Abstract
Exposure to certain pesticides induces oxidative stress and increases Parkinson’s disease (PD) risk. Mitochondrial DNA (mtDNA) damage is found in dopaminergic neurons in idiopathic PD and following pesticide exposure in experimental models thereof. Base excision repair (BER) is the major pathway responsible for repairing oxidative DNA damage in cells. Whether single nucleotide polymorphisms (SNPs) in BER genes alone or in combination with pesticide exposure influence PD risk is unknown. We investigated the contributions of functional SNPs in 2 BER genes (APEX1 and OGG1) and mitochondrial dysfunction- or oxidative stress-related pesticide exposure, including paraquat, to PD risk. We also studied the effect of paraquat on levels of mtDNA damage and mitochondrial bioenergetics. 619 PD patients and 854 population-based controls were analyzed for the 2 SNPs, APEX1 rs1130409 and OGG1 rs1052133. Ambient pesticide exposures were assessed with a geographic information system. Individually, or in combination, the BER SNPs did not influence PD risk. Mitochondrial-inhibiting (OR = 1.79, 95% CI [1.32, 2.42]), oxidative stress-inducing pesticides (OR = 1.61, 95% CI [1.22, 2.11]), and paraquat (OR = 1.54, 95% CI [1.23, 1.93]) were associated with PD. Statistical interactions were detected, including for a genetic risk score based on rs1130409 and rs1052133 and oxidative stress inducing pesticides, where highly exposed carriers of both risk genotypes were at the highest risk of PD (OR = 2.21, 95% CI [1.25, 3.86]); similar interactions were estimated for mitochondrial-inhibiting pesticides and paraquat alone. Additionally, paraquat exposure was found to impair mitochondrial respiration and increase mtDNA damage in in vivo and in vitro systems. Our findings provide insight into possible mechanisms involved in increased PD risk due to pesticide exposure in the context of BER genotype variants.
Keywords: Parkinson’s disease, base excision repair, pesticides, gene-environment interaction, mitochondrial DNA, paraquat
The most common neurodegenerative movement disorder is Parkinson’s disease (PD). The progressive loss of dopamine neurons in the substantia nigra pars compacta is a hallmark pathology. Since these dopaminergic neurons are required for proper motor function, the declining dopamine levels due to their loss is associated with tremor, rigidity and bradykinesia. Dopamine replacement treatments provide only a symptomatic benefit and are not a disease-modifying therapy. Current therapies also do not address the nonmotor symptoms of PD. In addition to the human suffering, the economic burden of PD exceeds $14.4 billion annually (Kowal et al., 2013).
PD is widely accepted as a multifactorial disease—with both genetic and environmental contributions (Gao and Hong, 2011). The discovery of the first genetic mutation responsible for familial PD occurred in 1996 (Polymeropoulos et al., 1996, 1997). Since then, only 6 highly penetrant Mendelian variants including SNCA, LRRK2, PARKIN, PINK1, and PARK7 cause monogenic PD (Verstraeten et al., 2015). Using genome-wide association studies (GWASs) 28 DNA variants at 24 loci were found to predict PD risk (Nalls et al., 2014). The identification of causative variants or risk alleles may be challenging due to the fact that heritability of PD is low and reported to be between 34% and 40% (Wirdefeldt et al., 2011).
The majority of cases are “idiopathic” and the multifactorial etiology is likely a complicated interplay between genetic risk factors and environmental exposures. PD has been associated with environmental exposures (Bronstein et al., 2009; Dick et al., 2007; Elbaz et al., 2007; Vance et al., 2010) and the best characterized environmental risk factor is occupational exposure to pesticides (Dhillon et al., 2008; Hancock et al., 2008; Kamel et al., 2007; Richardson et al., 2009). It is likely that interindividual differences in metabolizing specific substances or repairing damage induced by toxicants is genetically determined, and therefore effects of pesticide exposure on PD risk might be influenced by genetic variation.
For the widely used herbicide paraquat there is strong epidemiological evidence of a link to PD risk (Costello et al., 2009; Freire and Koifman, 2012), and for the pesticide rotenone, some human evidence is emerging (Tanner et al., 2011). Rotenone inhibits complex I of the mitochondrial electron transport chain and is a prototypical example of how an exogenous toxin can mimic clinical and pathological features of PD in an animal model (Martinez and Greenamyre, 2012). Paraquat, on the other hand, induces oxidative stress by redox cycling rather than by direct complex I inhibition; however, after prolonged exposure, it may inhibit mitochondrial function via oxidative mechanisms. In fact, there is evidence that paraquat redox cycling may involve complex I and generate superoxide in the mitochondrial matrix. Previous studies from several groups have shown paraquat to be a potent inducer of oxidative stress, alpha-synuclein aggregation, and selective nigral cell injury (Dinis-Oliveira et al., 2006; Kuter et al., 2010; McCormack et al., 2002). Long-term paraquat administration in rats leads to a slowly progressing dysfunction of the dopaminergic nigrostriatal system, a model which may resemble the early stages of PD (Ossowska et al., 2005).
Mitochondrial dysfunction and oxidative stress have been strongly associated with several age-related neurodegenerative diseases, including PD (Erlich et al., 2006; Sanders and Greenamyre, 2013). Oxidative stress and mitochondrial impairment can both cause and result from DNA damage. Furthermore, deficiency in the repair of nuclear and mitochondrial DNA (mtDNA) damage can contribute to neurodegeneration (Jeppesen et al., 2011). To protect against DNA damage, numerous cell mechanisms are devoted to DNA repair. The base excision repair (BER) pathway is responsible for repairing oxidative and alkylation base damage, apurinic/apyrimidinic (abasic sites) and single strand breaks. Little is known about BER in neurons or in the mitochondria (Krokan and Bjoras, 2013). Although expression of some BER proteins is increased in dopaminergic neurons in the substantia nigra of PD, the role of BER proteins in the development of PD is unclear (Arai et al., 2006; Fukae et al., 2005; Shimura-Miura et al., 1999).
BER is the major pathway for eliminating reactive oxygen species-induced DNA damage (Saki and Prakash, 2016). Oxidatively damaged DNA is formed as a byproduct of either normal cell metabolism or endogenous or exogenous exposures. One of the most common oxidative base modifications is 8-oxoguanine (8-oxo-dG). Oxoguanine DNA glycosylase (OGG1) is the primary enzyme responsible for the removal of the 8-oxo-dG lesion during BER. If OGG1-mediated BER does not remove the 8-oxo-dG, replication can still proceed but may lead to the accumulation of G → T mutations. During BER when the 8-oxo-dG lesion is removed, an abasic site is left as a DNA repair intermediate. Abasic sites are a type of DNA damage in which the DNA has neither a purine nor a pyrimidine. Abasic sites are potentially cytotoxic or genotoxic as they can block the polymerase during replication and transcription. In addition, an inability to effectively repair abasic sites can compromise mitochondrial function and contribute to neurodegeneration (Lauritzen et al., 2011). Abasic sites generated spontaneously, by exogenous stress, or as a DNA repair intermediate by monofunctional DNA glycosylases, are repaired by apurininc/apyrimidinic endonuclease I (APEX1). Polymorphisms in APEX1 and OGG1 have been studied extensively in human diseases and associations with several types of cancer have been found (Karahalil et al., 2012).
A recent study from our group showed that mtDNA damage, specifically in the form of abasic sites, accumulates in nigral neurons with rotenone exposure in vivo and in human postmortem PD brain tissue (Sanders et al., 2014c). These results suggest an ineffective BER response in PD. These findings led us to newly explore the effects of pesticide exposure on PD risk in the context of common genetic variants in BER. We hypothesized that pesticide exposure-induced mtDNA damage in individuals with genetically determined inefficiencies in the BER system may represent a synergism that increases the risk of developing PD. Therefore, we investigated the contributions of two single nucleotide polymorphisms (SNPs) in the APEX1 and OGG1 genes with pesticide exposure, including paraquat, and evaluate whether combinations of several BER risk SNPs together with pesticide exposure further modified PD risk. Last, we determined whether an increase in mtDNA damage and mitochondrial dysfunction was associated with exposure to the oxidative damage-inducing herbicide paraquat in vitro and in vivo.
