Abstract
Brain fiber pathways are presumed to follow smooth curves but recent high angular resolution diffusion MRI (dMRI) suggests that instead they follow 3 primary axes often nearly orthogonal. To investigate this, we analyzed axon pathways under monkey primary motor cortex with (1) dMRI tractography, (2) axon tract tracing, and (3) axon immunohistochemistry. dMRI tractography shows the predicted crossings of axons in mediolateral and dorsoventral orientations and does not show axon turns in this region. Axons labeled with tract tracer in the motor cortex dispersed in the centrum semiovale by microscopically sharp axonal turns and/or branches (radii ≤15 µm) into 2 sharply defined orientations, mediolateral and dorsoventral. Nearby sections processed with SMI-32 antibody to label projection axons and SMI-312 antibody to label all axons revealed axon distributions parallel to the tracer axons. All 3 histological methods confirmed preponderant axon distributions parallel with dMRI axes with few axons (<20%) following smooth curves or diagonal orientations. These findings indicate that axons navigate deep white matter via microscopic sharp turns and branches between primary axes. They support dMRI observations of primary fiber axes, as well as the prediction that fiber crossings include navigational events not yet directly resolved by dMRI. New methods will be needed to incorporate coherent microscopic navigation into dMRI of connectivity.
Keywords: confocal microscopy, connectome, rhesus monkey, tractography, white matter
Introduction
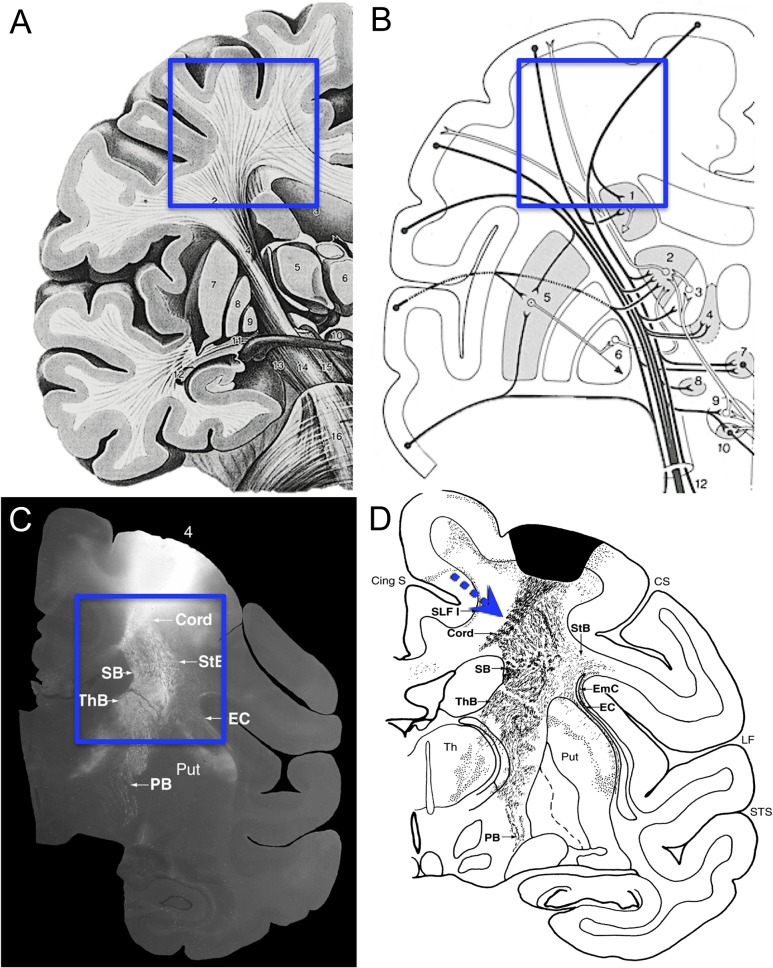
Efforts to identify cerebral connections date back to the 18th century with gross dissections of the brain and its white matter (for a historical review, see Schmahmann and Pandya 2006, 2007) and progressed to those of Klingler (1935) using freeze–thaw dissections to show hierarchic palisades of fiber structure. While these observations revealed some aspects of cerebral pathways, they were not reliable markers of point-to-point axonal connectivity. Instead, the study of axonal connectivity was launched by Santiago Ramon y Cajal's use of the Golgi stain in the late 19th century to microscopically follow the axons of individual neurons (Ramon y Cajal 1955, Figure 65; Ramon y Cajal 1995, Figure 519) and this approach has continued through the present with the addition of tract tracing in animals. Most recently, diffusion MRI (dMRI) tractography has been applied in animals and humans to explore questions of point-to-point connectivity (Schmahmann et al. 2007; Granziera et al. 2009). Common assumptions in studies of long-range connectivity are that connections may exist between any points in the cerebral hemispheres and that axons navigate between these points by following smooth, possibly spatially optimized, trajectories (Fig. 1A,B). Such paths are thought to be constrained mainly by curvature and sulci of the cerebral hemisphere although other factors, such as possible axon tension, have been postulated (Van Essen 1997). A counter example can be found in Krieg (1954), where degeneration studies in humans and monkeys with brain damage revealed efferents traveling smoothly for a distance but then dropping “sharply” via approximately right-angle turns (Krieg 1954, Fig. 96; reproduced in Wedeen et al. 2012b). Careful examination of tract tracing studies (e.g., Schmahmann and Pandya 2006) also shows data consistent with sharp turns in the efferents beneath motor cortex (Fig. 1C,D). Indeed, in that study using tracers to label entire axonal projections, sharp turning was noted in a variety of contexts but these were considered sporadic and of unclear significance (Schmahmann and Pandya 2006). In summary, there is reason to question the generality of tract tracer pathway reconstructions and textbook renderings that show smooth spatially optimized curvature.
Figure 1.
Fiber trajectories from published illustrations. Panel A is a typical artistic rendering of cerebral fibers originating from cortex. The superimposed box identifies the subcortical white matter, where fibers are shown descending from the cortex along smoothly curved geodesic paths into the internal capsule and corpus callosum (Fig. 21.8 from Nieuwenhuys et al. 2008). In B, the box identifies the same region, where connectional pathways are again shown following smooth arcs (Fig. 128, Nieuwenhuys et al. 1981). In contrast to these typical representations, Panel C is an autoradiograph of fiber projections from primary motor cortex with the box identifying subcortical white matter, where there are rather sharp breaks in the overall trajectories (Fig. 11-8B from Schmahmann and Pandya 2006). Panel D shows a “charting” of a nearby section from the same case as in C (Fig. 11-7(81) from Schmahmann and Pandya 2006). The superimposed dashed arrows to point out likely fiber trajectories at these sharp breaks. Whether axons actually branch or make sharp turns at these breaks cannot be established because individual fibers cannot be reliably followed in autoradiographs.
Recently, analysis of high angular resolution dMRI has suggested that cerebral pathways follow a geometric structure that appears incompatible with assumptions of a smoothly curving organization. Instead cerebral path orientations were found to conform to a geometric 3D coordinate system within which paths cross at approximately right-angles with little or no evidence of smooth or diagonal turns from one axis to another (Wedeen et al. 2012a) as long as high angular resolution dMRI is used (Wedeen et al. 2012b). It follows that the axons of cerebral pathways must navigate from one axis to another by making sharp right-angle turns not resolved by present dMRI. Support for this view can be found in studies of connectivity that traced individual labeled axons and noted examples of sharp turns and orthogonal branches (e.g., Rockland 1995, 2013) but these were not thought to be a general principle of axon navigation through white matter. To date, there have been no systematic studies of the presence or frequency of axons turning sharply or branching at right-angles to navigate cerebral pathways. This is a critical gap in our knowledge, particularly given the assumption inherent in dMRI tractography algorithms that the trajectories of axon bundles subserving cerebral connections are smooth and curvilinear. In fact, Catani et al. (2012) questioned the observation of geometric organization and asserted that the smooth arcs were indeed the rule. In this regard, we have noted (Wedeen et al. 2012b) that smooth curves may occur as artifacts when low angular resolution scans blur right-angle crossings (Wedeen et al. 2008).
To address these issues and determine if the structure observed with dMRI was valid at an axonal level, we took a multimodal approach to characterize the axons of white matter in the monkey brain. We used tract tracing with biotinylated dextran amine (BDA) to determine if corticofugal projections from primary motor cortex navigate as smooth curves or navigate along discrete primary axes. Since BDA labels only a sparse set of corticofugal fibers originating at the injection site, immunohistochemistry (IHC) was used in adjacent sections to label the larger subset of all long projection axons (SMI-32 positive fibers) and the total population of all axons (SMI-312 positive fibers) traversing the subcortical white matter beneath motor cortex. We quantified the orientations of BDA-labeled axons as well as the orientation and frequency of microscopically sharp right-angle turns and branches. We similarly quantified the orientations of the 2 populations of IHC labeled axons. Finally, we compared these histological and dMRI observations in the monkey brain with dMRI orientations in an equivalent region of human brain.
Materials and Methods
Subjects—Nonhuman Primates
Six adult rhesus monkeys were used in these experiments. All were obtained from national primate centers or domestic breeders and had known birthdates and health records, which were screened to ensure that they were free from disease or experimentation that might compromise the brain. All were part of other ongoing studies and were housed at the Laboratory Animal Science Center of the Boston University Medical Campus (BUMC). This facility is managed by a licensed veterinarian and is fully accredited by the Association for the Assessment and Accreditation of the Laboratory Animal Care. All procedures conformed to National Institutes of Health guidelines, the Institute of Laboratory Animal Resources Commission on Life Sciences’ Guide for the care and use of laboratory animals (1996) and were approved by the BUMC Institutional Animal Care and Use Committee.
Surgical Procedure and Tracer Injections
Three of the 6 subjects received injections of neuroanatomical tracers into the forebrain during a survival surgery carried out under aseptic conditions. Animals were sedated with ketamine hydrochloride (10 mg/kg) and anesthetized with isoflurane (1–3%) or sodium pentobarbital (25 mg/kg). Heart rate, respiration, oxygenation and muscle tonus were monitored to ensure a safe surgical level of anesthesia. The head was stabilized in a stereotactic apparatus, and a midline incision was made followed by reflection of the temporalis muscle. A bone flap was removed in one piece and the dura incised to expose the precentral gyrus. Using a 5 μL Hamilton syringe, 3–5 injections totaling 0.9–6.0 µL of BDA (MW 10 000; 10%) were placed 1.5 mm deep into the region of the hand representation of primary motor cortex (see Moore et al. 2012). Once injections were completed, the dura was closed, the bone flap sutured back in place and the muscle, fascia and skin closed in layers. Following surgery, monkeys were given antibiotics and analgesics and monitored for any signs of infection or complications.
