Abstract
Many genetic mutations have been identified as monogenic causes of nephrotic syndrome (NS), but important knowledge gaps exist in the roles of these genes in kidney cell biology and renal diseases. More animal models are needed to assess the functions of these genes in vivo, and to determine how they cause NS in a timely manner. Drosophila nephrocytes and human podocytes share striking similarities, but to what degree these known NS genes play conserved roles in nephrocytes remains unknown. Here we systematically studied 40 genes associated with NS, including 7 that have not previously been analysed for renal function in an animal model. We found that 85% of these genes are required for nephrocyte functions, suggesting that a majority of human genes known to be associated with NS play conserved roles in renal function from flies to humans. To investigate functional conservation in more detail, we focused on Cindr, the fly homolog of the human NS gene CD2AP. Silencing Cindr in nephrocytes led to dramatic nephrocyte functional impairment and shortened life span, as well as collapse of nephrocyte lacunar channels and effacement of nephrocyte slit diaphragms. These phenotypes could be rescued by expression of a wild-type human CD2AP gene, but not a mutant allele derived from a patient with CD2AP-associated NS. We conclude that the Drosophila nephrocyte can be used to elucidate clinically relevant molecular mechanisms underlying the pathogenesis of most monogenic forms of NS, and to efficiently generate personalized in vivo models of genetic renal diseases bearing patient-specific mutations.
Introduction
Advances in genomic analysis techniques have enabled the identification of a large number of genes, encoding products with diverse functions, that when mutated contribute to nephrotic syndrome (NS). Many cases of chronic kidney diseases (CKD) are associated with single gene mutations that affect the function of podocytes (1–5). These studies point to podocytes as a key renal target to precipitate CKD (6–9). For example, among patients with steroid-resistant nephrotic syndrome, several mutations have been found in podocyte genes that affect the structure and function of the slit diaphragm (SD), the regulation of the actin cytoskeleton proteins, basement membrane components, cell membrane, adhesion molecules, mitochondrial function, endocytosis, cation flux, Coenzyme Q10 biosynthesis, and the regulation of transcription (10–20). Our understanding of the molecular and cellular mechanisms underlying renal pathologies caused by these mutations, and our potential to develop new therapeutic treatment approaches based on that knowledge, have to date largely come from experimental studies performed in the mouse and zebrafish models. Although a relatively simple animal model system, Drosophila is unrivalled with respect to the abundance of highly sophisticated genetic approaches and well-established, available resources that can be exploited to rapidly and efficiently investigate specific molecular pathways and cellular targets implicated in abnormal cell physiologies relevant to disease (12,21–26). Drosophila can thus be used as a high-throughput experimental disease-model platform, to validate the large amount of data generated from clinical genomics.
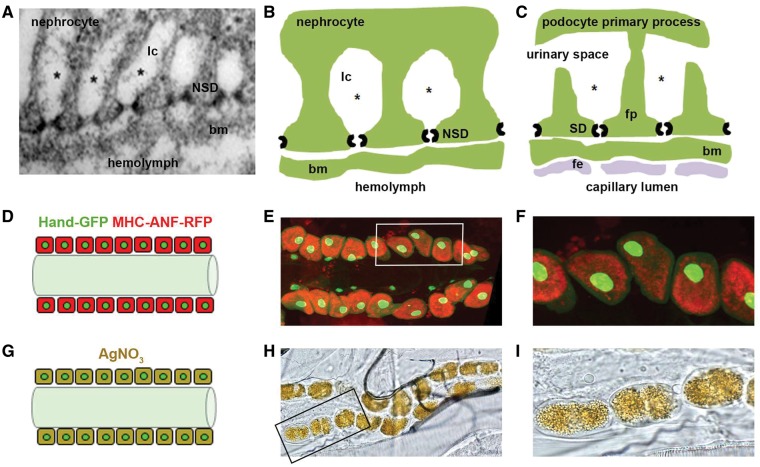
Overall, 75% of human disease associated genes are represented in the Drosophila genome by functional homologs (27,28). We have developed a fly model to investigate genetic, molecular, and cellular targets of cell injury in Drosophila nephrocytes, that is relevant to human podocytes (12,26,29,30), and provides new in vivo information to complement the excellent studies done in mice. Briefly, the Drosophila pericardial nephrocyte (hereafter, nephrocyte) is remarkably similar, both structurally and functionally, to the mammalian podocyte (Fig. 1A–C) (24,25,31,32). Nephrocytes are relatively large cells positioned beside the dorsal vessel (the fly heart). The dorsal vessel, by regular contractions, maintains hemolymph (insect "blood") circulation throughout the body cavity. The nephrocytes carry out hemolymph filtration and reabsorption functions homologous to mammalian podocytes and proximal tubule cells, respectively, by virtue of nephrocyte slit diaphragms (NSD) spanning the equivalents of foot processes and lacunar channels that expand the plasma membrane area to increase uptake of filtered hemolymph components (24,25,29). Nephrocytes are so positioned that filtered hemolymph enters the dorsal vessel for redistribution.
Figure 1.
Comparison of nephrocyte and podocyte basic features, and nephrocyte functional assays. (A–C). Insect nephrocytes and mammalian podocytes are structurally and functionally homologous. Panel A is a transmission electron micrograph of a Drosophila nephrocyte. Panels B and C compare structural and functional features of nephrocytes and podocytes, respectively. A basement membrane (bm) interposes a size and charge selective filtration barrier between the nephrocyte slit diaphragm (NSD) and the hemolymph of the insect open circulatory system, and the podocyte slit diaphragm (SD) and blood from the mammalian capillary lumen. In the latter case blood encounters the bm after exiting the capillary lumen through a fenestrated endothelial (fe) cell layer. The principal cellular structure in nephrocytes and podocytes mediating filtration is a slit diaphragm. In podocytes, SD proteins form intercellular filtration barriers between adjacent foot processes (fp), the intercalating termini of primary processes originating from different podocyte cells. In nephrocytes, NSD proteins form intracellular filtration barriers on either side of lacunar channel (lc) openings. In nephrocytes, filtered low molecular weight molecules (represented by *) are reabsorbed by endocytosis from the lacunar channel membrane. In podocytes the ultrafiltrate passing through the SD enters the urinary space, and is subsequently reabsorbed by proximal tubule cells. The nephrocyte therefore bears striking similarities not only to the mammalian podocyte, but is functionally homologous to proximal tubule cells as well. (D–F). Functional assay measuring nephrocyte uptake of hemolymph protein. Panel D is a schematic diagram showing pericardial nephrocytes aligned on either side of the heart tube. In this assay, muscle cells (not shown) express a MHC-ANF-RFP transgene in which a myosin heavy chain (MHC) promoter directs expression of an atrial natriuretic peptide (42) - red fluorescent protein (RFP) fusion protein. ANF-RFP is secreted into the fly hemolymph, from which it is filtered and endocytosed by nephrocytes leading to cytoplasmic red fluorescence. Hand-GFP transgene expression is visualized as green fluorescence concentrated in the nuclei of nephrocytes (shown) and cardiomyocytes (not shown). Panel E shows RFP fluorescence (54) in the cytoplasm of adult nephrocytes, reflecting endocytosis of ANF-RFP fusion protein filtered from the hemolymph. Cardiomyocytes lack red fluorescence. Green fluorescence in nephrocyte and cardiomyocyte nuclei is due to Hand-GFP expression. Panel F is a higher magnification image of cells (boxed) from panel E. (G–I). Functional assay measuring nephrocyte uptake of AgNO3. Panel G is a schematic diagram showing pericardial nephrocytes containing endocytosed and sequestered AgNO3. Panel H is a photomicrograph showing ingested AgNO3 sequestered in larval nephrocytes. Panel I is a higher magnification image of cells (boxed) from panel H.
