Here, Huang et al. applied domain-focused CRISPR screening to small cell lung cancer to identify a novel molecular subtype defined by the expression of and dependency on transcription factor POU2F3. They also demonstrate that POU2F3 is expressed exclusively in variant SCLC tumors that lack expression of neuroendocrine markers, which instead express markers of a chemosensory lineage known as tuft cells.
Keywords: POU2F3, small cell lung cancer, tuft cell, master regulator, enhancer
Abstract
Small cell lung cancer (SCLC) is widely considered to be a tumor of pulmonary neuroendocrine cells; however, a variant form of this disease has been described that lacks neuroendocrine features. Here, we applied domain-focused CRISPR screening to human cancer cell lines to identify the transcription factor (TF) POU2F3 (POU class 2 homeobox 3; also known as SKN-1a/OCT-11) as a powerful dependency in a subset of SCLC lines. An analysis of human SCLC specimens revealed that POU2F3 is expressed exclusively in variant SCLC tumors that lack expression of neuroendocrine markers and instead express markers of a chemosensory lineage known as tuft cells. Using chromatin- and RNA-profiling experiments, we provide evidence that POU2F3 is a master regulator of tuft cell identity in a variant form of SCLC. Moreover, we show that most SCLC tumors can be classified into one of three lineages based on the expression of POU2F3, ASCL1, or NEUROD1. Our CRISPR screens exposed other unique dependencies in POU2F3-expressing SCLC lines, including the lineage TFs SOX9 and ASCL2 and the receptor tyrosine kinase IGF1R (insulin-like growth factor 1 receptor). These data reveal POU2F3 as a cell identity determinant and a dependency in a tuft cell-like variant of SCLC, which may reflect a previously unrecognized cell of origin or a trans-differentiation event in this disease.
Small cell lung cancer (SCLC) is associated with rapid cell growth, early metastatic spread, and a complete lack of target-based therapies (Semenova et al. 2015). Consequently, the current treatment of SCLC with chemotherapy, radiotherapy, and surgery is associated with a dismal 5-yr survival rate of 6%. Comprehensive genome sequencing of SCLC tumors has revealed a high mutational load in this disease, with most tumors possessing inactivating mutations or deletions of the tumor suppressors RB1 and TP53 but few actionable oncogene targets (George et al. 2015). Thus, a much-needed personalized medicine paradigm has yet to be implemented in this disease.
Experiments in genetically engineered mice suggest a pulmonary neuroendocrine cell of origin for SCLC (Semenova et al. 2015), which is supported by the expression of neuroendocrine differentiation markers chromogranin A (CHGA) and insulinoma-associated protein 1 (INSM1) in human SCLC tumors (Gazdar et al. 2017). In addition, the neuroendocrine lineage master regulator ASCL1 is expressed and essential in SCLC tumors and cell lines (Augustyn et al. 2014; Borromeo et al. 2016).
However, it has long been recognized that a subset of SCLC cell lines exhibits a “variant” phenotype associated with low expression of neuroendocrine markers (Gazdar et al. 1985). A recent transcriptome analysis of human SCLC tumors revealed that ∼20% of samples expressed low levels of neuroendocrine markers CHGA and ASCL1 but still possessed the cell morphology and genetic profile of SCLC (George et al. 2015). The molecular basis of this variable neuroendocrine differentiation is not well understood. Studies in cell lines and mice suggest that the neurogenic transcription factor (TF) NEUROD1 can function as an alternative master regulator to ASCL1 in neurodendocrinelow SCLC (Borromeo et al. 2016; Mollaoglu et al. 2017). Elevated expression of MYC and REST as well as activation of NOTCH signaling are also known to inhibit neuroendocrine differentiation in SCLC (Lim et al. 2017; Mollaoglu et al. 2017). Nevertheless, the biological and clinical significance of variable neuroendocrine differentiation in SCLC remains unclear.
POU2F3 (POU class 2 homeobox 3; also known as SKN-1a/OCT-11) is a TF required for the generation of a rare chemosensory cell type found in the gastrointestinal and respiratory tracts (Matsumoto et al. 2011; Yamaguchi et al. 2014; Gerbe et al. 2016; Yamashita et al. 2017). These cells are known by a variety of names (including tuft, brush, microvillous, caveolated, or multivesicular cells), but, for simplicity, we refer to POU2F3-expressing chemosensory cells as a “tuft cell” lineage throughout this study. Like neuroendocrine cells, tuft cells respond to external stimuli by releasing bioactive substances to regulate local epithelial and immune cell functions (Howitt et al. 2016; von Moltke et al. 2016). Despite their similar functions and anatomical locations, tuft cells are distinct from the neuroendocrine cell lineage (Kaske et al. 2007). In addition to POU2F3, recent studies have exposed a variety of tuft cell lineage markers, including the TFs SOX9, GFI1B, and ASCL2; the ion channel TRPM5; the villin family protein AVIL; and choline O-acetyltransferase (CHAT) (Kaske et al. 2007; Bezencon et al. 2008; Bjerknes et al. 2012; Yamaguchi et al. 2014; Gerbe et al. 2016; Haber et al. 2017; Yan et al. 2017). Notably, an absence of tuft-like cells is the only known abnormality in Pou2f3-deficient mice (Andersen et al. 1997; Matsumoto et al. 2011; Yamaguchi et al. 2014; Gerbe et al. 2016). In this study we identify a link between the tuft cell lineage master regulator POU2F3 and the variant subtype of SCLC.
Results
CRISPR screen identifies POU2F3 as an essential TF in a subset of SCLC cell lines
We recently described domain-focused CRISPR screening as a strategy to identify essential transcriptional regulators in cancer (Shi et al. 2015). In this study, we applied this method to identify novel TF dependencies exhibiting exceptional potency and specificity for SCLC. To this end, we constructed a pooled library of 8658 single guide RNAs (sgRNAs) targeting 1427 DNA-binding domains of human TFs. We transduced this library via lentivirus into 19 heterologous Cas9-expressing cancer cell lines, including seven SCLC lines, and performed negative selection “dropout” screens (Supplemental Fig. S1). Importantly, we were unable to establish CRISPR screening in any neuroendocrinehigh/ASCL1high SCLC lines, which grew poorly in the presence of Cas9 (data not shown). Hence, our screening strategy was unintentionally biased toward identifying essential TFs in the neuroendocrinelow variant form of SCLC.
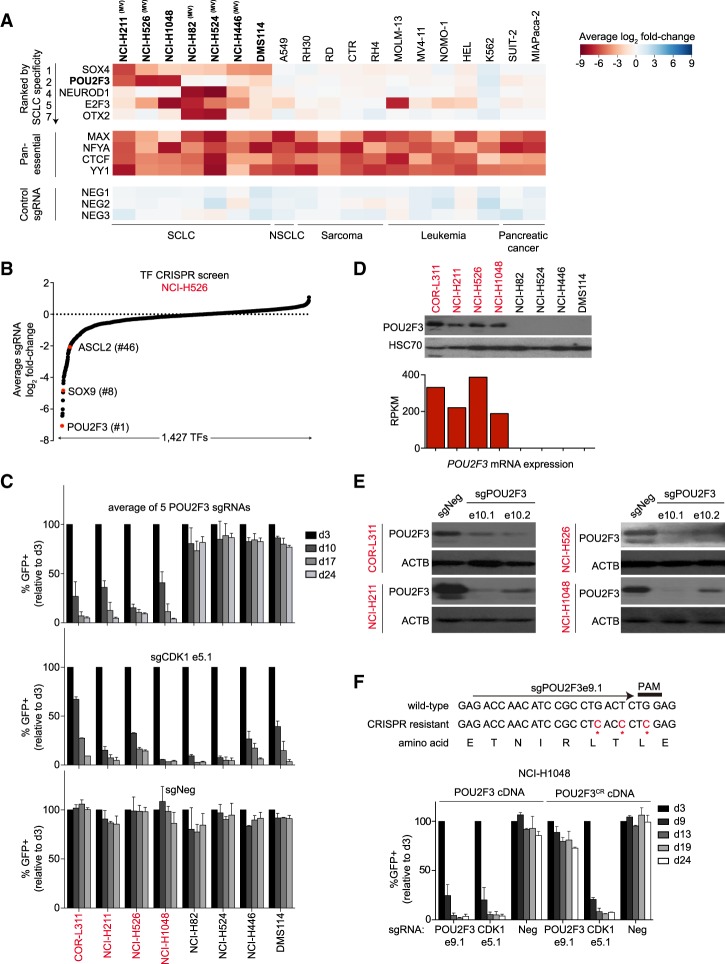
We performed deep sequencing analysis to quantify sgRNA abundance at an initial time point (3 d after infection with the library) and after 14 population doublings of each cell line. The log2 fold change of each sgRNA (averaging the effects of independent sgRNAs targeting each TF) was used to nominate essential TFs. While all 19 cell lines share a dependence on certain TFs (e.g., MAX, CTCF, and YY1), a subset of TFs was selectively essential in SCLC (Fig. 1A; Supplemental Table 1). This included SOX4, NEUROD1, E2F3, and OTX2, all of which have been implicated previously in SCLC (Cooper et al. 2006; Parisi et al. 2007; Castillo et al. 2012; Osborne et al. 2013; Christensen et al. 2014). Our screens identified POU2F3 as the second-ranked SCLC-specific TF dependency (Fig. 1A). POU2F3 was essential in only three SCLC lines (NCI-H211, NCI-H526, and NCI-H1048) but was dispensable in the other 16 cancer lines that we screened (Fig. 1A). In addition to its striking selectivity, POU2F3 was a potent dependency in each of these SCLC lines (Fig. 1B; Supplemental Fig. S2). We confirmed the pattern of POU2F3 dependence using competition-based assays of individual sgRNAs (Fig. 1C). To our knowledge, no prior study has demonstrated a role for POU2F3 as a cancer dependency, which prompted us to investigate its role in SCLC.
