family.3 The taxonomic position has not yet been defined, but it is likely that TTV will be named Torque Teno Virus and assigned to a new genus called Anellovirus, belonging to the Circoviridae family.4 The virus genome has a wide range of sequence divergence that has enabled the determination of several genotypes and subtypes.5–7 Viazov et al8 reported only two genotypes, while Okamoto et al9 and Muljono et al10 described the existence of 16 and 23 genotypes, respectively. However, genotypes 1 and 2 seem to be the most prevalent worldwide.11 Sequence analysis of the TTV genome of isolates from various countries showed a similar genomic organization, comprised of a coding region of 2.6 kb and a non-coding region of about 1.2 kb.12 Although TTV DNA was detected in the liver tissues of infected subjects in high titers,13 other studies have suggested that TTV is not necessarily hepatitis related, and it seems to be a widespread, non-pathogenic virus.14–16 The virus was reported to persist in patients with multiple transfusions and in renal transplant patients.17,18 These and other studies in hemodialysis patients and hemophiliacs indicate that TTV is probably a transmissible blood-borne virus. However, some investigators reported other possible routes of transmission, including vertical,19 sexual,20 and oral-fecal.21
Worldwide reports have described a TTV prevalence from 1% to 100%, depending on the area of the genomic DNA detected and on the geographical location.11 In Saudi Arabia, a report described a TTV prevalence of 19% among 48 blood donors residing in Riyadh.7 In a letter to the editor, Simmonds et al22 described a prevalence of 100% among the same blood donors using primers derived from the 5′UTR region. In this study, in which we used a reasonably high number of samples compared to previous observations,7,11,22 we report our findings on the rate of TTV infection among Saudi Arabian nationals, using the original primers derived from the ORF1 region of the virus genome for genotyping and for comparison with primers derived from the 5′UTR region. The study was performed on serum samples from adult Saudi Arabian nationals who were healthy blood donors and on hepatitis patients coming from various parts of the country for treatment in our tertiary care medical center. Through the determination of alanine aminotransferase (ALT) levels, we also investigated the effect of TTV positivity on the status of patients with hepatitis of various viral etiologies.
Subjects and Methods
Specimens
Two hundred serum samples were taken from healthy, male, volunteer blood donors who were negative for HIV, HBV, and HCV markers (Table 1). Forty-five serum samples taken from from HBV-positive patients (28 males, 17 females), as determined by HBsAg positivity and confirmed by polymerase chain reaction (PCR),23 were negative for all other viral hepatitis markers. One hundred serum samples from HCV-positive patients (65 males, 35 females), as determined by both antibodies and PCR,24 were negative for all other viral hepatitis markers. Nineteen serum samples from HGV-positive patients (13 males, 6 females), as determined by PCR,25 were negative for all other viral hepatitis markers. Fifty-six serum samples from non-A-G (cryptogenic) hepatitis patients (35 males, 21 females), as determined by serology and PCR, had elevated ALT. All samples were from adults and were stored at −80ºC until processed. All TTV-DNA-ORF1-positive samples that were sequenced were each termed KSA, followed by the sample number and the designation “-BD” if from a blood donor, “-C” if from an HCV-positive patient, “-Cr” if from a non-A-G hepatitis patient, and “-G” if from a HGV-positive patient. All samples were obtained with consent from Saudi Arabian nationals, coming from different geographical areas of Saudi Arabia. Our institutional review board approved the study.
Table 1.
