Abstract
Hus1, Rad1, and Rad9 are three evolutionarily conserved proteins required for checkpoint control in fission yeast. These proteins are known to form a stable complex in vivo. Recently, computational studies have predicted structural similarity between the individual proteins of Hus1-Rad1-Rad9 complex and the replication processivity factor proliferating cell nuclear antigen (PCNA). This has led to the proposal that the Hus1-Rad1-Rad9 complex may form a PCNA-like ring structure, and could function as a sliding clamp during checkpoint control. In the present study, we have attempted to test the predictions of this model by asking whether the PCNA alignment identifies functionally important residues or explains mutant phenotypes of hus1, rad1, or rad9 alleles. Although some of our results are consistent with the PCNA alignment, others indicate that the Hus1-Rad1-Rad9 complex possesses unique structural and functional features.
INTRODUCTION
Cell cycle checkpoints regulate the precise order of cell cycle events, thus ensuring the distribution of complete copies of the genome to daughter cells (Hartwell and Weinert, 1989). Defects in checkpoints lead to genomic instability, a major contributory factor in the development of cancer, because cells with faulty checkpoints proceed into mitosis with damaged or incompletely replicated DNA (Hartwell and Kastan, 1994; Nojima, 1997).
Molecular and genetic studies in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe have identified an intricate network of genes required for the S-G2/M checkpoint, which ensures cell cycle arrest in response to DNA damage and/or incomplete DNA replication (Sheldrick and Carr, 1993; Stewart and Enoch, 1996; Weinert, 1998). In fission yeast, a group of six nonessential checkpoint rad proteins, Hus1, Rad1, Rad3, Rad9, Rad17, and Rad26, constitute the sensor machinery of fission yeast checkpoint cascade (Boddy and Russell, 1999; Humphrey, 2000). These proteins are required for cell cycle arrest in response to the inhibition of DNA synthesis by compounds such as hydroxyurea (HU) or DNA damage caused by UV and gamma irradiation. Mutation in any of the genes encoding these proteins leads to virtually identical phenotypes, suggesting that they act in the same pathway (Boddy and Russell, 1999; Humphrey, 2000). These proteins have been highly conserved during evolution, and related proteins have been found in many other eukaryotes, including humans (Lieberman et al., 1996; Bluyssen et al., 1998; Weiss et al., 1999; Venclovas and Thelen, 2000).
The biochemical events underlying the checkpoint response are beginning to be understood. Rad3 is a phosphatidylinositol-3-related kinase that is likely to be activated in response to checkpoint signals (Bentley et al., 1996). The function of the other five checkpoint rad proteins in the checkpoint response has not been established. Hus1, Rad1, and Rad9 have been shown to form a stable complex and Hus1 is phosphorylated after DNA damage (Kostrub et al., 1998a; Caspari et al., 2000). Homologs of Hus1, Rad1, and Rad9 in other organisms also form a similar complex (Kondo et al., 1999; St Onge et al., 1999; Volkmer and Karnitz, 1999), suggesting a conserved role for this complex in response to genotoxic insults. These proteins do not show significant primary sequence similarity to proteins with known biochemical functions, although exonuclease activity associated with a Rad1-related protein in Ustilago maydis and with the human homolog of Rad1 has been reported (Onel et al., 1996; Parker et al., 1998).
Recently, several groups have used the sequence-structure threading method of Fisher and Eisenberg, sequence-structure alignments, and comparative molecular modeling to predict the presence of proliferating cell nuclear antigen (PCNA) folds in Hus1, Rad1, and Rad9 (Aravind et al., 1999; Thelen et al., 1999; Cai et al., 2000; Caspari et al., 2000; Venclovas and Thelen, 2000; Zuccola et al., 2000). PCNA is a processivity factor that functions in DNA replication and repair in all eukaryotic cells. The PCNA fold consists of two topologically similar domains, linked by a flexible interconnector domain (ICD) loop. The active complex consists of three such monomers that associate in a head-to-tail manner, forming a donut-shaped complex that encircles DNA (Krishna et al., 1994; Kelman, 1997). The ICD loops protrude from the surface of the complex, functioning as interacting sites for proteins such as DNA polymerase, p21, and FEN1 endonuclease (Tsurimoto, 1998).
Based on the potential presence of PCNA folds, it has been proposed that Hus1, Rad1, and Rad9 form a similar heterotrimeric, donut-shaped complex, which encircles DNA and acts as a sliding clamp (Aravind et al., 1999; Thelen et al., 1999; Cai et al., 2000; Caspari et al., 2000; Venclovas and Thelen, 2000; Zuccola et al., 2000). By sliding along the DNA, Hus1-Rad1-Rad9 could act as a sentinel complex scanning the genome for lesions and stalled replication forks. Alternatively, once the checkpoint signal is sensed by other checkpoint rad proteins, the Hus1-Rad1-Rad9 complex could act as a moving platform for the recruitment of other checkpoint proteins to the sites of lesions or stalled replication forks just as the PCNA complex acts to recruit replication and repair proteins (Zhang et al., 1999). In addition, another checkpoint rad protein, Rad17, shows structural and functional similarity to RF-C subunits, a complex that functions to load PCNA onto DNA (Mossi and Hubscher, 1998; Shimada et al., 1999; Naiki et al., 2000). The sliding clamp model thus also proposes a function for Rad17: it could act to load the Hus1-Rad1-Rad9 complex onto DNA (Rauen et al., 2000; Venclovas and Thelen, 2000).
Based on the PCNA model, a number of predictions can be made about the function of specific regions and residues in Hus1, Rad1, and Rad9. First, amino acids in Hus1, Rad1, or Rad9 that are conserved with PCNA should be functionally important. Second, the ICD loops of Hus1, Rad1, and Rad9 may interact with additional proteins involved in the checkpoint response. Third, the alignment makes predictions about interactions between Hus1, Rad1, and Rad9. For example, if Hus1, Rad1, and Rad9 form head-to-tail associations like those observed in the PCNA trimer, each protein should contain sites for interacting with both the other proteins. However, the two sites should be far way from each other, located on separate domains (Burtelow et al., 2001; Figures 4 and 7). The residues involved in each interaction have been predicted by combining information from the PCNA alignment with genetic studies of the equivalent S. cerevisiae proteins (Kondo et al., 1999; Venclovas and Thelen, 2000). Last, the model predicts that the C-terminal 126 amino acids of Rad9 should not be required for interactions with Hus1 and Rad1, because this region does not align with PCNA.
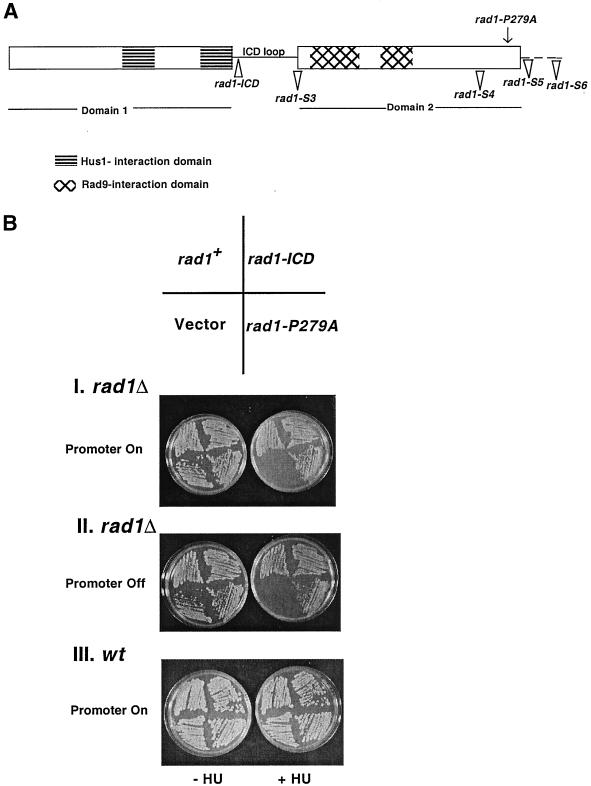
Figure 4.
rad1-ICD and rad1-P279A mutants complement the HU sensitivity of rad1Δ. (A) Schematic representation of the predicted structure of Rad1 (Venclovas and Thelen, 2000), indicating the position of various rad1 mutants on this structure. The dotted line represents the portion of the C-terminal region of Rad1, which does not align with PCNA. (B) rad1-ICD and rad1-P279A mutants rescue the HU sensitivity of rad1Δ and do not cause HU sensitivity when expressed in wt cells. rad1Δ cells transformed with pREP1 plasmid (pTE102), pREP1-rad1-ICD (pTE885), pREP1-P279A (pTE886), and pREP1-rad1+ (pTE567) were grown on EMM plates with or without 10 mM HU in the absence (I) or presence (II) of thiamine under inducing or noninducing conditions, respectively. (III) Growth of wt cells transformed with the same plasmids on EMM plates lacking or containing 10 mM HU.