Figure 1.
CRISPR screen reveals POU2F3 as a dependency in a subset of human SCLC lines. (A) Essential TFs identified via pooled DNA-binding domain-focused CRISPR–Cas9 screens performed in the indicated panel of cancer cell lines. Plotted is the log2 fold change of sgRNA abundance after ∼14 population doublings. The effect of individual sgRNAs targeting each DNA-binding domain was averaged. The TFs shown were ranked by specificity for SCLC or are examples of pan-essential TF dependencies. NEG1, NEG2, and NEG3 are spike-in negative control sgRNAs. (NSCLC) Non-small cell lung cancer; (MV) morphological variant form of SCLC defined previously (Gazdar et al. 1985). (B) TF dependencies in NCI-H526 cells ranked by the average sgRNA log2 fold change in the pooled CRISPR screen. (C) Competition-based proliferation assays of individual sgRNAs to validate the results from the pooled screen. Since sgRNA expression is linked to GFP, reductions in GFP percentage reflect the fitness disadvantage caused by CRISPR-based gene targeting. sgNeg is a nontargeting negative control. An sgRNA targeting CDK1 is a positive control, and “e” refers to the exon number that is targeted by each sgRNA. Plotted is the average effect of five independent sgRNAs targeting the POU2F3 DNA-binding domain. n = 3. (D, top) POU2F3 and HSC70 protein levels detected by Western blotting. (Bottom) POU2F3 mRNA levels (reads per kilobase of transcript per million mapped reads [RPKM] from RNA sequencing [RNA-seq] analysis) in the indicated SCLC cell lines. (E) Western blotting performed on day 3 after sgRNA vector transduction in the indicated cell lines. (F, top) The design of a CRISPR-resistant synonymous mutant of POU2F3. (Bottom) Competition-based proliferation assays in NCI-H1048 cells expressing wild-type or CRISPR-resistant POU2F3 cDNA. n = 3. All bar graphs represent the mean ± SEM.
By Western blotting and RNA sequencing (RNA-seq) analysis, we found that POU2F3 expression was limited to the subset of SCLC lines in which it was required for growth (Fig. 1D). Using the Cancer Cell Line Encyclopedia (Barretina et al. 2012), we identified a fourth POU2F3high SCLC line (COR-L311), which we found also depends on POU2F3 to proliferate (Fig. 1C,D). An RNA fluorescent in situ hybridization (FISH) analysis revealed that POU2F3 was expressed from both alleles in SCLC lines, suggesting that its expression is not driven by a cis-acting genetic alteration (Supplemental Fig. S3). We confirmed the on-target effects of POU2F3 sgRNAs via Western blotting and by cloning a CRISPR-resistant POU2F3 cDNA, which was able to rescue the growth arrest phenotype cause by a POU2F3 sgRNA (Fig. 1E,F). To rule out artifacts of using the CRISPR system, we also confirmed the POU2F3 dependency using shRNA-based knockdown (Supplemental Fig. S4). Taken together, these results validate that POU2F3 is essential in the subset of SCLC lines that express this TF at high levels.
POU2F3 is expressed in a variant form of SCLC that resembles the tuft cell lineage
We next evaluated the pattern of POU2F3 expression in human SCLC tumor samples. First, we performed immunohistochemical staining of POU2F3 using tissue microarrays of 204 SCLC samples, which revealed POU2F3 expression in 25 tumors (∼12%) (Fig. 2A). Using hematoxylin and eosin (H&E) staining, we observed that POU2F3low and POU2F3high samples each possessed the characteristic “oat cell” morphology that defines SCLC (Fig. 2A). To further validate the heterogeneous pattern of POU2F3 expression in SCLC, we analyzed two published transcriptome data sets obtained from human SCLC samples (Sato et al. 2013; George et al. 2015). In accord with our immunohistochemical analysis, POU2F3 was expressed at high levels in ∼18% of cases across these two studies: in 11 out of 79 samples in the George et al. (2015) data set and in seven out of 23 samples of the Sato et al. (2013) data set (Fig. 2B; Supplemental Fig. S5). These results confirm that POU2F3 is highly expressed in a subset of histologically confirmed SCLC.
Figure 2.
POU2F3 expression is associated with a neuroendocrinelow subtype of SCLC that expresses markers of the tuft cell lineage. (A) SCLC tissue microarray analysis by H&E and anti-POU2F3 immunohistochemical staining. (Left) Representative images of POU2F3high and POU2F3low samples. (Right) Summary of POU2F3 staining of 204 SCLC tumor samples. (B, top) Rederivation of an unsupervised clustering analysis of RNA-seq data obtained from 79 SCLC patient samples from a data set in George et al. (2015). Tumor samples are arranged in columns, and genes are arranged in rows, which were selected based on differential expression in these two groups of samples (George et al. 2015). (Bottom) Expression of POU2F3 and NEUROD1 across the 79 SCLC patient samples. (C) Gene set enrichment analysis (GSEA) of RNA-seq data, evaluating expression of “group 1 identity signature” genes after CRISPR-based targeting of POU2F3. RNA-seq was performed on cells at day 4 or 5 following lentiviral transduction of POU2F3 or control sgRNAs. (NES) Normalized enrichment score; (FWER P val) family-wise error rate P-value. (D) Expression of the indicated neuroendocrine cell lineage markers in POU2F3high and POU2F3low SCLC patient sample RNA-seq from George et al. (2015). Two-tailed t-test results comparing POU2F3high and POU2F3low samples were as follows: for INSM1, P < 0.0001; for CHGA, P = 0.0011; for GRP (gastrin-releasing peptide), P = 0.0046; for CALCA (calcitonin-related polypeptide), P = 0.1177; and for ASCL1, P < 0.0001. The horizontal line is the mean. (E) Expression of the indicated variant SCLC markers in POU2F3high and POU2F3low in SCLC patient sample RNA-seq data from George et al. (2015). Two-tailed t-test results comparing POU2F3high and POU2F3low samples were as follows: for REST, P < 0.0001; for MYC, P < 0.0001; and for NEUROD1, P = 0.0889. The horizontal line is the mean. (F) Expression of the indicated tuft cell markers in POU2F3high and POU2F3low in SCLC patient sample RNA-seq from George et al. (2015). Two-tailed t-test results were as follows: for POU2F3, P < 0.0001; for TRPM5, P < 0.0001; for SOX9, P < 0.0001; for GFI1B, P < 0.0001; for CHAT, P < 0.0001; and for AVIL, P < 0.0001. The horizontal line is the mean.
We next considered whether POU2F3 expression in SCLC tumors was associated with unique clinical, genetic, or molecular characteristics. In the cohort of 79 SCLC tumors analyzed by George et al. (2015), POU2F3 expression was not associated with a significant difference in overall survival (Supplemental Fig. S6). We also did not observe striking differences in the pattern of gene mutations in these two groups of patients (Supplemental Fig. S7). This led us to consider instead whether POU2F3 expression correlates with a unique gene expression profile in SCLC. It was shown previously that SCLC tumors can be classified into two major subgroups when performing an unsupervised clustering analysis of transcriptome data (George et al. 2015). The larger subgroup (group 2) is associated with high expression of the neuroendocrine markers CHGA and ASCL1, whereas the smaller subgroup (group 1) is characterized by low neuroendocrine marker expression (George et al. 2015). We reconstructed this clustering analysis from the investigators’ data set and evaluated POU2F3 expression within these two groups of samples (Fig. 2B). Remarkably, we found that POU2F3 expression was exclusive to group 1 tumors with low neuroendocrine marker expression (Fig. 2B). We applied this same clustering analysis to the microarray data produced by Sato et al. (2013) and confirmed the exclusive expression of POU2F3 to group 1 samples in this independent cohort (Supplemental Fig. S5).
The essentiality of POU2F3 and its association with a distinct transcriptional signature suggested that this TF might function as a master regulator that specifies cell identity in a subtype of SCLC. To investigate this, we constructed a “group 1 identity signature” and a “group 2 identity signature” by selecting ∼200 genes that were preferentially expressed in each set of tumor samples (Supplemental Fig. S8). Importantly, the group 1 identity genes were more highly expressed in POU2F3high SCLC cell lines, and the group 2 identity genes were expressed at higher levels in ASCL1high lines (Supplemental Fig. S8). This shows that SCLC lines retain the cell identity profile found in primary human tumors. We next performed RNA-seq analysis after POU2F3 or control sgRNA transduction in each of the four POU2F3high cell lines, which revealed that inactivating POU2F3 led to a global reduction in the expression of group 1 identity genes (Fig. 2C) and no significant effect on group 2 identity genes (data not shown). These findings suggest that POU2F3 specifies cell identity in the neuroendocrinelow subtype of SCLC in addition to its role as a dependency.
