Abstract
BACKGROUND AND OBJECTIVES
The tumor necrosis factor-alpha (TNF-α) and interleukin (IL)-6 gene polymorphism has a controversial role in the pathogenesis of cardiovascular disease among different populations. The effect of the cytokine’s gene polymorphism on idiopathic dilated cardiomyopathy (IDCM) is still unresolved. The current study aimed to evaluate the association of the TNF-α −308 G/A and IL-6 −174 G/C polymorphism with IDCM in a Pakistani population.
DESIGN AND SETTINGS
Blood samples for this case-control study were collected from the cardiology outpatient department of multiple cardiology centers of Rawalpindi/Islamabad, Pakistan, between July 2012 and December 2012.
PATIENTS AND METHODS
IDCM cases (number [n]=250) and healthy controls (n=300) were genotyped using polymerase chain reaction and restriction fragment length polymorphism.
RESULTS
The TNF-α −308 variant genotypes GA and AA were more prevalent in patients compared with the control group (P<.0001). Similarly, the IL-6 −174 variant genotypes GC and CC showed a high prevalence in patients with IDCM compared with healthy controls (P=.0019). IDCM cases had a higher prevalence of the TNF-α–308A (P<.0001) and the IL-6 −174C (P=.0008) mutant alleles than did the control group. The IDCM cases bearing the TNF-α−308 and IL-6 variant genotypes revealed elevated levels of high-sensitivity C-reactive protein (hs-CRP) when compared with the corresponding controls (P<.05).
CONCLUSION
The TNF-α −308 G/A and IL-6 −174 G/C gene polymorphisms and high levels of hs-CRP may be associated with the pathogenesis of IDCM in the study population.
Dilated cardiomyopathy (DCM) is characterized by dilatation and impaired contraction of either the left ventricle or both ventricles.1 It is an important cause of cardiac mortality and morbidity due to congestive heart failure or arrhythmias.2 The etiopathogenesis of DCM in about half of the patients is idiopathic, and among them 20% to 25% of cases are familial.3 These facts lead to the perception that genetic factors might contribute to disease susceptibility. This involvement of multiple genes in complex diseases that do not exhibit a clear pattern of familial aggregation, the candidate gene approach has not only been exceptional, but it is a strategy that is widely used to understand the underlying cause of the disease.4
A number of gene mutations, including troponin C,5 myosin-binding protein C,6 T cap,7 myosin light chain,8 and sarcomeric-associated proteins,9 have been associated with cardiomyopathy. In addition, proinflammatory cytokines have been ascribed as one of the contributors to the underlying cardiomyopathic process.10 Promoter region polymorphisms of cytokines may affect gene transcription, influencing both in vivo and in vitro cytokine production.11 Among these inflammatory cytokines, tumor necrosis factor-alpha (TNF-α) and interleukin (IL)-6 have been reported to be elevated in DCM patients and are considered as a part of the heart failure syndrome.12 Studies on xanthine-derived agents, which decrease the level of inflammatory mediators through the transcriptional blockade of inflammatory gene expression, have shown to increase ejection fraction and decrease ventricular size, thus improving the clinical status of idiopathic dilated cardiomyopathy (IDCM).13,14
It is widely accepted that genetic and environmental factors affect the pathogenesis of cardiac disease. In 2010, we published a pioneer report on the association of a novel cytokine resistin with cardiac hypertrophy in a Pakistani population.15 Molecular mechanisms underlying the pathology of IDCM are not fully understood. Variant mutations in more than 30 genes account for approximately 35% of the genetic factors involved in the pathogenesis of cardiomyopathy.16,17 Thus, a range of genetic variations that could contribute to the disease pathology are still unknown. Current attention is focused on the identification of novel risk factors of IDCM, which may improve the prognosis and management of the disease in susceptible populations.
Several studies on cytokine gene polymorphism have been conducted in patients with IDCM, which yielded conflicting results due to ethnic variations.18–20 Adamopoulos et al have reported that specific cytokine gene polymorphisms may be associated with a worse prognosis as well as with disease severity in patients with IDCM.20 South Asians are considered to be at an increased risk for cardiovascular complications, and it is probable that mutations in their genes may predispose them to the disease. In the present study, we investigated TNF-α −308 G/A and IL-6 −174 G/C gene polymorphisms in patients with IDCM and healthy controls from a Pakistani population. Furthermore, serum high-sensitivity C-reactive protein (hs-CRP) levels were determined in relation to the cytokines’ variant genotype from IDCM cases. We hypothesize that the TNF-α −308 G/A and IL-6 −174 G/C polymorphisms and elevated hs-CRP may be associated with the pathogenesis of IDCM in the study population.
PATIENTS AND METHODS
Study subjects
This study was reviewed and approved by the Institutional Review Board, Quaid-i-Azam University, Islamabad. Informed consent was obtained from all participants of the study in accordance with the Declaration of Helsinki of 1975, 1997 revision. We studied 250 unrelated patients with IDCM (mean [SD] age: 53 [14.7] years). The patients were recruited from the cardiology outpatient departments of multiple cardiology centers in Rawalpindi/Islamabad, Pakistan (Armed Forces Institute of Cardiology/National Institute of Heart Diseases, Rawalpindi; Pakistan Institute of Medical Sciences, Islamabad). The diagnosis of DCM was based on patient history, physical examination, electrocardiogram, and echocardiogram (Aplio-XV, Toshiba, Japan) according to World Health Organization guidelines.1 Chest X-ray demonstrated generalized ventricular enlargement. Electrocardiogram showed nonspecific ST segment and T-wave changes. IDCM patients were diagnosed on echocardiographic evidence of left ventricular enlargement, left ventricular systolic dysfunction (ejection fraction ≤40%), and end-diastolic diameter >34 mm/m2,21 excluding any known cause of myocardial disease. Patients with hypertension, coronary artery disease, atrial fibrillation, and ventricular arrhythmias were excluded from the study. Considering the genetic substructure in the Pakistani population, the control subjects (number [n]=300; mean [SD] age: 54.7 [13.4] years) were randomly selected from unrelated healthy individuals from the regions of the patients’ recruitment. Control subjects were clinically healthy, had normal electrocardiograms and echocardiography with no symptoms of any concomitant disease, and had no personal or familial history of cardiomyopathy or other heart-related diseases.