Rate of TTV infection among study groups as determined by PCR methods using primers derived from the ORF1 and 5′UTR regions.
| Groups | n | ALT (mean±SD) | Total TTV-positive (%) | |
|---|---|---|---|---|
| ORF1 Primers | 5′UTR Primers | |||
| Healthy blood donors | 200 | 33.6±10.7 | 11(5.5) | 101(50.5) |
| HBV-infected | 45 | 74.5±9.4 | 1(2.2) | 40(88.8) |
| HCV-infected | 100 | 71.8±9.3 | 2(2) | 70(70) |
| HGV-infected | 19 | 50.8±4.8 | 3(15.8) | 19(100) |
| Non-A-G hepatitis | 56 | 55.6±7.5 | 3(5.4) | 7(12.5) |
| Total | 420 | - | 20(4.8) | 237(56.4) |
Normal ALT range: 10–45 U/L
TTV DNA detection by PCR
Viral DNA from 100 μL of each serum sample was extracted by QIAmp System (Qiagen, Germany) according to the manufacturer’s instructions. Using the original primers derived from the ORF1 region (NG059, NG061 and NG063 as described by Nishizawa et al1), a semi-nested PCR was performed on all samples. Using primers derived from the 5′UTR (NG054/NG147 for the first round and NG133/NG132 for the second round as described by Okamoto et al26), nested PCR was performed on all samples. For both procedures, the first round of amplification was carried out in a total volume of 50 μL using 5 μL of extracted DNA and 45 μL of master mix (10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 0.2 mM of each of dNTPs, 20 pmole of each of sense [NG059 for ORF1 and NG054 for 5′UTR] and antisense [NG061 for ORF1 and NG147 for 5′UTR] primers, and 1 Unit of AmplitaqR DNA polymerase) for 35 cycles. Each cycle consisted of denaturation at 94°C for 1 min, annealing at 60ºC for 1 min, and extension at 72ºC for 1 min. The second round was performed with 20 pmoles of each of sense primer (NG061 for ORF1 and NG133 for 5′UTR) and antisense primer (NG063 for ORF1 and NG132 for 5′UTR) using 2.5 μL of the round 1 PCR product as a template for 25 cycles of the same cycling parameters as the first round except that the final extension step was increased to 7 minutes to allow complete formation of duplex molecules. Plasmid-cloned TTV DNA sequences were prepared in our laboratory and used as a positive control. Negative controls consisted of a negative serum and the PCR reaction mixture. Preparation of PCR mixture, viral DNA extraction, thermal cycling, and post-PCR analysis were done in separate areas. Amplified products (10 μL) were electrophoresed on 2% agarose, stained with 1 μg/mL ethidium bromide, and visualized under UV illumination.
Sequencing and phylogenetic analysis
PCR amplicons obtained from the amplification of the ORF1 region were subcloned into PCR 2.1 TA vector (Invitrogen, San Diego, USA). Two clones from each of 17 of the 20 positive samples were sequenced in both directions using the dideoxy nucleotide chain termination method (Taq Dye Deoxy Teminator Cycle Sequncing Kit, Applied Biosystems, Weiterstadt, Germany). Primers NG061 and NG063 were used for sequencing in an ABI 373A Autosequencer (Applied Biosystems). Nucleotide sequences of the positive amplicons and the published TTV prototype sequences were compared by multiple sequence alignment using the Lasergene Navigator computer program (DNA Star Sequence Analysis Software Package, Madison, WI). Nucleotide similarity and phylogenetic analysis were determined, using the CLUSTAL W algorithm.
Results were expressed as mean±standard deviation. Differences in proportions were tested by the chi-square test. Mean quantitative values were compared by Student’s t test. For all tests, P<0.05 was considered to have statistical significance.
Results
Prevalence of TTV DNA
When ORF1 region primers were used, TTV DNA was detected in 11 of 200 (5.5%) healthy volunteer blood donors, 1 of 45 (2.2%) HBV-positive patients, 2 of 100 (2.0%) HCV-positive patients, 3 of 19 (15.8%) HGV-positive patients, and 3 of 56 (5.4%) non-A-G hepatitis patients, with an overall rate of 4.8% (Table 1). When 5′UTR primers were used, TTV DNA was detected in 101 of 200 (50.5%) healthy volunteer blood donors, 40 of 45 (88.8%) HBV-positive patients, 70 of 100 (70.0%) HCV-positive patients, 19 of 19 (100%) HGV-positive patients, and 7 of 56 (12.5%) non-A-G hepatitis patients, with an overall rate of 56.4%.