Perfusion and Tissue Harvest
Postoperative survival times after the BDA tracer injection ranged from 13 to 38 days after which monkeys were euthanized. The other 3 monkeys, which did not receive tracer injections were euthanized at the end of their experimental protocols. In all cases, they were deeply anesthetized with intravenous sodium pentobarbital (25 mg/kg to effect) and euthanized by exsanguination during transcardial perfusion of the brain. Perfusion began with 4–6 L of cold (4°C) Krebs–Heinsleit buffer (pH 7.4) and was followed by 4–6 L of warm (37°C), buffered (pH 7.4) 4% paraformaldehyde containing up to 0.01% gadolinium. Following perfusion, the brain was removed, weighed, photographed and then postfixed in 4% paraformaldehyde with up to 0.01% gadolinium overnight at 4°C.
Ex Vivo MRI Acquisition
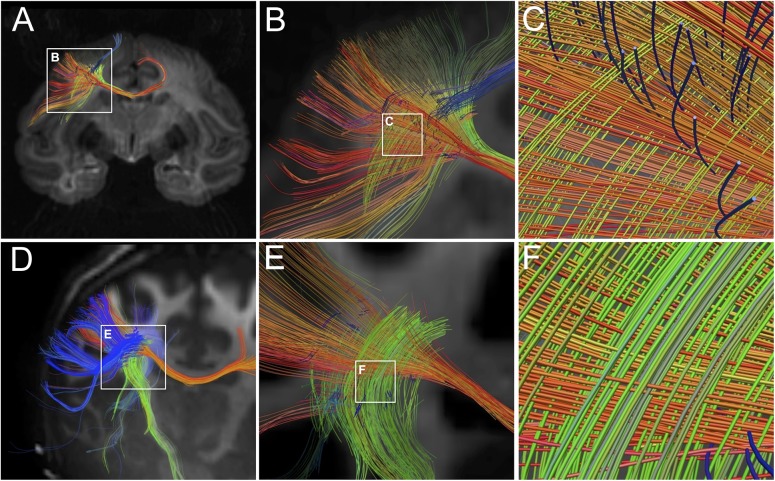
After 24 h of fixation, brains were rinsed in buffer and transferred to an MRI scanning solution of perfluorocarbon. Diffusion spectrum imaging (DSI) was acquired at 9.4 T (Biospec 20 cm, Bruker Corp.) as previously described (Wedeen et al. 2005). The acquisition pulse sequence was a spin-echo time repetition (TR)/time echo (TE) 1000/40 ms with hybrid 2D echo-planar–3D Fourier transform spatial encoding with isotropic resolution of 400–500 μm depending on brain size. A 3D volume image of the brain was acquired for each of the 515 diffusion-encoding q-vectors, with spin-echo diffusion-encoding gradient pulse-pair with peak intensity Gmax = 440 mT m−1, interpulse delay (mixing time) ∆ = 24 ms, and pulse durations d = 13 ms yielding maximum sensitivity bmax = 40 ms μm−2. Using 4 averages, the total acquisition times were 24–32 h. Orientation distribution functions (ODFs) were reconstructed, orientation maxima identified, and paths computed with streamline tractography. The pathways and their crossings (e.g., Fig. 2A–C) were visualized with MGH Diffusion Toolkit and TrackVis software, and crossing paths analyzed, as previously described (Wedeen 2012a). In addition, in 3 of the cases, crossing angles for fibers reconstructed in the white matter beneath primary motor cortex were computed and color coded for visualization (e.g., Supplementary Fig. 5).
Figure 2.
Fiber crossings in monkey and human forebrain with high angular resolution dMRI. The top row (Panels A–C) shows fiber reconstructions from ex vivo dMRI imaging of rhesus monkey brain using DSI protocol at 0.5 mm3 spatial resolution. The bottom row (Panels D–F) shows fiber reconstructions from in vivo dMRI images of human brain using QBI at 1.5 mm3 spatial resolution. Both images are approximately coronal sections that include the white matter of the centrum-semiovale beneath primary motor cortex. Panels A and D show at widefield similar geometric fiber crossings beneath primary motor cortex. Panels B,C and E,F show at higher magnification that these fibers are aligned in 3 approximately orthogonal directions forming a “grid-like” pattern. The in-plane mediolaterally and dorsoventrally oriented fibers are prominent. Some rostrocaudal fibers in the z-dimension can also be seen. Overall, this confirms at multiple scales the grid-like orientation of fibers in the subcortical white matter of both species.
Human Subjects and MRI Acquisition
Human subjects were healthy adult volunteers, recruited and scanned at Massachusetts General Hospital in the Siemens 3 T Connectom instrument with informed consent under an IRB-approved protocol. Q-ball dMRI (QBI) data were acquired with a spin-echo 2D echo-planar pulse sequence, 1.5 mm isotropic spatial resolution, TR/TE = 7000/52 ms, peak diffusion-encoding gradient intensity Gmax = 300 mT m−1, QBI encoding at diffusion sensitivity b = 10 ms μm−2 with 256 directions for a net scan time of 18 min. Image reconstruction and processing were done as for the ex vivo scans (e.g., Fig. 2D–F).
Nonhuman Primate Tissue Processing Procedures
Following dMRI scanning, all the monkey brains were blocked in the coronal stereotactic plane, photographed and then cryoprotected in 0.1 M phosphate buffer (pH 7.4) containing 10% glycerol and 2% DMSO for 2 days followed by the same buffer but with 20% glycerol and 2% DMSO for another 3 days. Each block was flash frozen in −75°C 2-methyl-butane (Rosene et al. 1986) and stored at −0°C until cut in the coronal plane on a sliding microtome into interrupted series of 30 µm thick sections, spaced at 300 µm intervals within each series. The coronal plane was chosen based on earlier tracer studies in this laboratory, which demonstrated that projections leave the primary motor cortex in the coronal plane where they can be followed as they disperse into the subcortical white matter of the centrum semiovale (Fig. 1C,D).
All cut sections were collected in buffer with 15% glycerol solution and stored at −80°C until further processed for BDA or IHC (e.g., Giannaris and Rosene 2012). It is important to note that glycerol cryoprotection does not shrink the tissue as other methods including cryoprotection with sucrose or embedding in paraffin do (Rosene et al. 1986). Moreover, processing tissue “free-floating” in isotonic buffers for IHC also preserves tissue proportions until the sections are mounted and dried onto gelatin subbed slides. While some minor distortions can occur in the mounting process, the x,y dimensions are generally preserved as the tissue adheres to the slide. In contrast, the z-axis of 30 µm thick sections can shrink over 50% if the sections are dehydrated for cover slipping with Permount, but z-axis thickness is preserved if the sections are covered in aqueous mounting media for confocal microscopy. The important thing for this study is that under the processing conditions used here, axon turns and branches in the x,y dimension cannot be an artifact of shrinkage.
Visualization of Corticofugal Axons with BDA Tract Tracer
To visualize the motor cortex projection fibers labeled in the BDA injection cases, one series of 30 µm thick sections from each case was removed from −80°C storage, thawed at room temperature, rinsed in 0.05 M TBS (pH 7.4) to remove glycerol and processed free-floating at room temperature (20°C). After rinses in 0.1 M phosphate buffer (pH 7.4) sections were incubated on a rocker with Vector ABC Elite solution (PK-6100) for 90 min, rinsed with 0.1 M phosphate buffer (pH 7.4), incubated in a nickel (0.05%) 3,3′-diaminobenzidene tetrachloride solution and 0.03% H2O2 for 20 min. Sections were then rinsed with 0.05 M sodium acetate (pH 7.4) followed by rinses in 0.1 M phosphate buffer (pH 7.4) before being mounted on gelatin-coated slides and dried at room temperature. The next day, they were counterstained with neutral red before being dehydrated, cleared and cover-slipped with Permount (Fisher Scientific).
IHC Visualization of Axons with SMI-32 and SMI-312 Antibodies
To label axon populations more densely than BDA, series of 30 µm sections adjacent to the BDA series from the 3 cases were thawed and rinsed in 0.1 M phosphate buffer (pH 7.4) to remove glycerol. They were then processed free-floating, at 20°C for IHC with either SMI-32 antibody to label all large projection fibers in the region or with SMI-312 pan-axonal neurofilament antibody to label all axons, both myelinated and unmyelinated. In both cases, sections were incubated in a blocking solution containing 10% normal goat serum with 0.5% Triton-X in 0.05 M TBS in a variable wattage microwave tissue processor (Biowave, Ted Pella) 3 times for 3 min at 150 W at 30° with each irradiation separated by 5 min at room temperature with gentle agitation. Sections remained in the blocking solution for an additional 2 h at room temperature and were then incubated in primary antibody solution (1:1000 of mouse monoclonal SMI-32 antibody or SMI-312 antibody, both Covance) and microwaved 3 times at 250 W for 3 min with intervening 10-min gentle agitation at room temperature. Sections were then incubated for an additional 36 h in the primary antibody solution at 4°C with gentle agitation.
After removal from primary antibody solution, sections were brought to room temperature and rinsed 10 times for a total of 100 min in 0.05 M TBS (pH 7.4). They were then incubated in a secondary solution containing 1:500 of goat antimouse Alexa 488 IgG1 (Invitrogen) and microwaved 3 times at 150 W for 3 min interspersed with 10-min gentle agitation at room temperature. Sections were kept at 4°C overnight in the secondary solution, after which, they were rinsed at room temperature in 0.05 M TBS (pH 7.4) for a total of 120 min and then mounted on gelatin-coated slides and cover-slipped with polyvinyl alcohol mounting medium (DABCO, Sigma).
Microscopic Analysis
For quantitative microscopic analysis of BDA, SMI-32, and SMI-312, 3 sections of each stain were selected to be equidistantly spaced across the volume of white matter beneath motor cortex (Bregma −5.85 mm to −13.05 in the atlas of Paxinos et al. (2009), the rostral section in the −5.85 to −6.03 range, the middle section between −7.65 and −8.5, and the caudal section between −13.05 and −13.5). BDA-stained sections were digitized using a 20× objective on a Nikon E600 microscope equipped with a motorized stage and Turboscan montaging system (Objective Imaging, Inc.). Immunofluroescent-stained sections (SMI-32 and SMI-312) were visualized and imaged using a 20× objective on a Zeiss LSM710 laser scanning confocal microscope with a motorized stage for collection of montages. Confocal images were acquired at a z-step of 1.0 µm and reconstructed using ImageJ (NIH version 1.47b).
Image Analysis for Fiber Orientations
We applied 3 different methods for quantifying the orientation distribution function of microscopic axon images from BDA and IHC experiments. All 3 were selected for their ability to measure orientations directly from digitized images, without manually selecting, marking or tracing individual axons and with minimal processing.
