Abstract
Purpose
Aerobactin is a critical factor for hypervirulent Klebsiella pneumoniae (hvKp) in genetic backgrounds, but data based on the genotype for the elderly is limited.
Materials and methods
A retrospective study was conducted on elderly patients from June 2008 to July 2017 in 2 teaching hospitals. The clinical and microbiological data, including antimicrobial susceptibility testing, string test, extended-spectrum β-lactamase (ESBL) production, virulence gene, and multilocus sequence typing, of the hvKp group defined as aerobactin positive were compared with those of classic K. pneumoniae isolates.
Results
A total of 45.7% of 202 K. pneumoniae isolates were hvKp.ST23, which were predominant in 2 hospitals, but they were not highly associated with hvKp in different hospitals. Hypermucoviscosity, K1, K2, magA, and rmpA/A2 genes were highly related to hvKp (P=0.000). With regard to the host, invasive infections (P=0.000), liver abscess (P=0.000), abdominal infection (P=0.000), pneumonia (P=0.037), and septic shock (P=0.045) were significantly higher in the elderly with hvKp. In the hvKp group, patients with better nutritional status were associated with a more severe sequential organ failure assessment score and a more serious inflammation reaction. Patients with diabetes (odds ratio [OR]=2.566) are more likely to be infected with hvKp. Previous hvKp is associated with hypermucoviscosity (OR=15.249) are often paralleled with hvKp. Importantly, 26% of hvKp isolates produced ESBLs, and most of them showed a carbapenems-resistant (CR) phenotype. Multivariate analysis implied that patients with a history of surgery within the last 1 month (OR=15.999) is an independent risk factor for CR-hvKp infection.
Conclusion
The prevalence of hvKP is high in the elderly. ESBL-hvKp, especially CR-hvKp, is emerging, which is a sign that clinical awareness and infection monitoring needs to improve.
Keywords: Klebsiella pneumoniae, hypervirulent, aerobactin, risk factor, ESBL-hvKp, CR-hvKp
Introduction
Klebsiella pneumoniae is a Gram-negative bacterium, causing various fatal infections. There are 2 pathotypes: hypervirulent (hvKp) and classical (cKp), which are detrimental to our health. Initially, a string test with a length >5 mm was defined as hypermucoviscosity, which is a traditional unique hvKp trait, triggering aggressive invasive infection, such as bloodstream infection and pyogenic liver abscesses (PLAs) for immunocompetent ambulatory younger individuals with non-underlying diseases.1–4 However, many studies do not agree with the definition of hvKp defined by hypermucoviscosity phenotype.5,6 The reason is that few hypermucoviscous K. pneumoniae (hmvKp) strains are associated with high virulence with in vitro and in vivo assays.5,6 Thus, using hvKp by hypermucoviscosity phenotype as the sole indicator of hvKp is not appropriate.7,8
Until recently, aerobactin, the dominant siderophore, was regarded as a critical virulence factor for hvKp genetic background.1,8,9 A multicenter study, focusing on middle-aged patients, first demonstrated the clinical and molecular characteristic of hvKp (defined as aerobactin positive) infection.8 But data are limited on the elderly who may have various underlying diseases, nutrition status, and atypical manifestations, along with being infected with genotype hvKp.
Many previous studies have illustrated that hvKp is sensitive to most antibiotics, which is not frequently present in infection with cKp strains. But emerging multidrug resistance (MDR) hvKp, especially resistant to colistin and carbapenems, has been reported in China.10–12 However, there are not enough adapted data on the elderly and the characteristics of antimicrobial-resistant hvKp infection.
Thus, for further investigation of the prevalence and antibiotic resistance of hvKp, we conducted a retrospective study in 2 teaching hospitals based on the genotype of hvKp, which was defined as aerobactin positive.
Materials and methods
Patients
A retrospective study was conducted on 202 K. pneumoniae culture-positive patients diagnosed at Beijing Tsinghua Changgung Hospital and Chinese PLA General Hospital from June 2008 to July 2017. The definition of elderly was if the patient was ≥65 years. Duplicate isolates from the same patient were excluded. The clinical characteristics, including underlying disease, infection type, nutritional status, mortality in 30 days, and sequential organ failure assessment (SOFA), were collected. To recognize the host responsibility and nutritional status between the 2 pathotypes, white blood cell count (WBC) and neutrophil percentage (NEU%) were used as primary and rough inflammatory factors. To evaluate the nutrition status, we used total protein (TP) and albumin (ALB) as markers. The study was approved by the Chinese PLA General Hospital Ethics Committee, and the Guidelines for Human Experimentation (China) were followed through the whole study. Informed consent was not needed due to the retrospective nature of the study; additionally, the patient data accessed in this research was anonymous. Therefore, the Chinese PLA General Hospital Ethics Committee waived the need for consent.
K. pneumoniae strains
All isolates were stored at −80°C and previously identified by the API 20 NE system and the Vitek II system. Additionally, species identification was further confirmed by 16S rRNA gene sequencing. HvKp was defined as aerobactin positive. Hypermucoviscous phenotype was confirmed by string test as described previously.13
Antimicrobial susceptibility testing and phenotypic detection of ESBLs
Antimicrobial susceptibility testing was conducted and the results were interpreted by 2017 Clinical and Laboratory Standards Institute (CLSI) guidelines. The antibiotics include amikacin, gentamicin, tobramycin, ampicillin/sulbactam, aztreonam, cefazolin, cefepime, ceftriaxone, ceftazidime, ciprofloxacin, levofloxacin, piperacillin/tazobactam, and trimethoprim/sulfamethoxazole. ESBL was confirmed by agar dilution test using ceftazidime and cefotaxime combined with clavulanate according to the CLSI guidelines.8 MDR strains were defined as resistant to ≥3 different antimicrobial categories.14 Isolates that are resistant to both imipenem and meropenem are defined as carbapenems-resistant (CR) isolates.
Detection of virulence gene
Genomic DNA of all K. pneumoniae isolates was extracted. Virulence-associated genes (rmpA, rmpA2, magA, and aerobactin) and capsular serotype-specific (cps) genes (K1, K2, K5, K20, K54, and K57) were amplified by polymerase chain reaction (PCR).8,15–17 The primers are listed in Table S1.