Effect of TTV infection on the liver
The effect of TTV infection (as determined by 5′UTR positivity) on the liver was investigated by measuring the liver enzyme, alanine aminotransferase (ALT) values for each patient in all groups positive and negative for TTV DNA (Table 2). Blood donors infected with TTV (n=101) had relatively lower levels of ALT (33.4±10.7) than those who were not infected (n=99, 37.4±10.3). In the case of HBV, HCV, and non-A-G patient groups, subjects positive for TTV infection had slightly lower levels of ALT than those with single HBV, HCV, or non-A-G infection. In the HGV group, all patients were positive for TTV DNA. Taken altogether but without HGV patients, hepatitis patients (all with ALT levels >45 U/L, n=201) who were positive for TTV DNA (n=117) had relatively lower levels of ALT (58.7±10.1) than those who were negative for TTV DNA (n=84, ALT 66.8±12.2). However, all these differences were not statistically significant.
Table 2.
Effect of TTV positivity (by using 5′UTR primers) on the outcome of hepatitis infection as measured by ALT levels.
| Groups | n | ALT (mean±SD) | |
|---|---|---|---|
|
| |||
| Blood donor | TTV(+) | 101 | 33.4±10.7 |
| TTV(−) | 99 | 37.4±10.3 | |
|
| |||
| HBV | TTV(+) | 40 | 74.2±7.9 |
| TTV(−) | 5 | 74.5±9.5 | |
|
| |||
| HCV | TTV(+) | 70 | 70±1.4 |
| TTV(−) | 30 | 71.9±9.4 | |
|
| |||
| HGV | TTV(+) | 19 | 52.6±4.7 |
| TTV(−) | - | - | |
|
| |||
| Non-A-G | TTV(+) | 7 | 52.3±4.9 |
| TTV(−) | 49 | 55.9±7.6 | |
Normal ALT range: 10–45 U/L
Nucleotide sequence analysis of ORF1 of the TTV genome
Based on availability, semi-nested PCR products, using N22 primers from ORF1, were sequenced from all but three TTV DNA-positive samples (all 11 positive blood donor samples and 2 samples of each HCV, HGV, and non-A-G). Gene sequences were deposited in the Gene Bank (GenBank accession numbers AY256663-AY256679). Nucleotide sequence alignments were performed on 222-bp segments of the amplicons (excluding sequencing primers) and the corresponding region of TTV sequences taken from GenBank, using the CLUSTAL W algorithm.27 The pair-wise distance matrices showed that TTV isolates from blood donors were closely related to each other with an overall nucleotide sequence homology of 83.2% to 100%. Sequence diversity was highest between the two HGV-positive patients with a sequence divergence of 55.9%. Patients positive for HCV showed a sequence divergence of 19.7%, whereas positive non-A-G cryptogenic hepatitis patients showed a sequence divergence of only 1.4% (unpublished, but available on request).