Sample collection
Venous blood samples were obtained from IDCM cases and healthy subjects. For biochemical analyses, blood samples were allowed to clot at room temperature and were then immediately centrifuged to separate serum and kept at −80°C. For the molecular study, blood samples were collected in ethylenediaminetetraacetic acid tubes to prevent the coagulation of blood samples. Genomic DNA was extracted from whole blood using the standard phenol-chloroform method.
Biochemical analyses
Biochemical analyses were carried out using AMP Diagnostics kits (AMEDA Laboradignostik GmbH, Graz, Austria) for the determination of total cholesterol (TC), triglycerides (TG), low-density cholesterol (LDL-C), and high-density cholesterol (HDL-C) from serum samples using Vitalab Selectra E Chemistry Analyzer (Vitalab, Hoogerheide, the Netherlands). The assays were performed according to the manufacturer’s instructions using standard enzymatic techniques. hs-CRP was measured by Tinaquant C-reactive protein (latex) high-sensitivity assay using a Roche/Hitachi-904 chemistry analyzer, Roche Diagnostics (Hoffmann-La Roche AG, Basel, Switzerland) according to the manufacturer’s recommendations.
Genotyping of the TNF-α−308 G/A polymorphism
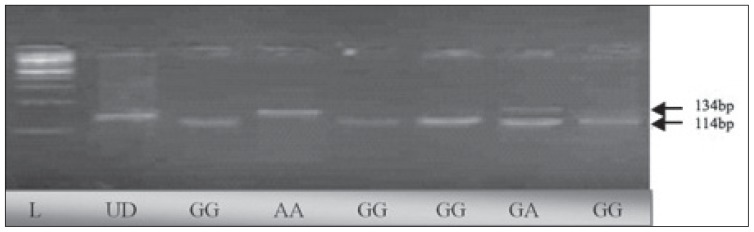
Polymorphism in the promoter region at position −308 G/A (rs 1800629)22 of the TNF-α gene was detected by polymerase chain reaction (PCR) using the forward primer 5′-AGG CAA TAG GTT TTG AGG GCC AT-3′ and reverse primer 5′-CAT CAA GGA TAC CCC TCA CAC TC-3′. Each 50 μL of PCR contained 5 μL of 10× reaction buffer, 1.5 μmol/L MgCl2 (MBI-Fermentas; Thermo Fisher Scientific, Waltham, MA, USA), 2.5 μL from 10 pmol of each primer, 1 μL of 10 mmol/L dNTPs, 1 unit of Taq DNA polymerase (MBI-Fermentas; Thermo Fisher Scientific), and 100 ng of genomic DNA. PCR was performed in the GeneAmp PCR System 9700 (Applied Biosystems Inc, Foster City, CA, USA). PCR conditions were as follows: initial denaturation at 95°C for 5 minutes, followed by 40 cycles of 1-minute denaturation at 95°C; 1-minute annealing at 60°C’; 1-minute elongation at 72°C; and the final extension for 10 minutes at 72°C. Amplified products were electrophoresed on 2% agarose gels to confirm the correct amplification using the appropriate DNA ladder (MBI-Fermentas; Thermo Fisher Scientific). PCR products were purified using JET quick PCR products purification spin/250 kit (GenoMed, Inc., Leesburg, FL, USA). An aliquot of 12 μL of PCR products was digested with restriction enzyme NcoI (MBI-Fermantas; Thermo Fisher Scientific) in 2 μL of 10× buffer G (10 mM Tris–HCl, 10 mM MgCl2, 50 mM NaCl, 0.1 mg/mL bovine serum albumin) and 10 U of NcoI enzyme and 5 μL of PCR water. The mixture was incubated at 37°C for 16 hours. The digested products were separated on 4% agarose gels stained with 10 mg/mL ethidium bromide and visualized under ultraviolet light. The GG genotype led to the formation of 2 fragments (114 bp and 20 bp); heterozygous variant GA showed 3 bands (134 bp, 114 bp, and 20 bp), while in the case of homozygous variant AA, the PCR-amplified product remained undigested, leaving a 134 bp fragment (Figure 1).
Figure 1.
Agarose gels showing the enzyme-digested patterns of PCR products of TNF-α −308 G/A. Electropherogram of ethidium bromide stained 4% agarose gel showing genotype pattern obtained with Ncol restriction digest at −308 G/A TNF-α polymorphism. Lane L represents 100-bp DNA ladder (Fermentas, Germany) while lane UD refers to the undigested PCR-amplified (134 bp) fragment. The other lanes refer to genotype pattern of samples. Genotypes of the samples are noted at the bottom of figure. Genotypes are characterized as GG (114 bp, 20 bp), GA (134 bp, 114 bp, and 20 bp), and AA (134 bp). On the gel, 20-bp fragments cannot be seen.