We next evaluated the expression of marker genes to clarify the lineage profile of POU2F3high SCLC. We began by examining neuroendocrine lineage markers INSM1, CHGA, GRP (gastrin-releasing peptide), and CALCA (calcitonin-related polypeptide) (Taneja and Sharma 2004; Gazdar et al. 2017). As expected, these genes were not expressed in POU2F3high tumors (Fig. 2D). The neuroendocrine lineage master regulator ASCL1 was similarly lacking in POU2F3high tumors (Fig. 2D). We then assessed the expression of TFs that had been linked previously with variant SCLC and found that POU2F3high tumors expressed higher levels of REST and MYC but lacked expression of NEUROD1 (Fig. 2E).
Recent studies have shown that POU2F3 functions as a master regulator of the tuft cell lineage (Gerbe et al. 2016; Yamashita et al. 2017). We therefore sought to determine whether POU2F3 drives the expression of tuft cell markers in this variant of SCLC. Remarkably, we found that several tuft cell markers were up-regulated in POU2F3high tumors, including TRPM5, SOX9, GFI1B, CHAT, ASCL2, and AVIL (Fig. 2F). We validated this overall pattern of marker gene expression in an independent SCLC transcriptome data set (Sato et al. 2013) and in our own RNA-seq analysis of human SCLC cell lines (Supplemental Figs. S9, S10). Of note, our TF CRISPR screens also identified SOX9 and ASCL2 as intermediate dependencies in POU2F3high cell lines (Fig. 1B), which we confirmed in competition-based assays of individual sgRNAs (Supplemental Fig. S11). Furthermore, inactivating POU2F3 led to diminished expression of several tuft cell marker genes in SCLC lines (Supplemental Fig. S12). Taken together, these findings suggest that POU2F3 specifies a tuft cell-like identity in a variant form of SCLC.
Enhancer landscapes classify SCLC lines based on POU2F3, ASCL1, or NEUROD1 expression
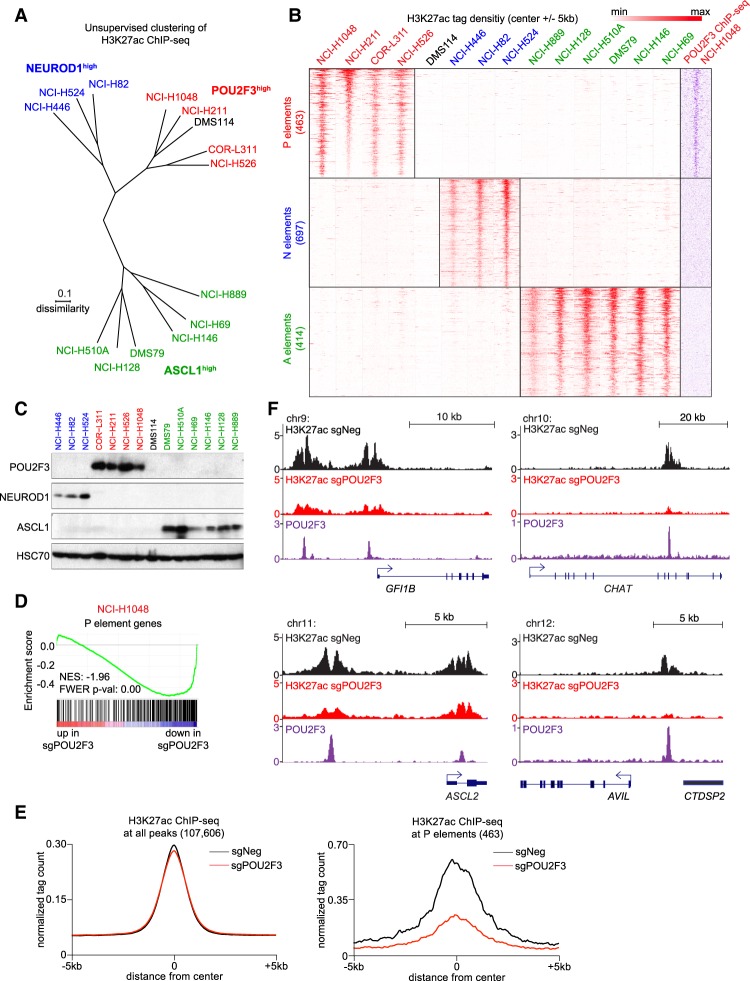
A prior study demonstrated that ASCL1high and NEUROD1high human SCLC lines harbor striking differences in their enhancer landscapes, suggesting that these two master regulators specify distinct chromatin states and lineage identities (Borromeo et al. 2016). This prompted us to consider whether POU2F3high cell lines possess a unique enhancer profile that differs from these two other SCLC subtypes. To investigate this, we performed chromatin immunoprecipitation (ChIP) followed by DNA sequencing (ChIP-seq) to profile histone H3K27 acetylation (H3K27ac) in SCLC lines. H3K27ac is enriched at active enhancers and promoter elements and is a validated mark for annotating such elements in a nonbiased manner (Rada-Iglesias et al. 2011). We chose 14 SCLC lines for our analysis: Four cell lines express POU2F3, three express NEUROD1, six express ASCL1, and one lacks all three TFs (Supplemental Fig. S10). We note that in both SCLC tumor samples and cell lines, these three TFs are expressed in a mutually exclusive manner (Fig. 2B; Supplemental Figs. S5, S9, S10, S13).
An unsupervised clustering analysis of the global H3K27ac enrichment pattern across these lines revealed three major groupings that corresponded to the expression of POU2F3, NEUROD1, or ASCL1 (Fig. 3A). One exception was the DMS114 line, which does not express any of these three TFs yet clustered with the POU2F3high lines (Fig. 3A). We extracted the subset of H3K27ac-enriched regions that correlated with expression of each of the three TFs, which we termed P, N, or A elements based on enrichment of H3K27ac in cell lines that express POU2F3, NEUROD1, or ASCL1, respectively (Fig. 3B). As expected, POU2F3, NEUROD1, and ASCL1 motifs were highly enriched at P, N, or A elements, respectively (Supplemental Fig. S14). We performed ChIP-seq analysis of POU2F3 occupancy in NCI-H1048 and NCI-H526 cells, which confirmed the selective association of this TF with P elements (Fig. 3B; Supplemental Fig. S15). Using published ChIP-seq data sets (Borromeo et al. 2016), we also confirmed the preferential association of NEUROD1 and ASCL1 with N and A elements, respectively (Supplemental Fig. S15). Taken together, this analysis identifies a unique chromatin landscape linked to POU2F3, which further implicates this TF as an alternative master regulator to ASCL1 and NEUROD1 in SCLC.
Figure 3.
Enhancer landscapes classify SCLC lines based on POU2F3, ASCL1, or NEUROD1 expression. (A) Unsupervised clustering (Euclidian) using dissimilarity level (1 = Pearson correlation coefficient) of global H3K27ac profiles from 14 human SCLC cell lines measured using ChIP-seq analysis. Global H3K27ac profiles were correlated (Pearson), and correlation coefficients were used to calculate distances in the tree representation. (B) ChIP-seq density plots of H3K27ac tag density (red) centered at the summit of 463 P elements, 697 N elements, and 414 A elements across the 14 SCLC cell lines. Density plot of POU2F3 tag density (purple) in NCI-H1048 cells at the indicated regions. (C) Western blotting of POU2F3, ASCL1, NEUROD1, and HSC70 (loading control) in lysates from SCLC cell lines. (D) GSEA of RNA-seq data obtained from NCI-H1048 cells on day 4 following transduction of POU2F3 or control sgRNA, evaluating an effect on genes located near P elements. (NES) Normalized enrichment score; (FWER P val) family-wise error rate P-value. (E) A metaprofile analysis of H3K27ac enrichment at all H3K27ac peaks (107,606 total) or at 463 P elements, evaluating effect of POU2F3 inactivation. NCI-H1048 cells were collected for ChIP-seq analysis on day 3 following transduction with POU2F3 (red) or control (black) sgRNAs. (F) ChIP-seq profiles of H3K27ac (black and red) and POU2F3 (purple) at the indicated of tuft cell marker gene loci.
POU2F3 establishes active enhancers at tuft cell identity genes in SCLC
We further characterized P elements as putative enhancer elements activated by POU2F3. The overwhelming majority of P elements was located in either intergenic or intronic regions, consistent with being distal enhancers (Supplemental Fig. S16). Our RNA-seq analysis revealed that genes located in the vicinity of P elements were suppressed upon inactivating POU2F3 in each of the four SCLC lines (Fig. 3D; Supplemental Fig. S17). We next performed H3K27ac ChIP-seq in NCI-H1048 cells after transduction with POU2F3 or control sgRNAs. Importantly, inactivating POU2F3 led to a loss of H3K27ac at P elements while having minimal effects on the global profile of H3K27ac in the cell (Fig. 3E). As examples, POU2F3 activates enhancers located near tuft cell marker genes GFI1B, CHAT, ASCL2, and AVIL (Fig. 3F). Taken together, these findings suggest that POU2F3 activates a set of enhancers to promote expression of tuft cell lineage genes in variant SCLC.
