Summary
By using meta-analysis and in silico functional analysis, we identified one functional SNP rs12587742 within the CpG island of DCAF4, associated with lung cancer risk, possibly by decreasing methylation status and up-regulating mRNA expression of DCAF4.
Abstract
Cullin-RING ubiquitin ligases (CRLs) responsible for substrate specificity of ubiquitination play a key role in cell-cycle control and DNA damage response. In this study, we assessed associations between 16 599 SNPs in 115 CRL genes and lung cancer risk by using summary data of six published genome-wide association studies (GWASs) of 12 160 cases and 16 838 cases of European ancestry. As a result, we identified three independent SNPs in DCAF4 (rs117781739, rs12587742 and rs2240980) associated with lung cancer risk (odds ratio = 0.91, 1.09 and 1.09, respectively; 95% confidence interval = 0.88–0.95, 1.05–1.14 and 1.05–1.13, respectively; and P = 3.99 × 10–6, 4.97 × 10–5 and 1.44 × 10–5, respectively) after multiple comparison correction by a false discovery rate <0.05. Since SNP rs12587742 is located within the promoter region and one CpG island of DCAF4, we further performed in silico functional analyses and found that the rs12587742 variant A allele was associated with an increased mRNA expression (P = 2.20 × 10−16, 1.79 × 10−13 and 0.001 in blood cells, normal lung tissues and tumor tissues of lung squamous carcinoma, respectively) and a decreased methylation status (P = 2.48 × 10−9 and 0.032 in adipose and lung tumor tissues, respectively). Moreover, evidence from differential expression analyses further supported an oncogenic effect of DCAF4 on lung cancer, with higher mRNA levels in both lung squamous carcinoma and adenocarcinoma (P = 4.48 × 10−11 and 1.22 × 10−9, respectively) than in adjacent normal tissues. Taken together, our results suggest that rs12587742 is associated with an increased lung cancer risk, possibly by up-regulating mRNA expression and decreasing methylation status of DCAF4.
Introduction
Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer deaths in the world. In USA, the estimated incidence of lung cancer in 2016 is 57.3 per 100 000 with an estimated mortality of 46 per 100 000 (1). Etiology studies have revealed several environmental risk factors for lung cancer, such as exposures to cigarette smoke, radon, asbestos and arsenic (2). Genetic factors such as heritable and somatic mutations are also involved in the etiology of lung cancer. Multiple genetic loci with moderate effects have also been reported by genome-wide association studies (GWASs) of lung cancer at chromosome regions of 3q28, 5p15.33, 6p21.33, 6p22.1, 13q13.1, 15q25.1 and 22q12.1 in European populations (3–8). However, most of the published GWASs had mainly focused on single nucleotide polymorphism (SNPs) that reached genome-wide significance, most of which did not have clear biological functions (9). In the post-GWAS era, identification of genetic variants with moderate but detectable effects and potential biological functions might provide additional insight about the complex mechanisms of cancer development. Currently, the availability of enormous genetic data made such studies feasible (8).
Carcinogenesis is a multiple-step process that often involves loss control of cell proliferation. The ubiquitin-proteasome system is a major player in the regulation of critical cellular processes, including cell proliferation, differentiation and apoptosis. Dysfunction of the system has been implicated in several clinical disorders including inflammation and cancer (10,11). There are three types of enzymes that specifically mediate ubiquitin attachment to the target proteins: ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s). In humans, there are only 2 E1s, at least 38 E2s and over 600 kinds of E3. Cullin-RING ubiquitin ligases (CRLs) represent one of the largest classes of E3 ubiquitin ligases mainly responsible for the substrate-specific ubiquitination. In addition, CRLs play a key role in cell-cycle control and DNA damage response (12), and deregulation of CRLs may lead to abnormal cell proliferation and genomic instability, which in turn could result in malignance transformation. Currently, several components of CRLs (e.g. SKP2, CUL4A, CUL1 and RBX1/2) have been found to behave as oncogenes and are frequently amplified or overexpressed in human cancers, while several others (e.g. FBXW7 and VHL) act as suppressor genes for they were often mutated or inactivated in cancers (13–15). Notably, as one of the most studied CRLs, SKP2 is found to be overexpressed and associated with aggressiveness and metastasis of non-small cell lung cancer, as a result of accelerated degradation of a cell-cycle inhibitor p27 (16,17). Moreover, large-scale somatic mutations of KEAP1, another well-studied CRL, occurred in multiple human cancers, including non-small cell lung cancer (18). According to the findings of these previous studies, we hypothesize that genetic variants with potential functions in genes encoding CRLs are associated with risk of lung cancer.
To test our hypothesis, we first performed a meta-analysis for SNPs in CRL-related genes by using summary statistics from six published lung cancer GWASs, including 12 160 cases and 16 838 controls from the TRICL–ILCCO Consortium (Transdisciplinary Research in Cancer of the Lung / the International Lung Cancer Consortium) (19). For those identified SNPs as significant, we further performed stratified analysis by smoking status and histological types and investigated their effects on gene expression and methylation in cell lines and tissues by using the available genomic and genetic data from multiple public databases (e.g. TCGA and GTEx).
Materials and methods
Study populations
The study populations included in this study have been detailed in previous publications from TRICL and ILCCO (8,19). Briefly, six published lung cancer GWASs were obtained from the TRICL–ILCCO consortium, which consists of 12 160 lung cancer cases and 16 838 controls of European descent. The GWAS participants included Institute of Cancer Research (ICR), The University of Texas MD Anderson Cancer Center (MDACC), International Agency for Research on Cancer (IARC), National Cancer Institute (NCI), Lunenfeld-Tanenbaum Research Institute study (Toronto) and German Lung Cancer Study (GLC). Two additional GWAS data sets were also requested from other independent GWASs of Caucasian populations: the Harvard Lung Cancer Study (984 cases and 970 controls) and Icelandic Lung Cancer Study (deCODE) (1319 cases and 26 380 controls) from the ILCCO (20,21). A written informed consent was obtained from each participant of each GWAS. The present study was approved by Duke University Health System Institutional Review Board and all methods performed in this study were in accordance with the relevant guidelines and regulations.
Genotyping platforms and quality controls
For all the GWAS datasets, multiple genotyping platforms were applied, including Illumina HumanHap 317, 317 + 240S, 370Duo, 550, 610 or 1M arrays (22). For the meta-analyses, imputation was performed based on the reference data from the 1000 Genomes Project (phase I integrated release 3, March 2012) by using IMPUTE2 v2.1.1 (23), MaCH v1.0 (24) or minimac (version 2012.10.3) software. Only SNPs with an information score ≥ 0.40 in IMPUTE2 or an r2 ≥ 0.30 in MaCH were included in the final analyses. Standard quality control on samples was performed on all scans, excluding individuals with a low call rate (<90%), extremely high or low heterozygosity (P < 1.0 × 10−4) and non-European ancestry (using the HapMap phase II CEU, JPT/CHB and YRI populations as a reference).
