Summary
Our study identifies a novel epigenetic factor that potentiates thyroid oncogenesis and raises the possibility that HMGN4 may serve as an additional diagnostic marker, or therapeutic target in certain thyroid cancers.
Abstract
Thyroid cancer originates from genetic and epigenetic changes that alter gene expression and cellular signaling pathways. Here, we report that altered expression of the nucleosome-binding protein HMGN4 potentiates thyroid tumorigenesis. Bioinformatics analyses reveal increased HMGN4 expression in thyroid cancer. We find that upregulation of HMGN4 expression in mouse and human cells, and in the thyroid of transgenic mice, alters the cellular transcription profile, downregulates the expression of the tumor suppressors Atm, Atrx and Brca2, and elevates the levels of the DNA damage marker γH2AX. Mouse and human cells overexpressing HMGN4 show increased tumorigenicity as measured by colony formation, by tumor generation in nude mice, and by the formation of preneoplastic lesions in the thyroid of transgenic mice. Our study identifies a novel epigenetic factor that potentiates thyroid oncogenesis and raises the possibility that HMGN4 may serve as an additional diagnostic marker, or therapeutic target in certain thyroid cancers.
Introduction
Thyroid cancer is one of the most common endocrine malignancies and its global incidence seems to be rising. Accumulating studies identified several genetic alterations including gene mutations, gene dosage effects, gene translocations and gene misexpression that are associated with the occurrence of various types of thyroid cancers (1,2). Here we show that misexpression of the high mobility group N4 (HMGN4) chromatin-binding protein can lead to thyroid tumorigenesis.
HMGN4 is a member of the high mobility group N (HMGN) protein family, which are nuclear proteins that are ubiquitously expressed in all vertebrate cells. The HMGN family contains five members, named HMGN1-5, all of which contain a highly conserved region through which they bind specifically to nucleosomes, the building block of the chromatin fiber, without known specificity for the underlying DNA sequence (3,4). Image analyses of living cells showed that the binding of HMGNs to nucleosomes is highly dynamic and that these proteins constantly roam throughout the entire nucleus, residing for only a short period at any specific site (5,6). Biochemical studies, including chromatin immunoprecipitation analyses, revealed that the binding of HMGNs to nucleosomes affects the structure and function of chromatin, and plays a role in epigenetic regulation of gene expression (7,8). HMGNs modulate the fidelity of the transcription profiles in a variant and tissue-specific manner; the functions of the variants are similar but not fully redundant (8,9). Each HMGN variant affects the expression of a limited number of genes, with only moderate overlap between the genes affected, and each tissue analyzed had a distinct set of genes affected by a specific HMGN variant. Misexpression of HMG proteins, including HMGN proteins has been linked to the etiology of certain cancers (reviews in 10,11).
HMGN4 is the latest discovered member of the HMGN protein family and its biological function has not been yet studied (12). Unlike other HMGN variants, HMGN4 is encoded by a retrogene. Usually, the intronless retrogenes do not encode for proteins; however, western analysis with antibodies specific to HMGN4 verified that this protein is expressed in human cells (12). In search of the biological properties of this protein, we queried several databases for its expression pattern and found that the thyroid expresses relatively high levels of HMGN4, and that these levels are further elevated in several cancers including thyroid cancers. We therefore explored the possibility that HMGN4 plays a role in thyroid carcinogenesis and examined the consequences of elevating HMGN4 in mouse and human cells, and in the thyroid of transgenic mice. Our studies indicate that expression of HMGN4 in mouse cells downregulates the expression of certain tumor suppressor genes, enhances the tumorigenic properties of cells, and can lead to preneoplastic lesions in the thyroids of transgenic mice.
Materials and methods
Construction of HMGN4 plasmids
The human HMGN4 gene (GenBank accession no. NM_006353.2) was amplified with primers containing EcoRI at the 5′ end and XhoI at the 3′ end and then ligated into pEGFP-N2 vector (Clontech), which was pre-cut by EcoRI at the 5′ end and by XhoI at the 3′ end. The HMGN4 sequences was also subcloned into retroviral vector pHAN-FLAG-HA using BamHI at the 5’ end and XhoI at the 3’ end. All clones were verified by DNA sequencing.
Cells and transfection
SV40-immortalized mouse embryonic fibroblast (MEF) and U2OS cell lines were obtained from the American Tissue Cell Collection (ATCC, Manassas, VA). The SV-40 cells which are tumorigenic are widely used to test for the tumorigenic potential of exogenous substances or specific treatments. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA). Follicular thyroid carcinoma-originated FTC-133 and FTC-238 cells were obtained from the European Collection of Cell Cultures (ECACC) through Sigma-Aldrich and cultured in DMEM/F12, supplemented with 10% fetal calf serum (FCS; Invitrogen), at 37°C and 5% CO2. All cell lines were authenticated by the vendor sources. Cultures were expanded, aliquots were frozen, and used within 4 weeks of receipt or resuscitation. Stable MEF cell lines were prepared using a retrovirus generated by transient transfection of amphotropic helper Phoenix cells with the retrovirus containing the HMGN4 gene or an empty vector which was used as a control. The retroviral supernatant was collected and added to MEF cells with 0.8 μg/ml Polybrene (Chemicon International). Stable clones were selected using 5 μg/ml puromycin (Sigma) for 1 week. Stable FTC-133 cell lines were obtained by transfecting with pHMGN4-EGFP or pEGFP using lipofectamine (Invitrogen) followed by selection with 700 μg/ml G418 (Invitrogen) for 1 week, followed by constant selection in 200 μg/ml G418 thereafter. All modified cell lines were aliquoted and frozen and used within 2 weeks or less of preparation or resuscitation.
Soft agar colony assay
One ml of 0.5% agar (42°C), suspended in DMEM with 10% fetal bovine serum, was poured into each 2 cm well of six-well Petri dishes and allowed to solidify. One-half ml of cells at different concentrations were mixed with 0.5 ml of the same agar suspension (42°C) and immediately layered over the hardened agar. The dishes were cultured at 37°C in 5% CO2, for 3 weeks until colonies were visible. Colonies were stained with crystal violet (Sigma), imaged, and counted using ImageJ.
Tumorigenesis by transfected cells in nude mice
Thirty immunodeficient nude (nu/nu) male mice, 8-week-old, were injected s.c. with 1 × 106 of MEFs, expressing either hHMGN4 (MEF-N4) or vector alone (MEF-V). The animals were monitored for tumor formation for 3 weeks, and then sent to necropsy. The developed tumors were excised and weighed. All animal procedures were performed according to approved ACUC protocols.
Transcription analyses and bioinformatics
RNA was prepared by TRIzol reagent (Invitrogen) according to the manufacturer’s protocol and cleaned by Qiagen RNeasy kit. Microarray expression analyses were performed using an Affymetrix Mouse Gene 1.0 ST array at the Laboratory of Molecular Technology (LMT), NCI, Frederick, MD. Data were analyzed by BRB Array Tools, GeneSpring GX 12.6 (Agilent Technologies) and Partek Genomics Suite 6.5 software. A P value < 0.0001 (FDR < 0.033) and fold change ≥2 was set as a threshold for significantly transcriptional changes. All microarray expression data have been deposited in GEO (GSE83313). For qPCR analysis, cDNA was prepared using iScript (Bio-Rad) with 0.3–1 μg of RNA as template. cDNA was used for real-time PCR in the presence of Power SYBR Green PCR Master Mix (Applied Biosystems). Reactions were analyzed on the Applied Biosystems 7900HT Fast Real-time PCR System. A comparative Ct method (2−ΔΔCt) was used to calculate the relative gene expression according to the formula ΔΔCt = [Ct target gene (wild type) – Ct control gene (wild type)] – [Ct target gene (genetically altered) – Ct control gene (genetically altered)]. Dhfr was used as a control gene. Assays were carried out in triplicates. Primer sequences are listed in Supplementary Table 1, available at Carcinogenesis Online. Multi-genome human HMGN4 alignment was performed using the VISTA browser (13) using the human HMGN4 locus as input. The cladogram was adapted from (14). Various databases: Cancer Genome Anatomy Project (NCI, NIH), Cancer Genomics Hub (CGHub, UCSC, CA); GEO datasets (NCBI, NIH) and Oncomine Cancer Genomics Data Analysis Tool (ThermoFisher Sci) were used to study expression profile of human HMGN4 gene in various tissues and tumors. Genomatix Software Suite was employed to perform GO pathway analysis with MEF-N4 gene expression outliers (GeneRanker module), and gene network generation (GePS module).
