Abstract
BACKGROUND
Resistance to beta-lactam antibiotics has become more common in Morganella morganii, which can cause of outbreaks of bacteremia and septicemia in postoperative patients.
OBJECTIVE
Investigate drug susceptibility of M morganii, identify the gene responsible for extended-spectrum beta-lactamase (ESBL) production and explore treatment options.
DESIGN
Descriptive study.
SETTING
Hospitals in An Najaf, Iraq.
METHODS
M morganii isolates were identified based on morphology, biochemical tests and VITEK® 2 compact system using (GN-ID) card. M morganii isolates were subjected to antibiotic resistance tests using the minimum inhibitory concentration (MIC) technique and an antibiogram was produced. Molecular studies were conducted using the polymerase chain reaction technique.
MAIN OUTCOME MEASURE (S)
Minimum inhibitory concentration.
RESULTS
From 395 gram-negative bacteria, only 17 isolates M morganii grew on MacConkey agar. M morganii isolates strongly resistant to several antibiotics were considered multidrug resistant. All M morganii isolates were ESBL producers. Four genes (CTX-M, SHV, TEM and OXA) encoding the β-lactamase enzyme were detected. Meropenem and imipenem were highly active against the M morganii isolates.
CONCLUSIONS
All isolates showed resistance to most common antibiotics, which limits options for treatment. This study provided useful information for selecting antibiotics to precisely target infections caused by M morganii.
LIMITATIONS
Limited to antibiotic susceptibility and genotype.
Morganella morganii is a rod-shaped gram-negative bacteria, a member of the tribe Proteeae of the family Enterobacteriaceae. The Proteeae, which consists of Morganella, Proteus and Providencia, are important opportunistic pathogens that cause a variety of nosocomial infections.1,2 Exposure of bacterial strains to β-lactam antibiotics has altered the dynamics of β-lactamase mutations, leading to an increase in activity against third- and fourth-generation cephalosporins such as cefepime, ceftazidime and cefotaxime as well as aztreonam.3 Like other Enterobacteriaceae, M morganii are naturally resistant to beta-lactam antibiotics, but resistance has become more common in M morganii, as demonstrated by the increased production of the so-called extended spectrum β-lactamases (ESBLs).4,5
M morganii has been implicated in outbreaks of bacteraemia and septicemia in humans that occur most commonly in postoperative patients.6,7 The majority of the known gene cassettes confer resistance to antibiotics.8 The integration of most β-lactamase genes in plasmids and transposons encourages rapid transferance of resistance genes between microbes.9 The β-lactamase genes are often found within the integrons with multi-drug resistance cassettes which are involved in resistance to chloramphenicol, aminoglycosides, sulphonamides and macrolides.10 Third-generation cephalosporins, referred to as extended-spectrum cephalosporins, which include ceftriaxone, ceftazidime, and cefotaxime, were effective against ampicillin hydrolyzing β-lactamases and gained wide-spread clinical use. However, within a few years of their introduction, hospital-acquired gram-negative bacilli (such as Klebsiella pneumonia) started producing a mutated version of β-lactamases called extended spectrum β-lactamases (ESBLs) and plasmid-mediated AmpC β-lactamases that conferred resistance to third-generation cephalosporins.11 The wide spectrum of β-lactamases represented by TEM-1 and SHV-1 gave rise to the name “extended spectrum” beta-lactamses (ESBL), which later involved CTX-M and OXA-type enzymes. 9 These enzymes are capable of hydrolyzing and inactivating a wide variety of therapeutic β-lactam antimicrobials. 12 The purpose of our study was to investigate drug susceptibility of M morganii at hospitals in the city of Najaf, Iraq, identify the gene responsible for extended-spectrum beta-lactamase (ESBL) production and explore treatment options.
PATIENTS AND METHODS
Samples collection
Clinical samples were collected from November 2014 to February 2015. These samples were collected from patients at different hospitals in An Najaf, Iraq (Alsader Teaching Hospital, Al Najaf General Hospital and Al-Furat Al-Awsat hospital). All samples were inoculated onto MacConkey agar plates and incubated at 37°C under aerobic conditions for 18 to 24 hours.