MATERIALS AND METHODS
Human subjects
The Parkinson’s Environment and Genes Study (PEG) has been enrolling new-onset PD patients initially through large medical groups, neurologists, and public service announcements (2001–2007). In addition, patient enrollment occurred through the pilot California Parkinson’s Disease Registry since its introduction in 2007. We also enrolled population controls through Medicare enrollee lists and residential tax-collector records from the same three highly agricultural counties in Central California in which the patients also resided: Kern, Fresno, and Tulare. Each PD patient, diagnosed on average 2.6 years (SD = 2.1) prior to enrollment, was seen by University of California at Los Angeles (UCLA) study movement disorder specialists (J.B.) and clinically confirmed as having idiopathic PD. A detailed discussion of subject recruitment methods and selection, case definition criteria, and clinical characteristics has previously been described in Costello et al. (2009) and Wang et al. (2011). Here, we included 619 PD patients and 854 population-based controls who were recruited between January 2001 and December 2013. All procedures were approved by the UCLA Human Subjects Committee and informed consent was obtained from all participants.
SNP selection and genotyping methods
Two SNPs, APEX1 rs1130409 (Asp148Glu, major allele T, minor allele G) and OGG1 rs1052133 (Ser326Cys, major allele C, minor allele G), functionally associated with oxidative DNA damage repair capacity, were selected. A single transition in the fifth APEX1 exon, from the base pair T allele to G allele induces the substitution of the 148th amino acid aspartate (Asp) to glutamate (Glu) (Asp148Glu, rs1130409). Functional analyses have found the Asp148Glu variant decreases repair of 8-oxo-dG, a widely accepted marker of oxidative DNA damage and oxidative stress (Breton et al., 2007). Similarly, for hOCCG1 a transition from the base pair C allele to G allele in exon 7 induces the substitution of the 326th amino acid serine (Ser) to cysteine (Cys) (Ser326Cys, rs1052133); with functional analyses finding the hOGG1-Cys326 protein having a significantly lower capacity to repair 8-oxo-dG than the hOGG1-Ser326 protein (Weiss et al., 2005). Both SNPs have been investigated in a wide range of health outcomes. Our recent findings suggest increased BER intermediates in PD models and postmortem brain tissue (Sanders et al., 2014c).
PEG participants provided blood or saliva samples from which we obtained DNA. EDTA preserved blood samples were collected by venipuncture; saliva samples with the Oragene kit. DNA was extracted at the UCLA Biologic Specimen Core Facility, using Autopure LSTM nucleic acid purification instrument from Gentra Systems or the best suitable Qiagen kit (ie, DNeasy). Primers and probes were designed based on the NCBI (National Center for Biotechnology Information) DNA sequence and purchased from Fluidigm Corporation (South San Francisco, California). Genotyping was done using the Fluidigm high throughput BioMark HD system (Fluidigm Corporation). Both SNPs were successfully genotyped, with call rates of 98.7% (APEX1 rs1130409) and 99.1% (OGG1 rs1052133).
Pesticide selection and exposure assessment
We selected specific pesticides considered mitochondrial complex I inhibitors and/or oxidative stressors based on a previous study (Tanner et al., 2011). To assess exposures to these specific pesticides, we employed a geographic information system (GIS)-based computer model which estimates ambient pesticide exposures from agricultural sources based on legally mandated pesticide use reports and land use in California. The exposure assessment model has been described in detail (Cockburn et al., 2011). Briefly, for each pesticide recorded (since 1974), we summed the pounds of pesticide applied per year per acre within a 500-m radius buffer of each participants’ geocoded addresses (both residential and workplace), then we calculated yearly averages for each subject and pesticide by summing the annual pounds of pesticide applied between 1974 and up to 10 years prior to the date of diagnosis (for patients) or interview (for controls) and divided this sum by the number of years in the relevant time period. We then dichotomized exposure to individual pesticides based on the pesticide-specific median level in exposed controls (at or above median). Participants could have received exposures at either residential or occupational addresses, or both. We counted the total number of oxidative stressor exposures and mitochondrial complex I inhibitor exposures for each participant and classified all participants as having either none/low or high exposure based on the distribution of the pesticide counts among exposed controls (high exposure: at or above the median number of chemicals in exposed controls, none/low exposure: exposure to less than the median number of chemicals in exposed controls). Additionally, we assessed exposure to paraquat alone, considering participants with high estimated ambient exposure (having a yearly average pounds of pesticides applied above the median found in exposed controls) at both their residence and occupation as highly exposed.
Quantification of mtDNA damage using a QPCR-based assay
Rat primary ventral midbrain (VMB) neurons were cultured as described in (Sanders et al., 2014c). All animals were treated humanely with regard to alleviation of suffering and followed the Guide for the Care and use of Laboratory Animals. Rat VMB neuronal cultures were treated with vehicle or paraquat, a common oxidative stress inducing pesticide, harvested in C1 buffer and H20 (QIAGEN), incubated on ice for 10 min and then centrifuged at 10 000 g for 20 min at 4 °C. The supernatant was removed and the remaining pellet was frozen and stored at −20 °C until analysis. Levels of mtDNA damage were measured using a quantitative polymerase chain reaction (QPCR)-based assay as described previously (Sanders et al., 2014b, c) with the following modifications. The KAPA Long Range Hot Start DNA Polymerase Kit (KAPABiosystems) was used since the availability of the GeneAmp XL PCR kit from Applied Biosystems has been discontinued (Rooney et al., 2015). The primers for the amplification of the rat mitochondrial fragment were described previously (Ayala-Torres et al., 2000).
We also evaluated paraquat treated Drosophila (see below for full description). The primers used for the quantification of Drosophila mtDNA damage were: large mitochondrial primer sequences were 5′-TTT AAA AAA TAA ATA TGA AAA TTT ATT CGC-3′ and 5′ATG AAG CGA TTG ATT GCA ATT AGT-3′, for the small mitochondrial PCR product, the primer sequences were 5′-GCT CCT GAT ATA GCA TTC CCA CGA-3′ and 5′-CAT GAG CAA TTC CAG CGG ATA AA-3′. All QPCR-based experiments were performed in technical triplicate for each biological replicate.
Measurement of cellular oxygen consumption rate
Cellular oxygen consumption rate (OCR) was measured using an extracellular flux analyzer (Seahorse Bioscience XF96) as described (Sanders et al., 2014c) with minor modifications. Intact primary VMB neurons were grown in XF96 plates seeded at 80 000 cells/well for 1 week in growth medium. Neuronal cultures were then treated with either vehicle (DMSO) or paraquat for 24 h. Plates were incubated in Calibrant medium (Seahorse Bioscience) for at least 30 min. Media containing final concentrations of 10 μM oligomycin, 300 nM carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone, and 1 μM rotenone and antimycin A were preloaded into the drug delivery system. Once the basal OCR was measured, the compounds were added sequentially and the effects on OCR measured every 8 min. OCR values were normalized by cell number.
Drosophila paraquat exposure
Wildtype Drosophila were maintained on standard cornmeal agar and incubated at 25°C and a 50% relative humidity. Paraquat exposure was performed as described previously in Chaudhuri et al. (2007) with some modifications. Briefly, 24–96 h old flies were sorted into groups of 600 flies per group (300 males and 300 females) and incubated on freshly prepared drug food comprising sucrose (Sigma-Aldrich, St Louis, Missouri) at a final concentration of 5% mixed into molten agar (Sigma-Aldrich) at a final concentration 1%; and paraquat (methyl viologen, Sigma Aldrich) at a final concentration of 20 mM. Bottles containing 5% sucrose only in 1% molten agar were used as control. A total of 5 groups were run in triplicate for each of the drug-treated and control paradigms. Both paraquat-treated and control flies were incubated at 25°C for 12 h after which fly heads were harvested by placing the flies in a glass vial; dipping the samples into liquid nitrogen; and vortexing the vial on a mechanical shaker to detach the heads of the flies. Severed fly heads were then quickly sorted using a sieve and shipped on dry-ice to the University of Pittsburgh for analysis of mtDNA damage.
Statistical methods
We tested Hardy-Weinberg equilibrium for both SNPs in control participants using a chi-square test and then evaluated marginal associations between the SNPs and PD using logistic regression to calculate odds ratios (ORs) and 95% CIs. The heterozygote and homozygote variant genotypes were compared with the homozygote wildtype genotype (general genetic model) for both SNPs, in addition we also compared GG/GC to CC (OGG1 rs1052133) and TT/TG to GG (APEX1 rs1130409) based on identifying pesticide-related “risk alleles” in gene-environment (GxE) analysis—T (APEX1) and G (OGG1), ie, for alleles linked to increased risk in conjunction with pesticide exposure. To assess statistical GxE interactions, we introduced a multiplicative interaction term into the logistic models; we grouped genotype carriers of risk alleles together. Based on risk allele counting we also created a 2-level combined BER genetic risk classification, comparing those with at least 1 risk allele at both APEX1 and OGG1 loci to those with risk alleles either at only one locus or at neither; eg, APEX1 GG and/or OGG1 CC versus APEX1 TG or TT and OGG1 GC or GG. We adjusted for potential confounders including age (continuous, at diagnosis for patients and interview for controls), sex, ever/never smoking status, and European ancestry (yes/no). We used SAS 9.4 (SAS Institute Inc., Cary, North Carolina) for these analyses.
