Abstract
Mutations in mitochondrial complex II (succinate dehydrogenase; SDH) genes predispose to paraganglioma tumors that show constitutive activation of hypoxia responses. We recently showed that SDHB mRNAs in hypoxic monocytes gain a stop codon mutation by APOBEC3A-mediated C-to-U RNA editing. Here, we test the hypothesis that inhibition of complex II facilitates hypoxic gene expression in monocytes using an integrative experimental approach. By RNA sequencing, we show that specific inhibition of complex II by atpenin A5 in normoxic conditions mimics hypoxia and induces hypoxic transcripts as well as APOBEC3A-mediated RNA editing in human monocytes. Myxothiazol, a complex III inhibitor, has similar effects in normoxic monocytes. Atpenin A5 partially inhibits oxygen consumption, and neither hypoxia nor atpenin A5 in normoxia robustly stabilizes hypoxia-inducible factor (HIF)-1α in primary monocytes. Several earlier studies in transformed cell lines suggested that normoxic stabilization of HIF-1α explains the persistent expression of hypoxic genes upon complex II inactivation. On the contrary, we find that atpenin A5 antagonizes the stabilization of HIF-1α and reduces hypoxic gene expression in transformed cell lines. Accordingly, compound germline heterozygosity of mouse Sdhb/Sdhc/Sdhd null alleles blunts chronic hypoxia-induced increases in hemoglobin levels, an adaptive response mainly regulated by HIF-2α. In contrast, atpenin A5 or myxothiazol does not reduce hypoxia-induced gene expression or RNA editing in monocytes. These results reveal a novel role for mitochondrial respiratory inhibition in induction of the hypoxic transcriptome in monocytes and suggest that inhibition of complex II activates a distinct hypoxia signaling pathway in a cell-type specific manner.
Introduction
Germline heterozygous mutations in SDH (mitochondrial complex II, MTCII) genes, primarily SDHB, SDHC, and SDHD, predispose to hereditary paragangliomas (PGLs) (1–3). MTCII is a heterotetrameric enzyme that has dual roles in the respiratory chain and TCA cycle (4). MTCII oxidizes succinate to fumarate and transfers the electrons from flavoprotein (SDHA) to ubiquinone at the interface between the iron-sulfur subunit (SDHB) and the membrane-spanning domain comprised of large and small cytochrome b subunits (SDHC/SDHD). SDH-mutated PGLs often develop in the oxygen-sensitive carotid body in the neck (1), and expression profiling shows normoxic induction of hypoxia-related genes (5–7). Persistent hypoxic stimulation, during high-altitude dwelling or chronic lung or cyanotic heart diseases increases the risk of sporadic carotid body PGLs (8,9). Higher altitudes also increase the severity of SDH-mutated PGLs (10,11). Collectively, these results suggest that MTCII is involved in oxygen sensing and signaling in paraganglionic cells and that persistent hypoxic stimulation may contribute to tumor formation in PGL.
Transcriptional responses to hypoxia are primarily mediated by hypoxia inducible factors (HIFs) (12). HIFs are comprised of an oxygen-labile α subunit and a constitutively expressed common β subunit. HIFα subunits are hydroxylated by prolyl or asparaginyl hydroxylases (PHD1, PHD2 and PHD3, FIH) in the presence of O2 and are subsequently degraded by ubiquitination where pVHL plays an adaptor role (13). O2 deprivation inhibits PHDs, stabilizes HIFαs and activates HIF-α/β dimer as a transcription factor. HIF-1α is expressed ubiquitously, whereas HIF-2α expression is confined to the heart, lung, endothelium, kidney, and small intestine (14). HIF-3α may negatively regulate the transcriptional activity of HIF-1 (15).
Several studies have suggested that normoxic RNA interference of SDH subunits in 293T or tumor cell lines stabilizes HIF-1α via increased reactive oxygen species or succinate levels and proposed this as the molecular basis of persistent hypoxic stimulation in SDH-mutated PGLs (16–18). However, in other studies, RNA interference of SDH subunits did not cause normoxic stabilization of HIF-1α in 293T, PC12 pheochromocytoma and C1 mouse ovarian cancer cell lines (19–21). Also, germline Sdh/SDH mutations did not stabilize HIF-1α in mouse primary cells or human neutrophils in normoxia (22,23). Sdhb or Sdhd homozygous null genotypes are lethal in utero; whereas heterozygous or conditional null genotypes do not develop PGLs in mice (24,25). Recently, somatic mutations in HIF2A (EPAS1) are described in PGL associated with erythrocytosis (26). Experimental, genetic and evolutionary studies have established that the PHD-HIF system, primarily through HIF2α, mediates hypoxia-induced erythrocytosis (27,28). In contrast, erythrocytosis is not known to be a feature of SDH-mutated paraganglioma syndrome.
We recently observed that hypoxia coordinately induces site-specific C-to-U (C > U) RNA editing of hundreds of genes in peripheral blood monocytes (29,30). Monocytes characteristically express the surface CD14 antigen, whereas the other mononuclear cells in peripheral blood (primarily B and T lymphocytes and natural killer cells) are negative for CD14. Widespread C > U RNA editing in monocytes is not observed in lymphocytes and is catalyzed by the innate viral restriction factor APOBEC3A (A3A) (29–31), which belongs to the seven member family of APOBEC3 cytidine deaminases (32). A3A inhibits retrotransposons and several exogenous viruses (e.g. hepadna and herpes) (33). A3A-mediated RNA editing is also induced by interferon type 1 (IFN1) which acts additively to hypoxia (30). Hypoxic induction of A3A-mediated RNA editing, which occurs without transcriptional induction of A3A, edits up to 20% of SDHB mRNAs in monocytes to introduce a pathogenic c.C136U/R46X mutation (34). SDHB c.136C > U RNA editing is associated with SDHB protein downregulation (30). The RNA seq analysis also revealed mRNA editing of SDHA (c.C562T/R188W) in hypoxic monocytes (30).
Monocytes circulate in highly oxygenated peripheral blood then exit to sites of inflammation, cancer, infection, atheroma plaques, which are characterized by micro-environmental hypoxia (35). Monocytes have direct antimicrobial roles and are precursors of macrophages and inflammatory dendritic cells (36). Therefore, hypoxia-sensing pathways in monocytes may define therapeutic targets in common diseases. Hypoxia induces substantial gene expression changes in monocytes by poorly understood mechanisms (37,38). Stabilization of HIF-1α, HIF-2α or HIF-3α subunits could not be determined in hypoxic monocytes (39). A recent study showed stabilization of HIF-1α in hypoxic monocytes, but HIF-1α was localized to cytoplasm not nucleus (39,40). Fangradt et al. suggested that NF-κB rather than HIFs mediate transcription of hypoxia-induced genes in monocytes (40).
SDHB mRNA editing in hypoxic monocytes raises the hypothesis that inactivation of MTCII may amplify hypoxia responses. In this study, we examined the role of MTCII in hypoxia responses in monocytes and transformed cell lines by pharmacologic inhibitors, and in Sdh knockout mouse model. Since HIF1A is ubiquitously expressed, including in monocytes, and stabilization of its protein product has been examined in multiple experimental models of MTCII (5,16–21), we studied HIF-1α in our cell culture models. We present evidence that inhibition of MTCII mimics the transcriptional effects of hypoxia in normoxic monocytes without robust stabilization of HIF-1α, but antagonizes (a) hypoxic stabilization of HIF-1α in transformed cell lines and (b) hypoxia-induced increases in hemoglobin levels in a heterozygous Sdh mouse model.