Gene and SNP selection
The CRL-related genes were collected from the category of ‘Cullin-RING ubiquitin ligase complex’ in the Gene Ontology database (http://amigo.geneontology.org/amigo/term/GO:0031461). In total, we retrieved 118 genes from the database, 115 of which were located in autosomes (listed in Supplementary Table 1, available at Carcinogenesis Online). We then mapped all the SNPs located within 2 KB up- and down-stream of the NCBI Reference sequence of those selected genes and extracted their summary data from the GWAS datasets. SNPs included in the final meta-analysis were those with call rate ≥90%, minor allele frequency ≥1%, and P value for the Hardy–Weinberg Equilibrium test ≥10–5. All remained SNPs also passed the quality control of imputation with info ≥0.40 in IMPUTE2 or an r2 ≥ 0.30 in MaCH.
In silico functional analysis as a biological validation
For those identified SNPs as significant, we first performed bioinformatic functional prediction by using three online tools: SNPinfo (http://snpinfo.niehs.nih.gov), RegulomeDB (http://www.regulomedb.org) and HaploReg (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php). We then performed expression quantitative trait loci analysis by using data from multiple sources: lymphoblastoid cell data of 373 European individuals from Genetic European Variation in Health and Disease Consortium (GEUVADIS) and the 1000 Genomes Project (phase I integrated release 3, March 2012) (25); lung tissues data from the genotype-tissue expression (GTEx) project (26); tumor tissues and adjacent normal tissue data from the Cancer Genome Atlas (TCGA) database (27,28). SNP-methylation correlation analysis was further performed by using the data from TCGA and the Multiple Tissue Human Expression Resource (MuTHER) project implemented in the Genevar software (29). Different expression analyzes between tumor and normal tissues were also performed for those identified genes using the data from TCGA and Oncomine (https://www.oncomine.org). The TCGA level 3 RNAseq data (LUSC_rnaseqv2_Level_3_RSEM_genes_normalized_data.2016012800.0.0.tar.gz and LUAD_Level_3_RSEM_genes_normalized_data_2016012800.0.0.tar.gz) and methylation data (gdac.broadinstitute.org_LUSC.Methylation_Preprocess.Level_3.2016012800.0.0.tar.gz and gdac.broadinstitute.org_LUAD.Methylation_Preprocess.Level_3.2016012800.0.0) were obtained from the Broad TCGA GDAC site (http://gdac.broadinstitute.org).
Statistical methods
For each GWAS data set, we performed an unconditional logistic regression to estimate odds ratios (ORs) and 95% confidence intervals (CIs) per effect allele by using R (v2.6), Stata (v10, State College, TX, USA) and PLINK (v1.06) software with adjustment for the top significant principle components (8). We performed meta-analysis by the inverse variance method using a fixed effects model (30). If the Cochran’s Q test P-value ≤ 0.100 or the heterogeneity statistic (I2) ≥ 25%, a random-effects model was employed. We used the linear step-up method of Benjamini and Hochberg to calculate false discovery rate (FDR) with a cut-off value of 0.05 to correct for multiple comparisons (31) and used linear regression for the expression quantitative trait loci analysis and paired t-test for the gene differential expression analysis between tumor and adjacent normal tissues. For the differential expression and mRNA-methylation correlation analyses, outliers were defined as those outside the interval (Q1 −3×IQR, Q3 + 3×IQR) and were removed in the final analysis. Q1 and Q3 denote the first and third quartiles, respectively, and IQR denotes the interquartile range. Based on the 1000 Genomes European (EUR) reference data (phase I integrated release 3, March 2012), we used LocusZoom (32) and Haploview v4.2 (33) to construct the regional association plots and linkage disequilibrium (LD) plots, respectively. SNP pruning was applied, and SNPs with paired-wise r2 < 0.30 were considered as independent. All other analyses were conducted with SAS (version 9.4; SAS Institute, Cary, NC, USA), if not mentioned specifically.
Results
Meta-analysis of the main effects
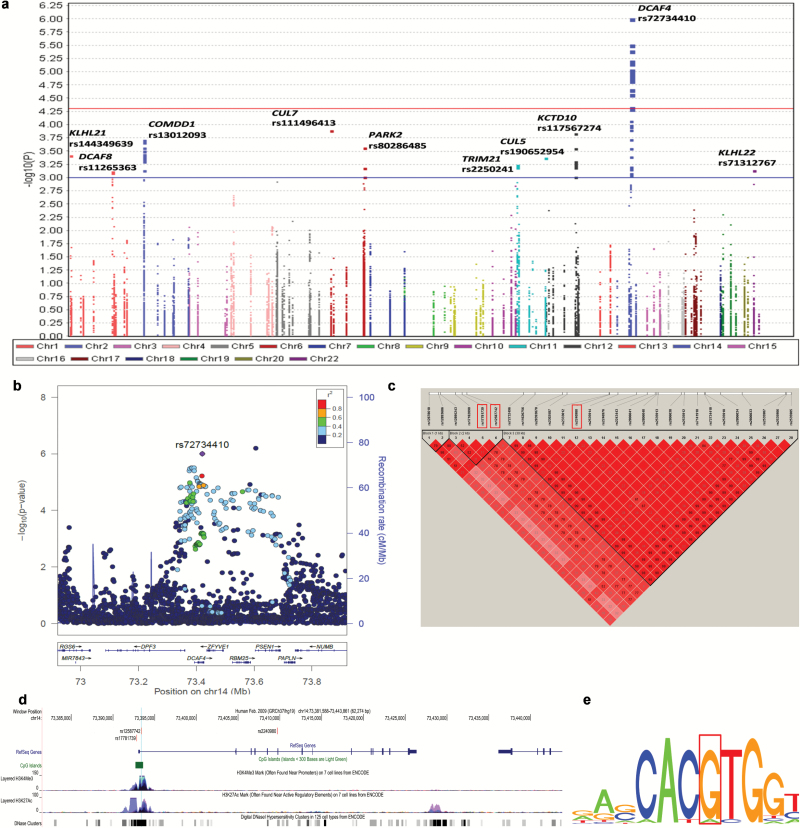
The sample sizes for the eight GWASs included in the present study are summarized in Supplementary Table 2, available at Carcinogenesis Online, and the workflow of this study is depicted in Figure 1. We first performed meta-analysis using summary statistics from six GWASs (i.e. ICR, MDACC, IARC, NCI, Toronto and GLC) including 12 160 lung cancer cases and 16 838 non-cancer controls. The overview of the overall association results is shown in the Manhattan plot (Figure 2a). We found there were 84 SNPs of 10 CRL-encoding genes with a nominal P < 0.001, 28 of which are in the DCAF4 gene with FDR < 0.05. More detailed information for each of the 84 SNPs (including position, effect allele, relative minor allelic frequency, effect sizes, unadjusted and FDR adjusted P-values, and heterogeneity test results) is summarized in Supplementary Table 3, available at Carcinogenesis Online. The regional association plots (Figure 2b) demonstrated that the top SNP rs72734410 of DCAF4 was in moderate to high LD with other SNPs of the same gene but in very low LD (r2 < 0.2) with the top SNP rs214278 in the neighboring gene PSEN1.
Figure 1.
Workflow of the study.
Figure 2.