Analysing the available literature up to the end of 2015, we identified 40 genes that are directly associated with NS and have clear Drosophila homologs with available RNAi targeting fly lines (33–36). We carried out this study to explore how the Drosophila orthologs of these 40 NS genes (Table 1) modulate the function of nephrocytes (34). More than half of the NS genes listed are associated with the development of steroid-resistant nephrotic syndrome in humans, which cause ∼15% of all CKD diagnosed before age 25 (1). We then used the Gal4-UAS system to direct expression of RNAi silencing transgenes in nephrocytes to systematically knockdown Drosophila orthologs of the NS genes listed in Table 1 (35–37). Subsequently, we quantitatively assessed the phenotypic severity of gene knockdowns using functional assays for protein uptake and sequestration of toxic silver nitrate (AgNO3), adult fly survival, and average lifespan. We have also validated our system, exploring the role of mutations present in key podocytes genes (i.e. the CD2AP gene) that have been linked to the pathogenesis of NS in humans and other experimental animal model systems (38–41). More specifically, we explored the role of Cindr (the fly ortholog of CD2AP) in Drosophila nephrocyte function (41). We also tested the ability of human patient-derived CD2AP alleles to rescue deficiencies associated with silencing endogenous Cindr gene expression. Our findings validate the efficacy and clinical relevance of the Drosophila experimental model system to study pathogenic mechanisms underlying most monogenic forms of NS, and reveal the possibility of using the fly to efficiently generate patient specific (i.e. personalized) in vivo models of monogenic renal disease.
Table 1.
Human monogenic Nephrotic Syndrome (NS) genes and their Drosophila orthologs
| Human Gene | Drosophila Ortholog | Function | Cellular Localization | Renal Pathology |
|---|---|---|---|---|
| NPHS1 | Sns | Filtration | SD | SRNS |
| NPHS2 | Mec2 | Linker | SRNS | |
| CD2AP | Cindr | Adaptor protein | SRNS | |
| PTPRO | Ptp10D | Receptor-type tyrosine phosphatase | SRNS | |
| TRPC6 | Trpgamma | Calcium channel | SRNS | |
| MYH9 | Zip | Myosin motor | Actin Cytoskeleton | SRNS |
| MYO1E | Myo61F | SRNS | ||
| ACTN4 | Actn | Actin binding | SRNS | |
| INF2 | Form3 | SRNS | ||
| SYNPO | CG1674 | NS | ||
| ANLN | Scra | SRNS | ||
| KANK1 | Kank | Actin regulation | SRNS | |
| KANK2 | SRNS | |||
| KANK4 | SRNS | |||
| ARHGAP24 | RhoGAP92B | Signaling | SRNS | |
| ARHGDIA | RhoGDI | SRNS | ||
| LMX1B | CG32105 | Transcription factor | Nucleus | SRNS |
| SMARCAL1 | Marcal1 | SRNS | ||
| PAX2 | Sv | NS | ||
| E2F3 | E2f | NS | ||
| NXF5 | Sbr | RNA export | NS | |
| ZMPSTE24 | Ste24a | Lamin A processing | NS | |
| LMNA | Lam | Lamina | NS | |
| CUBN | Cubn | Endocytosis | Plasma membrane | IGS |
| AMN | Amn | IGS | ||
| SCARB2 | Emp | Proteolysis | Lysosome | SRNS |
| PMM2 | CG10688 | Glycosylation | Cytosol | NS |
| ALG1 | CG18012 | ER | NS | |
| COQ2 | Coq2 | Coenzyme Q synthesis | Mitochondrion | SRNS |
| COQ6 | CG7277 | SRNS | ||
| PDSS2 | CG10585 | SRNS | ||
| ADCK4 | CG32649 | SRNS | ||
| GPC5 | Dally | Signaling | Plasma membrane | NS |
| ITGA3 | Mew | Adhesion | SRNS | |
| ITGB4 | Mys | SRNS | ||
| CD151 | Tsp74F | NS | ||
| LAMB2 | LanB1 | GBM Structure | ECM | SRNS |
| COL4A3 | Col4a1 | Alport | ||
| COL4A4 | Alport | |||
| COL4A5 | Alport |
Results
Silencing of most NS gene orthologs in nephrocytes reduced hemolymph protein uptake and AgNO3 sequestration
In order to assess the function of the Drosophila orthologous of human NS genes in nephrocytes, we employed two in vivo cellular assays that quantitatively assessed uptake of a fluorescent hemolymph fusion protein (ANF-RFP), and sequestration of ingested AgNO3. In the former, flies carry a transgenic construct in which a myosin heavy chain (MHC) gene promoter directs the expression of atrial natriuretic factor (ANF) (42) peptide - red fluorescent protein (RFP) fusion protein. Muscle cells expressing the MHC-ANF-RFP transgene secrete ANF-RFP fusion protein into the fly’s hemolymph, from which it is normally filtered and endocytosed by nephrocytes. The resulting intracellular red fluorescence is detected by fluorescence microscopy and can be quantitated. To confirm nephrocyte cell identity, flies also carry a transgene encoding green fluorescent protein (GFP) expressed under the control of the Drosophila Hand gene promoter (Hand-GFP) (Fig. 1D–F). MHC-ANF-RFP and Hand-GFP transgenes are combined in flies carrying a Dot-Gal4 driver directing nephrocyte-specific expression of a UAS-GeneX-RNAi transgene construct that knocks down expression of the endogenous NS gene ortholog (i.e. GeneX) target. Because off-target effects of a given RNAi silencing construct could potentially elicit a false-positive result, we tested two or more available, independent RNAi silencing lines for each gene of interest. In our over-all experience, discrepancies in induced phenotypes between lines are observed very rarely (data not shown), and such presumptive evidence of off-target effects was not encountered in this study. In the second in vivo functional assay, we add toxic silver nitrate (AgNO3) to the standard diet of developing larvae. Ingested AgNO3 is normally taken up by nephrocytes and sequestered intracellularly, where it can be detected and the metal levels quantitated using phase contrast microscopy (Fig. 1G–I).