Multilocus sequence typing (MLST) for K. pneumoniae
Seven housekeeping genes (gapA, mdh, phoE, tonB, infB, pgi, and rpoB) were amplified by PCR following the protocol (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html) (Table S1). Allelic profiling and sequence types (STs) determination were also confirmed on the aforementioned website. Moreover, to further analyze the relationship among different STs, phylogenetic analysis of spliced 7 housekeeping genes for frequency >1 isolates and strains contributing to invasive infection and mortality was performed by the neighbor-joining method (MEGA 7.0).
Statistical analysis
SPSS software (version 20.0; IBM Corporation, Armonk, NY, USA) was performed for data analysis. Measurement data were assessed as mean ± SD. The count data was analyzed as percentages. Continuous variables were analyzed by Student’s t-tests and the Wilcoxon rank-sum tests. Categorical variables were analyzed by χ2 or Fisher’s exact test. Univariate logistic regression analyses were performed for risk factor. To further analyze independent risk factors, a multivariable logistic regression analysis was conducted. All variables with P values of <0.05 were included in the multivariate model. All tests were 2-tailed. P-value <0.05 was considered significant.
Results
Patient characteristics
A total of 202 K. pneumoniae culture-positive patients were diagnosed at the 2 hospitals from June 2008 to July 2017. A total of 96 (47.5%) isolates were hvKp and 121 (59.9%) were hmvKp. All the PLA patients (10 cases) were infected with hvKp. The main infection type distribution in hospital was pneumonia (146, 72.3%), while other infection types included urinary infection (42, 20.8%), invasive infection (37, 18.3%), and abdominal infection (26, 12.9%). Moreover, almost half of the patients (98, 48.5%) presented with sepsis, and 24 (11.9%) were diagnosed as septic shock. A total of 181 (89.6%) were males and 21 (10.4%) were females; the mean age was 84.43±7.84 years.
Clinical features: hvKp vs. cKp
Clinical features are shown in Table 1. The mean age of the hvKp group was younger than the cKp group (83.24±7.35 vs. 85.5±8.14 years, P=0.039). Diabetes (72.9% vs. 48.1%; P=0.000) and digestive diseases (22.9% vs. 13.5%; P=0.046) were highly associated with the hvKp group as their underlying diseases. Compared with the cKp group, a significantly higher number of patients with the hvKp presented with invasive infections (30.2% vs. 7.5%; P=0.000), liver abscess (10.4% vs. 0%; P=0.000), other abscess (26.0% vs. 2.8%; P=0.035), septic shock (16.7% vs. 7.5%; P=0.045), pneumonia (79.2% vs. 66.0%; P=0.037), and abdominal infection (21.9% vs. 4.7%; P=0.000). However, the incidence rates of urinary infection (13.5% vs. 27.4%, P=0.016) and stomach tube indwelling (60.4% vs. 77.4%, P=0.009) were comparably lower in the hvKp group.
Table 1.
Clinical and microbiological features of hvKp
| Characteristic | hvKp (N=96) | cKp (N=106) | P-value |
|---|---|---|---|
| K serotype | |||
| K1 | 33 (34.4%) | 2 (1.9%) | 0.000 |
| K2 | 20 (20.8%) | 2 (1.9%) | 0.000 |
| K5 | 3 (3.1%) | 0 (0%) | 0.106 |
| K20 | 6 (6.3%) | 4 (3.8%) | 0.627 |
| K54 | 4 (3.8%) | 3 (2.8%) | 0.894 |
| K57 | 10 (10.4%) | 6 (6.3%) | 0.211 |
| rmpA | 76 (79.2%) | 12 (11.3%) | 0.000 |
| rmpA2 | 68 (70.8%) | 15 (14.2%) | 0.000 |
| magA | 77 (80.2%) | 50 (47.2%) | 0.000 |
| Hypermucoviscosity | 86 (89.6%) | 35 (33.0%) | 0.000 |
| Basic demographics | |||
| Age | 83.24±7.35 | 85.5±8.14 | 0.039 |
| Male | 93 (96.9%) | 88 (83.0%) | 0.361 |
| Underlying diseases | |||
| Pulmonary disease | 86 (89.6%) | 85 (80.2%) | 0.064 |
| Diabetes | 70 (72.9%) | 51 (48.1%) | 0.000 |
| Cardiovascular disease | 45 (46.9%) | 60 (56.6%) | 0.167 |
| Cerebrovascular disease | 15 (15.6%) | 27 (25.5%) | 0.085 |
| Cancer | 28 (29.2%) | 27 (25.5%) | 0.556 |
| Surgery within 1 month | 14 (14.6%) | 11 (10.4%) | 0.365 |
| Digestive disease | 22 (22.9%) | 13 (13.5%) | 0.046 |
| Catheter | |||
| Central intravenous catheter | 65 (67.7%) | 66 (62.3%) | 0. 418 |
| Urinary catheter | 73 (76.0%) | 87 (82.1%) | 0.291 |
| Tracheal catheter | 34 (35.4%) | 39 (36.8%) | 0.839 |
| Stomach tube | 58 (60.4%) | 82 (77.4%) | 0.009 |
| Infection type | |||
| Pneumonia | 76 (79.2%) | 70 (66.0%) | 0.037 |
| Urinary infection | 13 (13.5%) | 29 (27.4%) | 0.016 |
| Invasive infection | 29 (30.2%) | 8 (7.5%) | 0.000 |
| Bacteremia | 8 (8.3%) | 4 (3.8%) | 0.171 |
| Liver abscess | 10 (10.4%) | 0 (0%) | 0.000 |
| Other abscess | 25 (26.0%) | 3 (2.8%) | 0.000 |
| Abdominal infection | 21 (21.9%) | 5 (4.7%) | 0.000 |
| Sepsis | 48 (50.0%) | 50 (47.2%) | 0.688 |
| Septic shock | 16 (16.7%) | 8 (7.5%) | 0.045 |
| Host responsibility | |||
| WBC (109/L) | 12.74±3.94 | 10.59±3.48 | 0.000 |
| NEU% | 78.70±8.02 | 75.6±8.50 | 0.003 |
| Nutrition status | |||
| TP | 64.74±5.42 | 62.83±6.32 | 0.023 |
| ALB | 34.98±3.40 | 33.78±3.73 | 0.019 |
| SOFA score | 6.79±2.88 | 4.91±2.61 | 0.000 |
| Infection occurred in ICU | 19 (19.8%) | 16 (15.1%) | 0.378 |
| Mortality in 30 days | 16 (16.7%) | 23 (21.7%) | 0.366 |
Notes: Data presented as mean ± standard deviation, unless otherwise stated. Bold values indicate P<0.05.