Isolation and identification of the microorganism
M morganii isolates were recovered from clinical samples after culturing on MacConkey agar, which incubated for overnight at 37°C. Identification was based on morphology, gram staining, and biochemical tests (catalase, oxidase, methyl red, citrate, indole, Voges-Proskauer, Kligler iron agar, motility, urease, gelatin liquefaction and lactose). The VITEK® 2 compact system was used to carry out final identification.
Antibiogram testing
Antibiogram testing was carried out using the VITEK® 2 system according to manufacturer instructions using ASTN093 cards. The following antibiotics were tested by ASTN093: piperacillin-tazobactam, piperacillin, ticarcillin, ticarcillin-clavulanic acid, ceftazidime, cefepime, aztreonam, meropenem, imipenem, isepamicin, tobramycin, amikacin, gentamicin, pefloxacin, ciprofloxacin, trimethoprimsulfamethoxazole, minocycline, colistin and rifampicin.
Extended-spectrum β-lactamase production
All bacterial isolates were tested for extend spectrum-spectrum β-lactamase by an initial screening test depending on the MIC results for ceftazidime with the VITEK® 2 compact system. The isolate was considered a potential ESBL producer if the ceftazidime MIC was ≥2 μg/mL (CLSI, 2012). The disk approximation test was used to confirm ESBL production. The test involves 30 μg antibiotic disks of ceftazidime, ceftriaxione, cefotaxime and azetreonam placed 15 mm apart (edge to edge) around a central disk of amoxicillin-clavulanate disk (20:10μg) on Muller-Hinton agar plates inoculated with the organism being tested for ESBL production. The augmentation (increase in diameter of inhibition zone) between the central amoxicillin-clavulanate disk and β-lactam antibiotic disks reflects antibiotic resistance and the organism was recorded as an ESBL producer. 13
Isolation of plasmid DNA and polymerase chain reaction
Plasmid DNA was extracted by the alkaline method according to manufacturer instructions (G-Biosciences, USA). The polymerase chain reaction (PCR) test was carried out in a final volume of 25 μL containing a 4 μL template DNA, 12.5 μL of 2X KABA2G Robust HotStart ReadyMix, 1.25 μL of forward primer, 1.25 μL of reserve primer and 6 μL of PCR grade water for each sample. The DNA amplification was performed for the blaCTX-M gene by an initial denaturation at 94°C for 3 min, denaturation at 94°C for 45 sec, annealing 60°C for 30 sec, extension 72°C for 1 min and final extension 72°C for 3 min followed by 35 cycles of amplification. For the blaSHV gene an initial denaturation at 94°C for 3 min, denaturation at 94°C for 1 min, annealing 55 °C for 1 min, extension 72°C for 1 min and final extension 72°C for 7 min was followed by 40 cycles of amplification. For the blaTEM gene an initial denaturation at 94°C for 5 min, denaturation 94°C for 1 min, annealing 55°C for 1 min, extension 72°C for 30 sec and final extension 72°C for 5 min was followed by 35 cycles of amplification. For the blaOXA gene an initial denaturation at 94°C for 5 min, denaturation at 94°C for 45 sec, annealing 55°C for 45 sec, extension 72°C for 1 min and final extension 72°C for 5 min was followed by 30 cycles of amplification. PCR products were separated on 1 % agarose gel and visualized using a transilluminator (BIO-RAD, USA).
Statistical analysis
The Microsoft Excel (Package 2007) was used to compile the data.
RESULTS
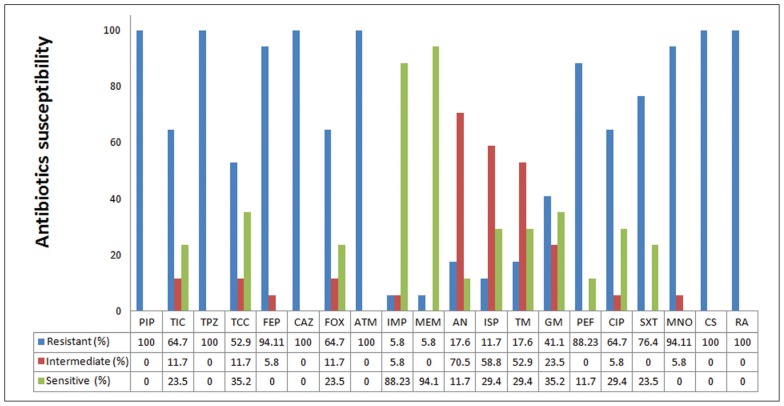
During the study interval from November 2014 to February 2015, clinical samples were collected from 800 patients (Table 1). All M morganii isolates were resistant to a minimum of three classes of antibiotics to which they were tested, and this were considered multidrug resistant (MDR) (Figure 1). All isolates were resistant to piperacillin, piperacillin-tazobactam, and aztreonam, with lower rates of resistance to ticarcillin and ticarcillin-clavulanic acid, with the lowest rates to imipenem and meropenem. Figure 1 shows the rates of resistance and sensitivity for other antibiotics. The aminoglycosides had a high-to-moderate effect against M morganii isolates, while most isolates were resistant to fluoroquinolones, colistin and rifampicin. Results of susceptibility revealed that none of the isolates was fully resistant or susceptible to antibiotics tested.