Association results and functional prediction of SNPs in 115 Cullin-ring ligase encoding genes. (a) Manhattan plot of the overall results. There were 84 SNPs on 10 CRLs genes with nominal P < 0.001 and 28 of them were on the DCAF4 gene with false discovery rate (FDR) < 0.05. The x-axis indicated the chromosome number and the y-axis showed the association P values with lung cancer risk (as –log10 P values). The horizontal blue line represents P values of 0.001 while the red line indicated the FDR threshold 0.05. (b) Regional association plot, which demonstrated that the linkage disequilibrium (LD) between the top SNP rs72734410 on DCAF4 and other SNPs in the region of 500kb up- or downstream of the top SNP. (c) Pair-wise LD plot between the 28 SNPs in DCAF4 with FDR < 0.05. Based on it, two tag SNPs (rs17781739 and rs2240980) together with one functional SNP rs12587742 were chosen for further analysis. (d) Locations and functional prediction of the three selected SNPs. Two SNPs (rs17781739 and rs12587742) are located within one CpG island and presented strong signals of active enhancer and promoter functions (indicated by DNase hypersensitivity, histone modification H3K27 acetylation and H3K4 methylation, respectively). (e) Position weight matrix (PWM) based Sequence Logo, which showed rs12587742 is located on the c-MYC motif.
We then performed functional prediction for these 28 significant SNPs by using three bioinformatics tools (SNPinfo, regulomDB and HaploReg) and selected those apparently independent SNPs (paired-wise r2 < 0.3) with potential effects on gene expression or functions for further analysis. As a result, two SNPs (rs17781739 and rs2240980) together with another functional SNP rs12587742 were chosen in further analysis (Figure 2c–e). As shown in Table 1, SNP rs17781739 G>T was associated with a significantly decreased risk of lung cancer [odds ratio (OR) = 0.91, 95% confidence interval (CI) = 0.88–0.95, P = 3.99 × 10–6], while two other SNPs in moderate LD (pair-wise r2 = 0.38) were associated with a significantly increased lung cancer risk (rs12587742 G>A: OR = 1.09, 95% CI = 1.05–1.14, P = 4.97 × 10–5; and rs2240980 C>G: OR = 1.09, 95% CI = 1.05–1.13, P = 1.44 × 10–5). There was no heterogeneity observed for the effect estimates of these three SNPs from the six GWASs (Table 1).
Table 1.
Results of the three tagSNPs in the DCAF4 gene with FDR < 0.05
| SNP | Position | Allelesa | Minor allele frequency | # Study | # Cases | # Controls | Effectsb | OR (95% CI) | P meta c | FDR | P Q-test | I 2 | Functional prediction | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPinfod | RegulomDB scoree | |||||||||||||
| rs17781739 | 14:73392839 | T/G | 0.30 | 6 | 12 160 | 16838 | ------ | 0.91 (0.88–0.95) | 3.99E−06 | 0.015 | 0.665 | 0 | SREBP | 4 |
| rs12587742 | 14:73393391 | A/G | 0.21 | 6 | 12 160 | 16838 | ++++-+ | 1.09 (1.05–1.14) | 4.97E−05 | 0.042 | 0.446 | 0 | MYC/MAX | 1a |
| rs2240980 | 14:73409683 | G/C | 0.32 | 6 | 12 160 | 16838 | ++++-+ | 1.09 (1.05–1.13) | 1.44E−05 | 0.015 | 0.715 | 0 | NA | 3a |
aEffect allele/reference allele.
b‘-’ indicated protective effect and ‘+’indicated risk effect of effect alleles in one of the six GWAS studies in the order of: ICR, MDACC, IARC, NCI, Toronto and GLC.
c P value from meta-analysis with fixed effects model.
dTranscription factors with the highest core match score and matrix match score in SNPinfo.
eThe scoring scheme refers to the available data type for the SNP position: ‘1a’ represents ‘eQTL + TF binding + matched TF motif + matched DNase Footprint + DNase peak’; ‘3a’ represents ‘TF binding + any motif + DNase peak’; ‘4’ represents ‘TF binding + DNase peak’.
We then expanded the meta-analysis for these three identified SNPs by including two additional GWASs with European descents from Harvard University (984 cases and 970 controls) and deCODE (1319 cases and 26 380 controls) as a population validation, and similar results were observed (Table 2).
Table 2.
Stratification analysis of the three identified SNPs by histological types and smoking status
| SNP | Impa | Overall | Adenocarcinoma | Squamous cell carcinoma | Never smoking | Ever smoking | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI)b | P b | OR (95% CI)b | P b | OR (95% CI)b | P b | OR (95% CI)b | P b | OR (95% CI)b | P b | ||
| rs17781739 | |||||||||||
| ICR | 0.97 | 0.96 (0.88–1.04) | 0.287 | 1.01 (0.87–1.17) | 0.880 | 0.94 (0.83–1.08) | 0.397 | NA | NA | NA | NA |
| MDACC | 0.92 | 0.95 (0.82–1.09) | 0.426 | 0.97 (0.82–1.15) | 0.745 | 0.86 (0.69–1.07) | 0.164 | NA | NA | 0.95 (0.82–1.09) | 0.426 |
| IARC | 0.92 | 0.93 (0.85–1.01) | 0.088 | 0.95 (0.82–1.11) | 0.524 | 0.98 (0.87–1.10) | 0.726 | 0.78 (0.59–1.04) | 0.089 | 0.93 (0.85–1.02) | 0.131 |
| NCI | 0.97 | 0.88 (0.83–0.93) | 2.00E−05 | 0.88 (0.81–0.96) | 2.60E−03 | 0.84 (0.77–0.92) | 2.14E−04 | 0.86 (0.69–1.05)4 | 0.1544 | 0.89 (0.83–0.95) | 5.31E−04 |