Results
Atpenin A5 (AtA5) in normoxia induces hypoxia-related RNA editing by A3A in monocytes
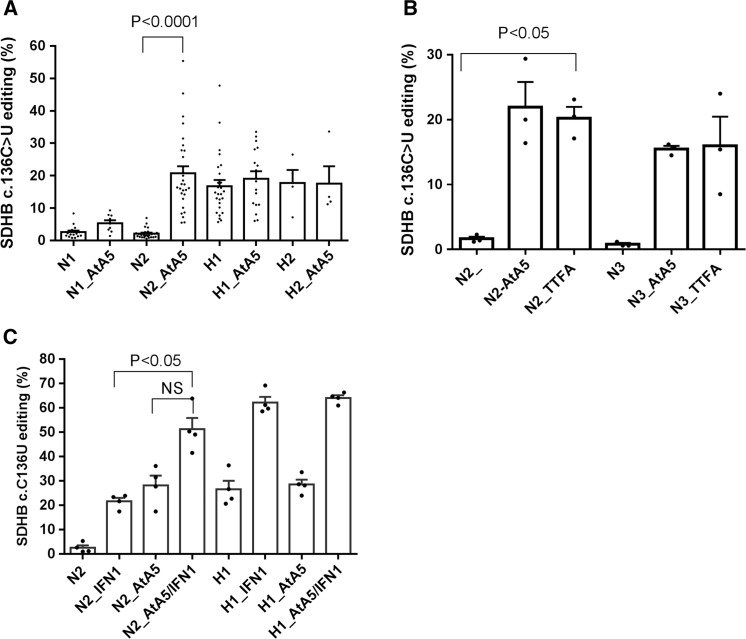
To test whether inactivation of MTCII triggers hypoxia responses in monocytes, we used AtA5, a ubiquinone homolog and a highly specific and potent inhibitor (41,42). AtA5 in normoxia (AtA5/normoxia) induced SDHB c.C136U RNA editing, especially on day 2 in cultures of monocyte-enriched PBMCs (MEPs) (Fig. 1A). RNA editing levels induced by hypoxia (day 1) versus AtA5/normoxia (day 2) were similar. Joint treatment by AtA5 and hypoxia did not further increase RNA editing levels. TTFA, another ubiquinone analog but a less potent inhibitor of MTCII, also induced RNA editing in normoxia (Fig. 1B). A3A-mediated RNA editing by hypoxia and IFN1 is additive (30). We find that RNA editing by AtA5 and IFN1 in normoxia is also additive (Fig. 1C), whereas no additional effect of AtA5 is seen in hypoxia with IFN1. These results demonstrate that normoxic inhibition of MTCII induces A3A-mediated RNA editing in monocytes in a manner similar to hypoxia.
Figure 1.
Normoxic inhibition of complex II triggers induction of A3A-mediated RNA editing observed in hypoxia. (A) Bar graph depicts percentage SDHB c.136 C > U RNA editing in monocyte-enriched PBMCs (MEPs), approximately 30 million/ml, when treated with Atpenin A5 (AtA5, 1 µM-2 µM) under normoxic (N) or hypoxic (H; 1% O2) conditions for 1 or 2 days (e.g. H2 = day 2 in hypoxia, minimum (n)=4 and maximum (n)=29 donors). (B) Bar graph depicts percentage SDHB c.136 C > U RNA editing upon treatment with TTFA in normoxia for 2 or 3 days. (C) Bar graph depicts percentage SDHB c.136 C > U RNA editing upon treatment with AtA5 and/or IFN1 when subjected to normoxia or hypoxia for 1 or 2 days. Mean and SEM are shown in scatter bar plot. NS: not significant.
AtA5 in normoxia induces hypoxia-related gene expression in monocytes
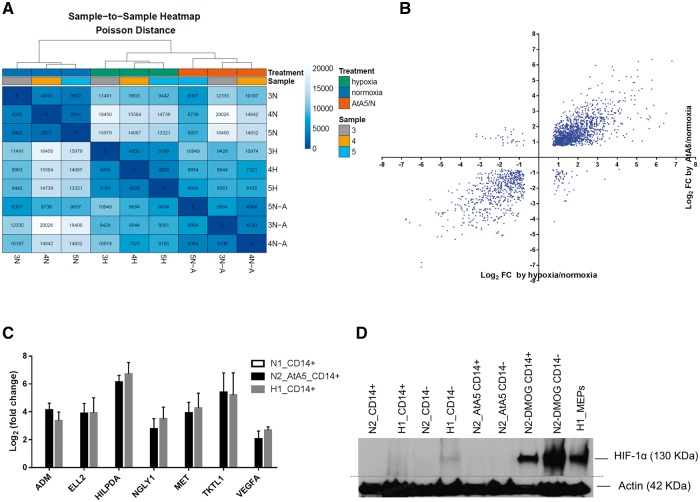
We examined whether MTCII also regulates induction of gene expression in primary monocytes under hypoxia or when inactivated, as observed in SDH-mutated paragangliomas. To test the impact of MTCII inhibition on monocyte gene expression, we cultured MEPs (n = 3 donors) in normoxia (1 day), hypoxia (1 day) and AtA5/normoxia (2 days), isolated CD14+ monocytes and performed RNA seq analysis. SDHB c.136C > U RNA editing increased in CD14+ cells in hypoxia (mean ± SEM = 29.9 ± 9.9%) and AtA5/normoxia (mean ± SEM = 32.1 ± 14.9%) relative to normoxic controls (mean ± SEM = 5.4 ± 1.7%). Gene expression data without making any assumptions on the experimental design revealed evidence of similar expression patterns with hypoxia and AtA5/normoxia treatment (Fig. 2A and Supplementary Material, Fig. S1). Genes that are expressed at high levels (RPKM > 0.5, n = 9,389) and statistically significantly affected by hypoxia or AtA5/normoxia (P-adjusted <0.05) showed a very strong positive correlation in fold changes (Fig. 2B). HIF1A but not EPAS1 (HIF2A) or HIF3A is expressed robustly (RPKM averages 21.5, 0.17 and 0.01, respectively) in monocytes. Hypoxia or AtA5/normoxia induced RNA editing without transcriptional upregulation of A3A (Supplementary Material, Table S2). RT-qPCR analyses confirmed the induction of selected highly-expressed genes (ADM, ELL2, HILPDA, NGLY1, MET, TKTL1 and VEGFA) both by hypoxia and AtA5/normoxia (Fig. 2C). RNA seq analysis showed induction of RNA editing in genes other than SDHB by both hypoxia and AtA5 (unpublished data). The induction of SDHB mRNA editing and hypoxic gene expression are confirmed in CD14+ monocytes obtained from three additional donors as well as in MEPs by hypoxia at day 1 and by AtA5/normoxia at day 2 but not in untreated normoxic controls at day 2 (Supplementary Material, Fig. S2). Western blot showed HIF-1α stabilization in hypoxic (1% O2) CD14– cells but not in hypoxic CD14+ monocytes nor in normoxic CD14+ or CD14– cells treated with AtA5 (Fig. 2D). DMOG, an inhibitor of PHD enzymes, caused robust normoxic stabilization of HIF-1α both in CD14+ and CD14– cells (day 2). These results suggest that hypoxia and AtA5/normoxia have similar signaling pathways to induce the gene expression changes in CD14+ monocytes independently of HIF-1α.
Figure 2.
Atpenin A5 (AtA5) in normoxia induces transcriptome-scale gene expression responses similar to hypoxia in monocytes (A) Unsupervised heat map shows clustering of hypoxic (day 1) and AtA5/normoxia (N-A) (day 2) samples. Samples 3, 4 and 5 represent CD14 positive monocytes from three donors, isolated after culture of MEPs. (B) Scatter plot shows a strong positive correlation between gene expression changes in CD14+ cells upon exposure to hypoxia (day 1) and AtA5/normoxia (day 2) (n = 2,131 genes, Pearson r = 0.8819, P < 0.0001). (C) Bar graph depicts validation of induced expression of selected genes from RNA seq analysis in CD14+ cells under AtA5/normoxia and hypoxic conditions, as determined by RT-qPCR. Inductions by AtA5/normoxia or hypoxia are statistically significant for each gene (P < 0.05, Dunnett’s multiple comparisons test). (D) Representative immunoblot shows the expression of HIF-1α in lysates (40 µl) of CD14+ and CD14– cells. The cells were isolated after culture for 1 or 2 days in normoxia or hypoxia upon treatment with AtA5 or DMOG. Hypoxia-exposed MEP cells were used to isolate CD14+ and CD14 – populations in hypoxia chamber. Actin was used as a loading control (n = 3). The immunoblot, performed on the same day, was cropped and merged as depicted by the dotted grey line.
