In this study, Okano-Uchida et al. describe the physiological role of ORC1 in mice by generating knock-in mice with LoxP sites flanking exons encoding the critical ATPase domain of ORC1. They show that ORC1 ablation in extraembryonic trophoblasts and hepatocytes failed to impede genome endoreduplication and organ development and function and conclude that ORC1 in mice is essential for mitotic cell divisions but dispensable for endoreduplication.
Keywords: endoreduplication, DNA replication, polyploidy, hepatocytes
Abstract
The largest subunit of the origin recognition complex (ORC1) is essential for assembly of the prereplicative complex, firing of DNA replication origins, and faithful duplication of the genome. Here, we generated knock-in mice with LoxP sites flanking exons encoding the critical ATPase domain of ORC1. Global or tissue-specific ablation of ORC1 function in mouse embryo fibroblasts and fetal and adult diploid tissues blocked DNA replication, cell lineage expansion, and organ development. Remarkably, ORC1 ablation in extraembryonic trophoblasts and hepatocytes, two polyploid cell types in mice, failed to impede genome endoreduplication and organ development and function. Thus, ORC1 in mice is essential for mitotic cell divisions but dispensable for endoreduplication. We propose that DNA replication of mammalian polyploid genomes uses a distinct ORC1-independent mechanism.
The loading of the origin recognition complex (ORC) is the first step in the assembly of origins of DNA replication. ORC loading limits the firing of each origin to once per cell division, ensuring faithful duplication of the genome. The largest subunit, ORC1, contains ATPase activity and is essential for mitotic cell divisions in yeast, flies, fish, and mammals (Grallert and Nurse 1996; Park and Asano 2008; Shu et al. 2008; Bicknell et al. 2011). ORC1 ATPase activity is required for recruitment of ORC2–6 and subsequent engagement of CDC6, CDT1, and MCMs to origins, completing the assembly of the prereplicative complex (pre-RC) (Chesnokov et al. 2001; Takehara et al. 2008). The pre-RC is the core machinery used to initiate DNA replication, and its assembly on origins is thought to be required for the accurate replication of all DNA genomes (DePamphilis et al. 2006). Hence, the molecular mechanisms regulating the initiation of DNA replication across cell types and species are presumed to be the same. Although deemed essential for DNA replication, two different groups have shown that certain cell types in flies and some human cancer cell lines can proliferate with significantly reduced levels of ORC1 (Park and Asano 2008; Shibata et al. 2016), raising the possibility that special conditions may support ORC1-independent DNA replication.
The archetypal cell division cycle culminates in the generation of genetically identical diploid cell progenies. However, a number of cell cycle variations exist in nature. The endocycle is one such variant characterized by alternating DNA synthesis (S) and gap (G) phases in the absence of an intervening cell division (Lee et al. 2009; Fox and Duronio 2013; Edgar et al. 2014), which can, in some cell types, result in ploidy levels that exceed 1000C. This remarkable cell cycle variant is used by a wide range of organisms to support organ function during development and in response to diverse environmental contexts. Flies and plants use endocycles extensively as a mechanism for cell and organ growth (Lee et al. 2009; Fox and Duronio 2013; Edgar et al. 2014). In mammals, however, endocycles are restricted to a few cell types, including trophoblast giant cells (TGCs) in the placenta, hepatocytes in the liver, and megakaryocytes (MKs) in the bone marrow. Polyploidy is proposed to provide these mammalian tissues with an augmented ability to respond to strenuous cellular metabolic demands, a dampened sensitivity to apoptotic or checkpoint stimuli, and a buffering capacity against deleterious loss-of-function mutations that may cause cancer (Lee et al. 2009; Fox and Duronio 2013; Edgar et al. 2014). However, altered ploidy levels have been noted in a number of gene knockout mouse models without any physiological consequence to organ function (Chen et al. 2012). Thus, the role of endoreduplication in mammalian organ physiology remains to be firmly established.
Here, we generated knock-in mice with LoxP sites flanking exons encoding the critical ATPase domain of ORC1. Evaluation of Orc1f/f mice with different tissue- and temporal-specific Cre drivers revealed that ORC1 function is essential for mitotic cell divisions and mouse embryonic development. Surprisingly, ORC1 was dispensable for the proliferation of extraembryonic trophoblasts and postnatal developing hepatocytes, two endoreduplicating cell types in mice. Remarkably, mutant Orc1Δ placentas could carry wild-type fetuses to term, and mutant Orc1Δ livers reached normal size and fully regenerated following acute injury. These findings reveal that mitotic cycles and endocycles have distinct requirements for ORC1, with variant cell cycle types using an ORC1-independent mechanism of DNA replication.
Results
ORC1 is essential for embryonic development
We used standard homologous recombination approaches to target the Orc1 allele in mice. LoxP sites were introduced flanking exons 9–14, which encode the critical ORC1 ATPase domain, and an Frt-flanked neomycin (Neo) selection cassette was introduced within intron 14 (Fig. 1A; Supplemental Fig. S1A). The targeting vector was electroporated into mouse embryonic stem (ES) cells, and correct integration was confirmed by Southern blot and PCR genotyping of genomic DNA (Fig. 1B,C; Supplemental Fig. S1B,C). Blastocysts injected with targeted ES cells were transplanted into pseudopregnant females, giving rise to founder Orc1+/neo knock-in mice. Breeding Orc1+/neo knock-in mice with the general deleter Actin-FLPe mice (Rodríguez et al. 2000) resulted in widespread excision of the Frt-flanked Neo and yielded germline conditional floxed (Orc1+/f) knock-in mice.
Figure 1.
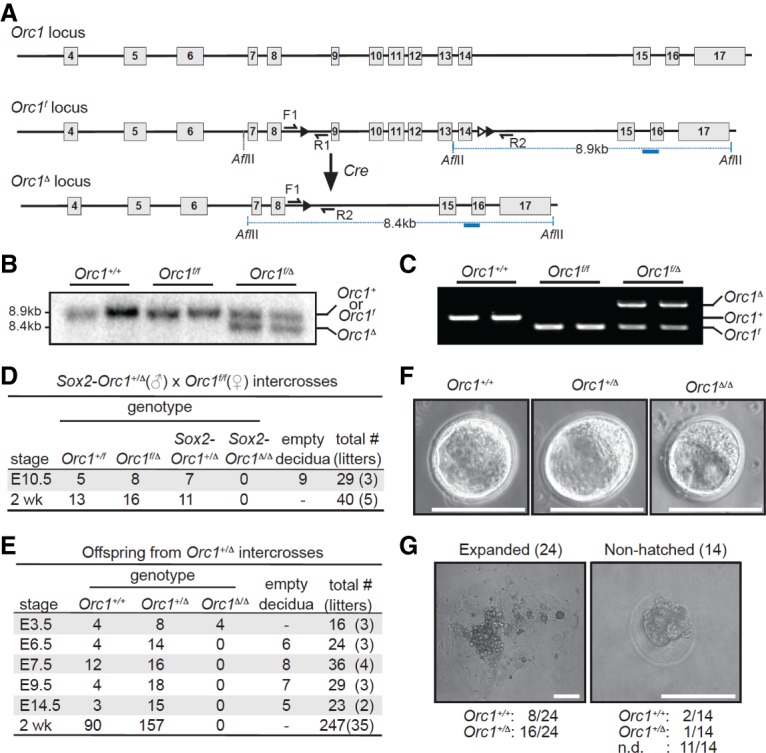
ORC1 is essential for early embryonic development. (A) Diagram of the mouse Orc1 locus. The floxed Orc1 (Orc1f) locus has exons 9–14 flanked with LoxP sites (black triangles), and the Orc1-deleted (Orc1Δ) locus has the targeted exons deleted. The blue line represents the Southern probe used to screen ES cell clones after AflII digestion. (B) Southern blotting of genomic liver DNA. The 8.9-kb wild-type (Orc1+) or Orc1f allele and the 8.4-kb Orc1Δ allele are indicated. (C) Genotyping PCR of genomic liver DNA from Orc1+/+, Orc1f/f, and Orc1f/Δ mice using the primers (F1, R1, and R2) shown as black arrows in A. (D,E) Genotypes of offspring from male Sox2-Cre;Orc1+/Δ and female Orc1f/f intercrosses (D) and Orc1+/Δ intercrosses (E) at various stages during gestation and after birth. (F) Representative pictures of embryonic day 3.5 (E3.5) blastocysts derived from the intercrosses in E. (G) Thirty-eight E3.5 blastocysts from Orc1+/Δ intercrosses (five litters) were individually cultured for 6–7 d in vitro, and examples of expanded and nonhatched blastocysts after in vitro culture are shown. Genotypes of the 24 expanded clones (left panel) and the 14 nonhatched blastocysts (right panel) are indicated below each sample picture. Bars: F,G, 100 µm.
