Abstract
Objectives
Manganese (Mn) is a known neurotoxicant, and given its health effects and ubiquitous nature in metal-working settings, identification of a valid and reproducible biomarker of Mn exposure is of interest. Here, global metabolomics is utilized to determine metabolites that differ between groups defined by Mn exposure status, with the goal being to help inform a potential metabolite biomarker of Mn exposure.
Methods
Mn exposed subjects were recruited from a Mn steel foundry and Mn unexposed subjects were recruited from crane operators at a metal recycling facility. Over the course of a work day, each subject wore a personal inhalable dust sampler (IOM), and provided an end of shift urine sample that underwent global metabolomics profiling. Both exposed and unexposed subjects were divided into a training set and demographically similar validation set. Using a two-sided adjusted t-test, relative abundances of all metabolites found were compared between Mn exposed and unexposed training sets, and those with a false discovery rates (FDR) <0.1 were further tested in the validation sets.
Results
Fifteen ions were found to be significantly different (FDR < 0.1) between the exposed and unexposed training sets, and nine of these ions remained significantly different between the exposed and unexposed validation set as well. When further dividing exposure status into ‘lower exposure’ and ‘higher exposure’, several of these nine ions exhibited an apparent exposure–response relationship.
Conclusions
This is the first time that metabolomics has been used to distinguish between Mn exposure status in an occupational cohort, though additional work should be done to replicate these findings with a larger cohort. With metabolite identification by name, empirical formula, or pathway, a better understanding of the relationship between Mn exposure and neurotoxic effects could be elucidated, and a potential metabolite biomarker of Mn exposure could be determined.
Keywords: biomarkers of exposure, exposome, exposure assessment, manganese, metabolomics
Introduction
Manganese (Mn) is an established neurotoxicant associated with deleterious cognitive (Mergler et al. 1994; Bouchard et al. 2007; Bowler et al. 2007) and motor (Roels et al. 1985; Roels et al. 1987; Lucchini et al. 1999) health outcomes. Environmental Mn exposures, from traffic or industrial sources, have been shown to increase the prevalence of Parkinsonian disorders (Finkelstein and Jerrett 2007; Lucchini et al. 2007). The American Conference of Governmental Industrial Hygienists (ACGIH) has recommended an 8-hour time weighted average threshold limit value (TWA TLV) of 100 µg/m3 for inhalable Mn based on neurological endpoints.
While Mn is ubiquitously found in ambient air, drinking water, and foods common to a Western diet, it is in occupational settings where chronic, elevated exposures are most frequently found. Elevated exposures, encountered in either community or occupational settings, make Mn a public health concern in need of further investigation.
Given the negative health effects related to Mn exposure, research has focused on finding both exposure and disease biomarkers for Mn and its related neurological disorders in readily available biofluids such as blood, plasma, and urine, or by using imaging techniques such as magnetic resonance imaging (MRI) or positron emission tomography (PET) scans (Apostoli, Lucchini, and Alessio 2000; Zheng et al. 2011; O’Neal and Zheng 2015; Baker, Criswell, et al. 2015; Baker et al. 2016). However, a lack of understanding of the timing between exposure and uptake (Roth 2006; Baker et al. 2014), natural human variability in Mn (Järvisalo et al. 1992; Baker, Simpson, et al. 2015), and costs or contraindications associated with MRI or PET scans (e.g. metal chips in eyes) make many biomarkers of Mn exposure and effect impractical for use in a field or clinical setting. Thus, the critical barrier to identifying a reliable, reproducible, and easily accessible biomarker of Mn exposure still remains largely unsolved. As such, there is a critical need in the fields of environmental health, neurology, toxicology, and public health to develop informative peripheral biomarkers of Mn exposure and its related diseases, and to better understand the underlying pathophysiology of Mn neurotoxicity.
Global metabolomics is one promising approach to identifying a biomarker of Mn exposure. Metabolomics is the study of the small molecules (<1000 daltons) in biological systems, often important in metabolic function and signaling, or representing a fingerprint of a specific cellular process (Wishart 2010). Global metabolomics is typically an exploratory analysis method and is a means of hypothesis generation because selection of putative biomarkers in biofluids using this approach is inherently statistical, based on comparisons between thousands of ions. Therefore, the false positives in the analysis represent ions that are statistically significant but are not related to the exposure in question. Results from metabolomics studies tend to have limited utility unless the selected putative ions can be replicated in an independent sample and the chemical nature and biological role of the ions identified (Broadhurst and Kell 2006).