AtA5 and myxothiazol inhibit oxygen consumption and induce hypoxia responses in monocytes without robust stabilization of HIF-1α
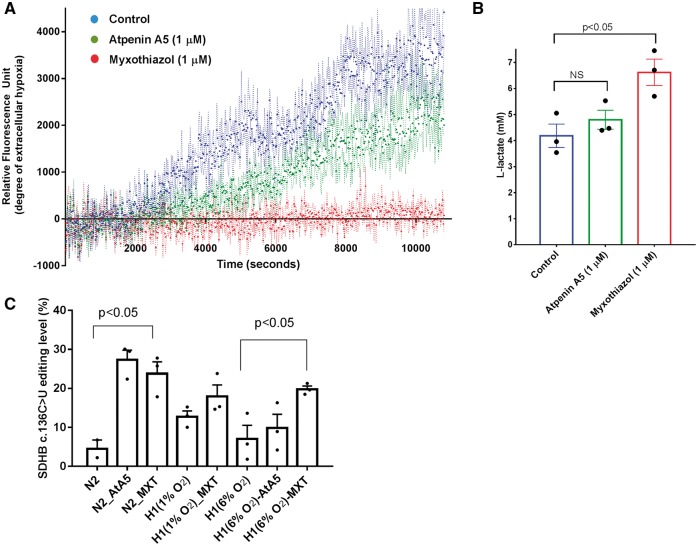
To examine whether hypoxia responses in monocytes are also induced by the inhibition of another mitochondrial complex, we used myxothiazol (MXT), a ubiquinole analog which inhibits complex III. We first measured oxygen consumption and L-lactate levels to confirm the effect of AtA5 and MXT on mitochondrial respiration in MEPs. We used a phosphorescent oxygen probe (MitoXpress-Xtra) which is quenched by oxygen. Cellular respiration in a closed system depletes oxygen and increases the fluorescence. We find that complex III inhibitor MXT completely suppressed oxygen consumption (Fig. 3A), whereas AtA5 reduced but did not abolish it in primary cells (MEPs). L-Lactate, the end product of glycolysis, increased relative to the degree of respiratory inhibition (Fig. 3B). MXT induced SDHB mRNA editing in normoxia and increased the editing level in mild hypoxia (6% O2), but had no statistically significant effect on it at 1% O2 when compared to hypoxia (1% O2) alone (Fig. 3C). Normoxic induction of RNA editing by MXT is also confirmed in a separate experiment from three additional donors (Supplementary Material, Fig. S3).
Figure 3.
AtA5 and myxothiazol (MXT) inhibit oxygen consumption and induce A3A-mediated RNA editing. (A) Graph depicts the relative fluorescence levels (mean and SEM with dashed lines), which reflect the degree of hypoxia, on treatment of monocyte-enriched PBMCs (MEPs) with AtA5 or MXT within approximately 3 h. Control indicates cells without any inhibitors. (B) Bar graph depicts L-Lactate levels in extra-cellular media from the samples analyzed in (A). NS: no significant (C) Bar graph depicts the percentage SDHB c.136 C > U RNA editing upon treatment of MEPs with MXT in normoxia or hypoxia (1 or 6% O2) for 1 or 2 days. All panels show mean and SEM in MEPs from n = 3 donors.
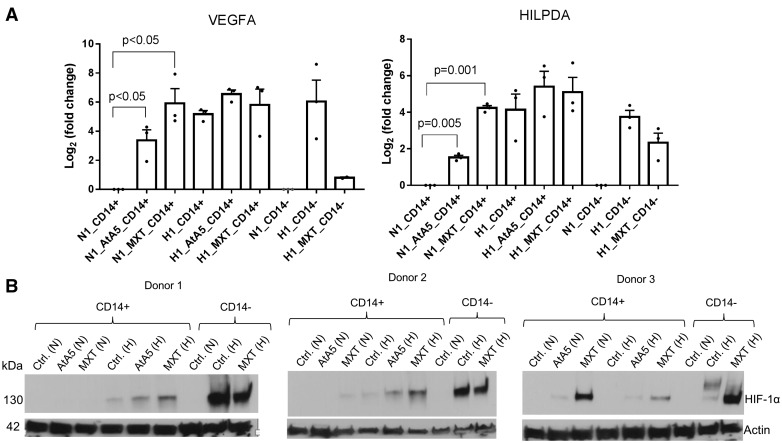
To further examine the effect of MXT and AtA5 on the expression of hypoxia regulated genes and HIF-1α protein expression, we first isolated CD14+ and CD14– cells from PBMCs of three additional donors and then exposed them for 1 day to normoxia or hypoxia (1% O2) with or without the inhibitors. MXT and AtA5 statistically significantly increased the mRNA expression of VEGFA and HILPDA in CD14+ monocytes in normoxia but not in hypoxia (Fig. 4A). Hypoxia showed robust stabilization of HIF-1α in CD14– but not in CD14+ cells (Fig. 4B). Similarly, AtA5 and MXT did not robustly stabilize HIF-1α in normoxia in monocytes. AtA5 and MXT appeared to enhance the stabilization of HIF-1α in hypoxic monocytes in two of three donors (Fig. 4B). Thus, hypoxia or mitochondrial inhibitors in normoxia induce hypoxic gene expression in monocytes without a consistent stabilization of HIF-1α when compared to its robust stabilization in hypoxic lymphocytes.
Figure 4.
AtA5 in normoxia induces hypoxic gene expression in monocytes without robust stabilization of HIF-1α. (A) Bar graph depicts fold changes in VEGF and HILPDA gene expression in normoxic and hypoxic CD14+ and CD14– cells upon treatment with AtA5 or MXT for 24 h (1 day) (n = 3 donors). (B) Immunoblot shows the expression of HIF-1α in lysates (40 µl) of CD14+ and CD14– cells examined in (A). The cells were isolated from PBMCs at room conditions followed by culture (5–7 million/ml) for 24 h in normoxia or hypoxia (1%) upon treatment with AtA5 or MXT. Actin was used as a loading control.
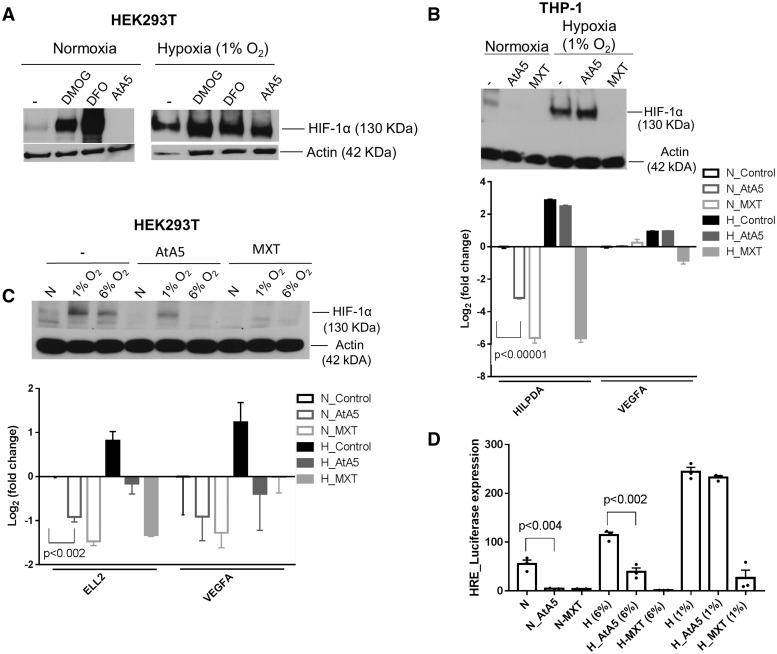
AtA5 and MXT suppress HIF-1α and hypoxia-induced gene expression in cell lines
Our results so far suggest that normoxic inhibition of complex II or complex III in monocytes induces hypoxia responses, both RNA editing and gene expression, without a consistent stabilization of HIF-1α. It is possible that HIF-1α may have degraded depending on cell type, time of analysis (24 h) or another factor. Therefore, we further examined the effect of AtA5 in HEK293T embryonic kidney cell line and THP-1 monocytic leukemia cell line over a 24-h period. Several studies in cell lines have reported normoxic stabilization of HIF-1α upon knocking down MTCII (16–18). We found that DMOG and DFO, but not AtA5 stabilized HIF-1α in normoxia in HEK293T cells. Moreover, AtA5 suppressed the weak HIF-1α expression in normoxia, which was possibly seen due to cellular crowding and peri-cellular hypoxia (Fig. 5A). Similarly, AtA5 antagonized weak normoxic stabilization of HIF-1α in THP-1 cells, whereas MXT inhibited it even at 1% O2 (Fig. 5B). Since AtA5 appeared to suppress HIF-1α in mild hypoxia, we exposed the cells to both 6 and 1% O2. A confirmatory experiment in 293T cells showed that AtA5 inhibited stabilization of HIF-1α only at 6% O2, whereas MXT inhibited it both at 6% O2 and 1% O2 (Fig. 5C). RT-qPCR analyses showed that expression of hypoxia-regulated genes is suppressed by AtA5 and MXT in parallel to their ability to suppress HIF-1α (Fig. 5B and C, lower panels). To confirm the suppression of HIF-regulated gene expression, we used a plasmid construct expressing firefly luciferase under the control of three hypoxia-response elements (HRE) from PGK1, a HIF-1α target gene. AtA5 statistically significantly suppressed HRE-driven expression in 6% O2 but not in 1% O2, whereas MXT inhibited HRE-driven expression both in 6% O2 and 1% O2 (Fig. 5D). These results demonstrate that inhibition of MTCII by AtA5 in transformed cell lines does not stabilize HIF-1α in normoxia, but rather blocks it in mild hypoxia.