To evaluate the role of ORC1 in mouse development, we used Sox2-Cre mice, which express the Sox2-Cre transgene in all cells of the inner cell mass following implantation (Hayashi et al. 2002). Crosses between Sox2-Orc1+/Δ and Orc1f/f mice failed to yield any Sox2-Orc1Δ/Δ offspring (Fig. 1D). Mice with germline deletion of floxed exons in one allele (Orc1+/Δ) were indistinguishable from wild-type littermates. Interbreeding Orc1+/Δ mice also failed to yield any live Orc1Δ/Δ offspring, and analysis of timed pregnancies revealed no viable embryos beyond embryonic day 6.5 (E6.5) (Fig. 1E,F). However, E3.5 Orc1Δ/Δ blastocysts were recovered at the expected frequency. We then cultured in vitro blastocysts derived from Orc1+/Δ intercrosses and found that all of the hatched embryos were wild type or heterozygous (eight Orc1+/+ and 16 Orc1+/Δ), whereas nonhatched blastocysts were either wild type (two Orc1+/+), heterozygous (one Orc1+/Δ), or dead and could not be genotyped (11 not determined) (Fig. 1G). From these observations, we conclude that ORC1 is essential for mouse development and that absence of intact ORC1 function leads to peri-implantation lethality.
Embryonic and adult mitotic cell divisions require ORC1
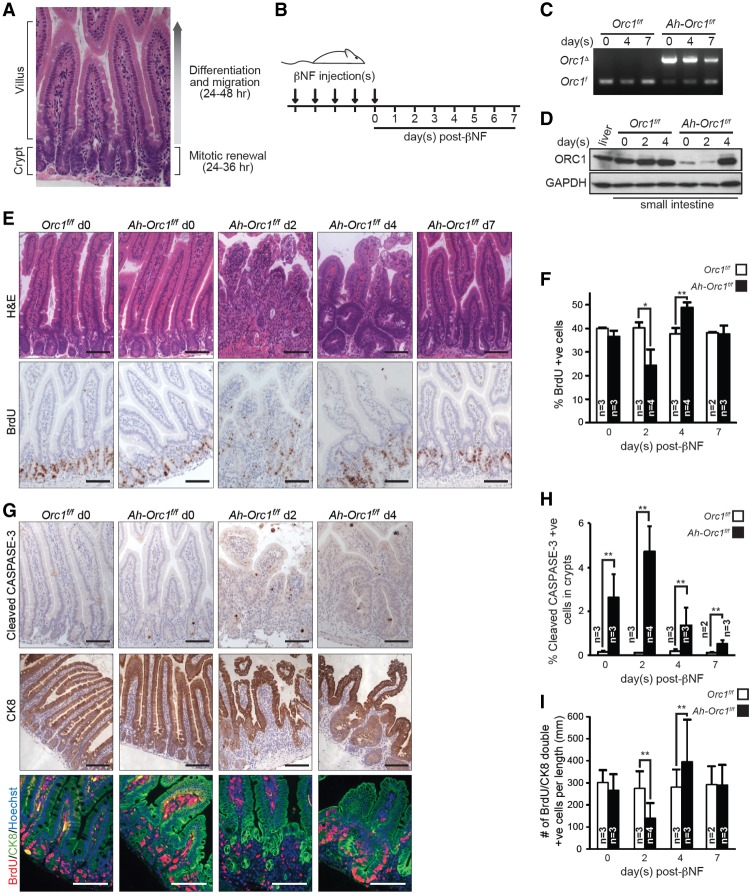
We then used the small intestine as a model organ to test whether ORC1 function is required for the expansion of diploid cells during adult development. Cell proliferation in the small intestine is compartmentalized within pouch-like invaginations called crypts (Fig. 2A). Here, intestinal stem cells divide asymmetrically to maintain the stem cell pool and generate progenitor cells that undergo several rounds of division before they exit the cell cycle, differentiate, and migrate into the finger-like projections called villi. As these differentiated enterocytes migrate into villi and encounter the harsh environment of the intestinal lumen, many of them die and are shed into the luminal space. The entire regenerative process—from asymmetric stem cell division within crypts to enterocyte shedding at the tips of villi—takes ∼4–5 d in mice and continues throughout the lifetime of the animal. Orc1 ablation in intestinal crypts was achieved by using an inducible Ah-Cre transgene, which can be activated in the intestinal tract by the administration of β-napthoflavone (βNF) (Ireland et al. 2004). Cohorts of Orc1f/f and Ah-Orc1f/f mice were injected with βNF for five consecutive days to recombine effectively the Orc1f allele in the entire crypt–villi unit (Fig. 2B). Treated mice were collected at various time points following the last day of treatment (days 0–7). Cre-mediated recombination of the floxed Orc1 allele was confirmed by PCR genotyping of genomic DNA derived from the small intestine and by immunohistochemistry (IHC) using CRE-specific antibodies (Fig. 2C; Supplemental Fig. S2A). Western blot analysis of small intestines showed that ORC1 protein was dramatically deceased by the end of the last βNF injection and that its depletion persisted for two to three additional days (Fig. 2D). Interestingly, by the fourth day following the last βNF injection, ORC1 protein was replenished to near-normal levels. Hematoxylin and eosin (H&E) staining of intestinal sections showed that by day 2 after βNF injection, Orc1 deficiency led to a progressive and severe disruption of intestinal architecture, with fewer and smaller crypts and shorter, hypocellular, and progressively disorganized villi (Fig. 2E). As expected, BrdU incorporation was decreased but not completely abolished, and there were marked levels of apoptosis in the crypts of Ah-Orc1f/f animals relative to controls (Fig. 2E–H; Supplemental Fig. S2B). IHC of intestinal sections from these animals using cytokeratin 8-specific antibodies confirmed the morphological disruption of crypt–villus architecture and decreased proliferation of progenitor cells (Fig. 2G,I).
Figure 2.
Orc1 deletion causes defects in DNA replication and cell renewal in the small intestine. (A) H&E-stained small intestine sections highlight the compartmentalized structure of the crypt–villus unit, consisting of the proliferative (crypt) and differentiated zones (villus). (B) Experimental scheme of Orc1 deletion in the small intestines of adult mice. Control and Ah-Cre 2-mo-old mice were injected with βNF for five consecutive days and harvested 0–7 d after the last βNF injection. BrdU was injected 1 h prior to dissection. (C) PCR genotyping detecting Orc1 deletion in the Ah-Orc1f/f small intestine. (D) Immunoblot showing depletion of the ORC1 protein in the Ah-Orc1f/f small intestine after various time points following βNF treatment. GAPDH was used as a loading control. (E) Representative images of H&E-stained sections (top) and IHC using BrdU antibody (bottom) of small intestine at 0–7 d after βNF treatment. (F) Quantification of BrdU-positive cells in crypt cells from E. At least 1600 cells in crypts were counted for each small intestine (total of 108,000 cells). n = 25 mice. (G) Representative images of IHC for cleaved CASPASE-3 (top), cytokeratin 8 (CK8; middle), and immunofluorescence (IF) for cytokeratin 8 (green)/BrdU (red)/Hoechst 33342 (blue) (bottom) in the small intestines of βNF-treated mice. (H) Quantification of cleaved CASPASE-3-positive cells in crypts. At least 1100 cells in crypts were counted for each section (total of 62,800 cells). n = 24 mice. (I) Quantification of BrdU/cytokeratin 8-double-positive crypt cells per the longitudinal axis of the small intestine. At least 1300 cytokeratin 8-positive cells were counted for each section (total of 56,900 cells). n = 24 mice. The results are shown as average + SD. (*) P ≤ 0.05; (**) P < 0.01, t-test. Bars: E,G, 50 µm. (n) Number of mice in each group.
Remarkably, the acute disruption of intestinal integrity observed in Ah-Orc1f/f animals was followed by a rapid repopulation of enterocytes, presumably derived from rare nondeleted intestinal stem cells (Fig. 2E, day 7 [d7]). During this 4- to 5-d repopulation period, we observed enhanced DNA replication, progenitor cell proliferation, and crypt hypertrophy, which followed the progressive restoration of ORC1 protein levels noted earlier (Fig. 2D). By 7 d after βNF treatment, the integrity of the crypt–villus was fully restored, and the small intestine was indistinguishable from littermate controls (Fig. 2E). βNF treatment had no impact on the small intestine of Orc1f/f mice, and additional control experiments showed that CRE expression and recombination activity, as measured by a Rosa26stop-EYFP reporter allele, had no adverse impact on cell proliferation, apoptosis, or intestinal architecture of animals containing at least one wild-type Orc1 allele (Supplemental Fig. S2C–E). We also evaluated animals containing one floxed Orc1 allele and one germline-deleted Orc1 allele (Ah-Orc1f/Δ). Treatment of Ah-Orc1f/Δ mice with βNF disrupted cell proliferation and intestinal architecture to the same extent as treatment of Ah-Orc1f/f mice (Supplemental Fig. S2F). Thus, based on the analysis of embryonic and adult tissues, we conclude that sustained proliferation of diploid cells in mice requires intact ORC1 protein.
