Here, Shimotohno et al. investigated how upstream factors that regulate WUS and WOX genes converge to position organizer cells during embryogenesis, initiation of new lateral organs, and regeneration after tissue damage in Arabodopsis. Here, they show that PLT and SCR genes genetically and physically interact with plant-specific teosinte-branched cycloidea PCNA (TCP) transcription factors to specify the stem cell niche during embryogenesis and maintain organizer cells post-embryonically.
Keywords: Arabidopsis, stem cell niche, organizer, quiescent center, PLT-TCP-SCR complexes, WUSCHEL-RELATED HOMEOBOX 5
Abstract
Continuous formation of somatic tissues in plants requires functional stem cell niches where undifferentiated cells are maintained. In Arabidopsis thaliana, PLETHORA (PLT) and SCARECROW (SCR) genes are outputs of apical–basal and radial patterning systems, and both are required for root stem cell specification and maintenance. The WUSCHEL-RELATED HOMEOBOX 5 (WOX5) gene is specifically expressed in and required for functions of a small group of root stem cell organizer cells, also called the quiescent center (QC). PLT and SCR are required for QC function, and their expression overlaps in the QC; however, how they specify the organizer has remained unknown. We show that PLT and SCR genetically and physically interact with plant-specific teosinte-branched cycloidea PCNA (TCP) transcription factors to specify the stem cell niche during embryogenesis and maintain organizer cells post-embryonically. PLT–TCP–SCR complexes converge on PLT-binding sites in the WOX5 promoter to induce expression.
In both the animal and plant kingdoms, stem cell niches contain specialized organizer cells that maintain surrounding stem cells (Dolan et al. 1993; Sablowski 2004, 2011; Scheres 2007; Dinneny and Benfey 2008). In Arabidopsis thaliana, root stem cells are maintained by a small group of slowly dividing organizer cells collectively called the quiescent center (QC). The QC cells have roles in the maintenance of abutting stem cells and the replenishment of a subset of these stem cells (Dolan et al. 1993; van den Berg et al. 1995; Cruz-Ramírez et al. 2013). The stem cells, in turn, are pluripotent cells whose progeny normally generate defined cell types of the root depending on their position but reveal developmental plasticity upon perturbation (Xu et al. 2006; Sena et al. 2009). The entire root stem cell niche is first laid down in the embryo but can be re-established during lateral root formation or during regeneration upon damage (Malamy and Benfey 1997; Sena et al. 2009; Efroni et al. 2016; Du and Scheres 2017).
In past decades, two different classes of plant-specific transcription factors that have key roles in root meristem and stem cell maintenance have been investigated in some detail. Double APETALA2 (AP2)/ethylene-responsive element-binding proteins (EREBPs) PLETHORA/ANTEGUMENTA-LIKE (PLT/AIL; PLT here) and the GRAS family transcription factor SCARECROW (SCR) play central roles for defining root stem cell niche identities (Wysocka-Diller et al. 2000; Sabatini et al. 2003; Aida et al. 2004; Heidstra et al. 2004; Galinha et al. 2007). In terms of maintenance of stem cell activities, most PLT genes act redundantly, and their ectopic induction results in ectopic stem cell niche formation and organogenesis (Boutilier et al. 2002; Galinha et al. 2007; Tsuwamoto et al. 2010). Besides their requirement for QC identity and stem cell maintenance, PLT proteins form a gradient that guides progression of cells from the stem cell state to the transit-amplifying cell state and finally to differentiation (Galinha et al. 2007; Mähönen et al. 2014; Santuari et al. 2016). SCR regulates root differentiation and is required to maintain stem cells by its cell-autonomous activity in organizer cells (Sabatini et al. 2003; Heidstra et al. 2004; Cruz-Ramírez et al. 2012, 2013; Moreno-Risueno et al. 2015). In addition, SCR (together with its partner protein, SHORT-ROOT [SHR]) is required for asymmetric cell division of the cortex/endodermis initial (CIE) daughter (CEID) cell in the ground tissue, which gives rise to cortex and endodermis cell lineages (Benfey et al. 1993; Scheres et al. 1994; Di Laurenzio et al. 1996; Scheres and Benfey 1999; Helariutta et al. 2000). Furthermore, SCR is required for the regulation of QC division rate, which determines how rapidly abutting stem cells are replenished (Cruz-Ramírez et al. 2013).
The embryonic initiation of the root stem cell niche in Arabidopsis is marked by a stereotypic transverse asymmetric cell division within the hypophyseal cell during the early to mid-globular embryo stages. The smaller lens-shaped apical daughter acquires QC cell identity, whereas the basal descendant cell becomes distal columella stem cells (Jürgens et al. 1994; Scheres and Benfey 1999; Jürgens 2001; Weigel and Jürgens 2002; Ten Hove and Heidstra 2008; Ten Hove et al. 2015). Although PLT and SCR genes are expressed in partially overlapping larger domains (of which the QC forms a subset), loss-of-function mutants of SCR and combinations of loss of function of PLT clade members lead to differentiation of the root stem cell niche and decrease the expression of different QC identity markers from embryogenesis onward (Sabatini et al. 2003; Aida et al. 2004; Galinha et al. 2007). Up to now, it has not been revealed how their activities might converge for QC specification in such a narrow domain. Reported target genes of the SHR/SCR pathway (Levesque et al. 2006; Sozzani et al. 2010; Moreno-Risueno et al. 2015) and of the root-expressed PLT genes (Santuari et al. 2016) do not show overall overlap, leaving it unclear whether SCR and PLT regulate identical target genes relevant for stem cell niche function.
In contrast to PLT and SCR (whose expression encompasses larger domains, including the QC), expression of the gene encoding homeodomain transcription factor WUSCHEL (WUS)-RELATED HOMEOBOX 5 (WOX5) is highly enriched in the QC (Sarkar et al. 2007). WOX5 is also required for QC division rate control and the maintenance of at least a subset of surrounding stem cells (Sarkar et al. 2007; Pi et al. 2015; Zhang et al. 2015). Both WOX5 and its shoot-expressed homolog, WUS, are required for the function of organizer cells of roots and shoots, respectively (Mayer et al. 1998; Sarkar et al. 2007). The mechanisms by which PLT and SCR converge in regulating the WOX5 root expression domain and how this links to specification of the QC have remained unknown.
Class I members of the teosinte-branched cycloidea PCNA (TCP) protein family encode plant-specific transcription factors (Li et al. 2005; Hervé et al. 2009; Martín-Trillo and Cubas 2010; Li 2015). Class I TCPs are implicated in the coordination of cell proliferation and development, especially during leaf development, lateral branching, and shoot apical meristem formation in several plant species (Aguilar-Martínez and Sinha 2013; Davière et al. 2014). Loss-of-function mutants in class I TCP genes or EREB factor-associated amphiphilic repression (EAR) motif-fused class I TCP proteins show developmental alterations, suggesting that they are positive regulators of meristem formation (Hervé et al. 2009; Kieffer et al. 2011; Aguilar-Martínez and Sinha 2013; Li 2015).
Here we investigate how the two major PLT and SCR pathways for root stem cell niche specification converge to specify QC cells within the root stem cell niche. We show that both PLTs and SCR interact with specific class I TCP proteins. We provide genetic and molecular evidence that PLT1, PLT3, SCR, TCP20, and TCP21 proteins cooperate for the specification of QC identity and the induction of WOX5 expression in at least four developmental contexts: embryogenesis, primary root formation, secondary root development, and the root regeneration process. Our data connect hitherto separated stem cell pathways through novel protein complexes that regulate the formation of the root stem cell organizer and expression of a key gene involved in its function.
Results
Class I TCP proteins interact with both PLTs and SCR protein through different motifs
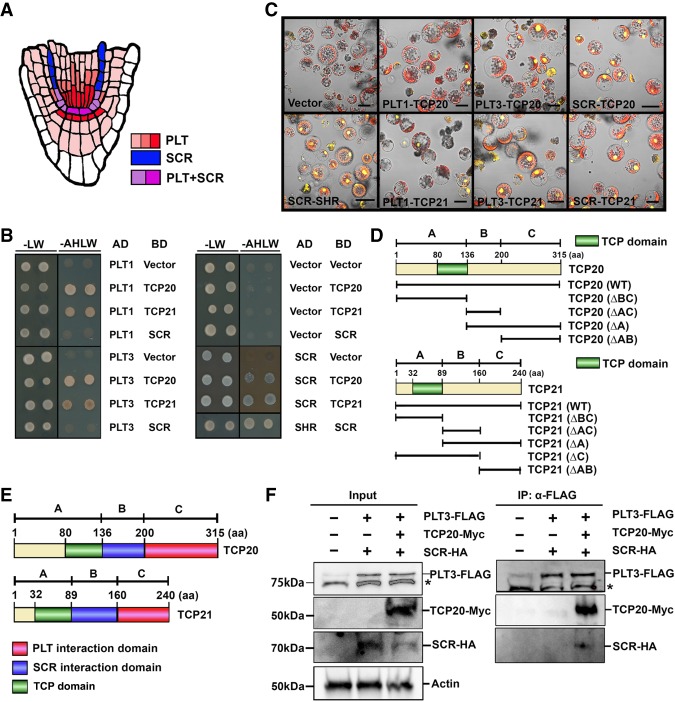
PLT1, PLT2, AIL6/PLT3 (referred to here as PLT3), and BBM/PLT4 (referred to here as PLT4) proteins form gradients across the root meristem with maximum levels in the stem cell niche domain, whereas SCR expression is restricted mainly to the QC, cortex/endodermis stem cell, and endodermis (Fig. 1A; Wysocka-Diller et al. 2000; Galinha et al. 2007). To investigate whether their functions in the stem cell niche might converge at the protein level, we performed yeast two-hybrid screening (Y2H) of an Arabidopsis root cDNA library to search for interacting factors of PLT1, PLT2, PLT3, PLT4, and SCR proteins. Since the full-length coding sequence of PLTs elicited strong autoactivation in yeast cells, we used C-terminal regions, including the double AP2 domain of PLT1 (395 amino acids), PLT2 (380 amino acids), PLT3 (338 amino acids), and PLT4 (376 amino acids), fused to the GAL4 activation domain. In total, 25 putative interactors were identified at least in triplicate for PLT1, 23 were identified for PLT2, 18 were identified for PLT3, 21 were identified for PLT4, and 25 were identified for SCR (Supplemental Table 1). We focused on two common interactors between the bait proteins. Retransformation full-length cDNAs for one-to-one assays with PLTs or SCR revealed that one of the plant-specific transcription factor TCP family members, TCP20, interacted with PLT1, PLT3, and SCR proteins (Fig. 1B). We did not find other TCP proteins in the SCR Y2H screening; however, like TCP20, TCP21 is highly expressed across the root meristem. Therefore, we tested the ability of TCP21 to interact with SCR in a one-to-one assay, which flagged TCP21 as a candidate SCR interactor. The Y2H tests did not reveal reproducible interactions between PLT2 and PLT4 with these two TCP proteins. We validated the shared interactors in plant cells using bimolecular fluorescent complementation (BiFC) analysis, where cDNAs driven by the 35S promoter were fused to the N-terminal or C-terminal half of YFP and cotransfected into Arabidopsis mesophyll protoplasts (Fig. 1C). TCP proteins possess a centrally located conserved common motif, the TCP domain, which has been implicated in DNA-binding sites as well as TCP–TCP dimerization but not in interactions with other proteins (Cubas et al. 1999; Broholm et al. 2008; Aggarwal et al. 2010; Shutian 2015). To assess whether the PLT and SCR proteins bound to separate or the same TCP interaction domains, we resolved which TCP protein regions are required for PLT and/or SCR interactions. We generated a truncation series of TCPs using three regions—for TCP20, domain A (1–408 base pairs [bp]), domain B (409–600 bp), and domain C (601–945 bp) and, for TCP21, domain A (1–267 bp), domain B (268–480 bp), and domain C (481–720 bp)—represented in Figure 1D. We investigated the interaction strengths of truncated TCPs with full-length PLT3 or SCR in the yeast system. As shown in Supplemental Figure 1, A and B, PLT3 interacted with domain C of TCP20 and TCP21, whereas SCR interacted with domain B of both TCP proteins. These data indicated that TCPs directly interact with PLTs and SCR through different regions C-terminal to the TCP domains. BiFC assays using these truncated TCP fragments confirmed these protein interactions in living plant cells, indicating that TCP20 and TCP21 bind with PLT proteins and SCR through different motifs (Fig. 1E; Supplemental Fig. 1C,D). To further test whether PLT and SCR could simultaneously interact with TCP proteins, we performed coimmunoprecipitation assays after transient expression in Arabidopsis mesophyll protoplasts. TCP20, PLT3, and SCR were fused with 10xMyc, 3xFlag, and 7xHA tags, respectively, driven by 35S promoters and cotransfected into Arabidopsis mesophyll protoplasts. Coimmunoprecipitation assays after anti-Flag pull-down showed that TCP20 interacted with PLT3 and SCR in planta and that TCP20 was required for SCR interaction with PLT3, suggesting that PLTs and SCR assemble in vivo into a TCP-containing complex (Fig. 1F; Supplemental Fig. 1E).