DNA damage analysis
Cells (1 × 104) were plated in six-well dishes a day before irradiation. The cells were washed in ice-cold 1× PBS and given 8, or 20 J/m2 of UVC radiation. Fresh medium was added to the plates immediately after irradiation, and cells were allowed to recover for 7 h before fixation or lysis for cell extracts.
Cell cycle analysis
Cells were fixed in 70% ethanol overnight at 4°C, washed twice in 1× PBS and stained with 10 µM propidium iodide for 30 min at room temperature in the dark. Cells were run through a Calibur sorter (Applied Biosystems), and the resulting data were analyzed using ModFit.
Antibodies, western blotting and immunofluorescence
Anti-HMGN4, elicited by a peptide spanning amino acids 67–82 of human HMGN4 and anti HMGN2 were as described (12). Rabbit polyclonal non-immune IgG (Millipore), rabbit anti-HA (#26183, Sigma-Aldrich), rabbit anti-BRCA2, (#ab27976, Abcam), rabbit anti-ATRX (#ab97508, Abcam) and rabbit anti-ATM (#ab32420, Abcam) were from the indicated commercial sources. For γH2AX westerns analyses, whole-cell lysates were prepared in 1× SDS-PAGE sample buffer (Bio-Rad) supplemented with protease inhibitors. The samples were fractionated in 12% Precast Mini-Protean-TGX gels (Bio-Rad), transferred by a semidry method to a polyvinylidene difluoride membrane, blocked with nonfat milk (5%) in 1× phosphate-buffered saline and probed with antibodies. Enhanced chemiluminescence detection was performed according to ThermoFisher recommendations for SuperSignal West kits. For HMGN4 and HMGN2 westerns, perchloric acid (PCA) extracts were prepared (15) and diluted with 2×SDS-PAGE sample buffer (Bio-Rad) supplemented with protease inhibitors. For ATM, ATRX and BRCA2 western blotting analyses, whole-cell lysates were prepared in 1× SDS-PAGE sample buffer (Bio-Rad) supplemented with protease inhibitors. The samples were fractionated in 4–15% precast gradient Mini-Protean-TGX gels (Bio-Rad), and transferred to polyvinylidene difluoride membrane. For immunofluorescence, stably transfected FTC-133 cells and MEFs were plated on poly-D-lysine-coated coverslips on six-well plates 24 h before fixation. Cells were fixed in 4% paraformaldehyde (Thermo, Rockford, IL) for 20 min at room temperature, permeabilized with 0.5% Triton X-100 (Sigma) for 10 min, and incubated with anti-γH2AX antibody for 2 h at room temperature. Cells were incubated with FITC-conjugated secondary antibody and Hoechst for 1 h at room temperature. Confocal images were collected using a Zeiss LSM 510 system mounted on a Zeiss Axiovert 200 M microscope (Carl Zeiss Inc., Thornwood, NY) using an oil-immersion Plan-Apochromat 63×/1.4 differential interference contrast (DIC) objective lens. Wide-field images were taken using the oil-immersion Plan-Apochromat 40×/1.4 DIC objective lens. Quantification of the γH2AX-positive cell numbers and the relative signal intensities was carried out using five fields of at least five independently prepared cells per construct and condition.
DNase I hypersensitivity
Mouse embryonic fibroblasts were harvested, nuclei were isolated and resuspended in digestion buffer (60 mM KCl, 15 mM NaCl, 15 mM Tris-HCl, pH 7.4, 0.25 M sucrose, 3 mM MgCl2, 0.5 mM DTT, 50% glycerol) at a concentration of 0.1 mg/ml DNA. Digestions were carried out at 37°C with 60 units of DNase I (Sigma Aldrich) per 100 µg DNA for 3 min. The samples were treated with Proteinase K 10 µg/ml, the DNA was purified with phenol/chloroform/isopropanol mixture and ethanol precipitation. Real-time quantitative PCR were performed using SYBR Green PCR (Roche) and SYBR Green RT-PCR reagent kits (Applied Biosystem) in triplicates. Each primer pair was designed by Primer 3 software to generate a 100–200 bp product. Only dissociation curves with single peaks, indicating specific amplification, were used in the analysis. ΔCt of the digested samples were determined by subtracting the Ct value of DNase I-digested sample from the non-DNase I digested sample. The 2−ΔΔCt method was used to calculate the differences among the various samples. ΔΔCt depicted a difference in DNAse I sensitivity between each primer pair (probe X) and control (Ctrl) primer pairs. ΔΔCtX-Ctrl = ΔCt digested probe X-ΔCt digested Ctrl, where ΔCt digested Ctrl was average of ΔCt digested for three control pairs, which represent ‘bulk’ intergenic chromatin. The sensitivity to DNase I is expressed as log2 ratio ΔΔCtX-Ctrl (MEF-N4)/ ΔΔCtX-Ctrl (MEF-V), pre-set for control primers as a zero (16). Primer sequences are listed in Supplementary Table 1, available at Carcinogenesis Online.
Generation of thyroid-specific hHMGN4-expressing mouse and induction of thyroid carcinogenesis
Mouse strain C57BL/6J was from Jackson Laboratories. All animal procedures were performed according to approved ACUC protocols. The plasmid used to insert the HMGN4 gene for generating the transgenic mice has been described before (17). A MfeI/NotI restriction fragment containing the HMGN4 cDNA sequence tagged with GFP was inserted downstream TPO (thyroid peroxidase promoter and enhancer). The linear DNA fragment was then microinjected into fertilized C57BL/6J embryos using standard procedures. To induce thyroid carcinogenesis, DHPN (N-nitrosodiisopropanol amine) were injected subcutaneously (5600 mg/kg, 200 µl in phosphate-buffered saline, 25G needle) at the age of 4 weeks. Two days after injection, mice were fed with iodine-deficient diet supplemented with 0.15% propylthiouracil (PTU) until euthanized for histological analysis.