Figure 5.
AtA5 or MXT antagonizes HIF-1α and reduces hypoxic gene expression in transformed cell lines. (A) Immunoblot shows the expression of HIF-1α in lysates (40 µl) of 293T cells upon treatment with DMOG (1 mM), DFO (0.5 mM) and AtA5 (1 µM) when subjected to normoxia or hypoxia (1%) for 24 h. (B) Immunoblot shows the expression of HIF-1α in lysates (40 µl) of THP-1 cells upon treatment with AtA5 or MXT when subjected to normoxia or hypoxia (1%) for 24 h (upper panel). Bar graph depicts the fold change in gene expression of HILPDA and VEGFA under the same treatment conditions (n = 3 replicates) (lower panel) (C) Immunoblot shows the expression of HIF-1α in lysates (40 µl) of 293T cells upon treatment with AtA5 or MXT when subjected to normoxia or hypoxia (1 or 6%) for 24 h (upper panel). Bar graph depicts the fold change in gene expression of ELL2 and VEGFA in normoxia and hypoxia (1% O2) (n = 3 replicates) (lower panel) (D) Bar graph depicts the expression of the plasmid expressing luciferase under the control of hypoxia response element (HRE) when 293T cells are treated with AtA5 or MXT in normoxia or hypoxia (1 or 6% O2) for 24 h. (n = 3 replicates, mean and SEM shown).
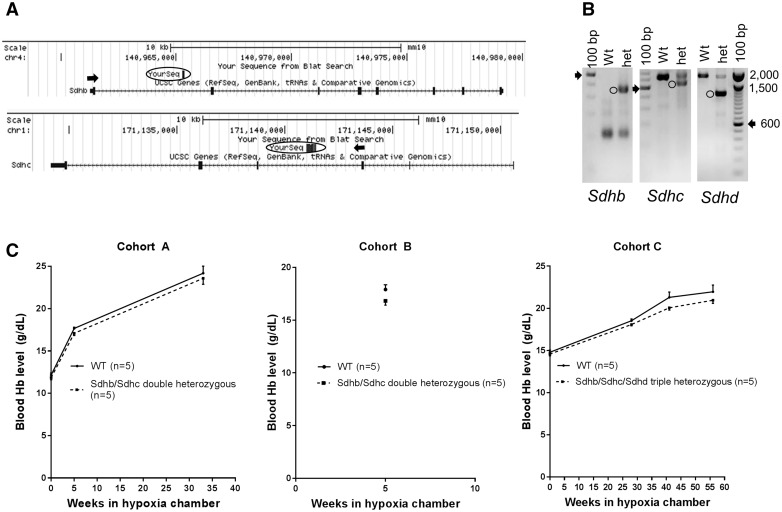
MTCII mutations reduce hemoglobin levels in chronically hypoxic mice
Since AtA5 does not induce HIF-1α in normoxia but appears to antagonize its hypoxic stabilization in 293T and THP-1 cell lines, we further studied the impact of MTCII inhibition on hypoxia response in vivo. Mice with Sdhb, Sdhc and Sdhd heterozygous knockout alleles were cross-bred to obtain Sdhb/Sdhc double heterozygous and Sdhb/Sdhc/Sdhd triple heterozygous mice. Sdhb, Sdhc, and Sdhd are located on mouse chromosomes 4, 1, and 9, respectively. Cross-mating of Sdhb/Sdhc double heterozygous mice did not give any viable progeny homozygous for Sdhb or Sdhc mutations (P < 0.0001, Chi-Square test), supporting that Sdhb and Sdhc alleles obtained by gene trapping are null (Fig. 6A and B). Mating of Sdhb/Sdhc double heterozygous mice with Sdhd heterozygous mice produced all possible genotypes including Sdhb/Shdc/Sdhd triple heterozygosity as expected from independent Mendelian segregation (P = 0.5161, Chi-Square test, Supplementary Material, Table S3). To examine the role of Sdh on hemoglobin levels, we exposed mice to chronic hypobaric hypoxia (9–10% O2). Hypoxia-induced increases in hemoglobin levels were less in Sdh mutant mice compared to the WT control in each six measurement performed in three cohorts (range = 2.4–6%) (Fig. 6C). Although the effect size was small in each measurement (P > 0.05), combined analysis was statistically significant to support the hypothesis that partial inactivation of Sdh antagonizes hypoxia-induced increases in Hb levels (P < 0.05, Fig. 6C and Supplementary Material, Table S3). Since hypoxia-induced increases in Hb levels are thought to be mediated by HIFs (27,28), primarily HIF-2, these in vivo results support the data obtained in cell lines treated with AtA5 to suggest that inactivation of MTCII antagonizes HIF-mediated responses to hypoxia.
Figure 6.
Compound heterozygosity for Sdh subunit null alleles in mice blunts hypoxia-induced increases in hemoglobin levels. (A) Diagram showing Sdhb and Sdhc upstream exons (arrows) splicing into the intronic gene traps (circled). (B) Transgene screening by genomic PCR detects wild-type (WT) homozygous or heterozygous alleles (circles). Arrow shows 600 bp band in 100 bp marker lane. (C) Hemoglobin levels in hypoxia in double (Sdhb/Sdhc) or triple (Sdhb/Sdhc/Sdhd) heterozygous mice are consistently lower than in wild-type controls (P < 0.05, Fisher's combined probability test (68), n = 4 or 5 males with similar age in each cohort, Supplementary Material, Table 3, Mean and SEM).
Discussion
We have shown here that pharmacologic inhibition of mitochondrial respiration in normoxia induces A3A-mediated RNA editing and the hypoxic transcriptome in primary monocytes. On the contrary, AtA5 and MXT reduce hypoxic gene expression in THP-1 monocytic leukemia and 293T embryonic kidney cell lines by antagonizing the stabilization of HIF-1α. Partial inactivation of MTCII by heterozygous gene knockouts of Sdh subunits blunts hypoxia-induced increases in hemoglobin levels in mice. Thus, inhibition of mitochondrial respiration activates the hypoxia responses in monocytes via a distinct mechanism.
Normoxic stabilization of HIF-1α upon knockdown of MTCII subunits in cell lines (16–18) cannot be confirmed by our data which show the opposite effect (i.e. less HIF-1α, Fig. 5). Of note, such studies did not compare transcriptome-wide responses in normoxic knockdown versus hypoxic wild-type cells as performed in the current study. This comparison is important because constitutive expression of hypoxia related genes is a defining feature of SDH-mutated paragangliomas. Cervera et al (18) performed global gene expression analysis in SDHB-silenced Hep3B hepatoma cell line which stabilized HIF-1α/2α but found enrichment of functionally diverse genes including a few hypoxia-regulated genes. Studies in fumarate hydratase-deficient mouse showed that stabilization of HIF-1α observed in fumarate hydratase-related human skin and kidney tumors (43) was neither necessary for the kidney cyst formation (44) nor sufficient for the induction of hypoxia-related transcriptome as observed in VHL-mutated kidney cancers (45). Thus, it remains uncertain whether reported stabilization of HIF-αs upon knockdown of MTCII subunits in cell lines plays a role in triggering transcriptomic responses to hypoxia or in the pathogenesis of SDH-mutated PGLs.