Genotyping of the IL-6 −174 G/C polymorphism
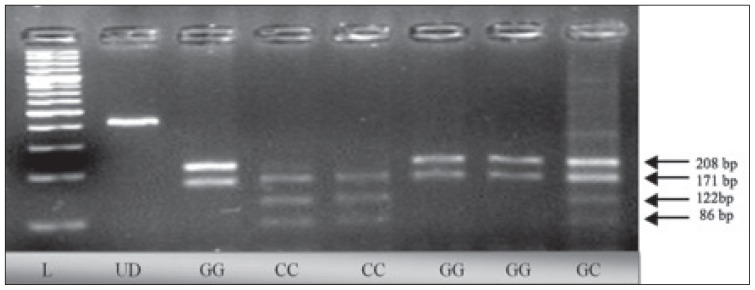
Amplification of the IL-6 promoter region −174 G/C (rs 1800795)22 was done by PCR using specific primers 5′-GCG ATG GAG TCA GAG GAA AC-3′(forward) and 5′-ATC TTT GTT GGA GGG TGA GG-3′ (reverse). A PCR protocol was adopted, as discussed previously. After purification, 12 μL of PCR products from each sample was digested by 10U of Nla III restriction enzyme (MBI-Fermentas; Thermo Fisher Scientific). The digested products were separated on 3% agarose gels stained with 10 mg/mL ethidium bromide and visualized under ultraviolet light. The GG genotype showed 3 bands of 208 bp, 171 bp, and 29 bp; the CC genotype showed bands of 171 bp, 122 bp, 86 bp, and 29 bp; whereas the heterozygous GC showed 208 bp, 171 bp, 122 bp, 86 bp, and 29 bp bands on the gels (Figure 2).
Figure 2.
Agarose gels showing the enzyme-digested patterns of PCR products of IL-6 −174 G/C polymorphism. Electropherogram of ethidium bromide stained 4% agarose gel showing genotype pattern obtained with Nla Ill restriction digest at −174 G/C IL-6 polymorphism. Lane L represents 100-bp DNA ladder (Fermentas, Germany) while lane UD refers to the undigested PCR-amplified (408 bp) fragment. Genotypes of the samples are noted at the bottom of figure. Genotypes are characterized as GG (208 bp, 171 bp, and 29 bp), GC (208 bp, 171 bp, 122 bp, 86 bp and 29 bp), and CC (171 bp, 122 bp, 86 bp, and 29 bp). On the gel, 29-bp fragments cannot be seen. tnf-a −308 g/a and il-6 −174 g/c gene polymo rphisms
Statistical analysis
Statistical analysis was performed by GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). The basic characteristics and clinical parameters (body mass index [BMI], TC, TG, LDL-C, HDL-C, and hs-CRP) of the study population are presented as the mean (SD). The statistical significance between the samples from the study groups was analyzed by the chi-square and Student t tests where indicated. Deviation from the Hardy–Weinberg equilibrium was evaluated by comparing observed and expected genotype frequencies via the chi-square test separately in cases and controls. The genotype and allele frequencies between the patient and control groups were compared by a chi-square test. The criterion for significance was P<.05.
RESULTS
Baseline and clinical characteristics of the study population
The baseline characteristics of the IDCM patients and control subjects are shown in Table 1. The patient group contained 181 (72%) and 69 (28%) males and females, respectively, whereas the control group contained 208 (69%) and 92 (31%) males and females, respectively. The IDCM cases had significantly lower BMI, TC, TG, LDL-C, and HDL-C, but higher hs-CRP when compared with the healthy individuals.
Table 1.
Baseline and clinical characteristics of control subjects and IDCM cases.
| Characteristics | Controls (n=300) | Patients (n=250) | P value |
|---|---|---|---|
|
| |||
| Age (years) | 53.7 (13.4) | 53 (14.7) | .5179a |
| Gender (M/F) | 208/92 | 181/69 | .4884b |
| BMI (kg/m2) | 25.1 (3.3) | 22.7 (2.9) | <.0001a |
| TC (mg/dL) | 167.5 (26.4) | 156.7 (44.7) | .0004a |
| TG (mg/dL) | 149.8 (58.6) | 138.1 (57.3) | .019a |
| LDL-C (mg/dL) | 101.5 (35.8) | 88.8 (37.1) | <.0001a |
| HDL-C (mg/dL) | 40.7 (8.7) | 35.2 (11.5) | <.0001a |
| hs-CRP (mg/L) | 1.11 (0.7) | 4.28 (4.6) | <.0001 |
Notes: Values are given as means (SD). A P value <.05 indicates statistical significance.
P value calculated by Student t test (unpaired).
P value calculated by the chi-square test.
Abbreviations: IDCM: Idiopathic dilated cardiomyopathy; n: number; M: male; F: female; BMI: body mass index; TC: total cholesterol; TG: triglycerides; LDL-C: low-density lipoprotein cholesterol; HDL-C: high-density lipoprotein cholesterol; hs-CRP: high-sensitivity C-reactive protein; SD: standard deviation.
Genotyping and haplotype analysis of the TNF-α −308 G/A and IL-6 −174 G/C polymorphism
Genotype frequencies were in Hardy–Weinberg equilibrium (P>.05) for both single nucleotide polymorphisms (SNPs) in controls; however, in some cases, significant deviation (P<.05) from Hardy–Weinberg equilibrium was observed in both SNPs. The −308 TNF-α variant genotype GA and AA was more prevalent in the patient group (71%) when compared with the control group at 26% (Table 2). A significant difference was found between the G and A allele from patients with IDCM versus healthy controls (odds ratio [OR]=4.0, 95% confidence interval [CI]3.0–5.3, P<.0001).
Table 2.