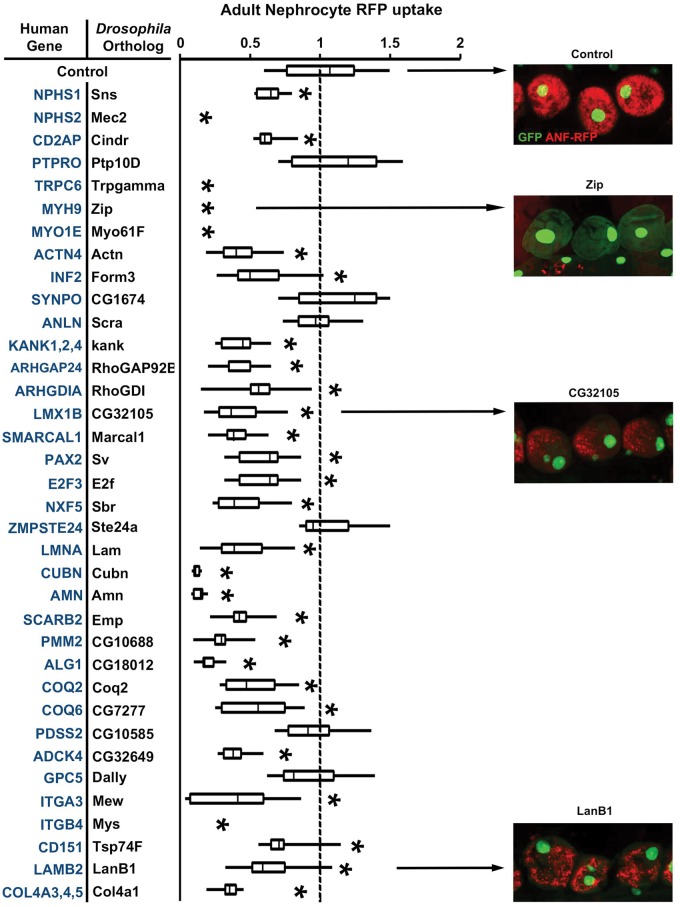
Table 1 lists 40 known human genes that are associated with NS. We identified Drosophila orthologs for all these 40 NS genes. In order to demonstrate the validity of the Drosophila nephrocyte model for in vivo investigations of mutation-associated renal disease, we conducted a systematic analysis of phenotypes induced by nephrocyte-specific RNAi based silencing of each Drosophila gene listed in Table 1. As shown in Figure 2, silencing of most of the genes reduced ANF-RFP fluorescence relative to wild type control nephrocytes. Zip and CG32105 knockdown illustrate the variable phenotypic severity as observed in this assay, while Ste24a knockdown illustrates a non-significant effect. Silencing of Mec2, Trpgamma, Zip, Myo61F, and Mys reduced RFP below the level of detection in our assay. By contrast, RFP levels were unaffected by silencing of Ptp10D, CG1674, Scra, CG10585, Ste24a, and Dally. We observed highly similar results when we assayed AgNO3 sequestration by nephrocytes in which NS gene orthologs were silenced (Fig. 3).
Figure 2.
Adult Nephrocyte RFP uptake. Human NS associated genes are listed with the corresponding Drosophila ortholog targeted by nephrocyte expression of inhibitory RNAi transgene. RFP fluorescence in nephrocytes was assessed in transgenic flies of genotype MHC-ANF-RFP; Hand-GFP; Dot-Gal4; UAS-GeneX-IR. ANF-RFP fusion protein uptake levels were determined from fluorescence micrographs and expressed relative to control nephrocytes of MHC-ANF-RFP; Hand-GFP; Dot-Gal4 transgenic flies. For quantification, ≥20 nephrocytes were analysed from each of three female flies per genotype. The results are presented as median plus interquartile range (IQR). Statistical significance (*) was defined as P < 0.05. Example fluorescence micrographs (merged GFP and RFP fluorescence) are shown to illustrate the range of effects on RFP levels induced by RNAi silencing of different genes.
Figure 3.
Adult Nephrocyte AgNO3 uptake. Human NS associated genes are listed with the corresponding Drosophila ortholog targeted by nephrocyte expression of inhibitory RNAi transgene. AgNO3 sequestration in nephrocytes was assessed in transgenic flies of genotype Dot-Gal4; UAS-GeneX-IR. AgNO3 levels were determined from photomicrographs and expressed relative to control nephrocytes in Dot-Gal4 transgenic flies. For quantification, ≥20 nephrocytes were analysed from each of three female flies per genotype. The results are presented as median plus interquartile range (IQR). Statistical significance (*) was defined as P < 0.05. Example photomicrographs are shown to illustrate the range of effects on AgNO3 levels induced by RNAi silencing of different genes.
Silencing of most NS gene orthologs in nephrocytes shortened adult fly life span
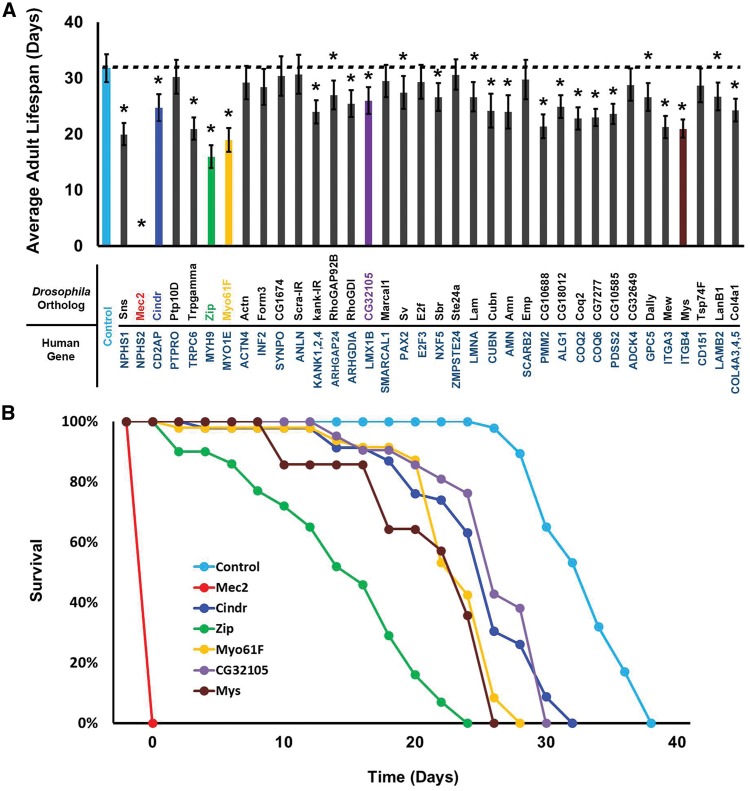
We observed that nephrocyte specific silencing of the majority of the NS gene orthologs shortened the average adult fly lifespan (Fig. 4A). Control wild type adult flies survived approximately 33 days on average. Significant reduction in average adult lifespan associated with nephrocyte gene silencing was observed for all genes except Ptp10D, Actn, Form3, CG1674, Scra, Marcal1, E2f, Ste24a, Emp, CG32649, and Tsp74F. Silencing of Ptp10D, CG1674, Scra, and Ste24a also did not reduce protein uptake or AgNO3 sequestration. Silencing of the CG10585 gene, by contrast, reduced fly lifespan without showing the effects in our nephrocyte functional assays. This may reflect a relatively high level of sensitivity to disruption of Coenzyme Q synthesis and increased susceptibility to ROS in aging adult flies. Alternatively, reduced lifespan may in this case be due to CG10585 knockdown in cells other than nephrocytes, as the Dot enhancer driving RNAi expression is also active in some neurons (43). Figure 4B shows selected survival curves that illustrate the range of phenotypes induced in gene knockdown experiments. Mec2 gene silencing in nephrocytes was associated with 100% developmental lethality (no adult flies were produced). Zip knockdown was associated with nearly continuous, progressive adult mortality over the course of approximately 25 days. In the case of CG32105 gene silencing, significant adult mortality was not observed until after day 15, followed by steady and then precipitous die-off through day 30.
Figure 4.
Nephrocyte specific gene silencing effects on adult fly survival. (A) Effect of gene silencing on average adult life span. NS associated genes are listed with the corresponding Drosophila ortholog targeted by nephrocyte expression of inhibitory RNAi transgene. Lifespan was assessed in transgenic flies of genotype Dot-Gal4; UAS-GeneX-IR and Control flies of genotype Dot-Gal4. 50 flies of each genotype were maintained at 29 °C (to increase Gal4 driven RNAi transgene expression) and mortality was recorded every 48 hours until all flies were dead. Experiments were performed in triplicate. The results are presented as average life span (in days). The results are presented as SEM. Statistical significance (*) was defined as P < 0.05. Statistical significance (*) was defined as P < 0.05. Colored histograms correspond to example survival curves shown in B. (B) Example survival curves for flies expressing the indicated gene silencing constructs. The curves correspond to the colored histograms in (A). Mec2 gene silencing was developmentally lethal, with no adult emergence from the pupa stage.