Targeted inactivation of the ATPase domain of ORC1 precludes proliferation of mouse embryonic fibroblasts (MEFs)
We also evaluated the consequence of ORC1 depletion in MEFs by standard cell culture methods. To this end, we generated Orc1f/f MEFs and infected them with either control (Cre−) or Cre-expressing (Cre+) retroviruses. Cre-mediated recombination of Orc1f/f alleles was confirmed by PCR-based genotyping of genomic DNA and quantitative RT–PCR analysis of mRNA (Fig. 3A,B; Supplemental Fig. S3A). Western blotting using an N terminus-specific antibody (Miyake et al. 2005) failed to detect any ORC1-specific products in Cre-treated Orc1f/f MEFs, indicating that deletion of exons 9–14 does not produce any appreciable level of truncated protein products (Fig. 3C; Supplemental Fig. S3B). Importantly, ORC1-deficient MEFs ceased to proliferate (Fig. 3D), had negligible BrdU incorporation (Fig. 3E), and accumulated ≥2C DNA content (Fig. 3F), whereas control MEFs proliferated normally. These observations suggested that loss of ORC1 function in MEFs is not permissive for DNA replication and leads to S-phase arrest.
Figure 3.
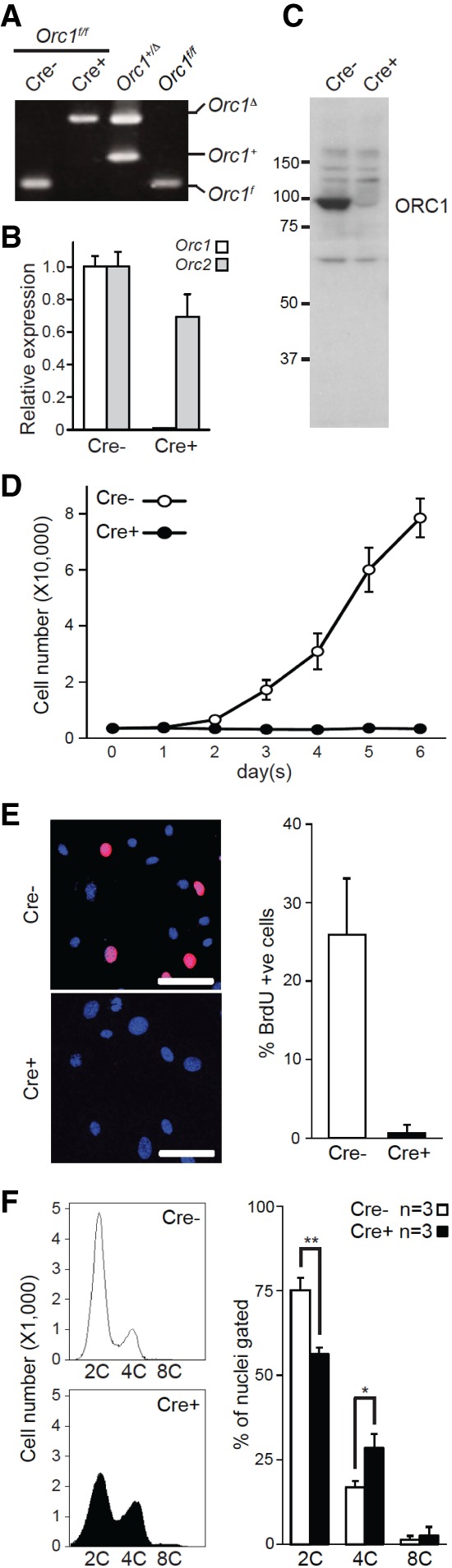
Ablation of Orc1 blocks DNA replication and cell proliferation in MEFs. Orc1f/f MEFs isolated from E13.5 embryo were infected with control (Cre−) or Cre-expressing (Cre+) retrovirus. The infected Orc1f/f MEFs were processed for PCR genotyping (A), quantitative RT–PCR for Orc1 and Orc2 (B), immunoblot using ORC1 antibody (C), and cell growth assay (D). (E) BrdU incorporation was detected in MEFs by IF (red), and Hoechst 33342 (blue) was used as a counterstain. Representative images (left) and quantification (right) of BrdU-incorporated MEFs are shown. At least 420 cells were counted for each sample. (F) Representative fluorescent-activated cell sorting (FACS) profiles of propidium iodide (PI)-stained Orc1f/f MEFs and quantification of DNA content. The results are shown as average + SD. n = 3. (*) P ≤ 0.05; (**) P ≤ 0.01, t-test. Bars: E, 100 µm.
Mutant Orc1Δ TGCs support placental development and a full pregnancy
Previous clonal analysis in Drosophila showed that endoreduplicating cells of the developing salivary gland expand into multicellular clones in the absence of ORC1 (Park and Asano 2008). Given the unique cell cycle nature of cells in the Drosophila tissue, we sought to determine whether ORC1-independent DNA replication might also be a feature of mammalian endoreduplicating cells. In mice, essentially all cells derived from the inner cell mass of embryos are uniformly diploid. However, critical extraembryonic-derived trophoblast lineages in the placenta are polyploid (Hu and Cross 2010). Secondary TGCs, a subpopulation that arises from trophoblast progenitors within the E6.5 ectoplacental cone and gives rise to the fetal–maternal boundary, acquire giant nuclei with genomes ranging in DNA content from 32C to 1024C. Thus, we first focused on the role of ORC1 in TGCs, the most exuberant endoreduplicating extraembryonic cell type of the mouse placenta. To this end, we used Plfcre/+ knock-in mice (Ouseph et al. 2012), which specifically express CRE in trophoblasts during the secondary wave of TGC expansion. Remarkably, intercrosses between Plfcre/+ and Orc1f/f mice yielded viable Plf-Orc1f/f E10.5 embryos and live offspring (Fig. 4A; Supplemental Fig. S4A). PCR genotyping demonstrated successful deletion of Orc1 in Plf-Orc1f/f placentae (Fig. 4B). Placentae from E10.5 embryos appeared histologically normal and supported the development of associated fetuses to term (Fig. 4A,C). Importantly, the nuclear size and genomic DNA content of E10.5 TGCs, as measured by Feulgen staining of placental sections, were unaffected by Orc1 deficiency (Fig. 4D,E). Coimmunofluorescence (co-IF) with CRE- and BrdU-specific antibodies confirmed that most CRE-positive TGCs incorporated BrdU (Fig. 4F). Moreover, IHC of Plf-Orc1f/f placental sections showed normal expression of the cell cycle marker Cyclin A2 in mutant Orc1Δ TGCs (Fig. 4G).
Figure 4.
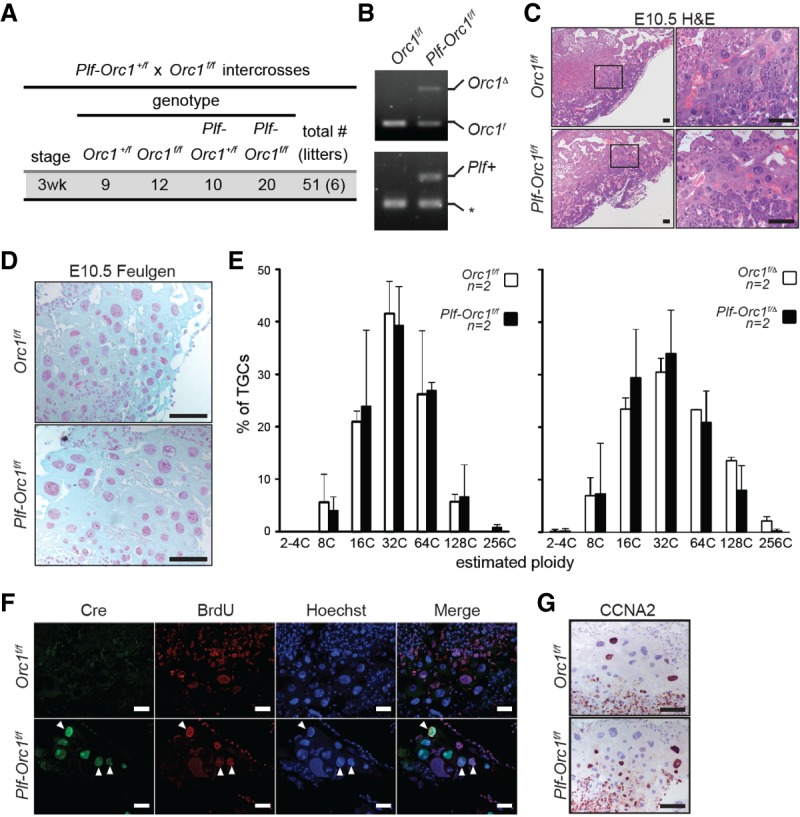
Deletion of Orc1 in TGCs has no effect on endoreduplication and placental development. (A) Genotypes of the offspring from Plf-Orc1+/f and Orc1f/f intercrosses. (B) PCR genotyping showing Orc1 deletion in E10.5 Plf-Orc1f/f placentae. Whole placenta sections (female genotype was Orc1f/f) were used to extract genomic DNA. (Bottom panel) The asterisk indicates generation of a PCR product used as an internal control. (C,D) Representative images of H&E-stained (C) and Feulgen-stained (D) E10.5 control (Orc1f/f) and Orc1-deleted (Plf-Orc1f/f) placentae. (E) Quantification of Feulgen-stained TGCs derived from E10.5 control (Orc1f/f and Orc1f/Δ) and Orc1-deleted (Plf-Orc1f/f and Plf-Orc1f/Δ) placentae. At least 85 TGCs were counted for each placenta. (F) IF images of placentae using CRE-specific (green) and BrdU-specific (red) antibodies; slides were counterstained with Hoechst 33342 (blue). BrdU was injected 1 h prior to dissection. Arrowheads indicate TGCs stained positive for both CRE and BrdU. (G) IHC for Cyclin A2 showing comparable expression in TGCs of E10.5 control (Orc1f/f) and Plf-Orc1f/f placentae. Bars: C,D,F,G, 50 µm. (n) Number of mice in each group.