RESULTS
To determine whether either the APEX1 rs1130409 or OGG1 rs1052133 SNP was marginally associated with PD, 619 incident PD patients were compared with 854 population controls enrolled in the PEG case-control study. Basic demographic characteristics are listed in (Table 1). PD patients were more likely to be male, have a family history of PD, and ambiently exposed to a higher number of pesticides over all, and less likely to have ever smoked, consistent with previous reports (Ritz et al., 2007). APEX1 rs1130409 has a minor allele frequency (MAF) of 44% in both PD patients and controls, and OGG1 rs1052133 has an MAF of 24% in PD patients and 26% in controls. The control population was in Hardy-Weinberg equilibrium for both SNPs, APEX1 rs1130409 (P = .22) and OGG1 rs1052133 (P = .07).
TABLE 1.
Demographic Characteristics of the PEG Case-Control Study Population (n = 1473)
| Characteristic | Patients (n = 619) | Controls (n = 854) | P-Value* |
|---|---|---|---|
| Agea (mean ± SEM) | 68.3 ± 0.42 | 66.1 ± 0.40 | .0001 |
| Male (%) | 62.5 | 48.8 | <.0001 |
| European ancestry (%) | 75.8 | 68.1 | .001 |
| Ever smokers (%) | 47.7 | 52.7 | .06 |
| Positive PD historyb (%) | 15.8 | 7.5 | <.0001 |
| Education in years (mean ± SEM) | 13.6 ± 0.18 | 13.6 ± 0.14 | .67 |
| Years with Disease prior to enrollment (mean ± SEM) | 2.62 ± 0.08 | – | – |
| UPDRS Score (mean ± SEM) | 21.9 ± 0.46 | – | – |
| Number of pesticides highly exposed (mean ± SEM) | 51.0 ± 2.25 | 38.4 ± 1.64 | <.0001 |
Age at PD diagnosis for patients and interview for controls.
First degree relative with PD.
P-value based on chi-square or t test, PD patients versus controls.
PD risk was not influenced by genotype alone: for APEX1 rs1130409 the OR was 0.98 (95% CI; 0.77, 1.36) for those with a GT and 1.01 (95% CI; 0.74, 1.36) for those with the GG genotype (Table 2). Results were similar for the OGG1 rs1052133 SNP with an OR of 1.01 (95% CI; 0.81, 1.27) for the CG and 0.80 (95% CI; 0.52, 1.23) for the GG genotype (Table 2). PD risk was also not influenced by the combined risk allele score (Table 2). All models controlled for age, sex, smoking status, and European ancestry.
TABLE 2.
Associations (Marginal) Between APEX1 and OGG1 SNP Genotypes and PD in PEG Participants
| Variant/Genotype | Patients |
Controls |
Adjusted ORa |
P-Value |
|---|---|---|---|---|
| Genotype n (%) | (95% CI) | |||
| APEX1 rs1130409 | ||||
| TT | 200 (33) | 276 (33) | 1.00 (ref) | – |
| GT | 284 (47) | 400 (47) | 0.98 (0.76, 1.26) | .88 |
| GG | 126 (21) | 172 (20) | 1.01 (0.74, 1.36) | .97 |
| TT/GT vs GG (ref) | 0.98 (0.76, 1.28) | .91 | ||
| OGG1 rs1052133 | ||||
| CC | 352 (58) | 480 (56) | 1.00 (ref) | – |
| CG | 221 (36) | 306 (36) | 1.01 (0.81, 1.27) | .90 |
| GG | 39 (6) | 67 (8) | 0.80 (0.52, 1.23) | .31 |
| CG/GG vs CC (ref) | 0.98 (0.78, 1.21) | .83 | ||
| Combined genetic risk score | ||||
| APEX1 GG and/or OGG1 CC | 405 (66) | 559 (65) | 1.00 (ref) | – |
| APEX1 TT or TG and OGG1 CG or GG | 208 (34) | 295 (35) | 1.01 (0.80, 1.26) | .96 |
Adjusted for age, sex, ever smoked, and European ancestry.
Mitochondrial complex I pesticide exposure (OR of 1.79, 95% CI; 1.32, 2.42), oxidative stress inducing pesticide exposure (OR of 1.61, 95% CI; 1.22, 2.11), and paraquat (OR of 1.54, 95% CI; 1.23, 1.93), were associated with an increased risk for PD (Table 3).
TABLE 3.
Associations Between Ambient Pesticide Exposures at Residences and Work Places Grouped by Presumed Function and PD in PEG Participants
| Pesticide | CA-DPR Chem Code | Cases (n = 545) | Controls (n = 854) | Adjusted ORa(95% CI) | P-Value |
|---|---|---|---|---|---|
| Mitochondrial complex I inhibitors | |||||
| Thiabendazole | 587 | 0 | 0 | – | |
| Carbendazim | 2176 | 0 | 0 | – | |
| Rotenone | 518 | 6 | 12 | 0.76 (0.28, 2.06) | .58 |
| Cyhalothrin | 2297 | 7 | 10 | 1.18 (0.44, 3.18) | .74 |
| Benomyl | 1552 | 177 | 207 | 1.51 (1.18, 1.93) | .0009 |
| Permethrin | 2008 | 82 | 82 | 1.70 (1.21, 2.37) | .002 |
| (S)-cypermethrin | 3866 | 5 | 6 | 1.64 (0.49, 5.51) | .43 |
| Cypermethrin | 2171 | 24 | 22 | 2.00 (1.09, 3.67) | .03 |
| Pyridaben | 3959 | 14 | 14 | 1.70 (0.79, 3.68) | .18 |
| Mitochondrial inhibitor combined | None/Low (0–2) | 437 | 749 | ref | – |
| High (>2) | 108 | 103 | 1.79 (1.32, 2.42) | .0002 | |
| Oxidative stressors | |||||
| Carbon disulfide | 108 | 1 | 2 | – | |
| Dichlone | 202 | 3 | 4 | – | |
| Mercury compounds | 454 | 0 | 0 | – | |
| Pybuthrin | 486 | 15 | 34 | 0.68 (0.36, 1.27) | .22 |
| Cyhalothrin | 2297 | 7 | 10 | 1.18 (0.44, 3.18) | .74 |
| Paraquat | 1601 | 245 | 296 | 1.54 (1.23-1.93) | .0002 |
| Permethrin | 2008 | 82 | 82 | 1.70 (1.21, 2.37) | .002 |
| (S)-cypermethrin | 3866 | 5 | 6 | 1.64 (0.49, 5.51) | .43 |
| Cypermethrin | 2171 | 24 | 22 | 2.00 (1.09, 3.67) | .03 |
| Oxidative stressor combined | None/Low (0–2) | 409 | 703 | ref | – |
| High (>2) | 136 | 149 | 1.61 (1.22, 2.11) | .0007 |
Chloranil considered an oxidative stressor, was not recorded in the CADPR.
Adjusted for age, sex, ever-smoked, and European ancestry; reference for each individual chemical is the chemical specific unexposed groups (not shown).
Statistical interaction results for APEX1 rs1130409 or OGG1 rs1052133 SNPs and pesticide classes are shown in the Supplementary Table 1. We detected statistically significant or near significant interactions for both APEX1 rs1130409 and OGG1 rs1052133 (respectively) and ambient residential and work place pesticide exposures in the class of oxidative stressors. At both loci, for pesticide exposed risk allele carriers we estimated more than multiplicative risk increases. Specifically, eg, for OGG1 rs1052133, unexposed variant allele (G) carriers experienced no increase in PD risk (OR = 0.85, 95% CI = 0.66–1.10). The homozygous carriers of the dominant allele (CC) who were exposed to oxidative stress inducing pesticides showed if anything, a slight but nonstatistically significant increase in risk (OR = 1.28, 95% CI = 0.89–1.85). In contrast, the risk was increased by almost 80% in exposed variant carriers (OR = 1.79, 95% CI = 1.22–2.64). Likewise, for APEX1 rs1130409, we did not see increases in risk for either the SNP or pesticide exposures alone. Yet, exposed T allele carriers experienced a 67% increase in risk (OR = 1.67, 95% CI = 1.13–2.47). Similar trends were estimated for paraquat alone. Neither SNP alone showed a significant statistical interaction with ambient mitochondrial inhibitor exposure, though effect estimate trends are similar to those we estimated for the oxidative stress inducing pesticides described earlier (Supplementary Table 1).