Drosophila cindr gene silencing rescued by normal human CD2AP gene expression but not by a patient-derived mutant CD2AP-K301M allele
The CD2AP adapter protein is critical for SD formation and maintenance, and CD2AP mutations are associated with nephrotic syndrome. CD2AP knockout mice exhibit podocyte foot process defects and die from renal failure. CD2AP associates with the NPHS1 encoded protein nephrin, the major SD filter component, and the NPHS2 gene product podocin (39,41,44).
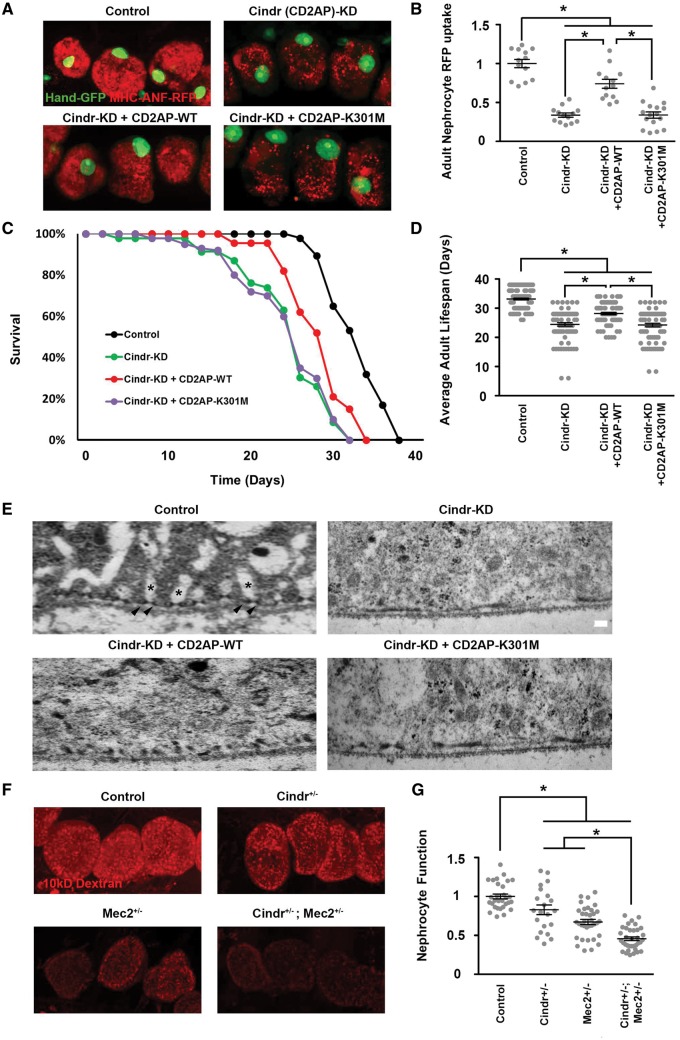
We tested the ability of a wild type (WT) human CD2AP transgene to rescue the ANF-RFP uptake deficiency induced by silencing of the Drosophila ortholog Cindr in nephrocytes. As shown in Figure 5A and B, the CD2AP-WT transgene expression in nephrocytes can significantly rescue the functional deficit in ANF-RFP uptake in a cell background of RNAi silenced endogenous Cindr gene expression. We extended our analysis to test CD2AP-K301M, a renal disease patient-derived mutant CD2AP allele (41), for the rescue of Cindr knockdown. As shown in Figure 5A and B the mutant allele did not rescue the ANF-RFP uptake defect. Furthermore, we showed that CD2AP-WT transgene expression (in Cindr silenced nephrocytes) significantly increased adult fly survival, but no rescue effect was observed with CD2AP-K301M mutant allele expression (Fig. 5C–D).
Figure 5.
Cindr is required for nephrocyte function, fly survival, and nephrocyte cell ultrastructure; Cindr gene silencing can be rescued by a normal but not a patient-derived mutant allele of human CD2AP; Cindr interacts with Mec2 in nephrocytes. (A). ANF-RFP uptake visualized by fluorescence microscopy in nephrocytes of Control flies (genotype Hand-GFP; Dot-Gal4), Cindr (CD2AP)-KD flies in which Cindr expression was silenced in nephrocytes with a Cindr targeting RNAi construct (genotype Hand-GFP; Dot-Gal4; UAS-Cindr-RNAi), Cindr-KD + CD2AP-WT flies in which a transgenic wild type allele of human CD2AP was expressed simultaneously with Cindr -targeting RNAi (genotype Hand-GFP; Dot-Gal4; UAS-Cindr-RNAi; UAS-CD2AP), and Cindr-KD + CD2AP-K301M flies in which a transgenic mutant allele of human CD2AP was expressed simultaneously with Cindr -targeting RNAi (genotype Hand-GFP; Dot-Gal4; UAS-Cindr-RNAi; UAS-CD2AP-K301M). The panels show merged ANF-RFP (54) and GFP (green, mostly nuclear) fluorescence. (B) Quantification of nephrocyte RFP levels relative to Control flies. For quantification, ≥20 nephrocytes were analysed from each of three female flies per genotype. The results are presented as mean ± s.e.m. Statistical significance (*) was defined as P < 0.05. (C) Adult fly survival curves illustrating effects on longevity of Cindr silencing and extent of rescue from expression of WT and mutant alleles of human CD2AP. 50 flies of each genotype were maintained at 29°C (to increase Gal4 driven RNAi transgene expression) and mortality was recorded every 48 hours until all flies were dead. Triplicate samples were analysed. (D) Average adult lifespan showing effects of Cindr silencing and rescue by WT and mutant alleles of human CD2AP. 50 flies of each genotype were maintained at 29°C (to increase Gal4 driven RNAi transgene expression) and mortality was recorded every 48 hours until all flies were dead. Experiments were performed in triplicate. The results are presented as mean ± s.e.m. Statistical significance (*) was defined as P < 0.05. (E) Transmission electron microscopy showing effects on nephrocyte NSD (arrowheads) and lacunar channel (*) ultrastructure of Cindr gene silencing and the extent of rescue from expression of WT and mutant alleles of human CD2AP. In Control nephrocytes NSDs were regularly and precisely spaced along the entire circumference of the cell and spanned the "“mouth”" of each lacunar channel. Silencing Cindr gene expression led to fusion of NSDs and the loss of lacunar channels. Expression of WT human CD2AP significantly rescued the ultrastructural defects, while the mutant CD2AP allele had very little effect. (F). Functional interaction between Cindr and Mec2 in nephrocytes demonstrated by Cindr+/-; Mec2+/- double heterozygote synergistic enhancement of deleterious effect on uptake of fluorescent Dextran particles in comparison to Cindr+/- or Mec2+/- single heterozygotes. (G). Quantitative analysis of fluorescent Dextran levels in nephrocytes of the indicated genotype, expressed relative to Control. For quantification, ≥20 nephrocytes were analysed from each of three female flies per genotype. The results are presented as mean ± s.e.m. Statistical significance (*) was defined as P < 0.05.
