Abstract
Filamins are a family of actin-binding proteins responsible for diverse biological functions in the context of regulating actin dynamics and vesicle trafficking. Disruption of these proteins has been implicated in multiple human developmental disorders. To investigate the roles of different filamin isoforms, we focused on FlnA and FlnB interactions in the cartilage growth plate, since mutations in both molecules cause chondrodysplasias. Current studies show that FlnA and FlnB share a common function in stabilizing the actin cytoskeleton, they physically interact in the cytoplasm of chondrocytes, and loss of FlnA enhances FlnB expression of chondrocytes in the growth plate (and vice versa), suggesting compensation. Prolonged FlnB loss, however, promotes actin-stress fiber formation following plating onto an integrin activating substrate whereas FlnA inhibition leads to decreased actin formation. FlnA more strongly binds RhoA, although both filamins overlap with RhoA expression in the cell cytoplasm. FlnA promotes RhoA activation whereas FlnB indirectly inhibits this pathway. Moreover, FlnA loss leads to diminished expression of β1-integrin, whereas FlnB loss promotes integrin expression. Finally, fibronectin mediated integrin activation has been shown to activate RhoA and activated RhoA leads to stress fiber formation and cell spreading. Fibronectin stimulation in null FlnA cells impairs enhanced spreading whereas FlnB inhibited cells show enhanced spreading. While filamins serve a primary static function in stabilization of the actin cytoskeleton, these studies are the first to demonstrate a dynamic and antagonistic relationship between different filamin isoforms in the dynamic regulation of integrin expression, RhoGTPase activity and actin stress fiber remodeling.
Introduction
Filamins comprise a family of actin-binding proteins responsible for diverse biological functions. In general, they are comprised of an N-terminal actin-binding domain, followed by immunoglobulin-like repeat domains that form a receptor binding region at the C-terminus. This structure allows for receptor activation and transduction of signals onto the actin cytoskeleton, thereby directing various cell functions including membrane stability, protrusion, and motility (1,2).The three members of the filamin family of proteins (Filamin A, B, and C) share a high degree of homology between the conserved exon/intron structure (3). Moreover, previous studies showed that Filamin A (FlnA) and Filamin B (FlnB) physically interact and heterodimerize, potentially suggesting a shared mechanism with which to regulate each other’s function (4). Disruption of these proteins has been shown to give rise to multiple human developmental disorders. Humans harboring mutations in the Filamin A are known to develop a wide variety of disorders, including periventricular heterotopia (malformation of brain development), otopalatodigital syndrome and Melnick-Needles syndrome. However, more recent work has also demonstrated problems in skeletal, cardiac, pulmonary, dermal, and gastrointestinal development (5,6). Recessively and dominantly inherited mutations in FLNB can result in dwarfism and skeletal dysplasia with joint dislocations, respectively (7). Filamin C (FlnC) defects lead to an underlying myopathy (8). These varied phenotypes reiterate the broad and important role that filamins and actin play in both development and maintenance of numerous cell types. A fundamental question exists as to whether the different filamins play similar roles in different organ systems or whether each filamin gene subserves specific functions in a shared pathway. In the current studies, we focus on FlnA and FlnB because loss of either protein results in skeletal defects, with both shared as well as distinct bone phenotypes (7,9). We find that FlnA and FlnB are broadly expressed in multiple organ systems although FlnA is more highly expressed in certain tissues, the two proteins physically interact to form heterodimers, and they share overlapping expression with cytoplasmic RhoA in chondrocytes. Both filamin proteins share overlapping static functions by stabilizing the actin cytoskeleton in unstimulated chondrocytes. Loss of expression of one filamin isoform leads to upregulation of the other, consistent with compensation. Actin assembly can be regulated by RhoGTPases and we find that FlnA more strongly binds RhoA GTPase than FlnB. While total RhoA levels are unchanged following FlnA/B inhibition, activated RhoA levels are increased with prolonged loss of FlnB and decreased with loss of FlnA. Moreover, loss of FlnA inhibits integrin expression and decreases stress fiber formation whereas FlnB knockdown promotes these processes. Finally, cell spreading (an indicator of RhoA activation and stress fiber formation) is impaired with loss of FlnA and promoted by loss of FlnB, after RhoA activation through fibronectin-integrin stimulation. Collectively, these findings suggest that FlnA physically binds to RhoA and upregulate its activity to affect downstream changes. While FlnB binds to RhoA to a lesser extent, it antagonizes RhoA activation though the formation of homo-dimers and hetero-dimers with FlnA. As a result, the dynamic interactions between FlnA and FlnB allow fine-tuning of integrin-dependent RhoA activation and subsequent cytoskeletal remodeling. These studies provide the first evidence for functional interactions between the two different filamin isoforms in regulating dynamic actin changes and giving rise to opposing cell spreading phenotypes after RhoA activation.
Results
FlnA and FlnB stabilize the actin cytoskeleton
Earlier studies have shown that FlnA/B, through their interactions with actin, anchor multiple cytoplasmic proteins and transmembrane receptors. Moreover, it has been suggested that their physical interactions with actin are essential for maintaining the mechanical stability of the actin cytoskeleton [1, 2]. To explore the functional significance of FlnA/B in chondrocytes, we inhibited FlnA/B expression in ATDC5 cells with shRNA (Supplementary Material, Fig. S1A). Acute simultaneous loss of both FlnA and FlnB resulted in smaller cell sizes with rounded morphologies, compared to control ATDC5 cells (Supplementary Material, Fig. S1B and C). These observations are consistent with the notion that FlnA/B share overlapping functions in the context of actin cytoskeleton stabilization, which is essential for maintenance of cell morphology (cortical actin) and locomotion.
FlnA and FlnB physically interact within the cytoplasm of chondrocytes
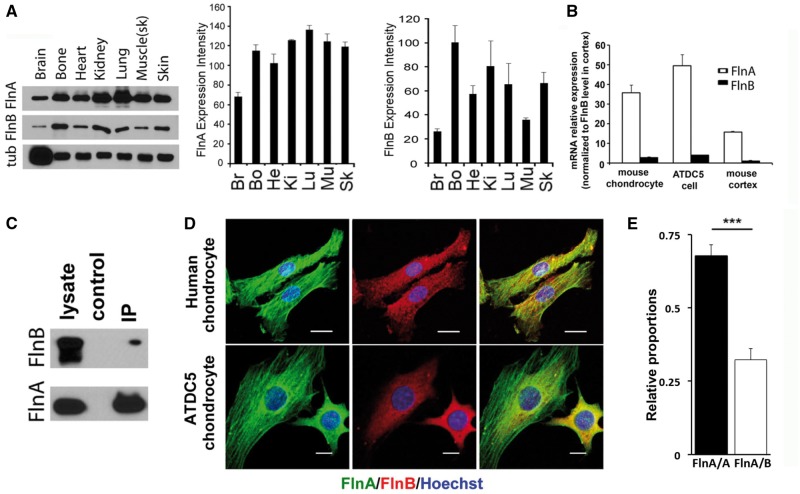
Our previous studies have shown that FlnA co-localizes with FlnB in murine brain and bone (4,10). However, FlnB levels are fairly low in the CNS and no gross neurological phenotype has been reported in humans, nor seen in the null FlnB mice. Endogenous expression levels in multiple tissues of postnatal day 1 wild type mice showed that FlnA appears to have relatively high levels of expression throughout various organ systems, whereas FlnB showed more variable expression, with highest levels seen in the bone and kidneys (Fig. 1A). To assess relative differences in expression between FlnA and FlnB within the same tissues, we compared expression levels between the two filamin genes by quantitative RT PCR. FlnA mRNA was more highly expressed relative to FlnB in mouse primary chondrocytes, ATDC5 chondrocytes, and cortex (Fig. 1B, Supplementary Material, Fig. S2). Using normalized protein standards for FlnA and FlnB, the FlnA protein expression is 1.35 fold that of FlnB in 293 cells. We had previously shown a direct interaction between FlnA and FlnB by yeast two hybrid, followed by co-immunoprecipitation in neural cells (4). Similar to these findings, endogenous FlnA could immunoprecipitate FlnB in ATDC5 mouse chondrocyte cells (Fig. 1C). To further characterize the interactions between FlnA and FlnB, we performed co-immunoprecipitation assays to evaluate levels of FlnA/A homodimers and FlnA/B heterodimers in 293 cells. Quantitative analysis showed that FlnA/A and FlnA/B exist at a ratio of approximately 2:1 (Fig. 1E, Supplementary Material, Fig. S3). Immunostaining of FlnA and FlnB in both human primary and mouse ATDC5 chondrocytes showed that the different filamin proteins had both overlapping and non-overlapping regions of expression (Fig. 1D). FlnA predominantly localized to both actin stress fibers and cell cytoplasm, while FlnB was mostly seen in the cytoplasm. Overall, these and prior findings suggest that the two filamin isoforms are found in the cytoplasm of chondrocytes, where the two potentially interact. Moreover, the more abundant and broader expression of FlnA, as well as its localization to the actin stress fibers, indicates that it might serve a more primary role in regulation of actin dynamics.