| Toronto | 0.94 | 0.88 (0.68–1.12) | 0.300 | 0.67 (0.46–0.99) | 0.043 | 1.05 (0.63–1.76) | 0.841 | 0.89 (0.58–1.36) | 0.579 | 0.85 (0.62–1.16) | 0.308 |
| GLC | 0.95 | 0.93 (0.76–1.13) | 0.455 | 0.86 (0.66–1.14) | 0.302 | 1.04 (0.74–1.46) | 0.836 | 0.81 (0.42–1.55) | 0.522 | 0.95 (0.74–1.23) | 0.714 |
| Harvard | 0.97 | 0.99 (0.91–1.07) | 0.752 | 0.93 (0.81–1.06) | 0.254 | 1.15 (0.95–1.39) | 0.141 | 1.49 (0.96–2.31) | 0.075 | 1.02 (0.88–1.20) | 0.767 |
| Decode | 0.95 | 1.11 (0.95–1.30) | 0.202 | 1.11 (0.93–1.32) | 0.256 | 1.17 (0.89–1.53) | 0.273 | NA | NA | NA | NA |
| Meta- analysisc | 0.94 (0.90–0.99) | 0.012 | 0.94 (0.88–1.01) | 0.070 | 0.96 (0.88–1.06) | 0.441 | 0.89 (0.77–1.03) | 0.112 | 0.92 (0.88–0.96) | 3.31E−04 | |
| rs12587742 | |||||||||||
| ICR | 0.93 | 1.12 (1.02–1.24) | 0.016 | 1.07 (0.9–1.28) | 0.423 | 1.06 (0.91–1.24) | 0.453 | NA | NA | NA | NA |
| MDACC | 0.89 | 1.18 (1.01–1.39) | 0.043 | 1.27 (1.05–1.53) | 0.015 | 1.05 (0.82–1.36) | 0.698 | NA | NA | 1.18 (1.01–1.39) | 0.043 |
| IARC | 0.90 | 1.11 (1.01–1.22) | 0.033 | 1.10 (0.93–1.31) | 0.273 | 1.18 (1.03–1.35) | 0.015 | 1.15 (0.85–1.56) | 0.356 | 1.14 (1.02–1.27) | 0.019 |
| NCI | 0.94 | 1.07 (1.00–1.14) | 0.050 | 1.04 (0.95–1.15) | 0.370 | 1.16 (1.05–1.29) | 0.005 | 0.88 (0.69–1.13)d | 0.331d | 1.03 (0.96–1.11) | 0.394 |
| Toronto | 0.90 | 0.88 (0.68–1.15) | 0.364 | 0.99 (0.65–1.50) | 0.965 | 0.79 (0.47–1.33) | 0.377 | 0.62 (0.38–1.01) | 0.054 | 1.06 (0.76–1.48) | 0.746 |
| GLC | 0.92 | 1.19 (0.94–1.51) | 0.143 | 1.24 (0.91–1.70) | 0.174 | 0.90 (0.59–1.37) | 0.631 | 1.46 (0.75–2.83) | 0.266 | 1.12 (0.82–1.53) | 0.467 |
| Harvard | 0.95 | 1.08 (0.98–1.19) | 0.137 | 1.15 (0.98–1.34) | 0.078 | 1.29 (1.03–1.61) | 0.026 | 1.14 (0.72–1.81) | 0.579 | 0.90 (0.76–1.07) | 0.246 |
| Decode | 0.91 | 0.90 (0.76–1.07) | 0.235 | 0.90 (0.74–1.10) | 0.310 | 0.84 (0.62–1.15) | 0.283 | NA | NA | NA | NA |
| Meta- analysisc | 1.08 (1.04–1.12) | 8.15E−05 | 1.08 (1.02–1.15) | 0.010 | 1.12 (1.05–1.20) | 4.16E−04 | 0.97 (0.76–1.25) | 0.840 | 1.05 (0.98–1.13) | 0.163 | |
| rs2240980 | |||||||||||
| ICR | 0.99 | 1.11 (1.02–1.20) | 0.012 | 1.16 (1.00–1.34) | 0.052 | 1.04 (0.91–1.18) | 0.599 | NA | NA | NA | NA |
| MDACC | 0.95 | 1.14 (1.00–1.31) | 0.048 | 1.20 (1.03–1.41) | 0.021 | 0.99 (0.80–1.22) | 0.932 | NA | NA | 1.14 (1.00–1.31) | 0.048 |
| IARC | 0.96 | 1.10 (1.02–1.20) | 0.017 | 1.09 (0.94–1.27) | 0.254 | 1.14 (1.02–1.29) | 0.026 | 1.32 (1.02–1.72) | 0.035 | 1.10 (1.00–1.20) | 0.046 |
| NCI | 0.99 | 1.07 (1.01–1.13) | 0.025 | 1.07 (0.99–1.16) | 0.101 | 1.11 (1.01–1.21) | 0.030 | 1.11 (0.90–1.35)4 | 0.325d | 1.03 (0.97–1.10) | 0.317 |
| Toronto | 0.97 | 0.94 (0.74–1.18) | 0.583 | 1.01 (0.70–1.44) | 0.968 | 0.77 (0.49–1.21) | 0.249 | 0.82 (0.54–1.23) | 0.329 | 1.00 (0.74–1.33) | 0.979 |
| GLC | 0.98 | 1.08 (0.88–1.33) | 0.455 | 1.10 (0.83–1.44) | 0.506 | 0.88 (0.61–1.27) | 0.496 | 1.17 (0.64–2.17) | 0.607 | 1.04 (0.80–1.36) | 0.756 |
| Harvard | 0.98 | 1.08 (0.99–1.17) | 0.087 | 1.15 (1.01–1.31) | 0.036 | 1.07 (0.89–1.29) | 0.481 | 0.87 (0.58–1.31) | 0.505 | 0.99 (0.86–1.15) | 0.933 |
| Decode | 0.98 | 0.96 (0.83–1.11) | 0.566 | 0.96 (0.82–1.13) | 0.640 | 0.92 (0.71–1.20) | 0.554 | NA | NA | NA | NA |
| Meta- analysisc | 1.08 (1.04–1.11) | 1.08E−05 | 1.10 (1.04–1.15) | 3.44E−04 | 1.07 (1.01–1.13) | 0.017 | 1.08 (0.87–1.34) | 0.481 | 1.05 (0.99–1.11) | 0.127 | |
Imp, imputation; Harvard, Harvard Lung Cancer Study, US; deCODE, Icelandic Lung Cancer Study, Iceland.
aImputation quality score: r-squared from MACH for the MDACC, IARC, GLC and Harvard studies; info values from IMPUTE2 were used in other studies.
bAdjusted for the top significant principle components for each study.
cFixed effects model was used in the meta-analysis if Q-test P > 0.1 and heterogeneity statistic I2 < 25%; otherwise random effects model.
dThe pooling results of the four NCI sub-studies: the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC), the Cancer Prevention Study II Nutrition Cohort (CPS-II), the Environment and Genetics in Lung Cancer Etiology (EAGLE) and the Prostate, Lung, Colon, Ovary Screening Trial (PLCO). The detailed results for each sub-study were presented in Figure 3.
Stratified analyses
As lung adenocarcinoma and squamous cell carcinoma may have different risk factors, we performed stratified analysis by these histological types. By using 4862 adenocarcinomas and 3897 squamous cell carcinomas from all the eight GWASs (Supplementary Table 1, available at Carcinogenesis Online), we found that the effect of rs1258772 was more significant in squamous cell carcinomas (OR = 1.12, 95% CI = 1.05–1.20, P = 4.16 × 10–4) than in adenocarcinomas (OR = 1.08, 95% CI = 1.02–1.15, P = 0.010), while SNP rs2240980 had more significant effects in adenocarcinomas (OR = 1.10, 95% CI = 1.04–1.15, P = 3.44 × 10–4) than in squamous cell carcinomas (OR = 1.07, 95% CI = 1.01–1.13, P = 0.017) (Table 2). However, heterogeneity test showed that the effect difference between two histological strata was non-significant for both SNPs.