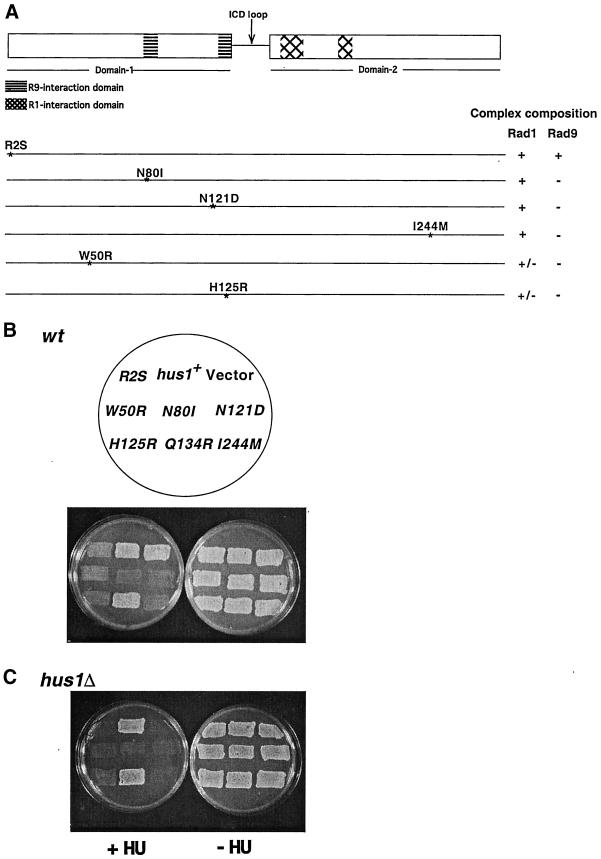
Figure 7.
hus1 dominant mutants disrupt checkpoint control in wt cells. (A) Schematic representation of single amino acid changes in hus1+ that are sufficient to create dominant negative (dn) alleles. Composition of the Hus1-Rad1-Rad9 complex in each hus1-dn mutant is shown on right side. (B) hus1-dominant alleles disrupt checkpoint control in wt cells. wt (TE237) cells transformed with pREP42 plasmid (pTE105), pREP42-hus1+ (pTE550), hus1-dominant mutants, pREP42-hus1-R2S (pTE601), pREP42-hus1-W50R (pTE823), pREP42-hus1-N80I (pTE592), pREP42-hus1-N121D (pTE599), pREP42-hus1-H125R (pTE887), pREP42-hus1-Q134R (pTE888), and pREP42-hus1-I244 M (pTE889) were grown on EMM plates in the presence or absence of 10 mM HU. (C) hus1-dn alleles do not rescue the HU sensitivity of hus1Δ. Growth of hus1Δ (TE484) cells transformed with the same plasmids as used above, on EMM plates with or without 10 mM HU.
Here, we describe experiments designed to test each of these predictions. While some of our results are consistent with the model, we have also found that the Hus1-Rad1-Rad9 complex has distinctive features that are not explained by the PCNA alignment.
MATERIALS AND METHODS
Schizosaccharomyces pombe Physiological Methods
The strains used in the present study are listed in Table 1. Strains were grown under standard conditions (Moreno et al., 1991). Transformation of S. pombe was carried out by the overnight lithium acetate procedure (Elble, 1992). To check the HU sensitivity of strains, cells were streaked on Edinburgh minimal medium (EMM; Bio 101, Vista, CA) plates containing thiamine (2 μM) and grown for 3 d at 29°C. They were then replica plated onto EMM plates to turn on the nmt1 promoter. After 24 h of promoter induction, cells were further replica plated onto EMM plates containing 5 mg/ml phloxine B or EMM plates containing 5 mg/ml phloxine B and 10 mM HU. Growth was scored after 3 d. HU (Sigma, St. Louis, MO) was prepared as a 200 mM stock solution in water, filter sterilized, and stored at 4°C.
Table 1.
S. pombe strains and plasmids
| Strain | Genotype | Reference |
|---|---|---|
| TE237 | leu1-32 ura4-D18 h+ | Unpublished |
| TE457 | rad9∷ura4+ ade6-704 leu1-32 ura4-D18 h+ | Gift of A. Carr |
| TE459 | rad1∷ura4+ leu1-32 ura4-D18 his− | Sunnerhagen et al. (1990) |
| TE484 | hus1∷LEU2 leu1-32 ura4-D18 h− | Kostrub et al. (1997) |
| TE992 | KLP8 (rad1-S5 his3- leu1-32 h−) | Kanter-Smoler et al. (1995) |
| TE993 | KLP9 (rad1-S6 his3-leu1-32 h−)− | Kanter-Smoler et al. (1995) |
| TE994 | KLP6 (rad1-S3 his3- leu1-32 h−) | Kanter-Smoler et al. (1995) |
| TE995 | KLP7 (rad1-S4 his3- leu1-32 h−) | Kanter-Smoler et al. (1995) |
| Plasmid | Construct | |
|---|---|---|
| pTE479 | pREP1-rad9+ | |
| pTE883 | pREP1-rad9N | |
| pTE884 | pREP1-rad9C | |
| pTE567 | pREP1-rad1+ | |
| pTE885 | pREP1-rad1-ICD | |
| pTE886 | pREP1-rad1-P279A | |
| pTE550 | pREP42-hus1+ | |
| pTE601 | pREP42-hus1-R2S | |
| pTE823 | pREP42-hus1-W50R | |
| pTE592 | pREP42-hus1-N80I | |
| pTE599 | pREP42-hus1-N121D | |
| pTE887 | pREP42-hus1-H125R | |
| pTE888 | pREP42-hus1Q134R | |
| pTE889 | pREP42-hus1-I244M |
To carry out immunoprecipitation experiments, cells were grown overnight in EMM liquid media containing thiamine, washed thrice with EMM media, and reinoculated into EMM media to induce the promoter. Cells were harvested after 20 h of growth in EMM media.
Identification of Functionally Important Regions of Hus1, Rad1, and Rad9
Venclovas and Thelen's (2000) sequence alignment of Hus1, Rad1, Rad9 with PCNA was used to determine the PCNA-like region of Rad9 and the residues of Rad1 and Hus1 that are involved in monomer–monomer interactions. According to this alignment, the N-terminal region of Rad9, comprising of residues 1–299, shows structural similarity to PCNA, whereas the C-terminal region of Rad9, which is composed of residues 299–426, bears no similarity to PCNA. This alignment also predicts that Rad1 has two binding sites for Hus1 in its N-terminal region, which span residues 75–81 and 134–140. The interaction sites for Rad9 are in the C-terminal region of Rad1 encompassing residues 165–176 and 204–209. The ICD loop connecting the N- and C-terminal region of Rad1 is formed by the residues 141–158. Based on Venclovas and Thelen's (2000) alignment, Hus1 is predicted to possess two Rad9 binding sites (residues 78–84 and 124–130) in its N-terminal region and two Rad1 interaction sites (residues 157–168 and 194–199) in its C-terminal region. The ICD loop of Hus1 is comprised of residues 131–149.
Construction of Plasmids
The plasmids used in this study are listed in Table 1. Standard molecular biology methods were used for all constructions. Polymerase chain reactions (PCRs) were carried out with the use of Pfu polymerase to minimize the rate of mutations during PCR amplification.
To construct pREP1-rad9-N (pTE883), which encodes 1–299 amino acids of Rad9, PCR was performed on pREP1-rad9+ plasmid (pTE479) with the use of the primers Rad9-N1, 5′-GGAGTTCCATATGGAATTCACTGTTTCAAATG-3′, and Rad9N2–2, 5′-CGCGGATCCCCAAGTTGCAAGAATGAACTGCGC-3′. These primers were designed to create an in-frame stop codon after the 299th amino acid (threonine) of Rad9 and also to create an NdeI and a BamHI site at the 5′ and 3′ end of the PCR product, respectively. The resulting PCR product was cloned with the use of the TA Cloning Vector (Invitrogen, Carlsbad, CA) and then subcloned into the pREP1 plasmid as NdeI-NdeI and NdeI-BamHI fragments with the use of a three-way ligation.