Typically, global metabolomics has been used to identify biomarkers or physiological alterations related to health effects, such as for neurologic diseases (Ibáñez et al. 2012; Hassan-Smith et al. 2012; Sato et al. 2012; Zhang, Sun, and Wang 2013), cancers (Zhang et al. 2012; Wang, Zhang, and Sun 2013), and, of particular relevance to Mn, Parkinson’s Disease (Bogdanov et al. 2008; Michell et al. 2008; Ahmed et al. 2009). While using metabolomics to distinguish between patients and controls is more common, using metabolomics to distinguish between exposed and non-exposed persons is a relatively novel concept (especially in occupational settings) and is a key approach in exploring the exposome—the biological signatures of lifelong environmental exposures within individual physiological histories (Wild 2005). Walker et al. (2016) used a metabolome-wide association study (MWAS) framework to investigate metabolic changes in plasma related to trichloroethylene exposure in an occupationally exposed cohort, and were able to link measured exposure to internal dose and measured endogenous metabolites. However, the other few human metabolomics studies investigating biomarkers of exposure have looked only at dietary exposure to vitamins or minerals, not chemical exposures in an environmental or occupational setting (Astle et al. 2007; Johansson-Persson et al. 2013). Few studies have investigated metabolomics related to Mn exposure, and those that have analyzed biofluids or tissues collected from model organisms, not biofluids collected from humans exposed in an occupational setting (Dorman et al. 2008; Fordahl et al. 2012; Kumar et al. 2015; Neth et al. 2015). Our study of occupational exposed workers is particularly important given the substantial limitations in available biomarkers of acute and chronic exposure to Mn. The objective of this work is to report on the use of a metabolomics analysis approach to investigate the differences between occupational groups classified by Mn exposure status, which could in turn inform a biomarker of exposure to Mn. Work presented here represents a novel method for exposure assessment in environmental and occupational health studies and the first time, to our knowledge, that metabolomics has been applied in an occupational setting to distinguish between exposed and unexposed workers.
Methods
During 1 week in October 2014, subjects were recruited from two Puget Sound worksites; Mn exposed subjects (n = 20) worked at a Mn foundry and Mn unexposed subjects worked as crane operators in a metal recycling facility (n = 17). Data were collected on worker demographics including age, race, and ethnicity. No exclusions were made on the basis of race, ethnicity, or gender, though all enrolled subjects identified as male. For each enrolled subject at both worksites, a personal, full-shift, inhalable dust sample was collected using an Institute of Occupational Medicine (IOM, Edinburgh, UK) sampler, and a single spot urine sample was collected at the end of the same day. The study protocols were approved by the University of Washington Institutional Review Board.
To increase the validity of our findings, both the Mn exposed and unexposed groups were divided into a training set (n = 12 in the Mn exposed group, n = 10 in the Mn unexposed group) and a demographically similar validation set (n = 8 in the exposed group, n = 7 in the unexposed group). Urine samples were processed and analyzed without regard to exposure status. After metabolomics data extraction and preprocessing (as described below), statistical analysis of the training data set yielded putative ions related to Mn exposure and a separate analysis was performed to compare the relative abundance of the selected significant ions in the validation data set.
Mn exposure assessment
Personal full-shift inhalable dust sampling was conducted at each location using IOM inhalable dust samplers according to the UK Health and Safety Executive’s Methods for the Determination of Hazardous Substances 14/4 (Mark and Vincent 1986; Health and Safety Executive 2014). The collected inhalable dust samples were analyzed at the University of Washington Environmental Health Laboratory for total Mn using inductively coupled plasma mass spectrometry and adjusted for field blanks handled in the same manner as the filters deployed in the field. Measured inhalable Mn concentration were normalized to 8-hour time weighted average concentrations. For the 37 subjects enrolled, three subjects were missing IOM samples (one from the foundry, and two from the scrap metal recycling yard). Missingness was due to pump failure in the field and is believed to be completely at random.
To explore exposure–response relationships, subjects were divided into three categorical exposure levels based on job title and results from inhalable dust sampling: No Exposure (operators at the scrap metal recycling yard, n = 10 in the training set and n = 7 in the validation set), Lower Exposure (forklift operators, foundry helpers, and molders at the foundry, n = 7 in the training set and n = 3 in the validation set), and Higher Exposure (melters or pourers at the foundry, n = 5 in the training set and n = 5 in the validation set).
Urine preparation and metabolomics analysis
After collection in the field, urine samples were immediately stored on dry ice for transport to a −80°C freezer at the University of Washington. All samples were frozen upon arrival to University of Washington and remained frozen in the −80°C freezer until being thawed on wet ice for sample preparation. All urine samples were prepared and analyzed on the same day, approximately 1 month after collection in the field. The procedure is described in full in Tay-Sontheimer et al. (2014). In brief, 200 µl urine was combined with 800 µl acetonitrile containing deuterated internal standards. After protein precipitation, samples were centrifuged and the supernatant was evaporated under nitrogen gas. Samples were reconstituted in methanol:water with 0.4% acetic acid, vortexed, centrifuged, and transferred to glass autosampler vials. A pooled quality control sample was made from 10 randomly selected urine samples and prepared as described above. Samples were analyzed on an Agilent (Santa Clara, CA, USA) 1200 HPLC coupled to Agilent 6520 Accurate Mass quadropole time-of-flight (Q-TOF) mass spectrometer, calibrated for accurate masses between m/z of 118 to 1700. A 3.5 μm, 2.1 × 30 mm Agilent Zorbax SB-C8 guard column and a 1.8 μm, 2.1 × 50 mm Agilent Zorbax SB-Aq analytical column heated to 60°C were used. The mobile phase consisted of 0.2% acetic acid in water (A) and 0.2% acetic acid in methanol (B) with the gradient starting at 2% B and increasing to 98% B in 13 min, held at 98% B for 6 min, followed by re-equilibration to 2% B for 6.5 min at a flow rate of 0.6 ml/min.