Cigarette smoking is one of the major risk factors for lung cancer and may interact with genetic factors. According to the currently available smoking data, study subjects were divided into two groups: ever smokers (defined as individuals having smoked at least 100 cigarettes in their lifetime) and never smokers. We performed stratified analysis by smoking status and found that only SNP rs17781739 had a significant effect in ever smokers (OR = 0.92, 95% CI = 0.88–0.96, P = 3.31 × 10–4) (Table 2). No significant association was observed in never smokers for all three SNPs, which might be due to a reduced sample size (731 never smokers). The forest plots of the overall and stratification results for these three SNPs are shown in Supplementary Figure 1a–c, available at Carcinogenesis Online.
In silico functional validation
The three SNPs were predicted with potentials to influence mRNA transcription (Table 1; Figure 2d and e). According to experimental data (e.g. histone modification, DNase cluster, transcription factor binding, RNAseq) from the ENCODE project (Figure 2d), we found that two SNPs (rs17781739 and rs12587742) are located within one CpG island with strong signals for active enhancer and promoter functions (indicated by DNase hypersensitivity and histone modification H3K27 acetylation, and H3K4 tri-methylation, respectively). Further transcription factor binding analysis (using the transcription factor ChIP-seq data) showed that rs12587742 is located at the c-MYC motif as shown by the position weight matrix (PWM) based Sequence Logo (Figure 2e), and the allele difference might influence the binding activity of the transcription factor. SNP rs2240980 was also predicted to be located at a regulatory region with evidence from DNase cluster and transcription factor CHIP-seq data (Figure 2d).
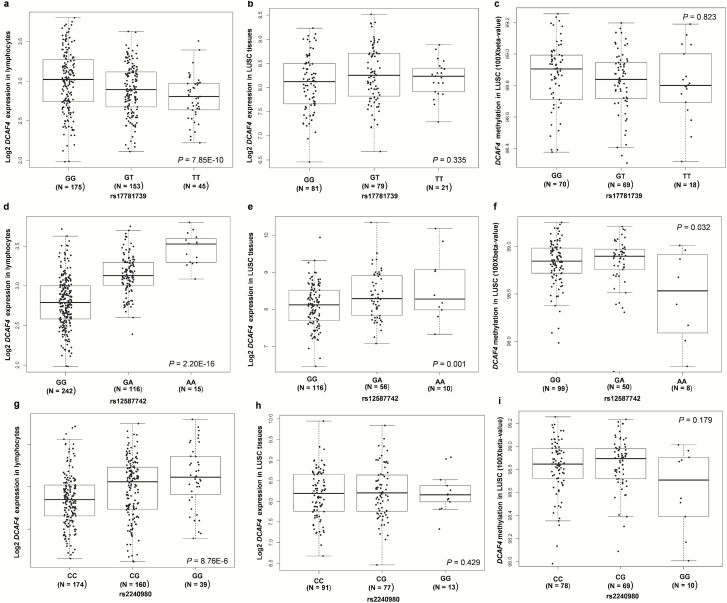
As genotyping data for the three identified SNPs were not available in the TCGA database, we performed imputation for them by using the reference data from the 1000 Genomes project. Further expression quantitative trait loci and meQTL analyses were conducted for SNPs with high quality imputation. Only SNPs from patients with lung squamous carcinoma passed the imputation quality control (imputation info > 0.9) and were used in further SNP-expression/methylation correlation analysis. As shown in Fig 3a, d and g, all of those three SNPs had a significant correlation with the mRNA expression of DCAF4 in the blood cells from 373 Europeans individuals (P = 7.85 × 10−10, 2.20 × 10−16 and 8.76 × 10−6 for rs17781739, rs12587742 and rs2240980, respectively). When put all these three SNPs into the same regression model, only SNP rs1258772 and rs2240980 remained significant (P =0.208, 5.86 × 10−25 and 0.003 for rs17781739, rs12587742 and rs2240980, respectively). These results suggest that two SNPs (rs1258772 and rs2240980) in DCAF4, particularly rs1258772, have an independent effect on the gene expression.
Figure 3.
Correlations of the three SNPs with DCAF4 mRNA expression and methylation status in blood cells and tumor tissues. Correlation between DCAF4 mRNA expression and (a) rs17781739; (d) rs12587742; (g) rs2240980 in 373 blood cells from 373 Europeans individuals in 1000 genomes project (P = 7.85 × 10−10, 2.20 × 10−16 and 8.76 × 10−6, respectively). Boxplots of DCAF4 mRNA expression and (b) rs17781739; (e) rs12587742; (h) rs2240980 in 182 lung squamous cell carcinomas (LUSC) tumor tissues from The Cancer Genome Atlas (TCGA) database (P = 0.335, 0.001 and 0.429, respectively). Boxplots of DCAF4 methylation status and (c) rs17781739; (f) rs12587742; (i) rs2240980 in 157 LUSC tumor tissues from the TCGA database (P = 0.823, 0.032 and 0.179, respectively).
We also performed SNP and mRNA expression correlation analysis by using the expression data in tumor tissues from 182 lung squamous cell carcinomas from TCGA database (Figure 3b, e and h). Once again, only SNP rs12587742 showed a significant correlation with increased mRNA expression of DCAF4 (P = 0.001). Such correlation was also supported by the results from normal lung tissues (P = 1.79 × 10−13) (Supplementary Figure 2a, available at Carcinogenesis Online) as well as multiple other tissues (e.g. testis, skin, colon, esophagus, subcutaneous adipose, stomach, pancreas, breast and thyroid) based on the data from the GTEx project (Supplementary Table 4, available at Carcinogenesis Online). Based on those results, the rs12587742 ‘A’ allele was associated with an increased mRNA expression of DCAF4 in most tissues except for testis. Considering this SNP is located within one CpG island, we further explored its influence on the methylation status of DCAF4 by using the data from TCGA and the MuTHER project. We observed that the ‘A’ allele was associated with a decreased methylation status (beta value, which is defined as the ratio of methylated probe intensity and the sum of methylated and unmethylated probe intensities) in the tumor tissues from 157 lung squamous cell carcinomas (Figure 3f, P = 0.032) and the adipose tissues from 428 female twin-pairs (Supplementary Figure 2b, available at Carcinogenesis Online, P = 2.48 × 10−9) (34). No significance was observed for two other SNPs (rs17781739 and rs2240980) to be associated with mRNA expression (Figure 3b and h) and methylation in the tumor tissues (Figure 3c and i). However, it should be noted that two other SNPs (rs2302587 and rs9788482) that had a moderate to relatively high LD with rs17781739 and rs2240980 (r2 = 0.73 and 0.43, respectively) showed a significant correlation with the methylation status in the adipose tissues from the female twin-pairs (Supplementary Figure 2c and d, available at Carcinogenesis Online).