We sought to rule out the unlikely but formal possibility that only one of the two floxed alleles might have been deleted in Plf-Orc1f/f placentae. To this end, we evaluated intercrosses between Plfcre/+ and Orc1f/Δ mice, which contain one floxed Orc1 allele and one germline-deleted Orc1 allele. This analysis showed that, like Plf-Orc1f/f embryos, Plf-Orc1f/Δ embryos could be carried to term and that all pups born were without any apparent abnormalities (Supplemental Fig. S4B,C). Together, these findings demonstrate that ORC1 is dispensable for TGC endoreduplication, development, and function.
Perinatal ORC1 deficient hepatocytes endoreduplicate prematurely
We then turned our attention to the role of ORC1 in hepatocytes of the liver, one of the few endoreduplicating cell types in adult animals. Endoreduplication of their genome begins at the time of weaning and continues through adulthood (Epstein 1967; Gentric and Desdouets 2014). By the age of 12 mo, mice acquire hepatocytes with polyploid genomes that range between 8C and 16C. We used the Albumin-Cre (Alb) transgene to target hepatocyte-specific recombination of the floxed Orc1 allele in perinatal livers (Postic and Magnuson 2000). Examination of newborn, infant, and adult Alb-Orc1f/f mice revealed livers with grossly normal appearance and weight at each age examined (Fig. 5A,B; Supplemental Fig. S5A). Quantification of H&E-stained Orc1-deficient liver sections showed fewer hepatocytes, but these cells were strikingly larger with correspondingly larger nuclei than controls (Fig. 5C,D; Supplemental Fig. S5F). Flow cytometry analysis revealed an increased proportion of hepatocytes with 4C, 8C, and 16C DNA content in Orc1-deficient livers when compared with controls (Fig. 5E; Supplemental Fig. S5B,C). Moreover, quantification of polyploidy (≥8C) showed that Orc1-deficient livers began to endoreduplicate hepatocyte genomes earlier in life (by 1 wk of age) and reached maximum ploidy by 6-mo of age, well in advance of control mice (Fig. 5F; Supplemental Fig. S5D). Consistent with precocious endoreduplication, the number of KI-67-, BrdU-, and P-H3-positive (Ser10 and Ser28) hepatocytes was dramatically increased in Orc1-deficient livers (Fig. 5G–J; Supplemental Fig. S5E). Thus, mutant Orc1Δ hepatocytes had the capacity to replicate their genomic DNA, accumulating ≥8C DNA content well before weaning.
Figure 5.
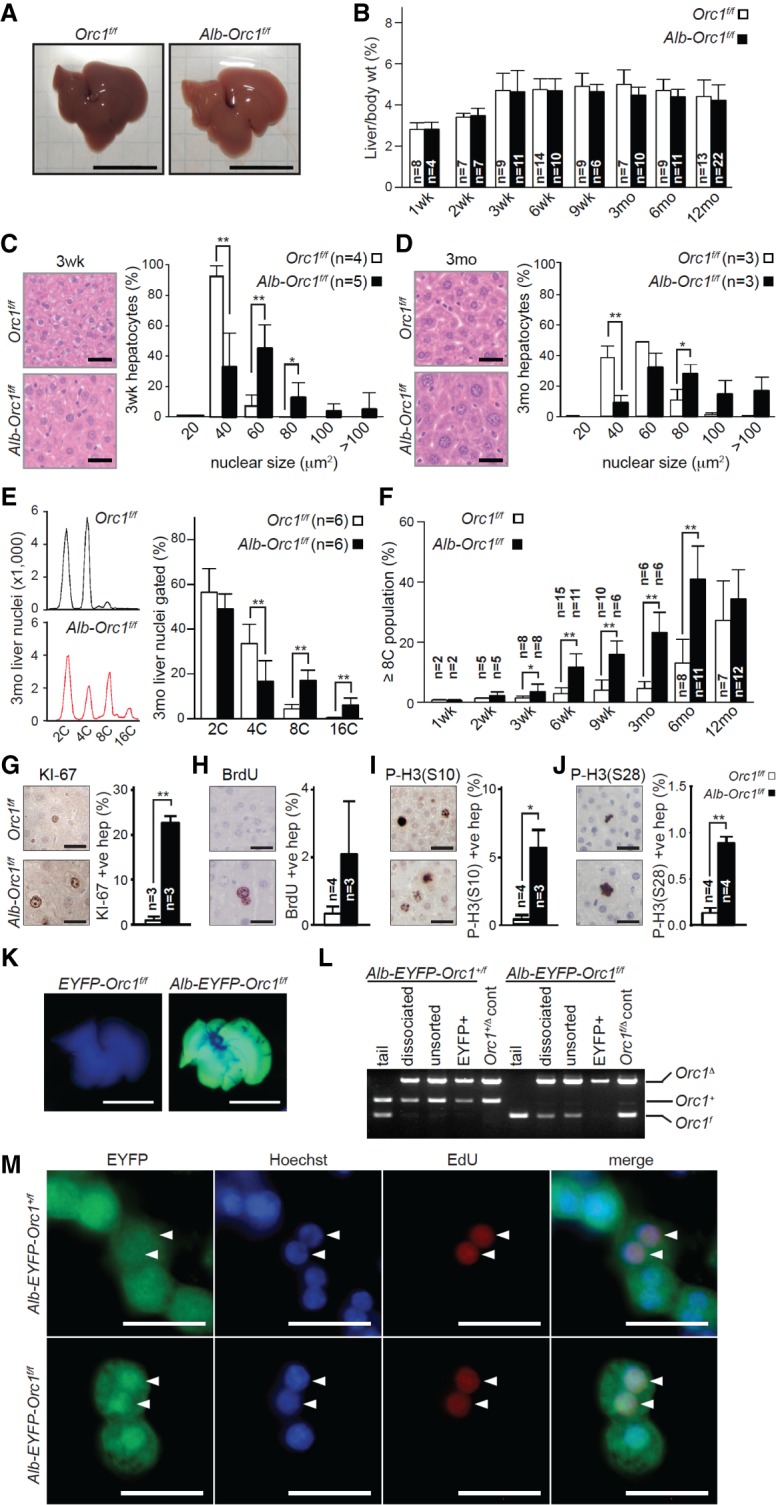
Orc1 ablation promotes polyploidization of hepatocytes. (A) Representative pictures of Orc1f/f (control) and Alb-Orc1f/f livers from 6-wk-old mice. (B) The ratio of liver to body weights of mice with the indicated ages. (C,D) Representative images of H&E-stained liver sections (left) and quantification of nuclear size in hepatocytes (right) from 3-wk-old (C) and 3-mo-old (D) mice. At least 300 hepatocyte nuclei were measured for each liver. (E) Representative FACS profiles of PI-stained liver nuclei from 3-mo-old mice (left panels) and quantification of DNA content (right panel). (F) The proportion of polyploid nuclei with ≥8C DNA content. (G–J) Representative IHC images and quantification results for KI-67-positive (G), BrdU-positive (H), Histone H3 phosphorylated at Ser 10 [P-H3 (S10)]-positive (I), and Histone H3 phosphorylated at Ser28 [P-H3 (S28)]-positive (J) hepatocytes in liver sections from 6-wk-old mice. At least 410 hepatocytes were counted for each liver. BrdU was injected 2 h prior to harvesting livers. (K) Ubiquitous EYFP expression in a 2-wk-old Alb-Orc1f/f liver carrying the ROSA26stop-EYFP reporter. (L) PCR genotyping in the FACS-sorted EYFP-expressing hepatocytes from 6-wk-old Alb-Orc1f/f mice carrying ROSA26stop-EYFP. (M) IF images showing DNA-replicating hepatocytes. Hepatocytes isolated from 3-wk-old Alb-Orc1+/f (control) and Alb-Orc1f/f mice carrying ROSA26stop-EYFP were cultured for 3 h in the presence of EdU. The fixed cells were stained with anti-GFP (EYFP; green) followed by EdU (red) detection. Hoechst 33342 (blue) was used as a counterstain. The results are shown as average + SD. (*) P ≤ 0.05; (**) P ≤ 0.01, t-test. Bars: A,K, 1 cm; C,D,G–J,M, 50 µm. (n) Number of mice in each group.