Autocorrelation
Autocorrelation (AC) is defined as:
| (1) |
The ODF of the AC is defined as
| (2) |
where θ is the polar angle and r dr is the polar area. Integration limits were from r = 5–50 μm, or just above the diameter of a large fiber to the length of a very long fiber. Integration limits were from r = 5–50 μm.
Structure Tensor
We applied the method of Budde and Annese (Knutsson et al. 2011; Budde and Annese 2012), where ST is computed from the spatial gradient of the images, indicating image intensity changes including boundaries. At each location (pixel) in the image, the structure tensor (ST) is defined as
| (3) |
a 2 × 2 matrix, where G = grad(S) is the smoothed discrete spatial gradient of S. STs were smoothed with a 2D Gaussian kernel with radius at 50% intensity equal to 3 pixels, and axon orientation at each location represented by the direction lesser eigenvector v2 of the ST, or perpendicular to greatest changes, which occur across the fiber. The ODF of the ST for a region is defined as density of orientation vectors in each angular interval.
Path Probability
To better differentiate neuronal fibers from nonaxonal structure and artifacts in BDA images, we developed “Path Probability” (PP). Here, we compute a cross-correlation between the log of the BDA image intensity B(x,y) and a template image, Rr,θ(x,y), modeling the 2D image of a connected linear segment of voxels of length r, orientation q, 0° ≤ q < 180°, and having the symmetry R(x,y) = R(–x,–y) as illustrated in Supplementary Fig. 1. Specifically, we define
| (4) |
where * is the 2D spatial convolution in (x,y), and ε > min(B) is a regularization constant. The use of log(B) identifies fractional changes in image intensities, to better accommodate variability of tracer uptake above a threshold intensity ε > 0 (Morrison, Boston University PhD Physics Dissertation 2015). In the present analyses, we used a constant template size of r = 20 µm = 30 pixels, corresponding to 3 times the maximum diameter of observed fiber (7 µm).
Peaks for Orientation Analyses
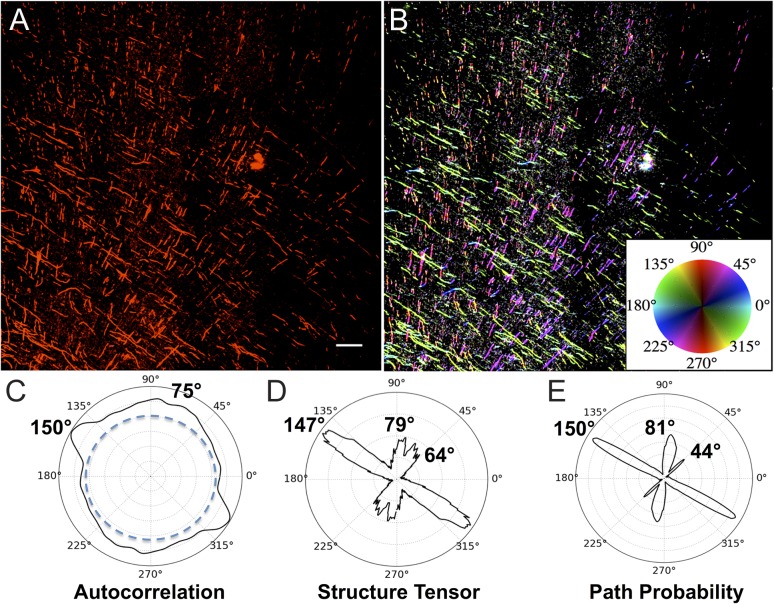
All orientation peaks were based on an ODF function f(θ), where the peak is defined as being centered at local maxima θ = M of f(θ), and we define the signal SM associated with this maximum as the area under the curve f~(θ) = f(θ)– b within the angle sector M ± d, where d is the angle to the local minimum nearest M and b = minθ(f) is the amplitude of the background. Comparison among ODFs derived from AC, ST, and PP showed good concordance between orientation maxima, with PP showing the highest rejection of nonoriented background signal. Thus, we generally use AC to compare ODF's across our 3 different histological labeling methods but use PP to obtain the sharpest maxima and pseudocolor illustrations.
Polar Plots of Orientation Analysis and Angle of Cut
All the orientation analyses were done on images acquired from slide mounted coronal sections, where the dorsoventral midline axis was imperfectly aligned to the axes of the rectangular slide so that image regions of interest (ROIs) were arbitrarily rotated different amounts. To remove this variability, the polar plots shown for all analyses were rotated to compensate for the amount by which the dorsoventral axis of the tissue sections was rotated. It is worth noting that visualization of axonal orientations also varies with the exact angle of cut through any region. Examples of how this varies in the sagittal stratum of the monkey brain are shown in Figure 17.6 of Mortazavi et al. (2015). Variation of the angle of cut across the widely spaced sections analyzed likely contributed to the small variations in orientation peaks present in the plots shown in Supplementary Figs 2–4.
Application of Pseudocolor to Visualize Fiber Orientation
To allow better visualization of the orientation of individual fibers in images, we pseudocolored fibers based on orientations obtained with the PP analysis. Specifically, we mapped orientation angle to color hue and the intensity of orientation contrast to color saturation (e.g., Fig. 3N inset). In some images, the brightness was inverted to better visualize these orientations relative to background (e.g., Fig. 5G inset). These details are reiterated in the figure captions.
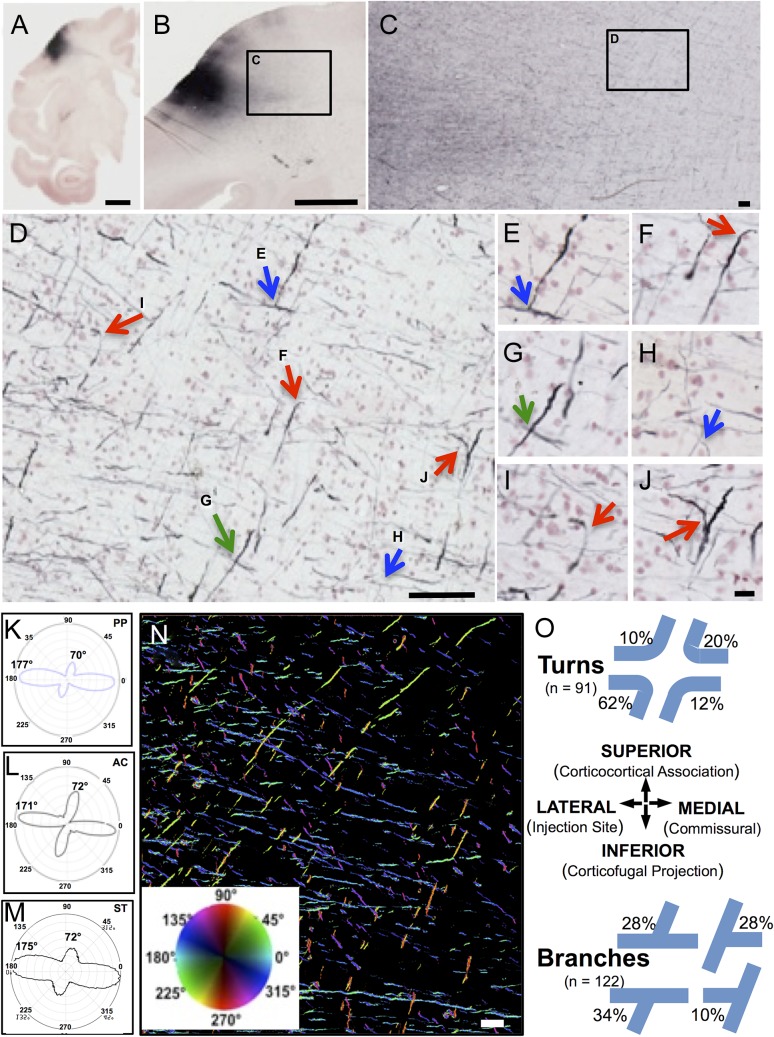
Figure 3.
Trajectories and quantitative orientations of BDA-labeled corticofugal axons. Panel A is a 30 µm whole hemisphere section though the BDA injection site in the precentral gyrus of rhesus monkey BM097, processed for BDA (black) and counterstained with neutral red to show cells. Panel B is a higher magnification view of a nearby section of the same case showing BDA-labeled fibers leaving the injection site (box C). These fibers are shown at successively higher magnifications in Panels C and D as they disburse in the white matter into mediolateral and dorsoventral directions. In D, fibers are identified as crossing (green arrows), branching (blue arrows), or making sharp turns (red arrows). Panels E–J show these and surrounding fibers at higher magnification. Panels K–M illustrate the orientations of the BDA fibers of D using quantified using (K) PP analysis, (L) AC, and (M) ST. Panel N is a field from the same case with BDA-labeled fibers pseudocolored for orientation using PP (K) and the color wheel inset where the fibers in the primary orientation at 177° are coded in reddish to green hues and those near the secondary peak at 70° are coded in aqua to blue hues. These 2 major directions account for about 80% of all fibers present in the image. Panel O graphically illustrates the frequency of different orientations observed as either sharp turns (top) or as T-branches (bottom) among the BDA fibers analyzed in all 9 sections across all 3 cases (e.g., Supplementary Fig. 2). Scale bars: A = 5 mm, B = 2 mm, C = 200 µm, D = 100 µm, J = 20 µm for E–J; N = 200 µm.
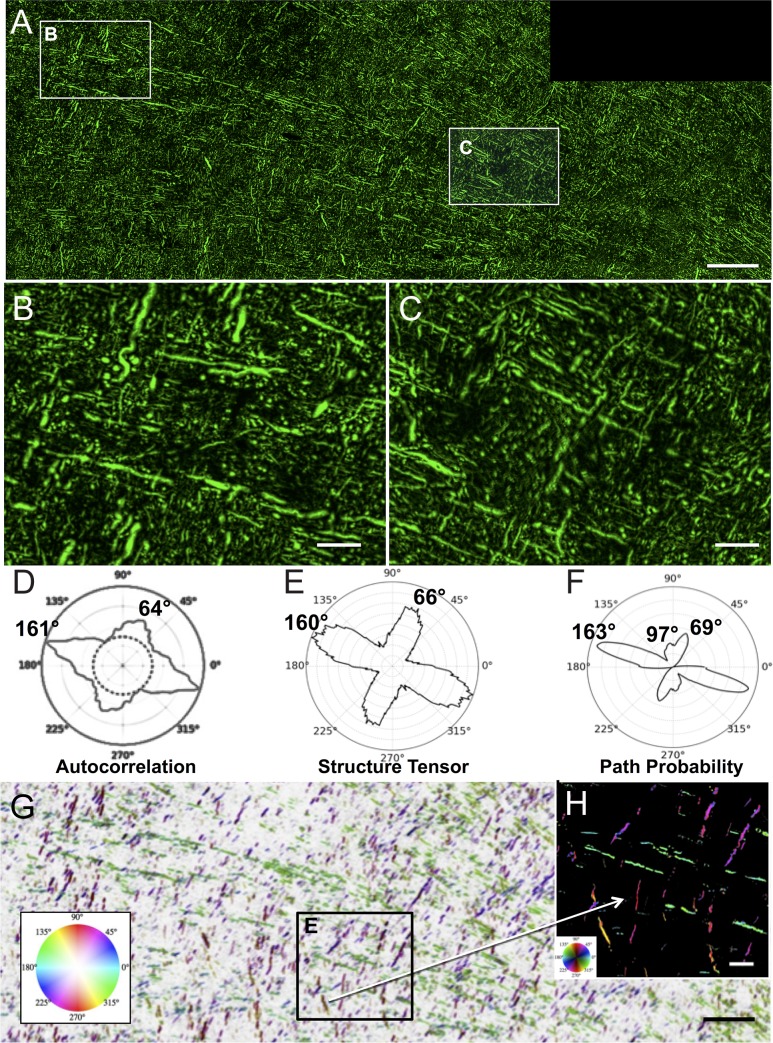
Figure 5.