The strongest statistical interactions we estimated were for the combined BER genetic risk allele score and pesticide exposures; we detected more than a multiplicative risk with oxidative stress exposure, mitochondrial inhibitor exposure, and paraquat exposure (Table 4). As reported for each individual SNP, we did not detect an increase in PD risk for those with risk alleles at both loci who were not exposed to these pesticides. We observed slight increases in risk with pesticide exposures in those with 1 or less risk alleles, mitochondrial inhibitor exposure: (OR = 1.43, 95% CI = 0.98–2.10); oxidative stressor exposure: (OR = 1.20, 95% CI = 0.85–1.69); and paraquat exposure (OR = 1.13, 95% CI = 0.75–1.70). The strongest risk was estimated for the joint effect of pesticide exposure and both risk alleles with mitochondrial inhibitor exposure: (OR = 2.32, 95% CI = 1.44–3.7); oxidative stressor exposure: (OR = 2.21, 95% CI = 1.45–3.38); and paraquat exposure: (OR = 2.38, 95% CI = 1.44–3.95, [Table 4]). Results were similar when we analyzed only participants who did not report a family history of PD (see Supplementary Tables 2 and 3).
TABLE 4.
Interaction and Main/Joint Effect Estimates for APEX1/OGG1 Combined Genetic Risk Score and Pesticide Exposure by Functional Group in PEG Participants
|
rs1130409 GG and/or rs1052133 CC |
rs1130409 GT or TT and rs1052133 CG or GG |
|||||||
|---|---|---|---|---|---|---|---|---|
|
0–1 Risk Genotypes |
2 Risk Genotypes |
|||||||
| Pesticide Exposure | Patients n (%) | Controls n (%) | Adj ORa (95% CI) | P-value | Patients n (%) | Controls n (%) | Adjusted ORa (95% CI) | P-Value |
| Ambient mitochondrial inhibitor exposure | ||||||||
| None/Low (0–2) | 298 (83) | 490 (88) | 1.00 (ref) | 139 (75) | 260 (88) | 0.91 (0.70–1.17) | .45 | |
| High (>2) | 62 (17) | 69 (12) | 1.43 (0.98, 2.10) | .07 | 46 (25) | 35 (12) | 2.32 (1.44–3.75) | .0006 |
| OR (95%) for interaction | 1.79 (0.96–3.35) | .07 | ||||||
| Ambient oxidative stressor exposure | ||||||||
| None/Low (0–2) | 283 (79) | 458 (82) | 1.00 (ref) | 126 (68) | 246 (83) | 0.84 (0.64–1.09) | .19 | |
| High (>2) | 77 (21) | 101 (18) | 1.20 (0.85, 1.69) | .29 | 59 (32) | 49 (17) | 2.21 (1.45–3.38) | .0003 |
| OR (95%) for interaction | 2.20 (1.25–3.86) | .006 | ||||||
| Paraquat exposure | ||||||||
| None/Low | 312 (87) | 496 (89) | 1.00 (ref) | 144 (78) | 264 (89) | 0.88 (0.68, 1.13) | .32 | |
| High | 48 (13) | 63 (11) | 1.13 (0.75, 1.70) | .57 | 41 (22) | 31 (11) | 2.38 (1.44, 3.95) | .0008 |
| OR (95%) for interaction | 2.40 (1.24, 4.67) | .01 | ||||||
Adjusted for age, sex, ever-smoking, and European ancestry.
Given the strong impact of paraquat alone on GxE interaction, next we investigated whether an increase in mtDNA damage was associated with paraquat exposure in vitro. Towards the goal of measuring mtDNA damage we used a QPCR-based assay, which has been applied to quantifying DNA damage in cells and tissues exposed to mutagens and pesticides (Furda et al., 2014; Sanders et al., 2014a). In brief, by primer design the assay involves amplification of a PCR fragment specific for the mitochondrial genome. When mtDNA damage or lesions interfere with the ability of the DNA polymerase to replicate, less PCR product will be produced. Therefore, mtDNA damage or mtDNA repair intermediates that slow down or block DNA polymerase progression will be detected. PCR products can then be compared from experimental and control conditions if equal amounts of DNA are amplified under identical conditions. The final amplified PCR product is inversely proportional to the number of mtDNA lesions.
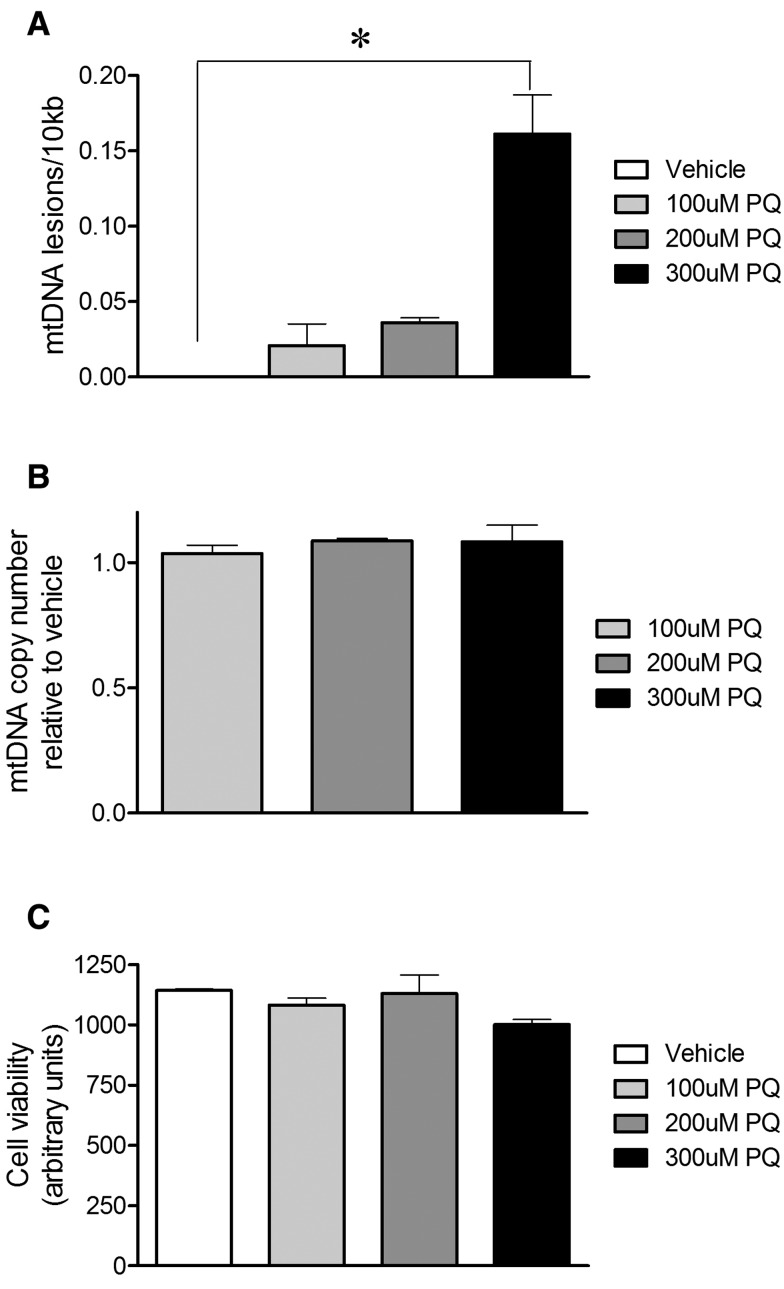
Rat primary VMB neuronal cultures were treated with a concentration range of paraquat (100–300 µM) or vehicle. After 24 h of paraquat treatment, DNA was purified and the QPCR-based assay performed. mtDNA damage was increased in primary VMB neuronal cultures after exposure to the highest concentration tested relative to vehicle-treated neuronal cultures (Figure 1a) . To control for a potential loss of mtDNA in response to paraquat exposure that could influence our interpretation of mtDNA damage, we amplified a short mtDNA fragment. Due to the small size of this mtDNA fragment it is less likely to contain a lesion, and this was used to assay for mitochondrial copy DNA number. The differences in levels of mtDNA damage in primary VMB neuronal cultures in response to paraquat were not attributable to changes in steady state mtDNA levels (Figure 1b). Cell viability was not affected at this time point by these concentrations of paraquat (Figure 1c).