In the absence of ORC1-specific antibodies suitable for IHC, we performed several control experiments to assess the recombination efficiency of the floxed alleles in Alb-Orc1f/f mice. First, we introduced the ROSA26stop-EYFP reporter allele into Alb-Orc1f/f and Orc1f/f control mice. Gross visualization of Alb-Orc1f/f; ROSA26stop-EYFP mice, in contrast to Orc1f/f; ROSA26stop-EYFP mice, displayed completely green fluorescent livers (Fig. 5K). IHC of livers showed that the vast majority of hepatocytes from Orc1-deficient livers expressed CRE as well as the reporter marker EYFP (Supplemental Fig. S5G,H), confirming efficient CRE-mediated recombination activity in these cells. Furthermore, sorted EYFP-positive hepatocytes from 6-wk-old Alb-Orc1f/f; ROSA26stop-EYFP livers contained the recombined Orc1Δ allele and lacked any detectable levels of the floxed Orc1f allele (Fig. 5L). Importantly, EYFP-positive hepatocytes from Alb-Orc1f/f liver incorporated EdU in vitro (3.3% of EdU/EYFP-double-positive hepatocytes in Alb-Orc1f/f, n = 241 EYFP+ cells; 3.8% in Alb-Orc1+/f, n = 160 EYFP+ cells) (Fig. 5M). Finally, we also analyzed Alb-Orc1f/Δ mice, which contain one floxed and one germline-deleted Orc1 allele, and observed that these livers had normal weight and appearance (Supplemental Fig. S5I,J). As anticipated, hepatocytes in Alb-Orc1f/Δ mice were fewer in number but bigger in size than controls, with larger nuclei containing ≥8C DNA content (Supplemental Fig. S5K–M). Hence, by all measures assessed, hepatocytes from Alb-Orc1f/Δ and Alb-Orc1f/f mice displayed similar phenotypes, with premature endoreduplication and highly polyploid genomes. Together, these additional lines of evidence demonstrate that endoreduplication of the genome in hepatocytes can proceed in the absence of functional ORC1.
Postnatal function of ORC1 in hepatocytes is confined to the first 2 wk of life
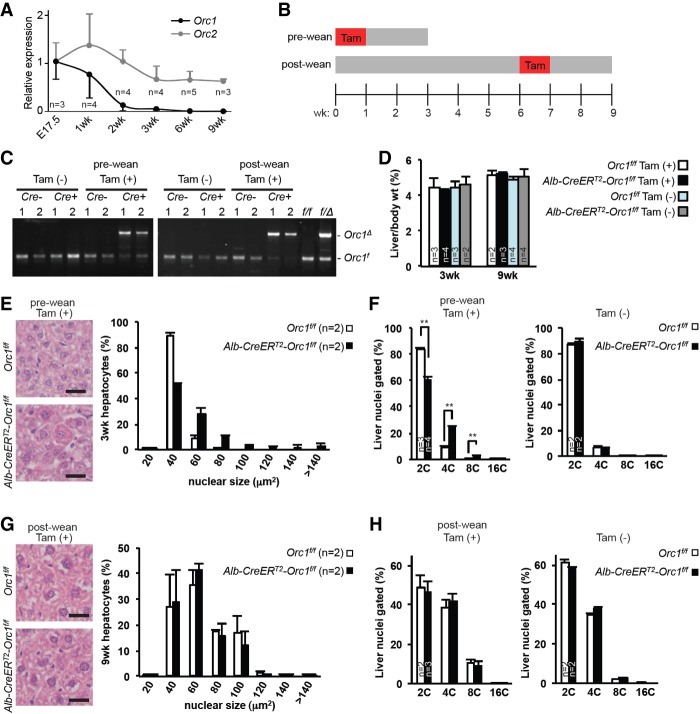
During the first 3 wk of life (prior to weaning), diploid hepatocytes divide vigorously in order to increase liver size proportionally to the rest of the body. After this initial rapid expansion, hepatocytes begin to transition from the use of mitotic cell divisions to endoreduplication as a mechanism of cell growth, which leads to a gradual increase in liver size and weight. Interestingly, we found that expression of Orc1 mRNA was relatively high in embryonic livers (E17.5), decreased after the first 2 wk of life, and was virtually undetectable by 3 wk of age (Fig. 6A). Expression of Orc2 was maintained throughout the life spans of the mice (Fig. 6A).
Figure 6.
Orc1 deletion promotes precocious polyploidization in neonatal mice. (A) Quantitative RT–PCR of Orc1 and Orc2 mRNA expression in the livers at various ages. Expression of Orc1 and Orc2 was normalized to Gapdh. (B) Experimental scheme of temporal Orc1 deletion using Alb-CreERT2. Orc1f/f (control) and Alb-CreERT2-Orc1f/f mice were fed tamoxifen chow (Tam) for 7 d starting at birth (fed to dam; pre-wean; harvested at 3 wk of age) or at 6 wk of age (post-wean; harvested at 9 wk of age). (C) Genotyping PCR detection of Orc1 deletion in the livers of Alb-CreERT2-Orc1f/f mice after feeding with tamoxifen chow. (D) The ratio of liver to body weights of 3-wk-old mice (prewean) and 9-wk-old mice (post-wean) with (+) or without (−) tamoxifen feeding. (E,G) Representative images of H&E-stained liver sections (left panels) and quantification of hepatocyte nuclear size (right panels) of prewean (E) and post-wean (G) tamoxifen-fed mice. At least 600 hepatocyte nuclei were measured for each liver. (F,H) Quantification of liver ploidy by FACS in prewean (F) and post-wean (H) mice with or without tamoxifen feeding. The results are shown as average + SD. (**) P < 0.01, t-test. Bars: E,G, 50 µm. (n) Number of mice in each group.
The observation that decreased Orc1 expression in hepatocytes coincided with the switch to endocycles prompted us to evaluate the temporal role of ORC1 in hepatocytes prior to and following weaning age. We used a tamoxifen-inducible Albumin-Cre transgene (Alb-CreERT2) (Schuler et al. 2004) and tamoxifen chow to delete Orc1 in hepatocytes within the first week of life (when diploid hepatocytes are rapidly dividing) or after 6 wk of age (when some hepatocytes are endoreduplicating) (see the diet regimen in Fig. 6B). To this end, Alb-CreERT2;Orc1f/f mice (Cre+) and control Orc1f/f littermates (Cre−) were fed the tamoxifen chow at either birth (preweaning) or 6 wk of age (post-weaning) for a period of 1 wk and then harvested 2 wk later. Deletion of Orc1 was confirmed by PCR analysis of genomic DNA (Fig. 6C). Liver weights were similar among all mouse groups regardless of genotype or diet regimen fed (Fig. 6D). Hepatocyte-specific ablation of Orc1 at birth resulted in livers with fewer hepatocytes containing visibly larger nuclei (Fig. 6E), accumulating ≥4C DNA content by the time mice were weaned (Fig. 6F). Interestingly, feeding Alb-CreERT2;Orc1f/f mice the tamoxifen chow at 6 wk of age did not impact hepatocyte size or ploidy levels beyond those normally observed in control mice (Fig. 6G,H). Together, these analyses temporally defined a role for ORC1 during the first 2 wk of life in replicating hepatocyte diploid genomes and in preventing premature endoreduplication prior to weaning.