To construct pREP1-rad9-C (pTE884), which encodes the 284–426 amino acid of Rad9, PCR was performed on pREP1-rad9+ (pTE479) plasmid with the use of the primers Rad9-C1, 5′-GGAGTTCCATATGGCAAAAGGGAAAAATTCC-3′, and Rad9-C2, 5′-CGCGGATCCCTAGTCTTCCTGAGAGAAAAT-3′. These primers were designed to create an in-frame stop codon after the 426th amino acid (aspartic acid) of Rad9 and also to create an NdeI and a BamHI site at the 5′ and 3′ end of the PCR product, respectively. The resulting PCR product was cloned with the use of the TA Cloning Vector (Invitrogen) and then subcloned into the pREP1 plasmid as a NdeI-BamHI fragment. The pREP1-rad9-N and pREP1-rad9-C constructs were sequenced to ensure that no mutation has been introduced during the PCR amplification.
To generate the pREP1-rad1-ICD (pTE885) and pREP1-rad1-P279A (pTE886) mutants, whole plasmid-based mutagenesis was performed on pREP1-rad1+ (pTE567) with the use of the QuickChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). For pREP1-rad1-ICD mutant, the primers Rad1-ICD1, 5′-GACTATGGAATGTGCGTCGGGCAACGA GGACGACGTTG-3′, and Rad1-ICD2, 5′-CAACGTCGTCCTCGTTGCCCGACGCACA TTCCATAGTC-3′, were used. For pREP1-rad1-P279A mutant, the primers Rad1-P279A1, 5′-GTAGACTTTTGTATTGTTGCTTTGGACCTTGTAAGTG-3′, and Rad1-P279A2, 5′-CACTTACAAGGTCCAAAGCAACAATACAAAAGTCTAC-3′, were used.
pREP42-hus1-H125R (pTE887), pREP42-hus1-Q134R (pTE888), and pREP42-hus1-I244 M (pTE889) were generated by performing PCR on pREP42-hus1+ (pTE550) plasmid with the use of the QuickChange Site-Directed Mutagenesis kit (Stratagene) as described above. The following sets of primers were used: Hus1-H125R1, 5′-GGTTCGAATATTGTGACTCGTAATATACCTGTTCGAG-3′, and Hus1-H125R2, 5′-CTCGAACAGGTATATTACGAGTCACAATATTCGAACC-3′, for pREP42-hus1-H125; Hus1-Q134R1, 5′-CCTGTTCGAGTACTATCACGATC ATACGTGTCAG-3′, and Hus1-Q134R2, 5′-CTGACACGTATGATCGTGATAGTAC TCGAACAGG-3′, for pREP42-hus1-Q134R; and Hus1-I244 M1, 5′-GTCAACATGTT AAAAATGTCCAGTGTTGCAAAGCG-3′, and Hus1-I244 M2, 5′-CGCTTTGCAACAC TGGACATTTTTAACATGTTGAC-3′, for pREP42-hus1-I244 M.
All the mutants generated by site-directed mutagenesis were sequenced to ensure that they contained the desired mutation and no other mutation has been introduced during the PCR amplification.
Screen for Dominant Negative Alleles of hus1
To carry out a screen for dominant negative alleles of hus1+, PCR reactions with the use of Taq polymerase in the presence of 50 mM MnCl2 were performed on pREP42-hus1+ plasmid (pTE550) with the use of the primers REP5, 5′-CCCACAATTTTGGGAATAGCGC AAG-3′ and REP3, 5′-CCTCCTTCCACATGCTGTAAACAAAG-3′. The mutated PCR fragments of hus1+ were cotransformed into the fission yeast wild-type cells along with the NdeI-BamHI digested, linearized pREP42-hus1+ plasmid whose ends shared homology to the ends of PCR-amplified hus1 fragments (Kostrub et al., 1998b). This places expression of the proteins under the control of a weakened version of the thiamine-repressible nmt1+ promoter (Mandrell, 1990; Basi et al., 1993). The mutant versions of hus1+ were obtained by homologous recombination through gap repair in vivo. Transformants were selected on EMM plates containing thiamine (to repress the expression of the hus1 mutants from the nmt1 promoter). Approximately 20,000 transformants were replica plated from EMM plates containing thiamine to EMM plates lacking thiamine. After 24 h of promoter induction, the cells were further replica plated onto EMM plates containing 10 mM HU to screen for dominant checkpoint defective mutants. Twenty-three transformants were found to be sensitive to HU only after thiamine induction, indicating the presence of dominant-negative alleles. Plasmids from these strains were recovered, retested, and subjected to DNA sequencing.
Protein Extracts and Immunoprecipitations
Antibodies against Rad9 were raised with the use of a histidine-tagged Rad9 protein expressed from pET16b-rad9-c (pTE795) plasmid. The fusion protein was purified from bacteria and injected into rabbits with the use of the standard methods (Harlow and Lane, 1988), resulting in the polyclonal antiserum that specifically recognized the fission yeast Rad9 protein. Production of Hus1 and Rad1 antibodies is described in Kostrub et al. (1997, 1998a).
Yeast protein extracts for immunoprecipitations (IPs) were prepared from log phase cultures, grown in the absence of thiamine for 20 h to induce the protein expression from nmt1 promoter. For the biochemical analysis of rad1-ICD and rad1-P279A mutants, extracts were made from the cells grown in EMM media containing thiamine (promoter repressed). The entire IP procedure was carried out at 4°C to minimize the protease activity. Cells (1 × 109) were harvested by centrifugation and washed with 1 ml of IP buffer (25 mM Tris, pH 7.5, 100 mM NaCl, 10% glycerol, 0.1% NP-40, 0.5 mM dithiothreitol, 15 mM MgCl2, 15 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 60 mM β-glycerophosphate, 15 mM p-nitrophenylphosphate, 1 mM sodium-orthovanadate, 0.1 mM sodium fluoride, and 1 mM 1,10-phenanthroline). Cell pellets were suspended in 100 μl of IP buffer and 0.6 g of glass beads (approximately equal to the volume of cell suspension) was added to the tubes. Cells were lysed with the use of a Fast-Prep FP120 (Bio 101 Savant) in two bursts of 30 s each with an intermittent cooling for 2 min on ice. Immediately after lysis, 1 ml of ice-cold IP buffer was added to the tube and samples were centrifuged for 30 s in a Microfuge to remove the cell debris. The supernatant was collected and centrifuged for another 15 min at 12K to remove any insoluble material. The resulting supernatant was added to 200 μl of antibody-coupled protein-A-Sepharose beads. Antibodies were coupled to protein A-Sepharose beads via the bifunctional reagent dimethylpimelimidate (Harlow and Lane, 1988). Cell extract was incubated with beads for 1 h at 4°C on a rotating wheel. The immunoprecipitates were washed three times with IP buffer to remove the unbound proteins and then boiled for 5 min in 2× SDS sample buffer.
Proteins were resolved on 10% SDS-PAGE and transferred onto Immobilon-P (Millipore, Bedford, MA) membrane with the use of semidry electrotransfer. Western blotting was performed with the use of the standard procedure and immunodetection was carried out with the use of the horseradish peroxidase-conjugated anti-rabbit secondary antibody and ECL chemiluminescence method (Amersham Pharmacia Biotech, Piscataway, NJ).