FIG. 1.
Paraquat-induced mtDNA damage in primary VMB neuronal cultures. Sublethal paraquat induced mtDNA damage in rat primary midbrain cultures. a, 24 h of exposure to 300 µM paraquat increased mtDNA lesions in rat primary VMB neuronal cultures (*P < .001, ANOVA). b, Paraquat exposure did not alter mtDNA copy number in rat primary VMB neuronal cultures relative to vehicle. c, In these cultures, paraquat exposure did not change viability relative to vehicle. Data are presented as mean ± SEM. The QPCR-based assay was performed in technical triplicate for each biological replicate (n = 3 vehicle, n = 3 paraquat-treated cultures).
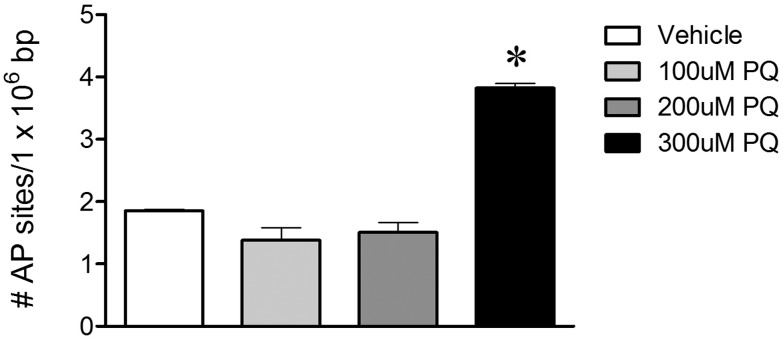
To examine whether abasic sites are specifically induced with paraquat exposure in vitro, we used a quantitative ELISA and found that 300 µM paraquat treatment resulted in an increase in abasic sites (Figure 2). The increase in abasic sites was consistent with the previously noted increase in mtDNA damage (Figure 1); however, because this assay used total cellular DNA, we could not determine whether abasic sites accumulated specifically in mtDNA and/or nuclear DNA.
FIG. 2.
Paraquat induced abasic sites in primary VMB neuronal cultures. Quantitative ELISA measurements using DNA from primary VMB neuronal culture extracts (black bar, *P < .0001, ANOVA) demonstrated increased levels of total abasic sites in VMB neurons treated with 300 µM paraquat compared with vehicle.
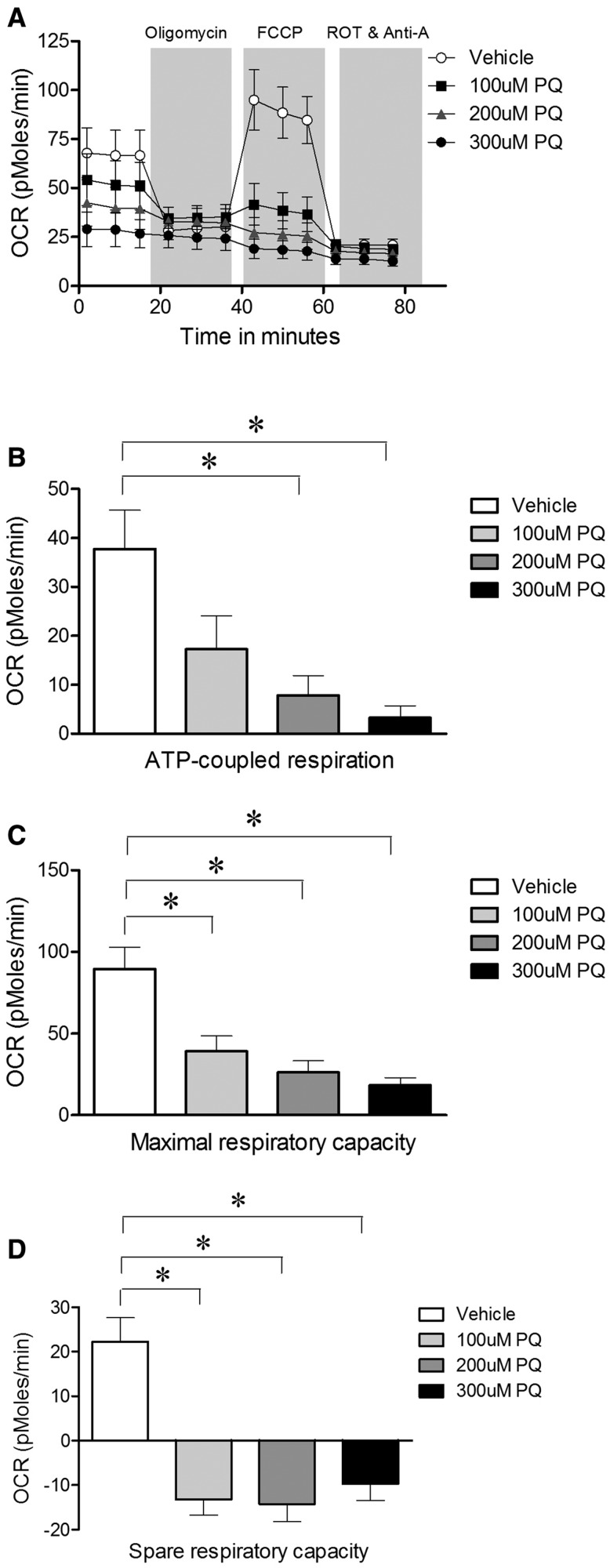
We then determined whether there were changes in mitochondrial respiration as assessed in intact neurons by measuring OCRs using a Seahorse respirometer. Treatment with 200 and 300 µM paraquat for 24 h (which does not affect neuronal viability or mtDNA copy number; Figs. 1b and c) reduced ATP-coupled respiration (Figure 3b). Both maximal and spare respiratory capacities were reduced at all paraquat concentrations tested (Figs. 3c and d).
FIG. 3.
Sublethal paraquat impaired mitochondrial respiration in rat primary VMB neuronal cultures. a, Treatment with 100, 200, and 300 µM paraquat for 24 h impaired OCR of rat primary midbrain neurons; b, the ATP-coupled respiration; c, maximal respiratory capacity; d, spare respiratory capacity was decreased with paraquat treatment. Seahorse experiments were performed with three biological replicates. Data are presented as mean ± SEM.
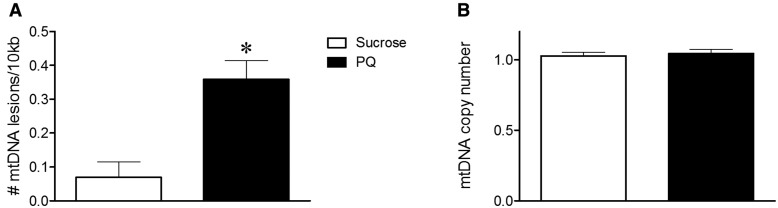
Last, we analyzed mtDNA damage with paraquat exposure in vivo. It is known that exposure to paraquat leads to parkinsonian symptoms in certain strains of Drosophila (Chaudhuri et al., 2007). Therefore, we used this model to study the effect of paraquat on mtDNA damage. Paraquat treated Drosophila heads had 5 times more mtDNA lesions than vehicle-treated flies (Figure 4, P-value < 0.001).
FIG. 4.
Paraquat-induced mtDNA damage in Drosophila. Sublethal paraquat-induced mtDNA damage in Drosophila heads. a, 12 h of exposure to 20 mM paraquat increased mtDNA lesions in Drosophila heads (*P < .001, Student t test). b, Paraquat exposure did not alter mtDNA copy number compared with sucrose-treated Drosophila. Data are presented as mean ± SEM. The QPCR-based assay was performed in technical triplicate for each biological replicate (n = 5 vehicle, n = 5 paraquat-treated Drosophila batches).
DISCUSSION
In this investigation, we identified novel APEX1- and OGG1-pesticide statistical interactions, providing support for the influence of BER in pesticide-exposed susceptible subpopulations. Importantly, we also demonstrated that paraquat exposure increased mtDNA damage and mitochondrial dysfunction, further substantiating a role for these mechanisms in the pathogenesis of PD.