Adult ORC1-deficient hepatocytes support liver regeneration
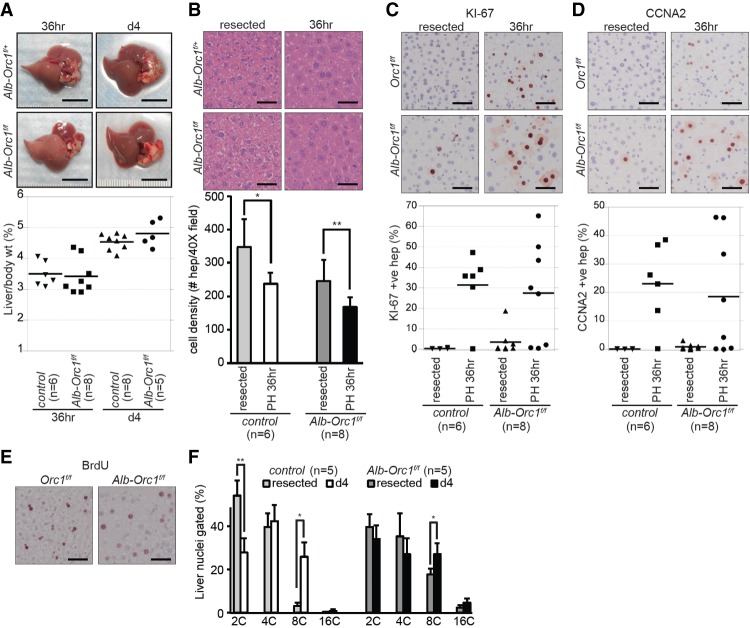
One of the remarkable features of mammalian livers is their capacity to regenerate rapidly after a major injury. Damaged livers engage a number of cellular response mechanisms, including cellular hypertrophy, proliferation, and endoreduplication in order to restore their mass and function (Gupta 2000; Mitchell and Willenbring 2008). We considered the possibility that ORC1 might provide damaged livers with an advantage to regenerate by better equipping one or more of these mechanisms. To this end, we assessed the ability of Orc1-deficient livers to regenerate following the surgical resection of the left lateral and median lobes. In both control (Alb-Orc1+/f or Orc1f/f) and Orc1-deficient (Alb-Orc1f/f) mice, liver weights increased during the first 36 h following the partial hepatectomy (Fig. 7A). Histological analysis showed hepatocyte hypertrophy characterized by increased cell size and significantly decreased cell density in both control and Orc1-deficient livers (Fig. 7B). Measurement of KI-67, BrdU incorporation, and Cyclin A2 expression showed overall marked hepatocyte proliferation in regenerating livers regardless of genotype (Fig. 7C–E). Fluorescent-activated cell sorting (FACS) analysis confirmed increased DNA content in hepatocytes within the regenerating lobe when compared with the surgically removed lobe (Fig. 7F). By 4 d after partial hepatectomy, all liver weights were restored to their original presurgery state (Fig. 7A). These results suggested that both control and Orc1-deficient livers regenerate equally well following a major injury and that regeneration involves the replication of the hepatocyte genome. We noted, however, that some Orc1-deficient livers lacked proliferating hepatocytes soon following the partial resection of the liver (36 h) even though liver mass was restored effectively after 4 d (Fig. 7C,D, 36 h). Hypertrophy of hepatocytes is known to be the initial response for liver regeneration following a partial hepatectomy (Miyaoka et al. 2012). Thus, we presume that Orc1-deficient hepatocytes may have an enhanced ability (over control animals) to support cellular hypertrophy, explaining the slight delay in some animals for mutant hepatocytes to enter S phase following an acute injury.
Figure 7.
Normal liver regeneration in Orc1-deficient livers. (A) Representative pictures of regenerating livers 36 h and 4 d after partial hepatectomy (PH). The restoration of liver size, as shown by the ratio of liver to body weight in both control (Alb-Orc1+/f or Orc1f/f) and Alb-Orc1f/f mice after partial hepatectomy. (B) Representative images of H&E-stained liver sections of the resected left lateral lobe (resected) and the regenerating right lobe 36 h after partial hepatectomy (36 h) and quantification of hepatocyte number per 40× image field. At least six images were counted for each liver. (C–E) IHC for KI-67 (C), Cyclin A2 (CCNA2) (D), and BrdU (E) showing comparable cell cycle entry of hepatocytes at 36 h after partial hepatectomy in control and Alb-Orc1f/f livers. Quantification results of KI-67- and CCNA2-positive hepatocytes are shown as dot plot graphs. At least 530 hepatocytes were counted for each liver. (F) Quantification of PI-stained liver nuclei showing significant ploidy increase at 4 d after partial hepatectomy in both control and Alb-Orc1f/f livers. The results are shown as average + SD. (*) P ≤ 0.05; (**) P ≤ 0.01, t-test in B and F. Bars: A, 1 cm; B–E, 50 µm. (n) Number of mice in each group.
Discussion
Precise duplication of the genome requires origins of DNA replication to fire once and only once per cell cycle. ORC1 is the only mammalian ORC subunit to function as an ATPase and thus is essential for assembly of the pre-RC, licensing origins, and preventing their rereplication. We generated mice with a conditional mutant allele of Orc1 lacking the ATPase domain and found that ORC1 function is required for DNA replication in diploid genomes but is dispensable for replication in polyploid genomes. These observations suggest that fundamentally different mechanisms regulate the initiation of DNA replication in mitotic cell cycles and endocycles.
Two previous studies indicating that some cells have the ability to replicate their DNA genomes with reduced ORC1 protein levels remain controversial (Park and Asano 2008; Shibata et al. 2016). We provide exhaustive in vivo evidence that mammalian cells with the capacity to endoreduplicate can replicate their DNA without ORC1 function. While feasible, these observations do not mean that endocycles normally proceed without ORC1. A hint that this may be the case, however, came from the temporal analysis of Orc1 expression in developing livers. While abundant during the first 2 wk of life, Orc1 mRNA levels precipitously decrease and are undetectable by 3 wk of age, coinciding with the transition to scheduled endoreduplication as a mechanism of liver growth. A second hint suggesting that ORC1-independent DNA replication may be a physiological response came from the unusual observation made here that ablation of ORC1 function in hepatocytes within the first week of life, but not later in adolescence, expedited precocious endoreduplication, yielding aberrantly large hepatocytes with polyploid genomes well before developmentally scheduled. Thus, we propose that developmentally scheduled endocycles proceed normally, at least in hepatocytes of mice, in the absence of ORC1. Clearly, additional work will be required to fully describe and expose the role and mechanisms of ORC1 and other ORC proteins in the replication of diploid and other variant mammalian genomes.
Why would cells need to use ORC1-dependent and ORC1-independent mechanisms for regulating DNA replication? Replication and segregation of diploid genomes are essential processes required for the accurate passage of genetic information from progenitor cells to their cell derivatives and from parents to progeny. We propose that while not strictly required for DNA replication per se, ORC1 in diploid cells likely plays a regulatory role in the engagement of the pre-RC and timely firing of origins, ensuring the accurate replication of the complete genome. Normally, in the absence of ORC1, the accumulation of genomic alterations in diploid cells would be expected to engage checkpoint controls that lead to either cell cycle arrest or apoptosis, ensuring the effective elimination of damaged cells from tissues and/or progeny. In some special cases, however, cells respond to developmental or external environmental cues by replicating their genome without an intervening mitosis, yielding polyploid cells. Interestingly, these variant cells often contain underreplicated or overreplicated regions of the genome. For example, in Drosophila salivary glands, cells with polytene chromosomes have vastly underreplicated regions of the genome (Nordman et al. 2011; Sher et al. 2013). In mice, certain regions of the placental TGC genome are underreplicated (6%), while other regions are amplified (0.2%) (Hannibal et al. 2014; Hannibal and Baker 2016). Similarly, mature hepatocytes of mouse livers have polyploid genomes that are aneuploid (Duncan et al. 2010; Duncan 2013). Remarkably, the accumulation of genetic alterations fails to engage checkpoint controls in these endoreduplicating cells, and thus polyploid cells avoid being eliminated and instead participate in the normal physiology of organ functions (Duncan et al. 2012). It would thus appear that some specialized endocycling cells in nature, perhaps in an attempt to effectively respond to heightened metabolic demands, have chosen an ORC1-independent mechanism to replicate their genome but at the cost of diminished genetic integrity.
A simple model could be that there is an endocycle-specific functional ORC1 homolog, protein X, that recruits the MCM helicase complex to replication origins. Arabidopsis has two ORC1 proteins (ORC1a and ORC1b), which are highly conserved at the amino acid level but differentially expressed in either proliferating cells or endoreduplicating cells with distinct promoter sequences (Diaz-Trivino et al. 2005). Another possibility is that Cdc6 could still bind to ORC2–5 in the absence of ORC1 and form a ring-shaped complex, which encircles the dsDNA to support MCM loading even if it is not like the completely closed structure shown by Tocilj et al. (2017). Our future task would be to elucidate how MCM helicase complexes are loaded onto DNA without ORC1 and whether ORC1 is involved in pre-RC formation in normal endoreduplicating cells in the presence of ORC1.
The observation that premature endoreduplication occurs when Orc1 is deleted in the hepatocytes of young mice was particularly intriguing. Premature endoreduplication was observed only when Orc1 was deleted in mitotic cells and not in cells already committed to or undergoing endoreduplication, such as TGCs (Plfcre) and hepatocytes in post-weaning age mice. In conjunction with this, Orc1 mRNA is highly expressed in mitotic populations such as in embryonic livers and during liver regeneration, while the expression of other Orc subunits is more static. Together these findings suggest that ORC1 functions as a suppressor to prevent cells from entering the endocycle and likely has novel and unique function distinct from other ORC subunits.