Results
Elevated HMGN4 expression in thyroid tumors
Querying of the Gene Expression Omnibus (GEO) and of the HUMAN Protein Atlas data bases showed that HMGN4 is expressed in numerous tissues with a relatively high expression in thyroid tissues. Among the normal tissues analyzed, the expression of HMGN4 in the thyroid ranked highest in the GEO database and fourth highest in the Human Protein Atlas database (Supplementary Figure 1, available at Carcinogenesis Online). Further analysis of the literature (18–21)and of several databases including GEO, Human Protein Atlas, Cancer Genome Anatomy Project (CGAP), Oncomine, and The Cancer Line Encyclopedia (CCLE), indicated that HMGN4 expression is elevated in several tumors, including thyroid cancers (Table 1, Supplementary Figure S2A–D, available at Carcinogenesis Online). In the GEO database, the average level of HMGN4 in the thyroid of seven individuals diagnosed with papillary thyroid carcinoma was 14% higher than that measured in the thyroid of seven healthy individuals (Supplementary Figure 2A, available at Carcinogenesis Online). Likewise, in the Cancer Genome Anatomy Project (CGAP) database, the average expression of HMGN4 in 18 libraries of follicular thyroid carcinoma tissues was twice that observed in 7 libraries constructed from normal thyroid tissues (Supplementary Figure 2B, available at Carcinogenesis Online). Further analysis of the available bioinformatics information reveals that HMGN4 is highly expressed in several types of thyroid cancers including papillary, follicular, and anaplastic carcinoma (Table 1 and Supplementary Figure 2C, available at Carcinogenesis Online) as well as in non-thyroid tumors (Supplementary Figure 2D, available at Carcinogenesis Online).
Table 1.
Bioinformatic data showing high HMGN4 expression in normal and malignant thyroid
| Database | Type of studies | Number of samples | HMGN4 expression | Reference |
|---|---|---|---|---|
| GEO | Expression array | 36 | Expression in thyroid is the highest among 36 types of normal tissues | https://www.ncbi.nlm.nih.gov/geoprofiles/? term=HMGN4+GSE2361 |
| GEO | Expression array | 14 | Elevated in papillary thyroid carcinoma | https://www.ncbi.nlm.nih.gov/geoprofiles/? term=HMGN4+GSE3678 |
| Human Protein Atlas | RNAseq | 122 | Expression in thyroid is fourth highest among 32 types of normal tissues | www.proteinatlas.org/ ENSG00000182952-HMGN4 |
| CGAP | SAGE | 25 | Elevated in follicular thyroid carcinoma | http://cgap.nci.nih.gov/SAGE/Viewer?TAG=AATATGGTAC&CELL=0&ORG=Hs&METHOD=SS10,LS10 |
| Oncomine | DNA-seq | 1,012 | Elevated in papillary thyroid carcinoma | https://www.oncomine.org/resource/login.html |
| CCLE | Expression array | 12 | Elevated in anaplastic and follicular thyroid carcinoma | http://www.broadinstitute.org/ccle/home |
CCLE, The cancer line encyclope; CGAP, Cancer Gene Anatomy Project; GEO, Gene Expression Omnibus.
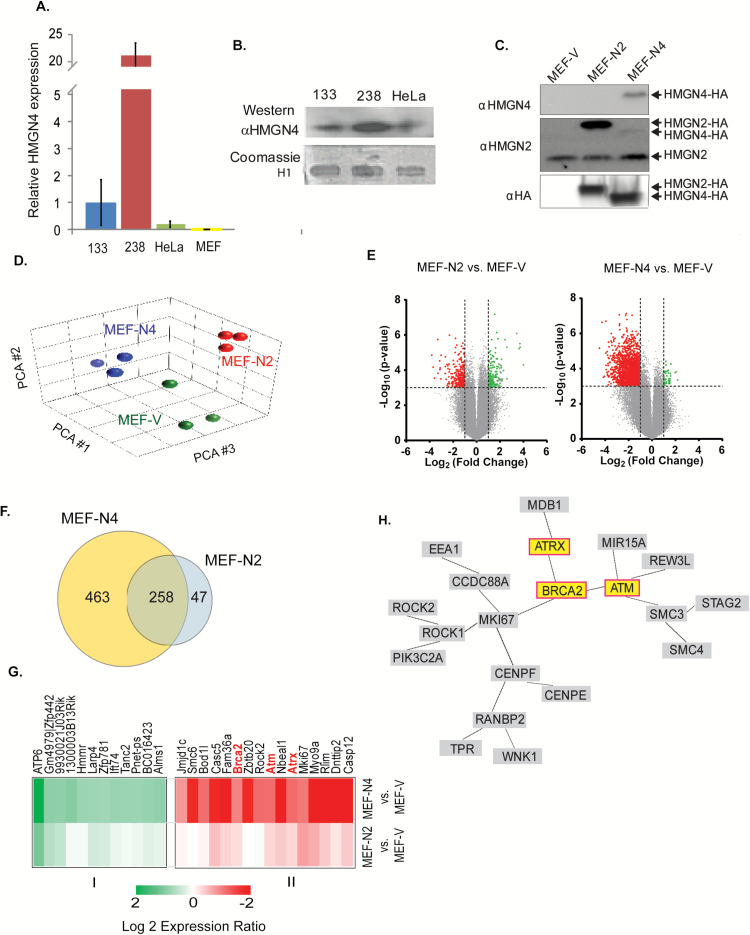
Since HMGN4 is highly expressed in thyroid tissues, we examined the expression of HMGN4 in the two follicular thyroid carcinoma cell lines FTC-133 and FTC-238 which differ in their malignant potential and in the levels of HMGN4 expression (Supplementary Figure 2C, available at Carcinogenesis Online). Quantitative RT-PCR and Western blots analyses of these thyroid carcinoma cell lines and of the non-thyroid human cancer cell line HeLa indicated that HMGN4 was significantly expressed in the thyroid cancer cell lines, with the highest level in the highly metastatic follicular thyroid carcinoma FTC-238 cell (Figure 1A and B), a finding that supports the link between HMGN4 expression and thyroid malignancy.
Figure 1.
Transcriptional Effects of HMGN4. (A) Increased HMGN4 expression in thyroid cancer cells. Shown is quantitative PCR data for HMGN4 expression in FTC-133, FTC-238, HeLa, and MEFs cells. (B) Western blot showing HMGN4 expression in FTC-133, FTC-238, and HeLa cells. Coomassie gel showing the histone H1 band in 5% perchloric acid extracts serves as a loading control. (C) Western blot of extracts from MEFs stably expressing FLAG-HA tagged HMGN4 (MEF-N4), HMGN2 (MEF-N2) or empty vector (MEF-V). (D) Principal component analysis of transcription profiles of the MEFs used in this study. Three biological replicates were used for each cell type. The data obtained from three technical replicates of each biological replicate were pooled and analyzed. (E) Volcano plots visualizing the transcription differences between MEF-V and either MEF-N2 or MEF-N4. (F) Venn diagram of the genes differentially expressed in MEF-N4 and MEF-N2 as compared to MEF-V cells. (G) Heat map of the most significantly differentially expressed genes. (H) Gene network generated from the “Genomic Instability” GO category using the Genomatix GeneRanker. Only interconnected genes are shown.
Interestingly, comparative genomics alignment by VISTA (13) revealed that the Hmgn4 locus is not present in mice and rat genomes (Supplementary Figure 3A, available at Carcinogenesis Online), an unexpected finding since the genes coding for the HMGN variants are present in all vertebrate genomes examined. Further analysis indicates that the gene coding for HMGN4 is present in the horse and dog genome, which split off the main mammalian lineage before the mouse and rat lineages. As visualized in the simplified version of the most recent mammalian cladogram (14), the data suggest that HMGN4 was lost from the rodent lineage (Supplementary Figure 3B, available at Carcinogenesis Online). The mouse cells lacking HMGN4 provide a test system where HMGN4 function can be examined without interference from endogenous protein.
HMGN4 downregulates tumor suppressor gene expression
To gain insight into the mechanisms that link HMGN4 expression to thyroid cancer, we generated MEFs stably expressing HMGN4-FLAG-HA (MEF-N4), HMGN2-FLAG-HA (MEF-N2) or empty FLAG-HA vector (MEF-V) and used microarray hybridization assays to examine the gene expression profiles of these cells. Western analysis with antibody to HA indicate that the cells expressed equal amounts of exogenous HMGN proteins, and westerns with an antibody to HMGN2 verify equal loadings of extracts (Figure 1C). The lack of HMGN4 in mice ensured that change in the properties of the cells can be attributed directly to the transgene rather than to interference with an endogenous protein, while the cells transfected with HMGN2-FLAG-HA serve as controls for HMGN4 variant-specific effects.