Trajectories and quantitative analysis of SMI-312 labeled axons. Panel A is a montage of 8 confocal images of a 30 µm thick section processed by IHC with the pan-axonal neurofilament antibody SMI-312 to label all axons. The section was nearly adjacent to the BDA section in Figure 3 and A shows a field corresponding to Figure 3C. Panels B and C are higher magnification images, showing that the majority of fibers within the plane of section are oriented in approximately orthogonal directions but also shows the small segments and round cross sections of axons running into the z-plane. The polar plots in D–F compare the orientation quantifications obtained with AC (D), ST (E), and PP (F) and all show similar major and minor orientation peaks. Panel G is the same image as A but with background inverted and pseudocolored according to the PP orientation analysis with hue of orientation according to the inset color wheel and brightness proportional to the anisotropy of PP (Pmax/Paverage). Thus, axons oriented near 163° are in green, those near 69° are in purple. PP also detected about 20% of the fibers oriented near 97° and these are colored in reddish-orange. An enlarged view is shown in H with background not inverted. Scale bar in A, and G = 200 µm and in B, C, and E = 50 µm.
Fiber Diameter and Turning Radius
To measure the diameter of BDA, SMI-32, and SMI-312 labeled axons, rectangular ROIs of 1 × 2 mm in size in white matter beneath the motor cortex were acquired as 40× montages using Turboscan (Objective Imaging, Inc.) image acquisition software on a Nikon E600 microscope for BDA-stained tissue sections or on the confocal microscope (40× 0.8 NA objective) for SMI-32 and SMI-312 stained sections. Using these calibrated high-resolution images, sampling grids consisting of squares 50 µm on a side were placed over these images and squares were randomly selected. Within these squares, diameter was measured by using the calibrated line tool of ImageJ (version 1.47b) placed orthogonally across the long axis at random locations on each axon. This resulted in diameters for the following number of axons: BDA: n = 102; SMI-312: n = 72; SMI-32: n = 45. To estimate the radius of each turn or branch, the calibrated BDA images were loaded into ImageJ and all turns or branches within the image were located and straight lines fit to each arm. Then, the intersection of these lines was identified and the radius of the turn computed trigonometrically from the angle of the intersection.
Turn and Branch Frequency
To estimate the number of turns and branches per fiber length for the BDA-labeled axons, rectangular ROIs 1.85 mm × 2.4 mm in size were acquired from the centrum semiovale with Turboscan as above. This was done from the 3 BDA-labeled sections analyzed for each of the 3 cases. Within each ROI, 80% of the field (avoiding edges) was examined and a total of 122 turns and 91 branches identified and marked. To compute a rate of turning or branching, the total length of labeled axons in the images was assessed by first thresholding the images to identify labeled axons with circularity Q > 3, defined as Q = perimeter2/(4π area) (Montero, 2009). An estimate of the mean width of the thresholded fibers was computed by placing sampling grids (80 squares of 200 × 200 µm) over the images and using the calibrated line tool in ImageJ to measure the width of the fiber nearest to the top left corner of each grid square. Under the assumption that the length of each fiber in the image is not dependent on the width, total length was calculated as the ratio of total fiber area divided by mean width. This produced a total estimated length for all fibers in this region of interest of 120 ± 10 mm. Comparison of this automated approach to lengths observed when the length was measured manually showed these to be within 5% of each other. These lengths were then used to compute the prevalence of fiber turning and branching across all 3 cases.
Results
Crossing Fibers Identified with dMRI Beneath Motor Cortex
High-resolution dMRI (DSI) acquired ex vivo from an intact monkey brain is shown in Figure 2A–C at a coronal level corresponding to primary motor cortex. This confirms that fibers in the centrum-semiovale beneath primary motor cortex have a geometric pattern of fiber crossing as previously reported (Wedeen et al. 2012a), even as they travel into the different cerebral paths described by Schmahmann et al. (2004) and Schmahmann and Pandya (2006). Figure 2D–F shows high-resolution dMRI of the human brain acquired in vivo at a coronal level beneath motor cortex and confirms a geometric organization within the white matter of the human brain congruent with that of the monkey brain. In both species, there are 2 distinct major orientations in the coronal plane: mediolateral (parallel to callosal fibers) and dorsoventral (parallel to fibers entering the internal capsule). Importantly, the coronal plane of the monkey dMRI is entirely congruent with the coronal sections used in the histological studies of this region of the centrum semiovale, facilitating comparison of neuroanatomical markers with observed dMRI.
Histological Analysis of BDA-Labeled Axon Trajectories from Motor Cortex
Axons labeled by BDA injections into the hand area of primary motor cortex of 3 monkeys (BM097, PIC, and AM244) were surveyed in the underlying centrum semiovale. Representative examples of labeled axons from 1 of the 3 BDA cases (BM097) are shown in Figure 3A–J. Images from 2 different 30 µm thick sections through the injection site (Fig. 3A,B) show fibers leaving the injection site and entering the underlying white matter where they disperse into discrete and approximately orthogonal directions within to the x,y plane of section (Fig. 3C), and confirmed at higher magnification in Figure 3D where the axons: (1) cross at approximately right-angles as predicted by dMRI, (2) form orthogonal “T-branches” as described by Ramon y Cajal (1955) for axons in cortical gray matter (e.g., Fig. 65 in Ramon y Cajal et al. 1955 and Zhang et al. 2010), and (3) make sharp turns at approximately right-angles. Examination at higher magnification (Fig. 3E–J) and with careful through focus confirms these features. Additional examples of turns, branches and crossings are visible (Fig. 3D–J) but not marked. In addition to in-plane fibers in the x,y axes, labeled axons also appear as many small dots in the third axis (z-axis, into the plane of section). However, the precise orientation of these z-axis fibers cannot be resolved because of the limited z-axis distance within the 30-µm thick sections where, at best, fibers turning into this axis may be identified as truncated segments (e.g., Fig. 3F). Notably, diagonally oriented axons were rare and axons following smooth curves were not observed.
Quantitative Image Analysis of Orientation of BDA-Labeled Axons
We applied the PP, AC, and ST algorithms to quantify the orientation of the BDA-labeled axons in image montages of 3 sections across the volume of white matter beneath primary motor cortex in each of the 3 BDA cases. Examples of the results of these analyses for this section of Case BM097 are presented as polar plots in Figure 3K–M. PP shows peaks at 177° and 70° (Fig. 3K) while the AC plot shows peaks at 171° and 72° (Fig. 3L) and ST plot shows peaks at 175° and 72° (Fig. 3M). The acute angles between these major and minor axis peaks obtained with all 3 methods were calculated and found to range from 73° to 87°. Figure 3N is taken from a representative field of the same BDA section analyzed for orientation (Fig. 3K–M), but with a pseudocolor map (inset) applied to visualize fiber segments according to orientation detected with the PP analysis. This illustrates that the projection axons from motor cortex labeled by BDA (Fig. 3A–C) disperse in the underlying subcortical white matter along 2 primary axes.
Supplementary Figure 2 shows polar plots derived from AC analysis of BDA images in 3 sections spaced along the volume of white matter beneath the motor cortex for each of the 3 cases (BM097, PIC, and AM254). The results of this quantitative analysis of fiber orientations are tabulated in Table 1. In all 3 cases, the polar plots from each of the sections analyzed show 2 major orientations for the BDA-labeled fibers, directions that are congruent with the peaks identified in Figure 3L. Taken together, these data provide strong evidence that the projection axons from motor cortex labeled by BDA (e.g., Fig. 3A,B) disperse in the underlying subcortical white matter along 2 major axes that are largely congruent to fiber orientations observed with dMRI in the same region (Fig. 2A–C). Additionally, the microscopic right-angle branches and turns shown in Figure 3D–J suggest that fibers navigate into and between the primary axis orientations by way of congruent microscopic, sharp, right-angle branches and turns.
Table 1.
Summary of quantitative analysis of fiber orientations observed for each of the histochemical markers
| Case | Values | BDA | SMI-312 | SMI-32 |
|---|---|---|---|---|
| BM097 | Average orientation offset (±SD) | 77° ± 4.5° | 78° ± 10.6° | 74° ± 9.5° |
| Fibers in major orientation | 50% | 67% | 59% | |
| Fibers in minor orientation | 29% | 30% | 36% | |
| Width of major peak | 14° | 78° | 13° | |
| Width of minor peak | 21° | 7° | 10° | |
| PIC | Average orientations offset (±SD) | 67° ± 12.0° | 61.5 ± 9.0 | 66.7 ± 11.0 |
| Fibers in major orientation | 64% | 71% | 55% | |
| Fibers in minor orientation | 35% | 6% | 23% | |
| Width of major peak | 16° | 21° | 22° | |
| Width of minor peak | 15° | 12° | 10° | |
| AM244 | Average orientation offset (±SD) o | 62 ± 17.0 | 81 ± 3.0 | 66 ± 11.0 |
| Fibers in major orientation | 56% | 75% | 83% | |
| Fibers in minor orientation | 21% | 23% | 9% | |
| Width of major peak | 13° | 14° | 14° | |
| Width of minor peak | 24° | 13° | 13° |
Values derived from the average of ACs from 3 sections of each case.
Orientation of Turns and Branches
Figure 3O provides a summary of the frequency of turns and branches with specific orientations detected in congruent rectangular ROIs placed in the centrum semiovale of all 3 sections of each of the 3 cases. A total of 91 turns and 122 branches were analyzed. It can be seen that the directions of turns and branches align to all 4 possible quadrants for turns and all 4 possible central axes for branches. However, the turns and branches are not equally distributed across these orientations but are most prevalent in the turn from lateral to ventral, toward the internal capsule, striatum and thalamus where corticofugal fibers such as those of the corticospinal, corticostriatal and other projections leave the forebrain.