Materials and methods
Mice
Mouse protocols were approved by the Institutional Animal Care and Use Committee at the Ohio State University and Medical University of South Carolina. The mouse BAC library was screened to identify the clones containing the Orc1 genomic region, and two clones (RP22-544B10 and RP22-574E14) were used to make the targeting vector. By retrieving from the BAC clones, the long homology arm (6.5 kb, introns 4–8) and floxed region (6.0 kb, exons 9–14) were introduced into PL451 plasmid containing the Neo cassette flanked by two FRT sites and one LoxP site, and the short homology arm (4.3 kb, intron 14) was introduced into PL253 plasmid containing the thymidine kinase cassette. The modified PL451 and PL253 plasmids were processed for restriction enzyme digestion and combined to obtain the Orc1 targeting vector. The linearized targeting vector was electroporated into ES cells. The ES cells were cultured in the presence of G418 and Ganciclovir and screened for identification of the correctly targeted clones by Southern blot using the 3′ probe amplified by PCR (561 base pairs [bp]; forward primer, 5′-TCACAGCTGCCTGTCAATTC-3′; reverse primer, 5′-GAACTCTGGACCCACCCATA-3′) and genotyping PCR using primers for 5′ solitary LoxP (wild-type allele [439 bp] and floxed allele [310 bp]; F1, 5′-GCTGCTTCAGTGTGGCAATA-3′; R1, 5′-CTCCAATTGTTCCCCAGCTA-3′) and the Neo cassette (wild-type allele [846 bp] and targeted Neo allele [2572 bp]; F2, 5′-TCTTTAGAAGGCCACGAGGA-3′; R2, 5′-CACCTGTCACTGGACCACAC-3′). The floxed PCR product is smaller than the wild-type one due to 175-bp deletion in intron 8 when the retrieval vector was made. Two correctly targeted clones were independently injected into blastocysts and then transplanted into pseudopregnant females to obtain chimeric mice. The chimeras were bred to C57BL/6 mice to obtain germline-transmitted mice (Orc1+/neo). The targeted Orc1+/neo mice were then bred to Actin-FLPe mice (Rodríguez et al. 2000) to obtain floxed Orc1 mice (Orc1+/f) after removal of the Neo cassette or Sox2-Cre mice (Hayashi et al. 2002) to obtain mice that have the Orc1-deleted allele (Orc1+/Δ). Floxed Orc1 mice were crossed to tissue-specific Cre mice (Ah-Cre, PlfCre, Alb-Cre, and Alb-CreERT2). The mice carrying ROSA26stop-EYFP were interbred to floxed Orc1 mice for usage of EYFP expression as a reporter for Cre recombinase activity and Orc1 deletion. For Orc1 deletion in the small intestine, 2-mo-old mice with or without inducible Ah-Cre (Ireland et al. 2004) received intraperitoneal injections of 80 mg/kg βNF (Sigma) dissolved in 80 mg/mL corn oil for five consecutive days. For Orc1 deletion using inducible Alb-CreERT2 (Schuler et al. 2004), mice were fed tamoxifen chow for 7 d at birth (administered to the dam) or at 6 wk of age. BrdU (Sigma) dissolved in 10 mg/mL sterile PBS was intraperitoneally injected at 100 mg per kilogram of body weight 1 or 2 h prior to harvesting tissues. All of the mice used in this study were maintained in a mixed background (129xC57BL/6x and FVB/N).
Fluorescence imaging of Orc1-deleted livers
The EYFP expression in the livers of Orc1f/f or Alb-Orc1f/f mice carrying the ROSA26stop-EYFP reporter was visualized, and images were taken via Cy2 channel using ChemiDoc MP (Bio-Rad). The far-red channel was used to visualize the livers.
Cell culture
E3.5 blastocysts were flushed from the uterus and individually cultured in 12-well plates in DMEM containing 10% FBS. After 6–7 d in vitro, expanded clones and unhatched embryos were lysed to isolate genomic DNA for genotyping PCR. MEFs were isolated from E13.5 Orc1f/f embryos and infected with either empty (pBP; Cre−) or Cre-expressing retrovirus (pBP-Cre; Cre+) vector. The infected Orc1f/f MEFs were cultured for 2–4 d in the presence of puromycin and replated for further experiments. For BrdU assay, MEFs were cultured for 3 h in the presence of 10 µM BrdU.
Isolation of hepatocytes
Alb-Orc1+/f (control) and Alb-Orc1f/f mice carrying ROSA26stop-EYFP were euthanized using CO2 and perfused with prewarmed (39°C) Hank's buffered salt solution (HBSS) containing EGTA for 10 min and then with prewarmed HBSS containing CaCl2 and 100 U/mL collagenase type II for 10 min. After perfusion, the livers were dissected out into a dish, minced with a razor blade, and further dissociated by gentle pipetting in the collagenase–HBSS solution. A single-cell suspension was obtained by filtering through a 100-µm cell strainer and then centrifuged at 50g for 3 min at 4°C. The cell pellet was resuspended in HBSS containing CaCl2 and washed four times. The final cell pellet was resuspended in ice-cold DMEM containing 20% FBS and processed for EYFP-positive cell sorting using Aria III (BD Biosciences).
Partial hepatectomy
Partial hepatectomy was performed as described previously (Mitchell and Willenbring 2008). In brief, two-thirds partial hepatectomy (resection of the left lateral and median lobes) was performed in 10- to 14-wk-old control (Alb-Orc1+/f or Orc1f/f) and Alb-Orc1f/f mice under anesthesia by isoflurane inhalation. The resected left lateral lobe was fixed in 10% formalin for histological analysis, and the median lobe was flash-frozen in liquid nitrogen for ploidy analysis. BrdU was injected 2 h prior to harvesting the regenerating liver after partial hepatectomy, and only the right lobes were used for further analysis.
PCR
Mice were genotyped using PCR primers for Orc1 (F1 forward, 5′-GCTGCTTCAGTGTGGCAATA-3′; R1 reverse, 5′-CTCCAATTGTTCCCCAGCTA-3; R2 reverse for deleted allele, 5′-CACCTGTCACTGGACCACAC-3′), Actin-FLPe (forward, 5′-CACTGATATTGTAAGTAGTTTGC-3′; reverse, 5′-CTAGTGCGAAGTAGTGATCAGG-3′), Sox2-Cre (forward, 5′-ATGCTTCTGTCCGTTTGCCG-3′; reverse, 5′-CCTGTTTTGCACGTTCACCG-3′), Ah-Cre (forward, 5′-CCTGACTAGCATGGCGATAC-3′; reverse, 5′-CCTGTTTTGCACGTTCACCG-3′), Plf-Cre (5′-CACAGCTAAGCCTGGGTAGG-3′; 5′-TTAATCAGTCTTCTTCATTCCTGA-3′; 5′-CGGTTATTCAACTTGCACCA-3′), Alb-Cre (wild-type forward, 5′-TGCAAACATCACATGCACAC-3′; mutant forward, 5′-GAAGCAGAAGCTTAGGAAGATGG-3′; common reverse, 5′-TTGGCCCCTTACCATAACTG-3′), Alb-CreERT2 (5′-GGAACCCAAACTGATGACCA-3′; 5′-TTAAACAAGCAAAACCAAAT-3′; 5′-ATCATTTCTT-TGTTTTCAGG-3′), and ROSA26 (common forward, 5′-AAAGTCGCTCTGAGTTGTTAT-3′; wild-type reverse, 5′-GGAGCGGGAGAAATGGATAT-3′; mutant reverse, 5′-GCGAAGAGTTTGTCCTCAACC-3′).
Quantitative RT–PCR
MEFs and frozen livers were processed for total RNA isolation using either TRIzol reagent (Invitrogen) or Nucleospin RNA II kit (Clontech). Quantitative RT–PCR was performed using iQ SYBR Green Supermix (Bio-Rad) and primers for Gapdh (forward, 5′-CGGTGTGAACGGATTTGGC-3′; reverse, 5′-TTTGATGTTAGTGGGGTCTCGC-3′), Orc1 (forward, 5′-TTTGAAGCGGATTAGGCAAC-3′; reverse, 5′-GTCTCGGATCTGAGGAGTGG-3′), Orc1-5′ (forward, 5′-TACGTGTTGTAGCCGTGGTC-3′; reverse, 5′-CTGCCAACCCATGAGAAAGT-3′), Orc1-3′ (forward, 5′-CGTCGGTCAGGACTAGAGGA-3′; reverse, 5′-CAGCACGTCATTCTGGCTAA-3′), and Orc2 (forward, 5′-GGAGAGCCAGAGTGGATCAG-3′; reverse, 5′-CCCAAGCCATAAAGCACAAT-3′) in triplicate samples. The results were normalized to Gapdh expression in each sample.
Ploidy analysis
Liver ploidy analysis was done as described previously (Mayhew et al. 2005). In brief, frozen liver was minced in ice-cold buffer A (25 mM Tris-HCl at pH 7.5, 50 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1 mM phenylmethanesulfonyl fluoride [PMSF]) followed by incubation for 5–10 min on ice. Liver nuclei were released using a glass Dounce homogenizer, washed twice with ice-cold buffer A and once with ice-cold PBS, and then resuspended in PBS containing 0.1% Triton X-100, 50 µg/mL propidium iodide (PI), and 100 µg/mL RNase A. PI-stained liver nucleus samples were processed to FACS using LSRII (BD Biosciences). For ploidy analysis of TGCs, paraffin-embedded E10.5 placenta sections were processed for Feulgen staining, and DNA content of TGCs was quantified using ImageJ as described previously (Chen et al. 2012).