For transcription analyses, three biological replicates of each MEF cell were analyzed using three technical replicates for each biological replicate. Principal component analysis of the transcriptomes shows distinct clustering of same cell lines indicating HMGN variant specific effects on the cellular transcription profile (Figure 1D). Further analysis of the pairwise differentially expressed genes between MEF-V and either MEF-N2 or MEF-N4, with ≥2.0 fold-change in expression and P ≤ 0.001 with false discovery rate <0.034, indicated that upregulation of HMGN2 altered the expression of 305 genes, while upregulation of HMGN4 altered the expression of 721 genes, with more genes downregulated than upregulated (Figure 1E and F). Of the total 768 affected genes, 463 were specific for MEF-N4 cells, 47 were specific for MEF-N2 cells and 258 were common to both MEFs (Figure 1F).
Of the 463 genes, changed in MEF-N4 but not in MEF-N2, we selected the group of differentially expressed genes with highest probability change between MEF-N4 and MEF-V cells (P > 0.01), and used Pearson’s correlation hierarchical analysis to construct a heat map (Figure 1G). The group revealed the presence of several cancer-related genes. Similar analysis with the genes that changes in MEF-N2 provided further evidence for specific effects of HMGN4 on the expression of this gene set. Categorical gene ontology (GO) analysis of the gene significantly changed in the cells overexpressing HMGN4 revealed downregulation of several categories including ‘Genomic Instability’, containing 62 genes and an adjusted P-value of 1.5 × 10−9 and ‘Chromosomal Instability’ containing 30 genes and an adjusted P-value of 4.2 × 10−6 (Supplementary Data File 1, available at Carcinogenesis Online). A network generated from the 62 genes in the ‘Genomic Instability’ category using the Genomatix GeneRanker program, identified the tumor suppressor genes Ataxia Telangiectasia Mutated (ATM), Breast Cancer Related 2 (BRCA2), and Alpha Thalassemia/Mental Retardation Syndrome X-Linked (ATRX) as some of the putative node elements in the downregulated pathway (Figure 1H). Given the known relation between these tumor suppressors and tumorigenesis we tested further the effect of HMGN4 on the expression of these genes.
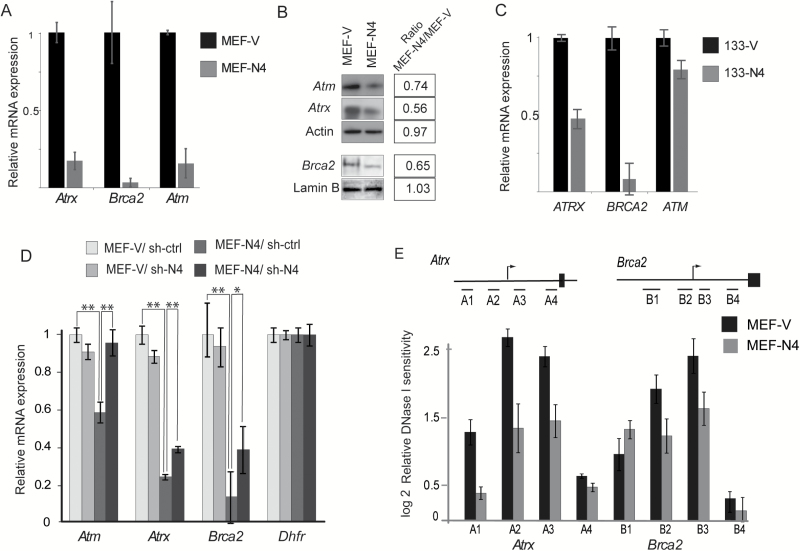
Quantitative RT-PCR of RNA extracted from MEF-V and MEF-N4 further verify that expression of HMGN4 downregulates the expression of these tumor suppressors. The qRT-PCR results indicate that the expression of Atm, Atrx and Brca2 is significantly more down regulated in cells expressing HMGN4 than in cells expressing control vector (Figure 2A). Western analyses of whole cell extracts verified that the protein levels of these tumor suppressors are indeed lower in MEF-N4 cells as compared to control MEF-V cells (Figure 2B). Furthermore, overexpression of HMGN4 downregulates ATM, ATRX and BRCA2 expression not only in MEFs but also in the human carcinoma cell line FTC133 (Figure 2C). In addition, rescue experiments in which the stably transformed cell lines MEF-V and MEF-N4 were treated with either control, or HMGN4 small hairpin RNA (sh-ctrl or sh-N4) verified that the downregulation of the three tumor suppressors are indeed due to HMGN4 overexpression, since the levels of Atm, Atrx and Brca2 transcripts in MEF-N4 treated with sh-N4 were higher than in MEF-N4 treated with sh-ctrl RNA (Figure 2D). Thus, downregulation of the exogenous HMGN4 expression in MEF-N4 cells upregulates Atm, Atrx and Brca2 expression. Neither the sh-ctrl nor the sh-N4 affected the levels of the tumor suppressors in the MEF-V cells which served as controls; further indication that indeed, HMGN4 expression affects the levels of the tumor suppressors (Figure 2D).
Figure 2.
HMGN4 expression downregulates ATRX, BRCA2 and ATM expression. (A) Quantitative PCR analyses of RNA extracted from MEF-V and MEF-N4 cells. Values are means from triplicates ± SD, normalized to the transcript levels in MEV-V. (B) Western blot analysis of ATRX, BRCA2 and ATM proteins in MEF-V and MEF-N4 MEFs. Actin and Lamin B serve as controls. Expression ratio of each protein, calculated from quantitative scans of the bands are shown on the right. (C) Quantitative PCR analyses of RNA extracted from human FTC-133-V and 133-N4 cells. Values are means from triplicates ± SD, normalized to the transcript levels in 133-V. (D) Rescue experiments. sh-4 mediated downregulation of Hmgn4 expression in MEF-N4 cells upregulates Atrx, Brca2 and Atm expression. MEFs were transfected with shRNA against HMGN4 (sh-N4) or scrambled control RNA (sh-ctrl), and 72h post-transfection the levels of the indicated genes were determined by qRT-PCR. Dhfr served as control gene. Values normalized to expression in MEF-V treated with sh-ctrl. *P > 0.01; **P > 0.001. (E) Decreased DNase I hypersensitivity in the promoter region of Atrx and Brca2 in MEF-N4, as compared to MEF-V. Schematic view of the promoter regions of mouse Atrx and Brca2 genes and the approximate position of the sites analyzed are illustrated by the lines on top of the bars (details in Supplementary Figure 4, available at Carcinogenesis Online). Data are means of three biological replicates ± SDs.
HMGN proteins have been shown to affect the sensitivity of chromatin regulatory sites to digestion by DNase I (8). A survey of the ENCODE DNase I sensitivity data for various mouse cells identified several DNase I hypersensitive sites at the promoter regions of Atrx and Brca2 genes that are conserved in several mouse cell types, including fibroblasts (Supplementary Figure 4, available at Carcinogenesis Online). Quantitative DNase I hypersensitivity assays revealed that these regulatory sites in the Atrx and Brca2 gene promoter regions were more resistant to digestion in MEF-N4 than in MEF-V cells, suggesting that HMGN4 downregulates the transcription of the tumor suppressors by altering the organization of regulatory sites in the promoter regions of these genes (Figure 2E).