Phylogenetic Analysis of TTV isolates
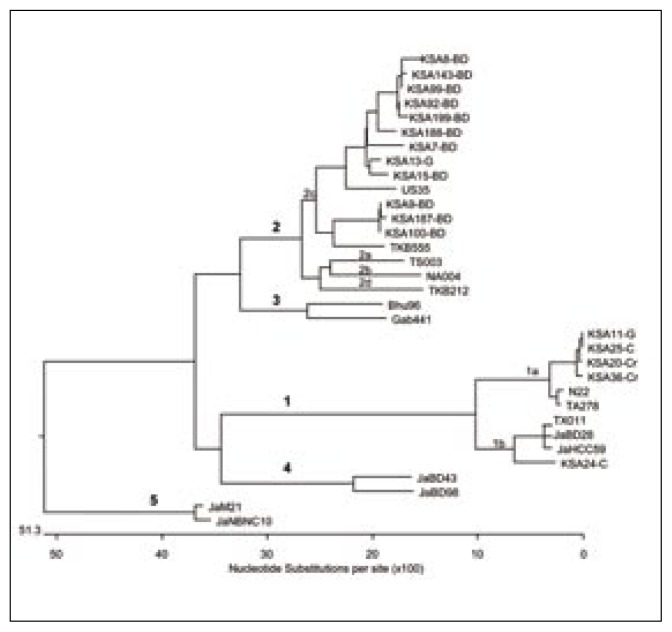
Sequences of local TTV isolates and 16 reference sequences taken from the GenBank database were subjected to phylogenetic analysis by the neighbor-joining method.28 The TTV isolates formed five distinct clades (Figure 1) which corresponded to 5 major genotypes separated by an evolutionary distance of >0.3. The genotypes were further divided into subtypes separated by an evolutionary distance of >0.15. It was found that all isolates from blood donors belonged to genotype 2c. Of the two TTV isolates from HCV-positive patients, one was found to be of genotype 1a and the other belonged to 1b. Of the two TTV isolates from HGV positive patients one was genotype 1a and the other was genotype 2c. Both TTV isolates from non-A-G patients were genotype 1a. No other genotype/subtype was found. The results are summarized in Table 3.
Figure 1.
Cladogram constructed with the nucleotide sequences derived from the ORF1 region of TTV isolates and reference strains by the neighbor-joining method [28]. For genotype 1a, the reference strains are TA278 (AB 0008394) and N22 (AB 017767). For genotype 1b, the reference strains are TX011 (AB 017769), JaBD28 (AB 018930), and JaHCC59 (AB 0188916). For genotypes 2a, 2b, 2c, and 2d, the reference strains are TS003 (AB 017770), NA004 (AB 017771), US35 (AF 124020), and TKB212 (AB 017772), respectively. For genotype 3, the reference strains are Bhu96 (AF 084107) and Gab441 (AF 084108). For genotype 4, the reference strains are JaBD43 (AB 018938) and JaBD98 (AB 018960). For genotype 5, the reference strains are JaM21 (AB 017887) and JaNBNC10 (AB 019861).
Table 3.
TTV genotypes/subtypes among the 17 Saudi Arabian isolates by ORF1 primers.
| Groups | TTV genotypes/subtypes | ||
|---|---|---|---|
| 1a | 1b | 2c | |
| Blood donors | - | - | 11 |
| HCV patients | 1 | 1 | - |
| HGV patients | 1 | - | 1 |
| Non-A-G patients | 2 | - | - |
| Total | 4/17 (23.5%) | 1/17 (5.9%) | 12/17 (70.6%) |
Other genotypes were not found. All genotypes/subtypes were deposited in GenBank (Accession numbers AY256663-AY256679)
Discussion
TTV is a novel single-stranded DNA virus that is transmitted both parenterally and non-parenterally. There is no clear association with liver disease or any other disease.14,15,29,30 TTV infection among adult Saudi Arabians was investigated in this study. Using primers derived from ORF1 and 5′UTR regions of the virus genome, respectively, a prevalence rate of 5.5% and 50.5% was determined among healthy blood donors, and 4.1% and 61.8% among patients with elevated ALT (hepatitis patients), for an overall prevalence of 4.8% and 56.4% (20 and 237 samples of 420). We decided to determine the prevalence of TTV infection by using the original primers derived from ORF1 region1 as well as primers derived from the 5′UTR since it was found that primers from this region detect more cases of TTV infection.26 We observed that this is true since the 5′UTR is a conserved region while the N22 region of ORF1 is variable. Prescott et al7 reported the prevalence of TTV in many countries, including Saudi Arabia, where a prevalence of 19% (9 samples of 48) was found. The primers they used were from the same ORF1 region that we used in our study. Simmonds et al22 reported a prevalence of 100% among Saudi Arabians, using primers derived from the 5′UTR region. However, a relatively small number of samples (n=48) in both reports were used and the samples were obtained only from blood donors living in Riyadh, which may have included non-Saudi Arabians. Early studies in Japan used primers based on the ORF1 region of the N22 clone.1,2 Later, it was found that primers deduced from the 5′UTR region showed a much higher prevalence in almost all geographical regions studied.11,12,31 For example, in neighboring Egypt the reported blood donor prevalence was 29% and 85% when primers from ORF1 and 5′UTR regions, respectively, were used.32,33 A recent study from Taiwan34 as well as a study of renal transplanted patients18 reported the same observation. A recent study on 449 serum samples from various populations in neighboring United Arab Emirates showed that nationals have a lower rate of infection than non-nationals, using 5′UTR-based primers.35 Like our study, they found a high TTV positivity in patients with hepatitis B and C.