Figure 1.
FlnA and FlnB expression pattern, interaction and co-localization. (A) Western blot analyses shows widespread endogenous expression levels for both FlnA in multiple tissues of postnatal day 1 wild type mice. FlnB protein shows relatively higher expression levels in bone and kidneys. (B) Quantitative RT-PCR shows that mRNA expression for FlnA is higher in primary mouse chondrocytes, ATDC5 chondrocytes, and murine cerebral cortex compared to FlnB. (C) Endogenous FlnA immunoprecipitation by FlnB in mouse ATDC5 cells indicate that these two proteins interact in proliferating chondrocytes. Mouse anti-FlnA antibody and rabbit anti-FlnB were used to detect the input and immunoprecipitated filamin proteins. The specificity for these antibodies has been previously demonstrated in the null FlnA/B mice (10,36). (D) Fluorescent photomicrographs show overlapping expression for both FlnA and FlnB in the cell cytoplasm for primary human and mouse ATDC5 chondrocytes. FlnA also localizes to the actin stress fibers. Scale bars = 20 μm for human chondrocytes and 10 μm for mouse ATDC5 cells in D. (E) FlnA/A homo-dimers and FlnA/B heterodimers exist at an approximate ratio of 2:1 in HEK293 cells. *P≤0.05,**P < 0.01, ***P <0.001.
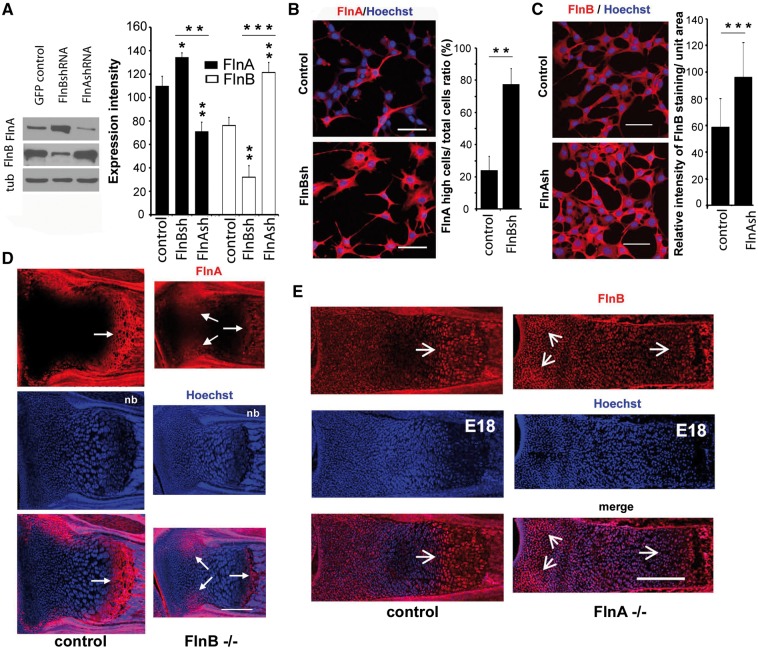
Figure 2.
FlnA regulates FlnB expression levels in chondrocytes and vice versa (A) Western blot and immunostaining analysis of FlnA and FlnB expression levels in FlnB knockdown and FlnA knockdown ATDC5 cells, respectively. FlnA protein levels are increased with FlnB inhibition. Conversely, FlnB levels are increased with FlnA inhibition. Levels are graphically quantified to the right. (B) Fluorescent photomicrograph demonstrates increased FlnA immunostaining (rhodamine) in stably transfected ATDC5 cells, lacking FlnB (FlnBsh), relative to control. Occasional control cells do show increased FlnA expression, with the vast majority exhibiting lower levels of staining. (C) Fluorescent photomicrograph demonstrates increased FlnA immunostaining (rhodamine) in stably transfected ATDC5 cells, lacking FlnB (FlnBsh), relative to control. (D) Fluorescent photomicrographs of the growth plate from the radius of newborn FlnB-/- mice. Immnostaining shows FlnA (rhodamine) expression is decreased in the hypertrophic zone (double arrows) but the increased in the proliferative zone (single arrow). (E) Similarly, FlnB expression (rhodamine) is decreased in the hypertrophic zone but increased in the proliferative zone of FlnA-/- mice. Scale bars in B and C =100 μm, D and E = 200 μm. Quantification is based on n ≥ 3 tissues sections/western blots per experiment and n > 50 cells.
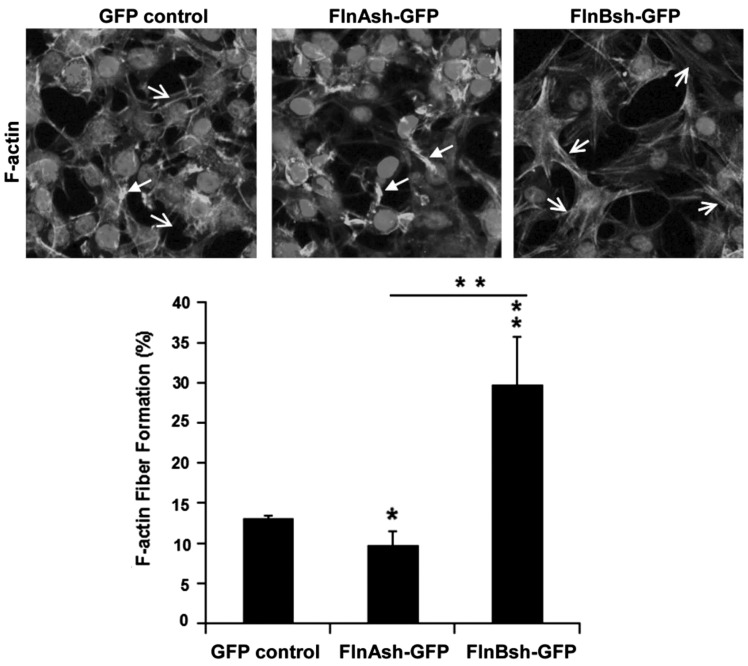
Figure 3.