Taken together, all the data are consistent with the notion that upregulation of HMGN4 down regulates the expression of the tumor suppressors Atm, Atrx and Brca2, thereby providing a possible clue to the mechanisms whereby upregulation of HMGN4 levels promotes thyroid tumorigenesis.
HMGN4 expression impairs DNA repair and perturbs the cell cycle
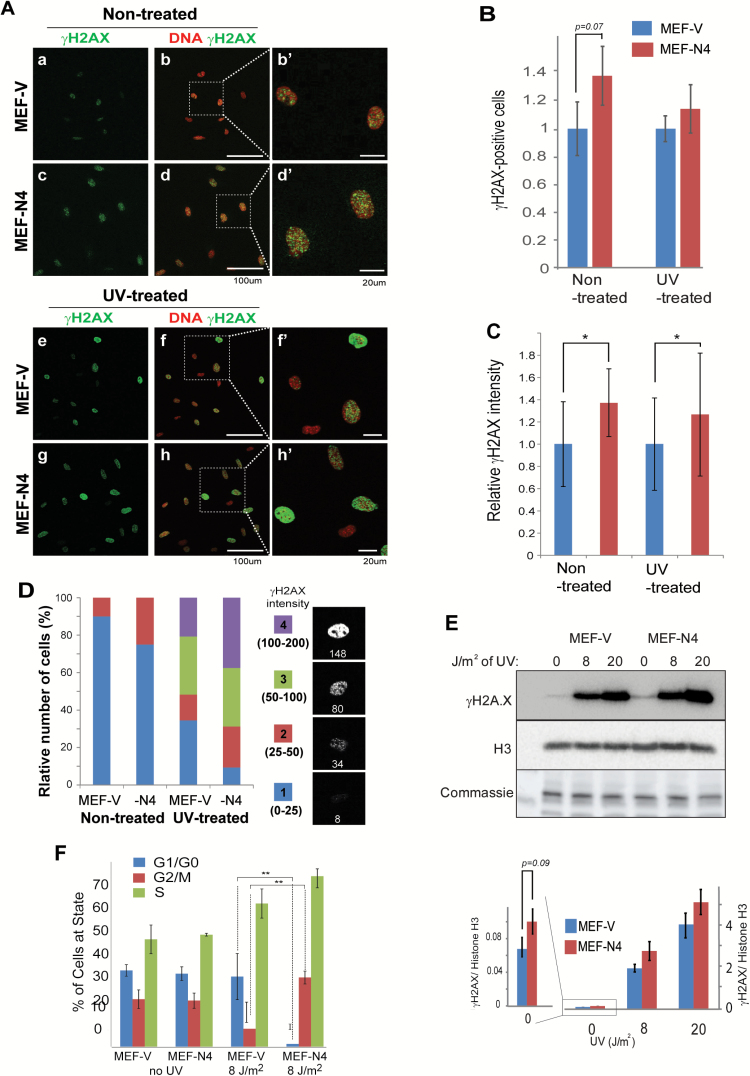
Decreased expression of Atm, Atrx and Brca2 is known to impair the repair of damaged DNA and to affect the progression of the cell cycle. To test whether ectopic HMGN4 expression downregulates these tumor suppressors to levels that are sufficient to affect DNA repair processes we used an antibody to gamma H2AX (γH2AX), a known marker of unrepaired DNA damage (22–25), to visualize the frequency and signal intensity of the γH2AX foci in MEF-V and MEF-N4, in both non-treated and in UV-treated cells. In these analyses, we quantified the number of γH2AX-positive cell and the relative γH2AX signal intensity in each cell, a measure of the level of this modification in a cell, using Nikon NIS-Elements program and classed the cells into four categories, according to their fluorescence intensity (Figure 3D). In non-treated growing cells, we find that the number of cells showing γH2AX is 30% higher in MEF-N4 than in MEF-V (Figure 3A and B). Likewise, the average γH2AX signal intensity in MEF-N4 is higher than in MEF-V cells (Figure 3A, C and D). Thus, the percentage of cells in category #2 signal intensity was two times higher in MEF-N4 (Figure 3D). Following exposure to UV, both the number and the signal intensity increased in both MEF-V and MEF-N4. MEF-N4 cells showed more foci with higher average signal intensity than MEF-V (Figure 3B–D) and 70% of the cells showed fluorescence intensity in categories 3 and 4 (Figure 3D). In addition, quantitative western analyses also indicated increased γH2AX levels in MEF-N4 both prior to, and following UV exposure (Figure 3E). Thus, all the tests consistently indicate increased γH2AX levels in cells expressing HMGN4.
Figure 3.
Increased DNA damage marker γH2AX in cells expressing HMGN4. (A) Fluorescence analysis of γH2AX in non-treated and UV-treated MEF cells. (B) Quantitation of γH2AX positive cells. (C) Quantitation of γH2AX signal intensity in cells. All quantifications were done with Nikon NIS-Elements. (D) Distribution of γH2AX cell intensity in non-treated and UV-treated MEF-V and MEF-N4 cells. The fluorescence intensities seen in the cells were subdivided into four categories as shown. Numbers below the cells show the fluorescence intensity of that cell. (E) Western blotting analysis for γH2AX foci in MEFs exposed to varying levels of UV radiation. Quantification of the western is shown below the gel images. Histone H3 and Coomassie stained bands are loading controls. (F) Perturbed cell cycle progression following UV irradiation. Shown are FACS analyses of cell cycle distributions of MEF-V and MEF-N4 that were either exposed or not exposed to 8 J/m2 UV irradiation. **P ≤ 0.05.
Faulty DNA repair due to altered levels of BRCA2 and ATM can lead to changes in cell cycle which can be more obvious in cells containing damaged DNA. To investigate possible effects of HMGN4 expression on the cell cycle, unsynchronized MEF-N4 and MEF-V cells that were either exposed or not exposed to UV irradiation, were subjected to flow-cytometry analysis. In untreated cells, no difference in cell cycle phase distribution between MEF-N4 cells and control MEF-V cells was observed. After exposure to UV radiation, the control MEF-v cells show the expected reduction of cells in G2M and increase of cells in S phase. Compared to these control cells under the same conditions, HMGN4-expressing cells displayed significant reduction of cells in G1 and a significant increase of cells in G2/M (Figure 3F). Taken together, our results indicate that ectopic expression of HMGN4 downregulates the expression of several tumor suppressors to levels that impairs the rate of repair of damaged DNA and cell cycle progression.