Figure 1.
Protein interactions between TCPs and PLTs/SCR. (A) Scheme of PLTs and SCR protein expression in the Arabidopsis root apical meristem. PLTs are expressed broadly across the stem cell niche in the root (red), whereas SCR expression is restricted in the endodermis and QC cells (blue). The PLT and SCR expression domains overlap predominantly in QC cells and CEIs (purple). (B) Y2H assays between TCP20 or TCP21 and PLT1 or PLT3 or SCR were performed on SD/−Leu−Trp− Ade−His (−AHLW) medium for 4 d. SD/−Leu−Trp (−LW) medium was used as a growth control. Vector combination was used as a negative control, whereas SCR–SHR interaction was used as a positive control. (C) BiFC assay of the interaction between full-length PLT1 or PLT3 or SCR and TCP20 or TCP21 in Arabidopsis mesophyll protoplasts. Vector combination (vector) was used as a negative control, whereas SCR–SHR was used as a positive control. Bar, 30 µm. (D) Schematic presentations of the protein domains for truncated TCP20 and TCP21 used in Supplemental Figure 1, A–C. TCP domains are indicated in green. The numbers at the top indicate amino acids (aa). (E) The deduced domains of TCP protein binding to PLT (red), SCR (blue), and TCP domain (green). (F) Coimmunoprecipitation assay showing the positive interaction of PLT3–TCP20–SCR. 35S::PLT3-3xFlag, 35S::SCR-7xHA, and 35S::TCP20-10xMyc constructs were cotransfected in Arabidopsis mesophyll protoplasts and subjected to immunoprecipitation with anti-Flag antibody (IP:α-Flag), shown in the right panel. The left panel shows total cell lysate (input) from Arabidopsis mesophyll protoplasts expressing PLT3-Flag, SCR-HA, and TCP20-Myc followed by Western blot analysis. The presence of PLT3, SCR, and TCP20 was determined by anti-Flag, anti-HA, and anti-Myc antibody, respectively. Anti-actin antibody was used as a loading control. The asterisks indicate nonspecific bands. The original gel blot source is available in Supplemental Figure 1E.
plt, tcp, and scr mutants display synergistic genetic interactions
To assess the potential relevance of PLT and SCR protein interaction mediated through TCP proteins, we investigated genetic interactions in different combinations of their T-DNA insertion alleles (Danisman et al. 2012). We selected plt1-3, plt3-1, tcp20-1, tcp21-1, and scr-3 mutant lines for the analysis (Supplemental Fig. 2A; Fukaki et al. 1998; Aida et al. 2004; Galinha et al. 2007). Quadruple plt1-3−/−plt3-1−/−tcp20-1−/−scr-3−/− mutants further enhanced the root phenotype of scr and blocked primary root, but not shoot, growth (Fig. 2A,B; Supplemental Fig. 2C). Specifically, primary root growth and meristem size were reduced in several combinations of loss-of-function alleles of PLT, TCP, and SCR genes (Fig. 2C; Supplemental Fig. 2B–D). We investigated the contribution of individual alleles further by varying plt1-3, plt3-1, and tcp20-1 mutant allele dosage in seedlings homozygous for scr-3−/− and determined primary root length in genotyped mutant backgrounds. The data revealed that primary root length was affected by PLT and TCP20 dosage in the absence of SCR (Supplemental Fig. 2C). Next, we investigated the dosage effect of SCR and TCP21 by analyzing the segregating lines of the scr-3−/− mutant crossed to the tcp21-1−/− mutant (Supplemental Fig. 2D). While the double-homozygous mutant caused severe primary root growth defects, the heteroallelic combination tcp21-1+/−scr-3+/−showed an intermediate root length, indicating that SCR function also contributes to root growth in a dose-dependent manner upon TCP21 reduction. Together, our data indicate that a reduced level of each of the four proteins creates dosage sensitivity for the others, which is consistent with their physical interactions being relevant for root growth.
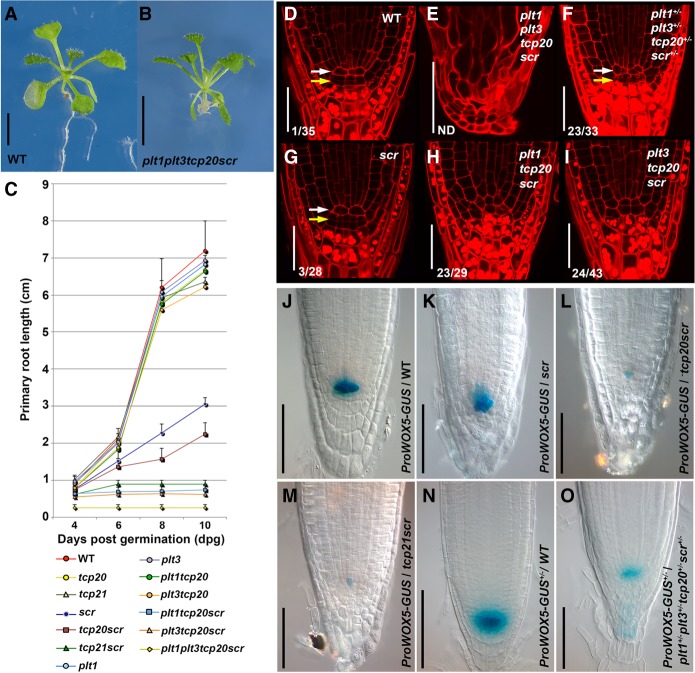
Figure 2.
The effect of plt, tcp, and scr genetic interaction on growth and stem cell maintenance in the Arabidopsis root. (A,B) Seedlings of wild-type (A) and the plt1-1−/−plt3-2−/−tcp20-1−/−scr-3−/− quadruple mutant (B) 10 d after germination (dag). (C) Primary root length measurements of indicated wild-type and mutant seedlings from 4 to 10 dag. Values are average lengths (means ± SD) of >25 seedling roots per genotype per time point. (D–I) The root apical meristem of 2-dag seedlings in wild-type (D), plt1−/−plt3−/−tcp20−/−scr−/− (E), plt1+/−plt3+/− tcp20+/−scr+/− (F), plt1+/+plt3+/+tcp20+/+scr−/− (G), plt1−/−plt3+/+tcp20−/−scr−/− (H), and plt1+/+plt3−/−tcp20−/−scr−/− (I). Arrows indicate the QC (white) and columella stem cells (CSCs; yellow), respectively. Numbers indicate roots with additional QC divisions of total roots examined. (ND) Not determined. (J–M) pWOX5-GUS expression levels in Columbia-0 (Col-0; J), scr-3−/− (K), tcp20-1−/−scr-3−/− (L), and tcp21-1−/−scr-3−/− (M) roots. (N,O) GUS expression levels in pWOX5-GUS+/− wild-type (N) and pWOX5-GUS+/−plt1+/−plt3+/− tcp20+/− scr-3+/− (O) roots. The images displayed in J–O are representative of at least three independent experiments with >10 seedlings examined that obtained similar results. Bars: A,B, 1 cm; D–I, 30 µm; J–O 40 µm.
We investigated whether the observed reduction of root growth and meristem cell number in plt, tcp, and scr mutant combinations was accompanied by defects in the root stem cell niche. In the wild type, only differentiated columella cells (CCs) contain large starch granules that are not observed in columella stem cells (CSCs) or in the QC (Fig. 2D). Two days after germination (dag) plt1-3−/− tcp20-1−/−scr-3−/− and plt3-1−/−tcp20-1−/−scr-3−/− triple-homozygous mutants displayed ectopic cell divisions in the QC and starch granule accumulation into CSCs (Fig. 2H,I), while the 2-dag scr-3−/− single mutant still maintains the wild-type stem cell niche anatomy (Fig. 2, cf. D and G). In addition, at 2 dag, the plt1-3−/−plt3-1−/−tcp20-1−/−scr-3−/− quadruple mutants no longer display the typical cell arrangement associated with a functional stem cell niche and meristem (Fig. 2, cf. D and E). To assess a potentially synergistic role in the stem cell niche for PLT, TCP, and SCR proteins, we generated plt1-3+/−plt3-1+/− tcp20-1+/−scr-3+/− trans-heterozygous mutant seedlings. Like in wild type, starch granule accumulation in plt1-3+/− plt3-1+/−tcp20-1+/−scr-3+/− mutants indicates the presence of starch-free CSCs below the QC. However, these mutants revealed extra cell divisions in the QC, which is generally associated with improper QC function (Fig. 2F; Capron et al. 2009; Bennett and Scheres 2010; Petricka et al. 2012; Forzani et al. 2014; Heyman et al. 2014). To test whether the extra QC cell divisions observed upon mutation of SCR and TCP genes correlated with incomplete QC specification, we introgressed the pWOX5-GUS promoter fusion. Five days after germination, WOX5 promoter activity was strongly reduced in tcp20−/−scr-3−/− and tcp21−/scr-3−/− double mutants, while it was much less affected in the scr-3−/− single-mutant root tip (Fig. 2J–M). Finally, we generated a plt1-3+/−plt3-1+/−tcp20-1+/− scr-3+/− trans-heterozygous mutant with one copy of the pWOX5-GUS promoter fusion and compared WOX5 promoter activity with the same single-gene copy in wild type. We consistently detected lower WOX5 expression in the quadruple trans-heterozygote (Fig. 2N,O). Together, our data indicate that the interacting PLT, TCP, and SCR proteins act in a dosage-dependent manner to maintain the QC and stem cells.