Abbreviations: ALB, albumin; cKp, classic Klebsiella pneumoniae; ESBLs, extended-spectrum β-lactamases; hvKp, hypervirulent Klebsiella pneumoniae; ICU, intensive care unit; NEU%, neutrophil percentage; SOFA, sequential organ failure assessment; TP, total protein; WBC, white blood cell count.
WBC (12.74±3.94 109/L vs. 10.59±3.48 109/L, P=0.000) and NEU% (78.70±8.02 vs. 75.60±8.50, P=0.003) of patients with hvKp, represented as host responsibility, were significantly higher than cKp group. However, patients infected with hvKp were more likely to have a poorer nutritional status in TP (64.74±5.42 vs. 62.83±6.32, P=0.023) and ALB (34.98±3.40 vs. 33.78±3.73, P=0.019). Moreover, although the mortality in 30 days (16.7% vs. 21.7%, P=0.366) was not significantly different, SOFA score in patients with hvKp was notably higher (6.79±2.88 vs. 4.93±2.59, P=0.000; Table 1).
Genetic and phenotype characteristics: hvKp vs. cKp
Previous studies confirmed that virulence-associated genes (rmpA, rmpA2, and magA) and cps genes (K1, K2, K5, K20, K54, and K57) are clustered in the hvKp group.18–20 A significant difference was that K1, K2, rmpA, rmpA2, and magA were highly clustered in hvKp (P=0.000, respectively), and K5, K20, K54, and K57 were not associated with hvKp (P=0.106, 0.627, 0.894, and 0.211, respectively). There was no strain in the cKp group with K5 (Table 1). It was strongly noted that hypermucoviscosity was highly associated with hvKp (P=0.000).
Antimicrobial resistance and prevalence of ESBL-producing K. pneumoniae isolates
Most of the hvKp isolates were sensitive to most of the antibiotics, with the exception of ampicillin, imipenem, and meropenem (Table 2). All Kp strains were resistant to ampicillin. In the hvKp group, 24 strains (25.0%) were MDR. A total of 25 hvKp isolates were identified as ESBL-producing K. pneumoniae isolates, which were more common in the cKp group (53.8% vs. 26.0%, P=0.001). One CR-hvKp isolate existed in one of these 2 teaching hospitals. In another referral center, 10 CR-hvKp strains were detected. The detailed information for the 11 CR-hvKp strains is shown in Table S2.
Table 2.
Antibiotic resistance: hvKp vs. cKp
| Antibiotic agent | hvKp (N=96) | cKp (N=106) | P-value |
|---|---|---|---|
| ESBLs | 25 (16.3%) | 57 (40.0%) | 0.000 |
| Amikacin | 10 (2.5%) | 22 (11.6%) | 0.044 |
| Gentamicin | 16 (8.8%) | 41 (29.5%) | 0.001 |
| Ampicillin/sulbactam | 28 (20.0%) | 60 (44.2%) | 0.000 |
| Aztreonam | 18 (8.8%) | 41 (23.2%) | 0.002 |
| Cefazolin | 28 (18.8%) | 59 (44.2%) | 0.000 |
| Cefotetan | 14 (14.6%) | 31 (29.2%) | 0.012 |
| Cefepime | 15 (5.0%) | 34 (14.7%) | 0.006 |
| Ceftriaxone | 25 (16.3%) | 51 (34.7%) | 0.001 |
| Ceftazidime | 17 (7.5%) | 43 (25.3%) | 0.000 |
| Ciprofloxacin | 18 (10.0%) | 43 (25.3%) | 0.001 |
| Levofloxacin | 14 (6.3%) | 40 (22.1%) | 0.000 |
| Trimethoprim/sulfamethoxazole | 15 (8.8%) | 44 (34.7%) | 0.000 |
| Piperacillin/tazobactam | 13 (3.8%) | 33 (13.7%) | 0.003 |
| Imipenem | 11 (1.3%) | 21 (2.1%) | 0.104 |
| Meropenem | 12 (2.5%) | 23 (2.1%) | 0.085 |
| Tobramycin | 17 (8.8%) | 39 (27.4%) | 0.002 |
Abbreviations: cKp, classic Klebsiella pneumoniae; ESBLs, extended spectrum β-lactamase; hvKp, hypervirulent Klebsiella pneumoniae.
Risk factors: hvKp vs. cKp
In this study, univariate regression analysis showed that diabetes (odds ratio [OR]=2.903), digestive diseases (OR=2.127), and hypermucoviscosity (OR=17.446) were notable risk factors for hvKp infection. However, indwelling stomach tube (OR=0.447) was a protective factor for hvKp infection. Moreover, multivariate analysis revealed that diabetes (OR=2.566) and hypermucoviscosity (OR=15.249) were independent risk factors for hvKp infections (Table 3).
Table 3.
Risk factors for hvKp vs. cKp
| Variable | Univariate OR (95% CI) | P-value | Multivariate OR (95% CI) | P-value |
|---|---|---|---|---|
| Infection occurred in ICU | 1.388 (0.668–2.884) | 0.380 | ||
| Male | 1.538 (0.608–3.888) | 0.363 | ||
| Hypermucoviscosity | 17.446 (8.079–37.673) | 0.000 | 15.249 (6.905–33.875) | 0.000 |
| Pulmonary diseases | 2.125 (0.945–4.779) | 0.068 | ||
| Diabetes | 2.903 (1.610–5.236) | 0.000 | 2.566 (1.258–5.235) | 0.010 |
| Cardiovascular disease | 0.676 (0.388–1.179) | 0.168 | ||
| Cerebrovascular disease | 0.542 (0.268–1.095) | 0.088 | ||
| Cancer | 1.205 (0.648–2.240) | 0.556 | ||
| Surgery within 1 month | 1.475 (0.635–3.426) | 0.367 | ||
| Digestive diseases | 2.127 (1.004–4.505) | 0.046 | ||
| Central intravenous catheter | 1.271 (0.711–2.271) | 0.419 | ||
| Urinary catheter | 0.693 (0.350–1.372) | 0.293 | ||
| Tracheal catheter | 0.942 (0.530–1.673) | 0.839 | ||
| Stomach tube | 0.447 (0.242–0.824) | 0.010 |
Note: Bold values indicate P<0.05.
Abbreviations: cKp, classic Klebsiella pneumoniae; hvKp, hypervirulent Klebsiella pneumoniae; ICU, intensive care unit; OR, odds ratio.