Table 1.
Sources and distribution of clinical samples according to type of infection.
| Infection type | Samples No. | Bacterial growth No. (%) |
Yielded no growth No. (%) |
|---|---|---|---|
|
| |||
| Urinary tract infection | 400 | 207 (51.8) | 193 (48.2) |
| Ear infection | 200 | 119 (59.5) | 81 (40.5) |
| Respiratory tract infection | 200 | 69 (34.5) | 131 (65.5) |
| Total (%) | 800 | 395 (49.4) | 405 (50.6) |
Figure 1.
Antibiogram testing of Morganella morganii isolates with the automated VITEK® 2 compact system by using AST-N093 cards (n=17). PIP, Piperacillin; TIC, Ticarcillin; TZP, Piperacillin-tazobactam; TCC, Ticarcillin-clavulanic acid; FEP, Cefepime; CAZ, Ceftazidime; FOX, Cefoxitin; ATM, Aztreonam; IPM, Imipenem; MEM, Meropenem; AN, Amikacin; ISP, Isepamicin; TM, Tobramycin; GN, Gentamicin; PEF, Pefloxacin; CIP, Ciprofloxacin; SXT, Trimethoprim-sulfamethoxazole; MNO, Minocycline; CS, Colistin and RA, Rifampicin.
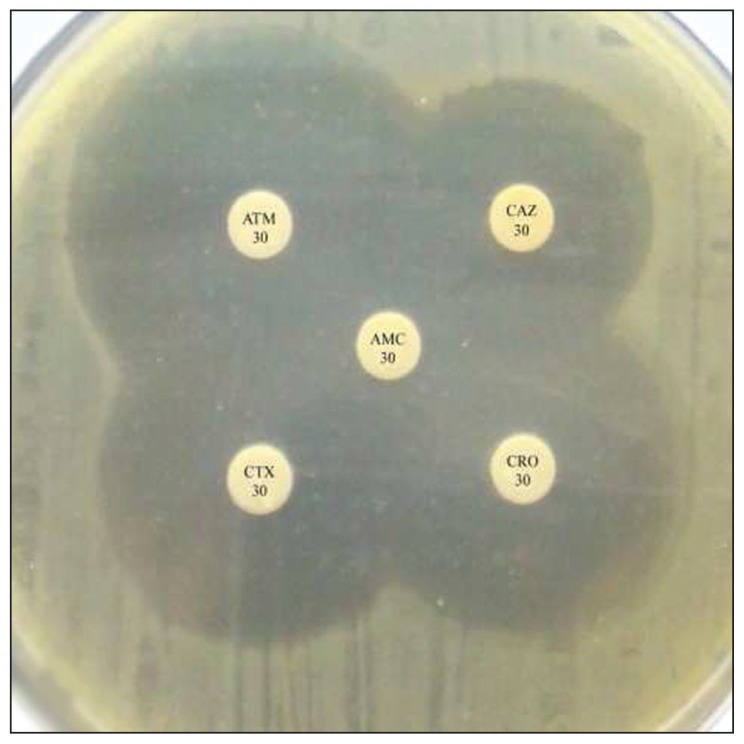
The ceftazidime MIC results from the VITEK® 2 compact system showed that all M morganii isolates (n=17) were ESBL producers during the initial screening (Table 2). The phenotypic confirmatory test using the disk approximation method confirmed the ability of M morganii isolates to produce ESBLs. All M morganii isolates (n=17) were ESBL positive on the phenotypic confirmatory test (Figure 2).
Table 2.