The discovery of TTV led to thoughts that this virus might be a cause of unexplained post-transfusion hepatitis. Accumulating reports, however, showed that the virus is ubiquitous, is found in a wide range of tissues, and is not clearly associated with any overt disease.36,37 By reporting ALT levels in this study, we have observed that TTV does not affect the outcome or the pathogenesis of other, well-characterized hepatotropic viruses. In fact, the average ALT levels of HBV, HCV, and non-A-G hepatitis patients with a TTV co-infection were slightly lower than the average ALT level of those with no TTV co-infection. We are therefore in agreement with previous observations that TTV infection does not seem to be related to abnormal ALT levels, and ALT abnormality was mainly attributable to HCV, HBV or HGV and not TTV infection.38,39 The presence or absence of serum TTV DNA does not seem to affect the clinical course of patients with chronic HBV or HCV infection and TTV viremia with any genotype was not associated with HBV, HCV or HGV infection and abnormal ALT levels.40,41
For sequencing and phylogenetic analysis, we used the semi-nested PCR products of the ORF1 region since these are employed for the characterization of major genotypes.31 Most of our TTV positive samples (17/20) were cloned, sequenced and subjected to phylogenetic analysis in our laboratories (all 11 blood donor positive samples, the 2 TTV positive samples from HCV-infected patients, 2/3 TTV positive samples from the HGV-infected patients, and 2/3 TTV positive samples from the non-A-G cryptogenic hepatitis patients). All isolates were found to be of TTV genotypes 1 and 2. The TTV positive blood donor samples (n=11) were all genotype 2, subtype c (2c). However, those samples that were TTV positive from hepatitis patients (n=6) were mostly genotype 1, subtype a (1a), with two that were 1b and 2c. Prescott et al7 reported the genotypes of 8/9 TTV-positive samples from Saudi Arabia. They found 4 samples of the 8 tested belonged to genotype 1a, 3 samples were genotype 1 but could not be subtyped, and 1 sample could not be typed. Although 28 genotypes have been described thus far,36 our results are in agreement with the literature that most of the reported genotypes from all over the world were G1 and G2, with occasional G3 and rarely other types, and that there was no significant differences in the distribution of these types based on gender or clinical manifestions.11
This study showed that the prevalence of TTV among Saudi Arabians using primers from the ORF1 region is rather low compared to studies from other countries. It also demonstrated that the major TTV genotype is 2c, especially among healthy blood donors, and illustrated that TTV co-infection did not aggravate other hepatitis infections. Further investigations are needed to augment these results with additional insights into the impact of TTV on the health of our population and to contribute to the global understanding of the unclear role of this virus in pathogenesis.
Acknowledgments
This study was partially supported by a grant from King Abdulaziz City for Science and Technology [LGP-5-45]. The authors are grateful to the Research Center Administration for providing the facilities, equipment and approvals. We thank Maria Cristina Rasing and Hanan Shaarawi for secretarial and logistic assistance.
Footnotes
The nucleotide sequences of ORF1 reported in this paper appear in GenBank database under accession numbers AY256663-AY256679 (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide).