Genotype and allele frequencies of the TNF-α −308 G/A and IL-6 −174 G/C polymorphism in the study population.
| TNF-α −308 G/A |
Controls n (%) 300 (100) |
Patients n (%) 250 (100) |
OR | 95% CI | P value |
|---|---|---|---|---|---|
|
| |||||
| GG | 223 (74) | 72 (29) | 7.16 | 4.91–10.44 | <.0001 |
| GA+AA | 77 (26) | 178 (71) | |||
| G | 510 (85) | 293 (59) | 4.00 | 3.00–5.33 | <.0001 |
| A | 90 (15) | 207 (41) | |||
| IL-6 –174 G/C | |||||
| GG | 252 (84) | 182 (73) | 1.96 | 1.29–2.97 | .0019 |
| GC+CC | 48 (16) | 68 (27) | |||
| G | 544 (91) | 419 (84) | 1.87 | 1.30–2.70 | .0008 |
| C | 56 (9) | 81 (16) | |||
Notes: P value calculated by chi-square test. A P value <.05 indicates statistical significance.
Abbreviations: TNF-α: Tumor necrosis factor-alpha; IL-6: interleukin-6; n: number; OR: odds ratio; CI: confidence interval.
The IL-6 −174 variant genotype GC and CC showed high prevalence in patients with IDCM (27%) compared with healthy controls at 16% (Table 2). The frequency of the C allele was significantly high in patients (16%) versus controls at 9% OR=1.8, 95% CI .3–2.7, P=.0008). Haplotypes A–G and A–C of the TNF-α (−308 G/A) and IL-6 (−174 G/C) genes showed higher frequency in the patient group compared with controls (P<.0001; Table 3).
Table 3.
Haplotype analysis of the TNF-α −308 G/A and IL-6 −174 G/C polymorphism in IDCM patients and control subjects.
| TNF-α −308/IL-6 −174 |
Controls n (%) 300 (100%) |
Patients n (%) 250 (100%) |
χ2 | 95% CI | P value |
|---|---|---|---|---|---|
|
| |||||
| G-G | 461 (77) | 263 (53) | Reference | ||
| G-C | 49 (8) | 30 (6) | 0.02 | 1.07 (0.66–1.7) | .868 |
| A-G | 83 (14) | 156 (31) | 60.8 | 3.29 (2.4–4.5) | <.0001 |
| A-C | 7 (1) | 51 (10) | 57.3 | 12.8 (5.7–28.5) | <.0001 |
Notes: P value calculated by chi-square test (Yates corrected). A P value <.05 indicates statistical significance.
Abbreviations: TNF-α: Tumor necrosis factor-alpha; IL-6: interleukin-6; IDCM: idiopathic dilated cardiomyopathy; n: number; CI: confidence interval.
Serum hs-CRP levels in relation to the TNF-α (−308 G/A) and IL-6 (−174 G/C) genotype in patients with IDCM
A comparison of the serum hs-CRP levels in relation to the TNF-α (−308 G/A) and IL-6 (−174 G/C) gene polymorphism is given in Table 4. The IDCM cases with the GG, GA, and AA variant genotypes showed significantly high hs-CRP levels compared with the corresponding genotypes at −308 of the TNF-α gene (P<.05) in the controls. Patients with the IL-6 GC variant genotype at −174 revealed significantly high hs-CRP levels when compared with the GG and GC genotype (P<.05) and when compared with the controls. Elevated levels of hs-CRP were observed from IDCM cases with the variant CC versus the wild control genotype, but this increase was insignificant; this may be due to the relatively lower number of CC genotype carriers and the high SD of the mean value.
Table 4.
The serum hs-CRP levels in relation to the TNF-α −308 G/A and IL-6 −174 G/C polymorphism in the study population.
| TNF-α −308 G/A |
(n=300) | CRP (mg/L) Controls | (n=250) | CRP (mg/L) Patients | P value |
|---|---|---|---|---|---|
|
| |||||
| GG | (n=223) | 1.08 (0.7) | (n=72) | 2.70 (3.4) | <.0001 |
| GA | (n=64) | 1.11 (0.6) | (n=149) | 4.57 (4.7) | <.0001 |
| AA | (n=13) | 1.59 (0.8) | (n=29) | 6.71 (5.9) | .0034 |
| IL-6 −174 G/C | |||||
| GG | (n=252) | 1.07 (0.7) | (n=182) | 3.69 (4.4) | <.0001 |
| GC | (n=40) | 1.24 (0.7) | (n=55) | 5.92 (4.9) | <.0001 |
| CC | (n=8) | 1.73 (1.1) | (n=13) | 5.57 (5.5) | .0671 |
Notes: Values are given as mean (SD). P value calculated by student t test (unpaired). P value (<.05) indicates statistical significance.
Abbreviations: hs-CRP: High-sensitivity C-reactive protein; TNF-α: tumor necrosis factor-alpha; IL-6: interleukin-6; n: number; CRP: C-reactive protein.
DISCUSSION
The genetic basis of IDCM has been understudied and is therefore unresolved as yet. Recently, a genomewide association study identified 2 loci associated with heart failure due to DCM;23 however, the role of inflammation in IDCM is still unclear. These findings instigated our interest in investigating the role of the TNF-α and IL-6 gene polymorphism in the pathogenesis of IDCM in a Pakistani population.