Kinase domain-focused CRISPR screening identifies an essential role for insulin-like growth factor 1 receptor (IGF1R) in POU2F3high SCLC
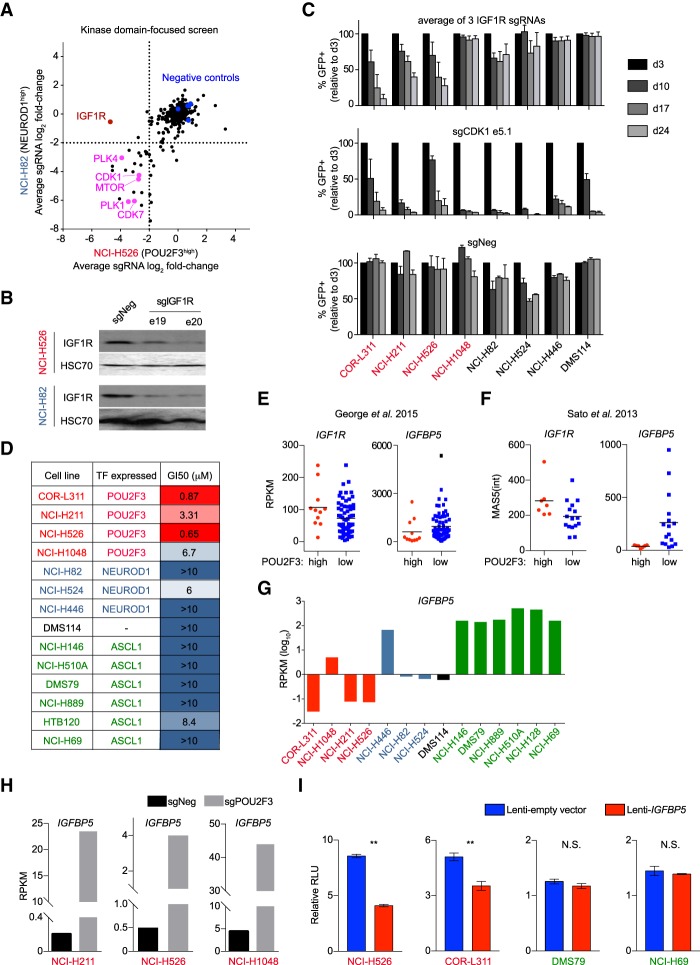
The practical utility of identifying novel subtypes of SCLC is in the potential to uncover an association with actionable dependencies. To investigate this possibility, we performed a CRISPR screen that compared the essentiality of 493 kinase domains in the POU2F3high line NCI-H526 with the NEUROD1high line NCI-H82 (Supplemental Table 6). While nearly all essential kinases were shared between these two cell lines, the IGF1R was identified as a unique dependency in NCI-H526 cells (Fig. 4A). By Western blotting, we confirmed that IGF1R sgRNAs led to depletion of IGF1R protein in both cell line contexts (Fig. 4B). A broader validation of this result in competition-based assays identified IGF1R dependence in three of the four POU2F3high lines but no growth arrest phenotype in the four SCLC lines that lacked POU2F3 expression (Fig. 4C). Of note, the POU2F3high SCLC line that grew independently of IGF1R (NCI-H1048) expressed IGF1R at low levels and had an activating mutation of PIK3CA (H1047R) (Supplemental Fig. S18; Bader et al. 2006; Barretina et al. 2012), which may bypass upstream receptor tyrosine kinases (Nisa et al. 2017). We next evaluated the sensitivity of 14 SCLC lines to linsitinib, a small molecule inhibitor of IGF1R kinase activity (Mulvihill et al. 2009). Using a compound titration and cell proliferation measurements, we observed that POU2F3high lines were consistently more sensitive to linsitinib than NEUROD1high or ASCL1high SCLC lines (Fig. 4D).
Figure 4.
POU2F3high SCLC lines are hypersensitive to targeting of IGF1R. (A) Kinase domain-focused CRISPR screening in NCI-H82 (NEUROD1high) and NCI-H526 (POU2F3high) cells. The averaged log2 fold change of sgRNA abundance after ∼14 population doublings for each kinase domain is plotted. (B) Western blot of IGF1R and HSC70 (loading control) in NCI-H526 and NCI-H82 cells on day 4 after sgRNA transduction. (C) Arrayed format competition-based proliferation assays to evaluate the effect of IGF1R and control sgRNAs on the indicated SCLC lines. n = 3. (D) Summary of proliferation assays following linisitinib treatment, showing the drug concentrations that cause a 50% suppression of cell growth (GI50) using CellTiter-Glo proliferation assays. (E) Expression of IGF1R and IGFBP5 in POU2F3high and POU2F3low SCLC patient sample RNA-seq data from George et al. (2015). Two-tailed t-test results comparing the RPKM (reads per kilobase per million mapped reads) values of POU2F3high and POU2F3low samples were as follows: for IGF1R, P = 0.0488, and for IGFBP5, P = 0.2525. The horizontal line is the mean. (F) Expression of IGF1R and IGFBP5 in POU2F3high and POU2F3low SCLC patient sample microarray data from Sato et al. (2013). Two-tailed t-test results comparing normalized MAS5 intensity value averages of all probes for each gene in POU2F3high and POU2F3low samples were as follows: for IGF1R, P = 0.042, and for IGFBP5, P = 0.0389. The horizontal line is the mean. (G) RNA-seq measurement of IGFBP5 expression (RPKM log10) in human SCLC cell lines. (H) IGFBP5 mRNA levels (RPKM) in the indicated cell lines following transduction with control (sgNeg) or POU2F3 sgRNA. (I) The effect of lentiviral expression of IGFBP5 on the relative proliferation rate of the indicated cell lines. Normalized relative luminescence unit (RLU) was measured using CellTiter-Glo after 2 d of culture of the indicated cells. n = 3. (**) P < 0.01; (N.S.) not significant. All bar graphs represent the mean ± SEM.
In human tumor transcriptome data (Sato et al. 2013; George et al. 2015), we found a modest elevation of IGF1R in POU2F3high relative to POU2F3low SCLC samples (Fig. 4E,F). However, IGF1R expression across the SCLC lines was not well correlated with IGF1R dependence (Supplemental Fig. S18), consistent with prior findings (Zinn et al. 2013). In addition, inactivating POU2F3 did not influence IGF1R expression (Supplemental Fig. S19). This prompted us to evaluate the expression of other IGF1R pathway components in SCLC. We found that IGFBP5, a negative regulator of IGF1R signaling (Ding et al. 2016), was expressed at low levels in POU2F3high patient samples and cell lines (Fig. 4E,F), and CRISPR-based targeting of POU2F3 in SCLC cell lines led to a marked increase in IGFPB5 expression (Fig. 4H). Furthermore, forced IGFBP5 expression via lentivirus in POU2F3high SCLC lines inhibited cell proliferation, while introducing IGFBP5 into ASCL1high cell lines had no effect (Fig. 4I). Taken together, these findings suggest that expression of IGF1R pathway genes and dependence on IGF1R differ between the POU2F3high and POU2F3low subtypes of SCLC.
Rare POU2F3-expressing cells are present in the mouse bronchial epithelium
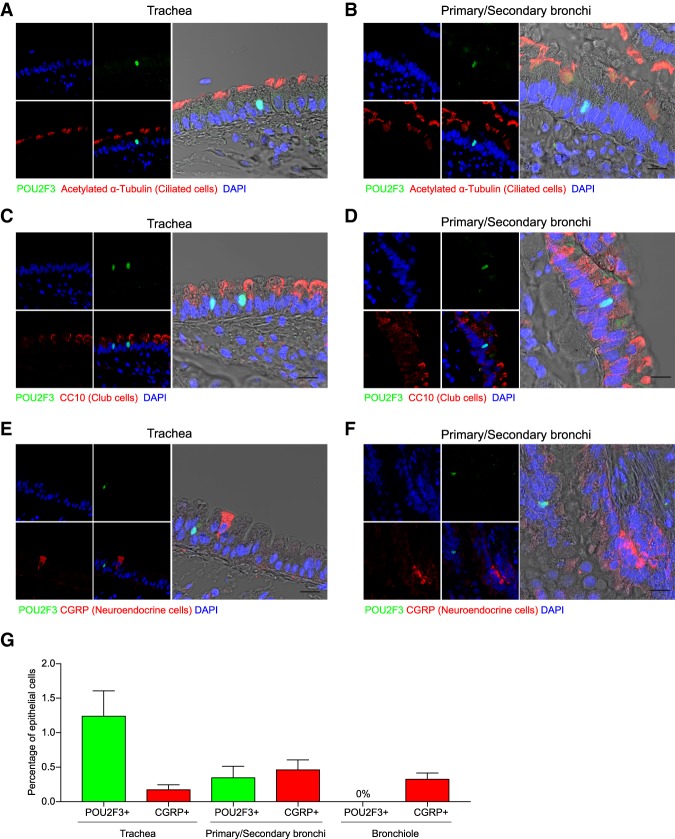
POU2F3-expressing tuft cells have been found previously in the mouse trachea (Yamashita et al. 2017), but their presence in the lung has not been demonstrated previously. To examine this, we performed immunofluorescence staining for POU2F3 in cross-sections of the mouse lung, which revealed rare POU2F3-expressing cells in the epithelial layer surrounding the primary/secondary bronchi but no detectable POU2F3 expression in the distal airways or alveoli (Fig. 5A–G; Supplemental Figs. S20, S21; data not shown). POU2F3-expressing cells lacked expression of the neuroendocrine cell marker CGRP, the ciliated cell marker acetylated α-tubulin, and the club cell marker CC10 (Fig. 5A–F; Supplemental Figs. S20, S21). We note that SCLC often presents as a mass in central airway locations, which is similar to the site in which we detected POU2F3 expression.