Data were acquired over the 25 minute run in both electrospray ionization positive (ESI+) and negative (ESI−) modes to detect cations and anions, respectively.
Data preprocessing
Data from the quality control samples were assessed for signal stability and retention time shifting over the course of the run. Preprocessing of the raw data from the Q-TOF was done using the open-source package xcms in R Studio, developed by the Scripps Center for Metabolomics (Smith et al. 2006; Tautenhahn, Böttcher, and Neumann 2008). We used xcms for feature detection, retention time alignment, and recursive filling of missing peaks. The relative abundance of each ion was normalized by dividing by the sum of the abundances of all ions detected. Signals were multiplied by 106 for convenience, were log10 transformed to better approximate Gaussian distributions, and are unitless. Features found in ESI+ and ESI− modes were combined, and further filtered into two data sets: the primary data set and the sensitivity analysis data set. When considering the percent of samples in which an ion was detected, 40 samples (37 subject and 3 pooled quality control samples) were included. The primary data set retained only those ions that were found in at least 50% of all samples (n = 20; P = 1736 ions), and the sensitivity data set retained those ions that were found in at least 25% of all samples (n = 10; P = 3380 ions). After filtering, the primary data and sensitivity analysis data sets were each split into a training and a validation data set as described above.
Statistical analyses
Discovery of biomarkers of exposure
The primary data set, split into training and validation data sets, was used for the discovery of biomarkers of exposure. Using the training data, relative abundances of all ions were compared between Mn exposed and unexposed workers using a 2-sided t-test. P values were adjusted for multiple comparisons using the Benjamini–Hochberg method to control false discovery rates (FDR). Given the exploratory nature of these analyses and the desire to find identify a larger set of potential ions that could distinguish between groups defined by exposure, an FDR <0.1 was considered to be significant. Ions found to be significantly different between the exposed and unexposed workers in the training group were subsequently tested in the validation set. Since the number of statistical comparisons was more limited in the validation set (n = 15), the unadjusted P values are reported. For those ions in the 50% (primary) data set that were found to be significantly different in the training group and were also significantly different in the validation group, the exposure–response relationship was explored categorically by comparing relative abundances in the three categories of exposure: No Exposure, Lower Exposure, and Higher Exposure.
Sensitivity analysis
The biomarker discovery process was repeated in the sensitivity data set with a 25% filtering step (i.e., less stringent) to see how changing the filtering criteria affected the number of ions found to be different between Mn exposed and Mn unexposed subjects. For the sensitivity analysis an FDR<0.05 and <0.01 in addition to an FDR <0.1 was considered to be the threshold for significance, to see how changing this criteria affected the number of ions found to be different between Mn exposed and unexposed subjects.
Results
Subject demographics for the Mn exposed and unexposed workers, stratified by training and validation sets, are presented in Table 1. Unexposed workers were slightly older than the exposed workers. The majority of exposed and unexposed workers self-identified as White, though there were more Hispanic workers in the Mn exposed group than the unexposed group. The unexposed workers were nearly equally divided between first and second shift, whereas the exposed workers were predominantly first shift workers. No Mn unexposed workers wore a respirator, but 12 (60%) of the Mn exposed workers self-reported to have worn an N95 dust mask for part or all of the workday. However, the foundry does not have a formal respiratory protection program, and overall respiratory hygiene at the site was observed to be poor. Thus, their use is unlikely to have a substantial effect on their received Mn dose.