PLT, TCP, and SCR are required for embryonic specification of the root stem cell niche
Our genetic analysis indicated that PLT–TCP–SCR complexes are required for stem cell niche function at the earliest stages of root emergence, raising the question of whether these proteins are jointly expressed and required during embryogenesis, where the root stem cell niche progenitors are established at the globular stage. In wild type, the extraembryonic hypophyseal cell is specified by a combination of auxin signaling and a mobile transcription factor (Schlereth et al. 2010). After this specification step, the hypophysis undergoes a stereotyped asymmetric cell division that creates the lens-shaped QC progenitor (Mansfield and Briarty 1991; Jürgens et al. 1994; Capron et al. 2009; Ten Hove et al. 2015). We traced back the origin of the abnormal root phenotype of the plt1-3plt3-1tcp20-1scr-3 mutant during embryogenesis. The quadruple mutant revealed atypical cell divisions in the hypophyseal cell at the dermatogen stage with a frequency of 38% at the globular stage, correlated with late embryo lethality (Fig. 3A; Supplemental Table 3). Lower-order plt tcp scr mutant combinations also revealed specific cell division orientation defects in hypophyseal cells but with lower frequency: 10.8% in plt1-3−/−tcp20-1−/−scr-3−/− and 18.3% in plt3-1−/−tcp20-1−/−scr-3−/− (Supplemental Table 3). Notably, the apical embryo region of triple and quadruple mutants at 16-cell and earlier stages revealed no morphological defects.
Figure 3.
Genetic interaction among PLT, TCP, and SCR during embryogenesis. (A) The wild-type (Col-0) and plt1-3−/−plt3-1−/−tcp20-1−/−scr-3−/− embryos at the dermatogen to globular stages. Original cleared images (left) and merged images with tracings of embryos (right) are shown. Bars, 20 µm. (B) Expression patterns of PLT3-YFP, SCR-YFP, TCP20-YFP, and pWOX5::H2B-YFP at the transition between the dermatogen and early globular stages (left and middle) and expression heat maps during the late globular/transition states (right) are shown. Bars, 10 µm. (C) YFP signal intensities of PLT1-YFP, PLT3-YFP, SCR-YFP, TCP20-YFP, TCP21-YFP, and pWOX5::H2B-YFP during the octant to transition/heart stages exemplified in B. Box length represents the range in which the central 50% of the values fall, with the box edges at the lower (orange) and upper (gray) quartiles. The whiskers indicate the highest and lowest values. YFP fluorescence intensities (n > 15) in the QC (or its precursor cells in the octant and dermatogen stages) were quantified using ImageJ. (D) Cellular anatomies of the radicle in wild-type and scr-3−/−, plt1-1−/−tcp20-1−/−scr-3−/−, plt3-1−/−tcp20-1−/−scr-3−/−, tcp20-1−/−scr-3−/−, and plt1-3−/−plt3-1−/−tcp20-1−/−scr-3−/− homozygous mature embryos. The numbers of embryos that showed improper cell divisions in the QC per examined total embryos are indicated in the respective panels. (Yellow arrows) Position of the QC; (dots) positions of the columella cell layers. Bars, 30 µm.
PLT1 mRNA accumulation in the preglobular stage embryo is restricted to basal cells, which will form the QC in later stage (Blilou et al. 2005). To assess whether the expression patterns of other PLT, TCP, and SCR proteins were consistent with a role in QC specification, we analyzed the expression level and distribution of PLT3, TCP20, TCP21, and SCR proteins during the relevant embryonic stages using functional fusion proteins. SCR, TCP20, and TCP21 protein fusions were specifically enriched in the hypophyseal cell at the dermatogen stage. Also at that stage, weak PLT3 expression was detected (Fig. 3B,C). At the early globular stage, the PLT3, TCP20, TCP21, and SCR proteins are enriched in the QC and neighboring cells at all embryonic stages and become more prominent in the apical region at the heart stage (Fig. 3B [right], C). It is at the early globular stage that WOX5 expression is initiated, consistent with the observed dependency of WOX5 expression on these factors in post-embryonic roots (Figs. 2J–O, 3B). It is of note that both TCP20 and TCP21 are expressed in (precursor) QC cells, but TCP21 expression is more restricted and retained in these cells from the dermatogen stage to the late globular stage (Fig. 3B). Taken together, the overlapping expression profiles of relevant PLT, TCP, and SCR fusion proteins coincide with the defects in the establishment of appropriate cell division planes in the hypophyseal cell that accompany the formation of the stem cell niche.
To test whether abnormal early stem cell progenitor formation interfered with the later development of the root stem cell niche during embryogenesis, we quantified the number of columella cells and frequencies of cell divisions in the QC in mature embryos of wild type, scr-3−/−, and four plt–tcp–scr-derived mutants: tcp20-1−/−scr-3−/−, plt1-3−/−tcp20-1−/−scr-3−/−, plt3-1−/−tcp20-1−/−scr-3−/−, and plt1-3−/−plt3-1−/−tcp20-1−/−scr-3−/− (Fig. 3D; Supplemental Table 4). All triple-mutant combinations showed an increase in embryonic QC divisions and fewer columella cell layers, whereas tcp20-1−/−scr-3−/− double and scr-3−/− single mutants were less affected (Fig. 3D; Supplemental Table 4).
Analysis of embryo development revealed that a stereotyped early embryonic induction pattern of PLT1, PLT3, SCR, TCP20, and TCP21 corresponded to an early requirement for these factors to specify the lens-shaped QC cell (Fig. 3C). However, QC specification in the primary root is not restricted to early embryonic stages, as manipulation of auxin distribution and regeneration after cell or tissue damage leads to respecification of QC identity in new regenerated tissues (Jiang and Feldman 2005; Weijers et al. 2005; Xu et al. 2006; Terpstra and Heidstra 2009; Pulianmackal et al. 2014; Efroni et al. 2016). To investigate how the expression patterns of the transcription factors involved in QC specification and WOX5 expression are set up during the re-establishment of a new stem cell niche during regeneration, we performed QC laser ablations and monitored expression recovery of PLT3, SCR, and TCP20 proteins as well as the QC marker pWOX5::H2B-YFP. Six hours after QC ablation, PLT3 already showed expression in the region of the presumptive regenerating stem cell niche when other regulatory factors (SCR and TCP20) were still undetectable (Supplemental Fig. 3). Six hours later, PLT3 and TCP20 showed pronounced expression in this area, and we detected SCR-YFP and pWOX5::H2B-YFP in the newly regenerated area (Supplemental Fig. 3). These observations and the reported requirement of stem cell niche regeneration on PLT and SCR genes (Xu et al. 2006) suggest a scenario in which the PLT–TCP–SCR expression pattern overlap triggers the selection of organizer cells also in the context of regeneration.
PLT–TCP–SCR genes are redeployed for QC specification during lateral root development
QC specification is reiterated in a different developmental context during the post-embryonic formation of lateral roots (Bennett and Scheres 2010; Hofhuis et al. 2013; Tian et al. 2014; Goh et al. 2016; Du and Scheres 2017). During lateral root initiation, two to three adjacent pericycle cell files undergo several rounds of anticlinal divisions to create a single layered stage I lateral root primordium (LRP) composed of up to eight to 10 cells of equal size. Two rounds of periclinal cell divisions in the central cells generate the stage III primordium. In the stage III LRP, the central cells act as QC progenitors based on fate mapping and the stable expression of WOX5 in these cells from that stage onward (Supplemental Fig. 4A; Ditengou et al. 2008; Goh et al. 2016; Rovere et al. 2016).
To investigate the potential relevance of PLT–TCP–SCR cooperative action during LRP formation, we monitored LRP development in 7-dag seedlings of wild type and three different plt tcp scr mutant combinations (Fig. 4; Supplemental Fig. 4). All compound mutant LRPs tested as well as the scr-3 single mutant itself were indistinguishable from wild type at stage I and early stage II (Fig. 4A-2,A-3,B-2,B-3). However, from late stage II onward, we observed aberrant cell sizes and division planes in the cell files that generate QC progenitor cells in wild type (Fig. 4B1–D; Supplemental Fig. 4A). To correlate the region of morphological abnormalities in compound mutant LRP-to-QC specification, we followed pWOX5-erGFP expression, which we consistently detected in the central cells of interposed cell layers at the emerging state, whereas consistent signals were undetectable in stage I and generally initiated in stage II (Fig. 4A-7,B-7,C-7). In contrast to the strong effects on LRP morphology in the compound mutants, we observed no strong reduction of LRP density in single, double, and triple plt tcp scr mutants (Fig. 4E) but rather an increase in the number of abnormal lateral roots with defective meristems, indicative of the failure to maintain a stem cell niche (Fig. 4D).
Figure 4.