Figure 5.
Rare POU2F3-expressing cells exist in the bronchial epithelium of the mouse lung. (A–F) Representative images of immunofluorescence staining for POU2F3 (green), CGRP (a neuroendocrine cell marker; red), acetylated α-tubulin (a ciliated cell marker; red), and CC10 (a club cell marker; red) in the mouse trachea or primary/secondary bronchi. (G) Quantification of the percentage of cells in the epithelial layer of the trachea, primary/secondary bronchi, and bronchiole that stain for POU2F3 or CGRP. The mean ± SEM for four independent mice is plotted.
Discussion
Our findings lead us to propose that POU2F3 is an essential master regulator of cell identity in the neuroendocrinelow variant of SCLC. One important line of evidence in support of this conclusion is that POU2F3high SCLC tumors and cell lines lack the expression of classical neuroendocrine lineage markers and instead express markers of the tuft cell lineage. In addition, our epigenomic analysis shows that POU2F3 establishes active enhancers at tuft cell identity genes in SCLC. The shared POU2F3 requirement in the normal tuft cell lineage and in the tuft cell-like variant of SCLC suggests a similar master regulator function for POU2F3 in normal and malignant contexts. Finally, we identify transcriptional and signaling dependencies that are linked to POU2F3 expression in SCLC. Together, these findings support consideration of POU2F3high SCLC as a disease entity that is distinct from the classical neuroendocrine form of this disease.
A key unanswered question in our study relates to the origin of POU2F3 expression in SCLC. We observed rare POU2F3-expressing cells in the epithelial layer of the primary and secondary bronchi, which is a common location for SCLC tumors to arise in humans. This raises a possibility that a POU2F3-expressing tuft cell is a cell of origin in a subset of SCLC cases, which will be evaluated in future studies by engineering Trp53 and Rb1 mutations in this cell population in mice. However, cell identity in lung cancer is known to be plastic, particularly for tumors under the selection pressure of therapy (Sequist et al. 2011). While POU2F3high tumors identified in the George et al. (2015) cohort were largely collected from SCLC patients before treatment, it is possible that POU2F3 expression was acquired during tumor evolution through a trans-differentiation mechanism or as a consequence of a yet to be identified genetic alteration. However, the biallelic expression pattern of POU2F3 in SCLC lines suggests that a cis-acting mutation at the POU2F3 locus is unlikely to be the source of its expression. It also remains unclear at present whether POU2F3high tumor cells can transition to a NEUROD1high or ASCL1high cell state (or vice versa) as an adaptive response to a selection pressure. In addition, the role of MYC overexpression in the POU2F3high variant of SCLC awaits further investigation. While POU2F3 and NEUROD1 expression accounts for the majority neuroendocrinelow variant SCLC tumors, we note that a small number of variant tumors lack expression of both TFs. This suggests that additional mechanisms await characterization within the variant form of this disease.
The current management of SCLC with chemotherapy leads to short-lived responses, and the prognosis of this disease has not changed over several decades. To improve outcomes for SCLC patients, substantial efforts have been made to evaluate targeted agents in the clinic, but with limited success (Joshi et al. 2013). However, these trials have been conducted largely in unselected SCLC patient populations. For example, a prior study demonstrated that a subset of SCLC lines is sensitive to linsitinib (Zinn et al. 2013), yet this drug failed to demonstrate efficacy in unselected SCLC patients in a phase II study (Chiappori et al. 2016). Our findings justify a reconsideration of agents such as linsitinib as well as new targeted agents in preclinical models to determine whether exceptional responses are associated with the expression of lineage-defining master regulators POU2F3, ASCL1, or NEUROD1. Such efforts might allow for the design of biomarker-driven clinical studies and the advancement of a personalized medicine strategy for this disease.
Finally, it has been shown that Pou2f3-null mice have a normal life span and only discrete chemosensory deficits due to the absence of the tuft cell lineage (Andersen et al. 1997; Gerbe et al. 2016; Yamashita et al. 2017). Hence, a therapeutic strategy aimed at blockade of POU2F3 function or ablation of the tuft cell lineage would be expected to have a wide therapeutic margin in patients with POU2F3high SCLC tumors. Chemical degradation approaches and targeting of transcriptional coactivator complexes are strategies for interfering with TFs in cancer (Bhagwat and Vakoc 2015). Thus, a deeper biochemical understanding of POU2F3 and its cofactors may expose strategies for targeting this dependency as a therapeutic approach in SCLC.
Materials and methods
Cell lines
All cell lines were obtained from American Type Culture Collection or Sigma. SCLC cell lines COR-L311, NCI-H211, NCI-H526, DMS114, NCI-H446, NCI-H82, NCI-H524, NCI-H889, DMS79, NCI-H146, and NCI-H69; leukemia cell lines MOLM-13, MV4-11, NOMO-1, HEL, and K562; and pancreatic cancer cell lines SUIT-2 and MIAPaca-2 were cultured in RPMI supplemented with 10% FBS. NCI-H510A was cultured in F12K supplemented with 10% FBS. NCI-H1048 was cultured in DMEM:F12 supplemented with 0.005 mg/mL insulin, 0.01 mg/mL transferrin, 30 nM sodium selenite, 10 nM hydrocortisone, 10 nM β-estradiol, 4.5 mM L-glutamine, and 5% FBS. NCI-H128 cells were cultured in RPMI supplemented with 20% FBS. HEK293T cells; sarcoma cell lines RD, RH30, CTR, and RH4; and non-small cell lung cancer cell line A549 were cultured in DMEM with 10% FBS. Penicillin/streptomycin was added to all media. All cell lines were cultured at 37°C with 5% CO2 and periodically tested to confirm the absence of mycoplasma.
Plasmid construction, sgRNA cloning, and shRNA cloning
For CRISPR screening, the optimized sgRNA lentiviral expression vector (LRG2.1T) and the lentiviral human codon-optimized Cas9 vector (LentiV_Cas9_puro) were used. For the arrayed format competition-based assays, sgRNAs were cloned into the LRG2.1 vector using a BsmBI restriction site. All sgRNA sequences in this study are listed in Supplemental Table 2. For the cDNA rescue experiment, a POU2F3 cDNA (DNASU, plasmid ID HsCD00329483) was cloned into the LentiV_Neo vector using the In-Fusion cloning system. The CRISPR-resistant synonymous mutant of POU2F3 was cloned using a standard PCR mutagenesis method. For the IGFBP5 overexpression experiment, IGFBP5 cDNA (Addgene, pcDNA3-IGFBP5-V5, plasmid no. 11608) was subcloned into LentiV_Puro vector using In-Fusion cloning. shRNAs targeting POU2F3 and PCNA were cloned into the mirE-based retroviral shRNA expression vector MLS-E. The sequences of each shRNA are summarized in Supplemental Table 2.
Construction of domain-focused sgRNA libraries
The human TF and kinase sgRNA pooled libraries were described in a previous study (Tarumoto et al. 2018). In brief, the TF DNA-binding domain and kinase domain annotations were retrieved from NCBI Conserved Domains Database. Five to six sgRNAs per domain were designed targeting 1427 TFs or 482 kinases in the human genome. sgRNAs with the predicted high off-target effect were excluded. Domain targeting and spike-in positive/negative control sgRNA oligonucleotides were synthesized in duplicate using an array platform (Twist Bioscience) and then PCR-cloned into the Bsmb1-digested LRG2.1 vector using a Gibson assembly kit (New England Biolabs, E2611). To verify the identity of and relative representative sgRNAs in the pooled plasmids, a deep-sequencing analysis was performed on a MiSeq instrument (Illumina), which confirmed that 100% of the designed sgRNAs were cloned in the LRG2.1 vector and that the abundance of >95% of individual sgRNA constructs was within fivefold of the mean.
Virus transduction
Lentivirus was produced in HEK293T cells by transfecting plasmid and helper packaging plasmid (pVSVG and psPAX2) with polyethylenimine (PEI 25000) transfection reagent. HEK293T cells were plated 1 d before transfection with 70%–80% confluency in a 10-cm tissue culture dish. For one 10-cm dish of HEK293T cells, 10 µg of plasmid DNA, 5 µg of VSVG, 7.5 µg of psPAX2, and 32 µL of 1 mg/mL PEI were mixed, incubated, and added to the cells. Fresh medium was changed 8 h after transfection, and lentivirus-containing supernatant was collected at 24, 48, and 72 h after transfection and pooled together.
For shRNA experiments, retrovirus was produced in Plat-E cells, which were transfected with retroviral DNA, VSVG, and Eco helper plasmids in a ratio of 10:1:1.5. Retrovirus-containing supernatant was collected at 24, 48, and 72 h after transfection and pooled together.