The TNF-α −308 G/A polymorphisms have been associated with increased serum TNF levels. It is considered that the −308 A allele is a powerful transcriptional activator when compared with the G allele.24 This is the reason behind the rationale of grouping GA and AA genotypes together, and for using the A allele as a dominant model in our study. Evidence shows increased TNF-α levels associated with a high prevalence of the A allele in patients with DCM compared with controls.18 In a recent meta-analysis, GA and AA were used as a dominant model to calculate the association between the TNF-α −308 gene variant and coronary heart disease. 25 Similarly, other studies have reported the association of the IL-6 −174 G/C polymorphism with increased serum IL-6 levels.26–28 We have used the IL-6 −174 GC+CC genotype model to investigate its association with IDCM in our study.
In this case-control study, we have observed a significant difference in the TNF-α −308 variant genotype (GA+AA) distribution between IDCM cases and healthy controls (P<.0001). The TNF-α −308 A allele was present in 41% of IDCM cases, which was significantly higher than controls at 15%. A similar trend was observed in the −308 A allele frequency from patients with DCM in the Han Chinese29 and Japanese populations. 18 A study from the African population also confirmed a positive association of the TNF-α −308 G/A polymorphism with IDCM.30 On the contrary, studies in French and Turkish populations showed contradictory results, showing no link between the TNF-α −308 G/A polymorphism and DCM.19,20 This controversy may be due to differences in the ethnic origin of the study populations. South Asians are considered to be at an increased risk for cardiovascular events compared to other ethnic populations.31 The high prevalence of the −308 A variant allele in patients with IDCM may indicate a link between the TNF-α SNP and cardiomyopathy in the study population.
Our study also demonstrated that the IL-6 −174 variant genotype GC and CC was more prevalent in patients with IDCM (27%) compared with healthy controls at 16% (P=.0019). The C allele frequency was significantly higher among IDCM cases than among controls in the study population. Evidence shows that the IL-6 −174 C allele carriers had higher levels of the cytokine compared with the −174 G allele individuals following coronary artery bypass surgery.11 Furthermore, patients with heart failure revealed significantly high IL-6 concentrations, which could be due to extracardiac production of the cytokine.32 Chang et al have reported that IL-6 levels were higher at the coronary sinus than that of the systemic artery in patients with IDCM.33 Enhanced IL-6 levels have been associated with the development34 and severity of coronary artery disease.35 Another study demonstrated that the IL-6 −174 C allele prevalence and the cytokine levels were significantly high in patients with aortic aneurysms.36 The study also demonstrated that the −174 CC genotype was associated with increased cardiovascular events. Some reports have presented a lack of association between the IL-6 −174 G/C polymorphism with DCM.20,37 Our findings show a high prevalence of the −174 C allele from patients with IDCM versus healthy controls. The controversy between our data and other reports may be due to differences in the ethnic populations studied.
Proinflammatory cytokines like TNF-α and IL-6 have been implicated in the pathogenesis of coronary artery disease and cardiomyopathy. Studies have shown that gene polymorphisms lead to increased cytokine concentrations in circulation, which affect the pathophysiology of the disease. TNF-α is considered to play a pivotal role in the development of CVD.38 The cardiac overexpression of TNF-α has been shown to induce the myocytes’ hypertrophy in transgenic mice.39 TNF-α also induces IL-6 gene expression in a variety of cells.40 It is widely accepted that the signaling cascade has an important role in mediating the effect of inflammatory cytokines in the pathogenesis of cardiac events. Increased profiles of TNF-α and its receptors (TNFRs) have been observed from patients with DCM and failing hearts.41 TNFRs transduce intracellular signals and activate nuclear factor-kappa B (NF-κB) by proteolytic breakdown of the transcriptional factor inhibitor.42 IL-6 decreases the contractility of ventricular myocytes via a nitric oxide-dependent pathway.43 This decrease in the myocytes’ contractility is mediated by the JAK2–STAT3 signaling pathway. Our data demonstrate that the TNF-α −308 G/A and IL-6 −174 G/C polymorphism is associated with IDCM in a Pakistani population. The TNF-α −308-containing region has been known to affect the transcriptional activity of TNF-α via multiple transcriptional factors.44 Some of the TNF-α promoter variants that have been implicated in the transcription include binding sites for NF-κB, c-Jun binding, and GCF2/LRRFIP1.45 The transcription of IL-6 is tightly regulated by the following transcription factors: NFIL6, Fos/Jun, NF-κB, CRBP, and the glucocorticoid receptor. The IL-6 −174 region is under the influence of the transcription factors NFIL-6 and CRE, specifically.46 The functional variants of the TNF-α and IL-6 gene may affect the cardiac contractility via receptors, signal transducers, and transcriptional activators, which could lead to heart failure.
In our study, the mean BMI, TC, TG, and LDL-C levels were significantly higher in controls than in the patient group. IDCM cases had lower BMIs; at this stage, we are not certain whether patients with IDCM suffered from some weight loss or if the control subjects selected for this study had relatively high BMIs. Regarding the lipid profile, it is probable that the use of lipid-lowering drugs led to decreased levels of TC, TG, LDL-C, and HDL-C in patients with IDCM. Similar results were observed in the Indian,47 Japanese,48 and Pakistani49 populations. Interestingly, we have observed high concentrations of hs-CRP from patients with variant genotypes of the TNF-α and IL-6 gene at −308 and −174, respectively. CRP is an acute phase protein; its concentrations reflect the inflammatory status of subjects. Evidence shows that CRP is associated with coronary heart disease.50,51 In healthy individuals, the TNF-α −308 variant genotype AA was linked with high concentrations of CRP.52 Similarly, the IL-6 −174 C allele appears to be associated with higher plasma CRP and IL-6 levels in patients undergoing coronary artery bypass graft surgery.53 CRP has been implicated in coronary heart disease; however, its potential involvement in IDCM is not well defined. Our data indicate that the TNF-α and IL-6 gene polymorphism at −308 and −174, respectively, is associated with higher concentrations of hs-CRP. Elevated levels of hs-CRP, in addition to other risk factors, may have a role in cardiovascular complications. At this stage, it is not certain whether hs-CRP is a causative factor of IDCM. Nevertheless, inflammatory polypeptides like TNF-α, IL-6, and CRP are now being recognized as additional risk factors that may contribute to cardiovascular events. The current investigation reveals that the TNF-α −308 A and IL-6 C allele-bearing haplotype was more frequent in patients versus the control group (P<.0001). The functional implications of variant alleles-bearing haplotypes may provide useful information regarding cardiovascular complications. Therefore, further in-depth studies with large sample sizes are required to determine the association and role of these haplotypes in IDCM.