HMGN4 expression increases the tumorigenic properties of MEFs
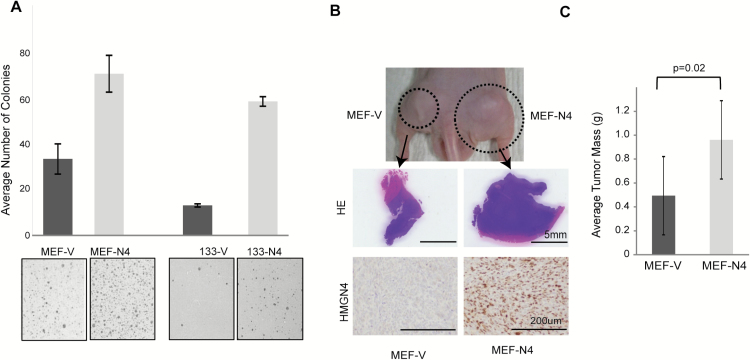
The elevated levels of HMGN4 in several tumors and the downregulated levels of tumor suppressors in MEF-N4 cells, raised the question of whether HMGN4 over-expression imparts tumorigenic properties to cell lines. As a first test, we used the soft agar colony formation assay to compare the tumorigenic potential of SV40 immortalized MEFs transfected with control vectors (MEF-V) to that of MEFs transfected with vectors expressing N4 (MEF-N4). We find that the MEF-N4 cells form two times more colonies than the control MEF-V cells (Figure 4A, left). Similarly, the human thyroid cancer cell line FTC-133, stably transfected with a plasmid expressing HMGN4-GFP, formed four times more colonies in soft agar than did the FTC-133 cells over-expressing only GFP (Figure 4A, right). Thus, by this assay, HMGN4 expression enhanced the tumorigenic potential of both mouse and human cells. As HMGN4 expression appeared to impart tumorigenic potential in vitro, we next studied its effect in vivo. A group of 30 nude mice were subcutaneously injected in opposite flanks with either MEF-N4 or with MEF-V cells. Three weeks after injection, the size and weight of the resulting tumors were measured. Tumors resulting from HMGN4-expressing MEFs weighed twice more and appeared larger by gross visual examination, than tumors initiated by the control MEF-V cells (Figure 4B and C). Immunostaining verified that the tumors generated from MEF-N4 cells, indeed express HMGN4 protein (Figure 4B bottom panels). Thus, both the agar colony formation assay and the nude mice tests indicate that HMGN4 expression increases the cellular tumorigenic potential.
Figure 4.
HMGN4 expression increases tumorigenicity. (A) Soft agar colony assay of MEFs and FTC-133 cells over-expressing either HMGN4 or empty vector. Three independent cell preparations were grown on agarose plates for 3 weeks, and colonies were stained with crystal violet, imaged and the numbers were counted using ImageJ. Significance was calculated using a one-sided t-test. (B) Nude mice assay. Nude mice were injected in opposite flanks with equal number of either MEF-N4 or MEF-V cells and tumors were excised 3 weeks after injection, weighed and analyzed for HMGN expression by immune-staining with anti HMGN4. (C) Quantification of tumor size. 60 tumors from 30 nude mice were weighed. Average mass per tumor for each group is shown.
HMGN4 expression in thyroid enhances focal hyperplasia
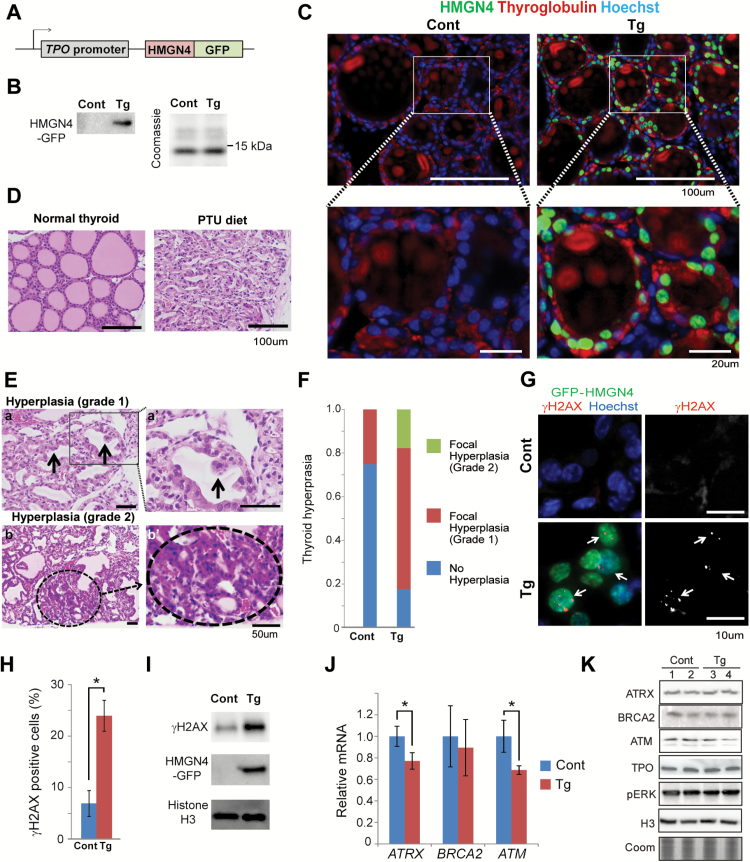
Previous studies reported that mice treated with DHPN, followed by administration of sulfadimethoxine (SDM) as a promoter in drinking water, developed goiter (92%), hyperplasia (25%), adenoma (8%) and carcinoma (8%) when examined 180 days after administration of DHPN (26). To test whether HMGN4 protein could play an oncogenic role in the pathogenesis of thyroid cancer in vivo, we generated transgenic mice expressing HMGN4-GFP protein under the control of the thyroid peroxidase (TPO) promoter (Figure 5A) which restricts exogenous protein expression to thyroid follicular cells (27), where the TPO promoter is specifically activated, mainly by the combined binding of the two transcription factors NKX2-1 and PAX8 (17). Western analysis with antibody to HMGN4 revealed prominent HMGN4 expression in the thyroid of these transgenic mice (Figure 5B) and fluorescence analysis of thyroids taken from the transgenic mice and their wild type littermates indicated that most of the thyroid follicular cells, but none of stromal cells express HMGN4, an indication of faithful HMGN4 expression under the follicular TPO gene promoter (Figure 5C).
Figure 5.
Increased DNA damage and hyperplasia in thyroid glands of HMGN4 over-expressing mice. (A) Outline of the vector used to generate transgenic mice ectopically expressing HMGN4 in thyroids. (B) Western blot showing HMGN4 expression in the thyroid of transgenic mice. (C) Fluorescence analysis showing HMGN4-GFP expression in the thyroid of transgenic mice. Note co-localization of GFP-HMGN4 (green) with thyroglobulin (red), a marker for thyroid follicular cells. (D) HE-stained cross-sections of thyroid glands of mice fed normally or with PTU diet. Note the shrunken follicles in mice fed with iodide-free PTU diet. (E) Evaluation of hyperplasia. Top, grade 1. Arrows point to grade 1 hyperplasia. Bottom, circle in bottom two image shows a grade 2 hyperplasia. (F) Comparison of severity of hyperplasia between control and HMGN4-Tg thyroids prepared from mice injected with DHPN and fed iodide-free PTU diet. Two microscopic fields per slide prepared from the thyroids of each of the 10 WT and 10 Transgenic mice were scored. (G) HMGN4 expression induces γH2AX. Immunofluorescence staining on thyroid section prepared from control (Cont) and hHMGN4-expressing mouse (Tg). Thyroid cells expressing HMGN4-GFP are detected by anti-GFP antibody (green) and nuclei are counterstained by Hoechst (blue). γH2AX is visualized with red (left panels) or white (right panels). (H) Relative amount of γH2AX positive cells in thyroid cross-sections from control (Cont) and HMGN4-expressing mice (Tg). Over 2000 cells from four slides were counted using Nikon Elements software. *P ≤ 0.01. (I) Western blot analysis of nuclear extracts of thyroid tissue from control (Cont) and HMGN4-expressing mouse (Tg). (J) Real-time qPCR analysis for Atrx, Brca2 and Atm mRNA in the thyroids prepared from control (Cont) and HMGN4 overexpressing (Tg) mice, injected with DHPN and fed iodide-free PTU diet. *P ≤ 0.05. (K) Western analysis of two biological replicates from control and HMG-4 expressing thyroids using the antibodies specific for the proteins listed on the left. Histone H3 and coomassie staining (Coom) are loading controls.