FlnA/B regulates FlnB/A expression in mouse long bone and actin stress fiber formation. Loss of FlnA and FlnB affects F-actin/stress fiber formation in ATDC5 cells. Compared to control cells, FlnA inhibition induces less F-actin formation whereas FlnB inhibition leads to more stress fiber formation following plating on fibronectin. Solid arrows in the fluorescent photomicrographs represent the non-stress fiber structure of F-actin, and open arrows represent stress fibers. Cultures were stained with phalloidin (rhodamine) and counterstained with Hoechst nuclear stain. The findings are quantitatively summarized in the graph below. Scale bars in A =50 μm. Quantification based on n > 50 cells.
Reciprocal regulation of FlnA/B expression, localization, and actin stress fiber formation
To determine any potential functional significance to the observed physical interaction between FlnA/B, we examined whether inhibition of one filamin isoform effected expression of the other. We generated stable FlnA/B ATDC5 chondrocytes and then determined the protein levels of and expression patterns for FlnA and FlnB (11). Western blot analyses demonstrated that loss of FlnA induced upregulation of FlnB expression, and vice versa (Fig. 2A). In addition, immunostaining for these proteins showed a similar compensatory regulation, with more intense staining seen for FlnB following FlnA inhibition and conversely, stronger FlnA staining observed with FlnB inhibition (Fig. 2B and C). These observations would be consistent for a shared static role in maintaining actin stability.
We have previously shown that FlnB has a broader expression across the growth plate of long bones, whereas FlnA is more restricted to the hypertrophic zone (10). Based on the in vitro chondrocyte data, we asked whether loss of FlnA/B altered the expression patterns of the different filamin isoforms in the long bones. Within the radius of newborn FlnB-/- mice, FlnA expression levels were increased in the proliferative zone of the growth plate, but decreased at the hypertrophic zone (Fig. 2D). Our prior work has shown that loss of FlnB in the mesenchymal progenitors undergo a delay in differentiation (10). Thus at a given age compared to control, there is a reduction in the size of the hypertrophic zone and consequent decrease in FlnA expression as well. Similarly, FlnB expression levels in the radius of embryonic day 18 FlnA-/- mice (null FlnA mice are embryonic lethal) were increased in the proliferative zone of the growth plate, but decreased in the hypertrophic zone (Fig. 2E). The changes in FlnA/B protein levels following loss of FlnB/A expression, respectively, occurs within proliferating and differentiating chondrocytes. The reciprocal changes (loss of one filamin isoform leading to upregulation of the other) seen in the proliferating chrondrocytes within the proliferative zone are consistent with the western blot and immunocytochemical staining seen with the ATDC5 FlnA and FlnB knockdowns. Interpretation of changes in the hypertrophic zone is more complex in the null FlnA and FlnB mice as the loss of these actin-binding proteins affects the subsequent differentiation of the chondroprogenitors. Thus, changes in FlnA/B expression in the hypertrophic zone may reflect an indirect effect from altered proliferating chondrocyte development and progression through the proliferating and prehypertrophic zones (10–12), as well as direct effects of FlnA/B on the hypertrophic chondrocytes.
Filamins regulate actin dynamics, such that a possible consequence of altered filamin expression would be changes in actin-stress fiber formation after integrin stimulation by plating on extracellular matrix (13). We therefore examined actin-stress fiber formation in null FlnA and FlnB ATDC5 cells by phalloidin staining following plating of cells on fibronectin. Compared with control chondrocytes, loss of FlnA impaired F-actin stress fiber formation, whereas FlnB inhibition resulted in the enhanced stress fiber formation (Fig. 3A). Surprisingly, these studies indicate that although FlnA/B physically bind and appear to compensate for each other’s expression (which likely reflect a general role for filamins in stabilizing receptors to the actin cytoskeleton), they have opposing effects on possibly dynamic changes in actin stress fibers.
FlnA shows stronger binding interactions to the small RhoGTPases than FlnB
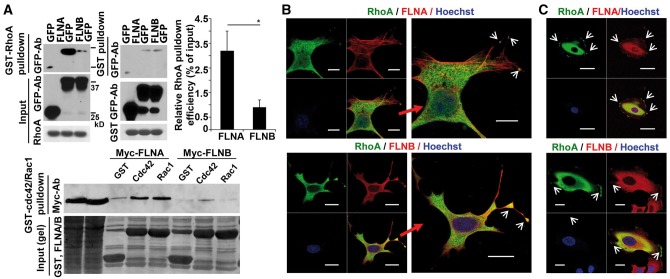
The small RhoGTPases (RhoA, Rac1 and Cdc42) physically bind to FlnA and direct actin stress fiber formation (14,15). To determine whether FlnA/B differentially regulate RhoGTPase function, we first performed immunoprecipitation studies to ask if FlnB shared similar physical interactions with the RhoGTPases, as seen with FlnA. Under identical conditions, no significant binding or very weak binding for any of the small RhoGTPases could be demonstrated with FlnB, whereas concurrently performed immunoprecipitations demonstrated interactions for RhoA, Cdc42 and Rac1 with FlnA (Fig. 4A). We then focused on FlnA/B interactions with RhoA, given that the small G protein has been specifically shown to guide chondrocyte differentiation (16–18). Activated RhoA but not activated Cdc42 or Rac1 also shows selective activation of Formin 1/2, novel FlnA/B effectors of actin nucleation (12). Immunostaining for FlnA/B and RhoA showed colocalization over the cell cytoplasm in ATDC5 chondrocytes (Fig. 4B) and human primary chondrocytes (Fig. 4C). The pattern of staining was similar to that seen with overlapping FlnA and FlnB expression in the cell cytoplasm and along the cell membrane. Collectively, these studies suggest that within chondrocytes, FlnA might serve as a primary regulator of RhoA activity given its stronger binding interaction with the small G protein. FlnB overlapping expression with both FlnA and RhoA, binding to FlnA but absence of strong binding to RhoA, suggest that it might modulate RhoA activity through FlnA.
Figure 4.
FlnA but not FlnB physically binds the small RhoGTPases. (A) Immunoprecipitation analyses between the small RhoGTPases and FLNA/B. Relative to FLNB, FLNA exhibits a much higher binding capacity for each of the small RhoGTPases (RhoA, Rac1, and Cdc42), as demonstrated by the levels of immunoprecipitated proteins. Levels are quantified graphically (n ≥ 3 independent experiments). (B,C) Fluorescent photomicrographs show that both filamins co-localize with RhoA in murine ATDC5 chondrocytes and human U2OS osteosarcoma cells, largely within the cytoplasm and peripheral cell membrane (arrowheads). Given the weak endogenous staining, RhoA localization and distribution was determined after transfection with GFP-RhoA. Molecular weight markers correspond to 25 and 30 kD. Scale bar = 10 μm.