For both lentivirus and retrovirus infections, corresponding target cells were mixed with viral supernatant supplemented with 4 µg/mL polybrene and then centrifuged at 1700 rpm for 30 min at room temperature. Fresh medium was changed 24 h after infection. Antibiotics (1 µg/mL puromycin and/or 1 mg/mL G418) were added 24 h after infection when selection was required.
Pooled CRISPR screening
CRISPR-based negative selection genetic screenings were performed in nonclonal cancer cell lines with stable Cas9 expression (LentiV-Cas9-Puro vector). Lentivirus of a pooled sgRNA library targeting DNA-binding domains or kinase domains was produced as described above. To ensure a single-copy sgRNA transduction per cell, multiplicity of infection (MOI) was set to 0.3–0.4. To maintain the representation of sgRNAs during the screen, the number of sgRNA-positive cells was kept at least 1000 times the number of sgRNAs in the library. Cells were harvested at day 3 after infection and were provided as a reference representation of the pooled sgRNA library. Cells were cultured for 14 population doublings and harvested as the final time point of the genetic screen. Genomic DNA was extracted using QIAamp DNA minikit (Qiagen) according to the manufacturer's instructions.
Sequencing libraries were constructed as described previously (Shi et al. 2015). Briefly, the sgRNA cassette was PCR-amplified from genomic DNA using high-fidelity polymerase (Phusion master mix, Theromo Fisher, F548L). The PCR product was end-repaired by T4 DNA polymerase (New England Biolabs, B02025), DNA polymerase I large (Klenow) fragment (New England Biolabs, M0210L), and T4 polynucleotide kinase (New England Biolabs, M0201L). A 3′ A overhang was then added to the ends of blunted DNA fragments with the Klenow fragment (3′–5′ exo; New England Biolabs, M0212L). The DNA fragments were ligated to diversity-increased custom barcodes with Quick ligation kit (New England Biolabs, M2200L). Illumina paired-end sequencing adaptors were attached to the barcoded ligated products through PCR reaction with high-fidelity polymerase (Phusion master mix, Thermo Fisher Scientific F548L). The final libraries were quantified using a bioanalyzer Agilent DNA 1000 (Agilent 5067-1504) and then pooled together in equal molar ratio followed by paired-end sequencing using MiSeq (Illumina) with MiSeq reagent kit version 3 (Illumina).
Pooled CRISPR screening data analysis
The sequencing data were demultiplexed and trimmed to contain only the sgRNA sequence cassettes. The read counts of each individual sgRNA were calculated, allowing no mismatches to the reference sgRNA sequence. Individual sgRNAs with a read count <50 in the initial time point were discarded. The total read counts were normalized between samples. The average log2 fold change in abundance of all sgRNA against a given domain/gene was calculated. SCLC-specificity was determined by subtracting the average of log2 fold change in non-SCLC cell lines from average log2 fold change in SCLC cell lines. The TF CRISPR screening data are shown in Supplemental Table 1. The kinase CRISPR screening data are shown in Supplemental Table 6.
Arrayed format competition-based assay to measure the effects of sgRNAs on cell proliferation
Cas9-expressing SCLC cell lines were lentivirally transduced with the LRG 2.1T sgRNA vector, which is linked to a GFP reporter, to achieve between 20% and 60% GFP-positive cells. The percentage of GFP-positive cell population was measured at various time points during culturing using a Guava Easycyte HT instrument (Millipore). The relative change in GFP percentage was used to assess the impact of individual sgRNAs on cellular proliferation, which reflects cells with a genetic knockout being outcompeted by nontransduced cells in the culture.
Human SCLC immunohistochemistry
The human SCLC tissue microarrays (TMAs) used in this study were OD-CT-RsLug01-003, LC818, and LC245, purchased from US Biomax. The TMA slides were stained with either H&E or a POU2F3 antibody (1:100; Santa Cruz Biotechnology, SC-330). The H&E-stained slides were used for SCLC morphology evaluation by pathologist Dr. John E. Wilkinson. POU2F3 positivity in the POU2F3 immunohistochemistry-stained slide was scored on a five-point scale as follows: 0 = negative, 1 = positive staining in <10% of cells, 2 = positive staining in >10% but <50% of cells, 3 = positive staining in >50% of cells, and 4 = almost all cells are positively stained.
RNA-FISH
RNA-FISH was carried out using Alexa 594 dUTP-labeled nick translated POU2F3 cDNA. The cells on coverslips were fixed in 4% freshly prepared PFA followed by permeablization with 0.5% Triton X-100 in PBS. Cells were washed in multiple changes of 2× SSC buffer for 2 h followed by hybridization with labeled nick translated probe in 10% formamide, 10 µg of transfer RNA (tRNA), and 2× SSC overnight at 37°C. Cells were washed in 2× SSC and 50% formamide followed by washes in 1× and 0.5× SSC, respectively. Finally, the cells were stained with DAPI, and coverslips were mounted on glass slides. Images were acquired and processed using a DeltaVision deconvolution microscope. Images were stacked, and the number of nuclear foci was manually counted across several random fields.
Tissue preparation for immunofluorescence and cell quantification
Freshly isolated trachea and lung tissue were fixed in 4% PFA for 2 h on ice, washed with PBS, and incubated in 30% sucrose in PBS overnight at 4°C. Tissue was mounted in OCT embedding compound and frozen at −80°C. Tissue sections (10 µm) were cut using a cryostat and incubated with 1× blocking buffer (5% goat serum, 2.5% bovine serum albumin in PBS) for 1 h followed by incubation in Fc receptor blocker (Innovex Biosciences) for 30 min. Sections were incubated with rabbit anti-POU2F3 (1:100; Sigma-Aldrich, HPA019652), mouse anti-CGRP (1:100; Abcam, ab81887), APC-conjugated rat anti-EpCAM (1:100; BioLegend), mouse anti-acetylated tubulin (1:400; Sigma-Aldrich, T7451), or mouse anti-CC10 (1:200; Santa Cruz Biotechnology, E-11) in 0.5× blocking buffer overnight at 4°C. After rinsing twice with PBS, tissue sections were incubated with secondary antibodies (goat anti-rabbit Alexa488 and goat anti-mouse Alexa568; 1:300 dilution; Life Technologies) for 2 h at room temperature. Nuclei were counterstained with DAPI (1:1000 dilution; Life Technologies). Fluorescent images were collected using a Leica TCS SP8 confocal laser scanning microscope (40× magnification) and an Olympus BX53 fluorescent microscope (5× and 10× magnification). Image acquisition and processing were conducted using the LAS AF Lite program (Leica). POU2F3- and CGRP-expressing cells were counted manually under a Zeiss fluorescent microscope (10× and 20× magnification). The DAPI channel of the slides was scanned by Aperio FL fluorescence slide scanner. The epithelial regions were drawn manually in each slide. The number of nuclei was counted using Aperio software.
Western blot
SCLC cells were counted before lysis with Laemmli sample buffer (Bio-Rad). One-million cells were lysed with 200 µL sample buffer containing 2-mercaptoethanol and boiled for 10 min. Equal volumes of samples were separated by SDS-PAGE gel followed by transfer to nitrocellulose membrane and immunoblotting. Primary antibodies used in this study included POU2F3 (1:1000; Sigma, HPA019652), ASCL1 (1:1000; BD Biosciences, clone 24B72D11.1 ), and NEUROD1 (1:1000; Abcam, ab60704).
CellTiter-Glo assay
To test cell growth upon Linsitinb (OSI-906) treatment, 2000∼6000 SCLC cells were plated in each well of an opaque-walled 96-well plate and mixed with a serially diluted concentration of Linsitinb (Selleckchem) or 0.1% DMSO as a control. After 72 h of incubation, the number of viable cells was measured using CellTiter-Glo luminescent cell viability assay kit (Promega) with SpectraMax plate reader (Molecular Devices) following the manufacturer's protocol. To test cell growth upon the overexpression IGFBP5 (after puromycin selection for 5 d), 2000∼6000 SCLC cells (infected with LentiV-Puro empty vector or LentiV-Puro-IGFBP5) were plated in each well of an opaque-walled 96-well plate. After 48 h of incubation, the number of viable cells was measured using CellTiter-Glo luminescent cell viability assay kit (Promega) with SpectraMax plate reader (Molecular Devices) following the manufacturer's protocol.
RNA-seq library construction
Total RNA of SCLC cell lines was extracted using TRIzol reagent (Thermo Fisher) according to the manufacturer's instructions. Briefly, ∼10 million cells were lysed with 1 mL of TRIzol and chloroform and incubated for 3 min at room temperature followed by centrifugation at 10,000g for 15 min at 4°C. The aqueous phase was added to 500 µL of isopropanol and incubated for 10 min at room temperature. RNA was precipitated at 10,000g for 10 min at 4°C, washed with 75% EtOH in DEPC-treated water, and eluted in RNase-free water. RNA-seq libraries were constructed using the TruSeq sample preparation kit version 2 (Illumina) according to the manufacturer's instructions. Briefly, 2 µg of purified RNA was poly-A-selected and fragmented with fragmentation enzyme. After first and second strand synthesis from a template of poly-A-selected/fragmented RNA, other procedures from end repair to PCR amplification were performed according to ChIP-seq library construction steps. The quantity of the RNA-seq library was determined by nanodrop, and the average quantity of RNA-seq libraries ranged from 40 to 80 ng/µL. Multiplexing for sequencing was performed as described for ChIP-seq library construction. RNA-seq libraries were sequenced using an Illumina HiSeq 2500 or NextSeq platform with single-end reads of 50 bases.