Thyroid cancer can be induced in mice by a single injection of the carcinogen, N-bis (2-hydroxypropyl) nitrosamine (DHPN) at the age of 4 weeks, followed by feeding with the iodide-free diet PTU (28). We exposed 10 HMGN4 transgenic mice and ten wild type littermates to this treatment, and after 6 months of feeding with the PTU diet we assessed their histological appearance and compared it to the thyroid of normal, non-treated mice of equal age.
Normal thyroid tissue is composed of round-shapes follicles (Figure 5C and D) which are filled with colloid containing iodinated thyroglobulin, the thyroid hormone precursor. In mice, iodide-free PTU diet administration leads to shrinkage of the thyroid follicles (Figure 5D). Combined DHPN injection and prolonged PTU diet administration can lead to focal hyperplasia, graded as 1 and 2 according to severity (Figure 5E), which eventually progress to form cancers (26,29).
Following the treatment regimen described, we detected focal hyperplasia grade 1 in 5 out of the 20 (25%) evaluations of the slides taken from the thyroids of 10 wild type mice, while the remaining 75% of the thyroid slides had no hyperplasias. In contrast, of the 21 evaluations of slides taken from the 10 HMGN4 transgenic mice, only 5 (24%) did not show hyperplasia, while 13 (62%) were scored as showing grade 1 hyperplasia and 3 (14%) were scored as hyperplasia grade 2. Thus, the incidence of pre-neoplastic lesions in the thyroid of HMGN4 expressing mice was significantly higher than in the thyroid of control mice that did not express HMGN4 (Figure 5F).
Immunofluorescence analyses of the thyroids of the mice showed significantly more γH2AX foci in the HMGN4 expressing cells, which could be identified by their GFP green fluorescence, than in cells that did not express HMGN4 (Figure 5G). For quantitative analysis we used the Nikon Elements program to count the number of positive cells in 4 slides prepared from the thyroid of either wild type or HMGN4 overexpressing mice fed 6 months with the PTU diet. Analysis of over 2000 cells from each genotype indicates that the overall level of γH2AX in HMGN4 expressing mice was 3 fold higher than in their control littermates (Figure 5H). Likewise, western analysis indicated a 2.5-fold higher content of γH2AX in the thyroid of HMGN4 transgenic mice (Figure 5I). Since in these mice only the follicular cells of the thyroid express HMGN4, we estimate that the occurrence of γH2AX in the HMGN4 expressing cells is approximately 5-fold higher than seen in cells that do not express HMGN4.
Given the link between HMGN4 overexpression and Atm, Atrx and Brca2 downregulation, we used qRT-PCR to examine the relative expression of these genes in the thyroid of the transgenic mice. We find decreased expression of these three genes in RNA derived from the HMGN4-Tg mice as compared to their non-transgenic littermates (Figure 5J). As expected, the small differences in transcription did not lead to measurable differences in protein expression levels, except with ATM which showed a 15% reduction, as measured by Western analyses (Figure 5K). Nevertheless, collectively, these results suggest that cell-specific HMGN4 overexpression leads to a decrease in the expression of several key tumor suppressor genes, thereby decreasing the rate of repair of damaged DNA, ultimately leading to increased incidence of thyroid tumor.
Discussion
Thyroid cancer variability and etiology is known to involve genetic and epigenetic changes and alteration in cellular signaling pathways (1,2). Our study identifies the nucleosome-binding protein HMGN4, encoded by an intronless retrogene, as an additional epigenetic factor that can promote thyroid tumorigenesis. Several types of observations support the link between HMGN4 and thyroid tumors. First, bioinformatics analyses indicate a relatively high expression of HMGN4 in thyroid tissues, and that the expression of this protein is further elevated in cells and tissues derived from thyroid tumors. Second, upregulation of HMGN4 expression in immortalized mouse fibroblasts and in human thyroid tumor cells increases the tumorigenic properties of the cells as measured by soft agar colony formation and tumor generation in nude mice. Third, in carcinogen treated transgenic mice, expression of HMGN4 in the thyroid increases the incidence of thyroid hyperplasia, an initial stage of tumorigenesis in rodents (26,29).
The upregulation of HMGN4 did not alter the expression of thyroid specific genes Nkx2-1 (Ttf1), Foxe1 (Ttf2), Pax8, Tpo and Slc5a5 (Nis) although there is a tendency for the reduced expression of all three transcription factors critical for the expression of thyroid specific genes, namely Nkx2-1 (Ttf1), Foex1 (Ttf2) and Pax8) (Supplementary Figure 5, available at Carcinogenesis Online, Figure 5K). Likely the lack of significant changes in the expression levels of these thyroid differentiation marker genes reflects the fact that the cells in hyperplasia are still in differentiated state, thus reduced expression of thyroid specific differentiation marker genes may not be expected. The results suggest that the HMGN4-dependent increase in tumorigenic potential is not directly due to altered expression of thyroid-specific genes. Likewise, examination of recent whole genome transcription data (30–32) did not suggest HMGN4 as a specific prognostic factor for thyroid tumorigenesis. We suggest that HMGN4 promotes thyroid tumorigenesis by altering the cellular transcription profile and down regulating the expression of Atm, Atrx and Brca2, three general tumor suppressors known to affect the cellular tumorigenic potential. The link between upregulation of HMGN4 and down regulation of these tumor suppressors is seen in both mouse and human thyroid cells expressing exogenous HMGN4, in rescue experiments showing that downregulation of HMGN4 elevated the level of these tumor suppressors, and in the thyroid of transgenic mice expressing HMGN4. Tissue cultured cells and thyroids of transgenic mice overexpressing HMGN4 show elevated levels of γH2AX, a hallmark of damaged, unrepaired DNA, which has been linked to mis-expression of tumor suppressors involved in DNA damage repair. These changes and thyroid hyperplasia, a hallmark of preneoplastic lesions (29), were seen already 6 months after administration of DPNH and continuous the PTU diet. Taken together, our studies indicate that HMGN4 enhances thyroid tumorigenesis by downregulating the expression of tumor suppressors and impairing the repair of damaged DNA. However, our studies do not fully exclude the possibility that changes in the expression of additional genes also contributes to the HMGN4-mediated enhancement of thyroid tumorigenesis. The widespread expression of HMGN4 in numerous tissues taken together with our finding that HMGN4 upregulation leads to downregulation of universal tumor suppressors, provides a partial explanation for the link between elevated HMGN4 levels and tumor occurrence which we noted by bioinformatics analysis (Supplementary Figure 2D, available at Carcinogenesis Online).
HMGN proteins have been shown to affect the fidelity of the cellular transcription profile and impair DNA repair processes. Thus, loss of HMGN1 impairs the rate of repair of DNA damaged by either UV or gamma irradiation, and increases the incidence of N-nitrosodiethylamine (DEN)-induced liver tumors (33–35), while upregulation of HMGN1 in trisomy 21 syndrome has been directly liked to increased risk of B cell acute lymphoblastic leukemia seen in Down syndrome (36). Since HMGN variants are not bona fide DNA repair factors it is likely that their effects are linked to their epigenetic functions; however, the precise molecular mechanisms whereby these abundant chromatin-binding proteins affect gene expression and impact the cellular phenotype are not fully understood. HMGNs affect the binding of histone H1 to chromatin (5), the level of histone modifications (37,38), and the landscape of regulatory sites in chromatin, in particular the maintenance and organization of enhancer sites (8). Chromatin regulatory sites are established by the dynamic binding of tissue specific transcription factors to chromatin (39). Changes in HMGN levels lead to both upregulation and downregulation of gene expression in a tissue and variant-specific manner (9). Thus, HMGNs may affect the cellular transcription profile by modulating and fine tuning the interaction of transcription factors with chromatin. We suggest that HMGN4 affects the expression of Atm, Atrx and Brca2 in thyroid cells, and perhaps in additional tissues, by altering the interaction of transcription factors with chromatin regions that regulate the expression of these genes.