Table 1.
Subject demographics by Mn exposure status.
| Mn exposed workers | Mn unexposed workers | |||||
|---|---|---|---|---|---|---|
| All (n = 20) | Training (n=12) | Validation (n=8) | All (n=17) | Training (n=10) | Validation (n=7) | |
| mean ± SD (range) | mean ± SD (range) | mean ± SD (range) | mean ± SD (range) | mean ± SD (range) | mean ± SD (range) | |
| Age | 41.9 ± 12.8 (21, 66) | 41.8 ± 12.6 (21, 62) | 42.1 ± 12.6 (29, 66) | 48.1 ± 11.3 (27, 60) | 48.3 ± 10.8 (31, 60) | 47.7 ± 10.8 (27, 60) |
| Ethnicity | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) |
| Hispanic | 12 (60.0) | 7 (58.3) | 5 (62.5) | 4 (23.5) | 3 (30.0) | 1 (14.3) |
| Not Hispanic | 8 (40.0) | 5 (41.7) | 3 (37.5) | 13 (76.5) | 7 (70.0) | 6 (85.7) |
| Race | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) |
| White | 15 (75.0) | 9 (75.0) | 6 (75.0) | 15 (88.2) | 9 (90.0) | 6 (85.7) |
| Other | 5 (25.0) | 3 (25.0) | 2 (25.0) | 2 (11.8) | 1 (10.0) | 1 (14.3) |
| Respirator | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) |
| Yes | 12 (60) | 7 (58.3) | 5 (62.5) | 0 (0) | 0 (0) | 0 (0) |
| No | 8 (40) | 5 (41.7) | 3 (37.5) | 17 (100) | 10 (100) | 7 (100) |
| Shift | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) |
| First | 17 (85.0) | 10 (83.3) | 7 (87.5) | 9 (52.9) | 5 (50.0) | 4 (57.1) |
| Second | 3 (15.0) | 2 (16.7) | 1 (12.5) | 8 (47.1) | 5 (50.0) | 3 (42.7) |
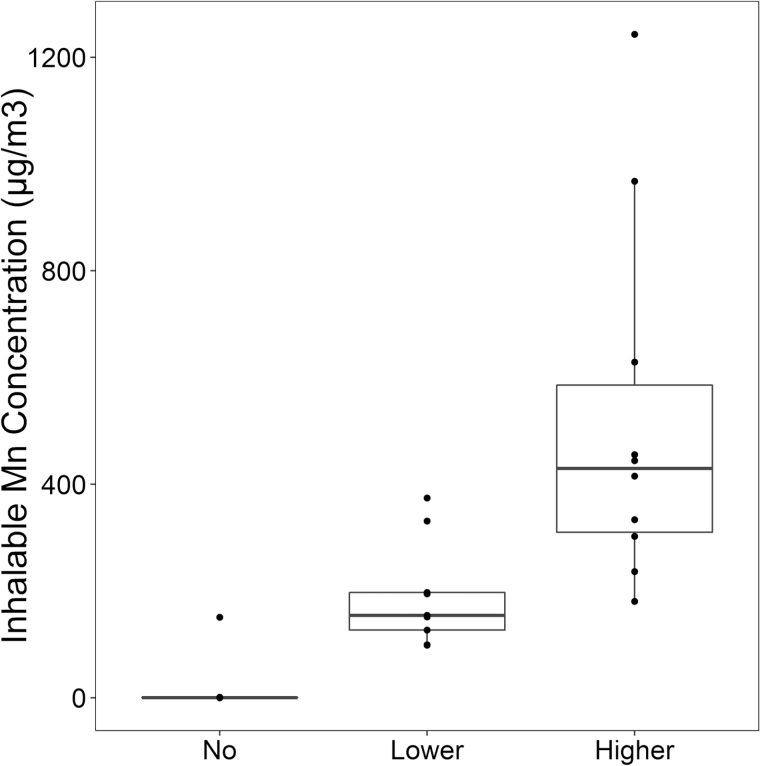
Results from personal inhalable dust monitoring are presented as box plots in Figure 1, with the training and validation sets combined, stratified into the three categories of exposure. The mean Mn exposure in the Lower Exposure group (n = 10) was 192.0 µg/m3 (standard deviation [SD]: 98.3, range: 98.5 to 374.3) and the mean Mn exposure in the Higher Exposure group (n = 10) was 520.7 µg/m3 (SD: 339.0, range: 180.4 to 1243.3). One worker in the No Exposure group had an inhalable dust measurement (150.8 µg/m3) that was substantially higher than the other workers in the No Exposure group. It is believed this is due to operating a vehicle with windows down near an area where welding was occurring, per worker report. However, this substantially higher exposure could have been due to another exposure source at the scrap yard that was not accounted for. Eighteen of the 34 inhalable Mn samples (53%) exceeded the ACGIH recommended TWA TLV of 100 µg/m3; all of which were collected at the foundry.
Figure 1.
Manganese exposure assessment by Mn exposure group (in µg/m3).
For each exposure group, the middle line that divides the box into two parts represents the median value while the top and bottom line of the box represent the 75th and 25th percentiles, respectively. The box represents the interquartile range (IQR) of scores for the group. The whiskers are extended to all values that are no >1.5 × IQR from the edge of the box.
Following metabolomics analysis of the urine samples, a total of 1736 ions were found in the primary data set (ions found in at least 50% of the samples). Nineteen ions were found to be significantly different between the Mn exposed and unexposed groups. However, in three instances, it was believed multiple isotopologues or fragments of another significant ion were classified as being significantly different between Mn exposed and unexposed groups. For these instances, only the ion with the largest relative abundance was retained and reported herein. Thus, a total of 15 unique ions were found to be significantly different between the Mn exposed and unexposed groups (Table 2). Nine of the 15 ions were replicated in the validation set.
Table 2.
Relative abundance of ions found to be significantly different (FDR < 0.1) between Mn exposed and unexposed subjects in the training group.