Multiple studies have demonstrated that an active mitochondrial respiration is required for HIF-α stabilization and activity (46–51). Genetic ablation or pharmacologic inhibitors of electron transport chain components oppose stabilization of HIF-α in hypoxia. In agreement with these studies, we have shown that pharmacologic inhibition or mutations of MTCII also antagonizes HIF-regulated activity in hypoxia in transformed cell lines and in mice, respectively (Figs 5 and 6). It has been suggested that the inhibition of mitochondrial respiration causes oxygen redistribution from mitochondria to cytoplasm, and blocks stabilization of HIF-1α by reactivating the PHD enzymes (52,53). An alternative model suggests that active mitochondrial respiration is required for the release of reactive oxygen species in hypoxia which then stabilizes HIF1α (46,54) Although we found reduced oxygen consumption by AtA5, these two models are inapplicable in monocytes due to fundamental differences in monocytes from the HIF-regulated systems. First, normoxic inhibition of mitochondrial respiration in monocytes induces transcriptomic responses to hypoxia, whereas no such effect is seen in HIF-regulated systems (Figs 1 and 2). Second, hypoxic inhibition of mitochondrial respiration in monocytes has no effect on transcriptomic responses to hypoxia (Figs 1 and 4), whereas it inhibits them in HIF-regulated systems (Figs 4 and 6). Finally, transcriptomic responses to hypoxia or AtA5/normoxia treatment in monocytes occur with only weak and variable stabilization of HIF-1α (Figs 2D and 4). Although we cannot completely rule out the involvement of HIF-1α without genetic studies, our findings demonstrate that a distinct hypoxia signaling pathway regulated by mitochondria is operational in monocytes.
Since EPAS1 is not expressed at any appreciable level in monocytes, we did not examine the stabilization of HIF-2α. EPAS1 mutations are linked to the development of paragangliomas in association with erythrocytosis (26); thus, a question arises whether HIF-2α, rather than HIF-1α, mediates hypoxic gene expression in SDH-mutated paragangliomas, even though erythrocytosis, which is mainly driven by HIF-2α, is not seen in SDH-related paraganglioma syndromes. We find that Sdh heterozygous knockout alleles suppress rather than increase hypoxia-induced hemoglobin levels in mice (Fig. 6). An earlier study in a chromaffin cell line found that inhibition of mitochondrial respiration suppresses hypoxic stabilization of HIF-2α (51). These results suggest that inhibition of mitochondrial respiration also opposes stabilization of HIF-2α in hypoxia. Therefore, a general model that posits that either HIF-1α or HIF-2α stabilizes when MTCII is mutated is not supported by our data. Whether inhibition of mitochondrial respiration could stabilize HIF-1α or HIF-2α in certain cell types requires further investigations.
We observe that mitochondrial inhibitors, especially MXT, may increase hypoxic stabilization of HIF-1α in monocytes (Fig. 4B), in contrast to cell lines where they suppress it (Fig. 5). This observation suggests that regulation of HIF-1α is different in monocytes compared to transformed cell lines. Perhaps intracellular oxygen tension is not the primary determinant of HIF-1α levels in monocytes and/or a downstream effector from mitochondria stabilizes HIF-1α. Regardless of the underlying mechanism, the functional significance of increased HIF-1α in hypoxic monocytes by mitochondrial inhibitors is uncertain, since we find no statistically significant increases in hypoxic (1% O2) gene expression or RNA editing levels with the addition of inhibitors.
Our findings support a novel oxygen sensing and signaling mechanism for hypoxic transcript induction that is triggered by the inhibition of mitochondrial respiration in a cell type specific manner. Primary monocytes and SDH-mutated paragangliomas may share this mechanism. Whether acute hypoxia responses triggered by normoxic inhibition of mitochondrial respiration in carotid body chief cells (55), pulmonary vascular smooth muscle cells (56) and astrocytes (57) are followed by induction of hypoxia-related transcriptomes is uncertain. To our knowledge, primary human monocytes are the first experimental model for SDH-mutated paragangliomas in mammals in which mitochondrial respiratory inhibition triggers transcriptome-scale responses to hypoxia. It is conceivable that other specialized cell types which depend on highly-oxygenated in vivo environments (e.g. arterial blood, alveolus) may utilize mitochondria, rather than the PHD-HIF system, for oxygen sensing to regulate hypoxic gene expression. Interestingly, mitochondrial inhibitors suppress rather than induce hypoxic gene expression in THP-1 monocytic leukemic cells suggesting that the hypoxia sensing apparatus switched from mitochondria to the PHD-HIF system in the THP-1 cell line. Downstream molecular actors that induce the hypoxic gene expression and A3A activity upon inhibition of mitochondrial respiration await future discoveries.
Many studies, some dating back several decades, show that inhibition of mitochondrial respiration mimics hypoxia and triggers neurotransmitter release by carotid body chief cells (references in (58)). Recently, a mouse model deficient in Ndufs2 subunit of complex I is shown to inhibit hypoxia responses by causing tonic stimulation in carotid body cells (59). Although interruption of the mitochondrial respiration at any complex may initiate hypoxia responses in experimental models, the precise mechanism of this inhibition, or whether such inhibition occurs in vivo in hypoxia is unknown. It is tempting to speculate that initial oxygen sensing mechanisms that trigger the acute responses in carotid body cells or gene expression changes in paraganglioma tumors and monocytes are similar. Several observations in humans including the association of complex II subunit mutations with hypoxia-associated carotid body paragangliomas (1), the role of altitude in determining clinical severity of SDH-mutated paragangliomas (10,11) and inactivation of SDHB mRNAs by hypoxia-induced RNA editing in monocytes (29,30), presumably an auto-inhibitory feedback loop for enhanced adaptation to hypoxia, suggest that complex II plays a role in this hypoxia sensing pathway in vivo. We note that in comparison to MXT, AtA5 has a weaker effect in normoxic induction of hypoxia-related RNA editing (Fig. 3C) and gene expression on day 1 (Fig. 4A). RNA editing and gene expression levels observed on day 1 at 1% O2 are reached on day 2 by AtA5/normoxia in our experimental conditions (Figs 1A and 2). These results support a model that physiological inactivation of MTCII by reduced oxygen tension activates a delayed but the complete transcriptomic response to hypoxia in monocytes. Perhaps, in vivo signaling in response to hypoxia occurs by inhibition of MTCII in addition to partial inhibition of another mitochondrial complex(es).
Finally, our findings reveal for the first time small molecule inducers of A3A’s cytidine deaminating activity in primary cells. The data presented here suggest that AtA5 activates the same physiological pathway as hypoxia to trigger RNA editing (Fig. 1). Since RNA editing induced by AtA5 is additive to that induced by IFN1, inhibitors of mitochondrial respiration provide tools to further investigate for therapy against viruses that A3A is known to inhibit.
Materials and Methods
Cells, cell lines and tissue culture
Leukoreduction filters (Terumo BCT, Lakewood, CO), waste products of the platelet donation process, were used to isolate PBMCs by an IRB-approved protocol. PBMCs were isolated using Histopaque-1077 (Sigma). Monocyte-enriched PBMCs (MEPs) were prepared using a cold-aggregation method with slight modifications (30,60). Monocytes were isolated from MEPs or PBMCs using EasySep Human CD14 Positive Selection Kit (STEMCELL Technologies). Flow cytometric verification of isolated CD14+ cells was performed using RPCI core facility services. The MEPs were cultured at an average density of 25–35 × 106/ml in 1 or 2 ml per well in 6- or 12-well standard tissue culture plates under standard conditions (37 °C/5% CO2) in RPMI-1640 medium with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin (Mediatech). Isolated CD14+ and CD14– cells were cultured at approximately 5×106 cells/ml and 7×106 cells per ml densities, respectively. THP-1 and TLA-HEK293T cell lines were purchased from ATCC, and Open Biosystems®, respectively, and cultured in recommended conditions. THP-1 cells were cultured in 106 cells per 100 µl in 96-well culture plates in ATCC-formulated-1640 medium (30-2001), whereas 293T cells were cultured in DMEM medium supplemented with 10% FBS.
Hypoxia, IFN-1 and inhibitor treatment
Cells were cultured under 1 or 6% O2, 5% CO2 and 94% N2 at 37 °C using Xvivo System (Biospherix). Human ‘universal’ type I IFN was obtained from the PBL Assay Science and used at 600 U/ml. Atpenin A5 (Cayman chemical #11898), myxothiazol (Sigma-Aldrich, #T5580) and 2-Thenoyltrifluoroacetone (TTFA) (Sigma-Aldrich, #T27006) were used at 1–2 µM, 1 µM and 400 µM final concentrations, respectively. DFO (Sigma-Aldrich # D9533) and DMOG (Sigma-Aldrich # D3695) were used in 0.5 mM and 1.0 mM final concentrations, respectively.