Radius of Axonal Turns or Branches in BDA-labeled Axons
The radius of curvature of BDA-labeled fibers was measured for random samples of turns and branches like those shown in Figure 3D–J. The observed radius ranged from 13 to 17 µm with a median of 14 µm. It is notable that this is well below the spatial resolution of high angular resolution dMRI in vivo (~1 mm) or ex vivo (~100 μm) (Wedeen et al. 2012a).
Frequency of Axonal Turns or Branches for BDA-labeled Axons
Finally, to further quantify the turning and branching, the frequency of BDA-labeled axonal turns and branches in the x,y plane of section was estimated by dividing the total number of branches and turns observed by the total length of all BDA-labeled axons in a rectangular ROI measuring 1.85 mm × 2.4 mm (ROI of about 4.4 mm2) within a field as illustrated in Figure 3C. It was found that there was one axon turn or branch every 1.75 (±0.4) mm of BDA-labeled axon length.
Axon Trajectories and Orientation of SMI-32 Labeled Large Projection Fibers
In order to determine how the observed orientations of motor cortex projection axons compares with the trajectories of projection axons that may originate in other areas but pass through the same part of the centrum semiovale, we processed sections through this same region from each of the 3 monkeys using IHC with SMI-32, an antibody that labels axons which originate from large projection neurons (Morrison et al. 1987; Bajo and Moore 2005). As shown in Figure 4A from case BM097, the fibers travel in 2 primary and approximately orthogonal orientations. This was quantified using all the same 3 analysis methods as in Figure 3K–M and identified 2 major orientation peaks—one ranging from 147° to 150° and the other from 75° to 81°. To further quantify this, the acute angles between the major and minor axis peaks were calculated and found to range from 68° to 75°. Supplementary Figure 3 shows the AC analysis of SMI-32 labeled axons in 3 sections from each of the 3 cases. These polar plots confirm the ubiquity and consistency of 2 primary axis orientations that are congruent with the axis orientations identified by dMRI (Fig. 2A–C) as well as the orientation of the BDA projections fibers (Fig. 3). These orientations can be further appreciated in Figure 4B, where pseudocolor coding of SMI-32 fibers based on PP analysis as in Figure 3N was applied and provides visual confirmation of the 2 major orientations. In comparing Figure 4B with Figure 3N, it is notable that there is a higher density of SMI-32 labeled fibers as expected and that the somewhat different offset angle of fibers analyzed in Figure 4 may reflect the somewhat more caudal location of this section in the centrum semiovale compared with the section in Figure 3N. The overall quantitative analysis of SMI-32 labeled axon orientations in all the sections analyzed with the 3 image analysis modalities is summarized in Table 1.
Figure 4.
Trajectories and quantitative orientations of SMI-32 labeled projection axons. Panel A shows axons labeled by IHC processing with the neurofilament H (heavy chain) antibody SMI-32. The image is taken in a region of subcortical white matter beneath motor cortex caudal to that of Figure 3D. Panel B shows the same image as A, with orientation encoded in hue according to PP analysis (E) and as per the inset color wheel. Panels C–E are polar plots comparing orientation analysis of fibers in Panel A using AC, ST, and PP with a major orientation between 147° and 150° and minor orientation between 75° and 81°. Notably ST and PP detect a smaller third peak between 44° and 64°. Scale bar in A = 200 µm.
Axon Trajectories and Orientation of Total Axon Population Labeled with SMI-312
To determine if the geometric organization observed by BDA labeling of corticofugal projection fibers (Fig. 3) and SMI-32 labeling of all large projection fibers (Fig. 4) was representative of all the axons that traverse this part of the centrum semiovale, we processed sections through this same region from each of the 3 monkeys using IHC with the pan-axonal neurofilament antibody SMI-312 (Lei et al. 2012; Zurashvili et al. 2013) to label the total population of axons in the white matter of the centrum-semiovale beneath motor cortex. Figure 5A is a large-scale montage of 8 confocal images of SMI-312 labeled fibers from one of the cases and shows that the population of all axons also appears to adhere to the primary axes in the x,y plane. Higher power images in Figure 5B,C show this approximately orthogonal organization of the 2 primary axes in the x,y plane and, as expected, also reveal fibers traveling in the z-axis as small dots or truncated segments traversing the short z-axis of the 30-µm thick sections. Similar to the analysis of the BDA and SMI-32, when orientation was quantified with AC, ST, and PP (Fig. 5D–F), it revealed one peak orientation ranging across all 3 methods from 160° to 163° and another from 64° to 69°. To further quantify this, the acute angles between the major and minor axis peaks were calculated and found to range from 83° to 86°. Interestingly, in contrast to AC and ST, the PP metric shows a shoulder of a third minor orientation centered at 97°. This minor peak represents <15% of fiber area but suggests that PP is more sensitive to fiber orientation in this dense field. Figure 5G shows the same image as Figure 5A but is pseudocolored for orientations quantified by PP with orientation angle mapped to hue, orientation contrast to saturation, but with the image brightness inverted. Inset Figure 5H is shown at a higher magnification but with image brightness not inverted. Supplementary Figure 4 shows AC polar plots for 3 sections from each of 3 cases and confirms that SMI-312 labeled fibers also adhere to 2 major orientations. The overall quantitative analyses of fiber orientations using the 3 image analysis modalities in all the sections analyzed are tabulated in Table 1. It is worth noting that the SMI-312 labeled fibers include axons from all sources that pass through this region of white matter regardless of their origin or destination and labels both myelinated and unmyelinated fibers. Examples of fiber populations would include thalamocortical fibers ascending to the cortex or corticocortical fibers coming from or going to other regions of the cortex (e.g., see Fig. 11-2 Sections 73, 81, 85 of Schmahmann and Pandya 2006).
Primary Axis Orientation Angles
The quantitative analysis of the histological data not only confirms the 2 primary axis orientations of dMRI but also reveals variability in the average angle of offset across cases (Supplementary Figs 2–4). To assess this variability, the 3 best highest angular resolution dMRI scan data sets were analyzed for the specific angle of offset at each fiber crossing within the centrum semiovale. One of these cases (BM097) is part of the histological analysis but the other 2 did not as their study origins could not incorporate BDA injections. Nevertheless, as shown in Supplementary Figure 5, all 3 cases not only showed the expected geometric organization, but also revealed variability in the exact angles that were color coded. This variability can be seen by comparing the offset angles between the 3 cases (Supplementary Fig. 5B–D) and likely reflects inevitable differences in plane of section and location of each region of interest across subjects. The variability is also evident within a case as one compares, for example, more dorsal and medial crossings with more ventral and lateral crossing. This variance likely reflects warping and shearing of these axes by the differential curvature of lobes, sulci, and gyri.
Fiber Diameters
We also measured the cross-sectional diameter of a sample of the x,y axis fibers detected in our orientation analyses of BDA, SMI-32, and SMI-312 labeled axons. As shown in Supplementary Table 1, the observed mean diameters of the different labeled axon populations ranged from about 5.21 µm for SMI-312 to 8.23 µm for SMI-32. This has several implications. First, the observed range of diameters is consistent with the presumed populations labeled and visualized by each method. The “projection fibers” labeled with SMI-32 that originate from this and other cortices have the largest average diameter (8.23 µm). In contrast, the total population of all axons labeled with SMI-312 had the smallest average diameter (5.21 µm) as this group should include smaller local circuit and unmyelinated fibers. Finally, the corticofugal fibers from motor cortex labeled with BDA had an intermediate average diameter of 6.12 µm. While this projection would include very large diameter fibers that could project all the way to the spinal cord, it would also include smaller diameter fibers that might only go short distances such as to adjacent somatosensory cortex. These measurements support the specificity and sensitivity of each label for their respective fiber populations. It also shows that the quantitative demonstration of orientations to grid axes adheres across these heterogeneous populations.
Discussion
Summary of Results
High angular resolution dMRI of the white matter beneath motor cortex in monkeys and humans shows that fibers in this region conform to axes of a geometric grid as previously described (Wedeen et al. 2012a). Labeling corticofugal projections from a limited region of primary motor cortex of monkeys using the tract tracer BDA revealed that axons observed in the coronal plane in this same white matter region disperse into 2 sharply defined primary orientations: mediolateral and dorsoventral. Congruent axonal orientations were found in the identical region using the SMI-32 antibody to label the population of large projection axons and using the SMI-312 pan-axonal antibody to label the entire population of axons. Quantitative analysis showed that with all 3 labeling methods over 80% of axons in the x,y coronal plane were oriented along 2 discrete, approximately orthogonal primary axes. Most importantly, these orientations conform to the pattern predicted by dMRI. Furthermore, it is notable that axons visualized with these methods and in these orientations likely include commissural projections to and from contralateral motor cortex, association projections with nearby premotor and somatosensory cortices, and subcortical projections to striatum, thalamus, brainstem (corticobulbar), and spinal cord (corticospinal). Notably, up to 20% of the fibers do not follow these 2 primary axes. These fibers may in part reflect axons oriented in the z-axis but unresolved by the 30 µm limit of section thickness, which does not allow computation of orientation as many of these fibers are cut in cross-section and appear as mere “dots” that are part of the isotropic background (e.g., Fig. 5A–C). Alternatively, in some areas, a part of this 20% can be axons that follow a slightly different axis (e.g., 44° peak in Fig. 4) which may reflect fibers that entered and left the region at a different point in development, and hence were subjected to different warping and shearing by the curvature of development. Finally, it is plausible that some of these fibers are not part of any major orientation but instead follow and independent course making them indeed a rare exception to the geometric organization.
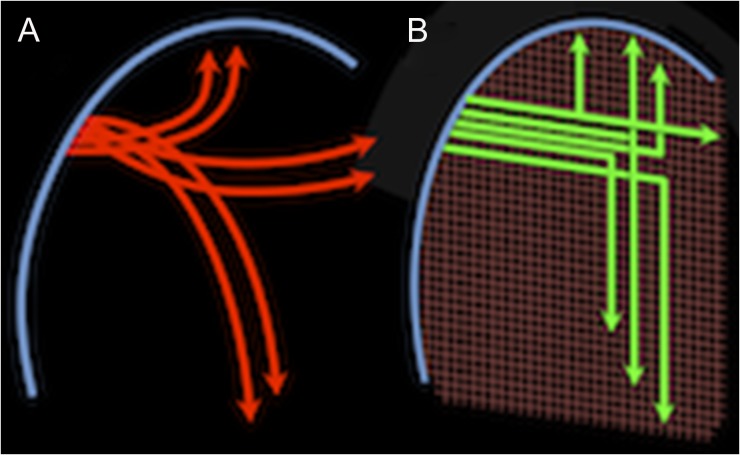
Careful microscopic analysis of BDA-labeled fibers shows that these corticofugal axons must navigate to these different targets by moving from one axis to another. Moreover, the data show that axons do this by making sharp right-angle turns and/or giving off right-angle branches. These data suggest that rather than traveling along smooth, distance minimizing arcs as traditionally assumed (e.g., Figs 1A,B and 6A), axons navigate from point to point along largely primary axes in a manner as simple as that shown in Figure 6B, or perhaps as complex as shown in Supplementary Figure 6. While high angular resolution dMRI shows fiber crossings as in Figure 2, dMRI cannot yet resolve the microscopic turns or branches (Fig. 3D–J and Supplementary Figs 7 and 8) nor their contributions to connectivity. These findings reinforce concerns expressed regarding the inability of dMRI to resolve “kissing fibers” (Basser et al. 2000) and other limitations on the accuracy of present dMRI tractography (Thomas et al. 2014).