| m/z | Retention time (mins) | Mode | Mn unexposed (n = 10) | Mn exposed (n = 12) | FDR traininga | P value validationb | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD | CV (%) | % Detected | Mean ± SD | CV (%) | % Detected | |||||
| 201.02c | 3.56 | ESI− | 2.05 ± 0.51 | 24.9 | 70 | 3.56 ± 0.40 | 12.4 | 100 | <0.01 | <0.01 |
| 297.10c | 3.98 | ESI− | 1.78 ± 0.29 | 16.3 | 10 | 3.23 ± 0.42 | 13.0 | 92 | <0.01 | <0.01 |
| 553.24 | 4.03 | ESI− | 0.76 ± 0.27 | 35.5 | 0 | 1.94 ± 0.58 | 29.9 | 75 | <0.01 | <0.01 |
| 160.08 | 4.21 | ESI− | 1.49 ± 0.36 | 24.2 | 40 | 2.14 ± 0.34 | 15.9 | 92 | 0.04 | 0.14 |
| 205.07 | 4.21 | ESI− | 1.78 ± 0.42 | 23.6 | 80 | 2.46 ± 0.35 | 14.2 | 92 | 0.08 | 0.13 |
| 246.01 | 4.68 | ESI− | 0.41 ± 0.76 | 185.4 | 0 | 1.99 ± 0.31 | 15.6 | 83 | 0.01 | 0.03 |
| 311.12 | 5.18 | ESI− | 1.45 ± 0.30 | 20.7 | 10 | 2.34 ± 0.34 | 14.5 | 92 | <0.01 | <0.01 |
| 415.22 | 8.95 | ESI− | 1.58 ± 0.16 | 10.1 | 20 | 2.04 ± 0.30 | 14.7 | 75 | 0.03 | 0.08 |
| 321.10 | 4.01 | ESI+ | 1.72 ± 0.26 | 15.1 | 0 | 2.47 ± 0.36 | 14.6 | 83 | 0.03 | <0.01 |
| 229.07 | 4.20 | ESI+ | 1.59 ± 0.27 | 17.0 | 10 | 2.07 ± 0.26 | 12.6 | 67 | 0.08 | 0.40 |
| 177.11c | 4.71 | ESI+ | 1.84 ± 0.37 | 20.1 | 30 | 2.46 ± 0.25 | 10.2 | 92 | 0.08 | <0.01 |
| 354.23 | 4.95 | ESI+ | 2.17 ± 0.27 | 12.4 | 50 | 2.62 ± 0.21 | 8.0 | 83 | 0.08 | 0.20 |
| 311.15 | 6.83 | ESI+ | 2.42 ± 0.22 | 9.1 | 80 | 1.97 ± 0.30 | 15.2 | 17 | 0.10 | 0.16 |
| 403.23 | 7.72 | ESI+ | 2.04 ± 0.18 | 8.8 | 60 | 1.69 ± 0.19 | 11.2 | 17 | 0.08 | 0.65 |
| 459.22 | 7.78 | ESI+ | 2.84 ± 0.21 | 7.4 | 100 | 2.41 ± 0.24 | 10.0 | 58 | 0.08 | 0.08 |
CV: coefficient of variation; m/z: mass to charge ratio.
Benjamini–Hochberg corrected P value between Mn unexposed (n = 10) and Mn exposed (n = 12 f) subjects in training group.
Two-sided t-test between exposed (n = 8) and unexposed (n = 7) in validation groups.
Had known isotopologues or fragments that were also found in the training group with FDR < 0.1.
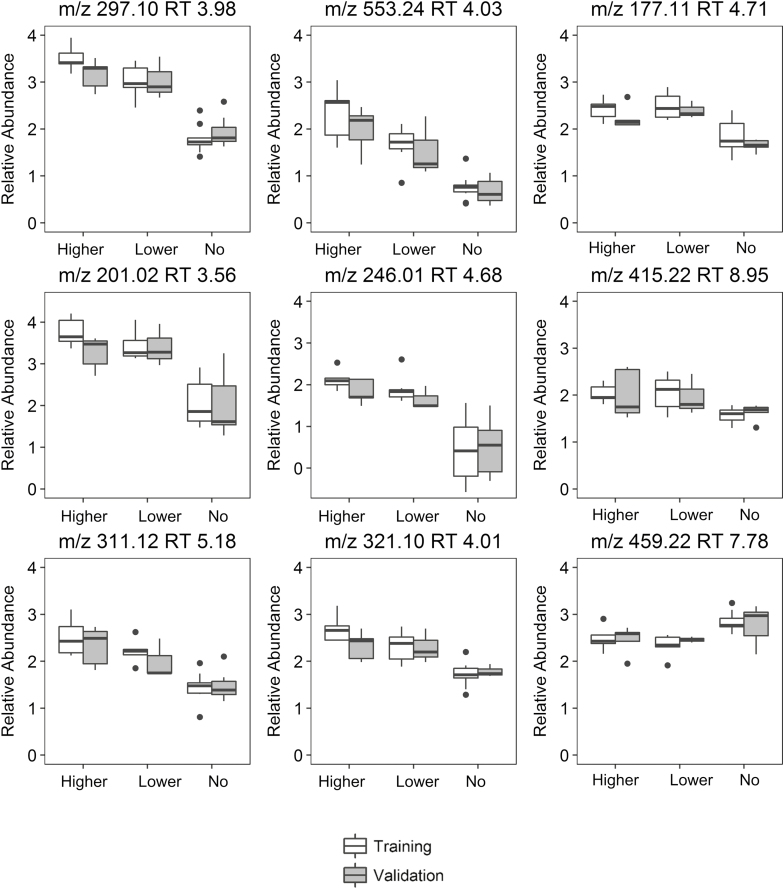
The exposure was stratified into three exposure categories (No Exposure, Lower Exposure, and Higher Exposure) to visualize possible exposure-response relationships. The box plots in Figure 2 illustrate the distribution of the abundance of these nine ions in the three exposure groups. Online Supplement 1 details the relative abundance of each of the ions stratified by the three exposure groups. Of the nine ions that replicated in the validation set, the abundance of all but one were increased in the exposed group, and were generally comparable in the training and validation sets. The ion with m/z 415.22 and retention time 8.95 minutes was the only ion of the nine to exhibit a lower abundance in the exposed group than in the unexposed group. Many of the ions exhibited an apparent exposure–response relationship across the three categorical exposure groups.
Figure 2.
