Abstract
Objectives
Melanoma is an aggressive skin cancer. Understanding the underlying mechanisms for melanomagenesis and identification of novel and effective melanoma treatment strategies are urgently necessary. The long-noncoding RNAs are considered as new essential players during cancer development, including the melanoma.
Materials and methods
In this study, we first determined the expression of LINC00888 in tumor tissues and adjacent normal tissues from 28 patients with melanoma using quantitative polymerase chain reaction, and the correlation between the expression level of LINC00888 and the survival months was also examined. Next, we investigated the effect of LINC00888 on the proliferation, apoptosis, and invasion in the melanoma cells. Moreover, LINC00888-specific miRNA and target gene were further confirmed using the dual-luciferase reporter assay and Western blotting. Last, the tumorigenesis role of LINC00888 was also explored using tumor xenografts mouse model.
Results
Elevated LINC00888 expression was found in melanoma specimens compared with adjacent normal tissues. The 4-year overall survival in melanoma patients with high expression of LINC00888 was substantially shorter than that in those with low expression of LINC00888. Knockdown of LINC00888 significantly inhibited the proliferation, apoptosis, epithelial–mesenchymal transition, and invasion of melanoma cells, while the overexpression of LINC00888 exerted opposite effect. Furthermore, we revealed that microRNA-126 (miR-126) was able to regulate LINC00888 expression and further influence the expression of CRK. Consistently, miR-126 inhibitor could rescue the expression of CRK in LINC00888-downregulated cells, while miR-126 mimics could reduce the CRK expression level in cells with the overexpression of LINC00888. Last, the animal experiment further demonstrated that the overexpression of LINC00888 enhanced the tumor development in vivo.
Conclusion
Our data showed that long-noncoding RNA LINC00888 functioned as an oncogene in melanoma tumorigenesis, it also regulated the cellular proliferation and invasion of melanoma via miR126/CRK signaling pathway and metastasis via miR-126/CRK signaling axis, which could be a promising molecular target for treating melanoma.
Keywords: long-noncoding RNAs, melanoma, melanomagenesis, proliferation, apoptosis
Introduction
Melanoma is the most common and aggressive skin cancer. According to an annual report, in the USA, around 76,380 cases were diagnosed with melanoma and there were 10,130 deaths by the end of 2016.1 Although primary melanoma could be cured via surgery with early diagnosis, the majority of melanoma, once clinically identified, is highly likely to be metastasized.2 Therefore, understanding the potential underlying mechanisms for both tumorigenesis and metastasis of melanoma and seeking novel and effective melanoma treatment strategies are urgently necessary.
Long non-coding RNAs (lncRNAs) are noncoding RNAs with various lengths, which range from 200 nucleotides to multiple kilobases.3 As defined, with lacking protein-coding function, lncRNAs are uniquely expressed in differentiated tissues or various cancer types.4 It modulates genes that do not code for proteins, accounting for >80% of genes, in both transcriptional and posttranscriptional levels, which is distinct from microRNAs (miRNAs) and other noncoding RNAs. It has been widely established that lncRNAs were involved in a large number of biological events, including proliferation, apoptosis, differentiation, tumorigenesis, and metastasis. Therefore, lncRNAs may be a new class of cancer biomarkers and therapeutic targets. A recent study showed that 48 lncRNAs were aberrantly expressed in melanoma.5 Among them, an upregulated lncRNA LINC00888 that was coexpressed with 15 mRNAs might play a vital role during the tumorigenesis and metastasis of melanoma. LINC00888, located on chromosome 3, had the most coexpressed RNAs among the unregulated lncRNAs in the tumorigenesis of melanoma.5 Specifically, 11 downregulated and two upregulated mRNAs were coregulated with LINC00888 between metastatic melanoma and normal tissues, suggesting that LINC00888 might be orchestra, a serial of key events to facilitate the tumor initiation, development, and metastasis. However, the biological function and the downstream signaling pathway of LINC00888 were rarely studied.
In this study, we attempted to determine LINC00888 expression in both tumor tissues and paired adjacent normal tissues in 28 patients with melanoma and identified any correlation between the level of LINC00888 and the survival months. In addition, we investigated the effect of LINC00888 on the proliferation, apoptosis, and invasion in human melanoma cells. Moreover, the potential downstream miRNA and target gene were identified using bioinformatics analysis and further confirmed using dual-luciferase reporter assay and Western blotting. Last, the tumorigenesis role of LINC00888 was also explored using tumor xenografts mouse model.
Materials and methods
Patients and clinical tissue samples
In total, 28 pairs of melanoma tumor samples and adjacent tissue samples were collected from specimens resected surgically from melanoma patients in the Zhejiang Provincial People’s Hospital; 10% formalin solution was used to fix the tissue specimens, and paraffins were used to be embedded in the specimens. All the collected cases were diagnosed as melanoma pathologically without any preoperative radiotherapy and/or chemotherapy. Patients who donated their tissues signed informed consent, which has been approved by the institutional review board of the hospital. The following information of patients was collected, including the survival months. The informed written consents were obtained from the patients, and the study was approved by the First Affiliated Hospital of Nanjing Medical University Institutional Review Board.
Experiment of siRNA
Invitrogen Block-iT RNAi Designer (http://rnaidesigner.thermofisher.com/rnaiexpress/) was used to design three LINC00888 siRNAs as follows: siRNA1, 5′-ACTTAATCCGATGAGAAATGC-3′; siRNA2, 5′-TGGACATGCCTCAGGTCTAGC-3′; and siRNA3, 5′-TAACTAGGGTACTAAGCCAGC-3′. The control siRNA 5′-TTTTAGCCTATGACCGTAATCA-3′ was used as a negative control (Thermo Fisher Scientific, Waltham, MA, USA). These siRNAs were provided by GenePharma (Shanghai, China). The A375-S2 cells were transfected with these siRNAs (50 nM) according to the protocol of Lipofectamine® 3000 DNA Transfection Reagent (Thermo Fisher Scientific).