Minimum inhibitory concentration (MIC) ranges for each antibiotic for Morganella morganii isolates.
| Antibiotic | MIC range (μg/ml) | Antibiotic | MIC range (μg/ml) |
|---|---|---|---|
|
| |||
| Cefepime | 4 – ≥64 | Gentamicin | 2 – ≥16 |
| Ceftazidime | 32 – ≥64 | Pefloxacin | 0.5 ≥16 |
| Aztreonam | 32 – ≥64 | Ciprofloxacin | 0.5 ≥4 |
| Imipenem | 2 – ≥16 | Trimethoprimsulfamethoxazole | ≤20 – ≥320 |
| Meropenem | 0.5 – ≥16 | Minocycline | 8 – ≥16 |
| Amikacin | 8 – ≥64 | Colistin | ≥16 |
| Isepamicin | 8 – ≥64 | Rifampicin | ≥32 |
| Tobramycin | 2 – ≥16 | ||
Figure 2.
Show positive result of phenotypic confirmatory test for ESBL.
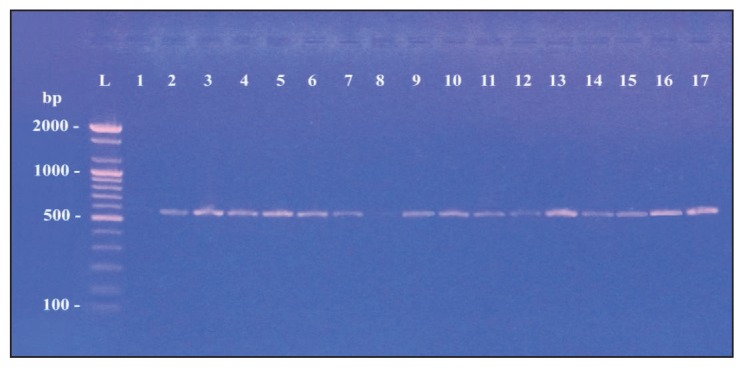
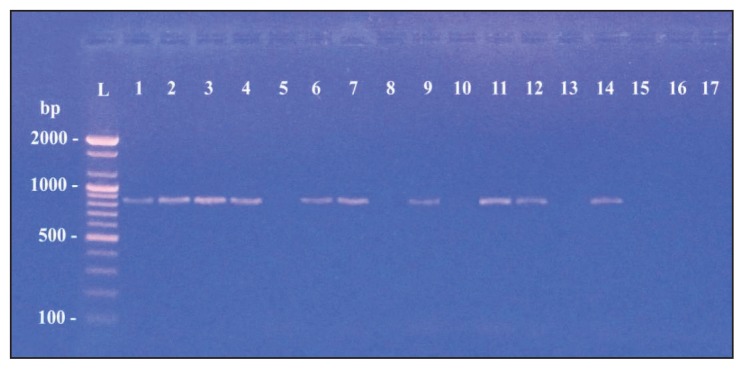
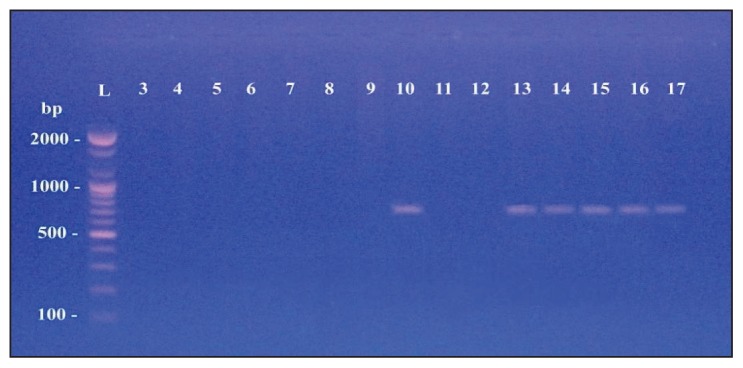
Table 3 shows the specific primers used to detect extended-spectrum β-lactamase genes (CTX-M, SHV, TEM and OXA) from the 17 isolates of M morganii.14–17 Of the 17 M morganii isolates, 16 carried at least one type of blagene (Figures 3, 4, 5 and 6). However, the most commonly identified ESBL gene was blaCTX-M type in 15 of the isolates (Figure 3). PCR amplification using specific primers of the blaSHV gene found the gene in 10 isolates (Figure 4). The blaTEM gene was detected in six isolates (Figure 5). Finally, only two of the examined isolates harbored a gene for the OXA type enzyme (Figure 6).