There are certain limitations of our study. The sample size in this study was relatively small. The investigation was carried out only in the Pakistani population, and therefore the findings from this study should be extrapolated to other populations with caution. More investigations are required to establish a link between the TNF-α −308 G/A and IL-6 −174 G/C polymorphism and the risk of IDCM.
In conclusion, this study demonstrates a statistically significant association between the TNF-α −308 G/A and IL-6 −174 G/C polymorphism and IDCM in a Pakistani population. Further studies are needed to signify the correlation of the functional variants of the inflammatory cytokines with IDCM and other risk factors of the disease.
REFERENCES
- 1.Richardson P, McKenna W, Bristow M, et al. Report of the 1995 WHO/International society and federation of cardiology task force on the definition and classification of cardiomyopathies. Circulation. 1996;93(5):841–842. doi: 10.1161/01.cir.93.5.841. [DOI] [PubMed] [Google Scholar]
- 2.Grimm W, Christ M, Bach J, Müller HH, Maisch B. Noninvasive arrhythmia risk stratification in idiopathic dilated cardiomyopathy: results of the Marburg cardiomyopathy study. Circulation. 2003;108(23):2883–2891. doi: 10.1161/01.CIR.0000100721.52503.85. [DOI] [PubMed] [Google Scholar]
- 3.Keeling PJ, Gang Y, Smith G, et al. Familial dilated cardiomyopathy in the United Kingdom. Br Heart J. 1995;73(5):417–421. doi: 10.1136/hrt.73.5.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motulsky AG. Genetics of complex diseases. J Zhejiang Univ Sci B. 2006;7(2):167–168. doi: 10.1631/jzus.2006.B0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann B, Schmidt-Traub H, Perrot A, Osterziel KJ, Gessner R. First mutation in cardiac troponin C, L29Q, in a patient with hypertrophic cardiomyopathy. Hum Mutat. 2001;17(6):524. doi: 10.1002/humu.1143. [DOI] [PubMed] [Google Scholar]
- 6.Niimura H, Bachinski LL, Sangwatanaroj S, et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med. 1998;338(18):1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi T, Arimura T, Itoh-Satoh M, et al. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J Am Coll Cardiol. 2004;44(11):2192–2201. doi: 10.1016/j.jacc.2004.08.058. [DOI] [PubMed] [Google Scholar]
- 8.Olson TM, Karst ML, Whitby FG, Driscoll D. Myosin light chain mutation causes autosomal recessive cardiomyopathy with mid-cavitary hypertrophy and restrictive physiology. Circulation. 2002;105(20):2337–2340. doi: 10.1161/01.cir.0000018444.47798.94. [DOI] [PubMed] [Google Scholar]
- 9.Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: An Intricate Web of Form and Function. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- 10.Pankuweit S, Ruppert V, Maisch B. Inflammation in dilated cardiomyopathy. Herz. 2004;29(8):788–793. doi: 10.1007/s00059-004-2626-9. [DOI] [PubMed] [Google Scholar]
- 11.Brull DJ, Montgomery HE, Sanders J, et al. Interleukin-6 gene −174g>c and −572g>c promoter polymorphisms are strong predictors of plasma interleukin-6 levels after coronary artery bypass surgery. Arterioscler Thromb Vasc Biol. 2001;21(9):1458–1463. doi: 10.1161/hq0901.094280. [DOI] [PubMed] [Google Scholar]
- 12.Aukrust P, Ueland T, Lien E, et al. Cytokine network in congestive heart failure secondary to ischemic or idiopathic dilated cardiomyopathy. Am J Cardiol. 1999;83(3):376–382. doi: 10.1016/s0002-9149(98)00872-8. [DOI] [PubMed] [Google Scholar]
- 13.Skudicky D, Bergemann A, Sliwa K, Candy G, Sareli P. Beneficial effects of pentoxifylline in patients with idiopathic dilated cardiomyopathy treated with angiotensin-converting enzyme inhibitors and carvedilol: results of a randomized study. Circulation. 2001;103(8):1083–1088. doi: 10.1161/01.cir.103.8.1083. [DOI] [PubMed] [Google Scholar]
- 14.Bahrmann P, Hengst UM, Richartz BM, Figulla HR. Pentoxifylline in ischemic, hypertensive and idiopathic-dilated cardiomyopathy: effects on left-ventricular function, inflammatory cytokines and symptoms. Eur J Heart Fail. 2004;6(2):195–201. doi: 10.1016/j.ejheart.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Hussain S, Asghar M, Javed Q. Resistin gene promoter region polymorphism and the risk of hypertrophic cardiomyopathy in patients. Transl Res. 2010;155(3):142–147. doi: 10.1016/j.trsl.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Hershberger RE, Lindenfeld J, Mestroni L, Seidman CE, Taylor MR, Towbin JA. Genetic evaluation of cardiomyopathy—a Heart Failure Society of America practice guideline. J Card Fail. 2009;15(2):83–97. doi: 10.1016/j.cardfail.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Hershberger RE, Norton N, Morales A, Li D, Siegfried JD, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3(2):155–161. doi: 10.1161/CIRCGENETICS.109.912345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito M, Takahashi H, Fuse K, et al. Polymorphisms of tumor necrosis factor-alpha and Interleukin-10 genes in Japanese patients with idiopathic dilated cardiomyopathy. Jpn Heart J. 2000;41(2):183–191. doi: 10.1536/jhj.41.183. [DOI] [PubMed] [Google Scholar]