Plasmid constructs and establishment of stable cell lines
LINC00888 transcript complementary DNA (cDNA) was amplified by polymerase chain reaction (PCR) and was inserted into pLenti6/V5-D-TOPO vector. To generate the lentivirus LINC00888 (lenti-LINC0088), the plasmid was further transfected into 293T cells using Lipofectamine 3000 DNA Transfection Reagent (Thermo Fisher Scientific). Later, the lentivirus particles were infected into A375-S2 cells, and the desired monoclonal stable cell line was further selected in selection medium.
Cell culture
Human melanoma cell lines, A375-S2 cell lines, were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA) and cultured in RPMI-1640 medium (Thermo Fisher Scientific) with 10% of fetal bovine serum (FBS; Thermo Fisher Scientific) in incubator (5% CO2 humidified) at 37°C. A375-S2 cells were plated into the 24-well plate and then transfected with designed siRNAs or LINC00888 overexpression particles following the manufacturer’s protocol.
Quantitative real-time PCR (qRT-PCR)
Total RNA was first extracted from cells or clinical tissue samples using TRIzol® reagent (Thermo Fisher Scientific) based on the standard protocol. Total cDNA was reversed-transcribed from isolated RNA using the Prime-Script RT Master Mix (Takara Biotechnology, Dalian, China). To detect the expression levels of interest target genes using the SYBR Premix Ex Taq (Takara), qRT-PCR was performed on the ABI Biosystems (Thermo Fisher Scientific). The relative expression levels of target genes were normalized by 2−ΔΔCt cycle threshold method to internal control (GAPDH). The reactions were performed at 95°C for 30 seconds, followed by 35 cycles of 95°C for 5 seconds, and 58°C for 30 seconds. The mean value of the LINC00888 expression was chosen as the cutoff between high and low LINC00888 expression groups in tumor tissues.
Cell proliferation analysis
The proliferation of A375-S2 cells was measured by Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Kumamoto, Japan) according to the manufacturer’s instructions. Briefly, the cells were washed with PBS buffer (pH =7.4), harvested by trypsinization, and reseeded into a 96-well plate. Then, the plate was preincubated in a humidified incubator (37°C, 5% CO2) for 0, 24, 48, and 72 hours. Next, 10 μL CCK-8 solution was added to each well of the plate, and the plate was incubated for 2 hours. The absorbance of each sample was measured at 450 nm using a microplate reader.
Cell invasion assay
Cell invasion assay was performed with transwell chambers (8 μm pore size; Corning Incorporated, Corning, NY, USA), which were precoated with 200 μg/mL Matrigel® (BD Biosciences, San Jose, CA, USA) and incubated overnight. Cells were pretreated with 1 μM mitomycin C for 1 hour and then harvested and suspended in serum-free medium in the upper chamber of a transwell plate. The upper channel was separated from the lower chambers with permeable 8.0-μm polycarbonate membranes. The lower chamber was added with 500 μL DMEM containing 10% FBS as a chemoattractant. After 24-hour incubation, cells remaining on the upper membrane which did not migrate were carefully removed. Later, the membranes were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The number of stained cells was manually counted under a phase-contrast inverted microscope from three randomly selected visual fields. Each experiment was conducted in triplicate.
Western blot
Total proteins were isolated from A375-S2 cells using protease inhibitor cocktail (Hoffman-La Roche Ltd., Basel, Switzerland). Protein concentrations were determined by bicinchoninic acid protein assay kit (Pierce, Rockford, IL, USA). Appropriate amounts of the protein were resolved by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel and transferred onto polyvinylidene fluoride membrane. The membrane was further blocked in Tris-buffered saline (pH =7.4) containing 0.05% Tween-20 and 5% nonfat dry milk. The following primary antibodies were used: GAPDH (#5174; Cell Signaling Technology, Danvers, MA, USA), CRK (610035; BD Biosciences), E-cadherin (#3195; Cell Signaling Technology), vimentin (#5741; Cell Signaling Technology), and PCNA (#13110; Cell Signaling Technology). GAPDH was served as the internal control. Chemiluminescence signals were detected incubated with horseradish peroxidase and secondary antibodies. The relative expressions of the proteins were valuated through the gray value ratio of each protein.
Flow cytometry analysis
The percentage of cell apoptosis was determined using flow cytometry. Briefly, cells were trypsinized without ethylenediaminetetraacetic acid followed by washing twice with PBS. The cells were then incubated with 5 μL of fluorescein isothiocyanate Annexin V (BD Pharmingen) and 5 μL of propidium iodide (BD Biosciences) in 400 μL of 1× binding buffer for 15 minutes at RT. FlowJo software (BD Biosciences) was used to analyze the data.
Colony formation
A375-S2 cells were seeded in 6-well plates at 500 or 1,000 cells per well overnight, and then, the cells were treated with control or LINC00888 overexpression particles for 72 hours. Later on, the cells were washed with PBS, fixed in ice-cold methanol, and stained with crystal violet solution (0.5% in 25% methanol) for 8 hours. Then, the pictures of colony format were taken using a digital camera. Finally, the violet crystals were dissolved with 70% ethanol, and absorbance (595 nm) was measured with microplate reader.
Target prediction
In order to predict the interactions between miRNAs and their target genes, the conserved miRNA target sites predicted by five algorithms (TargetScan, miRanda, Pictar2, PITA, and RNA22) from public databases were retrieved by starBase Version 2.0.6
Dual-luciferase reporter assay
The cells were cultured overnight until 60%–70% confluence. To determine the transcriptional level of LINC00888, A375-S2 cells were cotransfected with pmirGLO or pmirGLO-LINC00888. To detect the transcriptional level of CRK, the cells were cotransfected with LINC00888 siRNA2 with or without miR-126 inhibitor (20 nM). Similarly, the cells were cotransfected with lenti-LINC00888 with or without miR-126 mimics (20 nM). Lipofectamine 3000 (Thermo Fisher Scientific) was used as a transfection agent. The cells were harvested for luciferase detection using the dual-luciferase reporter assay (Promega, Madison, WI, USA) 48 hours posttransfection. The miR-126 mimics and inhibitor were provided by GenePharma (Shanghai, China).