RESULTS
Hus1, Rad1, and Rad9 Form a Complex Requiring the Presence of All Three Proteins
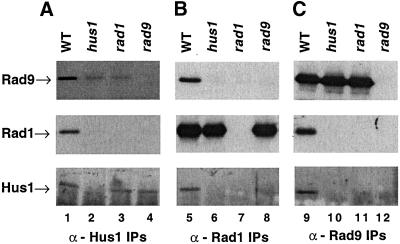
Previous work (Kostrub et al., 1998a; Caspari et al., 2000) has shown that Hus1, Rad1, and Rad9 interact to form a stable complex. In these studies, a direct association was detected between Hus1 and Rad1, which was dependent on Rad9 (Kostrub et al., 1998a). To determine directly whether Rad9 is present in the complex, we raised polyclonal antibodies against Rad9 and used them to probe for the presence of Rad9. The complex was examined by performing IPs with α-Rad9, α-Rad1, and α-Hus1 antibodies from wild-type (wt), hus1Δ, rad1Δ, and rad9Δ cells. As shown in Figure 1, Rad9 is detected only in the Hus1 and Rad1 immunoprecipitates prepared from wt cells (Figure 1A, top, lane 1; and B, top, lane 5). In contrast, significant amounts of Rad9 were not detected in the Hus1 or Rad1 immunoprecipitates from any of the deletion strains (Figure 1A, top, lanes 2–4; and B, top, lanes 6–8). Although Rad9 did not associate with Hus1 or Rad1 in any of the deletion strains, the protein was present and could be detected in the Rad9 immunoprecipitates from hus1Δ and rad1Δ strains (Figure 1C, top, lanes 10 and 11). In addition, Rad1 and Hus1 coprecipitated when wild-type extracts were precipitated with Rad9 antibodies (Figure 1C, middle and bottom, lane 9), although neither protein associated with Rad9 in extracts made from any of the deletion strains (Figure 1C, middle and bottom, lanes 10–12). Taken together, these results suggest that Hus1, Rad1, and Rad9 exist together in a complex in wild-type cells under normal growth conditions and the stability of the complex depends upon the presence of all three proteins.
Figure 1.
Hus1 and Rad1 physically interact with Rad9. Western blots of the IPs of the fission yeast protein extracts probed with anti-Rad9, anti-Rad1, or anti-Hus1 antibodies. (A) Hus1 immunoprecipitates immunoblotted with anti-Rad9 (top), anti-Rad1 (middle), or anti-Hus1 antibody (bottom) from wt (TE237, lane 1), hus1Δ::LEU2 (TE484, lane 2), rad1Δ::ura4+ (TE459, lane 3), and rad9Δ::ura4+ mutants (TE457, lane 4). (B) Rad1 immunoprecipitates prepared from the same strains immunoblotted with anti-Rad9 (top), anti-Rad1 (middle), and anti-Hus1 antibody (bottom). (C) Rad9 immunoprecipitates prepared from the same strains immunoblotted with anti-Rad9 (top), anti-Rad1 (middle), and anti-Hus1 antibody (bottom).
Interestingly, we occasionally observe low levels of band comigrating with Rad9 in the Hus1 immunoprecipitates in hus1Δ and rad1Δ cells (Figure 1A, top, lanes 2 and 3). We believe that this is Rad9, because it is not observed in the rad9Δ strain (Figure 1A, top, lane 4). We speculate that presence of this band is due to weak cross-reaction between Hus1 antibodies and the Rad9 protein, rather than the weak association between pairs of proteins, because we do not observe Hus1 or Rad1 in Rad9 immunoprecipitates from the same extracts (Figure 1C, lanes 10 and 11).
PCNA-like N-Terminal Domain of Rad9 Interacts with Hus1 and Rad1
The sequence alignment of Hus1, Rad1, Rad9 with PCNA predicts the structural similarity of Hus1 and Rad1 with PCNA to extend throughout the length of these proteins. In contrast, Rad9 has an additional, non-PCNA like domain at C terminus (Caspari et al., 2000; Venclovas and Thelen, 2000). This alignment suggests that these two regions of Rad9 could represent distinct domains with separate functions.
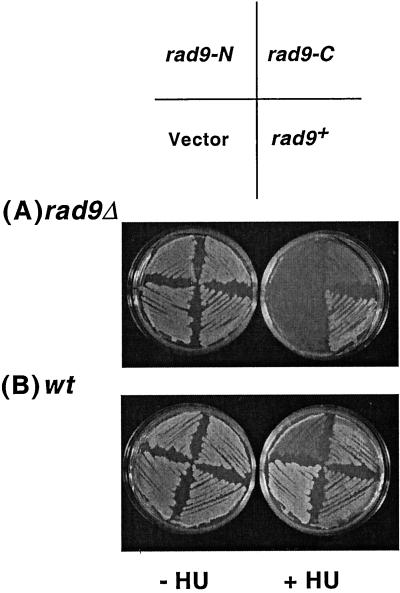
To investigate this possibility, we cloned the Rad9 N-terminal domain (Rad9-N, amino acids 1–299) and Rad9 C-terminal domain (Rad9-C, amino acids 284–426) independently under the nmt1+ promoter, which is repressed by thiamine (Maundrell, 1990). These constructs, as well as the full-length Rad9 and an empty vector control were introduced into wt (TE237) and rad9Δ (TE457) backgrounds. Neither Rad9-N nor Rad9-C expression rescued the HU sensitivity of rad9Δ (Figure 2A) establishing that neither peptide is sufficient to perform Rad9 function. Rad9-N and Rad9-C overexpression had no effect on the growth of wild-type cells under normal conditions (Figure 2B); however, cells expressing Rad9-N grew poorly on media containing the replication inhibitor HU. Microscopic examination revealed that many of the cells overexpressing Rad9-N were entering mitosis instead of arresting in HU because they were short, and in many cases, the septa was observed to cleave the nuclear material (our unpublished results). This phenotype is characteristic of cells with defects in the checkpoint that senses unreplicated DNA (Stewart and Enoch, 1996). In contrast, no growth inhibition was seen in cells expressing either Rad9-C or full-length Rad9 or empty vector (Figure 2B). Microscopic analysis of these cells suggested that they underwent cell cycle arrest normally because they were elongated and contained a single nucleus (our unpublished results).
Figure 2.
PCNA-like N-terminal domain of Rad9 disrupts checkpoint control in wild-type (wt) cells. (A) Expression of neither Rad9-N nor Rad9-C rescues HU sensitivity of rad9Δ (TE457). Growth of rad9Δ cells transformed with pREP1 plasmid (pTE102), pREP1-rad9-N (pTE883), pREP1-rad9-C (pTE884), and pREP1-rad9+ (pTE479) on EMM plates lacking or containing 10 mM HU. (B) Rad9-N expression causes HU sensitivity in wt (TE237) cells. wt cells transformed with the same plasmids as mentioned above were grown on EMM plates with or without 10 mM HU.
IP-Westerns on rad9Δ cells expressing Rad9-N and Rad9-C established that both Rad9-N and Rad9-C domains were stably expressed (Figure 3A, lanes 3–4, as can be seen in lane 1, the full-length Rad9 protein is somewhat degraded when expressed at these levels). Rad9-N levels appear somewhat lower (Figure 3A, lane 3), although this could also be due to the presence of fewer antigenic epitopes on this domain (note that the polyclonal sera used was raised against the full-length protein).
Figure 3.
PCNA-like N-terminal domain of Rad9 interacts with Hus1 and Rad1. (A) Rad9, Rad9-N, and Rad9-C protein expression. Western blot of Rad9 immunoprecipitates, probed with anti-Rad9 antibody, from rad9Δ cells expressing pREP1 plasmid (pTE102, lane 1), pREP1-rad9+ (pTE479, lane 2), pREP1-rad9-N (pTE883, lane 3), and pREP1-rad9-C (pTE884, lane 4). A weaker signal in the same position as Rad9 in lanes 1, 3, and 4 is probably due to IgG heavy chain background. Lower molecular weight bands in lane 2 are likely the degradation products of Rad9. (B) Western blots of anti-Rad1 immunoprecipitations probed with anti-Hus1 (top) or anti-Rad1 antibody (bottom) from cells transformed with the same plasmids as in A. (C) Western blots of anti-Hus1 immunoprecipitations probed with anti-Hus1 (top) or anti-Rad1 antibody (bottom) from the same strains as used in A.