Cindr gene silencing led to the effacement of normal NSD and lacunar channel ultrastructure
We used transmission electron microscopy to determine if defects in uptake and accumulation of ANF-RFP and AgNO3 due to Cindr gene silencing were associated with abnormalities in the characteristic nephrocyte NSD and lacunar channel ultrastructure (29,31). As shown in Figure 5E, in normal Control nephrocytes the lacunar channels and NSDs are regularly spaced along the circumference of the cell. A single NSD is located at the "mouth" of each lacunar channel. Silencing of Cindr expression led to loss of channels and dramatically altered NSD morphology. In place of regularly spaced, discreet pairs of opposing NSD structural units, we observed dysmorphic, flattened structures tending to fuse together into semi-continuous wavelike forms, and distinct lacunar channel ultrastructure was not evident. Expression of the CD2AP-WT transgene partially restored NSD pairing and regular spacing, while the mutant CD2AP-K301M allele failed to rescue the ultrastructural defects induced by silencing of Cindr (Fig. 5E).
Cindr interacts with Mec2 in drosophila nephrocytes
We used a genetic approach to investigate interactions between Cindr and other NSD proteins, informed by reported interactions between CD2AP and podocin in the podocyte SD (44). Figure 5F and G show that nephrocytes of Cindr+/- and Mec2+/- heterozygotes are each moderately though significantly impaired in uptake of fluorescent 10 kD Dextran particles. The nephrocyte function of Cindr+/-; Mec2+/- double heterozygotes are, in comparison to either single heterozygote, much more significantly impaired, providing in vivo evidence that Cindr and Mec2, like their human orthologs CD2AP and Podocin, interact and function together.
Discussion
Rapid advances in genomic sequencing technologies have led to the identification of large numbers of potential disease genes. To fully capitalize on these advances, a high-throughput animal model is required to provide disease-relevant functional data. As described here, we have established the Drosophila nephrocyte system as such a high-throughput functional validation system for nephrotic syndrome, an important type of renal disease. As proof-of-principle, we showed that 34 out of 40 known NS gene orthologs are required for nephrocyte function, using quantitative assays for protein uptake and sequestration of toxic AgNO3. Based on our observations approximately 85% of the identified NS genes have conserved roles in renal function from flies to humans, showing that Drosophila is a valuable simple animal model in which to investigate a majority of NS genes.
Of the 40 NS genes addressed in this study, 33 had previously been analysed for effects on kidney cell biology in mouse and/or zebrafish models. To our knowledge, the current study is the first published report of renal cell phenotypes induced by knockdown of NXF5, LMNA, PMM2, ALG1, COQ2, and ITGB4 gene homologs in an animal model system (ZMPSTE24 knockdown has also not previously been described in the context of model kidney studies, but we were unable to detect an Ste24a RNAi silencing induced phenotype in our assays). NXF5 mutations are associated with FSGS (42). Knockout of the mouse functional ortholog, Nxf7, was shown to impair memory but effects on kidney function were not examined (45). In mice, knockdown of Zmpste24 and Lmna caused a rapid aging phenotype (46) and deletion of Lmna led to growth delay and reduced body weight (47). Renal deficiencies, however, were not reported. Thus, a comprehensive functional analysis employing Drosophila nephrocytes has yielded novel observations on the physiological consequences of known NS gene silencing in vivo.
Furthermore, for 25 of the tested Drosophila genes, silencing in nephrocytes was associated with reduced adult fly lifespan, indicating that the compromised nephrocyte function was frequently associated with increased mortality. Effects on lifespan may be due to reduced nephrocyte function leading to deficiencies in reabsorption of proteins and recycling of metabolic resources and/or failure to sequester harmful metabolic waste products which eventually compromise fly health. Alternatively, nephrocyte physiological dysfunction may eventually render the nephrocyte itself harmful to the fly, for example through accumulation of ROS or other intermediaries of cellular stress. In either case, our observations suggest that Drosophila can indeed model pathogenic features of life threatening renal disease. Disparities between the severity of RFP and AgNO3 functional phenotypes and less severe effects on longevity may be explained by the optimal environmental conditions in which these flies were maintained, which are known to preserve their renal function. To test this hypothesis, we are currently testing the effects of gene silencing on adult survival for flies maintained under stress from toxin exposure.
In order to further confirm the function of a particular renal gene and exploit the nephrocyte model as a powerful in vivo experimental platform to explore the pathogenesis of NS in humans, we focused our attention on Cindr, the Drosophila ortholog of CD2AP, a well-studied monogenic risk factor for NS (39,41,48). We found that the diminished nephrocyte protein uptake induced by silencing Cindr was rescued by expression of a normal allele of human CD2AP, but not by a patient derived mutant allele. Differential rescue capabilities were also observed for adult fly lifespan, and nephrocyte cell ultrastructural features dependent upon CD2AP protein function. CD2AP has been reported to function as a linker protein associated with the podocyte SD where it has been shown to interact with both nephrin and podocin (44,48). We used genetics to show that in nephrocytes, Cindr interacts with Mec2, the Drosophila gene encoding the podocin homolog. This result confirms the similarities in organization of SD/NSD components at the molecular level in nephrocytes and podocytes, and extends earlier studies conducted in garland nephrocytes during Drosophila embryonic development (31).
Interestingly, six of the NS gene fly homologs tested by silencing in nephrocytes (Ptp10D, CG1674, Scra, Ste24a, CG10585, and Dally) did not induce functional deficiencies in our assays. In the case of Dally (the GPC5 homolog), this result was consistent with previous studies showing that GPC5 risk is associated with higher expression, and that podocyte-specific knockdown of gene expression actually confers resistance to cellular injury (49). Ste24a encodes a metalloprotease involved in the generation of mature lamin A, and mutations in the human homolog ZMPSTE24 are associated with FSGS (50). It has been reported that disease severity is correlated with levels of residual ZMPSTE24 enzyme activity, with complete loss-of-function alleles associated with the most severe disease manifestations (51). It is not unreasonable, therefore, to suggest that the degree of silencing of Ste24a by RNAi in nephrocytes may have been below the level required for detection in our assays. That lamin A production is important in nephrocytes as it is in podocytes, however, was clearly shown by the functional deficits induced by silencing of the LMNA homolog lam.
The complex cytoarchitecture of the podocyte and the cell’s ability to withstand the forces associated with blood filtration are critically supported by the actin cytoskeleton (52). Podocyte injury leads to cytoskeletal disruptions that commonly result in foot process effacement and proteinuria (53,54). Although the Drosophila nephrocyte is not subjected to the rigorous physical challenges associated with blood filtration under high pressure, we found that seven of nine homologs of NS genes associated with actin cytoskeletal dynamics displayed knock down phenotypes in our functional assays. RhoGDI and Kank gene silencing had previously been shown to impair nephrocyte function and disrupt NSD and the lacunar channel structure (12,26). These observations underscore the functional, structural, and regulatory linkages between slit diaphragm filtration and the actin cytoskeleton. The Drosophila nephrocyte model system can be a highly useful experimental platform to dissect in vivo and at high resolution the molecular interactions underlying these linkages.
Mutations in the genes COQ2, COQ6, PDSS2, and ADCK4 encoding enzymes involved in the biosynthesis of Coenzyme Q10 (CoQ10) are associated with NS (14,15,55,56). CoQ10 plays an essential role in the mitochondrial respiratory chain and protects against damage from reactive oxygen species (ROS) (56–58). Our observations indicate that nephrocyte functions are also susceptible to CoQ10 deficiency induced by knockdown of biosynthetic enzyme activity. Additional ongoing studies indicate that these phenotypes are associated with elevated ROS and cytotoxicity (data not shown).