Risk factors: CR-hvKp vs. non-CR-hvKp
Patients with surgery history within 1 month (OR=19.5) and catheterized tracheal catheter (OR=6.051) were closely associated with CR-hvKp. However, patients with diabetes (OR=0.256) were more likely to be infected with non-CR-hvKp. A history of surgery within the last 1 month is an independent risk factor for CR-hvKp infection (OR=15.999) (Table 4).
Table 4.
Risk factor for CR-hvKp vs. non-CR-hvKp
| Variable | Univariate OR (95% CI) | P-value | Multivariate OR (95% CI) | P-value |
|---|---|---|---|---|
| Hypermucoviscous | 0.468 (0.086–2.550) | 0.380 | ||
| Infection occurred in ICU | 2.667 (0.692–10.277) | 0.154 | ||
| Pulmonary diseases | 3.879 (0.052–1.112) | 0.162 | ||
| Cancer | 2.246 (0.625–8.077) | 0.215 | ||
| Diabetes | 0.256 (0.071–0.930) | 0.038 | ||
| Cardiovascular disease | 0.613 (0.167–2.250) | 0.461 | ||
| Cerebrovascular disease | 3.884 (0.965–15.313) | 0.056 | ||
| Surgery within 1 month | 19.5 (4.567–83.265) | 0.000 | 15.999 (3.412–75.026) | 0.000 |
| Digestive diseases | 2.386 (0.662–8.602) | 0.184 | ||
| Central intravenous catheter | 2.330 (0.472–11.501) | 0.299 | ||
| Tracheal catheter | 6.051 (1.485–24.658) | 0.012 | ||
| Stomach tube | 1.867 (0.462–7.535) | 0.381 | ||
| Urinary catheter | 3.492 (0.422–28.865) | 0.246 |
Note: Bold values indicate P<0.05.
Abbreviations: cKp, classic Klebsiella pneumoniae; CR, carbapenems-resistant; hvKp, hypervirulent Klebsiella pneumoniae; ICU, intensive care unit; OR, odds ratio.
MLST genotypic analysis
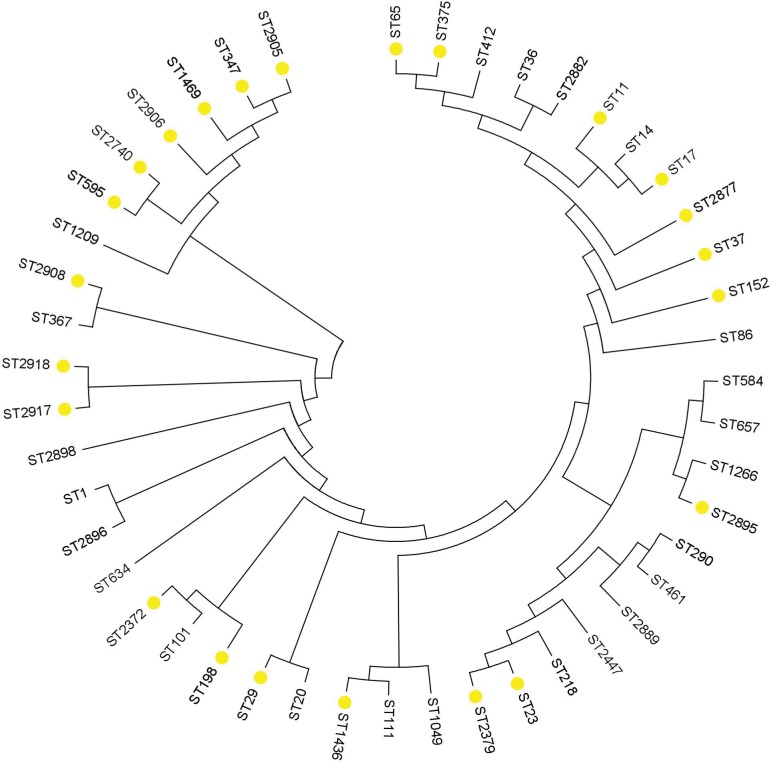
Among the 202 K. pneumoniae isolates, no new ST was identified in MLST database. The most prevalent ST in this study was ST23 (n=28; 13.9%), followed by ST412 (n=10; 5.0%), ST37 (n=7; 3.5%), ST65 (n=6; 3.0%), ST11 (n=5; 2.5%), ST17 (n=5; 2.5%), ST2905 (n=5; 2.5%), and ST2906 (n=5; 2.5%). The aforementioned STs accounted for 35.1% (43/202) of the total strains. Among the primary STs, ST23 (24/28), ST412 (6/9), ST17(5/5), and ST65(3/6) were strongly associated with hvKp, while ST11(0/5), ST2905(1/5), ST2906(1/5), and ST37(2/7) were more common in the cKp group. The more common clone complexes (CCs) of the CR-hvKp group were CC23 (n=3) and CC17 (n=3). There is an important phenomenon in the phylogenetic tree that a branch clustered with ST347, ST595, ST1469, ST2905, and ST2906 contributed to poor prognosis death in 30 days, which should be paid more attention (Figure 1).
Figure 1.
Neighbor-joining dendrogram showing concatenated sequences of 7 housekeeping genes from the MLST database for the frequency >1 STs and isolates contributing to invasive infection and mortality. Yellow solid rim represents death.
Abbreviations: MLST, multilocus sequence typing; ST, sequence type.
Discussion
To our knowledge, this is the first and biggest systematic study focusing on the elderly infected with hvKp in China. In this study, 59.9% of hvKp were identified as hypermucoviscous by string test, which was significantly different from a previous retrospective study conducted in a single center in China, with a prevalence of 33% in Beijing.13 In addition, the incidence of hvKp (47.5%) in genetic background is also higher than the figure in the previous multicenter studies (37.8%).8 It can be concluded that hvKp is emerging as the major pathotype for the elderly in the 2 hospitals, which should be paid more attention. The prevalent ST in 2 hospitals is the same: ST23. However, different hospitals isolated with ST23 are not highly associated with hvKp, which indicates that fully relying on STs to identify hvKp may be unreliable. Although the definition of hvKp is controversial, it is an objective marker, like plasmid type, biofilm producing, serotypes and the ability of trigger inflammatory factors that may be needed for further study to complement the real hypervirulence. Therefore, the prevalence of hvKp may be incorrectly estimated due to the lack of definite and objective diagnostic methods.