Figure 6.
Alternatives views of fiber navigation. This schematic compares 2 views of the trajectories taken by corticofugal axons leaving the cortical gray matter (blue arc on the left) and traveling to diverse cortical and subcortical targets. Association fibers are represented traveling vertically toward ipsilateral cortical targets, commissural fibers are represented traveling horizontally toward contralateral targets and projection fibers are represented traveling downward toward subcortical targets. In the classical model (A), these fibers (red) form groups, or bundles, that diverge smoothly from the cortex, possibly in geodesic arcs, as they interweave and disburse to different targets. In the geometric grid model (B), even as the grid deviates from perfectly orthogonal angles as it is warped by brain curvature, the different fibers (green) adhere to these coordinate axes as they make approximately right-angle sharp turns or branch as they travel to the different targets.
Neuroanatomical Results and High Angular Resolution dMRI Tractography
Our DSI results as shown in Figure 2A–C show that paths beneath primary motor cortex in the monkey conform to the large-scale grid-like organization previously reported (Wedeen et al. 2012a) even as they follow the different cerebral paths (commissural, association, and subcortical) described by Schmahmann et al. (2004) and Schmahmann and Pandya (2006). Figure 2D–F demonstrates a similar organization of cerebral white matter beneath human motor cortex observed by in vivo DSI of human brain. Considering the fiber pathway organization reported here using high angular resolution dMRI tractography, several issues are of note. First, the organization of axons observed at the microscopic level with tract tracers and IHC is congruent with dMRI of major fiber trajectories. Second, dMRI tractography showed no evidence of curved or diagonal paths within this region and microscopic analysis rarely detected curved or diagonally oriented axons. Third, dMRI tractography showed no evidence of the microscopic axonal turns and axonal branches seen with tractography, instead resolving these as fiber crossings. While several recent studies compared dMRI and microscopic findings in different systems, and with different approaches (e.g., Vasung et al. 2011; Budde and Annese 2012; Jbabdi et al. 2013), the question of resolving sharp axonal turns or branches has not been previously addressed.
Specific challenges for dMRI are the variety of axon turns and branches as observed in the current study as well as the overall geometry of fibers as identified in Figure 3D–J and confirmed in Supplementary Figure 7. How axons may navigate across axes deformed by brain curvature (angles different from 90°) to travel from one point to another is illustrated in Supplementary Figure 6, which is consistent with the observations presented here of angles ranging from 75° to 105° even as fibers make sharp turns or branches.
Relationship of Axon Orientations and Axon Turns and Branches to dMRI
dMRI has 2 kinds of resolution at 2 distinct spatial scales—(1) that of the diffusion of water molecules, which reflects the distributions of cellular components similar in size to diffusion distances of 1–10 µm and (2) the spatial resolution in the image in which these distributions are mapped positionally on a scale of millimeters. If axons were parallel arcs with moderate curvature (<1 radian of curvature per voxel), as the “standard model” of axons in cerebral pathways often assumes, then dMRI of axon orientations should be enough to infer the trajectories at larger scale and follow cerebral connectivity from point to point. Here, we show that this assumption is not valid for the corticofugal projection axons of primary motor cortex and the subcortical white matter in which they disperse. Whereas dMRI does indeed accurately reflect the predominant axon orientations, it is insensitive to microscopic turns and branches at angles of 75°−105° with turning radii of ≤15 µm, which appear to be the major means by which axons navigate across grid axes and travel from point to point. Depending on the angular resolution of the dMRI and the reconstruction algorithm employed dMRI tractography, may produce false-positive or false-negative maps of connectivity (Schmahmann et al. 2007; Wedeen et al. 2008). For example, at low angular resolution, fiber crossings are “averaged” and would be a false positive as they reconstruct all turning across smooth curves where none exists. Alternatively, at high resolutions, fiber tracks will appear to simply cross and would be a false negative as the sharp turns of axons that do change directions within the crossing go undetected. Possible instances of such inconsistency are described in detail by Thomas et al. (2014), and consistent with our interpretation of why the geometric organization was not seen by Catani et al. (2012) and Wedeen et al. (2012b). Thus, the phenomenon of microscopic right-angle axonal turns and branches poses a serious challenge for using dMRI to discover point-to-point cerebral connectivity.
Technical Considerations for Histological Analyses of Axons
First, while the BDA-labeled axons originating in motor cortex are clearly labeled, the proportion of the axons that leave primary motor cortex and were labeled by our BDA injections is unknown. Hence, one might argue that BDA selectively labeled only fibers that happen to disperse in a grid-like pattern (Fig. 3 and Supplementary Fig. 1). This possibility is unlikely since applying IHC to adjacent sections using SMI-32 to label additional projection axons passing through the same white matter shows that they conform (Fig. 4 and Supplementary Fig. 3) to orientations similar to the BDA-labeled fibers (Supplementary Fig. 2). The likely accuracy of both the BDA and SMI-32 observations was further confirmed using SMI-312 antibody to label the total population of axons in this region (Fig. 5 and Supplementary Fig. 4).
Second, the dMRI data predict that axons in the third axis (z-axis) should be approximately orthogonal to the x,y plane of section through white matter that is deep to primary motor cortex. However, because of limited depth in the z-axis (30 µm) for our histologic sections, z-axis fibers are truncated and either appear in cross-section as dots, or if they turn or branch, as extremely short truncated segments (Fig. 3F and Supplementary Fig. 8A). In both cases, their orientation in the z-axis cannot be mapped and their presence likely contributes to the isotropic background. So while high angular resolution tractography shows a 3D grid in x,y,z, 30-µm thick sections available for the present study were only able to confirm the orientations in the x,y plane.
Third, the sharp turns and branches we have observed in the x,y plane are not an artifact of tissue shrinkage due to the glycerol cryoprotection used as explained in the Methods. It is also noteworthy that turns are observed with confocal microscopy using water soluble mounting media that limits shrinkage in the z-axis (e.g., Supplementary Fig. 8A).
Fourth, our observation that there is one turn or branch per 1.75 mm of axon length raises the question of the overall prevalence of such turns or branches within forebrain white matter. Based on this rate and the 213 turns, we enumerated across the 3 cases (about 4 mm2 in each of 3 sections from each case), it can be estimated that there are 213 turns or branches per 390 mm of labeled corticofugal axon length. Since within our ROI axons the maximum axon length across the diagonal would be about 3 mm, then the minimum number of axons crossing the region would be 130 if all traveled the full diagonal. But the number of axons would range up to 780 if the average axon length within the field was 0.5 mm. While a true estimate of length and hence the number of individual axons that crossed our ROI's cannot be obtained because the thinness of our sections prevents tracing individual axons for such lengths, it seems unlikely that axon length traversing the 4.4 mm2 ROI would be much <0.5. So assuming a 0.5 mm length we would have 213 turns or branches for 780 axons or a probability of about 27% that any individual fiber would turn or branch within the ROI space analyzed. Additionally, our analysis of the direction of turns (Fig. 3O) shows that of the 91 fibers that turn, 56 turn ventrally and of the 122 that branch, 54 give off a ventrally oriented branch. This implies that nearly 50% of the fibers that do turn or branch, navigate ventrally toward the internal capsule, striatum, or other subcortical targets. Hence, while it appears that turns and branches are common for the corticofugal fibers from motor cortex, the actual frequency and specific targets cannot be established with the present 30 µm frozen sections and will require future experiments using 3D imaging to follow labeled fibers through thick sections (e.g., 3–5 mm).
Previous Neuroanatomical Studies: Tract Tracings
It is important to consider the reasons why prior tract tracing studies in large brain species like monkeys have not reported either the overall grid pattern or the right-angle turns and branches required to navigate between axes. One reason is that the majority of these studies from 1970s and on, utilized anterogradely transported, radioactively labeled amino acids to label connections across the entire monkey brain, where the emphasis was usually on finding terminal fields (e.g., Rosene and Van Hoesen 1977; Pandya and Yeterian 1996; Seltzer and Pandya 2009) rather than following fiber trajectories. When attention was directed to the fiber pathways themselves, the methodological approach using tritiated amino acids (autoradiogaphic studies; Cowan et al. 1972), made it possible to identify the trajectories of fiber aggregates, or bundles of fibers coursing through the white matter toward their destinations. These studies provided new insights into the overall structure of white matter connectivity at the mesoscale level, and often included reconstructions of the paths as artistic renderings that assumed smoothly curved trajectories (e.g., Pandya and Rosene 1985; Demeter et al. 1990; Schmahmann and Pandya 1992, 2006; Petrides and Pandya 1999). However, these histological investigations were unable to follow individual isotope-labeled axons for 2 pivotal technical reasons: First, labeled bundles were followed across nonadjacent spaced tissue sections. Second, the isotope which labels the fibers is detectable in the emulsion overlaying only the top ±3 µm of the section and scatters across up to 3 µm of emulsion. So, while sharp turns or branches of individual axons cannot be resolved, close inspection of these pathways reveals instances where entire fiber bundles make sharp turns, sometimes almost at right-angles (e.g., Fig. 1C,D). Thus, Schmahmann and Pandya (2006) reported that fibers leaving a longitudinal association bundle may turn abruptly at 90° to enter the cortex laterally or the corpus callosum medially. Similarly in a study of corticopontine pathways, Schmahmann and Pandya (1992) noted that fibers “descended sharply” as they turned into the cerebral peduncle toward the pons.