Box plots of exposure–response relationships for ions identified as different between no Mn exposure, lower Mn exposure, and higher Mn exposure, in both training and validation sets.
To determine whether the filtering criteria affected the number of ions found to be different between Mn exposed and Mn unexposed subjects, we compared our original results from the primary data (filtering criteria of ions found in at least 50% of the samples) to the ions identified in the sensitivity data (filtering criteria of ions found in at least 25% of the samples). Table 3 shows the results of the sensitivity analyses when less stringent filtering criteria were used (50% vs. 25% for the primary and sensitivity data sets, respectively) and when considering an FDR threshold of 0.01 and 0.05 as well as 0.1. As 0.1 was chosen a priori for the primary analysis, we wished to explore if more stringent FDR would also produce ions of interest. Using the sensitivity data set, the relative abundances of 22 unique ions were found to differ significantly between Mn exposed and unexposed workers in the training set when using a FDR <0.1, and 11 of these ions remained significantly different between the exposed and unexposed workers in the validation set. When considering an FDR threshold of 0.05, 12 ions were found to be different between the exposed and unexposed in the 25% data set and eight were found to be different in the 50% data set. In the 25% data set eight ions remained significant at the 0.05 level in the validation set, while seven remained significant in the 50% data set. Considering an even more stringent FDR of 0.01, eight ions were found to be different between the exposed and unexposed in the 25% data set and four were found to be different in the 50% data set. In the 25% data set six ions remained significant at the 0.01 level in the validation set while four remained significant in the 50% data set. One additional ion had a P value of <0.01 in the validation set, but had an FDR >0.01 (but <0.05) in the training set. All ions found to be significant in the primary data set were also found to be significant in the sensitivity data set.
Table 3.
Sensitivity analysis.
| Primary data set (50% filtering) | Sensitivity data set (25% filtering) | Overlapping ions | |
|---|---|---|---|
| Total ions | 1736 | 3380 | — |
| FDR < 0.1 | |||
| n ions found in training set | 15 | 22 | 15 |
| n ions found in validation set | 9 | 11 | 9 |
| FDR < 0.05 | |||
| n ions found in training set | 8 | 12 | 8 |
| n ions found in validation set | 7a | 8 | 7a |
| FDR < 0.01 | |||
| n ions found in training set | 4 | 8 | 4 |
| n ions found in validation set | 6b | 6 | 6b |
Ion m/z 177.11 RT 4.71 had an FDR <0.05 in the validation set, but had an FDR of 0.08 in the training set.
Ions m/z 177.11 RT 4.71 and m/z 321.10 RT 4.01 had FDR<0.01 in the validation set, but had FDRs of 0.08 and 0.03, respectively, in the training set.
Discussion
We found nine ions to be significantly different in the training and validation sets between groups defined by Mn exposure status (Table 3). An apparent exposure–response relationship emerged for most of the ions when looking at the relative abundances of these nine ions across three exposure groups, though no formal test of trend was done for the three groups. Despite the relatively modest number of workers participating in this study, these ions may in fact distinguish between groups defined by Mn exposure status in this occupational cohort and also provides evidence that metabolomics can be used as a method of exposure assessment (or in tandem with traditional exposure assessment techniques) in occupational settings. Exposure assessment using metabolomics methods is a novel and burgeoning field, with many as-yet unexplored applications for highly original and multidisciplinary environmental and occupational health research. The findings presented here represent an essential first step in the development and validation of a biomarker of Mn exposure using global metabolomics techniques.
Notably, work presented here does not include identification (by name or empirical formula) of any of the ions which we found to be significantly different between the Mn exposed and unexposed workers. Analytical work to determine the identities of these seven ions through fragmentation, isolation, and additional mass spectrometry and nuclear magnetic resonance methods is ongoing. However, as detailed by others, identification of unknown ions is a major bottleneck in metabolomics research. Knowing the chemical structure or formula of these promising ions would help to inform the biochemical pathways influenced by Mn exposure and allow us to better elucidate the relationship between exposure and health effect.
A challenge to using metabolomics methods for exposure assessment in occupational cohorts is that any metabolites believed to distinguish between Mn exposure status could be non-specific to Mn exposure. Instead, these metabolites could represent unmeasured confounders that differentiate the two groups, including other workplace exposures. In human cohorts, it is extremely challenging to comprehensively assess all confounding exposures that would be present in an occupational setting. While many of these workplace exposures (e.g. iron, chromium, nickel) would likely be correlated with Mn exposure, others may be related to specific industrial processes or base metals and not correlated with Mn exposure. It is always a limitation with metabolomics studies that ions found to differentiate between groups could actually relate to unmeasured confounders as opposed to the exposure in question. However, including separate training and validation groups minimizes the identification of false positive signals. We found nine ions to be different between groups defined by exposure in the training data set that were replicated in the validation data set. Additional studies in Mn exposed individuals and appropriate controls should be conducted to verify these results.
Additionally, the workplace is only one of many micro-environments in which these workers spend an appreciable amount of time, and while all such micro-environments have historically been thought of separately (including in this manuscript), it is the interaction of work and non-work factors that together contribute to overall human health (Schulte and Vainio 2010; Schulte et al. 2012). Despite classifying persons into groups defined by exposure or disease for metabolomics studies, at its very nature metabolomics is a reflection of exposures sustained in all micro-environments, and through multiple routes of exposure. While this can make it more challenging to investigate a single exposure, it does make metabolomics a powerful tool for exploring the human exposome, assuming that a life history of exposures can be assessed in a rigorous way.