Increasing evidence indicates that epigenetic mechanisms regulate the expression of cancer driver gene and play an important role in promoting oncogenesis (40,41). Our study identifies HMGN4 as an additional epigenetic regulator that modulates the expression of a subset of tumor suppressor genes in the thyroid. HMGN4 is encoded by an intronless retrogene; however, it remains to be seen if this unusual gene structure is related to its overexpression in thyroid cancer. Since both the human and mouse genome contain numerous HMGN-related retropseudogenes (42,43) it is conceivable that additional HMGN-derived retrogenes might be expressed in certain cells. Accumulating evidence suggest that HMGN variants affect gene expression by regulating the function of tissue specific transcription factors. Identification of regulatory factors affected by specific HMGN variants, including HMGN4 in the thyroid, remains a major experimental challenge. Nevertheless, our studies raise the possibility that HMGN4 could serve as an additional diagnostic marker or even therapeutic target in certain thyroid cancers.
Supplementary material
Supplementary data are available at Carcinogenesis online.
Funding
Research supported by the NIH Intramural Research Program, National Cancer Institute, Center for Cancer Research.
Supplementary Material
Acknowledgements
We thank Dr. Jerry Ward, Global VetPathology, Montgomery Village, MA 20886, USA, for pathological analysis of mice thyroids.
Conflict of Interest Statement: None declared.
Abbreviations
- DMEM
Dulbecco’s modified Eagle’s medium
- HMGN
high mobility group N
- MEF
mouse embryonic fibroblast
- PTU
propylthiouracil
References
- 1. Xing M. (2013) Molecular pathogenesis and mechanisms of thyroid cancer. Nat. Rev. Cancer, 13, 184–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kirschner L.S., et al. (2016) Mouse models of thyroid cancer: A 2015 update. Mol. Cell. Endocrinol., 421, 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Postnikov Y., et al. (2010) Regulation of chromatin structure and function by HMGN proteins. Biochim. Biophys. Acta, 1799, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ueda T., et al. (2008) Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol. Cell. Biol., 28, 2872–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Catez F., et al. (2002) Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep., 3, 760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Catez F., et al. (2010) Binding and interplay of HMG proteins on chromatin: lessons from live cell imaging. Biochim. Biophys. Acta, 1799, 15–27. [DOI] [PubMed] [Google Scholar]
- 7. Kugler J.E., et al. (2012) The HMGN family of chromatin-binding proteins: dynamic modulators of epigenetic processes. Biochim. Biophys. Acta, 1819, 652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deng T., et al. (2015) Functional compensation among HMGN variants modulates the DNase I hypersensitive sites at enhancers. Genome Res., 25, 1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kugler J.E., et al. (2013) HMGN proteins modulate the fidelity of the cellular transcriptional profile in a tissue and variant specific manner. J. Biol. Chem., 288, 16690–16703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fedele M., et al. (2010) HMGA and cancer. Biochim. Biophys. Acta, 1799, 48–54. [DOI] [PubMed] [Google Scholar]
- 11. Gerlitz G. (2010) HMGNs, DNA repair and cancer. Biochim. Biophys. Acta, 1799, 80–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Birger Y., et al. (2001) HMGN4, a newly discovered nucleosome-binding protein encoded by an intronless gene. DNA Cell Biol., 20, 257–264. [DOI] [PubMed] [Google Scholar]
- 13. Frazer K.A., et al. (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res., 32, W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lindblad-Toh K., et al. (2011) A high-resolution map of human evolutionary constraint using 29 mammals. Nature, 478, 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lim J.H., et al. (2004) Preparation and functional analysis of HMGN proteins. Methods Enzymol., 375, 323–342. [DOI] [PubMed] [Google Scholar]
- 16. McArthur M., et al. (2001) Quantification of DNaseI-sensitivity by real-time PCR: quantitative analysis of DNaseI-hypersensitivity of the mouse beta-globin LCR. J. Mol. Biol., 313, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Damante G., et al. (2001) A unique combination of transcription factors controls differentiation of thyroid cells. Prog. Nucleic Acid Res. Mol. Biol., 66, 307–356. [DOI] [PubMed] [Google Scholar]
- 18. Basso K., et al. (2005) Reverse engineering of regulatory networks in human B cells. Nat. Genet., 37, 382–390. [DOI] [PubMed] [Google Scholar]
- 19. D’Errico M., et al. (2009) Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur. J. Cancer, 45, 461–469. [DOI] [PubMed] [Google Scholar]
- 20. Roessler S., et al. (2010) A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res., 70, 10202–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. French P.J., et al. (2005) Gene expression profiles associated with treatment response in oligodendrogliomas. Cancer Res., 65, 11335–11344. [DOI] [PubMed] [Google Scholar]
- 22. Rogakou E.P., et al. (1999) Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol., 146, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Solier S., et al. (2014) The nuclear γ-H2AX apoptotic ring: implications for cancers and autoimmune diseases. Cell. Mol. Life Sci., 71, 2289–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ivashkevich A., et al. (2012) Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett., 327, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Godon C., et al. (2012) Generation of DNA single-strand displacement by compromised nucleotide excision repair. EMBO J., 31, 3550–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoshi S., et al. (2009) Role of NKX2-1 in N-bis(2-hydroxypropyl)-nitrosamine-induced thyroid adenoma in mice. Carcinogenesis, 30, 1614–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kusakabe T., et al. (2004) Thyrocyte-specific expression of Cre recombinase in transgenic mice. Genesis, 39, 212–216. [DOI] [PubMed] [Google Scholar]
- 28. Kitahori Y., et al. (1984) Effect of propylthiouracil on the thyroid tumorigenesis induced by N-bis(2-hydroxypropyl)nitrosamine in rats. Carcinogenesis, 5, 657–660. [DOI] [PubMed] [Google Scholar]
- 29. Lu C., et al. (2010) Growth activation alone is not sufficient to cause metastatic thyroid cancer in a mouse model of follicular thyroid carcinoma. Endocrinology, 151, 1929–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159: 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoo S.K., et al. (2016) Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. PLoS Genet., 12, e1006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Landa I., et al. (2016) Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J. Clin. Invest., 126, 1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Birger Y., et al. (2005) Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res., 65, 6711–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Birger Y., et al. (2003) Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J., 22, 1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Postnikov Y.V., et al. (2014) Loss of the nucleosome-binding protein HMGN1 affects the rate of N-nitrosodiethylamine-induced hepatocarcinogenesis in mice. Mol. Cancer Res., 12, 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lane A.A., et al. (2014) Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat. Genet., 46, 618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lim J.H., et al. (2004) Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol. Cell, 15, 573–584. [DOI] [PubMed] [Google Scholar]
- 38. Lim J.H., et al. (2005) Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J., 24, 3038–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Voss T.C., et al. (2014) Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet., 15, 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dawson M.A., et al. (2012) Cancer epigenetics: from mechanism to therapy. Cell, 150, 12–27. [DOI] [PubMed] [Google Scholar]
- 41. Garraway L.A., et al. (2013) Lessons from the cancer genome. Cell, 153, 17–37. [DOI] [PubMed] [Google Scholar]
- 42. Srikantha T., et al. (1987) Retropseudogenes for human chromosomal protein HMG-17. J. Mol. Biol., 197, 405–413. [DOI] [PubMed] [Google Scholar]
- 43. Strichman-Almashanu L.Z., et al. (2003) Retroposed copies of the HMG genes: a window to genome dynamics. Genome Res., 13, 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.