References
- 1.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 2.Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, et al. Molecular cloning and characterization of a novel DNA virus (TTV) associated with post transfusion hepatitis of unknown etiology. Hepatol Res. 1998;10:1–16. [Google Scholar]
- 3.Mushahwar IK, Erker JC, Muerhoff AS, Leary TP, Simons JN, Birkenmeyer LG, et al. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci USA. 1999;96:3177–3182. doi: 10.1073/pnas.96.6.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hino S. TTV, a new human virus with single stranded circular DNA genome. Rev Med Virol. 2002;12:151–158. doi: 10.1002/rmv.351. [DOI] [PubMed] [Google Scholar]
- 5.Simmonds P, Davidson F, Lycett C, Prescott LE, MacDonald DM, Ellender J, et al. Detection of a novel DNA virus (TT virus) in blood donors and blood products. Lancet. 1998;352:191–195. doi: 10.1016/s0140-6736(98)03056-6. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka Y, Mizokami M, Orito E, Ohno T, Nakano T, Kato T, et al. New genotypes of TT virus (TTV) and a genotyping assay based on restriction fragment length polymorphism. FEBS Lett. 1998;437:201–206. doi: 10.1016/s0014-5793(98)01231-9. [DOI] [PubMed] [Google Scholar]
- 7.Prescott LE, MacDonald DM, Davidson F, Mokili J, Pritchard DI, Arnot DE, et al. Sequence diversity of TT virus in geographically dispersed human populations. J Gen Virol. 1999;80:1751–1758. doi: 10.1099/0022-1317-80-7-1751. [DOI] [PubMed] [Google Scholar]
- 8.Viazov S, Ross RS, Niel C, de Oliveira JM, Varenholz C, Da Villa G, et al. Sequence variability in the putative coding region of TT virus: evidence for two rather than several major types. J Gen Virol. 1998;79:3085–3089. doi: 10.1099/0022-1317-79-12-3085. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto H, Takahashi M, Nishizawa T, Ukita M, Fukuda M, Tsuda F, et al. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology. 1999;259:428–436. doi: 10.1006/viro.1999.9770. [DOI] [PubMed] [Google Scholar]
- 10.Muljono DH, Nishizawa T, Tsuda F, Takahashi M, Okamoto H. Molecular epidemiology of TT virus (TTV) and characterization of two novel genotypes in Indonesia. Arch Virol. 2001;146:1249–1266. doi: 10.1007/s007050170089. [DOI] [PubMed] [Google Scholar]
- 11.Bindinelli M, Pistello M, Maggi F, Fornai C, Freer G, Vetteroni ML. Molecular properties, biology and clinical implications of TT virus, a recently identified widespread infectious agent of humans. Clin Microbiol Rev. 2001;14:98–113. doi: 10.1128/CMR.14.1.98-113.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biagini P. Human circoviruses. Vet Microbiol. 2004;98:95–101. doi: 10.1016/j.vetmic.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Alcorocho JM, Mariscal LF, de Lucas S, Rodriguez-Ingio E, Casqueiro M, Castillo I, et al. Presence of TTV DNA in serum, liver and peripheral blood mononuclear cells from patients with chronic hepatitis. J Viral Hepat. 2000;7:440–447. doi: 10.1046/j.1365-2893.2000.00252.x. [DOI] [PubMed] [Google Scholar]
- 14.Viazov S, Ross RS, Varenholz C, Lange R, Holtmann M, Niel C, et al. Lack of evidence for an association between TT infection and severe liver disease. J Clin Virol. 1998;11:183–187. doi: 10.1016/s0928-0197(98)00060-9. [DOI] [PubMed] [Google Scholar]