- 19.Alikasifoglu M, Tokgözoglu L, Acil T, et al. Tumor necrosis factor-alpha polymorphism in Turkish patients with dilated cardiomyopathy. Eur J Heart Fail. 2003;5(2):161–163. doi: 10.1016/s1388-9842(02)00238-6. [DOI] [PubMed] [Google Scholar]
- 20.Adamopoulos S, Kolokathis F, Gkouziouta A, et al. Cytokine gene polymorphisms are associated with markers of disease severity and prognosis in patients with idiopathic dilated cardiomyopathy. Cytokine. 2011;54(1):68–73. doi: 10.1016/j.cyto.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Tiret L, Mallet C, Poirier O, et al. Lack of association between polymorphisms of eight candidate genes and idiopathic dilated cardiomyopathy: The CARDIGENE study. J Am Coll Cardiol. 2000;35(1):29–35. doi: 10.1016/s0735-1097(99)00522-7. [DOI] [PubMed] [Google Scholar]
- 22.Bouhaha R, Baroudi T, Ennafaa H, et al. Study of TNF alpha −308G/A and IL6 −174G/C polymorphisms in type 2 diabetes and obesity risk in the Tunisian population. Clin Biochem. 2010;43(6):549–552. doi: 10.1016/j.clinbiochem.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Villard E, Perret C, Gary F, et al. A genomewide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32(9):1065–1076. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elahi MM, Asotra K, Matata BM, Mastana SS. Tumor necrosis factor alpha −308 gene locus promoter polymorphism: an analysis of association with health and disease. Biochim Biophys Acta. 2009;1792(3):163–172. doi: 10.1016/j.bbadis.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Zhang HF, Xie SL, Wang JF, Chen YX, Wang Y, Huang TC. Tumor necrosis factor-alpha G-308A gene polymorphism and coronary heart disease susceptibility: An updated meta-analysis. Thromb Res. 2011;127(5):400–405. doi: 10.1016/j.thromres.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 26.Bruunsgaard H, Christiansen L, Pedersen AN, Schroll M, Jørgensen T, Pedersen BK. The IL-6 −174G>C polymorphism is associated with cardiovascular diseases and mortality in 80-year-old humans. Exp Gerontol. 2004;39(2):255–261. doi: 10.1016/j.exger.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Ravaglia G, Forti P, Maioli F, et al. Associations of the −174 G/C interleukin-6 gene promoter polymorphism with serum interleukin 6 and mortality in the elderly. Biogerontology. 2005;6(6):415–423. doi: 10.1007/s10522-005-4908-x. [DOI] [PubMed] [Google Scholar]
- 28.Panoulas VF, Stavropoulos KA, Metsios GS, Smith JP, Milionis HJ, Douglas KM. Association of interleukin-6 (IL-6)-174G/C gene polymorphism with cardiovascular disease in patients with rheumatoid arthritis: the role of obesity and smoking. Atherosclerosis. 2009;204(1):178–183. doi: 10.1016/j.atherosclerosis.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 29.Liang WB, Lv ML, Su XW, et al. Association of tumor necrosis factor gene polymorphisms with susceptibility to dilated cardiomyopathy in a Han Chinese population. DNA Cell Biol. 2010;29(10):625–628. doi: 10.1089/dna.2010.1044. [DOI] [PubMed] [Google Scholar]
- 30.Brooksbank R, Badenhorst D, Sliwa K, Norton G, Woodiwiss A. The G-308A polymorphism of the TNF-α gene does not predict changes in cardiac function in response to medical therapy for idiopathic dilated cardiomyopathy. Cardiovasc J Afr. 2008;19(5):254–258. [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta M, Singh N, Verma S. South Asians and cardiovascular risk: what clinicians should know. Circulation. 2006;113(25):e924–e929. doi: 10.1161/CIRCULATIONAHA.105.583815. [DOI] [PubMed] [Google Scholar]
- 32.Deliargyris EN, Raymond RJ, Theoharides TC, Boucher WS, Tate DA, Dehmer GJ. Sites of interleukin-6 release in patients with acute coronary syndromes and in patients with congestive heart failure. Am J Cardiol. 2000;86(9):913–918. doi: 10.1016/s0002-9149(00)01121-8. [DOI] [PubMed] [Google Scholar]
- 33.Chang HJ, Chung J, Choi BJ, Choi TY, Choi SY, Yoon MH, et al. The origin of proinflammatorycytokines in patients with idiopathic dilated cardiomyopathy. J Korean Med Sci. 2003;18(6):791–796. doi: 10.3346/jkms.2003.18.6.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ridker PM. Are statins anti-inflammatory? Issues in the design and conduct of the pravastatin inflammation C-reactive protein evaluation. Curr Cardiol Rep. 2000;2(4):269–273. doi: 10.1007/s11886-000-0080-8. [DOI] [PubMed] [Google Scholar]