Tumor xenograft in vivo model
Next, 5- to 6-week-old BALB/c nude mice were purchased from the animal center at Vital River (Beijing, China) and maintained under pathogen-free conditions according to the protocols. The animals from this study were approved by the Institution of Animal Ethical and Welfare Committee. The effects of LINC00888 on tumor growth were determined using A375-S2 cells stably transfected with lentivirus LINC00888 or lentivirus vector. The mice were injected with 1×107 treated A375-S2 cells into posterior flank of each mouse subcutaneously. To examine the impact of LINC00888 on tumorigenesis, tumor weights were measured and pictured at day 28 after the cell injection. This animal study was approved by the Animal Ethics Committee of the First Affiliated Hospital of Nanjing Medical University. In addition, Guide for the Care and Use of Laboratory Animals (8th edition) was strictly followed by us for animal study.
Statistics
All values were described as the mean ± SD. Results were further analyzed using the statistical software SPSS 22.0. Student’s t-test or log-rank test was also performed when necessary. A two-sided p-value <0.05 was considered as statistically significant.
Results
LINC00888 was upregulated in melanoma tumor tissues compared with adjacent normal tissues
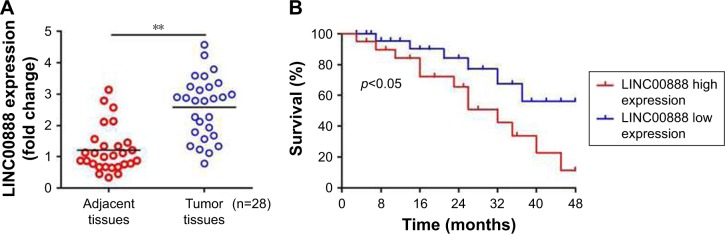
The qRT-PCR was performed to measure the expression level of LINC00888 in a total of 28 pairs of tumors and adjacent normal tissues. The results suggested a significantly higher level of LINC00888 in tumor tissues, compared with corresponding adjacent normal tissues (p<0.01; Figure 1A). In addition, the Kaplan–Meier analysis and the log-rank test revealed that the 4-year overall survival in melanoma patients with a high expression of LINC00888 was notably shorter than that in those with low expression of LINC00888 (p<0.05; Figure 1B). Moreover, as showed in Table 1, higher LINC00888 expression was observed more frequently in patients with bigger size melanoma and advanced lymphatic metastasis. The results showed that LINC00888 expression levels were significantly upregulated in melanoma tissues compared with normal tissues and strongly associated with the survival time of patients diagnosed with melanoma.
Figure 1.
Upregulation of LINC00888 in melanoma tumor tissues and its clinical significance. (A) Relative expression of LINC00888 in melanoma tumors and the paired adjacent normal tissues were detected by qRT-PCR (n=28; **p<0.01). (B) Kaplan–Meier analysis of the correlation between high and low LINC00888 expression in 28 melanoma patients and 4-year survival (log-rank test; p<0.05).
Abbreviation: qRT-PCR, quantitative real-time polymerase chain reaction.
Table 1.
Correlation of LINC00888 expression levels with clinicopathological parameters of patients with melanoma
| Parameters | n | LINC00888 level
|
p-value |
|---|---|---|---|
| qRT-PCR | |||
| Age (months) | 0.447 | ||
| <30 | 12 | 2.359±0.359 | |
| ≥30 | 16 | 2.777±0.404 | |
| Gender | 0.797 | ||
| Female | 11 | 2.501±0.381 | |
| Male | 17 | 2.641±0.391 | |
| Thickness | <0.01** | ||
| <2 mm | 15 | 1.707±0.267 | |
| >2 mm | 13 | 3.724±0.314 | |
| AJCC stage | <0.05* | ||
| I, II | 14 | 2.112±0.359 | |
| III, IV | 14 | 3.318±0.375 | |
| Lymphatic metastasis | <0.05* | ||
| Yes | 15 | 3.372±0.395 | |
| No | 13 | 2.147±0.342 |
Notes: Student’s t-test;
p<0.05,
p<0.01.
Abbreviations: AJCC, American Joint Committee on Cancer; qRT-PCR, quantitative real-time polymerase chain reaction.