Transfection of plasmid DNA
HEK293T cells were cotransfected with the 400 ng of HRE-luciferase (Addgene, plasmid # 26731), 1 ng of pRL-SV40 plasmid, 600 ng of pcDNA 3.1(+) (control empty vector) per well at ∼50–60% confluency using jetPRIME (Polyplus-transfection) in 12-well culture plates as per the manufacturer’s instructions. Transfection efficiency was 60–80% as assessed by fluorescent microscopy of cells co-transfected with the pLemiR plasmid DNA (Open Biosystems) for expression of a red fluorescent protein. Cells were harvested 2 days after transfection for measurement of their HRE and Renilla luciferase activities using Dual-Luciferase Reporter Assay System (Promega). HRE expression was quantified as a ratio of HRE/Renilla luciferase activities.
Immunoblotting of cell lysates
2X Laemmeli buffer (BIO-RAD) was used to prepare whole cell lysates. The lysate resuspended in the Laemmeli buffer was heated at 95 °C for 15 min, and 40 µl of the sample was used to perform gel electrophoresis on pre-cast, 4–15% gradient polyacrylamide gels (Mini-PROTEAN TGX, Bio-Rad) in Laemmli buffer system as described previously (30). Mouse monoclonal anti-HIF1α (product number GTX628480, GT10211; 1:1000 dilution, GeneTex, Inc.) and mouse monoclonal anti-β-actin (product number AM4302, AC-15; 1:15,000 dilution Life Technologies Corporation) was used to detect HIF-1α or actin, respectively, followed by incubation with HRP-conjugated goat anti-mouse antibodies (Life Technologies) at 1:2000 dilution (30). Bigger gel images of western blots of primary cells in Figures 1D and 4B are shown in Supplementary Material, Figure S4.
Oxygen consumption and L-lactate measurement
Oxygen consumption was measured using a phosphorescent oxygen probe, MitoXpress-Xtra (Cayman Dual Assay Kit, item no. 601060). Monocytes were enriched to >50% purity by short-term cold aggregation and first cultured in standard conditions for 24 h without treatment to stimulate metabolic activity. Cells were then centrifuged at 200×g for 7 min and resuspended in 1 ml RPMI/1% FBS with or without mitochondrial inhibitors. Cells are covered by mineral oil after addition of MitoXpress-Xtra following manufacturer’s protocol. The fluorescence was kinetically measured on a plate reader (Synergy H1) at 20 s intervals for approximately 3 h (delay 70 µs, collection time 30 µs). Supernatants of the oxygen consumption assay were used to measure L-lactate levels following manufacturer’s instructions.
Sdh transgenic mice and hypoxia exposure
Sdhb and Sdhc heterozygous mice in B6/129P2 background were gifts from Dr. Greg Cox (The Jackson Laboratory, Bar Harbor, Maine). The embryonic stem cell lines (Sdhb <6T(APO532)wtsi> and Sdhc <6T(BA0521)wtsi>) were generated by gene trapping (61). The gene trap vector insertion into Sdhb or Sdhc early introns creates fusion transcripts containing sequences from upstream gene exons joined to the β-geo marker, and interrupts the ORFs. Genetic verification of the knockout constructs was performed by genomic PCR and sequencing. A gene-specific intronic oligonucleotide PCR primer paired to either a vector-specific primer or another gene-specific intronic primer amplifies a knockout allele or a wild-type allele, respectively. We also re-derived a previously described Sdhd knockout mouse (62) in the C57BL/6J background at RPCI transgenic facilities using frozen sperm (mfd Diagnostics, Germany). Mouse genotyping was performed by tail DNA extraction using Allele-in-One Mouse Tail Direct PCR system (Allele Biotech) or by RPCI transgenic core facility.
Mice were exposed to chronic hypoxia (10%; range 9–11%) using a vacuum operated hypobaric chamber (Case Western Reserve University Design Fabrication Center, Cleveland, OH). Oxygen percentage is continuously monitored by a sensor. The chamber accommodates two standard cages, each for five mice. Mice (several weeks after weaning) were initially subjected to approximately 17–13% oxygen for six days and then chronically to 10% oxygen. The mice were exposed to room conditions for approximately 30 min twice a week during cage cleaning. Complete blood counts were obtained using automated cell counters Hemagen HC5 (cohorts A, B) or ProCyte Dx (cohort C) Hematology Analyzers. The mice were housed at RPCI core facility and studies were approved by IACUC.
RNA seq and bioinformatic analysis
RNAs extracted from CD14+ cells were purified using RNA clean-up and concentration kit (Norgen Biotek corp.). Illumina TruSeq paired stranded total RNA with RiboMinus Gold kit was used to obtain sequencing libraries. Paired 101 bp RNA sequencing was performed on an Illumina HiSeq 2500 system (all nine samples in one flow lane). Raw reads passed quality filter from Illumina RTA were first pre-processed by using FASTQC(v0.10.1) for sequencing base quality control, then mapped to the latest human reference genome (GRCh38.p7) and GENCODE annotation database (version 25) using Tophat2(v2.0.13) (63). Second round of QC using RSeQC (64) was applied to mapped bam files to identify potential RNA Seq library preparation problems. From the mapping results, the read counts for genes were obtained by HTSeq (65) using intersection-strict option. Differentially expressed genes were identified using DESeq2 (66), a variance-analysis package developed to infer the statically significant difference in RNA-seq data. Gene fold changes were calculated using regularized-log2 transformation in the DESeq2 R package. The raw RNA-seq data are submitted to the EMBL-EBI ENA archive under primary accession number PRJEB12121.
Other
RNA and plasmid DNA were isolated with commercial kits (TRIzol, Life Technologies and Qiagen, respectively). RNA/DNA was quantified by Nanodrop 2000 (Thermo Fisher). Proteins were quantified using Bio-Rad Dc assay with BSA standards. RNA was reverse transcribed with the Transcriptor First Strand cDNA Synthesis (Roche) kit. SDHB c.136C > U RNA editing was quantified by allele specific RT–qPCR (29). PCR oligonucleotide primers (Supplementary Material, Table S1) were obtained from Integrated DNA Technologies, Inc. ADM, ELL2, HILPDA, NGLY1, MET, TKTL1, VEGFA and B2M gene expression levels was assessed by qPCR using FastStart Taq DNA polymerase and SYBR Green I dye on a LightCycler 480 System (Roche). Quantification cycle values were calculated by the instrument software using the maximum second derivative method and the mean quantification cycle value of duplicate PCR reactions was used for analysis. Delta delta CT method is used to infer gene expression changes (67).
Statistical analysis
Effects of inhibitors and hypoxia on RNA editing in biological replicates (PBMCs and MEPs) were initially tested by ANOVA, then by multiple comparisons (Figs 1 and 3C). Effect of inhibitors on VEGFA and HILPDA expression in biological replicates (Fig. 4B and C) were tested by paired t-test (two-sided). Unpaired t-test (two-sided) was used to examine hemoglobin changes in mice and the effect of inhibitors on HRE expression in HEK293T cells (Fig. 5D). The false discovery rate approach was used to examine gene expression changes in THP-1 and HEK293T cells (Fig. 5). Statistical calculations were performed by GraphPad prism 7.00 software.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Drs G. Cox for providing Sdhb and Sdhc transgenic mice, S. Patnaik for initial help in bioinformatics analysis and providing HRE plasmid, S. Sexton and P. Wallace for helpful discussions and Ms. D. Tabaczynski in maintenance of mice.
Conflict of Interest statement. None declared.
Funding
National Cancer Institute (NCI) (P30CA016056) involving the use of Roswell Park Cancer Institute (RPCI)’s Laboratory Animal Shared Resources, Genomics Shared Resources, Bioinformatic Shared Resources, Flow Cytometry and Imaging Facilities.