Paraquat has been shown to have toxicity in both animals and humans (Baud et al., 1988; Peng et al., 2007; Wunnapuk et al., 2014). We previously reported in PEG an association of PD with ambient residential paraquat and maneb co-exposures (Costello et al., 2009) and a GxE interaction with dopamine transporter variants (Ritz et al., 2009). Building on this, in this study, now with nearly double the sample size, we provide further evidence in support of an increase in PD risk with paraquat exposure. However, in this study we were unable to replicate a previously reported association between rotenone exposure and PD (Tanner et al., 2011), most likely due to our study’s limited statistical power (only 6 patients and 12 controls with rotenone exposure). This is likely because rotenone is rarely used agriculturally, except in fisheries to control invasive species. Thus, while it is a household and home gardening pesticide, our exposure model only captured ambient exposures from agricultural uses. Lastly, our results implicate pyrethroids, specifically permethrin and cypermethrin, in increased PD risk, a finding which requires further replication. The potential link of pyrethroids and PD risk is concerning given the widespread use of these compounds in place of many organophosphates that are deemed to be acutely toxic.
Dysfunctional DNA repair and resulting DNA damage accumulation has been implicated in aging and age-related diseases (Jeppesen et al., 2013). Nevertheless, epidemiological data linking DNA repair genes and PD is currently limited. A small study previously reported a marginal association for the APEX1 rs1130409 GG genotype and PD relative to the TT genotype (Gencer et al., 2012). However, these findings were not replicated in this study. Furthermore, GWASs have not associated the APEX1 or OGG1 variants with PD.
Although it is widely accepted that genetic factors, including pesticide metabolism and transporter activities, may influence susceptibility to environmental exposures, GxE interaction investigations in PD still rely on candidate gene approaches. Investigators face many challenges in identifying and/or replicating interactions, including exposure assessment and sample size. There are few examples of specific GxE interactions which influence PD susceptibility that have been successfully replicated. We recently published a review highlighting GxE findings involving pesticide exposures in the PEG study over the last decade (Ritz et al., 2016). In this study, the strongest associations were seen among carriers of genotypes at both susceptibility loci we identified in the single SNP interaction analyses as pesticide associated risk genotypes. For example, participants who were highly exposed to mitochondrial-inhibiting pesticides who carried neither or only one risk genotype (APEX1 rs1130409 GG and/or OGG1 rs1052133 CC) were at a 43% increased risk of PD (95% CI = 0.98, 2.10), but those who were both highly exposed and who carried risk genotypes at both loci (APEX1 rs1130409 GT or TT and OGG1 rs1052133 CG or GG) experienced a 2.3-fold increased risk of PD (95% CI = 1.44, 3.75). Similar results were observed with oxidative stressor exposures and paraquat exposure. Thus, we have uncovered a novel GxE interaction such that humans with a genetically variant BER system seem to be particularly vulnerable to these types of pesticide exposure, with both SNP variants acting synergistically to increase risk for developing PD. Further research in an independent sample is required to re-examine and replicate findings.
The first genome-wide GxE interaction analysis for pesticides and PD was recently published using patients and sibling controls from the Mayo Clinic (Rochester, Minnesota, USA) and found that 2 SNPs in the ERCC6L2 gene showed the strongest statistical interactions (Biernacka et al., 2016). Although there is not much known about ERCC6L2, the proposed function of this gene links it to DNA repair and mitochondrial function (Tummala et al., 2014). In addition, associations between age at onset and genetic variants in DNA repair pathways have been reported in Huntington’s disease (Genetic Modifiers of Huntington’s Disease Consortium, 2015). The reported DNA repair genetic variants also modified age at onset in additional polyglutamine diseases, suggesting overlapping genetic mechanisms (Bettencourt et al., 2016). These findings, together with our results, highlight an important role for DNA repair in neurodegenerative diseases in potentially modulating age at onset, potentiating risk and suggest common pathogenic mechanisms.
Our PD case-control study population provides many advantages that allow us to pose and investigate such mechanistic hypotheses epidemiologically. Most epidemiologic studies rely on self-reported information for pesticide exposure assessment, a method prone to differential recall error, and they generally do not allow for the investigation of specific pesticides. We specifically assessed ambient oxidative stressor and mitochondrial complex I inhibitor pesticide exposures based on our GIS approach utilizing state-mandated pesticide use reports. Thus we do not rely on participant recall for pesticide use, and are able to investigate particular chemicals. Our ambient pesticide exposure method does not, however, account for factors such as wind patterns at the time of application, geographic features that may influence pesticide drift, and assumes that the participant was at the recorded location during the relevant time period or that the residents of nearby homes were exposed to the pesticides even after applications occurred such as through volatilization or dust etc. Another advantage of our study is that all PD patients were seen and well characterized by UCLA movement disorder specialists at least once, and often multiple times. This minimizes bias from disease misclassification, as PD is a commonly misdiagnosed disease (Meara et al., 1999; Wermuth et al., 2012). Finally, the population controls were drawn from the same region as the cases, likely providing adequate representativeness of the exposures in the source population. Although we estimated statistical interactions the confidence intervals for the high exposure groups across genetic risk status overlapped somewhat suggesting that while biologic relevance supports these findings, replication in an independent study would be preferable.
In experimental studies, we found that in rats treated with rotenone, at a dose that does not cause neurodegeneration, mtDNA damage was induced specifically in substantia nigra dopamine neurons (Sanders et al., 2014c). However, whether mtDNA damage is a common feature of pesticides that have been linked to an increased risk of PD is unknown. Recently, it was reported that paraquat caused mtDNA damage and dopaminergic neurodegeneration in Caenorhabditiselegans (Gonzalez-Hunt et al., 2014). Consistent with this, we found an increase in mtDNA damage following paraquat exposure in an in vitro model using VMB neurons. Similar results were obtained in an in vivo model with a striking 5-fold increase in mtDNA damage in paraquat-treated Drosophila relative to vehicle-treated flies. Using a different approach, oxidative DNA damage was also described by another group following paraquat treatment in flies; however, the authors evaluated total DNA and could not ascribe the effect to mtDNA specifically (Mehdi and Qamar, 2013). Increases in oxidative stress markers and impaired mitochondrial function have been reported in cell lines and in vivo with paraquat exposure (Czerniczyniec et al., 2011; de Oliveira et al., 2016; Hosamani and Muralidhara, 2013). We also found mitochondrial bioenergetic defects following paraquat exposure in VMB neuronal cultures, a cell type with clear-cut relevance for PD, which has not been examined previously. However, in VMB neuronal cultures we were able to detect a decrease in mitochondrial bioenergetics with paraquat exposures at lower concentrations that did not induce mtDNA damage. This suggests that mitochondrial respiration may be a more sensitive measure than mtDNA damage to acute paraquat exposure. Future experiments may include investigating whether following paraquat treatment there is an association between mitochondrial dysfunction and mtDNA damage. The mechanisms of action for other pesticides linked to PD, such as benomyl, permethrin, and pyridaben, are not known unambiguously and it will require further investigation to determine whether increased susceptibility is conferred through complex I inhibition, oxidative stress and/or other mechanisms (Fitzmaurice et al., 2013).
CONCLUSION
Disease-toxicant interactions likely play important roles in the etiology of PD (Kwakye et al., 2016). We found strong statistical interactions between exposures to relevant pesticides, including paraquat, with APEX1 and OGG1 genotypes and PD risk in our study participants. These epidemiologic findings are consistent with proposed pathophysiologic mechanisms implicated in experimental models for PD such as mitochondrial dysfunction, oxidative stress, and DNA repair deficiency (De Miranda, 2016; Ryan et al., 2015; Sanders and Greenamyre, 2013). Thus, our data support a role for DNA repair mechanisms and suggest that genetic risk variants in BER modify pesticide associations with PD.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
National Institute of Environmental Health Sciences at the National Institutes of Health (grant number R01ES010544, P01ES016732, U54ES12078), pilot funding from SCEHSC No. 5P30 ES07048 and The American Parkinson Disease Association (B.R. and J.T.G.); Burroughs Wellcome Fund Population and Laboratory Based Sciences Award (K.C.P.); Pittsburgh Claude D. Pepper OAIC (L.H.S.); William N. and Bernice E. Bumpus Foundation Innovation Award (L.H.S.); National Institute of Environmental Health Sciences at the National Institutes of Health (grant number R01ES020718) (J.T.G.).