Knockdown of LINC00888 inhibited melanoma cell proliferation and invasion
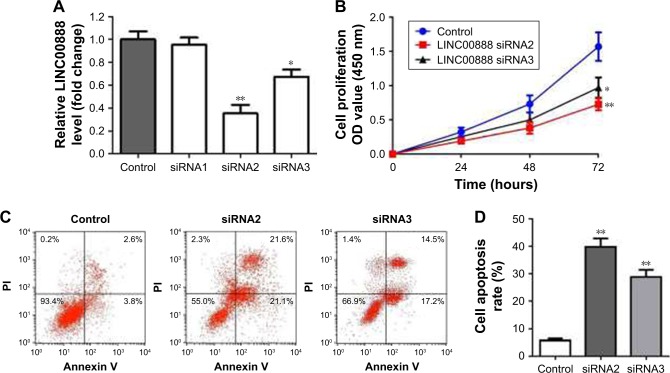
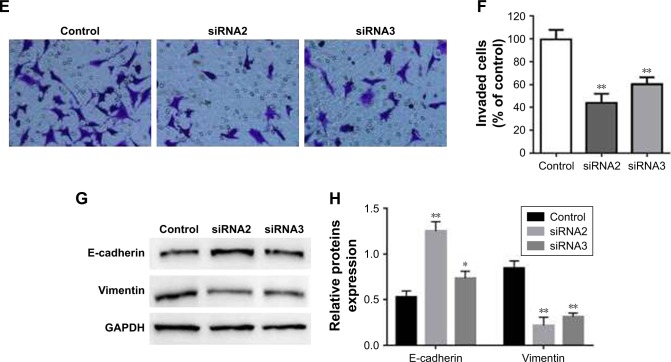
To assess the biological function of LINC00888 on melanoma cells, three siRNAs (siRNA1, siRNA2, and siRNA3) were designed to knockdown the expression of LINC00888. The qRT-PCR was performed to detect the knockdown efficiency of the LINC00888 siRNAs in A375-S2 melanoma cells. Figure 2A indicated that the knockdown efficiency of siRNA2 and siRNA3 was significantly higher than that of the negative control siRNA or siRNA1 (p<0.01 and <0.05, respectively). Since siRNA2 had the highest knockdown efficiency, which was selected for the following experiments. Next, the impact of LINC00888 on cell proliferation was investigated with CCK8 assay. As shown in Figure 2B, knockdown of LINC00888 in A375-S2 cells significantly inhibited cellular proliferation 72 hours post-transfection (p<0.01). In addition, knockdown of LINC00888 induced a remarkable cell apoptosis in 72 hours, determined by flow cytometric analysis (p<0.01; Figure 2C and D). Furthermore, it turned out that LINC00888 knockdown led to a notable reduction of cellular invasion in 24 hours (p<0.01; Figure 2E and F). Meanwhile, epithelial–mesenchymal transition (EMT) special markers E-cadherin and vimentin were detected with Western blot. Inhibition of LINC00888 inhibited EMT, while overexpression LINC00888 exerted the opposite effect (Figure 2G and H). All these observations suggested that knockdown of LINC00888 was able to inhibit the cellular proliferation and invasion.
Figure 2.
Knockdown of LINC00888 inhibited the cellular proliferation of melanoma. (A) LINC00888 expression in A375-S2 cells was detected by qRT-PCR after treatment with siRNA1, siRNA2, or siRNA3 for 72 hours. *p<0.05, **p<0.01. (B) The cell proliferation of A375-S2 after transfection with LINC00888 siRNAs or negative control for 24, 48, or 72 hours. *p<0.05, **p<0.01. (C) The apoptosis rate of A375-S2 cells with transfection of LINC00888 siRNAs (right panel) or negative control (left panel) was determined by Annexin V/PI staining in 72 hours. (D) The quantitative comparison of Annexin V/PI staining between positive cells and cells transfected with negative control or LIN00888 siRNA2 (n=3, **p<0.01 vs control). (E) Representative invasion assay readout of cells transfected with negative control or LIN00888 siRNA2 for 24 hours. (F) The cells which invaded through the membrane were fixed with paraformaldehyde, stained with crystal violet, and counted under an inverted microscope (normalized to control group, n=3, **p<0.01 vs control). (G) Protein expression of E-cadherin and vimentin in A375-S2 cells after transfection with LINC00888 siRNAs or negative control for 72 hours. (H) The quantification of E-cadherin and vimentin proteins expressions in the cells (n=3, *p<0.05, **p<0.01 vs control).
Abbreviations: PI, propidium iodide; qRT-PCR, quantitative real-time polymerase chain reaction.
Overexpression of LINC00888 enhanced A375-S2 cells invasion and colony formation
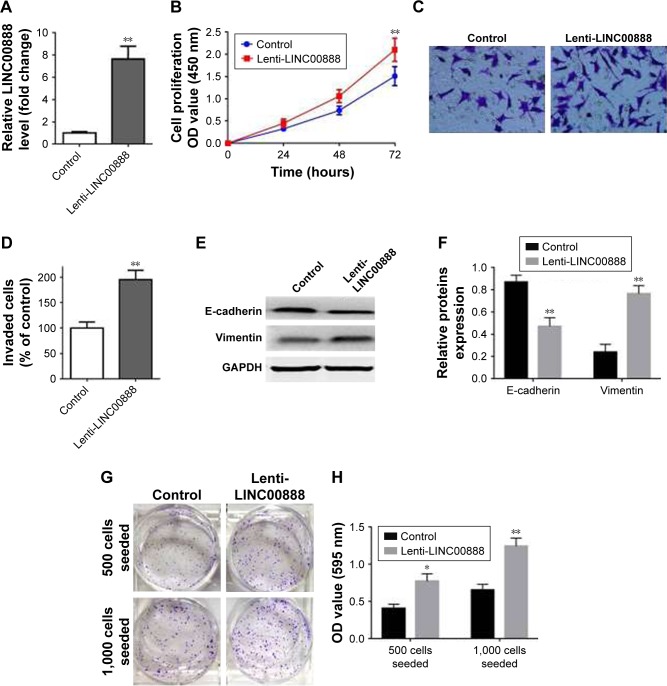
Having checked the impact of LINC00888 knockdown on A375-S2 cell proliferation and apoptosis, we further attempted to determine the biological effects of LINC00888 overexpression. We first checked the relative expression of LINC00888 in A375-S2 cells transfected with overexpression of lenti-LINC00888 or control. As shown in Figure 3A, transfection of lenti-LINC00888 induced a high level of LINC00888 in A375-S2 cells (p<0.01). We next determined the A375-S2 cell proliferation after transfection. The results indicated that lenti-LINC00888 time-dependently increased A375-S2 cell proliferation (p<0.01; Figure 3B). In addition, the results of Figure 3C and D suggested that overexpression of LINC00888 enhanced the cell invasion in the melanoma cells. Moreover, overexpression of LINC00888 not only increased EMT (Figure 3E and F), but promoted A375-S2 cell colony formation (Figure 3G and H). All these data were consistent with previous assays.
Figure 3.