Other examples of geometric organization with sharp turns can be found in the early tract tracing work of Wendell Krieg who followed degenerating myelinated axons from a lesion in motor cortex of the monkey and showed that bundles of axons made sharp turns into the internal capsule white matter (e.g., Figs 37 and 96, Krieg 1954). Most recently, radioisotope tracers have been replaced by anterograde tracers such as DiI, PHA-L, WGA, and BDA, which allow exquisite visualization of individual axons. These studies have also revealed fibers making sharp right-angle turns or branches (e.g., O'Leary and Terashima 1988; Rockland 1995, 2013). Nevertheless, in the absence of a 3D framework within which to view these observations, the sharp turns were regarded as interesting features rather than a reflection of an overall geometric organization that might be present across the brain. While the present results provide extensive data for corticofugal projections from primary motor cortex in the monkey, it is worth noting that this organization is also present in the sagittal stratum of the occipital lobe (Wedeen et al. 2012a Figure 2; Mortazavi et al. 2015 Figure 17.14) as well as in the cingulum bundle and superior temporal gyrus (Mortazavi et al. 2015 Figures 17.3 and 17.4, respectively). High-resolution confocal images of a sharp turn in the cingulum bundle and a branch in the white matter beneath area PG/Opt are shown in Supplementary Figure 8. This suggests that the geometric organization is as widespread at the histological level as recent dMRI observations of the 3D geometric pattern (Wedeen et al. 2012a, 2012b). The latter observations have provided both local and large-scale 3D context, allowing the significance of observations of turns and branches to be appreciated. It also suggests that re-analysis of earlier connectional studies such as those documented by Schmahmann and Pandya (2006) could reveal more evidence of fiber pathways making sharp turns.
Fiber Turning and Branching
Based on our original observations from dMRI of primary orientations of fiber pathways (Wedeen et al. 2012a, 2012b), this predicted that axons must make approximately right-angle turns congruent to the grid-like axes of a region to move from one direction to another. Indeed, this was observed in tissue sections following BDA injections into the motor cortex. Somewhat surprisingly, in addition to the predicted turns, we also found many right-angle branches within the white matter. When quantified, turns and branches were observed at a frequency of one turn per 1.75 mm of fiber length with branching occurring somewhat more often, suggests that the right-angle T-branches observed within cortical gray by Ramon y Cajal (1955) may also be a common feature of axons in subcortical white matter. Interestingly, if this branching is a common feature of many cortical pathways and the branching rate is consistent along a significant length of the fiber, it suggests that axons frequently branch and hence individual axons likely innervate multiple targets. This is surprising, as studies of multiple connectivity using discrete injections of different retrograde tracers to “double label” cells of origin of axon branches that terminate in multiple targets generally report “colabeling” from multiple branches of only around 10% with a report of about 20% being the high water mark (Rockland 2013). Finally, these branches have been described as a frequent feature of axon growth during development (O'Leary and Terashima 1988; Gallo 2011; Kalil and Dent 2014) but the present data suggest that such branches may be maintained in adulthood with both collaterals establishing effective connections. In contrast, for branches where connectivity is not effectively established in one arm, it is possible the axon of the poorly connected arm might “die back,” leaving behind the right-angle turns we observe in the adult brain.
Neurodevelopmental Studies
The notion of a geometric pattern of fiber trajectories and crossing has also been documented in studies of embryonic development, where a “checkerboard pattern” referred to as “migration areas” was identified as the underlying organization at early stages of development (Nieuuwenhuys and Puelles 2016). This is often considered to be the prosometric model which postulates that all vertebrate segmented brains structures develop along the same Bauplan (base-plan) and share a set of dorsoventral and anteroposterior axis coordinates (Puelles 2009). Hence, the present studies support 2 emerging views on the structure of the brain, and specifically, the relations between evolution, development, and connectivity. First, at the largest scales, it is suggested that an orthogonal structure may be a general feature of the nervous systems of vertebrates, a core feature of its base-plan of gene expression and of connectivity (Swanson 2007; Nieuwehuys and Puelles 2016). These patterns would be both 2D (Swanson 2007) and 3D (Nieuwenhuys and Puelles 2016), would be a rectilinear pattern modified by plastic deformation during development, as foreseen by d'Arcy Thompson (Katz et al. 1980; Nieuwenhuys 2009; Nieuwehuys and Puelles 2016), and would be strongly adhered to by the structure of axonal connectivity. Second, at a cellular level, axonal guidance via microscopic right-angle turns and branches has been noted in multiple brain systems, and governed by congruent guidance cues—dyads and tetrads of gene expression specifying local coordinate systems, which axons follow like the cursor of an etch-a-sketch. For example, this mechanism has been found to guide the genesis of the noted orthogonal wiring of the superior colliculus, in which sensory motor association results from 2D alignment of topographic sensory maps (retinotopic, somatotopic, acoustic, and others) to an parallel aligned motor output map directing visual gaze (Cang and Feldheim 2013). Our observations suggest that this geometric base-plan is characteristic of development across scales and remains evident in the adult primate forebrain. It provides evidence that the orthogonal morphology previously suggested (Wedeen et al. 2012a, 2012b) is observable from the scale of MRI tractography to that of the axons in cerebral white matter.
Conclusions and Future Directions
Observations of geometric structure with dMRI suggested that the pathways of the primate forebrain adhere to the cardinal axes of a 3D coordinate system (Wedeen et al. 2012a). The microscopic data reported here show that axon orientations of the corticofugal projections from primary motor cortex as well as all other axons in the same white matter volume also adhere to these dMRI directions and provide a histological basis for the dMRI observations. Furthermore, these observations show that axons navigate between these axes by way of microscopic sharp turns and branches, rather than along smooth curves or diagonals as previously supposed. While confirmation of the generality of these microscopic observations across the entire forebrain is necessary, the observations presented here provide quantitative microscopic data on axon branching, turning and trajectories that are entirely congruent with the pervasive geometric, grid-like structure of fibers revealed by high angular resolution dMRI (Wedeen et al. 2012a). Overall, these data suggest that a coherent grid structure is a possible feature of cerebral connectivity from the axonal scale to the entire forebrain. A recent review by Edwards et al. (2014) is instructive for the enumeration of the large number of signaling mechanisms involved in the normal and abnormal development of the corpus callosum and highlights the many unknown factors likely to affect the geometric structure.
Both in dMRI and in neuroanatomy, the idea of a “fiber crossing” has come to mean the situation where axons of distinct and unrelated pathways travel across the same volume. The question of the structure of these crossings has been a challenge. When MRI tractography of diffusion tensor imaging (DTI) was initially described (Wedeen 1995; Conturo et al. 1999) discussion ensued concerning whether DTI, which resolves only one orientation per MRI voxel, could provide enough information to infer connectivity. This limitation of DTI was addressed by the development of high angular resolution dMRI (Tuch et al. 2002; Wedeen et al. 2005), which revealed fiber crossings in multiple locations. The present studies suggest that rather than an epiphenomenon, the regions that appear to be simple fiber crossings in dMRI, in some cases can harbor sharp turns and branches as axons navigate between axes to establish cerebral connectivity as illustrated in Figure 6B.
These observations have notable implications for efforts to map the cerebral connectome. First, the conventional view of brain pathways organized as curved bundles like streamlines in fluid or strings in tension, needs to be revised. Rather, cerebral pathways as revealed in dMRI emerge from the statistics of axonal microstructure, specifically from coherent fibers that follow a grid-like orientation, crossing in approximately orthogonal directions that can be detected with dMRI but moving between axes by way of sharp turns and branches that dMRI does not resolve. Second, in brain mapping with dMRI, orientation contrast is averaged within voxels that are far larger than the microscopic axonal turns and branches by which axons are here shown to navigate. This further supports the previous suggestion (Wedeen et al. 2012b) that when tractography renders the pathways from cortex into white matter as a smooth arc (e.g., Catani et al. 2012), it is likely due to limited spatial resolution that fails to resolve the fiber orientations accurately. Third, while the axonal branches and turns documented here are too small to be spatially resolved with present dMRI, these structures may potentially be reflected and mapped using other features of diffusion contrast (e.g., Callaghan 1994; Lu et al. 2006; Assaf et al. 2008; White et al. 2013) including alternate analyses of crossings and of the time dependence of diffusion. In the interim, the present observations pose a notable challenge for and necessary caveat to the analysis and interpretation of dMRI mapping of the human brain connectome and to its validation.
Supplementary Material
Notes
We thank Wen-Yih Tseng, Krastan Blagoev, and Harvey Swadlow for helpful discussions and George Dai and Timothy G. Reese for assistance with MRI acquisitions. Conflict of Interest: None declared.
Supplementary Material
Supplementary material is available at Cerebral Cortex online.
Funding
NSF grants PHY-0855161 and PHY1505000 (D.L.R., H.E.S., and V.J.W.) and PHY-1444389 (W.Z.M.) as well as NIH grants R01-MH064044 (J.D.S. and V.J.W.), U01-MH093765 (V.J.W.), and R01-AG043640 (D.L.R.).
Author Contributions
F.M. wrote the manuscript, designed and conducted immunohistochemical experiments and acquired confocal images and prepared illustrations. A.L.O. designed and conducted the tract tracer experiments, acquired images for tract tracer figure and assisted with writing the manuscript. W.Z.M. designed and implemented the quantitative image analysis algorithms, analyzed turning and branching in histological images, and assisted in preparation of figures and in writing the manuscript. J.D.S. contributed to overall experimental design, designed the tract tracer experiments and assisted in writing the manuscript. H.E.S. oversaw design of image analysis algorithms and assisted in writing the manuscript. V.J.W. guided overall experimental design, conducted MRI experiments and participated in writing the manuscript. D.L.R. guided overall experimental design, conducted surgeries for tracer injections and participated in writing the manuscript.