Supplementary Material
REFERENCES
- Arai T., Fukae J., Hatano T., Kubo S., Ohtsubo T., Nakabeppu Y., Mori H., Mizuno Y., Hattori N. (2006). Up-regulation of hMUTYH, a DNA repair enzyme, in the mitochondria of substantia nigra in Parkinson's disease. Acta Neuropathol. 112, 139–145. [DOI] [PubMed] [Google Scholar]
- Ayala-Torres S., Chen Y., Svoboda T., Rosenblatt J., Van Houten B. (2000). Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods 22, 135–147. [DOI] [PubMed] [Google Scholar]
- Baud F. J., Houze P., Bismuth C., Scherrmann J. M., Jaeger A., Keyes C. (1988). Toxicokinetics of paraquat through the heart-lung block. Six cases of acute human poisoning. J. Toxicol. Clin. Toxicol. 26, 35–50. [DOI] [PubMed] [Google Scholar]
- Bettencourt C., Hensman-Moss D., Flower M., Wiethoff S., Brice A., Goizet C., Stevanin G., Koutsis G., Karadima G., Panas M., et al. (2016). DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases. Ann. Neurol. 79, 983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernacka J. M., Chung S. J., Armasu S. M., Anderson K. S., Lill C. M., Bertram L., Ahlskog J. E., Brighina L., Frigerio R., Maraganore D. M. (2016). Genome-wide gene-environment interaction analysis of pesticide exposure and risk of Parkinson’s disease. Parkinsonism Relat. Disord. 32, 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C. V., Kile M. L., Catalano P. J., Hoffman E., Quamruzzaman Q., Rahman M., Mahiuddin G., Christiani D. C. (2007). GSTM1 and APE1 genotypes affect arsenic-induced oxidative stress: A repeated measures study. Environ. Health 6, 39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronstein J., Carvey P., Chen H., Cory-Slechta D., DiMonte D., Duda J., English P., Goldman S., Grate S., Hansen J., et al. (2009). Meeting report: Consensus statement-Parkinson’s disease and the environment: Collaborative on health and the environment and Parkinson’s Action Network (CHE PAN) conference 26-28 June 2007. Environ. Health Perspect. 117, 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri A., Bowling K., Funderburk C., Lawal H., Inamdar A., Wang Z., O’Donnell J. M. (2007). Interaction of genetic and environmental factors in a Drosophila parkinsonism model. J. Neurosci. 27, 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn M., Mills P., Zhang X., Zadnick J., Goldberg D., Ritz B. (2011). Prostate cancer and ambient pesticide exposure in agriculturally intensive areas in California. Am. J. Epidemiol. 173, 1280–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello S., Cockburn M., Bronstein J., Zhang X., Ritz B. (2009). Parkinson’s disease and residential exposure to maneb and paraquat from agricultural applications in the central valley of California. Am. J. Epidemiol. 169, 919–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerniczyniec A., Karadayian A. G., Bustamante J., Cutrera R. A., Lores-Arnaiz S. (2011). Paraquat induces behavioral changes and cortical and striatal mitochondrial dysfunction. Free Radic. Biol. Med. 51, 1428–1436. [DOI] [PubMed] [Google Scholar]
- De Miranda B. R., Van Houten B., Sanders L. H. (2016). Mitochondrial Mechanisms of Degeneration and Repair in Parkinson’s Disease. doi: 10.1007/978-3-319-42139-1.
- de Oliveira M. R., Peres A., Gama C. S., Bosco S. M. (2016). Pinocembrin provides mitochondrial protection by the activation of the Erk1/2-Nrf2 signaling pathway in SH-SY5Y neuroblastoma cells exposed to paraquat. Mol. Neurobiol. doi: 10.1007/s12035-016-0135-5. [DOI] [PubMed] [Google Scholar]
- Dhillon A. S., Tarbutton G. L., Levin J. L., Plotkin G. M., Lowry L. K., Nalbone J. T., Shepherd S. (2008). Pesticide/environmental exposures and Parkinson’s disease in East Texas. J. Agromed. 13, 37–48. [DOI] [PubMed] [Google Scholar]
- Dick F. D., De Palma G., Ahmadi A., Osborne A., Scott N. W., Prescott G. J., Bennett J., Semple S., Dick S., Mozzoni P., et al. (2007). Gene-environment interactions in parkinsonism and Parkinson’s disease: The Geoparkinson study. Occup. Environ. Med. 64, 673–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinis-Oliveira R. J., Remiao F., Carmo H., Duarte J. A., Navarro A. S., Bastos M. L., Carvalho F. (2006). Paraquat exposure as an etiological factor of Parkinson’s disease. Neurotoxicology 27, 1110–1122. [DOI] [PubMed] [Google Scholar]
- Elbaz A., Tranchant C. (2007). Epidemiologic studies of environmental exposures in Parkinson’s disease. J. Neurol. Sci. 262, 37–44. [DOI] [PubMed] [Google Scholar]
- Erlich P. M., Lunetta K. L., Cupples L. A., Huyck M., Green R. C., Baldwin C. T., Farrer L. A., Group M. S., et al. and others. (2006). Polymorphisms in the PON gene cluster are associated with Alzheimer disease. Human Mol. Genet. 15, 77–85. [DOI] [PubMed] [Google Scholar]
- Fitzmaurice A. G., Rhodes S. L., Lulla A., Murphy N. P., Lam H. A., O’Donnell K. C., Barnhill L., Casida J. E., Cockburn M., Sagasti A., et al. (2013). Aldehyde dehydrogenase inhibition as a pathogenic mechanism in Parkinson disease. Proc. Natl. Acad. Sci. U. S. A. 110, 636–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire C., Koifman S. (2012). Pesticide exposure and Parkinson’s disease: Epidemiological evidence of association. Neurotoxicology 33, 947–971. [DOI] [PubMed] [Google Scholar]
- Fukae J., Takanashi M., Kubo S., Nishioka K., Nakabeppu Y., Mori H., Mizuno Y., Hattori N. (2005). Expression of 8-oxoguanine DNA glycosylase (OGG1) in Parkinson’s disease and related neurodegenerative disorders. Acta Neuropathol. 109, 256–262. [DOI] [PubMed] [Google Scholar]
- Furda A., Santos J. H., Meyer J. N., Van Houten B. (2014). Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 1105, 419–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H. M., Hong J. S. (2011). Gene-environment interactions: Key to unraveling the mystery of Parkinson’s disease. Progr. Neurobiol. 94, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gencer M., Dasdemir S., Cakmakoglu B., Cetinkaya Y., Varlibas F., Tireli H., Kucukali C. I., Ozkok E., Aydin M. (2012). DNA repair genes in Parkinson’s disease. Genet. Test. Mol. Biomarkers 16, 504–507. [DOI] [PubMed] [Google Scholar]
- Genetic Modifiers of Huntington’s Disease Consortium. (2015). Identification of genetic factors that modify clinical onset of Huntington’s disease. Cell 162, 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Hunt C. P., Leung M. C., Bodhicharla R. K., McKeever M. G., Arrant A. E., Margillo K. M., Ryde I. T., Cyr D. D., Kosmaczewski S. G., Hammarlund M., et al. (2014). Exposure to mitochondrial genotoxins and dopaminergic neurodegeneration in Caenorhabditis elegans. PloS One 9, e114459.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock D. B., Martin E. R., Mayhew G. M., Stajich J. M., Jewett R., Stacy M. A., Scott B. L., Vance J. M., Scott W. K. (2008). Pesticide exposure and risk of Parkinson’s disease: A family-based case-control study. BMC Neurol. 8, 6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosamani R., Muralidhara, (2013). Acute exposure of Drosophila melanogaster to paraquat causes oxidative stress and mitochondrial dysfunction. Arch. Insect Biochem. Physiol. 83, 25–40. [DOI] [PubMed] [Google Scholar]
- Jeppesen D. K., Bohr V. A., Stevnsner T. (2011). DNA repair deficiency in neurodegeneration. Progr. Neurobiol. 94, 166–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel F., Tanner C., Umbach D., Hoppin J., Alavanja M., Blair A., Comyns K., Goldman S., Korell M., Langston J., et al. (2007). Pesticide exposure and self-reported Parkinson’s disease in the agricultural health study. Am. J. Epidemiol. 165, 364–374. [DOI] [PubMed] [Google Scholar]
- Karahalil B., Bohr V. A., Wilson D. M. 3rd (2012). Impact of DNA polymorphisms in key DNA base excision repair proteins on cancer risk. Hum. Exp. Toxicology 31, 981–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal S. L., Dall T. M., Chakrabarti R., Storm M. V., Jain A. (2013). The current and projected economic burden of Parkinson’s disease in the United States. Mov. Disord. 28, 311–318. [DOI] [PubMed] [Google Scholar]