Overexpression of LINC00888 enhanced the A375-S2 cell proliferation and invasion. (A) Transfection of lenti-LINC00888 effectively overexpressed LINC00888 in the cells, revealed by qRT-PCR (n=3, **p<0.01 vs control). (B) The cell proliferation of cells transfected with lentivirus empty control or lenti-LINC00888 from 0 to 72 hours posttreatment (n=3, **p<0.01 vs control). (C) Representative invasion of cells transfected with negative control or lenti-LIN00888 was taken using a phase-contrast inverted microscope. (D) The quantitative comparison of invaded A375-S2 cells transfected with negative control or lenti-LIN00888 (n=3, **p<0.01 vs control). (E) Protein expression of E-cadherin and vimentin in A375-S2 cells after transfection with negative control or lenti-LIN00888 for 72 hours. (F) The quantification of E-cadherin and vimentin protein expression in the cells (n=3, **p<0.01 vs control). (G) A375-S2 cells were seeded in 6-well plates at 500 or 1,000 cells per well overnight, and then, the cells were treated with control or LINC00888 overexpression particles for 72 hours. And then, the pictures of colony format were taken using a digital camera. (H) The violet crystals were dissolved with 70% ethanol, and absorbance (595 nm) was measured with microplate reader (n=3, *p<0.05, **p<0.01 vs control).
Abbreviations: lenti-LINC0088, lentivirus LINC00888; qRT-PCR, quantitative real-time polymerase chain reaction.
LINC00888 regulated the melanoma cells via suppressing miR-126-5p level
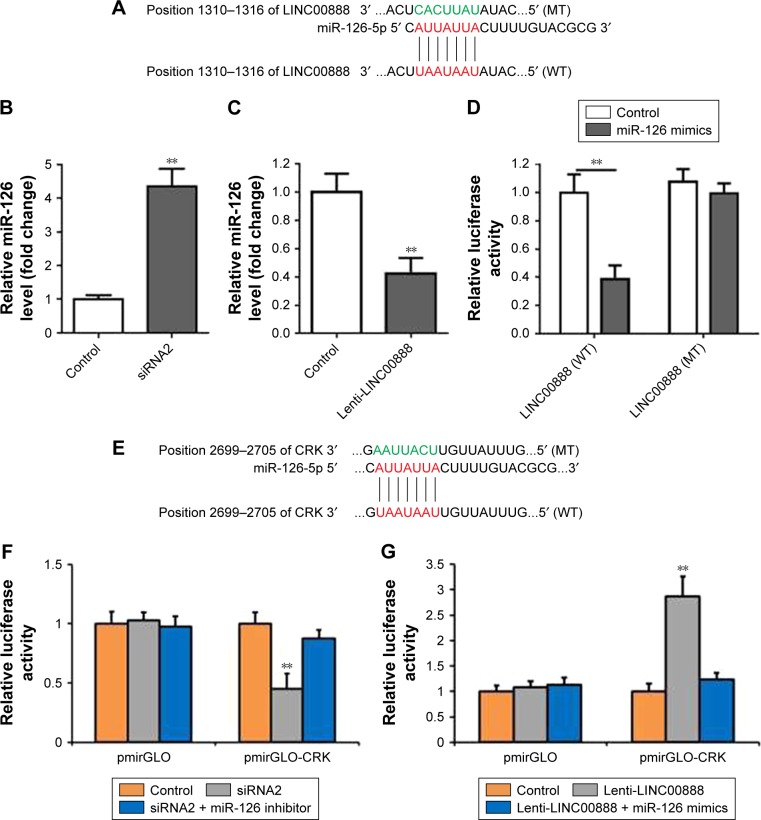
Recently, compelling evidence has demonstrated that lncRNAs contain the motif which is complementary to the sequence of miRNAs. In order to uncover whether LINC00888 could interact with miRNAs, starBase Version 2.0 was used to predict potential interact miRNAs.6,7 By searching this database, we found that miRNA-126 (miR-126) got the highest score and might interact with LINC00888 (Figure 4A). To experimentally confirm this result, qRT-PCR was performed to determine miR-126 expression in the condition of LINC00888 knockdown in A375-S2 cells. We found that miR-126 level was significantly elevated in the cells transfected with LINC00888 siRNA2, compared with control group (p<0.01; Figure 4B). In contrast, upregulation of LINC00888 exerted an inhibitory effect on miR-126 expression in the cells (p<0.01; Figure 4C). To further determine the direct interaction between miR-126 and LINC00888, dual-luciferase reporter assay was performed. The data showed that LINC00888 level was significantly decreased in the cells treated with miR-126 mimics (p<0.01), while there was little impact on the expression of LINC00888 mutant in cells with miR-126 mimics (Figure 4D). These results strongly suggested that LINC00888 promoted the melanoma tumorigenicity via directly interacting with miR-126.
Figure 4.
LINC00888 directly suppressed miR-126/CRK axis in A375-S2. (A) The predicted binding sites between LINC00888 and miR-126-5p through complementary base-pairs. The wild-type LINC00888 3′UTR (WT; containing the binding sites of miR-126-5p) or mutant-type LINC00888 3′UTR (MT) vectors were constructed. (B and C) A375-S2 cells were treated with LINC00888 siRNA2 or lenti-LINC00888 for 72 hours, and the expression of miR-126 were detected by qRT-PCR (n=3, **p<0.01 vs control). (D) miR-126 could directly regulate transcriptional level of LINC00888, demonstrated by dual-luciferase reporter assay (n=3, **p<0.01 vs control). (E) The predicted binding sites between targeted gene CRK and miR-126-5p through complementary base-pairs. The wild-type CRK 3′UTR (WT) or mutant-type CRK 3′UTR (MT) vectors containing the binding sites of miR-126-5p were constructed. (F) LINC00888 stable knockdown and control A375-S2 cells were cotransfected with miR-126 inhibitors and luciferase reporters containing CRK 3′UTR, or nothing. Luciferase activities were then detected by dual-luciferase reporter assay (n=3, **p<0.01 vs control). (G) LINC00888 stable overexpression and control A375-S2 cells were cotransfected with miR-126 inhibitors and luciferase reporters containing CRK 3′UTR, or nothing. Luciferase activities were then detected by dual-luciferase reporter assay (n=3, **p<0.01 vs control).