The PCNA alignment predicts that Rad9-N, but not Rad9-C, should interact with Hus1 and Rad1. The dominant negative activity of the overexpressed Rad9-N protein suggests that it may sequester Hus1 and Rad1 in nonfunctional complexes. To examine the interactions of the Rad9 domains, coimmunoprecipitation assays were carried out with the use of extracts from rad9Δ cells expressing either the empty vector, Rad9-N, Rad9-C, or full-length Rad9 protein. We were able to immunoprecipitate Hus1 with anti-Rad1 IPs from cells expressing either Rad9-N or full-length Rad9 (Figure 3B, top, lanes 2 and 4), suggesting that Rad9-N, like full-length Rad9, is able to associate with Rad1 and Hus1. In contrast, no Hus1 was precipitated with Rad1 antibodies in cells expressing empty vector or Rad9-C (Figure 3B, top, lanes 1 and 3), although the same amount of Rad1 was immunoprecipitated in all strains with the use of Rad1 antibody (Figure 3B, bottom, lanes 1–4). Similarly, Rad1 was found in the Hus1 immunoprecipitates from cells expressing either Rad9-N or full-length Rad9 (Figure 3C, bottom, lanes 2 and 4) but not from cells expressing empty vector or Rad9-C (Figure 3C, bottom, lanes 1 and 3). Equal amounts of Hus1 were present in each of the IPs (Figure 3C, top, lanes 1 and 4). Relatively lower amounts of Hus1 and Rad1 were found in the Rad1 and Hus1 immunoprecipitates, respectively, from cells expressing Rad9-N domain compared with the cells expressing full-length Rad9 (Figure 3, B and C, top and bottom, lanes 2 and 4). This difference could either be due to the lower expression of Rad9-N or alternatively, it may indicate that the Hus1-Rad1-Rad9-N complex is less stable than the wild-type complex. We were not able to use Rad9 IPs to determine directly whether Rad9-N or Rad9-C specifically associated with Rad1 and Hus1 because the overexpressed peptides interact nonspecifically with protein-A-Sepharose beads. However, because the interaction between Hus1 and Rad1 is observed in the presence of Rad9-N, we believe that Rad9-N forms a complex with Hus1 and Rad1. In contrast, Rad9-C does not appear to support the interaction between Hus1 and Rad1.
Use of PCNA Alignment to Identify Functionally Important Residues in Rad1
If Hus1, Rad1, and Rad9 are structurally similar to PCNA, it should be possible to use the PCNA alignment to identify important residues in checkpoint proteins especially considering that the overall degree of sequence conservation is low. To determine whether this was the case, we created a mutation that replaces a conserved proline in the Rad1-C terminus with an alanine (Figure 4A, rad1-P279A). According to the PCNA alignment, this proline residue is highly conserved between PCNA and Rad1 of many species (Venclovas and Thelen, 2000). Mutation of this residue in fission yeast PCNA makes cells sensitive to DNA damage and disrupts interactions with DNA polymerases, although it does not affect subunit–subunit interactions (Kelman et al., 1999). Thus, the PCNA alignment predicts that this mutation should disrupt the checkpoint function of Rad1, but might not alter its ability to interact with Hus1 and Rad9.
The PCNA alignment also predicts that disruption of the Rad1 region equivalent to the ICD loop in PCNA might interfere with interactions with other checkpoint proteins without affecting its interactions with Hus1 or Rad9. To test this idea, four amino acids (A-S-G-N) were inserted after the C-144 residue in the putative ICD loop of Rad1 by site-directed mutagenesis (Figure 4A, rad1-ICD). The addition of four amino acids is expected to perturb the overall binding surface of the loop thereby disrupting its interactions with other proteins (Venclovas and Thelen, 2000; Zuccola et al., 2000).
The mutants rad1-ICD and rad1-P279A, as well as wild-type rad1+ were expressed from nmt1 promoter and their phenotypes were scored. As shown in Figure 4B, I, both mutant proteins were able to complement the HU sensitivity of rad1Δ. They also rescued the UV sensitivity of the rad1Δ strain (our unpublished results). In addition, the mutant proteins had no effect on the growth of wt cells in the presence or absence of HU (Figure 4B, III). These results show that, contrary to the predictions of the PCNA alignment, both rad1-ICD and rad1-P279A mutants encode functional Rad1 proteins. To test whether rad1-ICD and rad1P279A mutants are partial loss of function mutants, we examined the ability of these mutants to rescue the HU sensitivity of rad1Δ under repressing conditions. nmt1 being a very strong promoter, basal level of protein expression is seen even when the promoter is repressed by thiamine, and this expression roughly equals the levels of expression of endogenous Rad1 (our unpublished results). As shown in Figure 4B, II, both the Rad1-ICD and Rad-P279A mutant proteins still complemented the HU sensitivity of rad1Δ, establishing that the mutant proteins are fully functional even when expressed at lower levels.
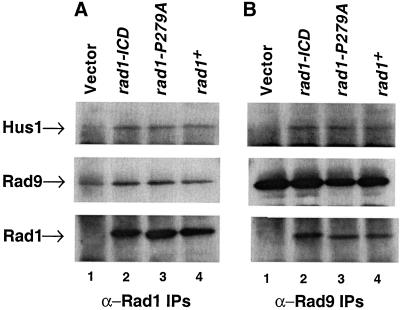
We next examined the interactions of Rad1-ICD and Rad1-P279A mutant proteins with Hus1 and Rad9. Hus1, Rad1, and Rad9 immunoprecipitates were prepared from rad1Δ cells expressing either empty vector, wild-type Rad1, or mutant Rad1 proteins, and the presence of Rad1, Rad9, and Hus1 in these immunoprecipitates was analyzed by Western blotting. Analysis of Rad1 IPs showed that both the mutant and wild-type Rad1 proteins were efficiently expressed (Figure 5A, bottom, lanes 2–4) and associated with equal amounts of Hus1 and Rad9 (Figure 5A, top and middle, respectively, lanes 2–4). None of the proteins were detected in the Rad1 immunoprecipitates from cells expressing empty vector (Figure 5A, top and middle, lane 1); however, a background band due to IgG heavy chain is seen in blots probed with Rad9 antibodies because the Rad9 protein comigrates with IgG heavy chain (Figure 5A, middle, lane 1). Similarly, Hus1 and Rad1 could be detected in the Rad9 immunoprecipitates from cells expressing either Rad1-ICD, Rad1-P279A, or wild-type Rad1 (Figure 5B, top and bottom, lanes 2–4, respectively) but not from cells with empty vector (Figure 5B, top and bottom, lane 1), although approximately the same amount of Rad9 was immunoprecipitated from all strains (Figure 5B, middle, lanes 1–4). The interactions of these two Rad1 mutant proteins, Rad1-ICD and Rad1-P279A, with Hus1 and Rad9 were also confirmed by anti-Hus1 IPs (our unpublished results). Collectively, these results suggest that both these Rad1 mutant proteins are able to form normal complexes with Hus1 and Rad9. Thus, these mutations do not alter Rad1 function, contrary to the predictions based on the PCNA alignment.
Figure 5.
Rad1-ICD and Rad1-P279 mutant proteins interact efficiently with Hus1 and Rad9. Western blots of IPs from fission yeast protein extracts probed with anti-Hus1, anti-Rad9, or anti-Rad1 antibody. (A) Anti-Rad1 IPs from rad1Δ cells expressing pREP1 plasmid (pTE102, lane 1), pREP1-rad1-ICD (pTE885, lane 2), pREP1-rad1-P279A (pTE886, lane 3), and pREP1-rad1+ (pTE567, lane 4) immunoblotted with anti-Hus1 (top), anti-Rad9 (middle), or anti-Rad1 antibody (bottom). (B) Anti-Rad9 IPs from rad1Δ cells expressing the same plasmids as mentioned above probed with anti-Hus1 (top), anti-Rad9 (middle), or anti-Rad1 antibody (bottom).
Analysis of Checkpoint-defective rad1 Alleles
We next examined four previously described rad1 mutant strains, rad1-S3, rad1-S4, rad1-S5, and rad1-S6 (Kanter-Smoler et al., 1995) to determine whether the biochemical defect of these mutant proteins can be explained with the use of the PCNA alignment. These strains contain the indicated rad1 alleles (Figure 4A) integrated at the rad1+ locus such that expression of mutant proteins is controlled by the rad1+ promoter (Kanter-Smoler et al., 1995). rad1-S3 and rad1-S4 are HU- and UV-sensitive (Kanter-Smoler et al., 1995; our unpublished results), indicating that the residues that are altered in these mutants are required for the checkpoint response. In contrast, rad1-S5 and rad1S-6 fully complement rad1Δ, establishing that the corresponding mutations do not affect important residues. rad1-S5 mutant has a deletion of 10 consecutive acidic amino acids (EDEEEDEEEE, 285–294) at the C terminus, whereas rad1-S6 mutant has a deletion of a stretch of seven amino acids (ETEDEDS, 317–323) at the C terminus of Rad1 (Kanter-Smoler et al., 1995; Figure 4A). This C-terminal sequence is not predicted to align with PCNA (Venclovas and Thelen, 2000). The PCNA alignment could thus explain the lack of effect of these mutations, because they lie in a less conserved predicted region that may not be required for Rad1 protein function.