Histology and immunostaining
Tissues were fixed in 10% formalin for 2 d and embedded in paraffin. Sections 5 μm thick were processed for H&E staining, IHC, or IF. The sections were incubated with primary antibodies against BrdU (Millipore, NA61), biotin-BrdU (Invitrogen, 03-3940), cytokeratin 8 (Developmental Studies Hybridoma Bank, TROMA-I), cleaved Caspase-3 (Cell Signaling, 9661), phospho-Histone H3 (Ser10; Millipore, 06-570), phospho-Histone H3 (Ser28; Sigma, H9908), KI-67 (Leica, NCL-Ki67p; BD Biosciences, 550609), Cyclin A2 (Santa Cruz Biotechnology, sc-596), Cre (Covance, PRB-1061C), or GFP (Abcam, ab6556). For detection of BrdU, the sections were treated in 2 M HCl for 10 min at 37°C prior to incubation with BrdU antibodies. Biotinylated secondary (Vector Laboratories) or fluorescent-conjugated secondary (Alexa fluor 488 or Alexa fluor 594, Invitrogen) antibodies were used for detection. Hoechst 33342 (10 µg/mL) was used as a counterstain for IF.
Immunoblotting
Whole-protein lysates from MEFs, small intestines, and livers were separated in either 10% or 9%–15% discontinuous acrylamide gel and transferred to PVDF membrane (Immobilon-FL, PDVF, Millipore). The membrane was blocked in 5% skim milk–TBST (20 mM Tris-HCl at pH 7.4, 150 mM NaCl, 0.1% Tween-20) for 30 min at room temperature and then incubated with primary antibodies against MCM3 (Santa Cruz Biotechnology, sc-9850), ORC1 (kindly provided by Dr. Hanaoka and Dr. Mizuno, RIKEN), α-Tubulin (Santa Cruz Biotechnology, sc-5286), GAPDH (Santa Cruz Biotechnology, sc-25778), PCNA (Cell Signaling, 13110), GFP (Cell Signaling, 2555), and phospho-Histone H3 (Ser10; Millipore, 06-570) overnight at 4°C. HRP-conjugated secondary antibodies and SuperSigmal WestPico chemiluminescent substrate (Thermo Scientific) were used for detection.
Supplementary Material
Acknowledgments
We thank J. Bice and D. Bryant for assistance with histology, and Dana LeMoine for assistance with the partial hepatectomies. This work was supported by the Ohio State University Comprehensive Cancer Center Genetically Engineered Mouse Modeling, Genomics, and Analytical Cytometry Core Shared Resources. We are grateful to R.P. Wharton for critical suggestions. We thank F. Hanaoka and T. Mizuno (RIKEN) for providing the mouse ORC1 antibodies. This work was funded by National Institutes of Health grants to G.L. (R01CA121275). T.O.-U. was a recipient of Pelotonia Post-doctoral Fellowship. This manuscript is dedicated in lasting memory of Dr. M. Asano, a wonderful colleague and mentor greatly missed.
Author contributions: T.O.-U., L.N.K., M.M.O., M.A., and G.L. designed the experiments. All authors performed the experiments and collected and analyzed data. M.A. and G.L. supervised the studies. T.O.-U., L.N.K., and G.L. wrote the manuscript with input from all authors.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.311910.118.
References
- Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C, Martin C-A, Yeyati P, Al Sanna N, Bober M, et al. 2011. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet 43: 350–355. [DOI] [PubMed] [Google Scholar]
- Chen H-Z, Ouseph MM, Li J, Pécot T, Chokshi V, Kent L, Bae S, Byrne M, Duran C, Comstock G, et al. 2012. Canonical and atypical E2Fs regulate the mammalian endocycle. Nat Cell Biol 14: 1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I, Remus D, Botchan M. 2001. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc Natl Acad Sci 98: 11997–12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A. 2006. Regulating the licensing of DNA replication origins in metazoa. Curr Opin Cell Biol 18: 231–239. [DOI] [PubMed] [Google Scholar]
- Diaz-Trivino S, del Mar Castellano M, de la Paz Sanchez M, Ramirez-Parra E, Desvoyes B, Gutierrez C. 2005. The genes encoding Arabidopsis ORC subunits are E2F targets and the two ORC1 genes are differently expressed in proliferating and endoreplicating cells. Nucleic Acids Res 33: 5404–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW. 2013. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol 24: 347–356. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, Grompe M. 2010. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467: 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Hanlon Newell AE, Bi W, Finegold MJ, Olson SB, Beaudet AL, Grompe M. 2012. Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest 122: 3307–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Zielke N, Gutierrez C. 2014. Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat Rev Mol Cell Biol 15: 197–210. [DOI] [PubMed] [Google Scholar]
- Epstein CJ. 1967. Cell size, nuclear content, and the development of polyploidy in the mammalian liver. Proc Natl Acad Sci 57: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ. 2013. Endoreplication and polyploidy: insights into development and disease. Development 140: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentric G, Desdouets C. 2014. Polyploidization in liver tissue. Am J Pathol 184: 322–331. [DOI] [PubMed] [Google Scholar]
- Grallert B, Nurse P. 1996. The ORCI homolog orpl in fission yeast plays a key role in regulating onset of S phase. Genes Dev 10: 2644–2654. [DOI] [PubMed] [Google Scholar]
- Gupta S. 2000. Hepatic polyploidy and liver growth control. Semin Cancer Biol 10: 161–171. [DOI] [PubMed] [Google Scholar]
- Hannibal RL, Baker JC. 2016. Selective amplification of the genome surrounding key placental genes in trophoblast giant cells. Curr Biol 26: 230–236. [DOI] [PubMed] [Google Scholar]
- Hannibal RL, Chuong EB, Rivera-Mulia JC, Gilbert DM, Valouev A, Baker JC. 2014. Copy number variation is a fundamental aspect of the placental genome. PLoS Genet 10: e1004290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP. 2002. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev 119S: S97–S101. [DOI] [PubMed] [Google Scholar]
- Hu D, Cross JC. 2010. Development and function of trophoblast giant cells in the rodent placenta. Int J Dev Biol 54: 341–354. [DOI] [PubMed] [Google Scholar]
- Ireland H, Kemp R, Houghton C, Howard L, Clarke AR, Sansom OJ, Winton DJ. 2004. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of β-catenin. Gastroenterology 126: 1236–1246. [DOI] [PubMed] [Google Scholar]
- Lee HO, Davidson JM, Duronio RJ. 2009. Endoreplication: polyploidy with purpose. Genes Dev 23: 2461–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, Babcock GF, Lentsch AB, Fukasawa K, Knudsen ES. 2005. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res 65: 4568–4577. [DOI] [PubMed] [Google Scholar]
- Mitchell C, Willenbring H. 2008. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc 3: 1167–1170. [DOI] [PubMed] [Google Scholar]
- Miyake Y, Mizuno T, Yanagi KI, Hanaoka F. 2005. Novel splicing variant of mouse Orc1 is deficient in nuclear translocation and resistant for proteasome-mediated degradation. J Biol Chem 280: 12643–12652. [DOI] [PubMed] [Google Scholar]
- Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. 2012. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol 22: 1166–1175. [DOI] [PubMed] [Google Scholar]
- Nordman J, Li S, Eng T, MacAlpine D, Orr-Weaver TL. 2011. Developmental control of the DNA replication and transcription programs. Genome Res 21: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouseph MM, Li J, Chen HZ, Pécot T, Wenzel P, Thompson JC, Comstock G, Chokshi V, Byrne M, Forde B, et al. 2012. Atypical E2F repressors and activators coordinate placental development. Dev Cell 22: 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Asano M. 2008. The origin recognition complex is dispensable for endoreplication in Drosophila. Proc Natl Acad Sci 105: 12343–12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C, Magnuson MA. 2000. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis 26: 149–150. [DOI] [PubMed] [Google Scholar]
- Rodríguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet 25: 139–140. [DOI] [PubMed] [Google Scholar]
- Schuler M, Dierich A, Chambon P, Metzger D. 2004. Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis 39: 167–172. [DOI] [PubMed] [Google Scholar]
- Sher N, Von Stetina JR, Bell GW, Matsuura S, Ravid K, Orr-Weaver TL. 2013. Fundamental differences in endoreplication in mammals and Drosophila revealed by analysis of endocycling and endomitotic cells. Proc Natl Acad Sci 110: 9368–9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata E, Kiran M, Shibata Y, Singh S, Kiran S, Dutta A. 2016. Two subunits of human ORC are dispensable for DNA replication and proliferation. Elife 5: e19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu M-Q, Qin Y-L, Jiang M-H. 2008. RNA interference targeting ORC1 gene suppresses the proliferation of vascular smooth muscle cells in rats. Exp Mol Pathol 84: 206–212. [DOI] [PubMed] [Google Scholar]
- Takehara M, Makise M, Takenaka H, Asano T, Mizushima T. 2008. Analysis of mutant origin recognition complex with reduced ATPase activity in vivo and in vitro. Biochem J 413: 535–543. [DOI] [PubMed] [Google Scholar]
- Tocilj A, On KF, Yuan Z, Sun J, Elkayam E, Li H, Stillman B, Joshua-Tor L. 2017. Structure of the active form of human origin recognition complex and its ATPase motor module. eLife 6: e20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.