Abbreviations: lenti-LINC0088, lentivirus LINC00888; miR-126, microRNA-126; qRT-PCR, quantitative real-time polymerase chain reaction; UTR, untranslated region.
MiR-126 restored LINC00888-induced cell proliferation in melanoma cells via targeting CRK
We next used starBase Version 2.0 to identify the possible targets that could be responsible for the impact of miR-126. The results indicated that v-crk sarcoma virus CT10 oncogene homolog (CRK) was the target of miR-126 (Figure 4E). CRK is a key adaptor molecule of migration and adhesion, which has been involved in various intracellular signal pathways related to cell adhesion, proliferation, and migration. Therefore, dual-luciferase reporter assay was used to further validate the interaction between miR-126 and CRK. As shown in Figure 4F, LINC00888 siRNA2 resulted in a significantly reduced CRK expression (p<0.01), which was dramatically reversed by miR-126 inhibitor. In contrast, overexpression of LINC00888 led to remarkable increased luciferase activity in A375-S2 cells (p<0.005), while miR-126 mimics attenuated this kind of increases (Figure 4G).
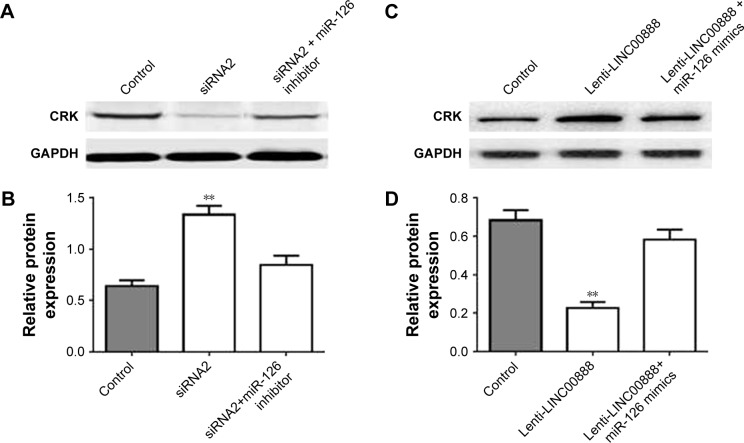
In consistent with dual-luciferase reporter assay, the results of Western blot demonstrated that miR-126 was able to rescue LINC00888-induced CRK protein upregulation (Figure 5A–D). Thus, these results further confirmed that miR-126 restored LINC00888-induced cell proliferation in melanoma cells via targeting CRK.
Figure 5.
Overexpression of LINC00888 promoted the CRK expression via suppressing miR-126. (A) CRK and GAPDH expressions in cells transfected with negative control, LINC00888 siRNA2, or LINC00888 siRNA2 plus miR-126 inhibitor were measured by Western blot. (B) The quantitation of CRK protein expression in cells transfected with control, LINC00888 siRNA2, or LINC00888 siRNA2 plus miR-126 inhibitor (n=3, **p<0.01 vs control). (C) CRK and GAPDH expression in the cells transfected with control, lenti-LINC00888, or lenti-LINC00888 plus miR-126 mimics were detected by Western blot. (D) The quantitation of CRK protein expression in cells transfected with control, lenti-LINC00888, or lenti-LINC00888 plus miR-126 mimics (n=3, **p<0.01 vs control).
Abbreviations: lenti-LINC0088, lentivirus LINC00888; miR-126, microRNA-126.
LINC00888 promoted A375-S2 tumor growth and EMT in vivo via increasing CRK protein expression
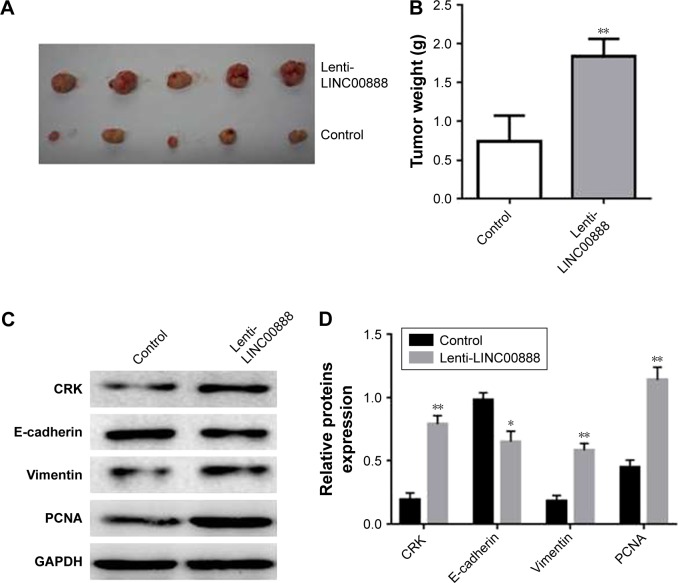
To determine whether LINC00888 could enhance tumor development and progression in vivo, A375-S2 cells transfected with lenti-LINC00888 or empty lentivirus were subcutaneously injected into nude mice, respectively. All the mice were sacrificed at day 28 after the cell injection. The results indicated that the tumors formed in lenti-LINC00888 group were significantly larger and heavier than those in control group (Figure 6A and B). In addition, LINC00888 overexpression promoted A375-S2 tumor growth and EMT in vivo via the upregulation of CRK protein (Figure 6C and D). All these data revealed that overexpression of LINC00888 was able to promote melanoma tumorigenesis in vivo via increasing CRK protein expression.
Figure 6.