The checkpoint defective rad1-S3 mutant has a deletion of five amino acids (157–161, LCTKI) that overlaps with two residues of the putative ICD loop and also spans the three residues of the first β-strand of the second domain of Rad1 lying near the predicted Rad9 interaction regions (Kanter-Smoler et al., 1995; Venclovas and Thelen, 2000; Figure 4A). According to the PCNA alignment of Rad1, deletion of the residues in the ICD loop might disrupt interactions with other proteins. Alternatively, the deletion could disrupt interactions with Rad9 but should not affect interactions with Hus1, because the Rad9 and Hus1 binding sites are in two separate domains of Rad1 (Venclovas and Thelen, 2000; Burtelow et al., 2001). The second checkpoint defective mutant, rad1-S4, has a deletion of five amino acids (235–239, RHALK) in the predicted α-β2 helix of Rad1 (Figure 4A). The PCNA alignment does not assign a specific function to this region (Venclovas and Thelen, 2000); however, this mutation might be expected to disrupt the overall structure of the C-terminal domain of Rad1, thus abolishing the interaction with Rad9, without affecting the interaction with Hus1 (Burtelow et al., 2001).
To examine the biochemical interactions of these Rad1 mutant proteins, coimmunoprecipitations were carried out on all four rad1 mutants and a wild-type (rad1+) strain. We were able to precipitate Rad1 with Rad1 antibodies from all mutant strains (Figure 6A, bottom, lanes 2–5), although Rad1 levels seem to be reduced for rad1-S3, rad1-S4, and rad1-S6 mutants (Figure 6A, bottom, lanes 2, 3, and 5; note the increased mobility of Rad1-S5 mutant protein). Rad9 was only found to interact with Rad1, Rad1-S5, and Rad1-S6 proteins (Figure 6A, middle, lanes 1, 4, and 5). No Rad9 was seen in the Rad1 immunoprecipitates from rad1S-3 and rad1-S4 mutants (Figure 6A, middle, lanes 2 and 3), indicating that these Rad1 proteins do not interact with Rad9 (the weak signal in Figure 6A, lanes 2 and 3, is probably due to IgG heavy chain and is also observed in vector only control, Figure 5A, middle, lane 1). Similarly, no Hus1 was detected in the Rad1 immunoprecipitates from rad1-S3 and rad1-S4 mutants (Figure 6A, top, lanes 2 and 3), indicating that these mutant proteins also fail to interact with Hus1. These results suggest that Rad1S-3 and Rad1-S4 mutant proteins cannot participate in the formation of a normal complex.
Figure 6.
Hus1–Rad1–Rad9 interactions are disrupted in the checkpoint defective rad1-S3 and rad1-S4 mutants. IPs were performed on fission yeast extracts with the use of anti-Rad1, anti-Hus1, or anti-Rad9 antibody. Immunoprecipitates were analyzed for the presence of Hus1, Rad9, or Rad1 from wt cells (TE237, lane 1), rad1-S3 (TE994, lane 2), rad1-S4 (TE995, lane 3), rad1-S5 (TE992, lane 4), and rad1-S6 (TE993, lane 5) mutants. (A) Western blot of anti-Rad1 IPs probed with anti-Hus1 (top), anti-Rad9 (middle), or anti-Rad1 antibody (bottom). (B) Western blot of anti-Rad9 IPs probed with anti-Hus1 (top), anti-Rad1 (middle), or anti-Rad9 antibody (bottom). (C) Western blot of anti-Hus1 IPs probed with anti-Rad1 (top), anti-Rad9 antibody (middle), or anti-Hus1 antibody (bottom).
This finding was also confirmed by performing reciprocal IPs with Rad9 and Hus1 antibodies. Neither Hus1 nor Rad1 were detected in the Rad9 immunoprecipitates from rad1-S3 and rad1-S4 mutants (Figure 6B, top and middle, lanes 2 and 3), although both proteins were readily detected in the Rad9 immunoprecipitates from rad1+, rad1-S5, and rad1-S6 (Figure 6B, top and middle panels, lanes 1, 4, and 5). The amount of Rad9 immunoprecipitated was the same in the Rad9 immunoprecipitates from all strains (Figure 6B, bottom, lanes 1–5), again suggesting that Rad1S-3 and Rad1-S4 mutant proteins do not form normal complexes. Similarly, although both Rad1 and Rad9 were found in the Hus1 immunoprecipitates from rad1+, rad1-S5, and rad1-S6 mutants (Figure 6C, top and middle, lanes 1, 4, and 5; note somewhat reduced amount of Rad1 is immunoprecipitated from rad1-S5 mutant in this experiment [Figure 6C, top, lane 4]; however, this was not observed consistently), neither Rad1 nor Rad9 was immunoprecipitated with Hus1 antibodies from rad1-S3 and rad1-S4 mutants (Figure 6C, top and middle panels, lanes 2 and 3; the weak signal in Figure 6C, middle, lanes 2 and 3, is probably due to IgG heavy chain), again indicating that Hus1-Rad1-Rad9 complex is disrupted in these two rad1 mutants.
These data suggest that Rad1-S3 and Rad1-S4 mutant proteins do not interact with either Hus1 or Rad9. This defect is not predicted by the PCNA alignment, because the alignment predicts that binding sites for Hus1 and Rad9 should be in separate domains of Rad1 (Venclavos and Thelen, 2000).
Identification of Dominant-Negative Alleles of hus1+
To test the predictions of the PCNA alignment as specifically as possible, we developed a screen for mutations that selectively disrupt subsets of protein–protein interactions. Such mutations should allow an unbiased identification of functionally important residues. To do this, we screened for alleles that disrupt checkpoint function when overexpressed in wild-type cells (dominant-negatives). We reasoned that proteins with selective defects in protein–protein interactions should sequester the proteins they can interact with in nonfunctional complexes (Herskowitz, 1987). Mutations that disrupt all protein–protein interactions by generally destabilizing the protein would not be detected in such a screen.
We carried out this screen with the use of hus1+, because we have previously identified dominant-negative alleles of this gene (Kostrub et al., 1998a,b). hus1+ was subjected to PCR mutagenesis, and a library of mutated constructs was introduced into fission yeast wt cells with the use of the method of gap-repair (Kostrub et al., 1998b). We screened 20,000 transformants and identified 23 hus1 dominant checkpoint defective mutants that disrupted checkpoint control under inducing conditions in wild-type cells (Figure 7B; our unpublished results). Microscopic analysis of these cells established that they were entering mitosis in HU, instead of undergoing normal cell cycle arrest (our unpublished results). Overexpressing these alleles did not have any effect on the growth of wild-type cells in the absence of HU (Figure 7B; our unpublished results). These alleles did not complement hus1Δ, indicating that all the mutant proteins were unable to perform the function of wild-type Hus1 (Figure 7C; and our unpublished results).
Sequence analysis of these 23 hus1-dn (dominant negative) alleles revealed that most of the alleles contained multiple amino acid substitutions that were scattered throughout the Hus1 protein (Table 2). As described below, by comparing the pattern of mutations among these alleles, we were able to identify six single amino acid changes (R2S, W50R, N80I, N121D, H125R, and I244 M) that were sufficient to cause dominant negative activity. A seventh substitution (Q134R), which did not have dominant negative activity, was also generated.
Table 2.
Summary of hus1-dominant negative alleles
| Allele | Presumed dominant-negative mutations | Other mutations |
|---|---|---|
| dn14 | R2S | |
| dn10 | R2Gb | Q134P, E203G |
| dn19 | R2Gb | E54D, D149Y, I190L, V286E |
| dn23 | W50R | |
| dn7 | W50R | D93G, F118L |
| dn8 | W50R | E54K, N67I |
| dn16 | W50R | K4E, Y264H |
| dn17 | W50R | Y285N |
| dn6 | N80I | |
| dn21 | N80Db | A19T, S246C |
| dn13 | N121D | |
| dn2 | H125R | S138T |
| dn4 | H125R | N189D |
| dn9 | H125R | E53G, SY135-136PF, H151R |
| dn12 | H125R | T5A, Q185L, D287E |
| dn15 | H125R | M176I, QU211-212HA |
| dn1 | I244M | F153L |
| dn18 | I244M | M46T, S108L, R161K, P209T, E273V |
| dn11a | I244Vb | A89T, S108L, N121Sb |
| dn3 | Unknown | A19T, F118I |
| dn5 | Unknown | Q104L, V262A, A276V |
| dn20 | Unknown | T124A, Q185R, I190V, M231V |
| dn22 | Unknown | N37I, T95A, Q134R, T222A |
hus1-dn11 allele contains two mutations, N121S and I244V, either of which could be responsible for the dominant negative phenotype. However, we favor I244V, because N121S is a conservative substitution compared with N121D, the change that causes dominant-negativity in dn13.