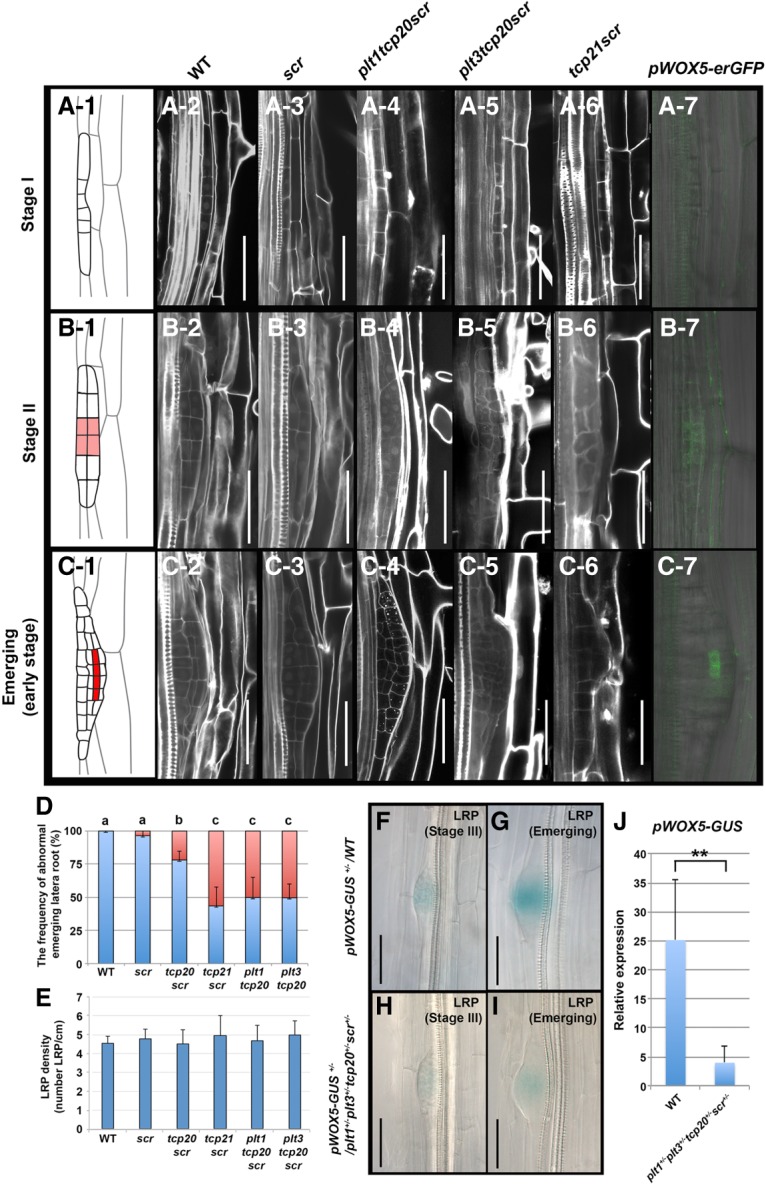
The lateral root formation in various plt–tcp–scr mutant combinations. (A-1–C-7) Early developmental stages during lateral root initiation in stage I (A-1–A-7), stage II (B-1–B-7), and emerging (C-1–C-7). Cell lineage maps are shown in A-1 (stage I), B-1 (stage II), and C-1 (emerging). (B-1,C-1) QC progenitor cells appear from late stage II onward (highlighted in red). Morphologies of stage I, stage II, and the emerging stage present in wild-type Col-0 (A-2,B-2,C-2), scr-3 (A-3,B-3,C-3), plt1-3−/−tcp20-1−/−scr-3−/− (A-4,B-4,C-4), plt3-1−/−tcp20-1−/− scr-3−/− (A-5,B-5,C-5), and tcp21-1−/− scr-3−/− (A-6,B-6,C-6), respectively. pWOX5-erGFP expression is observed in stage II (B-7) and the emerging stage (C-7) but is absent in stage I (A-7) wild-type primordium. Bars, 50 µm. (D) The frequencies (percentage) of the abnormal LRPs on the primary root from 8-dag wild-type and plt–tcp–scr mutant combination lines, as indicated. (Blue bar) Normal primordium; (red bar) abnormal primordium. Error bars show SDs. The letters above the bars (a, b, c) indicate significant differences (one-way ANOVA and Tukey's test, P < 0.01). Results are means ± SD, n = 15 per line. (E) Statistical analysis of the number of lateral roots per centimeter of the primary root from the 8-dag wild-type and the series of plt–tcp–scr mutant combinations. Results are means ± SD, n = 15, no significant differences by one-way ANOVA. (F–I) Expression levels of pWOX5-GUS+/− in LRPs of wild-type (F,G) and plt1-3+/− plt3-1+/−tcp20-1+/− scr-3+/− mutant (H,I) backgrounds. (F,H) Stage III. (G,I) Emerging LRP. Bar, 40 µm. (J) Statistical analysis of pWOX5-GUS+/− expression in F–I. Results are means ± SD. n = 46 in wild-type; n = 40 in plt1-3+/−plt3-1+/−tcp20-1+/−scr-3+/−. (**) P < 0.01, compared with the corresponding values of wild-type seedlings; two-tailed t-tests.
We asked whether PLT, TCP, and SCR proteins, such as in the context of the primary root, might cooperatively regulate WOX5 expression during lateral root formation. To this end, we monitored WOX5 promoter activity in pWOX5-GUS+/− and pWOX5-GUS+/−plt1-3+/−plt3-1+/− tcp20-1+/−scr-3+/− LRPs (Fig. 4F–I). In wild type, WOX5 expression was observed from stage II onward during lateral root formation and gradually increased in strength (Fig. 4B-7, C-7, F,G; Du and Scheres 2017). The trans-heterozygous pWOX5-GUS+/−plt1-3+/−plt3-1+/−tcp20-1+/−scr-3+/−, however, possessed weaker expression at stage II and later stages (Fig. 4H–J). Our results suggest that, like in primary roots, the induction of WOX5 expression during lateral root formation depends on joint activity of PLT, TCP, and SCR.
To determine whether PLT, TCP, SCR, and WOX5 expression patterns during LRP development coincide with the morphological and gene expression defects observed in their mutants, we compared the expression dynamics of five fusion proteins (PLT3, SCR, SHR, TCP20, TCP21) and WOX5 promoter activation during lateral root initiation (Supplemental Fig. 4B). PLT3, TCP20, and TCP21 fusion proteins were detected at stage I, whereas SCR expression was consistently observed in the stage II outer cell layer (Goh et al. 2016; Du and Scheres 2017). Notably, WOX5 promoter activity was detected from stage II onward in cells where PLT, SCR, and TCP proteins are all expressed. Together with our genetic data, these results indicate that joint activity of PLT–TCP–SCR proteins is essential for the specification of the QC and function of the stem cell niche in two very different developmental contexts: the early embryo and post-embryonic lateral root formation.
Expression overlap and synergistic activity of PLT and SCR proteins in the stem cell niche can limit WOX5 gene expression to the QC
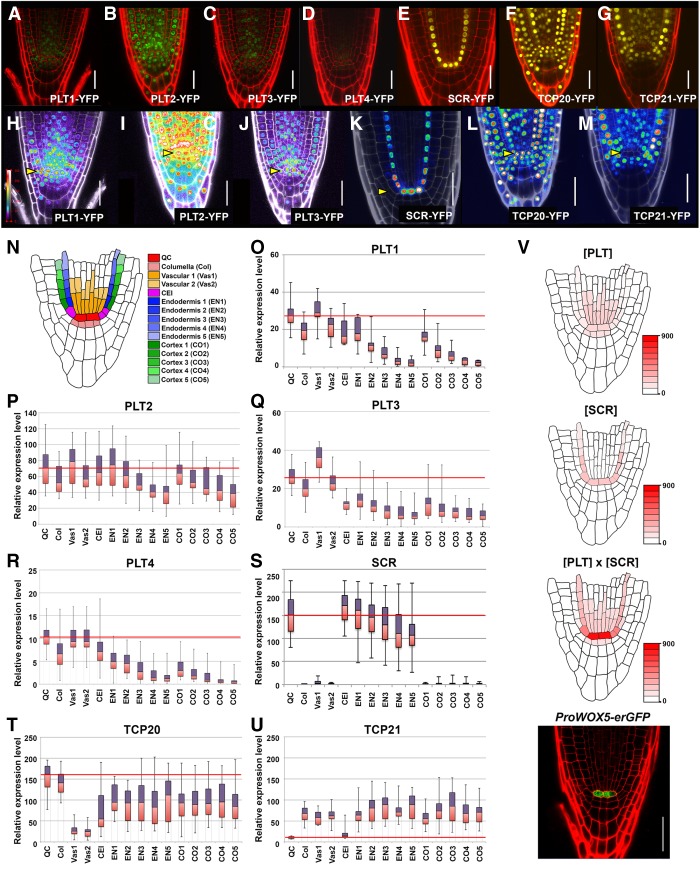
PLT proteins display a graded distribution and may assign stem cell niche states in a dose-dependent manner (Galinha et al. 2007; Mähönen et al. 2014). We asked whether the distribution of PLT–TCP–SCR proteins in the context of the primary root might explain the domain of QC specification. To this end, we quantified expression levels of the four PLTs (PLT1–PLT4), SCR, and two TCPs (TCP20 and TCP21) in the root meristem (Fig. 5). We classified 15 cell positions in and around the stem cell niche (Fig. 5N) and calculated fluorescence intensities in PLT-YFP, TCP-YFP, and SCR-GFP protein fusion lines pPLT1::PLT1-YFP, pPLT2::PLT2-YFP, pPLT3::PLT3-YFP, pPLT4::PLT4-YFP, pTCP20::TCP20-YFP, pTCP21::TCP21- YFP, and pSCR::SCR-YFP using ImageJ software (Fig. 5A–G,O–U).
Figure 5.
PLT1, PLT3, and SCR protein levels in and around the stem cell niche. (A–G) pPLT1::PLT1-YFP, pPLT2::PLT2-YFP, pPLT3::PLT3-YFP, pPLT4::PLT4-YFP, pSCR::SCR-YFP, pTCP20::TCP20-YFP, and pTCP21::TCP21-YFP expression in 2-dag root apical meristems. All fluorescence signals of PLT1-YFP (A), PLT2-YFP (B), PLT3-YFP (C), and PLT4-YFP (D) seedlings roots at 2 dag were imaged using identical laser settings. (H–M) Heat maps showing fluorescence intensities corresponding to A–G. PLT1-YFP (H), PLT2-YFP (I), PLT3-YFP (J), SCR-YFP (K), TCP20-YFP (L), and TCP21-YFP (M) expression patterns are shown. (Yellow arrowheads) Positions of the QC cells. Bar, 20 µm. (N) Schematic transverse section depicting Arabidopsis root stem cells and their progeny. (O–U) Box plot of average fluorescence intensities over the root stem cell niche of 2-dag seedlings for all four PLTs, SCR, and two TCPs according to the color code in N. The red lines across the middle of the boxes identify the median sample values in QC cells. The number of seedlings used in these experiments: pPLT1::PLT1-YFP (n = 19; O), pPLT2::PLT2-YFP (n = 13; P), pPLT3::PLT3-YFP (n = 45; Q), pPLT4::PLT4-YFP (n = 20; R), pSCR::SCR-YFP (n = 42; S), pTCP20::TCP20-YFP (n = 18; T), and pTCP21::TCP21-YFP (n = 20; U). (V) The overlap between PLT and SCR using expression levels from O, Q, and S and synergy data from Figure 6E ([PLT] × [SCR]) establishes a stronger enriched domain of QC specification and WOX5 expression in the stem cell niche than PLT or SCR alone ([PLT] or [SCR]). Red implies the highest expression of WOX5. The actual pWOX5-erGFP expression pattern in wild type (ProWOX5-erGFP). Bar, 30 µm.