HvKp causes various severe infections, posing a serious threat to health. Various types of K-antigen have been reported,17,21,22 with the more important elements in Asia being K1 and K2, which are the reason for severe infection. But they are not the unique trait for hvKp.23,24 MagA is not a specific virulence gene for hvKp isolates causing PLA,25 but it is highly associated with cps K1.22,26 Moreover, the mutant isolate (knockout magA) could not show hmv phenotype.27 RmpA/RmpA2 was proposed as a virulent factor in addition to magA and cps K1/K2.16 Although rmpA is not an independent factor contributing to pyogenic liver abscess, it promotes capsule synthesis, which is associated with hypermucoviscous.3,16 Our results are consistent with a previous study: invasive infection (especially liver abscess), hypermucoviscosity, and mainly virulence factors (K1, K2, rmpA, and magA genes) are highly presented in hvKp group.8 So, a better understanding of risk factors is essential to make interventions. Our results show that patients with diabetes are more likely to be infected with hvKp. Additionally, hypermucoviscosity is strongly associated with hvKp. In our study, patients with surgery history of <1 month are an independent risk factor for CR-hvKp infection, which should be focused more on how to prevent infection. A previous study concluded that major histocompatibility complex variants, nutritional status, and gut microbiota are essential host factors to improve the understanding of the hypervirulence phenomenon.7 Our results demonstrated that in the hvKp group, patients with better nutritional status are associated with a more severe SOFA score and more serious inflammation reaction. All aforementioned characteristics may be a potential marker for early and precise empirical interventions for the elderly with hvKp.
Previous studies have revealed that most hvKp and antimicrobial-resistant patterns were non-overlapping.8,13 In this study, most hvKp were sensitive to most of the aforementioned antibiotics. In the hvKp group, the number of MDR-hvKp (25.0%) and ESBL-hvKp (26.0%) is significantly higher compared to the previous study performed in the multicenter study, with a prevalence of 12.6%.8 It is alarming that the number of elderly with MDR-hvKp infection is increasing. Moreover, 1 CR-hvKp isolate was detected in 1 hospital, where long-term patients were hospitalized. However, 10 CR-hvKp strains were detected in the other hospital, which was a referral center receiving patients from other hospitals and the community. Therefore, the incidence of CR-hvKp may be underestimated in this region. Thus, these data revealed that MDR-hvKp is emerging among the elderly. However, to confirm this conclusion, further investigation using a larger population is needed.
The CR-hvKp was not detected in nosocomial environment by routine nosocomial infection surveillance. In addition, the 2 hospitals did not apply the use of anal swab for monitoring nosocomial infection. It is unclear whether gut microbiota composed of hvKp contributed to the infection. Thus, it is essential to enhance hospital infection surveillance for the elderly. A previous study suggested that wards previously infected with CR-hvKp should be disinfected and left unoccupied for >2 weeks.10 Otherwise it may be a good site for a fatal outbreak of the organism.
There are some limitations in our study. First, it was a retrospective study in 2 teaching hospitals for over 10 years. Most key inflammatory and nutrition status marker were not achieved. Second, in vitro and in vivo experiments as objective evidence, such as Galleria mellonella, mouse, or human neutrophil assay, may be needed to identify the real virulent Kp. Third, to further explore the pathogen genomic characteristics, especially for virulent and antibiotic-resistant environment, whole genome sequencing, transcriptomics, and proteomics may be needed. A larger prospective multicenter study, focusing on host, pathogen, and host–pathogen interaction (inflammatory factor), is needed to better defining the hvKp strains.
Conclusion
The prevalence of hvKp may be higher than expected in the elderly. The epidemiology for hvKp in different hospitals is different. Although the definition of hvKp is still controversial, hvKp (aerobactin positive) strains were more likely to cause serious infections, such as liver abscess and septic shock, and more severe inflammatory reaction in the host. To further understand hvKp, host, pathogen, and host–pathogen interaction may be taken into consideration. The emerging MDR-hvKp, especially CR-hvKp, will be a great challenge for treatment. It is essential to enhance clinical awareness and management for the different types of hvKp infections.
Supplementary materials
Table S1.
Primers
| Name | Sequence |
|---|---|
| rmpA | |
| Forward | 5-ACTGGGCTACCTCTGCTTCA-3 |
| Reverse | 5-CTTGCATGAGCCATCTTTCA-3 |
| rmpA2 | |
| Forward | 5-CTTTATGTGCAATAAG-GATGTT-3 |
| Reverse | 5-CCTCCTGGAGAGTAAGCATT-3 |
| magA | |
| Forward | 5-GGTGCTCTTTACATCATTGC-3 |
| Reverse | 5-GCAATGGCCATTTGCGTTAG-3 |
| aerobactin | |
| Forward | 5-GCATAGGCGGATACGAACAT-3 |
| Reverse | 5-CACAGGGCAATTGCTTACCT-3 |
| K1 | |
| Forward | 5-GTAGGTATTGCAAGCCATGC-3 |
| Reverse | 5-GCCCAGGTTAATGAATCCGT-3 |
| K2 | |
| Forward | 5-GGAGCCATTTGAATTCGGTG-3 |
| Reverse | 5-TCCCTAGCACTGGCTTAAGT-3 |
| K5 | |
| Forward | 5-GCCACCTCTAAGCATATAGC-3 |
| Reverse | 5-CGCACCAGTAATTCCAACAG-3 |
| K20 | |
| Forward | 5-CCGATTCGGTCAACTAGCTT-3 |
| Reverse | 5-GCACCTCTATGAACTTTCAG-3 |
| K54 | |
| Forward | 5-CATTAGCTCAGTGGTTGGCT-3 |
| Reverse | 5-GCTTGACAAACACCATAGCAG-3 |
| K57 | |
| Forward | 5-CGACAAATCTCTCCTGACGA-3 |
| Reverse | 5-CGCGACAAACATAACACTCG-3 |
| rpoB | |
| Forward | 5-GGCGAAATGGCWGAGAACCA-3 |
| Reverse | 5-GAGTCTTCGAAGTTGTAACC-3 |
| gapA | |
| Forward | 5-TGAAATATGACTCCACTCACGG-3 |
| Reverse | 5-CTTCAGAAGCGGCTTTGATGGCTT-3 |
| mdh | |
| Forward | 5-TGAAATATGACTCCACTCACGG-3 |
| Reverse | 5-CTTCAGAAGCGGCTTTGATGGCTT-3 |
| pgi | |
| Forward | 5-GAGAAAAACCTGCCTGTACTGCTGGC-3 |
| Reverse | 5-CGCGCCACGCTTTATAGCGGTTAAT-3 |
| phoE | |
| Forward | 5-ACCTACCGCAACACCGACTTCTTCGG-3 |
| Reverse | 5-TGATCAGAACTGGTAGGTGAT-3 |
| infB | |
| Forward | 5-CTCGCTGCTGGACTATATTCG-3 |
| Reverse | 5-CGCTTTCAGCTCAAGAACTTC-3 |
| tonB | |
| Forward | 5-CTTTATACCTCGGTACATCAGGTT-3 |
| Reverse | 5-ATTCGCCGGCTGRGCRGAGAG-3 |
Table S2.