Loss/gain of FlnA/B function causes opposite changes in RhoA activation
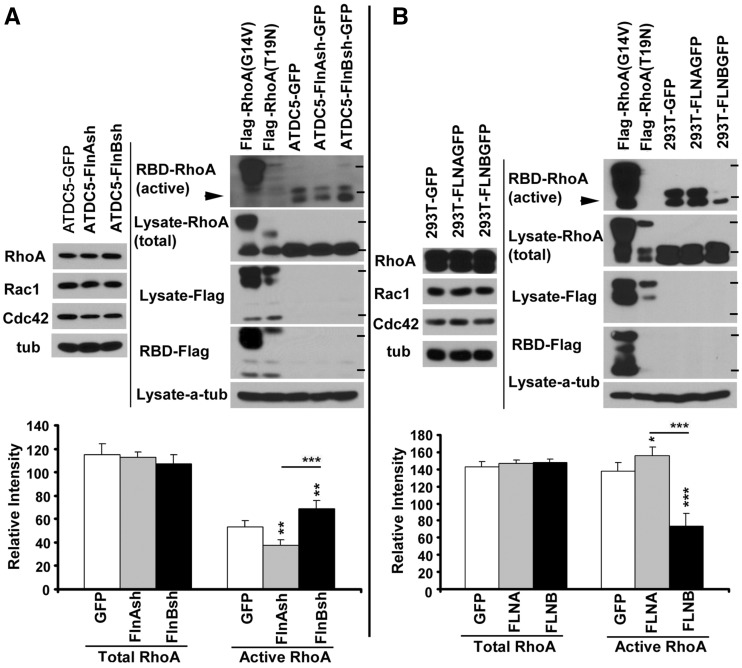
In order to characterize the effects of FlnA and FlnB function on RhoA activity, we quantified changes in total and active RhoA levels in filamin knockdown cells, as well as 293T cells, overexpressing these proteins. First, no significant changes in total levels of any of the Rho GTPases were seen with FlnA/B inhibition in ATDC5 chondrocytes or following overexpression of FlnA/B in 293T cells (Fig. 5A and B). Since RhoA activity is dependent on GDP to GTP conversion (19), we next examined changes in active RhoA GTP levels following FlnA/B inhibition and overexpression. Loss of FlnA caused decreased levels of active RhoA, whereas FlnB loss led to an opposite change with increased active RhoA levels (Fig. 5A). Similarly, overexpression of FlnA caused an increase in active RhoA levels, while FlnB overexpression decreased active RhoA (Fig. 5B). Taken in the context of the prior studies, these results are consistent with FlnA/B, having opposing effects on RhoA activation/inactivation.
Figure 5.
FlnA and FlnB differentially regulate RhoA activation. (A) Western blot analyses of total and activated RhoA following inhibition of FlnA/B. Prolonged loss of FlnA in stably transfected ATDC5 chondrocytes induces down-regulation of active RhoA levels (black arrowheads), while loss of FlnB induces up-regulation of active RhoA levels. Constitutively Flag-tagged active (RhoA G14V) and inactive (T19N) RhoA serve as controls. RhoA activity is assessed by pulldown using RBD (Rho binding domain), which interacts with RhoA-GTP. Following pulldown, blots are probed using RhoA antibody (or Flag antibody for the active/inactive RhoA) to assess activation levels. At times, two bands for activated RhoA are observed due to electrophoretic mobility. (B) In converse fashion, overexpression of FLNA causes up-regulation of active RhoA levels, while over-expression of FLNB causes down-regulation of active RhoA levels. HEK 293T cells are transfected with FLNA/B. 72 h post transfection, cells are harvested and RhoA activation levels assessed by RBD pulldown and blotting for RhoA. The statistical significance of the respective samples relative to control is indicated by asterisks on top of each bar. Asterisk localized above the line indicates statistical significance between samples. Molecular weight markers correspond to 25 and 30 kD. **P < 0.01, ***P <0.001 for n ≥ 3 independent experiments per variable.
Integrin dependent RhoA activation mediates cell spreading through FlnA/B
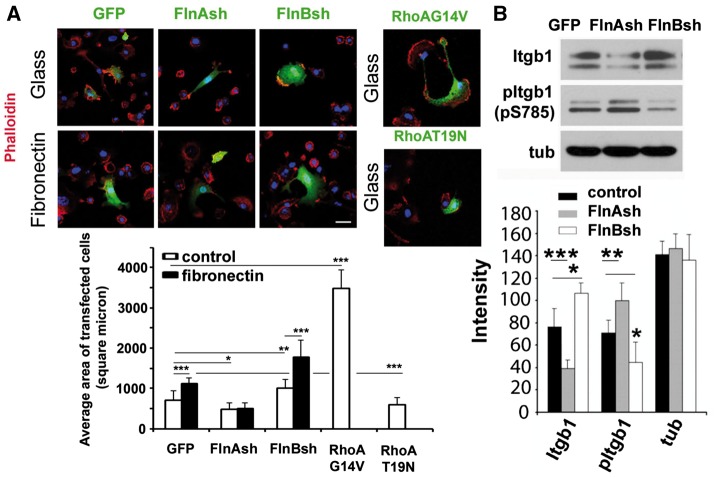
Integrin β1 binds FlnA/B, its activation occurs through extracellular matrix (ECM) signaling such as fibronectin, and it leads to RhoA activation. In general, RhoA activation leads to increased stress fiber formation and cell spreading (10,20,21). To address the dynamic effects on FlnA/B dependent RhoA activation on actin, we used fibronectin to activate integrin β1 and examine effects of FlnA/B loss on cell spreading. Consistent with prior reports (22), fibronectin stimulation of integrin (and presumptive RhoA activation leading to stress fiber formation) led to increased cell spreading of wild type mouse primary chondrocytes (n > 50 cells for in vitro studies, Fig. 6A and B). FlnA inhibited cells showed diminished spreading on either glass or fibronectin, much in the same manner as primary chondrocytes expressing constitutively inactive RhoA (T19N). Similarly, FlnB inhibited cells showed increased spreading on either glass or fibronectin, much in the same manner as primary chondrocytes expressing constitutively active RhoA (G14V). Collectively, these studies suggest that at a functional level, FlnA/B dynamically regulate RhoA activation and stress fiber formation, leading to opposite changes in cell spreading. Moreover, given that FlnB does not exhibit significant binding activity to RhoA, but does do so with FlnA, FlnB dependent changes in RhoA might potentially occur through FlnA.
Figure 6.
β1 integrin dependent activation of RhoA via FlnA/B regulates cell spreading. (A) Fluorescence photomicrographs demonstrate that fibronectin stimulation causes increased cell spreading in wildtype primary mouse chondrocytes. Cell spreading is diminished in FlnA knockdown primary mouse chondrocytes, regardless of fibronectin stimulation, consistent with impairment in RhoA activation. Conversely, increased cell spreading is seen with FlnB inhibited cells, following plating on either glass or fibronectin. FlnB knockdown cells display increased activated RhoA expression levels (see Fig. 5). Transient transfection of activated RhoA (G14V) leads to a similar pattern of spreading as seen with FlnB inhibition. Similar to the FlnA null cells and compared to constitutively active RhoA transfected cells, transfection of inactive RhoA (T19N) leads to impaired cell spreading. Changes in cell spreading area are quantified graphically for the different experimental variables. (B) Western blot demonstrates that loss of FlnA impairs β1-integrin expression levels but promotes expression of phospho- β1-integrin at serine 785. Phospho-integrin β1 (S785) promotes cell attachment but prevents spreading and migration (26). The opposite changes are seen with FlnB knockdown. *P≤0.05, **P≤ 0.01, ***P ≤ 0.001. Scale bar = 25 μm.