- Krokan H. E., Bjoras M. (2013). Base excision repair. Cold Spring Harb. Perspect. Biol. 5, a012583.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuter K., Nowak P., Golembiowska K., Ossowska K. (2010). Increased reactive oxygen species production in the brain after repeated low-dose pesticide paraquat exposure in rats. A comparison with peripheral tissues. Neurochem. Res. 35, 1121–1130. [DOI] [PubMed] [Google Scholar]
- Kwakye G. F., McMinimy R. A., Aschner M. (2016). Disease-toxicant interactions in Parkinson’s disease neuropathology. Neurochem. Res. doi: 10.1007/s11064-016-2052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen K. H., Cheng C., Wiksen H., Bergersen L. H., Klungland A. (2011). Mitochondrial DNA toxicity compromises mitochondrial dynamics and induces hippocampal antioxidant defenses. DNA Repair (Amst) 10, 639–653. [DOI] [PubMed] [Google Scholar]
- Martinez T. N., Greenamyre J. T. (2012). Toxin models of mitochondrial dysfunction in Parkinson’s disease. Antioxid. Redox. Signal. 16, 920–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack A. L., Thiruchelvam M., Manning-Bog A. B., Thiffault C., Langston J. W., Cory-Slechta D. A., Di Monte D. A. (2002). Environmental risk factors and Parkinson’s disease: Selective degeneration of nigral dopaminergic neurons caused by the herbicide paraquat. Neurobiol. Dis. 10, 119–127. [DOI] [PubMed] [Google Scholar]
- Meara J., Bhowmick B. K., Hobson P. (1999). Accuracy of diagnosis in patients with presumed Parkinson’s disease. Age Ageing 28, 99–102. [DOI] [PubMed] [Google Scholar]
- Mehdi S. H., Qamar A. (2013). Paraquat-induced ultrastructural changes and DNA damage in the nervous system is mediated via oxidative-stress-induced cytotoxicity in Drosophila melanogaster. Toxicol. Sci. 134, 355–365. [DOI] [PubMed] [Google Scholar]
- Nalls M. A., Pankratz N., Lill C. M., Do C. B., Hernandez D. G., Saad M., DeStefano A. L., Kara E., Bras J., Sharma M., et al. (2014). Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowska K., Wardas J., Smialowska M., Kuter K., Lenda T., Wieronska J. M., Zieba B., Nowak P., Dabrowska J., Bortel A., et al. (2005). A slowly developing dysfunction of dopaminergic nigrostriatal neurons induced by long-term paraquat administration in rats: An animal model of preclinical stages of Parkinson’s disease?. Eur. J. Neurosci. 22, 1294–1304. [DOI] [PubMed] [Google Scholar]
- Peng J., Peng L., Stevenson F. F., Doctrow S. R., Andersen J. K. (2007). Iron and paraquat as synergistic environmental risk factors in sporadic Parkinson’s disease accelerate age-related neurodegeneration. J. Neurosci. 27, 6914–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos M. H., Higgins J. J., Golbe L. I., Johnson W. G., Ide S. E., Di Iorio G., Sanges G., Stenroos E. S., Pho L. T., Schaffer A. A., et al. (1996). Mapping of a gene for Parkinson’s disease to chromosome 4q21-q23. Science 274, 1197–1199. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., et al. (1997). Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047. [DOI] [PubMed] [Google Scholar]
- Richardson J. R., Shalat S. L., Buckley B., Winnik B., O’Suilleabhain P., Diaz-Arrastia R., Reisch J., German D. C. (2009). Elevated serum pesticide levels and risk of Parkinson disease. Arch. Neurol. 66, 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz B. R., Ascherio A., Checkoway H., Marder K. S., Nelson L. M., Rocca W. A., Ross G. W., Strickland D., Van Den Eeden S. K., Gorell J. (2007). Pooled Analysis Of Tobacco Use And Risk Of Parkinson Disease. Arch Neurol. 647, 960–997. [DOI] [PubMed] [Google Scholar]
- Ritz B. R., Manthripragada A. D., Costello S., Lincoln S. J., Farrer M. J., Cockburn M., Bronstein J. (2009). Dopamine transporter genetic variants and pesticides in Parkinson’s disease. Environ. Health Perspect. 117, 964–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz B. R., Paul K. C., Bronstein J. M. (2016). Of Pesticides and Men: A California Story of Genes and Environment in Parkinson’s Disease. Curr. Environ. Health Rep. 3, 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney J. P., Ryde I. T., Sanders L. H., Howlett E. H., Colton M. D., Germ K. E., Mayer G. D., Greenamyre J. T., Meyer J. N. (2015). PCR based determination of mitochondrial DNA copy number in multiple species. Methods Mol. Biol. 1241, 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan B. J., Hoek S., Fon E. A., Wade-Martins R. (2015). Mitochondrial dysfunction and mitophagy in Parkinson’s: From familial to sporadic disease. Trends Biochem. Sci. 40, 200–210. [DOI] [PubMed] [Google Scholar]
- Saki M., Prakash A. (2016). DNA damage related crosstalk between the nucleus and mitochondria. Free Radic. Biol. Med. doi: 10.1016/j.freeradbiomed.2016.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders L. H., Greenamyre J. T. (2013). Oxidative damage to macromolecules in human Parkinson disease and the rotenone model. Free Radic. Biol. Med. 62, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders L. H., Howlett E. H., McCoy J., Greenamyre J. T. (2014a). Mitochondrial DNA damage as a peripheral biomarker for mitochondrial toxin exposure in rats. Toxicol. Sci. 142, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders L. H., Laganiere J., Cooper O., Mak S. K., Vu B. J., Huang Y. A., Paschon D. E., Vangipuram M., Sundararajan R., Urnov F. D., et al. (2014b). LRRK2 mutations cause mitochondrial DNA damage in iPSC-derived neural cells from Parkinson’s disease patients: Reversal by gene correction. Neurobiol. Dis. 62, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders L. H., McCoy J., Hu X., Mastroberardino P. G., Dickinson B. C., Chang C. J., Chu C. T., Van Houten B., Greenamyre J. T. (2014c). Mitochondrial DNA damage: Molecular marker of vulnerable nigral neurons in Parkinson’s disease. Neurobiol. Dis. 70, 214–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura-Miura H., Hattori N., Kang D., Miyako K., Nakabeppu Y., Mizuno Y. (1999). Increased 8-oxo-dGTPase in the mitochondria of substantia nigral neurons in Parkinson’s disease. Ann. Neurol. 46, 920–924. [PubMed] [Google Scholar]
- Tanner C. M., Kamel F., Ross G. W., Hoppin J. A., Goldman S. M., Korell M., Marras C., Bhudhikanok G. S., Kasten M., Chade A. R., et al. (2011). Rotenone, paraquat, and Parkinson’s disease. Environ. Health Perspect. 119, 866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummala H., Kirwan M., Walne A. J., Hossain U., Jackson N., Pondarre C., Plagnol V., Vulliamy T., Dokal I. (2014). ERCC6L2 mutations link a distinct bone-marrow-failure syndrome to DNA repair and mitochondrial function. Am. J. Hum. Genet. 94, 246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance J. M., Ali S., Bradley W. G., Singer C., Di Monte D. A. (2010). Gene-environment interactions in Parkinson’s disease and other forms of parkinsonism. Neurotoxicology 31, 598–602. [DOI] [PubMed] [Google Scholar]
- Verstraeten A., Theuns J., Van Broeckhoven C. (2015). Progress in unraveling the genetic etiology of Parkinson disease in a genomic era. Trends Genet. 31, 140–149. [DOI] [PubMed] [Google Scholar]
- Wang A., Costello S., Cockburn M., Zhang X., Bronstein J., Ritz B. (2011). Parkinson's Disease risk from ambient exposure to pesticides. Eur J Epidemiol. 267, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J. M., Goode E. L., Ladiges W. C., Ulrich C. M. (2005). Polymorphic variation in hOGG1 and risk of cancer: A review of the functional and epidemiologic literature. Mol. Carcinog. 42, 127–141. [DOI] [PubMed] [Google Scholar]
- Wermuth L., Lassen C. F., Himmerslev L., Olsen J., Ritz B. (2012). Validation of hospital register-based diagnosis of Parkinson’s disease. Dan. Med. J. 59, A4391.. [PMC free article] [PubMed] [Google Scholar]
- Wirdefeldt K., Gatz M., Reynolds C. A., Prescott C. A., Pedersen N. L. (2011). Heritability of Parkinson disease in Swedish twins: A longitudinal study. Neurobiol. Aging 32, 1923 e1–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunnapuk K., Mohammed F., Gawarammana I., Liu X., Verbeeck R. K., Buckley N. A., Roberts M. S., Musuamba F. T. (2014). Prediction of paraquat exposure and toxicity in clinically ill poisoned patients: A model based approach. Br. J. Clin. Pharmacol. 78, 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.