References
- 1. Baysal B.E., Ferrell R.E., Willett-Brozick J.E., Lawrence E.C., Myssiorek D., Bosch A., van der Mey A., Taschner P.E., Rubinstein W.S., Myers E.N., et al. (2000) Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science, 287, 848–851. [DOI] [PubMed] [Google Scholar]
- 2. Niemann S., Muller U. (2000) Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat. Genet., 26, 268–270. [DOI] [PubMed] [Google Scholar]
- 3. Astuti D., Latif F., Dallol A., Dahia P.L., Douglas F., George E., Skoldberg F., Husebye E.S., Eng C., Maher E.R. (2001) Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am. J. Hum. Genet., 69, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cecchini G. (2003) Function and structure of complex II of the respiratory chain*. Annu. Rev. Biochem., 72, 77–109. [DOI] [PubMed] [Google Scholar]
- 5. Dahia P.L., Ross K.N., Wright M.E., Hayashida C.Y., Santagata S., Barontini M., Kung A.L., Sanso G., Powers J.F., Tischler A.S., et al. (2005) A HIF1alpha regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet., 1, 72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lopez-Jimenez E., Gomez-Lopez G., Leandro-Garcia L.J., Munoz I., Schiavi F., Montero-Conde C., de Cubas A.A., Ramires R., Landa I., Leskela S., et al. (2010) Research resource: Transcriptional profiling reveals different pseudohypoxic signatures in SDHB and VHL-related pheochromocytomas. Mol. Endocrinol., 24, 2382–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castro-Vega L.J., Letouzé E., Burnichon N., Buffet A., Disderot P., Khalifa E., Loriot C., Elarouci N., Morin A., Menara M., et al. (2015) Multi-omics analysis defines core genomic alterations in pheochromocytomas and paragangliomas. Nat. Comm., 6, 6044.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saldana M.J., Salem L.E., Travezan R. (1973) High altitude hypoxia and chemodectomas. Hum. Pathol., 4, 251–263. [DOI] [PubMed] [Google Scholar]
- 9. Opotowsky A.R., Moko L., Ginns J., Rosenbaum M., Greutmann M., Aboulhosn J., Hageman A., Kim Y., Deng L.X., Grewal J. (2015) Pheochromocytoma and paraganglioma in cyanotic congenital heart disease. J. Clin. Endocrinol. Metab., 100, 1325–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Astrom K., Cohen J.E., Willett-Brozick J.E., Aston C.E., Baysal B.E. (2003) Altitude is a phenotypic modifier in hereditary paraganglioma type 1: Evidence for an oxygen-sensing defect. Hum. Genet., 113, 228–237. [DOI] [PubMed] [Google Scholar]
- 11. Cerecer-Gil N.Y., Figuera L.E., Llamas F.J., Lara M., Escamilla J.G., Ramos R., Estrada G., Hussain A.K., Gaal J., Korpershoek E., et al. (2010) Mutation of SDHB is a cause of hypoxia-related high-altitude paraganglioma. Clin. Cancer Res., 16, 4148–4154. [DOI] [PubMed] [Google Scholar]
- 12. Semenza G.L. (2012) Hypoxia-inducible factors in physiology and medicine. Cell, 148, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaelin W.G. Jr., Ratcliffe P.J. (2008) Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell, 30, 393–402. [DOI] [PubMed] [Google Scholar]
- 14. Gordan J.D., Bertout J.A., Hu C., Diehl J.A., Simon M.C. (2007) HIF-2α promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell, 11, 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Makino Y., Uenishi R., Okamoto K., Isoe T., Hosono O., Tanaka H., Kanopka A., Poellinger L., Haneda M., Morimoto C. (2007) Transcriptional up-regulation of inhibitory PAS domain protein gene expression by hypoxia-inducible factor 1 (HIF-1): A negative feedback regulatory circuit in HIF-1-mediated signaling in hypoxic cells. J. Biol. Chem., 282, 14073–14082. [DOI] [PubMed] [Google Scholar]
- 16. Selak M.A., Armour S.M., MacKenzie E.D., Boulahbel H., Watson D.G., Mansfield K.D., Pan Y., Simon M.C., Thompson C.B., Gottlieb E. (2005) Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell, 7, 77–85. [DOI] [PubMed] [Google Scholar]
- 17. Guzy R.D., Sharma B., Bell E., Chandel N.S., Schumacker P.T. (2008) Loss of the SdhB, but not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol. Cell. Biol., 28, 718–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cervera A.M., Apostolova N., Crespo F.L., Mata M., McCreath K.J. (2008) Cells silenced for SDHB expression display characteristic features of the tumor phenotype. Cancer Res., 68, 4058–4067. [DOI] [PubMed] [Google Scholar]
- 19. Lee S., Nakamura E., Yang H., Wei W., Linggi M.S., Sajan M.P., Farese R.V., Freeman R.S., Carter B.D., Kaelin W.G. (2005) Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: Developmental culling and cancer. Cancer Cell, 8, 155–167. [DOI] [PubMed] [Google Scholar]
- 20. Aspuria P.P., Lunt S.Y., Väremo L., Vergnes L., Gozo M., Beach J.A., Salumbides B., Reue K., Wiedemeyer W.R., Nielsen J. (2014) Succinate dehydrogenase inhibition leads to epithelial-mesenchymal transition and reprogrammed carbon metabolism. Cancer Metab., 2, 21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Her Y.F., Nelson-Holte M., Maher L.J. III(2015) Oxygen concentration controls epigenetic effects in models of familial paraganglioma. PloS One, 10, e0127471.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Millán-Uclés Á., Díaz-Castro B., García-Flores P., Báez A., Pérez-Simón J.A., López-Barneo J., Piruat J.I. (2014) A conditional mouse mutant in the tumor suppressor SdhD gene unveils a link between p21WAF1/Cip1 induction and mitochondrial dysfunction. PloS One, 9, e85528.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones R., McDonald K.E., Willson J.A., Ghesquière B., Sammut D., Daniel E., Harris A.J., Lewis A., Thompson A., Dickinson R.S. (2016) Mutations in succinate dehydrogenase B (SDHB) enhance neutrophil survival independent of HIF-1α expression. Blood, 127, 2641–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maher L.J. III, Smith E.H., Rueter E.M., van Deursen J., Bida J.P., Palomo J.I.P., López-Barneo J., Nelson-Holte M., Becker N.A., García-Flores P. (2011) Mouse Models of Human Familial Paraganglioma. Chapter 3 in Pheochromocytoma - A New View of the Old Problem. Publisher: INTECH.