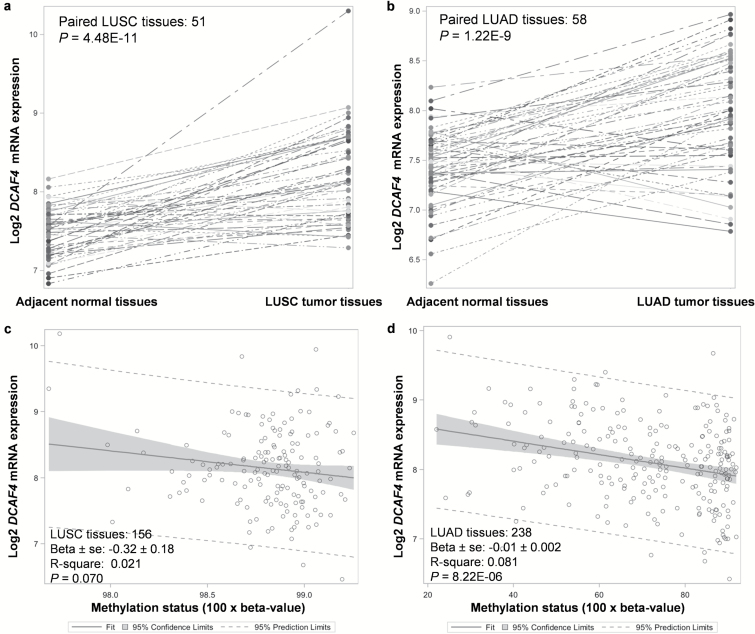
Differential expression analyses revealed that the DCAF4 gene had higher mRNA expression in tumor tissues from 156 lung squamous cell carcinomas and 238 adenocarcinomas (P = 4.48 × 10–11 and 1.22 × 10–9) than in adjacent normal tissues (Figure 4a and b). Results from other studies collected in the cancer microarray database Oncomine also showed some evidence for a high expression level of DCAF4 in lung adenocarcinomas than in the normal tissues (Supplementary Figure 3a and b, available at Carcinogenesis Online). We also observed a significantly negative correlation between the DCAF4 methylation status and mRNA expression levels in tumor tissues from both lung squamous cell carcinomas and adenocarcinomas (P = 0.070 and 8.22 × 10–6, respectively) (Figure 4c and d), which suggests that a high methylation status may lead to a decrease in mRNA expression of DCAF4 in the target tissues.
Figure 4.
Differential mRNA expression and methylation analysis by using the data generated by The Cancer Genome Atlas (TCGA). Higher DCAF4 mRNA expression were found in the tumor tissues of (a) 156 lung squamous cell carcinomas (LUSC) and (b) 238 adenocarcinomas (LUAD) than in the adjacent normal tissues (P = 4.48 × 10–11 and 1.22 × 10–9, respectively). Negative correlations were found between DCAF4 methylation and mRNA expression in both the (c) 156 lung squamous cell carcinomas and (d) 238 adenocarcinomas (P = 0.070 and 8.22 × 10–6, respectively).
We finally investigated the mutations of DCAF4 in lung tumor tissues by using the public available data from the database of the cBioPortal for Cancer Genomics (http://www.cbioportal.org). As shown in Supplementary Figure 4, available at Carcinogenesis Online, this gene had low somatic mutation rates in both the lung adenocarcinoma (LUAD; mutation rate = 0.5% [1/183], 5.9% [2/34], 0.4% [1/230] and 0% [0/163] in the Broad, MSKCC, TCGA and TSP studies, respectively) and squamous cell carcinoma (LUSC; mutation rate = 1.1% [2/178] in the TCGA study). Such results suggested the functional SNPs in DCAF4 might play more important role in the dysregulation of mRNA expression and methylation than mutations in tumor tissues.
Discussion
In this study, we performed an extensive analysis for associations between SNPs in 115 CRL-related genes and lung cancer risk by combining the summary data of six GWASs from the TRICL–ILLCO consortium including 12 160 cases and 16 838 cases. Such a large sample size allowed us to identify novel susceptibility loci with some moderate effects, which would have been often omitted in previous single GWAS. As a result, we identified three independent, potentially functional DCAF4 SNPs (rs117781739, rs12587742 and rs2240980) that were significantly associated with lung cancer risk in European populations. Further functional prediction analyses using data from blood cells and tumor tissues from the LUSC database revealed that the rs12587742 variant A allele was associated with an increased mRNA expression and a decreased methylation status of DCAF4. In addition, higher mRNA expression level of DCAF4 was also observed in tumor tissues than in adjacent normal tissues from patients with lung squamous cell carcinoma and adenocarcinoma. Moreover, significantly negative correlations were also observed between methylation status and mRNA expression levels in both sub-types of lung cancer. Taken together, our results provide a strong case that this novel genetic variant in DCAF4 was associated with lung cancer risk possibly by decreasing gene methylation status that had led to an increased mRNA expression of DCAF4.
DCAF4, also known as WDR21, is located on chromosome region 14q24.3 and encodes a WD40 repeat protein that interacts with the CUL4 and DDB1 to form the CUL4A–DDB1–DCAF complex. This interaction suggests that DCAF4 may be involved in nucleotide excision repair (NER), since DDB1 is one key component of the NER pathway, and that the CUL4A–DDBs complex may regulate NER activity through ubiquitination of several NER components, e.g. DDB2, XPC and histone H2A at the damaged DNA sites (35,36). Considering that smoking is the major risk factor for lung cancer and that smoking caused DNA damage is mainly repaired by the NER pathway, the increased DCAF4 expression as a compensation to a high level of damage to DNA may not be sufficient for the NER activity and thus result in high risk of lung cancer. This may partly explain the underlying biological and molecular mechanisms for the observed associations. In addition, DCAF4 may also be involved in the regulation of the telomere pathway and influence the telomere length, which is associated with risk of many cancers (37). Indeed, SNP rs2535913 in the DCAF4 gene was recently reported to be associated with a shorter leucocyte telomere length (38). A shorter telomere length had been found to be associated with an increased risk of lung squamous carcinoma and a decreased risk of lung adenocarcinoma in one large population study (39) and one recent meta-analysis (40). In this study, we found that the rs2535913 minor A allele (38) also showed a significant association with a decreased lung cancer risk (Supplementary Table 3, available at Carcinogenesis Online) and a decreased DCAF4 expression in adipose tissue and blood cells (GTEx data not shown). This SNP was also in a high LD (r2 = 0.78) with one identified functional SNP rs17781739. Although there is still no report about functions of DCAF4 on telomerase activity and telomere length, it is known that the DDB1 is involved in the regulation of telomerase expression via E2f1 (41,42) and the telomerase inhibition through ubiquitination-mediated TERT protein degradation (43). Thus, DCAF4 might indirectly influence telomerase activity and telomere length through interaction with DDB1 to inhibit the formation of other DDB1 complexes.
CRLs mediate the substrate-specific binding in the ubiquitination and play important roles in maintaining cellular protein homeostasis, which is especially critical for the lung, as the lung often experiences chronic or acute inflammation and frequent immune responses as well as DNA damage-repair responses induced by toxic or pathogenic exposures (24,44,45). Previous studies have reported that multiple CRL-related genes have been associated with inflammatory response and lung cancer. In the present study, in addition to DCAF4, we also found genetic variants in nine other CRL-related genes (i.e. COMMD1, CUL5, CUL7, DCAF8, KCTD10, KLHL21, KLHL22, PARK2 and TRIM21) to be associated with lung cancer risk with FDR < 0.2. Most of these genes were reported as tumor suppressor genes and also involved in the inflammation regulation (46–49). Notably, PARK2 is well studied as a Parkinson disease gene located at a fragile region of chromosome 6, which is prone to breakage and rearrangement. Genetic changes in this region have been found in several types of tumors, including glioma, lung cancer, colorectal and ovarian cancer (49). We also observed that SNPs in PARK2 are associated with lung cancer risk, which might provide some additional biological support for the connection between risks of cancer and Parkinson (49,50).