A different mutation of this residue, when present as a single mutation, causes dominant-negativity.
Four of these amino acid changes (R2S, W50R, N80I, and N121D) were present as single mutants in the initial collection of 23 dominant-negative alleles. Seven of the other multiply mutated alleles contained one of these substitutions, and interestingly, no allele contained more than one of these changes. Thus, it seems that mutation of one of these four residues is sufficient to explain the dominant-negative phenotype of 10 of our alleles (Table 2; note that in some cases, the amino acid change is different from the single mutation prototype).
We further noted that the mutation H125R occurred in five alleles and never occurred in combination with any of the other four amino acids changes that we already knew caused dominant-negativity. Site-directed mutagenesis was used to construct an allele with only this alteration, and as shown in Figure 7C, this single mutation abolished Hus1 function and caused dominant-negativity in wt cells (Figure 7B). Therefore, it seems that the H125R mutation explains the phenotype of a further five dominant-negative alleles (Table 2).
Only one mutation, I244 M, was altered in more than one of the remaining seven alleles (dn1 and dn18). In addition, the similar mutation I244V occurs in combination with N121S in the dn11 allele. We were particularly interested in I244, because it lies in the C terminus, and is far away from the other five residues that cause dominant-negativity (Figure 7A). We therefore used site-directed mutagenesis to construct an allele with only the I244 M mutation. As shown in Figure 7, B and C, this alteration on its own is also sufficient to abolish Hus1 function and to cause a dominant-negative phenotype. We also constructed the Q134R single mutation (mutated in dn10 and dn22) because it was the only mutation we obtained more than once in the putative ICD loop of Hus1 (Venclavos and Thelen, 2000). However, an allele bearing just this substitution did not act as a dominant-negative (Figure 7B) and was fully functional (Figure 7C).
Overall, mutation of any of six amino acids (R2, W50, N80, N121, H125, and I244) is sufficient to explain the dominant-negative phenotypes of 19 of the 23 (82.6%) alleles we isolated, suggesting that our screen was fairly comprehensive (for summary, see Table 2). It is of course possible that some of the alterations we did not analyze make further contributions to the phenotypes. None of the mutations identified are in residues that are conserved between PCNA and Hus1 (Venclavos and Thelen, 2000); however, three of the altered residues (W50, N80, and H125) are conserved between the human, mouse, fly, worm, and fission yeast Hus1 proteins (Venclavos and Thelen, 2000). A fourth residue, R2, is also significantly conserved as the human, mouse, fly, and worm proteins have lysine, the other basic amino acids, at this position.
Biochemical Analysis of hus1-dn Mutants
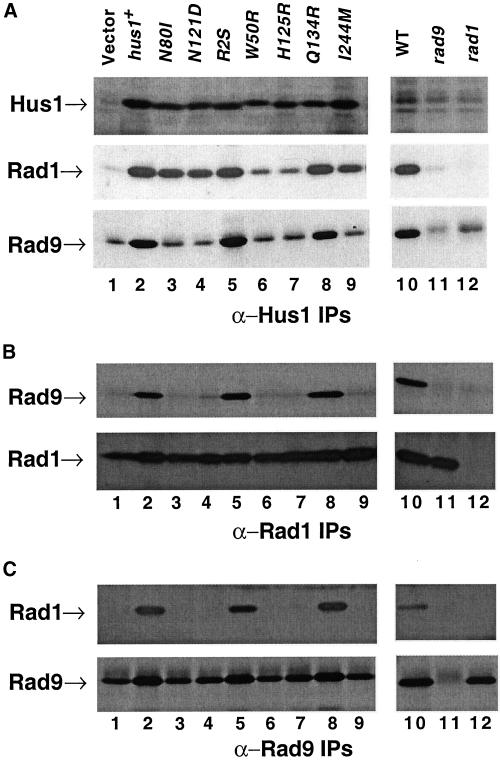
To investigate the molecular basis of the dominant-negativity of these alleles, we examined the interactions of the mutant Hus1 proteins with Rad1 and Rad9. The dominant negative alleles hus1-R2S, hus1-W50R, hus1-N80I, hus1-N121D, hus1-H125R, and hus1-I244 M as well as hus1-Q134R (which is not dominant negative), wild-type hus1+, and an empty vector control were expressed in a hus1Δ strain under the control of a weakened REP42 nmt1 promoter (Maundrell, 1990; Basi et al., 1993) for this analysis. IPs with the use of Hus1, Rad1, and Rad9 antibodies were performed on all the transformant strains. Analysis of Hus1-IP Western showed that all the Hus1 mutant proteins were expressed at approximately the same levels (Figure 8A, top, lanes 3–9; small differences in the mobility of some of these proteins were observed, e.g., lane 6). To investigate the interactions of these Hus1 mutant proteins with Rad1 and Rad9, we probed the same blot with Rad1 and Rad9 antibodies. Hus1-R2S, Hus1-N80I, Hus1-N121D, Hus1-Q134R, Hus1-I244 M mutant proteins interact with Rad1 approximately as efficiently as the wild-type Hus1 protein (Figure 8A, middle, compare lanes 3–5 and 8–9 with lane 2). In contrast, lower levels of Rad1 are associated with the Hus1-W50R and Hus1-H125R mutant proteins (Figure 8A, middle panel, lanes 6 and 7), indicating that the corresponding mutations destabilize the Hus1–Rad1 interactions.
Figure 8.
Biochemical analysis of hus1-dominant negative mutants. Western blots of the immunoprecipitations of fission yeast protein extracts probed with anti-Hus1, anti-Rad1, or anti-Rad9 antibody. (A) Hus1 immunoprecipitates prepared from hus1Δ (TE484) cells expressing the indicated plasmids, wt (TE237, lane 10), rad9Δ::ura4+ (TE457, lane 11), and rad1Δ::ura4+ (TE459, lane 12) strains probed with anti-Hus1 antibody (top), anti-Rad1 antibody (middle), or anti-Rad9 antibody (bottom). (B) Lanes 1–12 show the anti-Rad1 immunoprecipitations from the same strains as used above and probed with anti-Rad9 antibody (top) or anti-Rad1 antibody (bottom). (C) Lanes 1–12 show the anti-Rad9 immunoprecipitations from the same strains as used above and probed with anti-Rad1 antibody (top) or anti-Rad9 antibody (bottom).
In contrast to Rad1, wild-type levels of Rad9 were only detected in cells expressing wild-type Hus1 protein and Hus1-R2S and Hus1-Q134R mutant proteins (Figure 8A, bottom, lanes 2, 5 and 8). The remaining mutant proteins probably did not interact with Rad9 (Figure 8A, bottom, lanes 3, 4, 6, 7, and 9), although the presence of a background signal, which we believe is due to the residual amounts of IgG heavy chain, makes it difficult to rule out a weak association. Note that equivalent amounts of this background band are found in cells expressing the empty vector (Figure 8A, bottom, compare lane 1 with lanes 3, 4, 6, 7, and 9).