RNA-seq data analysis
Sequencing reads were mapped into reference genome hg19 using TopHat. Differentially expressed genes were analyzed using Cufflinks and Cuffdiff with masking of all noncoding RNAs. Genes with RPKM (reads per kilobase per million mapped reads) of more than three in the control were considered as expressed genes in subsequent analyses.
Gene set enrichment analysis (GSEA)
The fold change of RPKM was calculated by the ratio of the average RPKM of two independent sgRNA samples (sgPOU2F3 [e10.1 and e10.2]) to the RPKM of control sgRNA samples (sgNeg). The ranked list was prepared using the log2 fold change of all expressed genes. Group 1 gene signatures were derived from patient sample RNA-seq from George et al. (2015) with an expression ratio of average group 1 RPKM versus average group 2 RPKM of >5 (gene lists are in Supplemental Tables 3, 4). Custom P-element genes were obtained by the association of each P element to its nearest expressed gene using annotatePeaks from the Homer suite (the gene list is in Supplemental Table 5). GSEA was performed using the custom gene signatures.
Unsupervised expression clustering and gene expression level comparison
We took all of the 81 SCLC patient sample RNA-seq data (George et al. 2015) and computed sample correlations using the top 1000 genes that show the highest MAD (median absolute deviation) across all samples to check whether there were any outliers. Since samples S02297 and S02353 had a Spearman correlation coefficient of <0.7 across all of the other samples, we omitted these samples from all downstream analysis. Unsupervised expression clustering was reconstructed as described by George et al. (2015). We first obtained the “differentially expressed genes in SCLC patient samples” gene set from this prior study and separated samples into two groups for a high and low expression using hierarchical clustering of gene expression (Euclidean distance and complete linkage) on the log2(1 + RPKM values) as described (George et al. 2015).
Using the same “differentially expressed genes in SCLC patient samples” gene set and clustering strategy, we performed the unsupervised expression clustering using the RNA-seq data of 14 SCLC cell lines used in this study, the data of 42 SCLC cell lines from the cancer cell line encyclopedia (CCLE) (PMID:22460905), and the microarray data of 23 SCLC patient samples from Sato et al. (2013). For the cell line RNA-seq data, we used gene expression levels instead of considering isoform expression levels. For the microarray data set, the normalized MAS5 intensity values of the individual gene probes were averaged for the gene expression values. Hierarchical clustering (Euclidean distance and complete linkage) was performed on the log2 transformed raw expression values.
Gene expression analysis of SCLC patient samples
For the patient cohort with RNA-seq data (George et al. 2015), the available data contain RPKM values of isoforms only. In order to estimate gene-level expression, we reversed the step of normalizing by feature lengths in the calculation of RPKM to get fragment counts of each isoform with actual size. To do this, we divided isoform RPKM value by 1000 and multiplied it by its length. Next, for each gene, we added up the resulting counts of all isoforms and then normalized to the length of 1000 base pairs (bp). Gene expression level comparisons were performed using raw RPKM values in RNA-seq data. For the 23 SCLC patient sample cohort with microarray data (Sato et al. 2013), there were 11,037 genes with MAS5(int) > 200 in at least one sample. Only these genes were used in further analysis. All probes of each given gene were averaged for further analysis.
ChIP assay
For ChIP experiment with SCLC cell lines, 5 × 106 cells were cross-linked for 15 min at room temperature with formaldehyde (1% final) with constant shaking and then quenched with 0.125 M glycine for 15 min. Cells were pelleted and washed with PBS. Cells were then lysed with 1 mL of cell lysis buffer (10 mM Tris-Cl at pH 8.0, 10 mM NaCl, 0.2% NP-40) with protease inhibitor and incubated for 10 min at 4°C. Chromatin was isolated by centrifugation at 4200 rpm for 30 sec and lysed with 500 µL of nuclear lysis buffer (50 mM Tris-Cl at pH 8.0, 10 mM EDTA, 1% SDS) with protease inhibitor followed by incubation for 10 min at 4°C. Lysed chromatin was then sonicated for 15 min at low intensity for 0.5 sec using a Bioruptor (Diagenode). Chromatin was centrifuged at 14,000 rpm for 15 min at 4°C, pellets were discarded, the supernatants were preincubated for 2 h with 3.5 mL of immunoprecipitation dilution buffer (20 mM Tris-Cl at pH 8.0, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, 0.01% SDS) and 1 µg of rabbit antibody (H3K27ac [Abcam, ab4729] and POU2F3 [Santa Cruz Biotechnology, SC-330]), and 20 µL of protein A magnetic beads (Invitrogen) was added for incubation overnight with rotation at 4°C.
The next day, immunocomplexes were washed once with immunoprecipitation wash I buffer (20 mM Tris-Cl at pH 8.0, 2 mM EDTA, 50 mM NaCl, 1% Triton X-100, 0.1% SDS), twice with high-salt buffer (20 mM Tris-Cl at pH 8.0, 2 mM EDTA, 500 mM NaCl, 1% Triton X-100, 0.01% SDS), once with immunoprecipitation wash II buffer (10 mM Tris-Cl at pH 8.0, 1 mM EDTA, 0.25 M LiCl, 1% NP-40, 1% Na-deoxycholate), and twice with TE (pH 8.0). The washed immunocomplexes were eluted with 200 µL of elution buffer (50 mM Tris 8.0, 10 mM EDTA, 1% SDS) for 15 min at 65°C with constant shaking and reverse cross-linking with addition of 1 µg/µL RNase A and 0.25 M NaCl overnight at 65°C in a water bath. The next day, immunoprecipitated DNA was treated with 0.2 mg/mL proteinase K for 2 h at 42°C and purified with QIAquick PCR purification kit in 55 µL of distilled water.
ChIP-seq library construction
ChIP-seq libraries were constructed using the TruSeq ChIP sample preparation kit (Illumina). Briefly, 50 µL of purified ChIP DNA was end-repaired for dA tailing followed by adaptor ligation. Adaptor-ligated ChIP DNA was size-selected (250–300 bp) via agarose gel electrophoresis, gel-extracted with QIAquick gel extraction kit, and used for 15 cycles of PCR amplification. Amplified DNA was finally purified with SPRI cleanup by AMPure XP beads (Invitrogen). The quality of the ChIP-seq library was determined by bioanalyzer using the high-sensitivity chip (Agilent), and the average size of the ChIP-seq libraries ranged from 250 to 350 bp. Equal molar quantities of libraries were combined and sequenced using an Illumina NextSeq platform with single-end reads of 75 bases.
ChIP-seq peak calling method and annotation
All ChIP-seq data sets were mapped to the build version GRCh37/HG19 of the human genome using Bowtie2. Peak calling was done using a MACS2 false discovery rate cutoff of 5% with the broad peak and narrow peak option for H3K27ac and TF ChIP-seq data sets, respectively. Peaks were annotated to the closest expressed gene using annotatePeaks from the Homer suite. To compare the enhancer landscape of SCLC cell lines, all called peaks across the H3K27ac ChIP-seq data sets were combined into a single peak interval set, while overlapping peaks were merged into a single interval using BEDtools. For global correlation analysis, peak intensities (50-bin average normalized tag count) were calculated for each H3K27ac data set using a combined interval set followed by Pearson correlation. Correlation coefficients were used to calculate a dissimilarity value (1 = correlation coefficient) that was subsequently used to generate unsupervised clustering (Ward/Euclidean distance) of the epigenetic landscape between the cell lines. Distance in the phylogenetic tree represents the dissimilarity. A normalized tag count was calculated using the Bamliquidator package (https://github.com/BradnerLab/pipeline).
Identification of P, N, and A elements
P, N, and A groups of cis elements were identified by calculating the H3K27ac peak intensities using the single peak interval set. The peak intensities of POU2F3-expressing (P), NEUROD1-expressing (N), or ASCL1-expressing (A) cell lines were compared with the average peak intensity of the corresponding nonexpressing cell lines identifying 463 regions (P), 697 regions (N), or 414 regions (A) of increased H3K27ac marks, respectively. Regions with a uniform peak intensity gain of at least fourfold and a minimum average peak intensity of 0.1 were considered.
Motif enrichment analysis
For transcriptional factor-binding motif analysis of 500 bp around the summit of the gained H3K27ac regions, AME (MEME suite and the JASPER and Homo sapiens Comprehensive Model Collection [HOCOMOCO] database) was used. For motifs for enrichment at P, N, and A elements, de novo motifs for NEUROD1 (GSE69398) or ASCL1 (GSE69398) (MEME-ChIP) were added to the JASPER and HOCOMOCO merged databases.
Generation of ChIP-seq tracks and heat maps
ChIP-seq tracks of individual loci were generated using the University of California at Santa Cruz genome browser. Heat map density plots and metaplots were generated using ±5 kb around each center of the H3K27ac peak intervals with 50-bp binning size. Metaplots for POU2F3, NEUROD1 (GSE69398), or ASCL1 (GSE69398) occupancy were generated from ChIP-seq data sets in NCI-H526, NCI-H82, or NCI-H128 cells, respectively.