- 15.Kanda T, Yokosuka O, Ikeuchi T, Seta T, Kawai S, Imazeki F, et al. The role of TT virus infection in acute viral hepatitis. Hepatology. 1999;29:1905–1908. doi: 10.1002/hep.510290613. [DOI] [PubMed] [Google Scholar]
- 16.Das K, Kar P, Gupta RK, Das BC. Role of transfusion-transmitted virus in acute viral hepatitis and fulminant hepatic failure of unknown etiology. J Gastroenterol Hepatol. 2004;19:406–412. doi: 10.1111/j.1440-1746.2003.03308.x. [DOI] [PubMed] [Google Scholar]
- 17.Lefrere JJ, Roudot-Thoraval F, Lefrere F, Kanfer A, Mariotti M, Lerable J, et al. Natural history of the TT virus unfection through follow-up of TTV DNA-positive multiple-transfused patients. Blood. 2000;95:347–351. [PubMed] [Google Scholar]
- 18.Szladek G, Juhasz A, Asztalos L, Szoke K, Murvai M, Szarka K, et al. Persisting TT virus (TTV) genogroup 1 variants in renal transplant recipients. Arch Virol. 2003;148:841–845. doi: 10.1007/s00705-002-0001-9. [DOI] [PubMed] [Google Scholar]
- 19.Bagaglio S, Sitia G, Prati D, Cella D, Hasson H, Novati R, et al. Mother-to-child transmission of TT virus: sequence analysis of non-coding region of TT virus in infected mother-infant pairs. Arch Virol. 2002;147:803–812. doi: 10.1007/s007050200027. [DOI] [PubMed] [Google Scholar]
- 20.Krekulova L, Rehak V, Killoran P, Madrigal N, Riley LW. Genotypic distribution of TT virus (TTV) in a Czech population: evidence for sexual transmission of the virus. J Clin Virol. 2001;23:31–41. doi: 10.1016/s1386-6532(01)00185-8. [DOI] [PubMed] [Google Scholar]
- 21.Ross RS, Viazov S, Runde V, Schaefer UW, Roggendorf M. Detection of TT virus DNA in specimens other than blood. J Clin Virol. 1999;13:181–184. doi: 10.1016/s1386-6532(99)00015-3. [DOI] [PubMed] [Google Scholar]
- 22.Simmonds P, Prescott LE, Logue C, Davidson F, Thomas AE, Ludlam CA. TT virus - Part of the normal human flora? J Infect Dis. 1999;180:1748–1749. doi: 10.1086/315103. [DOI] [PubMed] [Google Scholar]
- 23.Zeldis JB, Lee JH, Mamish D, Finegold DJ, Sircar R, Ling Q, et al. Direct method for detecting small quantities of hepatitis B virus DNA in serum and plasma using the polymerase chain reaction. J Clin Invest. 1989;84:1503–1508. doi: 10.1172/JCI114326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Ahdal MN, Kessie G. Serological diagnosis of hepatitis C virus in patients with liver disease in Saudi Arabia. Evaluation of antibody determination by recombinant immunoblot assays in relation to detection by polymerase chain reaction. Diagn Microbiol Infect Dis. 1997;27:69–73. doi: 10.1016/s0732-8893(97)00001-1. [DOI] [PubMed] [Google Scholar]
- 25.Al-Ahdal MN, Kessie G, Chaudhary F, Rezeig MA, Al-Shammary FJ. GB virus C/Hepatitis G virus infection in Saudi Arabian blood donors and patients with cryptogenic hepatitis. Arch Virol. 2000;145:73–84. doi: 10.1007/s007050050006. [DOI] [PubMed] [Google Scholar]
- 26.Okamoto H, Nishizawa T, Ukita M, Takahashi M, Fukuda M, Iizuka H, et al. The entire nucleotide sequence of a TT virus isolate from the United States (TUS01): comparison with reported isolates and phylogenetic analysis. Virology. 1999;259:437–48. doi: 10.1006/viro.1999.9769. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acid Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Toniutto P, Fabris C, Falleti E, Lombardelli T, Gasparini V, Barillari G, et al. Evidence against a direct role played by transfusion-transmitted virus infection in causing hepatic or hematologic manifestations. Blood. 1999;93:2426–2427. [PubMed] [Google Scholar]