In this study, although we revealed associations between multiple genetic variants in DCAF4 and lung cancer risk and also provided functional evidence to support these associations, the exact biochemical and molecular mechanisms of the effects of those variants on DNA methylation and expression as well as possibly inflammation, DNA repair and telomere functions are still unclear. The associations between DCAF4 expression levels with telomerase activity and telomere length warrant additional experimental validation. Further biochemical studies are also required to reveal the hidden mechanisms, such as the role of DCAF4 in DNA repair. Although these identified variants only had a moderate effect on lung cancer risk, their joint effect might have driven the risk higher, which needs to be further explored in future association studies. In addition, as shown in the supplementary data, rs12587742 is significantly associated with DCAF4 mRNA levels in multiple tumor tissues, which implies this SNP might have a pleiotropic effect on cancer risk. This also needs to be clarified by future population studies across cancers.
In conclusion, the present study revealed one novel functional genetic variant rs12587742 in DCAF4, which is associated with a moderately increased lung cancer risk possibly by influencing its gene expression in normal and tumor tissues. We also provided multiple levels of evidence to support possible oncogenic effect of DCAF4. Our findings have provided new clues for future functional studies to investigate the roles of CRL-related genes in lung carcinogenesis.
Funding
TRICL-ILCCO
This work was supported by the Transdisciplinary Research in Cancer of the Lung (TRICL) Study, U19-CA148127 on behalf of the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network. The Toronto study was supported by Canadian Cancer Society Research Institute (020214), Ontario Institute of Cancer and Cancer Care Ontario Chair Award to RH The ICR study was supported by Cancer Research UK (C1298/A8780 andC1298/A8362—Bobby Moore Fund for Cancer Research UK) and NCRN, HEAL and Sanofi-Aventis. Additional funding was obtained from NIH grants (5R01CA055769, 5R01CA127219, 5R01CA133996 and 5R01CA121197). The Liverpool Lung Project (LLP) was supported by The Roy Castle Lung Cancer Foundation, UK. The ICR and LLP studies made use of genotyping data from the Wellcome Trust Case Control Consortium 2 (WTCCC2); a full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Sample collection for the Heidelberg lung cancer study was in part supported by a grant (70–2919) from the Deutsche Krebshilfe. The work was additionally supported by a Helmholtz-DAAD fellowship (A/07/97379 to MNT) and by the NIH (U19CA148127). The KORA Surveys were financed by the GSF, which is funded by the German Federal Ministry of Education, Science, Research and Technology and the State of Bavaria. The Lung Cancer in the Young study (LUCY) was funded in part by the National Genome Research Network (NGFN), the DFG (BI576/2-1; BI 576/2-2), the Helmholtzgemeinschaft (HGF) and the Federal office for Radiation Protection (BfS: STSch4454). Genotyping was performed in the Genome Analysis Center (GAC) of the Helmholtz Zentrum Muenchen. Support for the Central Europe, HUNT2/Tromsø and CARET genome-wide studies was provided by Institute National du Cancer, France. Support for the HUNT2/Tromsø genome-wide study was also provided by the European Community
(Integrated Project DNA repair, LSHG-CT- 2005–512113), the Norwegian Cancer Association and the Functional Genomics Programme of Research Council of Norway. Support for the Central Europe study, Czech Republic, was also provided by the European Regional Development Fund and the State Budget of the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101). Support for the CARET genome-wide study was also provided by grants from the US National Cancer Institute, NIH (R01 CA111703 and UO1 CA63673), and by funds from the Fred Hutchinson Cancer Research Center. Additional funding for study coordination, genotyping of replication studies and statistical analysis was provided by the US National Cancer Institute (R01 CA092039). The lung cancer GWAS from Estonia was partly supported by a FP7 grant (REGPOT245536), by the Estonian Government (SF0180142s08), by EU RDF in the frame of Centre of Excellence in Genomics and Estoinian Research Infrastructure’s Roadmap and by University of Tartu (SP1GVARENG). The work reported in this paper was partly undertaken during the tenure of a Postdoctoral Fellowship from the IARC (for MNT). The Environment and Genetics in Lung Cancer Etiology (EAGLE), the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC) and the Prostate, Lung, Colon, Ovary Screening Trial (PLCO)studies and the genotyping of ATBC, the Cancer Prevention Study II Nutrition Cohort (CPS-II) and part of PLCO were supported by the Intramural Research Program of NIH, NCI, Division of Cancer Epidemiology and Genetics. ATBC was also supported by US Public Health Service contracts (N01-CN-45165, N01-RC-45035 and N01-RC-37004) from the NCI. PLCO was also supported by individual contracts from the NCI to the University of Colorado Denver (NO1-CN-25514), Georgetown University(NO1-CN-25522), Pacific Health Research Institute (NO1-CN-25515),Henry Ford Health System (NO1-CN-25512), University of Minnesota(NO1-CN-25513), Washington University(NO1-CN-25516), University of Pittsburgh (NO1-CN-25511), University of Utah (NO1-CN-25524), Marshfield Clinic Research Foundation (NO1-CN-25518), University of Alabama at Birmingham (NO1-CN-75022, Westat, Inc. NO1-CN-25476), University of California, Los Angeles (NO1-CN-25404). The Cancer Prevention Study II Nutrition Cohort was supported by the American Cancer Society. The NIH Genes, Environment and Health Initiative (GEI) partly funded DNA extraction and statistical analyses (HG-06-033-NCI-01 andRO1HL091172-01), genotyping at the Johns Hopkins University Center for Inherited Disease Research (U01HG004438 and NIH HHSN268200782096C) and study coordination at the GENEVA Coordination Center (U01 HG004446) for EAGLE and part of PLCO studies. Funding for the MD Anderson Cancer Study was provided by NIH grants (P50 CA70907, R01CA121197, R01CA127219, U19 CA148127, R01 CA55769, and K07CA160753) and CPRIT grant (RP100443). Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is funded through a federal contract from the NIH to The Johns Hopkins University (HHSN268200782096C). The Harvard Lung Cancer Study was supported by the NIH (National Cancer Institute) grants CA092824, CA090578 and CA074386.
deCODE
The project was funded in part by GENADDICT: LSHMCT-2004–005166), the National Institutes of Health (R01-DA017932)
TCGA
The results published here are in whole or part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute The Cancer Genome Atlas (TCGA) Research Network can be found at ‘http://cancergenome.nih.gov’. The TCGA SNP data analyzed here are requested through dbGAP (accession#: phs000178.v1.p1).
Supplementary material
Supplementary data are available at Carcinogenesis online.
Supplementary Material
Acknowledgements
As Duke Cancer Institute members, Q.W. and K.O. acknowledges support from the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (Grant ID: NIH CA014236). Q.W. was also supported by a start-up fund from Duke Cancer Institute, Duke University Medical Center.