- 35.Gabriel AS, Ahnve S, Wretlind B, Martinsson A. IL-6 and IL-1 receptor antagonist in stable angina pectoris and relation of IL-6 to clinical findings in acute myocardial infarction. J Intern Med. 2000;248(1):61–66. doi: 10.1046/j.1365-2796.2000.00701.x. [DOI] [PubMed] [Google Scholar]
- 36.Jones KG, Brull DJ, Brown LC, et al. Interleukin-6 (IL-6) and the prognosis of abdominal aortic aneurysms. Circulation. 2001;103(18):2260–2265. doi: 10.1161/01.cir.103.18.2260. [DOI] [PubMed] [Google Scholar]
- 37.Spiroska V, Kedev S, Antov S, et al. Association between 22 cytokine gene polymorphisms and dilated cardiomyopathy in Macedonian patients. Kardiol Pol. 2009;67(11):1237–1247. [PubMed] [Google Scholar]
- 38.Levine B, Kalman J, Mayer L, Fillit H, Packer M. Elevated circulating levels of tumor necrosis factor in severe chronic heart failure. N Engl J Med. 1990;323(4):236–241. doi: 10.1056/NEJM199007263230405. [DOI] [PubMed] [Google Scholar]
- 39.Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296(5573):1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 40.Feldman AM, Combes A, Wagner D, et al. The role of tumor necrosis factor in the pathophysiology of heart failure. J Am Coll Cardiol. 2000;35(3):537–544. doi: 10.1016/s0735-1097(99)00600-2. [DOI] [PubMed] [Google Scholar]
- 41.Torre-Amione G, Kapadia S, Lee J, et al. Tumor necrosis factor-alpha and tumor necrosis factor receptors in the failing human heart. Circulation. 1996;93(4):704–711. doi: 10.1161/01.cir.93.4.704. [DOI] [PubMed] [Google Scholar]
- 42.Kuwano K, Hara N. Signal transduction pathways of apoptosis and inflammation induced by the tumor necrosis factor receptor family. Am J Respir Cell Mol Biol. 2000;22(2):147–149. doi: 10.1165/ajrcmb.22.2.f178. [DOI] [PubMed] [Google Scholar]
- 43.Yu X, Kennedy RH, Liu SJ. JAK2, STAT3, not ERK1/2, mediates Interleukin 6 induced activation of inducible nitric-oxide synthase and decrease in contractility of adult ventricular myocytes. J Biol Chem. 2003;278(18):16304–16309. doi: 10.1074/jbc.M212321200. [DOI] [PubMed] [Google Scholar]
- 44.Abraham LJ, Kroeger KM. Impact of the −308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: relevance to disease. J Leukoc Biol. 1999;66(4):562–566. doi: 10.1002/jlb.66.4.562. [DOI] [PubMed] [Google Scholar]
- 45.Suriano AR, Sanford AN, Kim N, Oh M, Kennedy S, Henderson MJ, et al. GCF2/LRRFIP1 represses tumor necrosis factor alpha expression. Mol Cell Biol. 2005;25(20):9073–9081. doi: 10.1128/MCB.25.20.9073-9081.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terry CF, Loukaci V, Green FR. Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem. 2000;275(24):18138–18144. doi: 10.1074/jbc.M000379200. [DOI] [PubMed] [Google Scholar]
- 47.Banerjee I, Pandey U, Hasan OM, Parihar R, Tripathi V, et al. Association between inflammatory gene polymorphisms and coronary artery disease in an Indian population. J Thromb Thrombolysis. 2009;27(1):88–94. doi: 10.1007/s11239-007-0184-8. [DOI] [PubMed] [Google Scholar]
- 48.Nakayama T, Asai S, Sato N, Soma M. Genotype and haplotype association study of the STRK1 region on 5q12 among Japanese: a case-control study. Stroke. 2006;37(1):69–76. doi: 10.1161/01.STR.0000194961.17292.33. [DOI] [PubMed] [Google Scholar]
- 49.Saleheen D, Bukhari S, Haider SR, et al. Association of phosphodiesterase 4D gene with ischemic stroke in a Pakistani population. Stroke. 2005;36(10):2275–2277. doi: 10.1161/01.STR.0000182242.59466.ee. [DOI] [PubMed] [Google Scholar]
- 50.Sabatine MS, Morrow DA, Jablonski KA, Rice MM, Warnica JW, Domanski MJ, et al. Prognostic significance of high-sensitivity C-reactive protein cut points for cardiovascular and other outcomes in patients with stable coronary artery disease. Circulation. 2007;115(12):1528–1536. doi: 10.1161/CIRCULATIONAHA.106.649939. [DOI] [PubMed] [Google Scholar]
- 51.Torzewski J, Torzewski M, Bowyer DE, et al. C-reactive protein frequently colocalizes with the terminal complement complex in the intima of early atherosclerotic lesions of human coronary arteries. Arteroscler Thromb Vasc Biol. 1998;18(9):1386–1392. doi: 10.1161/01.atv.18.9.1386. [DOI] [PubMed] [Google Scholar]
- 52.Lakka HM, Lakka TA, Rankinen T, et al. The TNF-α G-308A polymorphism is associated with C-reactive protein levels: The Heritage Family Study. Vascul Pharmacol. 2006;44(5):377–383. doi: 10.1016/j.vph.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Wypasek E, Undas A, Sniezek MM, et al. The increased plasma C-reactive protein and interleukin-6 levels in patients undergoing coronary artery bypass grafting surgery are associated with the interleukin-6 −174 G/C gene polymorphism. Ann Clin Biochem. 2010;47(Pt 4):343–349. doi: 10.1258/acb.2010.090305. [DOI] [PubMed] [Google Scholar]