- 30.Suzuki F, Kazuaki C, Tsubota A, Akuta N, Someya T, Kobayashi M, et al. Pathogenic significance and organic virus levels in patients infected with TT virus. Intervirology. 2001;44:291–297. doi: 10.1159/000050060. [DOI] [PubMed] [Google Scholar]
- 31.Lemey P, Salemi M, Bassit L, Vandamme AM. Phylogenetic classification of TT virus groups based on the N22 region is unreliable. Virus Res. 2002;85:47–59. doi: 10.1016/s0168-1702(02)00017-5. [DOI] [PubMed] [Google Scholar]
- 32.Abe K, Inami T, Asano K, Miyoshi C, Masaki N, Hayashi S, et al. TT Virus infection is widespread in the general populations from different geographic regions. J Clin Microbiol. 1999;37:2703–2705. doi: 10.1128/jcm.37.8.2703-2705.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gad A, Tanaka E, Orii K, Kafumi T, Serwah AE, El-Sherif A, et al. Clinical significance of TT virus infection in patients with chronic liver disease and volunteer blood donors in Egypt. J Med Virol. 2000;60:177–181. [PubMed] [Google Scholar]
- 34.Hsu HY, Ni YH, Chen HL, Kao JH, Chang MH. TT virus infection in healthy children after blood transfusion, and children with non-A to E hepatitis or other liver disease in Taiwan. J Med Virol. 2003;69:66–71. doi: 10.1002/jmv.10249. [DOI] [PubMed] [Google Scholar]
- 35.Al-Moslih MI, Abuodeh RO, Hu YW. Detection and genotyping of TT virus in healthy and subjects with HBV or HCV in different populations in the United Arab Emirates. J Med Virol. 2004;72:502–508. doi: 10.1002/jmv.20017. [DOI] [PubMed] [Google Scholar]
- 36.Simmonds P. TT virus infection: a novel virus-host relationship. J Med Microbiol. 2002;51:455–458. doi: 10.1099/0022-1317-51-6-455. [DOI] [PubMed] [Google Scholar]
- 37.Pollicino T, Raffa G, Squadrito G, Costantino L, Cacciola I, Brancatelli S, et al. TT virus has a ubiquitous diffusion in human body tissues: analyses of paired serum and tissue samples. J Viral Hepat. 2003;10:95–102. doi: 10.1046/j.1365-2893.2003.00417.x. [DOI] [PubMed] [Google Scholar]
- 38.Moriyama M, Longren W, Zi-Yi Z, Oshiro S, Matsumura H, Aoki H, et al. TT virus infection does not affect the clinical profiles of patients with hepatitis B and C in Yanbian City, China. Intervirology. 2003;46:214–221. doi: 10.1159/000072430. [DOI] [PubMed] [Google Scholar]
- 39.Kao JH, Chen W, Chen PJ, Lai MY, Chen DS. TT virus infection in patients with chronic hepatitis B or C: influence on clinical, histological and virological features. J Med Virol. 2000;60:387–392. doi: 10.1002/(sici)1096-9071(200004)60:4<387::aid-jmv4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 40.Dai CY, Yu ML, Hou C, Lu SN, Wang JH, Huang JF, et al. Clinical characteristics and distribution of genotypes of TT virus infection in a hepatitis C virus-hyperendemic township of a hepatitis B virus-endemic country (Taiwan) J Gastroenterol Hepatol. 2002;17:1192–1197. doi: 10.1046/j.1440-1746.2002.02878.x. [DOI] [PubMed] [Google Scholar]
- 41.Lai YC, Hu RT, Yang SS, Wu CH. Coinfection of TT virus and response to interferon therapy in patients with chronic hepatitis B or C. World J Gastroenterol. 2002;8:567–570. doi: 10.3748/wjg.v8.i3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]