Overexpression of LINC00888 promoted A375-S2 tumor growth via increasing CRK, expression, and EMT in vivo. (A) The comparison of tumor sizes from five nude mice inoculated with LINC00888 stable overexpression or control A375-S2 cells, respectively. (B) Tumor weights of nude mice inoculated with LINC00888 stable overexpression or control A375-S2 cells were recorded in 28 days, respectively (n=5, **p<0.01 vs control). (C) CRK, E-cadherin, vimentin, and PCNA protein expressions in these two groups were detected by Western blot on day 28. (D) The quantification of CRK, E-cadherin, vimentin, and PCNA proteins expressions in tumor tissues (n=3, *p<0.05, **p<0.01 vs control).
Discussion
In this study, we demonstrated the oncogenic role of LINC00888 in both clinical melanoma samples and melanoma cell lines. First, we identified a remarkable elevated level of LINC00888 expression in melanoma specimens compared with adjacent normal tissues. Clinical observation further showed that 4-year overall survival in patients with low level of LINC00888 was improved compared with patients with a high level of LINC00888. LINC00888 enhanced cellular proliferation, invasion, and EMT but inhibited the cellular apoptosis in melanoma cells. Furthermore, LINC00888 regulated the melanoma cells via suppressing miR-126-5/CRK axis, as LINC00888 was able to regulate miR-126 expression and further modulate the transcriptional and translational levels of CRK. Last, the tumor xenografts study further demonstrated that LINC00888 enhanced the tumor development in vivo. Thus, given the above results, we reasoned that lncRNA LINC00888 functioned as an oncogene in melanoma tumorigenesis and metastasis, which could be a promising molecular target for treating melanoma.
Dysregulation of miRNAs has been linked with suppression or progression of initiation, differentiation, development, or prognostic biomarkers for melanoma.8–10 Derived from a common precursor at chromosome 9q34.3, miR-126 is derived from epidermal growth factor-like domain 7 (egfl7) genes, which is important for vascular system development and highly expressed in vascularized tissues.11 The expression of miR-126 is produced by EGFL7 pre-mRNA splicing and modulated by gene promoter of EGFL7.12 Although we did not determine the miR-126 expression in melanoma, substantial different expression of miR-126 has been reported to be downregulated in various types of cancer,13–16 including lung cancer,15 gastric cancer,16 leukemia,17 thyroid cancer,18 and melanoma,19 possibly through the induction of cell arrest in G0/G1 phase.20 This is consistent with our hypothesis that elevated LINC00888 expression facilitates melanoma development via direct downregulation of miR-126 expression. A previous study also suggested that in melanoma cell lines, miR-126 may serve as a tumor suppressor via direct or indirect regulation of several important oncogenic molecules, such as ADAM9 and MMP7.19 Therefore, miR-126 might suppress melanoma development through inhibiting angiogenesis.21 However, the detailed mechanisms of miR-126 as a tumor suppressor in melanoma remained unclear, and more investigations are needed.
CRK is the predicted and further validated target gene for miR-126 in previous and present studies.15,16 As a member of a family of master modulators of migration and adhesion in many cell types, CRK was involved in various intracellular signal pathways that modulating cell adhesion, proliferation, and migration. In lung cancer cells, CRK knockdown induced an evident decreased cancer cell adhesion, migration, and invasion, suggesting a critical role in tumorigenesis. Recently, it was shown that CRK proteins can selectively regulate T-cell adhesion and migration into effector sites, which is consistent with our observation in melanoma cells.22 In addition, CRK also plays an important role in various types of cancer, including gastric cancer,16 breast cancer,23 and lung cancer.24 For example, CRK expression could be functioned as an independent prognostic factor for patients with gastric cancer.16 It was suggested that CRK received oncogenic signals from PRAS, and therefore, PARK1/CRK axis is a prominent prooncogenic pathway in nonsmall cell lung cancer.24 Therefore, CRK protein could be also essential in melanoma development and progression, while the underlying molecular mechanisms in the melanoma need further investigation.
MiR-126/CRK axis has been highlighted in gastric cancer.16,25,26 It was suggested that miR-126 can suppress the gastric cancer cell by targeting three genes simultaneously, PI3KR2, CRK, and the PLK2.25,26 The combined panel of miRNA-126 and CRK expression may function as an independent prognostic biomarker for patients with advanced gastric cancer. In addition to CRK protein tested in this study, previous bioinformatics analysis highlighted that a simultaneously downregulated gene of LINC00888 is basonuclin 1 (BNC1),5 a zinc finger transcription factor that was able to activate many known targets, and the absence of BNC1 enhances the metastasis of breast cancer.27 Although we did not test BNC1 protein expression in this study, it will be interesting to determine any direct association of LINC00888, BNC1, and CRK protein in the context of melanoma in future.
It is well established that lncRNAs can modulate the downstream target genes by various approaches at both transcriptional and posttranscriptional levels.28 Indeed, it has been widely established that lncRNAs are linked with clinicopathogenesis and prognosis of malignant tumors, including tumorigenesis, metastasis, migration, and drug resistance.29–31 The lncRNAs can orchestrate complex mechanisms on many important cancer phenotypes, including epigenetic regulation of protein-coding genes and dysregulated noncoding RNAs. Indeed, in our study, LINC00888 was found to enhance the melanoma development and metastasis by downregulating the expression of miRNA-126 and further modulating target CRK protein. These findings indicated that LINC00888 may serve as a new prognostic biomarker for melanoma and might be also functioned as a potential target for cancer therapy.
Conclusion
For the first time, the present study highlighted the potential clinical utility of LINC00888 as a novel prognostic biomarker for melanoma patients. We validated the clinical significance of LINC00888 not only from the clinical samples but also from cellular and molecular level. We concluded that LINC00888 could regulate the cellular proliferation and invasion via miR126/CRK signaling pathway and thus play an essential role in melanoma initiation, development, and metastasis.