Integrin β1 mediated activation of the RhoA/Rock pathway mediates endocytosis (23,24). Conversely, endocytosis regulates the stability and subsequent degradation of cell membrane adhesion receptors such as integrins (25). Our recent work also suggests that FlnA and Formin regulate endocytosis and degradation of cell receptors (unpublished observations). We therefore examined the expression levels of integrin β1 following FlnA/B knockdown. Loss of FlnA leads to decreased overall levels of integrin β1 but increased phosphorylated integrin β1 at Ser785. Phospho-integrin β1 (S785) promotes cell attachment but prevents spreading and migration (26). FlnB knockdown leads to the opposite changes. Impaired RhoA activation, diminished integrin β1expression levels, and enhanced integrin β1 phosphorylation at (S785) are all consistent with the observed decrease in cell spreading and associated with FlnA loss. That FlnB loss is associated with the exact opposite cellular and molecular changes suggests that the two filamin proteins serve complementary (scaffolding support) but antagonistic (RhoA specific pathways) roles.
Discussion
Filamins are actin-binding proteins which transduce signals from their receptor binding domains to the actin cytoskeleton. Many studies have described various intracellular signaling molecules, receptors, ion channels, transcription factors, and cytoskeletal and adhesion proteins which bind filamins (particularly FlnA) and disrupt some aspect of development (27). These works have provided a generalized role for filamins in regulating mechanosensing and serving as a scaffolding for various molecules. However, little is known about the functional differences between the filamin isoforms. The current studies demonstrate that FlnA and FlnB both are essential in maintaining cell shape and morphology through stabilization of the actin cytoskeleton in the static state. Loss of either protein leads to reciprocal upregulation of the other protein to compensate and assist in this essential function. The two proteins, however, also bind and interact in chondrocytes, they have opposing effects on RhoA activation, and FlnA more strongly binds RhoA. Moreover, FlnA/B exhibit opposing effects on β1integrin receptor expression levels. Integrin stimulation has been shown to promote RhoA activation and cell spreading. This effect is diminished with loss of FlnA but enhanced with FlnB inhibition. The antagonistic roles for these two filamin isoforms in directing integrin and RhoA activation provide a mechanism for greater refinement and regulation of actin dependent mechanisms in the dynamic state.
Although FlnA/B lead to opposite effects in RhoA activation and integrin expression, all the filamin proteins likely serve a broader and a more generalized function as scaffolding proteins for the actin cytoskeleton in their static state. FlnB shares greater than 90% homology with FlnA including the actin-binding domain. Their interactions with the actin cytoskeleton should thereby stabilize various cell membrane receptors, channels or molecules. Consistent with this possibility, we observe a compensatory response where loss of FlnA promotes FlnB expression and vice versa within the developmental growth plate. That said, other studies have shown no change in FLNA levels following loss of FLNB in lymphoblastoid cells, which may relate to cell type specific differences (28). Our current studies also show that both proteins, in their static state, fundamentally stabilize the actin cytoskeleton and cell morphology, presumably through their binding of the cortical actin. Reciprocal, compensatory expression would maintain this essential function, inherent to all filamin proteins.
The current work now shows a novel functional difference between the filamin proteins. Different filamin isoforms show varying interactions with the small RhoGTPases, suggesting functional differences between the various filamins. Consistent with other studies, RhoA expression is localized to the cell membrane and cytoplasm (29). We see a similar distribution for FlnA and FlnB, which suggested initially that both proteins might interact with the RhoGTPases. However, the current studies show that binding of the RhoGTPases is significantly stronger for FlnA than FlnB. These observations are largely consistent with prior reports that have observed FlnB binding with Rac1 but not RhoA or Cdc42 (30,31). We did observe faint bands with pulldowns for each of the small RhoGTPases by FlnB but they were not significantly different than GST alone. The reported Rac1 binding for FlnB might reflect this type of interaction. These discrepancies in binding might also relate to different tissues or cells used for the immunoprecipitation or even indirect binding through FlnA. Perhaps more importantly, the binding between the small RhoGTPases and FlnA are significantly stronger under similar conditions than that seen with FlnB. We also observe that FlnA is more broadly expressed across different organ systems and along actin stress fibers within cells. These observations would be more consistent with a primary role for FlnA in the regulation of RhoGTPase activation than FlnB. Finally, we show that FlnA and FlnB co-exist as hetero- and homodimers in chondrocytes, leading to the possibility that FlnA/B heterodimerization or FlnB homodimerization would impair RhoA activation in the dynamic state (as FlnB does not bind RhoA or does so weakly and would therefore impair FlnA dependent RhoA activation through integrins). Thus, loss of FlnB would thereby promote stress fiber formation and cell spreading. In this context, dynamic changes observed with integrin-dependent RhoA activation would serve as means of fine-tuning actin cytoskeletal turnover through FlnA and FlnB interactions.
The current studies refine our understanding of FlnA/B function. While all the filamins might serve as scaffolding to associate receptors with the actin cytoskeleton and also stabilize the actin cytoskeleton in the static state, activation of Rho GTPases may occur primarily through FlnA. We have observed that RhoA localization is dependent on FlnA function (unpublished observations, Lian and Sheen), perhaps in a manner similar to our previous reports of FlnA phosphorylation directing localization of guanine exchange factors (GEFs) involved in vesicle trafficking (32,33). The GEFs similarly promote GDP to GTP conversion of the ARFs, thereby initiating vesicle coat formation. In this respect, FlnA might serve to localize Rho GTPases toward the membrane periphery to allow for activation from various signaling molecules. The current work does show that RhoA activation can be dependent on the filamins after integrin activation. Given its relatively weak binding to RhoGTPases, FlnB may prevent localization of RhoGTPases to the required membrane subcompartment through its heterodimerization with FlnA or homodimerization alone and thereby inhibit Rho GTPase activation. Alternatively, FlnA/B may differentially stabilize integrin receptors at the cell membrane and changes in integrin activation levels could also alter RhoA activation.
Interactions between the filamin isoforms and Rho GTPases allow for differential regulation of shared pathways. The current studies suggest that both FlnA and FlnB can interact within chondrocytes to alter RhoA activation. In theory, different degrees of FlnA/B homo and heterodimerization will provide a secondary level of regulation over integrin β1 mediated functions. The current studies also suggest that FlnA can bind other small Rho GTPases such as Rac1 and Cdc42, whereas FlnB either does not or does so very weakly. Recent work has suggested that FlnA repeats 1 to 4 alter the balance between RhoA and Rac1 GTPase activities (34). These differences in binding affinities for the different Rho GTPases between different filamins and even within specific filamin isoforms suggest additional means of regulating actin dynamics. Lastly, we recently have identified formins as downstream filamin effectors of actin nucleation (12). Formins possess an autoinhibitory domain that is under specific Rho GTPase regulation. For example, Formin2 autoinhibition is regulated by activated RhoA but not activated Rac1 or Cdc42 (unpublished observations, Lian and Sheen). Taken in this context, several layers of complexity (from different filamin binding receptors, interactions between different filamin isoforms, activation of different Rho GTPases, and different RhoGTPase responsive formins) begin to provide a means for differential regulation of multiple actin dependent processes.