Detailed clinical and microbiological features of CR-hvKp strains
| Clinical features | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | P11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (years) | 87 | 87 | 87 | 88 | 79 | 71 | 65 | 65 | 77 | 84 | 94 |
| Gender | F | M | M | F | M | F | F | M | M | M | M |
| Clinical department | Internal medicine | Emergency | General surgery | ICU | General surgery | ICU | ICU | Thoracic surgery | ICU | Thoracic surgery | Respiratory |
| Main underlying diseases | Bone fracture; Cardiovascular diseases | Diabetes | Surgery with in 1 month | Cerebrovascular disease | Cancer; Surgery with in 1 month | Cancer; Surgery with in 1 month | Diabetes; Cerebrovascular disease | Cancer | Cancer; Surgery with in 1 month | Surgery with in 1 month | Cancer; Cerebrovascular disease; diabetes |
| Tube | CVC; Ureter | Ureter; Stomach tube | None | CVC; Ureter; Stomach tube; Tracheal catheter | CVC; Ureter; Stomach tube; Tracheal catheter | CVC; Ureter; Tracheal catheter | CVC; Ureter; Stomach tube; Tracheal catheter | CVC; Ureter; Stomach tube; Tracheal catheter | CVC; Ureter; Stomach tube; Tracheal catheter | CVC; Ureter; Stomach tube; Tracheal catheter | CVC; Ureter; Stomach tube; Tracheal catheter |
| Specimen type | Sputum | Sputum | Pyogenic fluids | Bile | Sputum +Blood +Wound | Bile+Blood | Sputum+Urine +Feces | Sputum | Pyogenic fluids | Sputum+Urine | Drainage liquid |
| Infection type | Pneumonia | Pneumonia | Abscess | PLA | Sepsis | Septic shock | Septic shock | Sepsis | Septic shock | Septic shock | Sepsis |
| WBC(109/L) | 12.21 | 10.85 | 13.76 | 16.44 | 8.80 | 15.49 | 10.0 | 18.32 | 11.66 | 9.51 | 4.64 |
| NEU (%) | 75.1 | 86.6 | 92.5 | 82.2 | 56.8 | 83.4 | 69.9 | 87.4 | 88.0 | 65.2 | 68.3 |
| TP (g/L) | 79.4 | 60.5 | 69.4 | 52.8 | 54.7 | 49.6 | 59 | 70.9 | 62 | 67.4 | 61.2 |
| ALB (g/L) | 40.3 | 27.7 | 34.6 | 28.2 | 20.6 | 30.0 | 32.6 | 36.4 | 27.4 | 31.7 | 35.0 |
| Sensitive antibiotics | N | N | N | N | GEN; LEV; TOB | N | SMZ; TOB; AMK | SMZ | N | N | SMZ; AMK |
| SOFA score | 6 | 10 | 2 | 13 | 7 | 9 | 7 | 4 | 5 | 6 | 7 |
| Empiric therapy | CIP+CAZ | CIP+CAZ | IPM | IPM+ISE | IPM+MXF | CIP+CAZ | IPM+ISE | IPM | IPM | MXF | MEM |
| Switched therapy | MEM+TGC | MEM+TGC | TGC | TGC | GEN+LEV | TGC+MEM | TGC+MEM+FOS | TGC+MEM | TGC+MEM | TGC+MEM+FOS | TGC |
| Clinical outcome | Survived | Survived | Survived | Survived | Survived | Survived | Survived | Died | Survived | Survived | Survived |
| String test | + | + | + | + | + | + | + | − | − | + | + |
| Virulence-associated genes | |||||||||||
| rmpA | + | + | + | + | + | + | + | − | − | + | + |
| rmpA2 | + | + | + | + | + | + | + | − | − | + | + |
| magA | + | + | + | + | + | + | + | − | + | + | + |
| cps genes | |||||||||||
| K1 | + | − | − | + | + | + | + | − | − | + | − |
| K2 | + | + | + | + | + | + | + | − | − | + | − |
| K5 | − | − | − | − | − | − | − | + | + | − | − |
| K20 | − | + | − | − | + | − | − | − | + | − | − |
| K54 | − | − | − | − | − | − | + | − | − | − | − |
| K57 | + | − | − | − | − | + | − | − | − | − | − |
| MLST genotyping | 23 | 23 | 17 | 17 | 347 | 17 | 2905 | 2906 | 23 | 412 | 2874 |
Abbreviations: ALB, albumin; AMK, amikacin; CAZ, ceftazidime; CIP, ciprofloxacin; CMZ, cefmetazole; CVC, central venous catheter; F, female; FOS, fosfomycin; GEN, gentamicin; ICU, intensive care unit; IPM, imipenem; ISE, isepamicin; LEV, levofloxacin; MLST, multilocus sequence typing; M, male; MEM, meropenem; Mo, month; MXF, moxifloxacin; NEU, neutrophils; N, no; PLA; pyogenic liver abscess; SMZ, sulfamethoxazole; SOFA, sequential organ failure assessment; TGC, tigecycline; TOB, tobramycin; TP, total protein; TZP, piperacillin tazobactam; WBC, white blood cell; Y, yes; +, positive; −, negative.