References
- Assaf Y, Blumenfeld-Katzir T, Yovel Y, Basser PJ. 2008. AxCaliber: a method for measuring axon diameter distribution from diffusion MRI. Magn Reson Med. 59:1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajo VM, Moore DR. 2005. Descending projections from the auditory cortex to the inferior colliculus in the gerbil, Meriones unguiculatus. J Comp Neurol. 486(2):101–116. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A. 2000. In vivo fiber tractography using DT-MRI data. Magn Reson Med. 44:526–532. [DOI] [PubMed] [Google Scholar]
- Budde MD, Annese J. 2012. Quantification of anisotropy and fiber orientation in human brain histological sections. Front Integ Neurosci. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan PT. 1994. Principles of nuclear magnetic resonance microscopy. Oxford: Oxford University Press. [Google Scholar]
- Cang J, Feldheim DA. 2013. Developmental mechanisms of topographic map formation and alignment. Annu Rev Neurosci. 36:51–77. [DOI] [PubMed] [Google Scholar]
- Catani M, Bodi I, Dell'Acqua F. 2012. Comment on “The geometric structure of the brain fiber pathways”. Science. 337(6102):1605. [DOI] [PubMed] [Google Scholar]
- Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder AZ, Shimony JS, McKinstry RC, Burton H, Raichle ME. 1999. Tracking neuronal fiber pathways in the living human brain. Proc Natl Acad Sci. 96(18):10422–10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan WM, Gottlieb DI, Hendrickson AE, Price JL, Woosely TA. 1972. The autoradiographic demonstration of axonal connections in the central nervous system. Brain Res. 37(1):21–51. [DOI] [PubMed] [Google Scholar]
- Demeter S, Rosene DL, Van Hoesen GW. 1990. Fields of origin and pathways of the interhemispheric commissures in the temporal lobe of macaques. J Comp Neurol. 302:29–53. [DOI] [PubMed] [Google Scholar]
- Edwards TJ, Sherr EH, Barkovich AJ, Richards LJ. 2014. Clinical, genetic and imaging findings identify new causes for corpus callosum development syndromes. Brain. 137:1579–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo G. 2011. The cytoskeletal and signaling mechanisms of axon collateral branching. Dev Neurobiol. 71:201–220. [DOI] [PubMed] [Google Scholar]
- Giannaris EL, Rosene DL. 2012. A stereological study of the numbers of neurons and glia in the primary visual cortex across the lifespan of male and female rhesus monkeys. J Comp Neurol. 520:3492–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granziera C, Schmahmann JD, Hadjikhani N, Myer H, Meuli R, Wedeen VJ, Krueger G. 2009. Diffusion spectrum imaging shows the structural basis of functional cerebellar circuits in the human cerebellum in vivo. PLoS One. 4(4):e5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Laboratory Animal Resources Commission of Life Sciences NRC 1996. Guide for the care and use of laboratory animals. Washington, DC: National Academy Press. [Google Scholar]
- Jbabdi S, Lehman JF, Haber SN, Behrens TE. 2013. Human and monkey ventral prefrontal fibers use the same organizational principles to reach their targets: tracing versus tractography. J Neurosci. 33:3190–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil K, Dent EW. 2014. Branch management: mechanisms of axon branching in the developing vertebrate CNS. Nat Rev Neurosci. 15:7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz MJ, Lasek RJ, Nauta HJ. 1980. Ontogeny of substrate pathways and the origin of the neural circuit pattern. Neuroscience. 5(5):821–833. [DOI] [PubMed] [Google Scholar]
- Klingler J. 1935. Erleichterung der makroskopischen Praeparation des Gehirns durch den Gefrierprozess. Schweiz Arch Neurol Psychiatr. 36:247–256. [Google Scholar]
- Knutsson H, Westin C-F, Andersson M. 2011. Representing local structure using tensors II. SCIA, Berlin-Heidelberg: Springer-Verlag. [Google Scholar]
- Krieg WJS. 1954. Connections of the frontal cortex of the monkey. Springfield, IL: Charles C. Thomas., 1954, Chapter 2, Fig. 38. [Google Scholar]
- Lei WL, Xing SG, Deng CY, Ju XC, Jiang XY, Luo ZG. 2012. Laminin/β1 integrin signal triggers axon formation by promoting microtubule assembly and stabilization. Cell Res. 22(6):954–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Jensen JH, Ramani A, Helpern JA. 2006. Three-dimensional characterization of non-Gaussian water diffusion in humans using diffusion kurtosis imaging. NMR Biomed. 19(2):236–247. [DOI] [PubMed] [Google Scholar]
- Montero RS. 2009. State of the art of compactness and circularity measures. Int Math Forum. 4(27):1305–1335. [Google Scholar]
- Moore TL, Killiany RJ, Pessina MA, Moss MB, Finklestein SP, Rosene DL. 2012. Recovery from ischemia in the middle-aged brain: a non-human primate model. Neurobiol Aging. 33(3):619.e9–619.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JH, Lewis DA, Campbell MJ, Huntley GW, Benson DL, Boura C. 1987. A monoclonal antibody to non-phosphorylated neurofilament protein marks the vulnerable cortical neurons in Alzheimer's disease. Brain Res. 416:331–336. [DOI] [PubMed] [Google Scholar]
- Morrison WZ. 2015. Understanding the brain through its spatial structure (Order No. 3684846). Available from ProQuest Dissertations & Theses Global. (1660976115). Retrieved from http://search.proquest.com/docview/1660976115?accountid=967
- Mortazavi F, Wedeen VJ, Rosene DL. 2015. Neuroanatomical techniques for analysis of axonal trajectories in the cerebral cortex of the rhesus monkey In: Rockland KS, editor. Axons and brain architecture. Oxford: Elsevier Publishing. Chapter 17. [Google Scholar]
- Nieuwenhuys R, Voogd J, van Huijzen C. 1981. The human central nervous system a synopsis and atlas. New York: Springer-Verlag. [Google Scholar]
- Nieuwenhuys R, Voogd J, van Huijzen C. 2008. The human central nervous system. 4th ed New York: Springer. [Google Scholar]
- Nieuwenhuys R. 2009. Analysis of the structure of the brain stem of mammals by means of a modified D'Arcy Thompson procedure. Brain Struct Funct. 214:79–85. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuys R, Puelles L. 2016. Towards a new neuromorphology. 1st ed New York: Springer. [Google Scholar]
- O'Leary DDM, Terashima T. 1988. Cortical axons branch to multiple subcortical targets by interstitial axon budding: implications for target recognition and “waiting periods”. Neuron. 1:901–910. [DOI] [PubMed] [Google Scholar]
- Pandya DN, Rosene DL. 1985. Some observations on trajectories and topography of commissural fibers In: Reeves AG, editor. Epilepsy and the corpus callosum. New York: Plenum; p. 21–39. [Google Scholar]
- Pandya DN, Yeterian EH. 1996. Comparison of prefrontal architecture and connections. Philos Trans R Soc Lond B Biol Sci. 351(1346):1423–1432. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Huan X-F, Petrides M, Toga AW. 2009. The rhesus monkey brain in stereotaxic coordinates. 2nd ed London: Academic Press. [Google Scholar]
- Petrides M, Pandya DN. 1999. Dorsolateral prefrontal cortex: comparative cytoarchitectonic analysis in the human and the macaque brain and corticocortical connection patterns. Eur J Neurosci. 11(3):1011–1036. [DOI] [PubMed] [Google Scholar]
- Puelles L. 2009. Forebrain development: prosomere model In: Squire LR, editor. Encyclopedia of neuroscience. Oxford: Academic Press, pp. 315–319. [Google Scholar]
- Ramon y Cajal S. 1995. Histology of the nervous system of man and vertebrates, Vol. 2; Translated from French (1909 & 1911) by Neely Swanson and Larry W. Swanson, Oxford, Oxford University Press. [Google Scholar]
- Ramon y Cajal S. 1955. Studies on the cerebral cortex (limbic structures) Translated from the Spanish (1901-1902) by Lisbeth M Kraft, Chicago, Year Book Publishers. [Google Scholar]
- Rockland KS. 1995. Morphology of individual axons projecting from area V2 to MT in the Macaque. J Comp Neurol. 355:15–26. [DOI] [PubMed] [Google Scholar]
- Rockland KS. 2013. Collateral branching of long-distance cortical projections in monkey. J Comp Neurol. 521(18):4112–4123. [DOI] [PubMed] [Google Scholar]
- Rosene DL, Van Hoesen GW. 1977. Hippocampal efferents reach widespread areas of the cerebral cortex and amygdala in the rhesus monkey. Science. 198:315–317. [DOI] [PubMed] [Google Scholar]
- Rosene DL, Roy NJ, Davis BJ. 1986. A cryoprotection method that facilitates cutting frozen sections of whole monkey brains for histological and histochemical processing without freezing artifact. J Histochem Cytochem. 34:1301–1315. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Pandya DN. 1992. Course of the fiber pathways to pons from parasensory association areas in the rhesus monkey. J Comp Neurol. 326:159–179. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Rosene DL, Pandya DN. 2004. Motor projections to the basis pontis in rhesus monkey. J Comp Neurol. 478(3):248–268. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Pandya DN. 2006. Fiber pathways of the brain. New York: Oxford University Press. [Google Scholar]
- Schmahmann JD, Pandya DN. 2007. Cerebral white matter – evolution of facts and notions concerning the organization of the fiber pathways of the brain. J Hist Neurosci. 16:237–267. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Pandya DN, Wang R, Dai G, D'Arceuil HE, de Crespigny AJ, Wedeen VJ. 2007. Association fiber pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain. 130(Pt 3):630–653. [DOI] [PubMed] [Google Scholar]
- Seltzer B, Pandya DN. 2009. Posterior cingulate and retrosplenial cortex connections of the caudal superior temporal region in the rhesus monkey. Exp Brain Res. 195:325–334. [DOI] [PubMed] [Google Scholar]
- Swanson LW. 2007. Quest for the basic plan of nervous system circuitry. Brain Res Rev. 55(2):356–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C. 2014. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci. 111(46):16574–16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch DS, Reese TG, Wiegell MR, Makris N, Belliveau JW, Wedeen VJ. 2002. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med. 48(4):577–582. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. 1997. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 385:313–318. [DOI] [PubMed] [Google Scholar]
- Vasung L, Jovanov-Milošević N, Pletikos M, Mori S, Judaš M, Kostović I. 2011. Prominent periventricular fiber system related to ganglionic eminence and striatum in the human fetal cerebrum. Brain Struct Funct. 215(3-4):237–253. [DOI] [PubMed] [Google Scholar]
- Wedeen VJ, Davis TL, Weisskoff RM, Tootell R, Rosen BR, Belliveau JW. 1995. White matter connectivity explored by MRI. Proceedings of the First International Conference for Functional Mapping of the Human Brain, Paris; P1.69.
- Wedeen VJ, Hagmann P, Tseng WY, Reese TG, Weisskoff RM. 2005. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn Reson Med. 54(6):1377–1386. [DOI] [PubMed] [Google Scholar]
- Wedeen VJ, Wang RP, Schmahmann JD, Benner T, Tseng WYI, Dai D, Pandya DN, Hagmann P, D'Arceuil H, de Crespigny AJ. 2008. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage. 41(4):1267–1277. [DOI] [PubMed] [Google Scholar]
- Wedeen VJ, Rosene DL, Wang R, Dai G, Mortazavi F, Hagmann P, Kaas JH, Tseng WYI. 2012. a. The Geometric structure of the brain fiber pathways. Science. 335:1628–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeen VJ, Rosene DL, Wang R, Dai G, Mortazavi F, Hagmann P, Kaas JH, Tseng WYI. 2012. b. Response to comment on “the geometric structure of the brain fiber pathways”. Science. 337:6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NS, Leergaard TB, D-Arceuil H, Bjaalie JG, Dale AM. 2013. Probing tissue microstructure with restriction spectrum imaging: histological and theoretical validation. Human Brain Map. 34(2):327–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Guo Y, Slater BJ, Miller NR, Bernstein SL. 2010. Axonal degeneration, regeneration and ganglion cell death in a rodent model of anterior ischemic optic neuropathy (rAION). Exp Eye Res. 91(2):286–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurashvili T, Cordón-Barris L, Ruiz-Babot G, Zhou X, Lizcano JM, Gómez N, Giménez-Llort L, Bayascasa JR. 2013. Interaction of PDK1 with phosphoinositides is essential for neuronal differentiation but dispensable for neuronal survival. Mol Cell Biol. 33(5):1027–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.