Despite these inherent limitations and considerations, the novelty of the work presented here should not be minimized. As limits on occupational exposures are becoming increasingly stringent and chronic diseases are being linked to low-level exposures, a new set of exposure assessment tools is necessary to assess how subtle biochemical changes in workers influence their health. Indeed, for an exposure such as Mn, it is likely that the majority of exposure would be sustained in a work environment relative to exposures through dietary or ambient sources, making the workplace a logical setting for investigation of Mn. Research presented here shows that metabolomics is one tool that can be used to find subtle biochemical differences between groups classified by exposure, and would have increased utility in well-characterized prospective longitudinal cohort studies with repeat measures as suggested by Wild, where both work and non-work exposures can be simultaneously assessed, and the contribution of each micro-environment characterized (Wild 2005).
In conclusion, this research presents the first time, to our knowledge, that metabolomics methods have been used in an occupational setting to investigate biochemical differences between Mn exposed and unexposed workers. We hope that, with identification of all or some of these seven promising ions and subsequent interpretation of their biological relevance, a greater understanding of the mechanisms behind manganese’s neurotoxic effects. Additionally, we hope that work presented here can be used by others to expand the use of metabolomics-based analyses in occupational exposure assessment. Doing so will inform the use of metabolomics in exposome studies looking to integrate work and non-work factors in interpreting human exposure and health.
Conflict of interest
The authors have no conflicts of interest to report.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under the Center for Ecogenetics and Environmental Health, Award Number P30ES007033. Marissa Baker was further supported by a National Institute of Environmental Health Sciences Biostatistics, Epidemiologic, and Bioinformatics Training in Environmental Health, Award Number T32ES015459 and Ruth L. Kirschstein National Research Service Award F31ES027304. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Thanks to Tauri Senn, University of Washington Department of Medicinal Chemistry for help with sample analysis and interpretation.
References
- Ahmed SS, Santosh W, Kumar S, et al. (2009) Metabolic profiling of Parkinson’s disease: evidence of biomarker from gene expression analysis and rapid neural network detection. J Biomed Sci; 16: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostoli P, Lucchini R, Alessio L. (2000) Are current biomarkers suitable for the assessment of manganese exposure in individual workers? Am J Ind Med; 37: 283–90. [DOI] [PubMed] [Google Scholar]
- Astle J, Ferguson JT, German JB, et al. (2007) Characterization of proteomic and metabolomic responses to dietary factors and supplements. J Nutr; 137: 2787–93. [DOI] [PubMed] [Google Scholar]
- Baker MG, Criswell SR, Racette BA, et al. (2015) Neurological outcomes associated with low-level manganese exposure in an inception cohort of asymptomatic welding trainees. Scand J Work Environ Health; 41: 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MG, Simpson CD, Sheppard L, et al. (2015) Variance components of short-term biomarkers of manganese exposure in an inception cohort of welding trainees. J Trace Elem Med Biol; 29: 123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MG, Simpson CD, Stover B, et al. (2014) Blood manganese as an exposure biomarker: state of the evidence. J Occup Environ Hyg; 11: 210–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MG, Stover B, Simpson CD, et al. (2016) Using exposure windows to explore an elusive biomarker: blood manganese. Int Arch Occup Environ Health; 89: 679–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Matson WR, Wang L, et al. (2008) Metabolomic profiling to develop blood biomarkers for Parkinson’s disease. Brain; 131: 389–96. [DOI] [PubMed] [Google Scholar]
- Bouchard M, Mergler D, Baldwin M, et al. (2007) Neuropsychiatric symptoms and past manganese exposure in a ferro-alloy plant. Neurotoxicology; 28: 290–7. [DOI] [PubMed] [Google Scholar]
- Bowler RM, Roels HA, Nakagawa S, et al. (2007) Dose-effect relationships between manganese exposure and neurological, neuropsychological and pulmonary function in confined space bridge welders. Occup Environ Med; 64: 167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst DI, Kell DB. (2006) Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics; 2: 171–96. [Google Scholar]
- Dorman DC, Struve MF, Norris A, et al. (2008) Metabolomic analyses of body fluids after subchronic manganese inhalation in rhesus monkeys. Toxicol Sci; 106: 46–54. [DOI] [PubMed] [Google Scholar]
- Finkelstein MM, Jerrett M. (2007) A study of the relationships between Parkinson’s disease and markers of traffic-derived and environmental manganese air pollution in two Canadian cities. Environ Res; 104: 420–32. [DOI] [PubMed] [Google Scholar]
- Fordahl S, Cooney P, Qiu Y, et al. (2012) Waterborne manganese exposure alters plasma, brain, and liver metabolites accompanied by changes in stereotypic behaviors. Neurotoxicol Teratol; 34: 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan-Smith G, Wallace GR, Douglas MR, et al. (2012) The role of metabolomics in neurological disease. J Neuroimmunol; 248: 48–52. [DOI] [PubMed] [Google Scholar]