Acknowledgments
We thank the team of curators of the Institut Pasteur MLST and whole genome MLST databases for curating the data and making them publicly available at http://bigsdb.pasteur.fr. Additionally, we thank Huicheng Wang for statistics, who works in China Food and Drug Administration. This work was supported by the China Postdoctoral Science Foundation (grant number 2014M562610) and the Excellent Young Program of the Organization Department of Beijing Municipal Party Committee (grant number 2016000057592G258).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun. 2015;83(8):3325–3333. doi: 10.1128/IAI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12(11):881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 4.Pomakova DK, Hsiao CB, Beanan JM, et al. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis. 2012;31(6):981–989. doi: 10.1007/s10096-011-1396-6. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Zeng J, Liu W, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71(5):553–560. doi: 10.1016/j.jinf.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Lin YC, Lu MC, Tang HL, et al. Assessment of hypermucoviscosity as a virulence factor for experimental Klebsiella pneumoniae infections: comparative virulence analysis with hypermucoviscosity-negative strain. BMC Microbiol. 2011;11:50. doi: 10.1186/1471-2180-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catalan-Najera JC, Garza-Ramos U, Barrios-Camacho H. Hyper-virulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence. 2017:1–13. doi: 10.1080/21505594.2017.1317412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Zhao C, Wang Q, et al. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60(10):6115–6120. doi: 10.1128/AAC.01127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russo TA, Olson R, Macdonald U, et al. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun. 2014;82(6):2356–2367. doi: 10.1128/IAI.01667-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2017;18(1):37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 11.Gu DX, Huang YL, Ma JH, et al. Detection of colistin resistance gene mcr-1 in hypervirulent Klebsiella pneumoniae and Escherichia coli isolates from an infant with diarrhea in China. Antimicrob Agents Chemother. 2016;60(8):5099–5100. doi: 10.1128/AAC.00476-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2015;60(1):709–711. doi: 10.1128/AAC.02173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Sun G, Yu Y, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58(2):225–232. doi: 10.1093/cid/cit675. [DOI] [PubMed] [Google Scholar]
- 14.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 15.Choi MJ, Ko KS. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother. 2015;59(11):6763–6773. doi: 10.1128/AAC.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu WL, Ko WC, Cheng KC, et al. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42(10):1351–1358. doi: 10.1086/503420. [DOI] [PubMed] [Google Scholar]
- 17.Cheng NC, Yu YC, Tai HC, et al. Recent trend of necrotizing fasciitis in Taiwan: focus on monomicrobial Klebsiella pneumoniae necrotizing fasciitis. Clin Infect Dis. 2012;55(7):930–939. doi: 10.1093/cid/cis565. [DOI] [PubMed] [Google Scholar]
- 18.Jung SW, Chae HJ, Park YJ, et al. Microbiological and clinical characteristics of bacteraemia caused by the hypermucoviscosity phenotype of Klebsiella pneumoniae in Korea. Epidemiol Infect. 2013;141(2):334–340. doi: 10.1017/S0950268812000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin JC, Yeh KM, Chang FY. The distant metastasis of pyogenic liver abscess caused by Klebsiella pneumoniae serotype K2 and the underlying disease of diabetes mellitus should be carefully interpreted. Clin Infect Dis. 2007;45(11):1531–1532. doi: 10.1086/523008. author reply 1532–1533. [DOI] [PubMed] [Google Scholar]
- 20.Lin WH, Wang MC, Tseng CC, et al. Clinical and microbiological characteristics of Klebsiella pneumoniae isolates causing community-acquired urinary tract infections. Infection. 2010;38(6):459–464. doi: 10.1007/s15010-010-0049-5. [DOI] [PubMed] [Google Scholar]
- 21.Pan YJ, Fang HC, Yang HC, et al. Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J Clin Microbiol. 2008;46(7):2231–2240. doi: 10.1128/JCM.01716-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuang YP, Fang CT, Lai SY, Chang SC, Wang JT. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis. 2006;193(5):645–654. doi: 10.1086/499968. [DOI] [PubMed] [Google Scholar]
- 23.Yeh KM, Kurup A, Siu LK, et al. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol. 2007;45(2):466–471. doi: 10.1128/JCM.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brisse S, Fevre C, Passet V, et al. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One. 2009;4(3):e4982. doi: 10.1371/journal.pone.0004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeh KM, Chang FY, Fung CP, Lin JC, Siu LK. magA is not a specific virulence gene for Klebsiella pneumoniae strains causing liver abscess but is part of the capsular polysaccharide gene cluster of K. pneumoniae serotype K1. J Med Microbiol. 2006;55(Pt 6):803–804. doi: 10.1099/jmm.0.46368-0. [DOI] [PubMed] [Google Scholar]
- 26.Struve C, Bojer M, Nielsen EM, Hansen DS, Krogfelt KA. Investigation of the putative virulence gene magA in a worldwide collection of 495 Klebsiella isolates: magA is restricted to the gene cluster of Klebsiella pneumoniae capsule serotype K1. J Med Microbiol. 2005;54(Pt 11):1111–1113. doi: 10.1099/jmm.0.46165-0. [DOI] [PubMed] [Google Scholar]
- 27.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Primers
| Name | Sequence |
|---|---|
| rmpA | |
| Forward | 5-ACTGGGCTACCTCTGCTTCA-3 |
| Reverse | 5-CTTGCATGAGCCATCTTTCA-3 |
| rmpA2 | |
| Forward | 5-CTTTATGTGCAATAAG-GATGTT-3 |
| Reverse | 5-CCTCCTGGAGAGTAAGCATT-3 |
| magA | |
| Forward | 5-GGTGCTCTTTACATCATTGC-3 |
| Reverse | 5-GCAATGGCCATTTGCGTTAG-3 |
| aerobactin | |
| Forward | 5-GCATAGGCGGATACGAACAT-3 |
| Reverse | 5-CACAGGGCAATTGCTTACCT-3 |
| K1 | |
| Forward | 5-GTAGGTATTGCAAGCCATGC-3 |
| Reverse | 5-GCCCAGGTTAATGAATCCGT-3 |
| K2 | |
| Forward | 5-GGAGCCATTTGAATTCGGTG-3 |
| Reverse | 5-TCCCTAGCACTGGCTTAAGT-3 |
| K5 | |
| Forward | 5-GCCACCTCTAAGCATATAGC-3 |
| Reverse | 5-CGCACCAGTAATTCCAACAG-3 |
| K20 | |
| Forward | 5-CCGATTCGGTCAACTAGCTT-3 |
| Reverse | 5-GCACCTCTATGAACTTTCAG-3 |
| K54 | |
| Forward | 5-CATTAGCTCAGTGGTTGGCT-3 |
| Reverse | 5-GCTTGACAAACACCATAGCAG-3 |
| K57 | |
| Forward | 5-CGACAAATCTCTCCTGACGA-3 |
| Reverse | 5-CGCGACAAACATAACACTCG-3 |
| rpoB | |
| Forward | 5-GGCGAAATGGCWGAGAACCA-3 |
| Reverse | 5-GAGTCTTCGAAGTTGTAACC-3 |
| gapA | |
| Forward | 5-TGAAATATGACTCCACTCACGG-3 |
| Reverse | 5-CTTCAGAAGCGGCTTTGATGGCTT-3 |
| mdh | |
| Forward | 5-TGAAATATGACTCCACTCACGG-3 |
| Reverse | 5-CTTCAGAAGCGGCTTTGATGGCTT-3 |
| pgi | |
| Forward | 5-GAGAAAAACCTGCCTGTACTGCTGGC-3 |
| Reverse | 5-CGCGCCACGCTTTATAGCGGTTAAT-3 |
| phoE | |
| Forward | 5-ACCTACCGCAACACCGACTTCTTCGG-3 |
| Reverse | 5-TGATCAGAACTGGTAGGTGAT-3 |
| infB | |
| Forward | 5-CTCGCTGCTGGACTATATTCG-3 |
| Reverse | 5-CGCTTTCAGCTCAAGAACTTC-3 |
| tonB | |
| Forward | 5-CTTTATACCTCGGTACATCAGGTT-3 |
| Reverse | 5-ATTCGCCGGCTGRGCRGAGAG-3 |
Table S2.