The effects of FlnA/B interactions on chondrocyte development are likely complex given the multiple interacting molecules that regulate actin dependent processes. Prior work suggests that RhoA/ROCK pathways are important regulators of chondrocyte proliferation and differentiation. Hence, in the context of cartilage development, our results suggest that FlnA and FlnB serve as important upstream modulators of RhoA activation to effect normal chondrocytes development. Additionally, both FlnA and FlnB can bind Formin1 (Fmn1) (12), and mutations in Fmn1 give rise to skeletal defects. Activated Rho GTPases have been shown to mediate formin activation by competing with formin autoinhibition domains. Thus, varying combinations of FlnA/B hetero- and homo-dimers would have differential effects on multiple Rho GTPases dependent pathways involved in chondrogenesis.
Filamin pathways are regulated by their binding to many different intracellular, cell membrane and extracellular signaling, adhesion, and receptor proteins. These many interactions likely explain the increasingly broad and varied phenotypes seen across various organ systems with loss of FlnA function. The current studies provide a potentially important functional difference between filamin isoforms that direct the activation and/or function of the filamin interacting proteins.
Materials and Methods
Ethics statement
All mouse studies were performed under approval from the Institutional Animal Care and Use Committees of Harvard Medical School and Beth Israel Deaconess Medical Center in accordance with The National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Human primary chondrocytes were obtained through a discarded tissue protocol. The study has been approved by the Institutional Review Board (IRB) at the Beth Israel Deaconess Medical Center and Brigham and Women's Hospital. The identified human discarded tissues were obtained from pathological samples obtained during planned, induced abortions.
FlnA-/- and FlnB-/- mice breeding, tissue isolation and bone decalcification
FlnB-/- mice were generated and bred as previously reported (10). The wild type allele was detected by PCR amplification using the primer pair 5’-agattattcacccggacgtg-3’ and 5’-cctgggctaataatggcaga-3’, and the mutated allele by using 5’-ctgtgctcgacgttgtcactg-3’ and 5’- gatcccctcagaagaactcgt-3’. FlnA null mice (Dilp2 mice) were obtained from the Comparative and Developmental Genetics Department Of MRC Human Genetics Unit, Edinburgh EH4 2XU, UK. The wild type and targeted allele was detected by PCR amplification using the primer pair Forward: 5-GCAGGCATTTTGCTTGTTATTCC and Reverse: 5-ACCTACCTGTGACACCACCTTCC, and the mutation site was confirmed by sequencing. Ketamine/Xylazine combination was used for anesthesia and euthanization (100mg/kg and 10mg/kg, 400mg/kg and 40mg/kg, respectively, injected intraperitoneally). Tissues were isolated after euthanization and fixed with 4% paraformaldehyde or 10% trichloroacetic acid (TCA). Additional samples were frozen, sectioned, and fixed prior to staining.
Immunostaining and imaging
Immunostaining of decalcified bone sections was previously performed in this laboratory (10). In brief, mice were euthanized as above, and then forearms dissected. Fixed bone tissues were further decalcified by 5% trichloroacetic acid solution containing 1% HCl and 1% acetic acid for 7 days, and then placed in 1:1 mixed OCT/30% sucrose mixture solution at least 24 h. 18 micron frozen sections were placed into 1X PBS for 5 min, before undergoing conventional immunostaining. For FlnA and FlnB staining, 10% (w/V) TCA fixed sections and cultured cells were used. For staining of other antibodies, 4% paraformaldehyde fixed sections and cultured cells were used. After washing in PBS, fixed samples were permeabilized with 0.5% Triton X-100 and blocked with 5% normal horse serum for 2 h. Tissue was incubated with the primary antibodies for 1 h at room temperature or overnight at 4 °C. The Dylight488- and Dylight594-conjugated secondary antibodies (Jackson Immunoresearch, West Grove PA, USA) were incubated for 1 h at room temperature. Samples were further counterstained with 100 ng/ml Hoechst33342 (Life Technologies, Grand Island, NY, USA). Images were obtained with an LSM5 Pascal confocal microscope (Zeiss, Germany). Given the relatively weak immunostaining using antibodies against RhoA, GFP-tagged RhoA was used for FLNA-RhoA and FLNB-RhoA double staining. The primary antibodies (for immunostaining and some also for western blotting) were: rabbit anti-FLNA monoclonal antibody (1:300, Cat.# 2242, Epitomics, Burlingame, CA, USA); rabbit anti-FlnB polyclonal antibody (Gifted by Dr. Kao, CWRU).
FlnA/a and FlnA/B dimerization analysis
Since the different binding affinities of FlnA and FlnB antibodies could potentially bias the comparison of protein expression levels, we first transfected 293 cells with FlnA-GFP and FlnB-GFP. Western blot analyses probing for GFP was then performed to establish a standard between FlnA and FlnB. Next, endogenous FlnA/A and FlnA/B dimers were precipitated from 293 cell lysate supernatant using FlnA specific antibody. Then, the co-immunoprecipitated FlnA, along with FlnA-GFP and FlnB-GFP controls, were detected on Western blots using FlnA and FlnB antibodies respectively. The amount of FlnA and FlnB were extrapolated based on their measured band intensities relative to that of FlnA-GFP and FlnB-GFP standards, respectively. The ratio of FlnA/A to FlnA/B was calculated using the following equation: .
ATDC5 and U-2 OS cell culture, primary chondrocyte isolation and cell culture
Mouse ATDC5 chondrocyte cells (Cat# 99072806, Sigma), shRNA expressing ATDC5 cells, and U-2 OS cells (Cat.# ATCC HTB-96) were cultured in DMEM medium containing 10% fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin (Cat.# 15140, Invitrogen). Unless otherwise specified, ATDC5 cells were passaged every two days. Primary chondrocytes from the growth plates of newborn mice were prepared according to the Lefebvre’s protocol (35) with mild modifications (10). In brief, the growth plates of radius and ulna were dissected from P7 newborn mice, or from the cartilage of discarded human fetal tissue limbs. Tissues were minced and washed in cold Hanks buffered saline solution, and placed in 0.2% collagenase type I (Cat.# 4196, Worthington, Lakewood, NJ, USA) solution in DMEM at 37 °C for 3 h. Soft tissues were detached from cartilage by repeated pipetting; the sediment cartilage was further digested with 0.2% collagenase I for 6 h. The dissociated cells were then passed through a cell strainer to isolate single cells and were plated at a feasible density in DMEM with 10% FBS. Primary human chondrocytes from aborted on long bone tissue were cultured following the same protocol as in mice.
shRNA vector construction, lentivirus preparation and infection
The shRNA target sequence used for FlnA was 5'-GCA TCC TAGTTAAGTACAA-3' and for FlnB: 5’-GCCCA AATCA AGACT CTT AAT-3’. The shRNA target sequences were cloned into pSicoR lentivirus vector. In brief, after heating to 98 °C for 10 min, the oligomers were reannealed in boiling water that gradually cooled back to room temperature, followed by gradual cooling back to room temperature. Then the annealed products were ligated into the HpaI/XhoI sites of the EGFP tagged pSicoR lentivirus vector. The helper vectors, pSPAX2 and pMD2G, were transfected together with the lentivirus vector into 293T cells. For the control group, empty pSicoR(GFP) vector was used. The TransFectin Lipid reagent was used according to manufacturer’s instructions (Cat.# 170-3352, BIO-RAD). The lentivirus-containing medium was collected by centrifugation at 2000xg for 10 min. Then the supernatant was passed through a 0.45um filter. The filtered medium was directly added to the pre-seeded ATDC5 cells. The infected ATDC5 cells were visualized by the microscope and were verified by confocal scanning. After two rounds of infection, nearly all the ATDC5 cells were EGFP positive. The EGFP positive ATDC5 cells were used for further in vitro experiments.