Conflict of Interest Statement: None declared.
Abbreviations
- CI
confidence interval
- CRL
cullin-RING ubiquitin ligases
- FDR
false discovery rate
- GWAS
genome-wide association study
- LD
linkage disequilibrium
- NER
nucleotide excision repair
- OR
odds ratio
- SNP
single nucleotide polymorphism
References
- 1. Howlader N., et al. (2016) SEER Cancer Statistics Review, 1975–2013, National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2013/, based on November 2015 SEER data submission, posted to the SEER web site, April 2016. [Google Scholar]
- 2. Molina J.R., et al. (2008) Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc., 83, 584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amos C.I., et al. (2008) Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet., 40, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hung R.J., et al. (2008) A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature, 452, 633–637. [DOI] [PubMed] [Google Scholar]
- 5. McKay J.D., et al. ; EPIC Study (2008) Lung cancer susceptibility locus at 5p15.33. Nat. Genet., 40, 1404–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y., et al. (2008) Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet., 40, 1407–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Landi M.T., et al. (2009) A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet., 85, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y., et al. (2014) Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat. Genet., 46, 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Freedman M.L., et al. (2011) Principles for the post-GWAS functional characterization of cancer risk loci. Nat. Genet., 43, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mani A., et al. (2005) The ubiquitin-proteasome pathway and its role in cancer. J. Clin. Oncol., 23, 4776–4789. [DOI] [PubMed] [Google Scholar]
- 11. Zhao Y., et al. (2012) Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis., 3, e386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bassermann F., et al. (2014) The ubiquitin proteasome system - implications for cell cycle control and the targeted treatment of cancer. Biochim. Biophys. Acta, 1843, 150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frescas D., et al. (2008) Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer, 8, 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Salon C., et al. (2007) Altered pattern of Cul-1 protein expression and neddylation in human lung tumours: relationships with CAND1 and cyclin E protein levels. J. Pathol., 213, 303–310. [DOI] [PubMed] [Google Scholar]
- 15. Welcker M., et al. (2008) FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer, 8, 83–93. [DOI] [PubMed] [Google Scholar]
- 16. Osoegawa A., et al. (2004) Regulation of p27 by S-phase kinase-associated protein 2 is associated with aggressiveness in non-small-cell lung cancer. J. Clin. Oncol., 22, 4165–4173. [DOI] [PubMed] [Google Scholar]
- 17. Yokoi S., et al. (2004) Amplification and overexpression of SKP2 are associated with metastasis of non-small-cell lung cancers to lymph nodes. Am. J. Pathol., 165, 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kandoth C., et al. (2013) Mutational landscape and significance across 12 major cancer types. Nature, 502, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Timofeeva M.N., et al. ; Transdisciplinary Research in Cancer of the Lung (TRICL) Research Team (2012) Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum. Mol. Genet., 21, 4980–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Su L., et al. (2006) Genotypes and haplotypes of matrix metalloproteinase 1, 3 and 12 genes and the risk of lung cancer. Carcinogenesis, 27, 1024–1029. [DOI] [PubMed] [Google Scholar]
- 21. Thorgeirsson T.E., et al. (2008) A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature, 452, 638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Y., et al. (2015) Deciphering associations for lung cancer risk through imputation and analysis of 12 316 cases and 16 831 controls. Eur. J. Hum. Genet., 23, 1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Howie B.N., et al. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tan Y.H., et al. (2010) CBL is frequently altered in lung cancers: its relationship to mutations in MET and EGFR tyrosine kinases. PLoS One, 5, e8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lappalainen T., et al. ; Geuvadis Consortium (2013) Transcriptome and genome sequencing uncovers functional variation in humans. Nature, 501, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Consortium G.T. (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cancer Genome Atlas Research, N. (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature, 511, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cancer Genome Atlas Research, N. (2012) Comprehensive genomic characterization of squamous cell lung cancers. Nature, 489, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang T.P., et al. (2010) Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics, 26, 2474–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Begum F., et al. (2012) Comprehensive literature review and statistical considerations for GWAS meta-analysis. Nucleic Acids Res., 40, 3777–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benjamini Y., et al. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B (Methodological), 57, 289–300. [Google Scholar]
- 32. Pruim R.J., et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barrett J.C., et al. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. [DOI] [PubMed] [Google Scholar]
- 34. Bell J.T., et al. ; MuTHER Consortium (2012) Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet., 8, e1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li J., et al. (2006) DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity. Cancer Res., 66, 8590–8597. [DOI] [PubMed] [Google Scholar]
- 36. Guerrero-Santoro J., et al. (2008) The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res., 68, 5014–5022. [DOI] [PubMed] [Google Scholar]
- 37. Wentzensen I.M., et al. (2011) The association of telomere length and cancer: a meta-analysis. Cancer Epidemiol. Biomarkers Prev., 20, 1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mangino M., et al. (2015) DCAF4, a novel gene associated with leucocyte telomere length. J. Med. Genet., 52, 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sanchez-Espiridion B., et al. (2014) Telomere length in peripheral blood leukocytes and lung cancer risk: a large case-control study in Caucasians. Cancer Res., 74, 2476–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang C., et al. ; GECCO and GAME-ON Network: CORECT, DRIVE, ELLIPSE, FOCI, and TRICL (2015) Genetic determinants of telomere length and risk of common cancers: a Mendelian randomization study. Hum. Mol. Genet., 24, 5356–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alonso M.M., et al. (2006) E2F1 and telomerase: alliance in the dark side. Cell Cycle, 5, 930–935. [DOI] [PubMed] [Google Scholar]
- 42. Hayes S., et al. (1998) DDB, a putative DNA repair protein, can function as a transcriptional partner of E2F1. Mol. Cell. Biol., 18, 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jung H.Y., et al. (2013) Dyrk2-associated EDD-DDB1-VprBP E3 ligase inhibits telomerase by TERT degradation. J. Biol. Chem., 288, 7252–7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bachmaier K., et al. (2007) E3 ubiquitin ligase Cblb regulates the acute inflammatory response underlying lung injury. Nat. Med., 13, 920–926. [DOI] [PubMed] [Google Scholar]
- 45. Weathington N.M., et al. (2013) New insights on the function of SCF ubiquitin E3 ligases in the lung. Cell. Signal., 25, 1792–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van de Sluis B., et al. (2010) COMMD1 disrupts HIF-1alpha/beta dimerization and inhibits human tumor cell invasion. J. Clin. Invest., 120, 2119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Samant R.S., et al. (2014) E3 ubiquitin ligase Cullin-5 modulates multiple molecular and cellular responses to heat shock protein 90 inhibition in human cancer cells. Proc. Natl. Acad. Sci. USA, 111, 6834–6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Y., et al. (2009) KCTD10 interacts with proliferating cell nuclear antigen and its down-regulation could inhibit cell proliferation. J. Cell. Biochem., 106, 409–413. [DOI] [PubMed] [Google Scholar]
- 49. Veeriah S., et al. (2010) Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat. Genet., 42, 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Garber K. (2010) Parkinson’s disease and cancer: the unexplored connection. J. Natl. Cancer Inst., 102, 371–374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.