Detailed clinical and microbiological features of CR-hvKp strains
| Clinical features | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | P11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (years) | 87 | 87 | 87 | 88 | 79 | 71 | 65 | 65 | 77 | 84 | 94 |
| Gender | F | M | M | F | M | F | F | M | M | M | M |
| Clinical department | Internal medicine | Emergency | General surgery | ICU | General surgery | ICU | ICU | Thoracic surgery | ICU | Thoracic surgery | Respiratory |
| Main underlying diseases | Bone fracture; Cardiovascular diseases | Diabetes | Surgery with in 1 month | Cerebrovascular disease | Cancer; Surgery with in 1 month | Cancer; Surgery with in 1 month | Diabetes; Cerebrovascular disease | Cancer | Cancer; Surgery with in 1 month | Surgery with in 1 month | Cancer; Cerebrovascular disease; diabetes |
| Tube | CVC; Ureter | Ureter; Stomach tube | None | CVC; Ureter; Stomach tube; Tracheal catheter | CVC; Ureter; Stomach tube; Tracheal catheter | CVC; Ureter; Tracheal catheter | CVC; Ureter; Stomach tube; Tracheal catheter | CVC; Ureter; Stomach tube; Tracheal catheter | CVC; Ureter; Stomach tube; Tracheal catheter | CVC; Ureter; Stomach tube; Tracheal catheter | CVC; Ureter; Stomach tube; Tracheal catheter |
| Specimen type | Sputum | Sputum | Pyogenic fluids | Bile | Sputum +Blood +Wound | Bile+Blood | Sputum+Urine +Feces | Sputum | Pyogenic fluids | Sputum+Urine | Drainage liquid |
| Infection type | Pneumonia | Pneumonia | Abscess | PLA | Sepsis | Septic shock | Septic shock | Sepsis | Septic shock | Septic shock | Sepsis |
| WBC(109/L) | 12.21 | 10.85 | 13.76 | 16.44 | 8.80 | 15.49 | 10.0 | 18.32 | 11.66 | 9.51 | 4.64 |
| NEU (%) | 75.1 | 86.6 | 92.5 | 82.2 | 56.8 | 83.4 | 69.9 | 87.4 | 88.0 | 65.2 | 68.3 |
| TP (g/L) | 79.4 | 60.5 | 69.4 | 52.8 | 54.7 | 49.6 | 59 | 70.9 | 62 | 67.4 | 61.2 |
| ALB (g/L) | 40.3 | 27.7 | 34.6 | 28.2 | 20.6 | 30.0 | 32.6 | 36.4 | 27.4 | 31.7 | 35.0 |
| Sensitive antibiotics | N | N | N | N | GEN; LEV; TOB | N | SMZ; TOB; AMK | SMZ | N | N | SMZ; AMK |
| SOFA score | 6 | 10 | 2 | 13 | 7 | 9 | 7 | 4 | 5 | 6 | 7 |
| Empiric therapy | CIP+CAZ | CIP+CAZ | IPM | IPM+ISE | IPM+MXF | CIP+CAZ | IPM+ISE | IPM | IPM | MXF | MEM |
| Switched therapy | MEM+TGC | MEM+TGC | TGC | TGC | GEN+LEV | TGC+MEM | TGC+MEM+FOS | TGC+MEM | TGC+MEM | TGC+MEM+FOS | TGC |
| Clinical outcome | Survived | Survived | Survived | Survived | Survived | Survived | Survived | Died | Survived | Survived | Survived |
| String test | + | + | + | + | + | + | + | − | − | + | + |
| Virulence-associated genes | |||||||||||
| rmpA | + | + | + | + | + | + | + | − | − | + | + |
| rmpA2 | + | + | + | + | + | + | + | − | − | + | + |
| magA | + | + | + | + | + | + | + | − | + | + | + |
| cps genes | |||||||||||
| K1 | + | − | − | + | + | + | + | − | − | + | − |
| K2 | + | + | + | + | + | + | + | − | − | + | − |
| K5 | − | − | − | − | − | − | − | + | + | − | − |
| K20 | − | + | − | − | + | − | − | − | + | − | − |
| K54 | − | − | − | − | − | − | + | − | − | − | − |
| K57 | + | − | − | − | − | + | − | − | − | − | − |
| MLST genotyping | 23 | 23 | 17 | 17 | 347 | 17 | 2905 | 2906 | 23 | 412 | 2874 |
Abbreviations: ALB, albumin; AMK, amikacin; CAZ, ceftazidime; CIP, ciprofloxacin; CMZ, cefmetazole; CVC, central venous catheter; F, female; FOS, fosfomycin; GEN, gentamicin; ICU, intensive care unit; IPM, imipenem; ISE, isepamicin; LEV, levofloxacin; MLST, multilocus sequence typing; M, male; MEM, meropenem; Mo, month; MXF, moxifloxacin; NEU, neutrophils; N, no; PLA; pyogenic liver abscess; SMZ, sulfamethoxazole; SOFA, sequential organ failure assessment; TGC, tigecycline; TOB, tobramycin; TP, total protein; TZP, piperacillin tazobactam; WBC, white blood cell; Y, yes; +, positive; −, negative.