Acknowledgments
We would like to thank Ke Wu for bioinformatics analysis.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.National Cancer Institute [webpage on the Internet] Surveillance, epidemiology, and end results. 2017. [Accessed May 8, 2017]. Available from: http://www.seercancergov/statfacts/
- 2.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 3.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 4.Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, Fan W, Wan B, et al. Characterization of long noncoding RNA and messenger RNA signatures in melanoma tumorigenesis and metastasis. PLoS One. 2017;12(2):e0172498. doi: 10.1371/journal.pone.0172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dweep H, Gretz N, Sticht C. miRWalk database for miRNA-target interactions. Methods Mol Biol. 2014;1182:289–305. doi: 10.1007/978-1-4939-1062-5_25. [DOI] [PubMed] [Google Scholar]
- 8.Thyagarajan A, Shaban A, Sahu RP. miRNA directed cancer therapies: implications in melanoma intervention. J Pharmacol Exp Ther. 2018;364(1):1–12. doi: 10.1124/jpet.117.242636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011;286(19):16606–16614. doi: 10.1074/jbc.M111.227611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mannavola F, Tucci M, Felici C, Stucci S, Silvestris F. miRNAs in melanoma: a defined role in tumor progression and metastasis. Expert Rev Clin Immunol. 2016;12(1):79–89. doi: 10.1586/1744666X.2016.1100965. [DOI] [PubMed] [Google Scholar]
- 11.Hansen TF, Christensen Rd, Andersen RF, Sørensen FB, Johnsson A, Jakobsen A. microRNA-126 and epidermal growth factor-like domain 7-an angiogenic couple of importance in metastatic colorectal cancer. Results from the Nordic ACT trial. Br J Cancer. 2013;109(5):1243–1251. doi: 10.1038/bjc.2013.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15(2):261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu LY, Wang W, Zhao LY, et al. miR-126 inhibits growth of SGC-7901 cells by synergistically targeting the oncogenes PI3KR2 and Crk, and the tumor suppressor PLK2. Int J Oncol. 2014;45(3):1257–1265. doi: 10.3892/ijo.2014.2516. [DOI] [PubMed] [Google Scholar]
- 14.Liu B, Peng XC, Zheng XL, Wang J, Qin YW. miR-126 restoration downregulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66(2):169–175. doi: 10.1016/j.lungcan.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Crawford M, Brawner E, Batte K, et al. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373(4):607–612. doi: 10.1016/j.bbrc.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 16.Yue S, Shi H, Han J, Zhang T, Zhu W, Zhang D. Prognostic value of microRNA-126 and CRK expression in gastric cancer. Onco Targets Ther. 2016;9:6127–6135. doi: 10.2147/OTT.S87778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibayama Y, Kondo T, Ohya H, Fujisawa S, Teshima T, Iseki K. Upregulation of microRNA-126-5p is associated with drug resistance to cytarabine and poor prognosis in AML patients. Oncol Rep. 2015;33(5):2176–2182. doi: 10.3892/or.2015.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian Y, Wang X, Lv Z, et al. MicroRNA126 is downregulated in thyroid cancer cells, and regulates proliferation, migration and invasion by targeting CXCR4. Mol Med Rep. 2016;14(1):453–459. doi: 10.3892/mmr.2016.5276. [DOI] [PubMed] [Google Scholar]
- 19.Felli N, Felicetti F, Lustri AM, et al. miR-126&126* restored expressions play a tumor suppressor role by directly regulating ADAM9 and MMP7 in melanoma. PLoS One. 2013;8(2):e56824. doi: 10.1371/journal.pone.0056824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng R, Chen X, Yu Y, et al. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298(1):50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Hu MH, Ma CY, Wang XM, et al. MicroRNA-126 inhibits tumor proliferation and angiogenesis of hepatocellular carcinoma by down-regulating EGFL7 expression. Oncotarget. 2016;7(41):66922–66934. doi: 10.18632/oncotarget.11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Clarke F, Karimi M, et al. CRK proteins selectively regulate T cell migration into inflamed tissues. J Clin Invest. 2015;125(3):1019–1032. doi: 10.1172/JCI77278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S, Lu B, Davra V, Hornbeck P, Machida K, Birge RB. Crk tyrosine phosphorylation regulates pdgf-bb-inducible src activation and breast tumorigenicity and metastasis. Mol Cancer Res. 2018;16(1):173–183. doi: 10.1158/1541-7786.MCR-17-0242. [DOI] [PubMed] [Google Scholar]
- 24.Mortazavi F, Lu J, Phan R, et al. Significance of KRAS/PAK1/Crk pathway in non-small cell lung cancer oncogenesis. BMC Cancer. 2015;15:381. doi: 10.1186/s12885-015-1360-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Wang F, Qi Y. miR-126 inhibits the invasion of gastric cancer cell in part by targeting Crk. Eur Rev Med Pharmacol Sci. 2014;18(14):2031–2037. [PubMed] [Google Scholar]
- 26.Liu LY, Wang W, Zhao LY, et al. miR-126 inhibits growth of SGC-7901 cells by synergistically targeting the oncogenes PI3KR2 and Crk, and the tumor suppressor PLK2. Int J Oncol. 2014;45(3):1257–1265. doi: 10.3892/ijo.2014.2516. [DOI] [PubMed] [Google Scholar]
- 27.Pangeni RP, Channathodiyil P, Huen DS, et al. The GALNT9, BNC1 and CCDC8 genes are frequently epigenetically dysregulated in breast tumours that metastasise to the brain. Clin Epigenetics. 2015;7:57. doi: 10.1186/s13148-015-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30(8):348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Liu X, Yan C, et al. LEF1-AS1, a long-noncoding RNA, promotes malignancy in glioblastoma. Onco Targets Ther. 2017;10:4251–4260. doi: 10.2147/OTT.S130365. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Zhang C, Liu C, Wu J, et al. Upregulation of long noncoding RNA LOC440040 promotes tumor progression and predicts poor prognosis in patients with prostate cancer. Onco Targets Ther. 2017;10:4945–4954. doi: 10.2147/OTT.S138354. [DOI] [PMC free article] [PubMed] [Google Scholar]