β1 integrin activation by fibronectin and F-actin staining
For β1 integrin activation, culture dishes or cover slips were pre-coated with fibronectin (F0895, Sigma) following manufacturer’s instructions. Cells were plated and incubated for 0.5, 1, 2, and 3 h, respectively and were fixed by 4% PFA for 20 min after PBS washing. After further washing in PBS, fixed samples were permeabilized with 0.5% Triton X-100 and blocked with 5% normal horse serum for 2 h. Samples were incubated with the 1:40 diluted Rhodamine-Phalloidin for f-actin staining for 1 h at room temperature or overnight at 4 °C. Samples were further counterstained with 100 ng/ml Hoechst33342 (Life Technologies, Grand Island, NY, USA) for 10 min. Images were obtained with an LSM5 Pascal confocal microscope (Zeiss, Germany). The primary mouse chondrocytes were transfected with pSicoR-GFP, pSicoR-GFP-FlnashRNA, pSicoR-GFP-FlnbshRNA, pSicoR-GFP-RhoA-G14V, or pSico-GFP-RhoA-T19N for 3 days before dissociation and replated on fibronectin-coated or uncoated coverslips.
Immunoprecipitation and western blotting
Samples were collected directly for endogenous interaction (for FlnA-FlnB interaction), or 36 h after transfection of the FlnA/B modifying constructs. Modified RIPA buffer (50 mM Tris-HCl, pH7.5; 150mM NaCl, 1.0% Triton x-100; 0.5% sodium deoxycholate and 0.1% SDS), with proteinase inhibitor cocktail and protein phosphotase inhibitor cocktail, as well as additional PMSF (1 mM), NaF (10mM) and Na3VO4 (1mM), were used for cell lysis. Conventional immunoprecipitations were performed by using Protein-A/G ultralink resin beads according to the protocol provided by the manufacturer (Cat.# 53132, Thermo Scientific, Rockford, IL, USA Proteins were separated in 8 or 10% SDS-polyacrylamide gels and transblotted to PVDF membrane. Immunoblot analysis was performed with primary antibodies based on manufactures’ guides or suggested dilutions. Blots were detected by using LumiGOLD ECL western blotting detection kit (Cat.# SL100309, SignaGen, Rockville, MD, USA).
Active RhoA pulldown and western blot detection of RhoA
GST-Rhotekin-RBD (Rho binding domain) beads were used for active RhoA pulldown. Samples were prepared and procedures were used following the manufacturer’s instructions (Cat. # RT01, Cytoskeleton, USA) with slight modifications. In brief, proteins were extracted from cultured cells by using the cell lysis buffer for RBD-RhoA pulldown (50 mM Tris pH 7.5, 30 mM MgCl2, 0.3 M NaCl, 1% triton X-100). After centrifugation at 13000gx10min, the supernatants containing the same amount of cells or total proteins were added into 1.5 ml eppendorf tubes each containing 60 μg Rhotekin-RBD beads. After gentle resuspension of the beads, the mixtures were incubated at 4 °C for 1 h with gentle rotating. 10. Gently rotate the tubes at 4 °C for 1 h. The beads were pelleted by centrifugation at 5k rpm, 4 °C for 1 min and the supernatants were removed. After washing the beads twice in 500 μl of Wash Buffer (25 mM Tris pH 7.5, 30 mM MgCl2, 40 mM NaCl), the beads were pelleted by centrifugation and resuspended from each tube in 100 μl of SDS sample buffer. After boiling at 100 °C for 5 min, the supernatant samples were collected and analyzed by conventional western blot procedure using a RhoA specific mono-clonal antibody (Cat. # ARH03, Cytoskeleton, USA).
Statistical analyses
Results were expressed as the mean ± s.d. of n experiments (n ≥ 3). For section staining, unless otherwise specified, 3 animals (except the section staining from E18.5 FlnA mouse, which is extremely hard to be obtained) were used for each experiment and 3–4 sections were stained per animal. For cultured cell staining, cells were plated and growing on 3 coverslips per sample. For cell growth curve analysis, three independent experiments were performed, with 3–4 wells were collected from each sample at each time point. Statistical analysis was performed with Student’s t-test, with P <0.05 considered significant. For quantitative analyses of immunostaining results, luminosity (histogram value, Adobe photoshop) values were obtained and based on the threshold (luminosity value 20) cells (n > 50 cells) were grouped into highly stained and weakly/negatively stained subpopulations. For quantitative analyses of western blotting results, band intensity was obtained by subtracting background luminosity from the total luminosity of each band (histogram value, Adobe photoshop).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Dr. Hung-Ying Kao (Case Western Reserve University, Cleveland, OH, USA) for gifting the FLNB antibody, Dr. Tyler Jacks (MIT, Cambridge, MA, USA) for providing the pSicoR lentivirus vector, and Dr. Fumihiko Nakamura (Harvard, Boston, MA, USA) for kindly providing the FLNA-EGFP vector.
Conflict of Interest statement. None declared.
Funding
National Institutes of Health [1 R01 NS092062-01 to VLS].