- 25. Diaz-Castro B., Pintado C.O., Garcia-Flores P., Lopez-Barneo J., Piruat J.I. (2012) Differential impairment of catecholaminergic cell maturation and survival by genetic mitochondrial complex II dysfunction. Mol. Cell. Biol., 32, 3347–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhuang Z., Yang C., Lorenzo F., Merino M., Fojo T., Kebebew E., Popovic V., Stratakis C.A., Prchal J.T., Pacak K. (2012) Somatic HIF2A gain-of-function mutations in paraganglioma with polycythemia. N. Engl. J. Med., 367, 922–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Franke K., Gassmann M., Wielockx B. (2013) Erythrocytosis: The HIF pathway in control. Blood, 122, 1122–1128. [DOI] [PubMed] [Google Scholar]
- 28. Haase V.H. (2013) Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev., 27, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baysal B.E., De Jong K., Liu B., Wang J., Patnaik S.K., Wallace P.K., Taggart R.T. (2013) Hypoxia-inducible C-to-U coding RNA editing downregulates SDHB in monocytes. Peer J., 1, e152.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharma S., Patnaik S.K., Taggart R.T., Kannisto E.D., Enriquez S.M., Gollnick P., Baysal B.E. (2015) APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Comm., 6, 6881.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sharma S., Patnaik S.K., Kemer Z., Baysal B.E. (2016) Transient overexpression of exogenous APOBEC3A causes C-to-U RNA editing of thousands of genes. RNA Biol, 5, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Salter J.D., Bennett R.P., Smith H.C. (2016) The APOBEC protein family: United by structure, divergent in function. Trends Biochem. Sci., 41, 578–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Willems L., Gillet N.A. (2015) APOBEC3 interference during replication of viral genomes. Viruses, 7, 2999–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bayley J.P., Devilee P., Taschner P.E. (2005) The SDH mutation database: An online resource for succinate dehydrogenase sequence variants involved in pheochromocytoma, paraganglioma and mitochondrial complex II deficiency. BMC Med. Genet., 6, 39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jaipersad A.S., Lip G.Y., Silverman S., Shantsila E. (2014) The role of monocytes in angiogenesis and atherosclerosis. J. Am. Coll. Cardiol., 63, 1–11. [DOI] [PubMed] [Google Scholar]
- 36. Geissmann F., Manz M.G., Jung S., Sieweke M.H., Merad M., Ley K. (2010) Development of monocytes, macrophages, and dendritic cells. Science, 327, 656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bosco M.C., Puppo M., Santangelo C., Anfosso L., Pfeffer U., Fardin P., Battaglia F., Varesio L. (2006) Hypoxia modifies the transcriptome of primary human monocytes: Modulation of novel immune-related genes and identification of CC-chemokine ligand 20 as a new hypoxia-inducible gene. J. Immunol., 177, 1941–1955. [DOI] [PubMed] [Google Scholar]
- 38. Strehl C., Fangradt M., Fearon U., Gaber T., Buttgereit F., Veale D.J. (2014) Hypoxia: How does the monocyte-macrophage system respond to changes in oxygen availability? J. Leukoc. Biol., 95, 233–241. [DOI] [PubMed] [Google Scholar]
- 39. Elbarghati L., Murdoch C., Lewis C.E. (2008) Effects of hypoxia on transcription factor expression in human monocytes and macrophages. Immunobiology, 213, 899–908. [DOI] [PubMed] [Google Scholar]
- 40. Fangradt M., Hahne M., Gaber T., Strehl C., Rauch R., Hoff P., Lohning M., Burmester G.R., Buttgereit F. (2012) Human monocytes and macrophages differ in their mechanisms of adaptation to hypoxia. Arthritis Res. Ther., 14, R181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miyadera H., Shiomi K., Ui H., Yamaguchi Y., Masuma R., Tomoda H., Miyoshi H., Osanai A., Kita K., Omura S. (2003) Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase). Proc. Natl. Acad. Sci. U. S. A, 100, 473–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Horsefield R., Yankovskaya V., Sexton G., Whittingham W., Shiomi K., Omura S., Byrne B., Cecchini G., Iwata S. (2006) Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): A mechanism of electron transfer and proton conduction during ubiquinone reduction. J. Biol. Chem., 281, 7309–7316. [DOI] [PubMed] [Google Scholar]
- 43. Pollard P.J., Briere J.J., Alam N.A., Barwell J., Barclay E., Wortham N.C., Hunt T., Mitchell M., Olpin S., Moat S.J., et al. (2005) Accumulation of krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum. Mol. Genet., 14, 2231–2239. [DOI] [PubMed] [Google Scholar]
- 44. Adam J., Hatipoglu E., O'Flaherty L., Ternette N., Sahgal N., Lockstone H., Baban D., Nye E., Stamp G.W., Wolhuter K.. et al. (2011) Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: Roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell, 20, 524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ooi A., Wong J., Petillo D., Roossien D., Perrier-Trudova V., Whitten D., Min B.W.H., Tan M., Zhang Z., Yang X.J. (2011) An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell, 20, 511–523. [DOI] [PubMed] [Google Scholar]
- 46. Chandel N.S., Maltepe E., Goldwasser E., Mathieu C.E., Simon M.C., Schumacker P.T. (1998) Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. U. S. A, 95, 11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brunelle J.K., Bell E.L., Quesada N.M., Vercauteren K., Tiranti V., Zeviani M., Scarpulla R.C., Chandel N.S. (2005) Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab., 1, 409–414. [DOI] [PubMed] [Google Scholar]
- 48. Guzy R.D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K.D., Simon M.C., Hammerling U., Schumacker P.T. (2005) Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab., 1, 401–408. [DOI] [PubMed] [Google Scholar]
- 49. Mansfield K.D., Guzy R.D., Pan Y., Young R.M., Cash T.P., Schumacker P.T., Simon M.C. (2005) Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-α activation. Cell Metab., 1, 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin X., David C.A., Donnelly J.B., Michaelides M., Chandel N.S., Huang X., Warrior U., Weinberg F., Tormos K.V., Fesik S.W., et al. (2008) A chemical genomics screen highlights the essential role of mitochondria in HIF-1 regulation. Proc. Natl. Acad. Sci. U. S. A, 105, 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brown S.T., Nurse C.A. (2008) Induction of HIF-2alpha is dependent on mitochondrial O2 consumption in an O2-sensitive adrenomedullary chromaffin cell line. Am. J. Physiol. Cell. Physiol., 294, C1305–C1312. [DOI] [PubMed] [Google Scholar]
- 52. Hagen T., Taylor C.T., Lam F., Moncada S. (2003) Redistribution of intracellular oxygen in hypoxia by nitric oxide: Effect on HIF1alpha. Science, 302, 1975–1978. [DOI] [PubMed] [Google Scholar]
- 53. Chua Y.L., Dufour E., Dassa E.P., Rustin P., Jacobs H.T., Taylor C.T., Hagen T. (2010) Stabilization of hypoxia-inducible factor-1alpha protein in hypoxia occurs independently of mitochondrial reactive oxygen species production. J. Biol. Chem., 285, 31277–31284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Waypa G.B., Smith K.A., Schumacker P.T. (2016) O2 sensing, mitochondria and ROS signaling: The fog is lifting. Mol. Aspects Med., 47, 76–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lopez-Barneo J., Gonzalez-Rodriguez P., Gao L., Fernandez-Aguera M.C., Pardal R., Ortega-Saenz P. (2016) Oxygen sensing by the carotid body: Mechanisms and role in adaptation to hypoxia. Am. J. Physiol. Cell. Physiol., 310, C629–C642. [DOI] [PubMed] [Google Scholar]
- 56. Michelakis E.D., Thébaud B., Weir E.K., Archer S.L. (2004) Hypoxic pulmonary vasoconstriction: Redox regulation of O2-sensitive K channels by a mitochondrial O2-sensor in resistance artery smooth muscle cells. J. Mol. Cell. Cardiol., 37, 1119–1136. [DOI] [PubMed] [Google Scholar]
- 57. Angelova P.R., Kasymov V., Christie I., Sheikhbahaei S., Turovsky E., Marina N., Korsak A., Zwicker J., Teschemacher A.G., Ackland G.L., et al. (2015) Functional oxygen sensitivity of astrocytes. J. Neurosci., 35, 10460–10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Buckler K.J., Turner P.J. (2013) Oxygen sensitivity of mitochondrial function in rat arterial chemoreceptor cells. J. Physiol. (Lond.), 591, 3549–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fernández-Agüera M.C., Gao L., González-Rodríguez P., Pintado C.O., Arias-Mayenco I., García-Flores P., García-Pergañeda A., Pascual A., Ortega-Sáenz P., López-Barneo J. (2015) Oxygen sensing by arterial chemoreceptors depends on mitochondrial complex I signaling. Cell Metab., 22, 825–837. [DOI] [PubMed] [Google Scholar]
- 60. Mentzer S.J., Guyre P.M., Burakoff S.J., Faller D.V. (1986) Spontaneous aggregation as a mechanism for human monocyte purification. Cell. Immunol., 101, 312–319. [DOI] [PubMed] [Google Scholar]
- 61. Nord A.S., Chang P.J., Conklin B.R., Cox A.V., Harper C.A., Hicks G.G., Huang C.C., Johns S.J., Kawamoto M., Liu S., et al. (2006) The international gene trap consortium website: A portal to all publicly available gene trap cell lines in mouse. Nucleic Acids Res., 34, D642–D648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Piruat J.I., Pintado C.O., Ortega-Saenz P., Roche M., Lopez-Barneo J. (2004) The mitochondrial SDHD gene is required for early embryogenesis, and its partial deficiency results in persistent carotid body glomus cell activation with full responsiveness to hypoxia. Mol. Cell. Biol., 24, 10933–10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Trapnell C., Pachter L., Salzberg S.L. (2009) TopHat: Discovering splice junctions with RNA-seq. Bioinformatics, 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang L., Wang S., Li W. (2012) RSeQC: Quality control of RNA-seq experiments. Bioinformatics, 28, 2184–2185. [DOI] [PubMed] [Google Scholar]
- 65. Anders S., Pyl P.T., Huber W. (2015) HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics, 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Love M.I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Livak K.J., Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 68. Fisher R.A. (1925) Statistical Methods for Research Workers. Genesis Publishing Pvt Ltd. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.