It has recently been reported that collagen IV mutations are frequently associated with FSGS and SRNS (59). We observed that nephrocyte specific silencing of Drosophila Col4A1 induced functional defects and shortened fly average lifespan. The critical importance of extracellular matrix and actin cytoskeleton in both nephrocyte and podocyte function was further emphasized by our finding that silencing of the Drosophila integrin genes Mew and Mys (homologs of human ITGA3 and ITGB4, respectively) led to severe functional defects and lifespan reduction. Integrins are known to be essential in podocytes, and integrin gene mutations are associated with nephrotic syndrome (60).
A number of genes associated with NS play roles in the nucleus, including transcription and RNA export. We observed that Drosophila homologs of these genes are essential for nephrocyte function. For example, the homeobox transcription factor LMX1B is involved in the maintenance of the actin cytoskeleton and regulation of the SD, and LMX1B haploinsufficiency is associated with NS (61,62). We observed strong functional phenotypes and reduced fly lifespan induced by nephrocyte specific silencing of the Drosophila homolog CG32105.
Overall, our findings show that of human genes known to be associated with NS, a majority of them play conserved roles in renal function from flies to humans. We conclude that the Drosophila nephrocyte can be used both to elucidate clinically relevant molecular mechanisms underlying the pathogenesis of most monogenic forms of NS, and efficiently generate patient-specific in vivo models of genetic renal diseases caused by specific mutations.
Materials and Methods
Fly strains
Flies were reared on standard fly food at room temperature, or at 29 °C for experiments involving Gal4 (to boost the activity of the yeast-derived Gal4 protein). Flies carrying Hand-GFP, Dot-Gal4, MHC-ANF-RFP transgenes were described previously (30,63). All RNAi based gene silencing lines were obtained from the Bloomington Drosophila Stock Center except for the Sns targeting line, which was from the Vienna Drosophila Resource Center. Flies with multiple transgenes were generated using standard genetic methods.
DNA cloning and generation of transgenic fly strains
A normal human CD2AP cDNA was obtained from OriGene, which encodes the common 639 a.a. isoform with NCBI reference sequence NP_036252. The cDNA of CD2AP-K301M mutation identified from a NS patient (41) was generated using QuikChange Site-Directed Mutagenesis Kit (Clontech) from a wild-type CD2AP cDNA. This wild-type and mutant CD2AP cDNA are only different at the lysine 301. To generate UAS-CD2AP and UAS-CD2AP-K301M constructs, the above cDNAs were cloned into the pUAST vector and introduced into the germ cells of flies by standard P element-mediated germ line transformation.
RFP uptake assay
Flies of the appropriate genotypes were allowed to lay eggs at 25 °C. One day after egg laying, embryos were shifted to 29 °C. RFP uptake by pericardial nephrocytes was assessed in adult flies one day post-emergence by dissecting heart tissues into Drosophila Schneider’s Medium (ThermoFisher) and examining the cells by fluorescence microscopy. For quantification, ≥20 nephrocytes were analysed from each of three flies per genotype.
AgNO3 uptake assay
Flies of the appropriate genotype were allowed to lay eggs on standard apple juice plates for 24 hours. Freshly emerged first instar larvae were transferred to agar-only plates supplemented with a regular yeast paste containing AgNO3 (2.0 g yeast in 3.5 ml 0.0005% AgNO3 solution) and allowed to develop at 29 °C until adulthood. AgNO3 uptake by pericardial nephrocytes was assessed in adult flies one day post-emergence by dissecting heart tissues into Drosophila Schneider’s Medium (ThermoFisher) and examining the cells by phase contrast microscopy. For quantification, ≥20 nephrocytes were analysed from each of three female flies per genotype.
Survival assay
Flies of the appropriate genotype were allowed to lay eggs at 25 °C. One day after egg collection, embryos were shifted to 29 °C and animals were maintained at that temperature throughout subsequent development and adulthood. Within one day of emergence, a total of 50 adult male flies of a given genotype were transferred to fresh vials at ≤15 per vial. Triplicate samples were analysed. Vials were checked and mortality recorded every 48 hours until all flies were dead.
Dextran uptake assay
Flies of the appropriate genotypes were allowed to lay eggs at 25 °C. One day after egg laying embryos were shifted to 29 °C. Dextran uptake by pericardial nephrocytes was assessed ex vivo in adult flies one-day post-emergence by dissecting heart tissues into Drosophila Schneider’s Medium (Thermo Fisher) and examining the cells by fluorescence microscopy after 20 min incubation with AlexaFluo568-Dextran (10 kD, 0.05 mg/ml). For quantification, ≥20 nephrocytes were analysed from each of three flies per genotype.
Light microscopy and confocal imaging
Drosophila tissues were dissected and fixed for 10 minutes in 4% paraformaldehyde in phosphate-buffered saline (PBS). Confocal imaging was performed with a Zeiss ApoTome.2 microscope using a 20× Plan-Apochromat 0.8 N.A. air objective. For quantitative image comparisons common settings were chosen to avoid oversaturation. ImageJ Software Version 1.49 was used for image processing.
Statistical analysis
Statistical tests were performed using PAST.exe software (http://�folk.uio.no/ohammer/past/index.html) unless otherwise noted. Sample errors are given as standard error of the mean (s.e.m). Data were first tested for normality by using the Shapiro-Wilk test (α = 0.05). Normally distributed data were analysed either by Student's t-test (two groups) and Bonferroni comparison to adjust the P value or by a one-way analysis of variance followed by a Tukey-Kramer post-test for comparing multiple groups. Non-normal distributed data were analysed by either a Mann–Whitney test (two groups) and Bonferroni comparison to adjust P value or a Kruskal-Wallis H-test followed by a Dunn's test for comparisons between multiple groups. Statistical significance was defined as P < 0.05.
TEM
TEM was carried out using established, standard procedures (29). Briefly, flies of the indicated genotype were fixed with Sorensen phosphate buffer containing 4% paraformaldehyde and 2.5% glutaraldehyde. The processed samples were analysed using a Philips CM100 TEM.
Acknowledgements
We thank the Bloomington Stock Center and the Vienna Drosophila Resource Center for providing fly stocks.
Conflict of Interest statement. None declared.
Funding
Z.H. is supported by the National Institute of Health (NIH) R01 grant DK098410. P.E.R is supported by NIH R01 grants DK49419, DK103564, DK108368.