pPLT1::PLT1-YFP, pPLT2::PLT2-YFP, pPLT3::PLT3-YFP, pPLT4::PLT4-YFP, and pSCR::SCR-YFP lines were selected previously for (near) wild-type function activities by complementation tests, and we used these homozygous lines for analysis (Galinha et al. 2007; Cruz-Ramírez et al. 2012). Both TCP20 and TCP21 are broadly expressed across the root stem cell niche at 2 dag, coinciding with the reactivation of the post-embryonic stem cell niche after seed germination (Fig. 5F,G,L,M,T,U). Consistent with previous reports, all of the PLT proteins measured revealed graded distributions that peak within the stem cell niche. However, the expression levels among PLTs throughout the root meristem were diverse (Fig. 5A–D). PLT1 and PLT3 fusions, which have the most prominent TCP interaction, peak in QC cells and vascular initial cells (Fig. 5A,C,H,J,O,Q). PLT2-YFP was more broadly expressed and revealed no significant differences among neighboring cells within the stem cell niches (Fig. 5B,I,P). PLT4-YFP expression peaked in the QC and vasculature, but its level was low overall (Fig. 5C,R). SCR-YFP was expressed mainly in the QC and endodermis cell lineages, and its expression level revealed a shallow proximodistal gradient with a peak in the QC, cortex/endodermis stem cells, and their immediate daughters (Fig. 5E,I,O).
With quantitative data on the PLT and SCR protein accumulation levels at hand, we asked whether the synergistic activity of TCP20- and TCP21-interacting PLT proteins with SCR could effectively limit WOX5 gene expression to the QC. Indeed, when fluorescence intensities of PLT3-YFP and SCR-YFP were combined by multiplication, maximal synergy was achieved in the QC, where pWOX5-erGFP is most abundantly expressed (Fig. 5V).
PLTs directly induce WOX5 gene expression by cooperative interaction with TCP and SCR
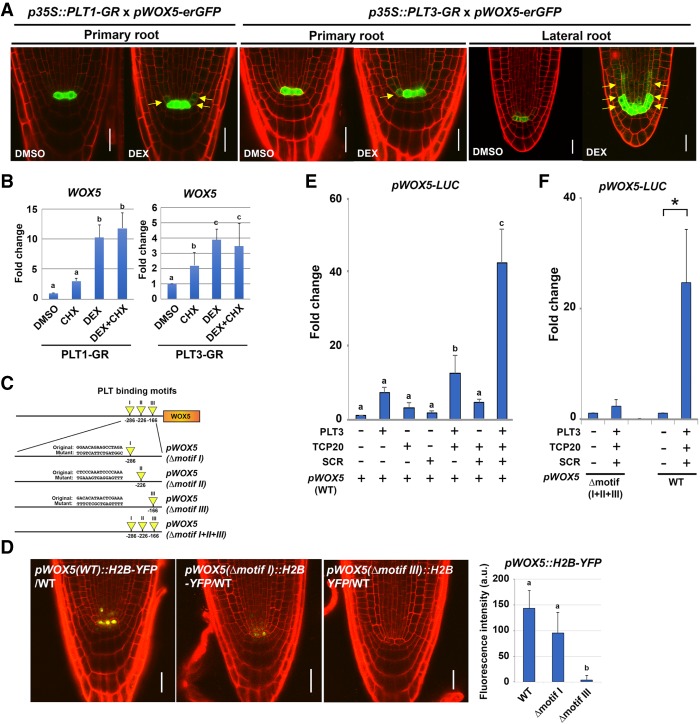
Our findings implied that PLT–TCP–SCR transcription factors function together in QC function and specification as measured by WOX5 gene expression. wox5-1 mutants display extra cell divisions in the QC and lack a columella stem cell layer (Sarkar et al. 2007), which is similar to the late embryogenesis phenotypes observed in compound plt tcp scr mutant combinations. WOX5 may therefore be a significant downstream effector of PLT–TCP–SCR activity. To uncover the mechanism involved, we asked whether WOX5 could be directly regulated by these transcription factors. Previously, in vivo binding sites for PLT transcription factors have been identified on the WOX5 promoter, and, in addition, WOX5 gene expression is induced upon ectopic dexamethasone (DEX) induction of glucocorticoid receptor (GR)-fused PLT2 (Santuari et al. 2016). We examined whether the WOX5 promoter could also be (ectopically) activated in a DEX-inducible PLT1-GR and PLT3-GR activation system. After 3 h of DEX induction in both PLT1-GR and PLT3-GR lines, pWOX5-erGFP expression slightly expanded into the CEI and CEID in primary root apical meristems within the domain where SCR and TCP proteins reside (Fig. 6A, left and middle panels). Induced ectopic WOX5 promoter activity in the SCR expression domain was more conspicuous after PLT3-GR activation in lateral root meristems (Fig. 6A, right panel). To assess whether PLT1 and PLT3 proteins directly activate WOX5 gene expression, we performed quantitative RT–PCR (qRT–PCR) analysis on 3-h DEX- and cyclohexamide (CHX)-treated seedlings of 35S::PLT1-GR and 35S::PLT3-GR. WOX5 mRNA levels were increased by DEX and combined DEX/CHX treatment compared with the DMSO mock and CHX control, respectively, consistent with direct activation of WOX5 by PLT1 and PLT3 (Fig. 6B). The accumulation of WOX5 transcript in 35S::PLT1-GR and 35S::PLT3-GR seedlings upon induction suggested that PLT1 and PLT3 act as direct transcriptional activators, consistent with the reported occurrence of PLT2 ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) peaks on the WOX5 promoter (Santuari et al. 2016). To further test this, we identified three putative PLT-binding motifs (I–III) in the ChIP-seq data on a 1.6-kb WOX5 promoter fragment (Fig. 6C). Like other reported WOX5 gene constructs, the 1.6-kb WOX5 promoter is expressed most abundantly in the QC. Each motif was replaced with random sequences by site-directed mutagenesis, fused to YFP, and transformed into Arabidopsis wild-type plants. Motif I substitution slightly decreased expression, and motif III substitution fully abolished detectable gene expression (Fig. 6D), suggesting that these PLT-binding motifs are crucial for in vivo WOX5 expression in the QC cells of primary roots. To understand whether PLT–TCP–SCR complex function underlies WOX5 induction, we coexpressed PLT3 together with TCP20 and SCR proteins in Arabidopsis mesophyll protoplasts, commonly applied to investigate transcription factor interactions with promoters (Yoo et al. 2007; Long et al. 2015; Díaz-Triviño et al. 2017), and tested their potential to activate the WOX5 promoter. Protoplasts were transfected with reporter constructs harboring endogenous promoter sequences (wild type) or the variant where the predicted PLT-binding motifs were substituted (Δmotif [I + II + III]) (Fig. 6E,F). PLT3, TCP20, and SCR genes were cotransfected, and WOX5 promoter activities were determined by measuring luciferase (LUC) intensities. We noted that PLT3 alone could activate the WOX5 promoter (Fig. 6E). Single transfection of SCR and cotransfection of TCP20 and SCR were unable to raise WOX5 wild-type promoter activity above that observed with PLT3 transfection alone (Fig. 6E). However, combining PLT3 with TCP20 and SCR expression drastically enhanced WOX5 promoter activity sixfold over the induction observed with PLT3 alone (Fig. 6E). In addition, the WOX5 promoter with disrupted PLT-binding sites could only be activated around threefold over the vector control when cotransfected with PLT3, TCP20, and SCR, whereas the intact WOX5 promoter was enhanced >20 times over the vector control (Fig. 6F). In addition, when driving WOX5 gene expression, the 1.6-kb promoter could partially complement the wox5-1 stem cell maintenance phenotype, whereas the version with disrupted motif III-binding sites could not (Supplemental Fig. 5). Collectively, our data show that predicted PLT protein-binding sites are critical for WOX5 induction and that TCP20 and SCR can act as positive regulatory factors mainly through these PLT-binding sites.
Figure 6.
PLTs directly induce WOX5 expression in cooperation with SCR and TCP proteins. (A) pWOX5-erGFP expression alterations by short-time PLT induction. Confocal images were taken after 3 h of 10 µM dexamethasone (DEX) treatment of 35S::PLT1-GR or 35S::PLT3-GR. (Yellow arrows) Ectopically expressed GFP. Seedlings at 2 dag and 5 dag were used for primary root primordium and LRP observations, respectively. Bars, 20 µm. (B) Results of quantitative RT–PCR assays showing WOX5 mRNA levels in Arabidopsis 4-dag seedling roots after 35S::PLT1-GR (left) or 35S::PLT3-GR (right) induction, respectively. The Arabidopsis roots were first treated with mock or 10 µM cycloheximide (CHX) for 15 min and then transferred and induced for 3 h on 10 µM DEX or 10 µM DEX plus CHX plates, respectively. Results are means ± SD. n ≥ 3. One-way ANOVA (Tukey's-Kramer test) was performed. Statistically significant differences are marked by lowercase letters. P < 0.01. (C) Diagrams of mutation analysis of the PLT-binding sites in the 1.6-kb WOX5 promoter. Yellow arrowheads indicate the positions of the indicated sequences of predicted PLT-binding motifs, and the numbers below the promoter represent nucleotide positions upstream of the transcription start site. For the mutated versions (Δmotif), the PLT binding motifs (Original) were replaced with the indicated random oligonucleotides (Mutant). (D) Expression of pWOX5::H2B-YFP variants from C in 5-dag wild-type seedlings. Bars, 20 µm. (Bottom right) Relative YFP expression levels in the QC are quantified. Results are means ± SD. n ≥ 9. One-way ANOVA (Tukey's-Kramer test) was performed. Statistically significant differences are marked by lowercase letters. P < 0.01. (E) Transient luciferase expression analysis in Arabidopsis mesophyll protoplasts normalized by vector control showing enhanced WOX5 promoter activity in 35S::PLT3-3xFlag, 35S::TCP20-10xMyc, and 35S::SCR-7xHA triple-infected protoplasts. Results are means ± SD. n ≥ 12. One-way ANOVA (Tukey's-Kramer test) was performed. Statistically significant differences are marked by lowercase letters. P < 0.01. (F) Transient luciferase expression analysis in Arabidopsis mesophyll protoplasts showing that PLT-binding motifs in the WOX5 promoter are crucial for activation by the PLT3, TCP20, and SCR protein combination. Protoplasts were prepared from 4-wk-old plants transfected with a pWOX5::LUC or pWOX5(Δmotif I+II+III)::LUC. All measurements were normalized to the cotransfected p35S::renilla luciferase (rLUC) activity. Results are means ± SD. n ≥ 3. Two-tailed t test, P < 0.05.