- Health and Safety Executive (2014) MDHS 14/4 General methods for sampling and gravimetric analysis of respirable, thoracic and inhalable aerosols. UK: Health and Safety Executive. [Google Scholar]
- Ibáñez C, Simó C, Martín-Álvarez PJ, et al. (2012) Toward a predictive model of Alzheimer’s disease progression using capillary electrophoresis-mass spectrometry metabolomics. Anal Chem; 84: 8532–40. [DOI] [PubMed] [Google Scholar]
- Järvisalo J, Olkinuora M, Kiilunen M, et al. (1992) Urinary and blood manganese in occupationally nonexposed populations and in manual metal arc welders of mild steel. Int Arch Occup Environ Health; 63: 495–501. [DOI] [PubMed] [Google Scholar]
- Johansson-Persson A, Barri T, Ulmius M, et al. (2013) LC-QTOF/MS metabolomic profiles in human plasma after a 5-week high dietary fiber intake. Anal Bioanal Chem; 405: 4799–809. [DOI] [PubMed] [Google Scholar]
- Kumar KK, Goodwin CR, Uhouse MA, et al. (2015) Untargeted metabolic profiling identifies interactions between Huntington’s disease and neuronal manganese status. Metallomics; 7: 363–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini R, Apostoli P, Perrone C, et al. (1999) Long-term exposure to “low levels” of manganese oxides and neurofunctional changes in ferroalloy workers. Neurotoxicology; 20: 287–97. [PubMed] [Google Scholar]
- Lucchini RG, Albini E, Benedetti L, et al. (2007) High prevalence of Parkinsonian disorders associated to manganese exposure in the vicinities of ferroalloy industries. Am J Ind Med; 50: 788–800. [DOI] [PubMed] [Google Scholar]
- Mark D, Vincent JH. (1986) A new personal sampler for airborne total dust in workplaces. Ann Occup Hyg; 30: 89–102. [DOI] [PubMed] [Google Scholar]
- Mergler D, Huel G, Bowler R, et al. (1994) Nervous system dysfunction among workers with long-term exposure to manganese. Environ Res; 64: 151–80. [DOI] [PubMed] [Google Scholar]
- Michell AW, Mosedale D, Grainger DJ, et al. (2008) Metabolomic analysis of urine and serum in Parkinson’s disease. Metabolomics; 4191–201. [Google Scholar]
- Neth K, Lucio M, Walker A, et al. (2015) Changes in brain metallome/metabolome pattern due to a single i.v. injection of manganese in rats. PLOS ONE; 10: e0138270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neal SL, Zheng W. (2015) Manganese toxicity upon overexposure: a decade in review. Curr Environ Health Rep; 2: 315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roels H, Lauwerys R, Buchet JP, et al. (1987) Epidemiological survey among workers exposed to manganese: effects on lung, central nervous system, and some biological indices. Am J Ind Med; 11: 307–27. [DOI] [PubMed] [Google Scholar]
- Roels H, Sarhan MJ, Hanotiau I, et al. (1985) Preclinical toxic effects of manganese in workers from a Mn salts and oxides producing plant. Sci Total Environ; 42: 201–6. [DOI] [PubMed] [Google Scholar]
- Roth JA. (2006) Homeostatic and toxic mechanisms regulating manganese uptake, retention, and elimination. Biol Res; 39: 45–57. [DOI] [PubMed] [Google Scholar]
- Sato Y, Suzuki I, Nakamura T, et al. (2012) Identification of a new plasma biomarker of Alzheimer’s disease using metabolomics technology. J Lipid Res; 53: 567–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte P, Vainio H. (2010) Well-being at work–overview and perspective. Scand J Work Environ Health; 36: 422–9. [DOI] [PubMed] [Google Scholar]
- Schulte PA, Pandalai S, Wulsin V, et al. (2012) Interaction of occupational and personal risk factors in workforce health and safety. Am J Public Health; 102: 434–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, et al. (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem; 78: 779–87. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, Böttcher C, Neumann S. (2008) Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics; 9: 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay-Sontheimer J, Shireman LM, Beyer RP, et al. (2014) Detection of an endogenous urinary biomarker associated with CYP2D6 activity using global metabolomics. Pharmacogenomics; 15: 1947–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DI, Uppal K, Zhang L, et al. (2016) High-resolution metabolomics of occupational exposure to trichloroethylene. Int J Epidemiol; 45: 1517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Zhang AH, Sun H. (2013) Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology; 572072–77. [DOI] [PubMed] [Google Scholar]
- Wild CP. (2005) Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev; 14: 1847–50. [DOI] [PubMed] [Google Scholar]
- Wishart DS. (2010) Computational approaches to metabolomics. New York, NY: Bioinformatics Methods in Clinical Research, Springer. [DOI] [PubMed] [Google Scholar]
- Zhang AH, Sun H, Wang XJ. (2013) Recent advances in metabolomics in neurological disease, and future perspectives. Anal Bioanal Chem; 405: 8143–50. [DOI] [PubMed] [Google Scholar]
- Zhang J, Bowers J, Liu L, et al. (2012) Esophageal cancer metabolite biomarkers detected by LC-MS and NMR methods. PLOS ONE; 7: e30181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Fu SX, Dydak U, et al. (2011) Biomarkers of manganese intoxication. Neurotoxicology; 32: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.