References
- 1. Sadowski C.E., Lovric S., Ashraf S., Pabst W.L., Gee H.Y., Kohl S., Engelmann S., Vega-Warner V., Fang H., Halbritter J., et al. (2015) A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J. Am. Soc. Nephrol., 26, 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hildebrandt F. (2010) Genetic kidney diseases. Lancet, 375, 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vivante A., Kohl S., Hwang D.Y., Dworschak G.C., Hildebrandt F. (2014) Single-gene causes of congenital anomalies of the kidney and urinary tract (CAKUT) in humans. Pediatr. Nephrol., 29, 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Devuyst O., Knoers N.V., Remuzzi G., Schaefer F.. and Board of the Working Group for Inherited Kidney Diseases of the European Renal Association and European Dialysis and Transplant Association (2014) Rare inherited kidney diseases: challenges, opportunities, and perspectives. Lancet, 383, 1844–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vivante A., Hildebrandt F. (2016) Exploring the genetic basis of early-onset chronic kidney disease. Nat. Rev. Nephrol., 12, 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smoyer W.E., Mundel P. (1998) Regulation of podocyte structure during the development of nephrotic syndrome. J. Mol. Med., 76, 172–183. [DOI] [PubMed] [Google Scholar]
- 7. Lovric S., Ashraf S., Tan W., Hildebrandt F. (2015) Genetic testing in steroid-resistant nephrotic syndrome: when and how?. Nephrol. Dial. Transplant, 31, 1802–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Greka A., Mundel P. (2012) Cell biology and pathology of podocytes. Ann. Rev. Physiol., 74, 299–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Greka A. (2016) Human genetics of nephrotic syndrome and the quest for precision medicine. Curr. Opin. Nephrol. Hypertens, 25, 138–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boute N., Gribouval O., Roselli S., Benessy F., Lee H., Fuchshuber A., Dahan K., Gubler M.C., Niaudet P., Antignac C. (2000) NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat. Gen., 24, 349–354. [DOI] [PubMed] [Google Scholar]
- 11. Kestila M., Lenkkeri U., Mannikko M., Lamerdin J., McCready P., Putaala H., Ruotsalainen V., Morita T., Nissinen M., Herva R., et al. (1998) Positionally cloned gene for a novel glomerular protein–nephrin–is mutated in congenital nephrotic syndrome. Mol. Cell, 1, 575–582. [DOI] [PubMed] [Google Scholar]
- 12. Gee H.Y., Saisawat P., Ashraf S., Hurd T.W., Vega-Warner V., Fang H., Beck B.B., Gribouval O., Zhou W., Diaz K.A., et al. (2013) ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J. Clin. Invest., 123, 3243–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kambham N., Tanji N., Seigle R.L., Markowitz G.S., Pulkkinen L., Uitto J., D’Agati V.D. (2000) Congenital focal segmental glomerulosclerosis associated with beta4 integrin mutation and epidermolysis bullosa. Am. J. Kidney Dis., 36, 190–196. [DOI] [PubMed] [Google Scholar]
- 14. Ashraf S., Gee H.Y., Woerner S., Xie L.X., Vega-Warner V., Lovric S., Fang H., Song X., Cattran D.C., Avila-Casado C., et al. (2013) ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J. Clin. Invest., 123, 5179–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heeringa S.F., Chernin G., Chaki M., Zhou W., Sloan A.J., Ji Z., Xie L.X., Salviati L., Hurd T.W., Vega-Warner V. (2011) COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J. Clin. Invest., 121, 2013–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ovunc B., Otto E.A., Vega-Warner V., Saisawat P., Ashraf S., Ramaswami G., Fathy H.M., Schoeb D., Chernin G., Lyons R.H., et al. (2011) Exome sequencing reveals cubilin mutation as a single-gene cause of proteinuria. J. Am. Soc. Nephrol., 22, 1815–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Montgomery E., Sayer J.A., Baines L.A., Hynes A.M., Vega-Warner V., Johnson S., Goodship J.A., Otto E.A. (2015) Novel compound heterozygous mutations in AMN cause Imerslund-Grasbeck syndrome in two half-sisters: a case report. BMC Med. Genet., 16, 35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Inoue K., Ishibe S. (2015) Podocyte endocytosis in the regulation of the glomerular filtration barrier. Am. J. Physiol. Renal Physiol., 309, F398–F405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Winn M.P., Conlon P.J., Lynn K.L., Farrington M.K., Creazzo T., Hawkins A.F., Daskalakis N., Kwan S.Y., Ebersviller S., Burchette J.L., et al. (2005) A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science, 308, 1801–1804. [DOI] [PubMed] [Google Scholar]
- 20. He B., Ebarasi L., Zhao Z., Guo J., Ojala J.R., Hultenby K., De Val S., Betsholtz C., Tryggvason K. (2014) Lmx1b and FoxC combinatorially regulate podocin expression in podocytes. J. Am. Soc. Nephrol., 25, 2764–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bier E. (2005) Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet., 6, 9–23. [DOI] [PubMed] [Google Scholar]
- 22. Denholm B., Skaer H. (2009) Bringing together components of the fly renal system. Curr. Opin. Genet. Dev., 19, 526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dow J.A., Romero M.F. (2010) Drosophila provides rapid modeling of renal development, function, and disease. Am. J. Physiol. Renal Physiol., 299, F1237–F1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simons M., Huber T.B. (2009) Flying podocytes. Kidney Int., 75, 455–457. [DOI] [PubMed] [Google Scholar]
- 25. Na J., Cagan R. (2013) The Drosophila nephrocyte: back on stage. J. Am. Soc. Nephrol., 24, 161–163. [DOI] [PubMed] [Google Scholar]
- 26. Gee H.Y., Zhang F., Ashraf S., Kohl S., Sadowski C.E., Vega-Warner V., Zhou W., Lovric S., Fang H., Nettleton M., et al. (2015) KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J. Clin. Invest., 125, 2375–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reiter L.T., Potocki L., Chien S., Gribskov M., Bier E. (2001) A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res., 11, 1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chien S., Reiter L.T., Bier E., Gribskov M. (2002) Homophila: human disease gene cognates in Drosophila. Nucleic Acids Res., 30, 149–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang F., Zhao Y., Chao Y., Muir K., Han Z. (2013) Cubilin and amnionless mediate protein reabsorption in Drosophila nephrocytes. J. Am. Soc. Nephrol., 24, 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang F., Zhao Y., Han Z. (2013) An in vivo functional analysis system for renal gene discovery in Drosophila pericardial nephrocytes. J. Am. Soc. Nephrol., 24, 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weavers H., Prieto-Sanchez S., Grawe F., Garcia-Lopez A., Artero R., Wilsch-Brauninger M., Ruiz-Gomez M., Skaer H., Denholm B. (2009) The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature, 457, 322–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhuang S., Shao H., Guo F., Trimble R., Pearce E., Abmayr S. (2009) Sns and Kirre, the Drosophila orthologs of Nephrin and Neph1, direct adhesion, fusion and formation of a slit diaphragm-like structure in insect nephrocytes. Development, 136, 2335–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E. (2011) An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics, 12, 357.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bierzynska A., Soderquest K., Koziell A. (2014) Genes and podocytes - new insights into mechanisms of podocytopathy. Front. Endocrinol., 5, 226.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ni J.Q., Markstein M., Binari R., Pfeiffer B., Liu L.P., Villalta C., Booker M., Perkins L., Perrimon N. (2008) Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat. Methods, 5, 49–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ni J.Q., Zhou R., Czech B., Liu L.P., Holderbaum L., Yang-Zhou D., Shim H.S., Tao R., Handler D., Karpowicz P. (2011) A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods, 8, 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brand A.H., Perrimon N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 38. Kim J.M., Wu H., Green G., Winkler C.A., Kopp J.B., Miner J.H., Unanue E.R., Shaw A.S. (2003) CD2-associated protein haploinsufficiency is linked to glomerular disease susceptibility. Science, 300, 1298–1300. [DOI] [PubMed] [Google Scholar]