Discussion
A dosage-dependent combinatorial model for stem cell organizer specification
Here we provide evidence of how two major transcription factor modules required for maintenance of the root stem cell niche—the PLT pathway and the SCR pathway—interact at the molecular level with distinct domains of specific members of the TCP protein family. Reduced levels of the relevant PLT, SCR, and TCP proteins in primary roots and their embryonic progenitors, regenerating roots, and LRPs reveal dose-dependent defects in the specification and activity of organizer cells within the root stem cell niche: the QC cells. Our genetic and protein interaction data indicate that dosage-sensitive combinatorial interactions among PLT–TCP–SCR directly regulate promoter activity of the WOX5 gene, which is the best-studied specific molecular marker for QC activity (Sarkar et al. 2007; Pi et al. 2015; Zhang et al. 2017b). Consistent with this scenario, overexpressed PLTs can only effectively expand the WOX5 expression domain where SCR and TCP also are highly expressed.
Our data reveal how the broader roles of PLT and SCR pathways in meristem function are combined for QC specification in a highly specific subdomain, which had been postulated more than a decade ago (Sabatini et al. 2003; Aida et al. 2004; Bennett and Scheres 2010).
Quantification of expression domains indicate that cooperative interaction between PLT1, PLT3, and SCR at specific gene promoters is sufficient to restrict a molecular response to a handful of cells in a single layer (Fig. 5V). The existence of cell-specific complexes that encompass only a part of the, and not the entire, domain of overlap between transcription factors has been corroborated recently by direct visualization of different SCR complexes in the Arabidopsis root using fluorescence resonance energy transfer-fluorescence lifetime imaging (FRET-FLIM) (Long et al. 2017). Note that our cooperative combinatorial model does not exclude the existence of either additional shared targets or many unshared PLT and SCR target genes. Unshared targets can serve broader roles in root development, such as the progression of cell division and differentiation in the root PLT gradient (Mähönen et al. 2014) and the regulation of division and differentiation of the cortical–endodermal lineage by SCR and its binding partner, SHR (Di Laurenzio et al. 1996; Helariutta et al. 2000; Sabatini et al. 2003; Cui et al. 2007; Cruz-Ramírez et al. 2012; Clark et al. 2016; Long et al. 2017). The many additional nonoverlapping functions are consistent with the observation that the published regulated targets of the PLT and SCR pathways are quite distinct (Levesque et al. 2006; Moreno-Risueno et al. 2015; Santuari et al. 2016).
In the shoot apical meristem, the WOX5 sister protein WUS defines the organizing center for the overlying stem cells, and its expression domain is generated by different genetic programs to maintain the shoot stem cell niche (Meng et al. 2017; Zhang et al. 2017a,; Zubo et al. 2017). During initiation of the embryonic shoot stem cells, a WOX2 module promotes the expression of HD-ZIPIII genes to ensure the appropriate ratio of cytokinin and auxin pathways (Zhang et al. 2017b). During de novo shoot regeneration, cytokinin B-type Arabidopsis response regulators (ARRs) combine with HD-ZIP III coactivators for region-specific activation of the WUS promoter (Zhang et al. 2017a), whereas, in the SAM, ARRs appear to maintain mainly WUS expression (Meng et al. 2017; Zhang et al. 2017a). WUS expression is restricted by the stem cell-expressed CLAVATA3 (CLV3) peptide and interacting receptor kinase pathways (Clark et al. 1997; Brand et al. 2000; Schoof et al. 2000; Müller et al. 2008; Ogawa et al. 2008; Nimchuk et al. 2011). Notably, the root WOX5 expression domain is also restricted by signaling through the CLV3/EMBRYO-SURROUNDING REGION40 (CLE40) peptide acting through partially overlapping receptor kinases (Stahl et al. 2009, 2013). Thus, while positive regulators of the expression domain of WUS/WOX5 organizer domain proteins in roots and shoots may be quite distinct, the signaling systems that maintain homeostasis of organizer and (subsets of) stem cells show similarities.
Developmental plasticity of stem cell specification
Somatic stem cells are commonly preserved by signals from “organizer cells,” which provide a microenvironment to maintain the undifferentiated state (Scheres 2007; Biteau et al. 2011; Heidstra and Sabatini 2014; Gaillochet and Lohmann 2015). Over the last two decades, research in animals and plants has revealed examples of considerable plasticity in the specification of organizer cells (Boyer et al. 2006; Wilson et al. 2008; Tian et al. 2011; Dejana et al. 2017; Hoeck et al. 2017). A dramatic demonstration of this plasticity in plants is the regeneration of an entirely new stem cell niche upon surgical dissection (Xu et al. 2006; Efroni et al. 2016; Zhang et al. 2017a). In both roots and shoots, this leads to the re-establishment of a new domain of expression of WOX5/WUS. Relocation of WOX5 expression at the center of the new stem cell niche during root regeneration correlates with shifts in the expression domains of PLT and SCR proteins (Xu et al. 2006). While it has not yet been clarified exactly how these “organizer upstream” expression domains are established, clues for this are suggested from the known root regulatory networks. First, PLT transcription is a slow response to persistent high auxin and therefore can respond to organ injuries that impart auxin flux and thereby change auxin distribution. Second, SCR regulation requires SHR protein movement from vascular to nonvascular cells and subsequent SHR nuclear retention (Sabatini et al. 1999; Dolan 2001; Nakajima et al. 2001; Long et al. 2015). This nuclear retention is restricted to a single layer through a dynamic developmental control mechanism (Nakajima et al. 2001; Long et al. 2015) and can be envisaged to be reinstated in the regenerating cells that surround intact vascular cells. Therefore, the normal mechanisms that guide PLT and SCR expression are dynamic in that they continue to depend on mobile signals provided by their cellular context. In this way, tissue severing or de novo stem cell niche establishment can create new regions of PLT–SCR overlap, which—as we show in three different contexts (embryogenesis, lateral root formation, and regeneration)—can lead to combinatorial activation of WOX5. The activity of WOX5 then contributes to the specification of new organizer cells, which maintain the undifferentiated state of neighboring stem cells (Sarkar et al. 2007; Forzani et al. 2014; Pi et al. 2015).
Materials and methods
Arabidopsis genetics, transformation, and growth conditions
All of the A. thaliana lines used in this study were in the Columbia-0 (Col-0) background. Arabidopsis mutants plt1-3, plt3-1, and scr-3 have been described previously (Fukaki et al. 1998; Aida et al. 2004; Galinha et al. 2007). tcp20-1 (SALK_016203) and tcp21-1 (SALK_106694) mutants were obtained from the Arabidopsis Biological Resource Center (ABRC). plt1-3+/−plt3-1+/− tcp20-1+/−scr-3+/− trans-heterozygous mutant seedlings were generated by crossing the quadruple homozygote of plt1-3−/− plt3-1−/−tcp20-1−/−scr-3−/− with Col-0 and harvesting these siliques (F1 seeds). Combinations of stacked mutant lines with heterozygous alleles in Supplemental Figure 2 were F1 seedlings and were generated as follows: tcp20+/−scr−/− was generated by crossing tcp20scr with scr. plt3+/−tcp20−/−scr−/− was generated by crossing plt3tcp20scr with tcp20scr. plt1+/−tcp20−/−scr−/− was generated by crossing plt1tcp20scr with tcp20scr. plt1+/−plt3−/− tcp20+/−scr−/− was generated by crossing plt1plt3tcp20scr with plt3scr. plt1−/−plt3+/−tcp20−/−scr−/− was generated by crossing plt1plt3tcp20scr with plt1tcp20scr. plt1+/−plt3−/−tcp20−/−scr−/− was generated by crossing plt1plt3tcp20scr with plt3tcp20scr. tcp21+/−scr+/− was generated was by crossing tcp21scr with Col-0. tcp21+/−scr−/− was generated by crossing tcp21scr with scr. Obtained F1 seedling genotypes were confirmed by PCR using proper primer combinations in Supplemental Table 5. All plants used in this study were grown on soil or 0.5× Murashige-Skoog (MS) medium plate under long-day conditions (16-h light/8-h dark period) at 22°C. Seeds were fume-sterilized (100 mL of bleach supplemented with 3 mL of hydrochloric acid), soaked into 0.1% (w/v) agarose, and plated on 0.5× MS plate (pH 5.8). Floral dip transformation methods were described previously (Clough and Bent 1998).
Construction of promoter and protein fusions
MultiSite Gateway cloning (Invitrogen, Thermo Fisher Scientific, Inc.) was performed to generate transgenic plants expressing TCP20 and TCP21 protein fusions. Promoter sequences upstream of the ATG start codon of TCP20 (2.56 kb) and TCP21 (2.15 kb) were cloned into pDONR P4-P1R. The coding sequences (CDSs) of TCP20 and TCP21 were amplified and cloned into a Gateway-adapted pGEMTeasy221. A YFP-NOS terminator cassette was cloned into pGEMTeasyR2R3. The three types of entry vectors were recombined into pGreenII (Hellens et al. 2000) expression constructs adapted for Gateway cloning that generate basta resistance in plants. These constructs were transformed into Col-0 and also into tcp20-1 for pTCP20::TCP20-YFP and into tcp21-1 mutants for pTCP21::TCP21-YFP, of which the extra cell divisions in the QC at 5 dag of these genotypes were complemented. To introduce specific motif mutants into the WOX5 promoter, we applied site-directed mutagenesis (Kunkel 1985), for which sequence information is available in Supplemental Table 5. The resultant WOX5 promoters were subcloned into pDONR P4-P1R and fused with H2B-YFP in-frame using the MultiSite Gateway system. The pWOX5-erGFP line was published previously (Xu et al. 2006). For BiFC analysis in Arabidopsis mesophyll protoplasts, all full-length CDSs without stop codons were cloned into pGEMTeasy221 vectors and recombined with the N-terminal half YFP (pARC235) or C-terminal half YFP (pARC236) and driven under the constitutive cauliflower mosaic virus 35S promoter. For transient expression in Arabidopsis mesophyll cells, CDS cassettes of PLT3, SCR, and TCP20 genes were fused in-frame with synthesized 3xFlag, 7xHA, and 10xMyc tags, respectively, and recombined into the plant binary vector pGII227 (hygromycin resistance in planta) for the generation of in-frame fusion with the 35S promoter by Multisite Gateway LR reactions (Invitrogen). Truncated coding sequences of PLT1, PLT3, TCP20, and TCP21 were amplified by PCR, cloned into pGEMTeasy221 vector, and recombined to pDEST22 or pDEST32 for binding assay in a Y2H system. Truncated TCP fragments for BiFC assays were created in p35S-N-terminal fusion YFP or p35S-C-terminal fusion YFP vectors based on pARC235 and pARC236, respectively. Detailed information of primers used for genotyping of T-DNA insertion lines and the cloning procedures used in this study are in Supplemental Table 5.