Table 3.
Primer sequences for detection of extended-spectrum beta-lactamase genes.
| No. | Target gene | Primer sequence | Amplicaon Size (bP) | References |
|---|---|---|---|---|
|
| ||||
| 1 | blaCTX-M | F CGCTGTTGTTAGGAAGTGTG R GGCTGGGTGAAGTAAGTGAC |
569 | 14 |
| 2 | blaOXA | F ATATCTCTACTGTTGCATCTCC R AAACCCTTCAAACCATCC |
619 | 15 |
| 3 | blaTEM | F TGCAACAGTGCCTCTCGATA R CTCGTGCACCCAACTGATCT |
717 | 16 |
| 4 | blaSHV | F GGTTATGCGTTATATTCGCC R GGTTAGCGTTGCCAGTGCTC |
867 | 17 |
Figure 3.
blaCTX-M gene with amplified product 569bp.
Figure 4.
blaSHV gene with amplified product 867 bp.
Figure 5.
blaTEM gene with amplified product 717 bp.
Figure 6.
blaOXA gene with amplified product 619 bp.
DISCUSSION
Much evidence supports the hypotheses that two factors largely contribute to the development and spread of antimicrobial resistance: the transmission of plasmids between microbes and poor selection of antimicrobials for treatment of infections. Plasmid-mediated antimicrobial resistance from horizontal transmission of plasmids is a global threat.18
In the present study, M morganii isolates were highly resistant to many antimicrobials (cephalosporins, ceftazidime and cefepime). However, resistance to third-generation cephalosporins occurred mainly by mutations in the common group of class A β-lactamases that include TEM, SHV and CTX-M.19 We found high resistance to third-generation cephalosporins such as ceftazidime that may be due to the production of cefotaximases encoded by the CTX-M gene. These CTX-Ms appear to have more activity toward cefotaxime than ceftazidime.20
We found intermediate resistance (47%) toward ticarcillin/clavulanic acid. Although β-lactamase inhibitors prevent the inactivation of the β-lactam antibiotics, they have low antimicrobial activity themselves.21 Since high susceptibility rates to imipenem and meropenem (88.2% and 94.1%, respectively) were found among M morganii isolates, this study shows that those two β-lactam antibiotics are the most effective against M morganii, a finding consistent with previous data by researchers from Greece and China.2,22
In this study, ceftazidime resistance was chosen to detect ESBL producers because it is the best third-generation cephalosporin substrate for most TEM, SHV and CTX-M derived ESBLs.23,24 The CTX-M genes are a recent family of plasmid-mediated ESBLs; some of them are part of transposons or constitute gene cassettes in integrons.25,26 The blaSHV gene was detected in 10 isolates. The SHV β-lactamases enzymes are mainly found in gram-negative bacteria.27 These enzymes possess variants of substituted serine instead of glycine at position 238 and lysine instead of glutamate at position 240.28 There are more than 125 SHV varieties described worldwide.29 The SHV β-lactamases were the predominant ESBL types in Europe and the United States.30 The blaTEM gene was detected in six of the tested M morganii isolates. The TEM-ESBLs are the first plasmid-mediated β-lactamase that are detected in many genera of Enterobacteriaceae such as Proteus mirabilis and Klebsiella pneumonia, E coli; and also in non-Enterobacteriaceae like Pseudomonas aeruginosa.31 Finally, two of the M morganii isolates harbored the blaOXA gene. However, most of the OXA derivative genes are located in plasmids and integrons,32 while some OXA-type β-lactamases are encoded by chromosomal genes that appear to be resident in some microbial genomes such as those in Pseudomonas aeruginosa.33 The integrons that harbor co-resistance genes make it a useful tool for facilitating dissemination.34,35
Our study was limited to antibiotic susceptibility and genotype testing. further study using sequencing technology and proteomic analysis is recommended.
In summary, M morganii harbors several beta-lactamases genes and shows resistance to most common antibiotics, which limit options for treatment. This study provided useful information for seleccting antibiotics to precisely target infections caused by M morganii.