- 39. Shih N.Y., Li J., Karpitskii V., Nguyen A., Dustin M.L., Kanagawa O., Miner J.H., Shaw A.S. (1999) Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science, 286, 312–315. [DOI] [PubMed] [Google Scholar]
- 40. Lowik M., Levtchenko E., Westra D., Groenen P., Steenbergen E., Weening J., Lilien M., Monnens L., van den Heuvel L. (2008) Bigenic heterozygosity and the development of steroid-resistant focal segmental glomerulosclerosis. Nephrol. Dial. Transplant, 23, 3146–3151. [DOI] [PubMed] [Google Scholar]
- 41. Gigante M., Pontrelli P., Montemurno E., Roca L., Aucella F., Penza R., Caridi G., Ranieri E., Ghiggeri G.M., Gesualdo L. (2009) CD2AP mutations are associated with sporadic nephrotic syndrome and focal segmental glomerulosclerosis (FSGS). Nephrol. Dial. Transplant, 24, 1858–1864. [DOI] [PubMed] [Google Scholar]
- 42. Esposito T., Lea R.A., Maher B.H., Moses D., Cox H.C., Magliocca S., Angius A., Nyholt D.R., Titus T., Kay T., et al. (2013) Unique X-linked familial FSGS with co-segregating heart block disorder is associated with a mutation in the NXF5 gene. Hum. Mol. Genet., 22, 3654–3666. [DOI] [PubMed] [Google Scholar]
- 43. Krupp J.J., Billeter J.C., Wong A., Choi C., Nitabach M.N., Levine J.D. (2013) Pigment-dispersing factor modulates pheromone production in clock cells that influence mating in drosophila. Neuron, 79, 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwarz K., Simons M., Reiser J., Saleem M.A., Faul C., Kriz W., Shaw A.S., Holzman L.B., Mundel P. (2001) Podocin, a raft-associated component of the glomerular slit diaphragm, interacts with CD2AP and nephrin. J. Clin. Invest., 108, 1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vanmarsenille L., Verbeeck J., Belet S., Roebroek A.J., Van de Putte T., Nevelsteen J., Callaerts-Vegh Z., D’Hooge R., Marynen P., Froyen G. (2013) Generation and characterization of an Nxf7 knockout mouse to study NXF5 deficiency in a patient with intellectual disability. PloS One, 8, e64144.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Varela I., Cadinanos J., Pendas A.M., Gutierrez-Fernandez A., Folgueras A.R., Sanchez L.M., Zhou Z., Rodriguez F.J., Stewart C.L., Vega J.A., et al. (2005) Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature, 437, 564–568. [DOI] [PubMed] [Google Scholar]
- 47. Ruan J., Liu X.G., Zheng H.L., Li J.B., Xiong X.D., Zhang C.L., Luo C.Y., Zhou Z.J., Shi Q., Weng Y.G. (2014) Deletion of the lmna gene induces growth delay and serum biochemical changes in C57BL/6 mice. Asian-Australas. J. Anim. Sci., 27, 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shih N.Y., Li J., Cotran R., Mundel P., Miner J.H., Shaw A.S. (2001) CD2AP localizes to the slit diaphragm and binds to nephrin via a novel C-terminal domain. Am. J. Pathol., 159, 2303–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Okamoto K., Tokunaga K., Doi K., Fujita T., Suzuki H., Katoh T., Watanabe T., Nishida N., Mabuchi A., Takahashi A. (2011) Common variation in GPC5 is associated with acquired nephrotic syndrome. Nat. Genet., 43, 459–463. [DOI] [PubMed] [Google Scholar]
- 50. Agarwal A.K., Zhou X.J., Hall R.K., Nicholls K., Bankier A., Van Esch H., Fryns J.P., Garg A. (2006) Focal segmental glomerulosclerosis in patients with mandibuloacral dysplasia owing to ZMPSTE24 deficiency. J. Investig. Med., 54, 208–213. [DOI] [PubMed] [Google Scholar]
- 51. Barrowman J., Wiley P.A., Hudon-Miller S.E., Hrycyna C.A., Michaelis S. (2012) Human ZMPSTE24 disease mutations: residual proteolytic activity correlates with disease severity. Hum. Mol. Genet., 21, 4084–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saleem M.A., Zavadil J., Bailly M., McGee K., Witherden I.R., Pavenstadt H., Hsu H., Sanday J., Satchell S.C., Lennon R., et al. (2008) The molecular and functional phenotype of glomerular podocytes reveals key features of contractile smooth muscle cells. Am. J. Physiol Renal Physiol., 295, F959–F970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang L., Ellis M.J., Gomez J.A., Eisner W., Fennell W., Howell D.N., Ruiz P., Fields T.A., Spurney R.F. (2012) Mechanisms of the proteinuria induced by Rho GTPases. Kidney Int., 81, 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schell C., Baumhakl L., Salou S., Conzelmann A.C., Meyer C., Helmstadter M., Wrede C., Grahammer F., Eimer S., Kerjaschki D., et al. (2013) N-wasp is required for stabilization of podocyte foot processes. J. Am. Soc. Nephrol., 24, 713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lopez L.C., Schuelke M., Quinzii C.M., Kanki T., Rodenburg R.J., Naini A., Dimauro S., Hirano M. (2006) Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am. J. Hum. Genet., 79, 1125–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Diomedi-Camassei F., Di Giandomenico S., Santorelli F.M., Caridi G., Piemonte F., Montini G., Ghiggeri G.M., Murer L., Barisoni L., Pastore A., et al. (2007) COQ2 nephropathy: a newly described inherited mitochondriopathy with primary renal involvement. J. Am. Soc. Nephrol., 18, 2773–2780. [DOI] [PubMed] [Google Scholar]
- 57. Quinzii C.M., Lopez L.C., Gilkerson R.W., Dorado B., Coku J., Naini A.B., Lagier-Tourenne C., Schuelke M., Salviati L., Carrozzo R., et al. (2010) Reactive oxygen species, oxidative stress, and cell death correlate with level of CoQ10 deficiency. Faseb J., 24, 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ozaltin F. (2014) Primary coenzyme Q10 (CoQ 10) deficiencies and related nephropathies. Pediatr. Nephrol,, 29, 961–969. [DOI] [PubMed] [Google Scholar]
- 59. Gast C., Pengelly R.J., Lyon M., Bunyan D.J., Seaby E.G., Graham N., Venkat-Raman G., Ennis S. (2016) Collagen (COL4A) mutations are the most frequent mutations underlying adult focal segmental glomerulosclerosis. Nephrol. Dial. Transplant, 31, 961–970. [DOI] [PubMed] [Google Scholar]
- 60. He Y., Balasubramanian M., Humphreys N., Waruiru C., Brauner M., Kohlhase J., O’Reilly R., Has C. (2016) Intronic ITGA3 Mutation Impacts Splicing Regulation and Causes Interstitial Lung Disease, Nephrotic Syndrome, and Epidermolysis Bullosa. J. Invest. Dermatol., 136, 1056–1059. [DOI] [PubMed] [Google Scholar]
- 61. Boyer O., Woerner S., Yang F., Oakeley E.J., Linghu B., Gribouval O., Tete M.J., Duca J.S., Klickstein L., Damask A.J., et al. (2013) LMX1B mutations cause hereditary FSGS without extrarenal involvement. J. Am. Soc. Nephrol., 24, 1216–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Konomoto T., Imamura H., Orita M., Tanaka E., Moritake H., Sato Y., Fujimoto S., Harita Y., Hisano S., Yoshiura K., et al. (2016) Clinical and histological findings of autosomal dominant renal-limited disease with LMX1B mutation. Nephrology, 21, 765–773. [DOI] [PubMed] [Google Scholar]
- 63. Han Z., Olson E.N. (2005) Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development, 132, 3525–3536. [DOI] [PubMed] [Google Scholar]