Data availability
All next-generation sequencing data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE115124.
Supplementary Material
Acknowledgments
We thank Rafaella Sordella and members of the Vakoc laboratory for helpful discussions and suggestions throughout the course of this study. We also thank the staff at the DNA Sequencing Shared Resource and the Bioinformatics Shared Resource of the Cold Spring Harbor Laboratory Cancer Center (National Cancer Institute 5P30CA045508). C.R.V. received funding from the Pershing Square Sohn Cancer Research Alliance, Boehringer Ingelheim, the Cold Spring Harbor Laboratory and Northwell Health Affiliation, and National Institutes of Health grant National Cancer Institute RO1 CA174793. This study was also funded by National Cancer Institute 5P01CA013106 Project 3 (D.L.S), Project 4 (C.R.V), and Core C (M.E.). T.D.D.S. and X.Y.H. are supported by a grant from the state of New York (C150158).
Author contributions: Y.-H.H., X.W., X.-Y.H., G.A., B.L., T.D.D.S., and J.P.M. performed the experiments. Y.-H.H., J.S., and C.R.V. designed the study and wrote the manuscript. O.E.D. and O.K. performed bioinformatics analyses. J.E.W. performed histology analysis. J.S., D.L.S., M.E., and C.R.V. supervised the research.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.314815.118.
References
- Andersen B, Weinberg WC, Rennekampff O, McEvilly RJ, Bermingham JR Jr, Hooshmand F, Vasilyev V, Hansbrough JF, Pittelkow MR, Yuspa SH, et al. 1997. Functions of the POU domain genes Skn-1a/i and Tst-1/Oct-6/SCIP in epidermal differentiation. Genes Dev 11: 1873–1884. [DOI] [PubMed] [Google Scholar]
- Augustyn A, Borromeo M, Wang T, Fujimoto J, Shao C, Dospoy PD, Lee V, Tan C, Sullivan JP, Larsen JE, et al. 2014. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc Natl Acad Sci 111: 14788–14793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Kang S, Vogt PK. 2006. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci 103: 1475–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezencon C, Furholz A, Raymond F, Mansourian R, Metairon S, Le Coutre J, Damak S. 2008. Murine intestinal cells expressing Trpm5 are mostly brush cells and express markers of neuronal and inflammatory cells. J Comp Neurol 509: 514–525. [DOI] [PubMed] [Google Scholar]
- Bhagwat AS, Vakoc CR. 2015. Targeting transcription factors in cancer. Trends Cancer 1: 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerknes M, Khandanpour C, Moroy T, Fujiyama T, Hoshino M, Klisch TJ, Ding Q, Gan L, Wang J, Martin MG, et al. 2012. Origin of the brush cell lineage in the mouse intestinal epithelium. Dev Biol 362: 194–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, Girard L, Minna JD, Gazdar AF, Cobb MH, et al. 2016. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep 16: 1259–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo SD, Matheu A, Mariani N, Carretero J, Lopez-Rios F, Lovell-Badge R, Sanchez-Cespedes M. 2012. Novel transcriptional targets of the SRY-HMG box transcription factor SOX4 link its expression to the development of small cell lung cancer. Cancer Res 72: 176–186. [DOI] [PubMed] [Google Scholar]
- Chiappori AA, Otterson GA, Dowlati A, Traynor AM, Horn L, Owonikoko TK, Ross HJ, Hann CL, Abu Hejleh T, Nieva J, et al. 2016. A randomized phase II study of linsitinib (OSI-906) versus topotecan in patients with relapsed small-cell lung cancer. Oncologist 21: 1163–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang T, Chipumuro E, Herter-Sprie GS, Akbay EA, Altabef A, et al. 2014. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 26: 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CS, Nicholson AG, Foster C, Dodson A, Edwards S, Fletcher A, Roe T, Clark J, Joshi A, Norman A, et al. 2006. Nuclear overexpression of the E2F3 transcription factor in human lung cancer. Lung Cancer 54: 155–162. [DOI] [PubMed] [Google Scholar]
- Ding M, Bruick RK, Yu Y. 2016. Secreted IGFBP5 mediates mTORC1-dependent feedback inhibition of IGF-1 signalling. Nat Cell Biol 18: 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Carney DN, Nau MM, Minna JD. 1985. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res 45: 2924–2930. [PubMed] [Google Scholar]
- Gazdar AF, Bunn PA, Minna JD. 2017. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer 17: 725–737. [DOI] [PubMed] [Google Scholar]
- George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, Leenders F, Lu X, Fernandez-Cuesta L, Bosco G, et al. 2015. Comprehensive genomic profiles of small cell lung cancer. Nature 524: 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, et al. 2016. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529: 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et al. 2017. A single-cell survey of the small intestinal epithelium. Nature 551: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, et al. 2016. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351: 1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi M, Ayoola A, Belani CP. 2013. Small-cell lung cancer: an update on targeted therapies. Adv Exp Med Biol 779: 385–404. [DOI] [PubMed] [Google Scholar]
- Kaske S, Krasteva G, Konig P, Kummer W, Hofmann T, Gudermann T, Chubanov V. 2007. TRPM5, a taste-signaling transient receptor potential ion-channel, is a ubiquitous signaling component in chemosensory cells. BMC Neurosci 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JS, Ibaseta A, Fischer MM, Cancilla B, O'Young G, Cristea S, Luca VC, Yang D, Jahchan NS, Hamard C, et al. 2017. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 545: 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto I, Ohmoto M, Narukawa M, Yoshihara Y, Abe K. 2011. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat Neurosci 14: 685–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollaoglu G, Guthrie MR, Bohm S, Bragelmann J, Can I, Ballieu PM, Marx A, George J, Heinen C, Chalishazar MD, et al. 2017. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell 31: 270–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill MJ, Cooke A, Rosenfeld-Franklin M, Buck E, Foreman K, Landfair D, O'Connor M, Pirritt C, Sun Y, Yao Y, et al. 2009. Discovery of OSI-906: a selective and orally efficacious dual inhibitor of the IGF-1 receptor and insulin receptor. Future Med Chem 1: 1153–1171. [DOI] [PubMed] [Google Scholar]
- Nisa L, Hafliger P, Poliakova M, Giger R, Francica P, Aebersold DM, Charles RP, Zimmer Y, Medova M. 2017. PIK3CA hotspot mutations differentially impact responses to MET targeting in MET-driven and non-driven preclinical cancer models. Mol Cancer 16: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne JK, Larsen JE, Shields MD, Gonzales JX, Shames DS, Sato M, Kulkarni A, Wistuba II, Girard L, Minna JD, et al. 2013. NeuroD1 regulates survival and migration of neuroendocrine lung carcinomas via signaling molecules TrkB and NCAM. Proc Natl Acad Sci 110: 6524–6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi T, Yuan TL, Faust AM, Caron AM, Bronson R, Lees JA. 2007. Selective requirements for E2f3 in the development and tumorigenicity of Rb-deficient chimeric tissues. Mol Cell Biol 27: 2283–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Kaneda A, Tsuji S, Isagawa T, Yamamoto S, Fujita T, Yamanaka R, Tanaka Y, Nukiwa T, Marquez VE, et al. 2013. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci Rep 3: 1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova EA, Nagel R, Berns A. 2015. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev 29: 1447–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. 2011. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 3: 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. 2015. Discovery of cancer drug targets by CRISPR–Cas9 screening of protein domains. Nat Biotechnol 33: 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja TK, Sharma SK. 2004. Markers of small cell lung cancer. World J Surg Oncol 2: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarumoto Y, Lu B, Somerville TDD, Huang YH, Milazzo JP, Wu XS, Klingbeil O, El Demerdash O, Shi J, Vakoc CR. 2018. LKB1, salt-inducible kinases, and MEF2C are linked dependencies in acute myeloid leukemia. Mol Cell 69: 1017–1027.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J, Ji M, Liang HE, Locksley RM. 2016. Tuft-cell-derived IL-25 regulates an intestinal ILC2–epithelial response circuit. Nature 529: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Yamashita J, Ohmoto M, Aoude I, Ogura T, Luo W, Bachmanov AA, Lin W, Matsumoto I, Hirota J. 2014. Skn-1a/Pou2f3 is required for the generation of Trpm5-expressing microvillous cells in the mouse main olfactory epithelium. BMC Neurosci 15: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita J, Ohmoto M, Yamaguchi T, Matsumoto I, Hirota J. 2017. Skn-1a/Pou2f3 functions as a master regulator to generate Trpm5-expressing chemosensory cells in mice. PLoS One 12: e0189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Gevaert O, Zheng GXY, Anchang B, Probert CS, Larkin KA, Davies PS, Cheng ZF, Kaddis JS, Han A, et al. 2017. Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell 21: 78–90 e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn RL, Gardner EE, Marchionni L, Murphy SC, Dobromilskaya I, Hann CL, Rudin CM. 2013. ERK phosphorylation is predictive of resistance to IGF-1R inhibition in small cell lung cancer. Mol Cancer Ther 12: 1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All next-generation sequencing data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE115124.