REFERENCES
- 1.Chen Y, Peng H, Shia W, Hsu F, Ken C, Tsao Y, Chen C, et al. Wholegenome sequencing and identification of Morganella morganii KT pathogenicity-related genes. BMC Genomics. 2012;13:4–11. doi: 10.1186/1471-2164-13-S7-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falagas ME, Kavvadia PK, Mantadakis E, Kofteridis DP, Bliziotis IA, Saloustros E, Maraki S, Samonis G. Morganella morganii infections in a general tertiary hospital. Infection. 2006;34:315–321. doi: 10.1007/s15010-006-6682-3. [DOI] [PubMed] [Google Scholar]
- 3.Samaha-Kfoury JN, Araj GF. Recent developments in b-lactamases and extended spectrum b-lactamases. BMJ. 2003;327:1209–1213. doi: 10.1136/bmj.327.7425.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Jasser AM. Extended-specrum betalactamases (ESBLs): a global problem. Kuwait Medical Journal. 2006;38(3):171–185. [Google Scholar]
- 5.Poirel L, Guibert M, Girlich D, Naas T, Nordmann P. Cloning sequence analyses expression and distribution of amp-CampR from Morganella morganii clinical isolates. Antimicrob Agents Chemother. 1999;43:769–776. doi: 10.1128/aac.43.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowen JL, Lopez SM. Morganella morganii early onset sepsis. Pediatr Infect Dis J. 1998;17:1176–1177. doi: 10.1097/00006454-199812000-00021. [DOI] [PubMed] [Google Scholar]
- 7.Manos J, Belas R. The Genera Proteus Providencia and Morganella. Prokaryotes. 2006;6:245–269. [Google Scholar]
- 8.Fluit AC, Schmitz FJ. Class 1 integrons gene cassettes mobility and epidemiology. Eur J Clin Microbiol Infect Dis. 1999;18:761–770. doi: 10.1007/s100960050398. [DOI] [PubMed] [Google Scholar]
- 9.Bradford PA. Extended-spectrum b-lactamases in the 21st century: characterization epidemiology and detection of this important resistance threat. Clinical Microbiology Reviews. 2001;14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eldhagen GF. Integrons and b-lactamases-a novel perspective on resistance. Int J Antimicrob AG. 2004;23:556–562. doi: 10.1016/j.ijantimicag.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Mocktar C, Govinden U, Sturm AW, Essack SY. CMY- 20 a novel AmpC-type beta-lactamase from South African clinical Escherichia coli isolates. Diagn Micr Infec Dis. 2008;60:405–408. doi: 10.1016/j.diagmicrobio.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhary U, Aggarwal R. Extended-spectrum b-lactamases (ESBL) An emerging threat to clinical therapeutics. Indian Journal Med Microb. 2004;22:75–80. [PubMed] [Google Scholar]
- 13.Murray PR, Baron EJ, Pfaller MA, et al., editors. Manual of Clinical Microbiology. 7th edition. American Society for Microbiology; Washington, DC, USA: 1999. [Google Scholar]
- 14.Bali Elif Burcu, Acik Leyla, Sultan Nedim. Phenotypic and molecular characterization of SHV, TEM, CTX-M and extended-spectrum beta-lactamase produced by Escherichia coli, Acinobacter baumannii and Klebsiella isolates in a Turkish hospital. Afr J Microbiol Res. 2010;4(8):650–654. [Google Scholar]
- 15.Colom, Karmele, Pérez J, Alonso R, Fernández-Aranguiz A, Lariño E, Cisterna R. Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and bla-OXA–1 genes in Enterobacteriaceae. FEMS Microbiol Lett. 2003;223(2):147–151. doi: 10.1016/S0378-1097(03)00306-9. [DOI] [PubMed] [Google Scholar]
- 16.Sompolinsky D, Nitzan Y, Tetry S, Wolk M, Vulikh I, Kerrn MB, Katcoff DJ. Integron-mediated ESBL resistance in rare serotypes of Escherichia coli causing infections in an elderly population of Israel. J Antimicrob Chemother. 2005;55(1):119–122. doi: 10.1093/jac/dkh517. [DOI] [PubMed] [Google Scholar]
- 17.Wang C, Cai P, Chang D, Mi Z. A Pseudomonas aeruginosa isolate producing the GES-5 extended-spectrum β-lactamase. J Antimicrob Chemother. 2006;57(6):1261–1262. doi: 10.1093/jac/dkl116. [DOI] [PubMed] [Google Scholar]
- 18.Carattoli A. Plasmids in clinically significant Gram-negative bacteria. report in European Infectious Disease Istituto Superiore di Sanità Rome. 2008;7:116–118. [Google Scholar]