Y2H screening and binding test
Y2H screenings of PLT and SCR cointeractors were performed using truncated PLT (C-terminal regions of PLT1 [395 amino acids], PLT2 [380 amino acids], PLT3 [338 amino acids], and PLT4 [376 amino acids], including the double AP2 domain) and full-length SCR CDSs into pDEST32 (Invitrogen) in-frame fused with the Gal4-DNA-binding domain. Each construct was cotransformed with an Arabidopsis root cDNA library cloned in pDEST22 (Invitrogen) into yeast strain PJ69A (James et al. 1996). Screens were performed on SD medium without His, Leu, and Trp and plus 30 mM 3-aminotriazole (Sigma-Aldrich). Interaction studies were performed using the recommended protocols for the ProQuest two-hybrid system (Invitrogen). For one-to-one interaction studies, at least three experimental and three technical replicates were performed. Primers used for cloning are listed in Supplemental Table 5.
Transient BiFC assay and transcriptional assay in Arabidopsis mesophyll protoplasts
For BiFC assays, plasmids were constructed as indicated above. The transfection protocols for Arabidopsis mesophyll protoplasts, Renilla luciferase transcriptional analysis, and microscopy have been described previously (Cruz-Ramírez et al. 2012; Díaz-Triviño et al. 2017). More than 200 cells per combination were observed, and at least two independent biological replicates were accomplished. For transcription assays, pWOX5 was first cloned into pGEMTeasy221 and subcloned by Gateway LR recombination into pGWB35 (luciferase) vector and cotransfected with 35S::PLT3-3xFlag, 35S::TCP20-10xMyc, and 35S::SCR-7xHA constructs. The protoplast transformation method was described previously (Yoo et al. 2007). Protoplasts were transfected by 10 µg of DNA in 10 µL of water. Transfected protoplasts were resuspended with W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES at pH 5.7) and incubated overnight at room temperature. Extracted protein mixtures were scored for firefly and Renilla luciferase activity with the dual-luciferase reporter assay system (Promega). At least three technical replicates for Figure 6, E and F, were accomplished. P-values were calculated by Tukey's honest significant difference (HSD) test (Fig. 6E) and two-tailed t-test (Fig. 6F).
Microscopy analysis, image processing, and GUS staining
Arabidopsis roots were mounted in water containing 10 µM propidium iodide (Sigma-Aldrich) and imaged with a confocal laser-scanning microscope. For the embryos, whole-mount immature seeds were dissected from siliques and transferred to a solution containing 5% glucose. Stained embryos and primary and lateral roots by Schiff reagent were mounted in chloral hydrate solution (chloral hydrate:water:glycerol, ratio [w/v/v] 8:3:1) and visualized by confocal optics (Leica SP2 and Zeiss LSM710). Details of the Schiff stain method are described in Truernit et al. (2008). LRPs and embryos were observed for >20 seedlings and 50 embryos per line, respectively, and at least three independent experiments were performed. YFP fluorescence intensities of pPLT1::PLT1-YFP, pPLT2::PLT2-YFP, pPLT3::PLT3-YFP, pPLT4::PLT4-YFP, pSCR::SCR-YFP, pTCP20::TCP20-YFP, and pTCP21::TCP21-YFP lines in primary roots and embryos were observed using equal laser power, detector gain, and pinhole, respectively. A region of interest (ROI) was set around the nucleus, and mean values were scored. The background signal was substracted by selecting a ROI from nonfluorescent cells within the same root. Heat maps of protein expression profiles were generated by Zen 2012 software. pWOX5-erGFP-induced signals by 35S::PLT1-GR or 35S::PLT3-GR induction were also tested using equal laser power and other settings among samples. For background subtraction, a ROI was selected where fluorescence signal was minimal. For GUS expression analysis, Arabidopsis 5-dag seedlings were incubated in 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (X-Gluc) staining buffer solution [1 mg/mL X-Gluc, 0.1% Triton X-100, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 50 mM NaH2PO4 at pH 7.0] for appropriate time periods. All quantifications were conducted with ImageJ software (http://rsbweb.nih.gov/ij). Root meristem length was estimated by measuring the distance between the QC and the first elongated cortex cell (Perilli and Sabatini 2010). Images were cropped to obtain properly sized images and rotated to have vertical orientations using Adobe Photoshop CS6 software.
Laser ablations and regeneration experiments
Laser ablations of QC cells were performed on a Leica TCS SP2 confocal microscope using 4-dag seedlings of pPLT3::PLT3-YFP-, pSCR::SCR-YFP-, pTCP20::TCP20-YFP-, or pWOX5::H2B-YFP-expressing lines (n = 5) as described previously (van den Berg et al. 1995; Xu et al. 2006). Briefly, roots were prestained with 10 µg/mL propidium iodide on slides to visualize the cellular structure of stem cells and target the QC for laser ablation. After ablation, seedlings were rinsed with distilled water and put back on 0.5× MS plates. Regeneration was monitored every 6 h for 24 h.
RNA isolation and qRT–PCR analysis of PLT-GR
PLT1-GR- and PLT3-GR-induced activation of the WOX5 promoter by qRT–PCR analysis was performed as follows: For DEX + CHX tests, p35S::PLT1-GR and p35S::PLT3-GR seedlings were germinated on 0.5× MS agar plates for 3 d and subsequently transferred to 0.5× MS agar plates containing 10 µM CHX (Sigma) for 15 min. Next, they were immediately transferred to 0.5× MS agar plates containing 10 µM DEX (Sigma) plus 10 µM CHX or 10 µM CHX control for another 3 h. For DEX induction tests, seedlings were similarly germinated on 0.5× MS agar plates for 3 d. Next, they were all transferred to 0.5× MS agar plates containing 10 µM DEX or DMSO for mock control for another 3 h. Total root RNA isolation was performed with the plant RNA purification kit (Qiagen). For each RNA sample, 1 µg of total RNA was treated with DNase I (Fermentas) and reverse-transcribed using SuperScript III first strand synthesis system (Invitrogen) according to the manufacturer's instructions. Quantitative real-time PCR was performed using LightCycler 480 II (Roche Diagnostics GmbH). Obtained results were normalized against UBQ10 expression. Primer information is in Supplemental Table 5. At least two technical replicates with two biological replicates were performed for each time period and sample. P-values were calculated by Tukey's HSD test. The study of spatio–temporal induction of the WOX5 promoter by PLT1 and PLT3 induction was performed using p35S::PLT1-GR and p35S::PLT3-GR seedlings harboring homozygous pWOX5-erGFP, respectively. For each line, seeds were germinated on 0.5× MS agar plates for 2 or 5 d for analysis of primary root or lateral root meristems, respectively. For induction, the seedlings were treated with 10 µM DEX for 3 h followed by the fluorescence imaging by confocal microscopy. Seedlings treated with DMSO for 3 h were used as controls.
Coimmunoprecipitation and Western blotting
For transient expression analysis in Arabidopsis mesophyll protoplasts, cells were ground in liquid nitrogen using a mortar and pestle. Powdered materials were mixed with extraction buffer (50 mM Tris-HCl at pH 7.5, 100 mM NaCl, 1% Triton X-100, protease inhibitors). After thawing, samples were centrifuged at 14,000 rpm for 10 min at 4°C. The resulting supernatant was incubated with anti-Flag antibodies under rotation for 4 h at 4°C. Bound fractions were immobilized on protein G beads (GE healthcare) and washed twice with 200 µL of extraction buffer. Precipitates were eluted from beads with 40 µL of SDS-PAGE loading buffer (1% SDS, 0.1 M dithithreitol, 0.08 M Tris-HCl at pH 6.8, 5% glycerol, bromophenol blue). Eluted proteins were separated on 10% polyacrylamide SDS gels and transferred to nitrocellulose membranes (Hybond, GE Healthcare) by blotting. All of the primary monoclonal antibodies produced in mice were incubated for 1 h in 3% nonfat milk in PBS-T (0.1% Tween), and subsequent anti-mouse secondary antibodies marked HRP were incubated for another 1 h in PBS-T (0.1% Tween). The following antibodies were used: anti-Myc antibody (Santa Cruz Biotechnology), anti-Flag antibody (Sigma), anti-HA antibody (Santa Cruz Biotechnology), anti-actin antibody (Sigma), and goat anti-mouse HRP-conjugated antibody (Abcam). Signals were detected using the Amersham ECL start kit (GE Healthcare, RPN3244). At least three biological replicates were performed and obtained similar results.
Supplementary Material
Acknowledgments
We thank Du Yujuan for technical support on imaging LRPs. This work was supported by an EMBO long-term fellowship to A.S., European Research Council Advanced Grant SysArc and Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) Spinoza grants to B.S., an NWO Vidi grant to I.B., and an NWO Horizon grant to R.H.
Author contributions: A.S., R.H., and B.S. conceived the study. A.S., R.H., I.B., and B.S. designed the methodology. A.S. and R.H. performed the investigation. A.S., R.H., and I.B. supplied the resources. A.S., R.H., I.B., and B.S. wrote the original draft of the manuscript. A.S., R.H., I.B., and B.S. acquired the funding.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.314096.118.
Freely available online through the Genes & Development Open Access option.