References
- 1. Gorlin J.B., Yamin R., Egan S., Stewart M., Stossel T.P., Kwiatkowski D.J., Hartwig J.H. (1990) Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J. Cell. Biol., 111, 1089–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cunningham C.C., Gorlin J.B., Kwiatkowski D.J., Hartwig J.H., Janmey P.A., Byers H.R., Stossel T.P. (1992) Actin-binding protein requirement for cortical stability and efficient locomotion. Science, 255, 325–327. [DOI] [PubMed] [Google Scholar]
- 3. Chakarova C., Wehnert M.S., Uhl K., Sakthivel S., Vosberg H.P., van der Ven P.F., Furst D.O. (2000) Genomic structure and fine mapping of the two human filamin gene paralogues FLNB and FLNC and comparative analysis of the filamin gene family. Hum. Genet., 107, 597–611. [DOI] [PubMed] [Google Scholar]
- 4. Sheen V.L., Feng Y., Graham D., Takafuta T., Shapiro S.S., Walsh C.A. (2002) Filamin A and Filamin B are co-expressed within neurons during periods of neuronal migration and can physically interact. Hum. Mol. Genet., 11, 2845–2854. [DOI] [PubMed] [Google Scholar]
- 5. Sheen V.L., Jansen A., Chen M.H., Parrini E., Morgan T., Ravenscroft R., Ganesh V., Underwood T., Wiley J., Leventer R., et al. (2005) Filamin A mutations cause periventricular heterotopia with Ehlers-Danlos syndrome. Neurology, 64, 254–262. [DOI] [PubMed] [Google Scholar]
- 6. Sheen V.L. (2012) Periventricular Heterotopia: Shuttling of Proteins through Vesicles and Actin in Cortical Development and Disease. Scientifica, 2012, 480129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krakow D., Robertson S.P., King L.M., Morgan T., Sebald E.T., Bertolotto C., Wachsmann-Hogiu S., Acuna D., Shapiro S.S., Takafuta T., et al. (2004) Mutations in the gene encoding filamin B disrupt vertebral segmentation, joint formation and skeletogenesis. Nat. Genet., 36, 405–410. [DOI] [PubMed] [Google Scholar]
- 8. Vorgerd M., van der Ven P.F., Bruchertseifer V., Lowe T., Kley R.A., Schroder R., Lochmuller H., Himmel M., Koehler K., Furst D.O., et al. (2005) A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am. J. Hum. Genet., 77, 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parrini E., Rivas I.L., Toral J.F., Pucatti D., Giglio S., Mei D., Guerrini R. (2011) In-frame deletion in FLNA causing familial periventricular heterotopia with skeletal dysplasia in males. Am. J. Med. Genet. A, 155A, 1140–1146. [DOI] [PubMed] [Google Scholar]
- 10. Lu J., Lian G., Lenkinski R., De Grand A., Vaid R.R., Bryce T., Stasenko M., Boskey A., Walsh C., Sheen V. (2007) Filamin B mutations cause chondrocyte defects in skeletal development. Hum. Mol. Genet., 16, 1661–1675. [DOI] [PubMed] [Google Scholar]
- 11. Hu J., Lu J., Lian G., Zhang J., Hecht J.L., Sheen V.L. (2014) Filamin B regulates chondrocyte proliferation and differentiation through Cdk1 signaling. PLoS One, 9, e89352.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu J., Lu J., Lian G., Ferland R.J., Dettenhofer M., Sheen V.L. (2014) Formin 1 and filamin B physically interact to coordinate chondrocyte proliferation and differentiation in the growth plate. Hum. Mol. Genet., 23, 4663–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stossel T.P., Condeelis J., Cooley L., Hartwig J.H., Noegel A., Schleicher M., Shapiro S.S. (2001) Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell. Biol., 2, 138–145. [DOI] [PubMed] [Google Scholar]
- 14. Ohta Y., Suzuki N., Nakamura S., Hartwig J.H., Stossel T.P. (1999) The small GTPase RalA targets filamin to induce filopodia. Proc. Natl. Acad. Sci. U. S. A, 96, 2122–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chrzanowska-Wodnicka M., Burridge K. (1996) Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol., 133, 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang G., Woods A., Sabari S., Pagnotta L., Stanton L.A., Beier F. (2004) RhoA/ROCK signaling suppresses hypertrophic chondrocyte differentiation. J. Biol. Chem., 279, 13205–13214. [DOI] [PubMed] [Google Scholar]
- 17. Woods A., Beier F. (2006) RhoA/ROCK signaling regulates chondrogenesis in a context-dependent manner. J. Biol. Chem., 281, 13134–13140. [DOI] [PubMed] [Google Scholar]
- 18. Woods A., Wang G., Beier F. (2005) RhoA/ROCK signaling regulates Sox9 expression and actin organization during chondrogenesis. J. Biol. Chem., 280, 11626–11634. [DOI] [PubMed] [Google Scholar]
- 19. Cherfils J., Zeghouf M.. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev., 93, 269–309. [DOI] [PubMed] [Google Scholar]
- 20. Arthur W.T., Petch L.A., Burridge K. (2000) Integrin engagement suppresses RhoA activity via a c-Src-dependent mechanism. Curr. Biol., 10, 719–722. [DOI] [PubMed] [Google Scholar]
- 21. Calderwood D.A., Huttenlocher A., Kiosses W.B., Rose D.M., Woodside D.G., Schwartz M.A., Ginsberg M.H. (2001) Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat. Cell. Biol., 3, 1060–1068. [DOI] [PubMed] [Google Scholar]
- 22. Bourdoulous S., Orend G., MacKenna D.A., Pasqualini R., Ruoslahti E. (1998) Fibronectin matrix regulates activation of RHO and CDC42 GTPases and cell cycle progression. J. Cell Biol., 143, 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Khelfaoui M., Pavlowsky A., Powell A.D., Valnegri P., Cheong K.W., Blandin Y., Passafaro M., Jefferys J.G., Chelly J., Billuart P. (2009) Inhibition of RhoA pathway rescues the endocytosis defects in Oligophrenin1 mouse model of mental retardation. Hum. Mol. Genet., 18, 2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ellis S., Mellor H. (2000) Regulation of endocytic traffic by rho family GTPases. Trends Cell Biol., 10, 85–88. [DOI] [PubMed] [Google Scholar]
- 25. Caswell P.T., Vadrevu S., Norman J.C. (2009) Integrins: masters and slaves of endocytic transport. Nat. Rev. Mol. Cell Biol., 10, 843–853. [DOI] [PubMed] [Google Scholar]
- 26. Mulrooney J.P., Hong T., Grabel L.B. (2001) Serine 785 phosphorylation of the beta1 cytoplasmic domain modulates beta1A-integrin-dependent functions. J. Cell Sci., 114, 2525–2533. [DOI] [PubMed] [Google Scholar]
- 27. Razinia Z., Makela T., Ylanne J., Calderwood D.A. (2012) Filamins in mechanosensing and signaling. Annu. Rev. Biophys., 41, 227–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Farrington-Rock C., Kirilova V., Dillard-Telm L., Borowsky A.D., Chalk S., Rock M.J., Cohn D.H., Krakow D. (2008) Disruption of the Flnb gene in mice phenocopies the human disease spondylocarpotarsal synostosis syndrome. Hum. Mol. Genet., 17, 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Michaelson D., Silletti J., Murphy G., D'Eustachio P., Rush M., Philips M.R. (2001) Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol., 152, 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Del Valle-Perez B., Martinez V.G., Lacasa-Salavert C., Figueras A., Shapiro S.S., Takafuta T., Casanovas O., Capella G., Ventura F., Vinals F.. Filamin B plays a key role in vascular endothelial growth factor-induced endothelial cell motility through its interaction with Rac-1 and Vav-2. J. Biol. Chem., 285, 10748–10760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jeon Y.J., Choi J.S., Lee J.Y., Yu K.R., Ka S.H., Cho Y., Choi E.J., Baek S.H., Seol J.H., Park D., et al. (2008) Filamin B serves as a molecular scaffold for type I interferon-induced c-Jun NH2-terminal kinase signaling pathway. Mol. Biol. Cell, 19, 5116–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang J., Neal J., Lian G., Hu J., Lu J., Sheen V. (2013) Filamin A regulates neuronal migration through brefeldin A-inhibited guanine exchange factor 2-dependent Arf1 activation. J. Neurosci., 33, 15735–15746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J., Neal J., Lian G., Shi B., Ferland R.J., Sheen V. (2012) Brefeldin A-inhibited guanine exchange factor 2 regulates filamin A phosphorylation and neuronal migration. J. Neurosci., 32, 12619–12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Duval D., Lardeux A., Tourneau T.L., Norris R.A., Markwald R.R., Sauzeau V., Probst V., Marec H.L., Levine R., Schott J.J., et al. (2014) Valvular dystrophy associated filamin A mutations reveal a new role of its first repeats in small-GTPase regulation. Biochim. Biophys. Acta, 1843, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lefebvre V., Garofalo S., Zhou G., Metsaranta M., Vuorio E., De Crombrugghe B. (1994) Characterization of primary cultures of chondrocytes from type II collagen/beta-galactosidase transgenic mice. Matrix Biol., 14, 329–335. [DOI] [PubMed] [Google Scholar]
- 36. Lian G., Lu J., Hu J., Zhang J., Cross S.H., Ferland R.J., Sheen V.L. (2012) Filamin a regulates neural progenitor proliferation and cortical size through Wee1-dependent Cdk1 phosphorylation. J. Neurosci., 32, 7672–7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.