- 19.Bush K, Jacoby G, Medeiros A. A functional classification scheme for b-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walther-Rasmussen J, Hoiby N. Cefotaximases (CTX-Mases) an expanding family of extended-spectrum b-lactamases. Can J Microbiol. 2004;50:137–165. doi: 10.1139/w03-111. [DOI] [PubMed] [Google Scholar]
- 21.Marti MS. PhD Thesis. Faculty of Medicine University of Barcelona; 2008. Molecular bases of antimicrobial resistance in Acinetobacter spp. clinical isolates. [Google Scholar]
- 22.Cai JC, Yang W, Hu YY, Zhang R, Zhou HW, Chen GX. Detection of KPC-2 and qnrS1 in clinical isolates of Morganella morganii from China. Diagn Micr Infec Dis. 2012;73:207–209. doi: 10.1016/j.diagmicrobio.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Livermore DM, Brown DFJ. Detection of b-lactamasemediated resistance. Antimicrob Chemother. 2001;48:59–64. doi: 10.1093/jac/48.suppl_1.59. [DOI] [PubMed] [Google Scholar]
- 24.Aggarwal R, Chaudhary U. Extended spectrum b-lactamases (ESBL) an emerging threat to clinical therapeutics. Indian J Med Microbiol. 2004;22:75–80. [PubMed] [Google Scholar]
- 25.Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, et al. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother. 2007;59:165–174. doi: 10.1093/jac/dkl483. [DOI] [PubMed] [Google Scholar]
- 26.Bonnet R. Growing group of extended-spectrum b-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang ZM, Mao PH, Chen Y, Wu L, Wu J. Study on molecular epidemiology of SHV type beta-lactamase encoding genes of multiple-drug- resistant Acinetobacter baumannii. Zhonghua Liu Xing Bing Xue Za Zhi. 2004;25:425–427. [PubMed] [Google Scholar]
- 28.Poole K. Resistance to b-lactam antibiotics. Cell Mol Life Sci. 2004;61:2200–2223. doi: 10.1007/s00018-004-4060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heritage John, M’Zali Fatima H, Gascoyne-Binzi Deborah, Hawkey Peter M. Evolution and spread of SHV extended-spectrum b-lactamases in Gram-negative bacteria. J Antimicrob Chemother. 1999;44(3):309–318. doi: 10.1093/jac/44.3.309. [DOI] [PubMed] [Google Scholar]
- 30.Paterson DL, Hujer KM, Hujer AM, Yeiser B, Bonomo MD, Rice LB, Bonomo RA the International Klebsiella Study Group. Extended-spectrum b-lactamases in Klebsiella pneumoniae blood stream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type b-lactamases. Antimicrob Agents Chemother. 2003;47:3554–3560. doi: 10.1128/AAC.47.11.3554-3560.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah AA, Hasan F, Ahmed S, Hameed A. Characteristics epidemiology and clinical importance of emerging strains of Gram-negative bacilli producing extendedspectrum betalactamases. Res Microbiol. 2004;155:409–421. doi: 10.1016/j.resmic.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Poirel L, Weldhagen GF, Naas T, De Champs C, Dove MG, Nordmann P. GES-2 a class A beta-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob Agents Chemother. 2001;45:2598–2603. doi: 10.1128/AAC.45.9.2598-2603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giuliani F, Docquier JD, Riccio ML, Pagani L, Rossolini GM. OXA-46 a new class D b-lactamase of narrow substrate specificity encoded by a blaVIM-1 containing integron from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 2005;49:1973–1980. doi: 10.1128/AAC.49.5.1973-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirel L, Weldhagen GF, De Champs C, Nordmann PA. Nosocomial outbreak of Pseudomonas aeruginosa isolates expressing the extended-spectrum beta-lactamase GES-2 in South Africa. J Antimicrob Chemother. 2002;49:561–565. doi: 10.1093/jac/49.3.561. [DOI] [PubMed] [Google Scholar]
- 35.Toleman MA, Rolston K, Jones RN, Walsh TR. Molecular and biochemical characterization of OXA-45 an extended-spectrum Class 2d? b-lactamase in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2003;47:2859–2863. doi: 10.1128/AAC.47.9.2859-2863.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]