References
- Aggarwal P, Gupta MD, Joseph AP, Chatterjee N, Srinivasan N, Nath U. 2010. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22: 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martínez JA, Sinha N. 2013. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front Plant Sci 4: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. 1993. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70. [DOI] [PubMed] [Google Scholar]
- Bennett T, Scheres B. 2010. Root development-two meristems for the price of one? Curr Top Dev Biol 91: 67–102. [DOI] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. 2011. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell 9: 402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AA, Miki BL, et al. 2002. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14: 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353. [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. 2000. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619. [DOI] [PubMed] [Google Scholar]
- Broholm SK, Tähtiharju S, Laitinen RA, Albert VA, Teeri TH, Elomaa P. 2008. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc Natl Acad Sci 105: 9117–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron A, Chatfield S, Provart N, Berleth T. 2009. Embryogenesis: pattern formation from a single cell. Arabidopsis Book 7: e0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585. [DOI] [PubMed] [Google Scholar]
- Clark NM, Hinde E, Winter CM, Fisher AP, Crosti G, Blilou I, Gratton E, Benfey PN, Sozzani R. 2016. Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy. eLife 5: e14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Blilou I, Grieneisen VA, Sozzani R, Zamioudis C, Miskolczi P, Nieuwland J, Benjamins R, Dhonukshe P, et al. 2012. A bistable circuit involving SCARECROW–RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150: 1002–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Wachsman G, Du Y, Arteága-Vázquez M, Zhang H, Benjamins R, Blilou I, Neef AB, Chandler V, et al. 2013. A SCARECROW–RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol 11: e1001724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E. 1999. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222. [DOI] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. 2007. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425. [DOI] [PubMed] [Google Scholar]
- Danisman S, van der Wal F, Dhondt S, Waites R, de Folter S, Bimbo A, van Dijk AD, Muino JM, Cutri L, Dornelas MC, et al. 2012. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol 159: 1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P. 2014. Class I TCP–DELLA interactions in inflorescence shoot apex determine plant height. Curr Biol 24: 1923–1928. [DOI] [PubMed] [Google Scholar]
- Dejana E, Hirschi KK, Simons M. 2017. The molecular basis of endothelial cell plasticity. Nat Commun 8: 14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Triviño S, Long Y, Scheres B, Bliou I. 2017. Analysis of a plant transcriptional regulatory network using transient expression systems. Methods Mol Biol 1629: 83–103. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. 1996. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Benfey PN. 2008. Plant stem cell niches: standing the test of time. Cell 132: 553–557. [DOI] [PubMed] [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, Flittner KA, Kneuper I, van der Graaff E, Nziengui H, Pinosa F, Li X, Nitschke R, et al. 2008. Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc Natl Acad Sci 105: 18818–18823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L. 2001. Root patterning: SHORT ROOT on the move. Curr Biol 11: R983–R985. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84. [DOI] [PubMed] [Google Scholar]
- Du Y, Scheres B. 2017. PLETHORA transcription factors orchestrate de novo organ patterning during Arabidopsis lateral root outgrowth. Proc Natl Acad Sci 114: 11709–11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip PL, Rahni R, DelRose N, Powers A, Satija R, Birnbaum KD. 2016. Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165: 1721–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forzani C, Aichinger E, Sornay E, Willemsen V, Laux T, Dewitte W, Murray JAH. 2014. WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr Biol 24: 1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey PN, Tasaka M. 1998. Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J 14: 425–430. [DOI] [PubMed] [Google Scholar]
- Gaillochet C, Lohmann JU. 2015. The never-ending story: from pluripotency to plant developmental plasticity. Development 142: 2237–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Goh T, Toyokura K, Wells DM, Swarup K, Yamamoto M, Mimura T, Weijers D, Fukaki H, Laplaze L, Bennett MJ, et al. 2016. Quiescent center initiation in the Arabidopsis lateral root primordia is dependent on the SCARECROW transcription factor. Development 143: 3363–3371. [DOI] [PubMed] [Google Scholar]
- Heidstra R, Sabatini S. 2014. Plant and animal stem cells: similar yet different. Nat Rev Mol Cell Biol 15: 301–312. [DOI] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B. 2004. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev 18: 1964–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. 2000. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. 2000. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Hervé C, Dabos P, Bardet C, Jaunean A, Auriac MC, Ramboer A, Lacout F, Tremousaygue D. 2009. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol 149: 1462–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J, Kumpf RP, De Veylder L. 2014. A quiescent path to plant longevity. Trends Cell Biol 24: 443–448. [DOI] [PubMed] [Google Scholar]
- Hoeck JD, Biehs B, Kurtova AV, Kljavin NM, de Sousa E, Melo F, Alicke B, Koeppen H, Modrusan Z, Piskol R, et al. 2017. Stem cell plasticity enables hair regeneration following Lgr5+ cell loss. Nat Cell Biol 19: 666–676. [DOI] [PubMed] [Google Scholar]
- Hofhuis H, Laskowski M, Du Y, Prasad K, Grigg S, Pinon V, Scheres B. 2013. Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Curr Biol 23: 956–962. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Feldman LJ. 2005. Regulation of root apical meristem development. Annu Rev Cell Dev Biol 21: 485–509. [DOI] [PubMed] [Google Scholar]
- Jürgens G. 2001. Apical–basal pattern formation in Arabidopsis embryogenesis. EMBO J 20: 3609–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G, Ruiz RAT, Laux T, Mayer U, Berleth T. 1994. Early events in apical-basal pattern formation in Arabidopsis. In Plant molecular biology: Molecular-genetic analysis of plant development and metabolism (ed. Coruzzi G, Puigdomenech P), pp. 95–103. Springer, Berlin. [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B. 2011. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68: 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci 82: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. 2006. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. 2015. The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signal Behav 10: e1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P. 2005. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci 102: 12978–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Smet W, Cruz-Ramírez A, Castelijns B, de Jonge W, Mähönen AP, Bouchet BP, Perez GS, Akhmanova A, Scheres B, et al. 2015. Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell 27: 1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Stahl Y, Weidtkamp-Peters S, Postma M, Zhou W, Goedhart J, Sánchez-Pérez MI, Gadella TWJ, Simon R, Scheres B, et al. 2017. In vivo FRET-FLIM reveals cell-type-specific protein interactions in Arabidopsis roots. Nature 548: 97–102. [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B. 2014. PLETHORA gradient formation mechanism separates auxin responses. Nature 515: 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44. [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. 1991. Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot 69: 461–476. [Google Scholar]
- Martín-Trillo M, Cubas P. 2010. TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815. [DOI] [PubMed] [Google Scholar]
- Meng WJ, Cheng ZJ, Sang YL, Zhang MM, Rong XF, Wang ZW, Tang YY, Zhang XS. 2017. Type-B Arabidopsis response regulators specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29: 1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Sozzani R, Yardımcı GG, Petricka JJ, Vernoux T, Blilou I, Alonso J, Winter CM, Ohler U, Scheres B, et al. 2015. Transcriptional control of tissue formation throughout root development. Science 350: 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Bleckmann A, Simon R. 2008. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. 2001. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311. [DOI] [PubMed] [Google Scholar]
- Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM. 2011. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr Biol 21: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. 2008. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Perilli S, Sabatini S. 2010. Analysis of root meristem size development. Methods Mol Biol 655: 177–187. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN. 2012. Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T. 2015. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev Cell 33: 576–588. [DOI] [PubMed] [Google Scholar]
- Pulianmackal AJ, Kareem AV, Durgaprasad K, Trivedi ZB, Prasad K. 2014. Competence and regulatory interactions during regeneration in plants. Front Plant Sci 5: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovere FD, Fattorini L, Ronzan M, Falasca G, Altamura MM. 2016. The quiescent center and the stem cell niche in the adventitious roots of Arabidopsis thaliana. Plant Signal Behav 11: e1176660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey PN, Leyser O, Bechtold N, Weisbeek P, et al. 1999. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. 2003. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R. 2004. Plant and animal stem cells: conceptually similar, molecularly distinct? Trends Cell Biol 14: 605–611. [DOI] [PubMed] [Google Scholar]
- Sablowski R. 2011. Plant stem cell niches: from signalling to execution. Curr Opin Plant Biol 14: 4–9. [DOI] [PubMed] [Google Scholar]
- Santuari L, Sanchez-Perez GF, Luijten M, Rutjens B, Terpstra I, Berke L, Gorte M, Prasad K, Bao D, Timmermans-Hereijgers JL, et al. 2016. The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell 28: 2937–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. 2007. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814. [DOI] [PubMed] [Google Scholar]
- Scheres B. 2007. Stem-cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol 8: 345–354. [DOI] [PubMed] [Google Scholar]
- Scheres B, Benfey PN. 1999. Asymmetric cell division in plants. Annu Rev Plant Physiol Plant Mol Biol 50: 505–537. [DOI] [PubMed] [Google Scholar]
- Scheres BJG, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P. 1994. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120: 2475–2487. [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. 2010. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. 2000. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644. [DOI] [PubMed] [Google Scholar]
- Sena G, Wang X, Liu HY, Hofhuis H, Birnbaum KD. 2009. Organ regeneration does not require a functional stem cell niche in plants. Nature 457: 1150–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutian L. 2015. The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signal Behav 10: e1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN. 2010. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R. 2009. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 19: 909–914. [DOI] [PubMed] [Google Scholar]
- Stahl Y, Grabowski S, Bleckmann A, Kühnemuth R, Weidtkamp-Peters S, Pinto KG, Kirschner GK, Schmid JB, Wink RH, Hülsewede A, et al. 2013. Moderation of Arabidopsis root stemness by CLAVATA1 and Arabidopsis CRINKLY4 receptor kinase complexes. Curr Biol 23: 362–371. [DOI] [PubMed] [Google Scholar]
- Ten Hove CA, Heidstra R. 2008. Who begets whom? Plant cell fate determination by asymmetric cell division. Curr Opin Plant Biol 11: 34–41. [DOI] [PubMed] [Google Scholar]
- Ten Hove CA, Lu K-J, Weijers D. 2015. Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142: 420–430. [DOI] [PubMed] [Google Scholar]
- Terpstra I, Heidstra R. 2009. Stem cells: the root of all cells. Semin Cell Dev Biol 20: 1089–1096. [DOI] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. 2011. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478: 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Jia Y, Niu T, Yu Q, Ding Z. 2014. The key players of the primary root growth and development also function in lateral roots in Arabidopsis. Plant Cell Rep 33: 745–753. [DOI] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui JC. 2008. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuwamoto R, Yokoi S, Takahata Y. 2010. Arabidopsis EMBRYOMAKER encoding an AP2 domain transcription factor plays a key role in developmental change from vegetative to embryonic phase. Plant Mol Biol 73: 481–492. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. 1995. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378: 62–65. [DOI] [PubMed] [Google Scholar]
- Weigel D, Jürgens G. 2002. Stem cells that make stems. Nature 415: 751–754. [DOI] [PubMed] [Google Scholar]
- Weijers D, Sauer M, Meurette O, Friml J, Ljung K, Sandberg G, Hooykaas P, Offringa R. 2005. Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell 17: 2517–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Laurenti E, Oser G, Van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135: 1118–1129. [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. 2000. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603. [DOI] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B. 2006. A molecular framework for plant regeneration. Science 311: 385–388. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jiao Y, Liu Z, Zhu Y. 2015. ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat Commun 6: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TQ, Lian H, Zhou CM, Xu L, Jiao Y, Wang JW. 2017a. A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29: 1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tucker E, Hermann M, Laux T. 2017b. A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev Cell 40: 264–277. [DOI] [PubMed] [Google Scholar]
- Zubo YO, Blakley IC, Yamburenko MV, Worthen JM, Street IH, Franco-Zorrilla JM, Zhang W, Hill K, Raines T, Solano R, et al. 2017. Cytokinin induces genome-wide binding of the type B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc Natl Acad Sci 114: E5995–E6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.