To confirm these results, we also examined the composition of Rad1 and Rad9 immunoprecipitates prepared from cells expressing either empty vector or Hus1 or Hus1 mutant proteins. As shown in Figure 8B, Rad9 was immunoprecipitated with Rad1 antibodies in cells expressing either the wild-type Hus1 protein, Hus1-R2S mutant protein, or Hus1-Q134R mutant protein (Figure 8B, top, lanes 2, 5, and 8). In contrast, no Rad9 was seen in the Rad1 immunoprecipitates from cells expressing either empty vector or other Hus1 mutant proteins (Figure 8B, top, lanes 1, 3, 4, 6, 7, and 9), although again background IgG makes it difficult to completely rule out some interaction. In contrast, the same amount of Rad1 was precipitated from all samples (Figure 8B, bottom, lanes 1–9). Similarly, Rad1 was detected in the Rad9 immunoprecipitates from cells expressing either the wild-type Hus1, Hus1-R2S, or Hus1-Q134R mutant (Figure 8C, top, lanes 2, 5, and 8, respectively), whereas no Rad1 was seen in the Rad9 immunoprecipitates from cells transformed with the vector alone or any of the other Hus1 mutant protein (Figure 8C, top, lanes 1, 3, 4, 6, 7, and 9). Equivalent amounts of Rad9 were precipitated with Rad9 antibodies from all strains (Figure 8C, bottom, lanes 1–9). Interestingly, the levels of Rad9 seem to be slightly reduced where the complex is disrupted (Figure 8C, bottom, compare lanes 1, 3, 4, 6, 7, and 9 with lanes 2, 5, and 8), suggesting that the protein may be less stable when it is not a part of the complex. These results confirm the absence of Rad9 in the Hus1-Rad1-Rad9 complexes from hus1-W50R, hus1-N80I, hus1-N121D, hus1-H125R, and hus1-I244 M mutants.
We also probed the Rad1 and Rad9 immunoprecipitates for Hus1; however, these results could not be interpreted because the overexpressed Hus1 protein bound nonspecifically to protein-A-Sepharose beads (our unpublished results). Despite this problem, we believe that we are detecting specific interactions between Hus1, Rad1, and Rad9 because Rad1 and Rad9 do not bind in the absence of Hus1 (Figure 8A, middle and bottom, lane 1), and we also observe differences in the interactions between mutant proteins that are confirmed by our analysis of Rad1 and Rad9 immunoprecipitates.
The results of our biochemical analysis are summarized in Figure 7A. As shown, the dominant-negative proteins fall into three classes. One class (Hus1-R2S) interacts normally with both Rad1 and Rad9. A second class of mutant proteins interacts inefficiently with Rad9, and normally with Rad1 (Hus1-N80I, Hus1-N121D, Hus1-I244 M). The last class of mutants shows reduced interaction with Rad1 and no interaction with Rad9 (Hus1-W50R and Hus1-H125R).
DISCUSSION
We have used a variety of approaches to examine what structural and functional features of the Hus1-Rad1-Rad9 complex are explained by its proposed similarity to the PCNA homotrimer.
First, we have used site-directed mutagenesis to construct mutations, and asked whether the PCNA model can explain the phenotypes of the resulting alleles. With this approach, we found that the region of Rad9 predicted to form a PCNA fold is necessary and sufficient for formation of a stable Hus1-Rad1-Rad9 complex, whereas the region that is not predicted to be PCNA-like plays no role in complex formation (Figure 3). In contrast, we had less success with the use of the PCNA alignment to predict functionally important residues in Rad1. Based on the alignment, we mutated a proline residue that is conserved between Rad1 and PCNA and is required for PCNA function. We also made an insertion into the putative ICD loop of Rad1, which, according to the model, should disrupt interactions of the Hus1-Rad1-Rad9 complex with other proteins. As shown in Figures 4 and 5, neither of these mutations affected Rad1 function. Thus, the PCNA alignment was not useful for predicting the function of these residues.
As a second approach, we analyzed rad1 alleles known to be defective for checkpoint function to see whether their phenotypes could be explained by the PCNA alignment. One of the mutations we analyzed, rad1-S3, overlaps with the putative ICD loop, and therefore might be expected to abolish interactions between the Hus1-Rad1-Rad9 complex and other proteins. The other mutation, rad1-S4, lies in a conserved helix in the C terminus, and might be expected to disrupt the conformation of the C-terminal lobe of Rad1. The PCNA model predicts that such an alteration might be expected to abolish Rad9 interaction, without affecting the interaction with Hus1 (Venclovas and Thelen, 2000; Burtelow et al., 2001). In contrast to these predictions, we found that the mutant proteins were unable to interact with either Hus1 or Rad9 (Figure 6). This finding could suggest that interaction between any pair of proteins requires the third protein, a finding that is not consistent with recent studies of the human proteins (Burtelow et al., 2001) and is also not explained by the PCNA alignment, which predicts that binding sites for Hus1 and Rad9 should lie on different domains of the Rad1 protein (Venclovas and Thelen, 2000). However, it is difficult to interpret these results conclusively, because the mutations could be having a general effect on Rad1 protein conformation.
As a third approach, we carried out a genetic screen for dominant-negative hus1 alleles to identify mutations that selectively disrupt specific protein–protein interactions. We reasoned that such mutants would allow us to rigorously test the predictions of the PCNA model, because they should identify specific residues involved in protein–protein interactions. With the use of this approach, we identified six single amino acid substitutions that selectively disrupted interactions of Hus1 with other proteins. Only one of the mutations (R2S) did not disrupt the interactions of Hus1 with Rad1 or Rad9. This mutation is presumed to identify a region of Hus1 that is involved in interactions with other proteins such as Rad3, Rad17, or other components of the checkpoint response pathway. PCNA mutants with alterations near the mutated residue, which do not interact normally with DNA polymerase δ, have been described (Arroyo et al., 1996; Henderson et al., 2000). Thus, this mutation may identify a domain that is functionally conserved between Hus1 and PCNA.
Surprisingly, we did not find any alleles in this category with mutations in the putative ICD loop of Hus1. In fact, we specifically created an allele with a mutation in the ICD loop (Q134R), because this alteration was found in two of the dominant-negative alleles that we identified (Table 2). However, this mutation had no effect on Hus1 function, or on its interactions with Rad1, Rad9, or other proteins (Figures 7 and 8). This, combined with the lack of a phenotype in the rad1-ICD allele (Figures 4 and 5), argues that ICD region does not have an important function in the checkpoint proteins. In PCNA structure, the ICD loop protrudes from the surface of the protein and plays a critical role in interactions with other proteins (Kelman and Hurwitz, 1998; Tsurimoto, 1998). A domain equivalent to the ICD loop is involved in protein–protein interactions for herpes simplex virus protein UL-42, a protein that consists of a PCNA fold, although it does not trimerize or interact with clamp loaders (Zuccola et al., 2000). These studies suggest that function of the ICD region of the PCNA fold can be highly conserved. Thus, it is surprising that it appears to play no role in Hus1 or Rad1 function.
We found three mutations (N80I, N121D, I244 M) that disrupted Hus1–Rad9 interactions without affecting Hus1–Rad1 interactions. The existence of these mutants argues that binding to Rad1 and Rad9 are separable functions of Hus1. This is in agreement with the PCNA model, which predicts these binding sites to lie in different lobes of the protein (Venclovas and Thelen, 2000). It is also consistent with recent studies of the human Hus1-Rad1-Rad9 complex (Burtelow et al., 2001). The PCNA alignment combined with genetic and biochemical data from S. cerevisiae (Kondo et al., 1999), predicts two Rad9 interaction sites for Hus1, one between amino acids 78–84, and another between amino acids 124–130 (Venclovas and Thelen, 2000). Interestingly, hus1-N80I contains an altered residue in the first domain, whereas hus1-N121D alters a residue that is very close to the second region (Figure 7A). Thus, the PCNA alignment explains the phenotypes of these mutants with remarkable accuracy.
In contrast, the third mutation that disrupts Hus1–Rad9 interactions, hus1-I244 M, lies in a region equivalent to the C-terminal domain of Hus1. Because the PCNA alignment predicts that Hus1, Rad1, and Rad9 are arranged in the head-to-tail configuration, residues involved in Rad9 interactions should lie in either the N- or the C-terminal domain, but not in both (Burtelow et al., 2001; Venclovas and Thelen, 2000). Thus, the location of this mutation suggests that not all aspects of the Hus1–Rad9 interaction can be modeled by analogy with the PCNA structure.
The detection of a Hus1-Rad1 complex in these mutants is surprising given that Hus1 and Rad1 do not interact in rad9Δ mutant (Figure 1; Kostrub et al., 1998a). It is possible that we observe these interactions because the Hus1 mutant proteins are overexpressed in the present study. Alternatively, it is possible that these mutant proteins interact transiently with Rad9. This transient association may not be detectable in immunoprecipitates, but may be sufficient to stabilize the Rad1-Hus1 interaction. Finally, it is possible that these alleles represent neomorphs, which have acquired novel functions as a result of the mutation. Experiments to distinguish between these possibilities are currently underway.
A third class of mutant proteins interacted weakly with Rad1 and was disrupted for their interactions with Rad9. Mutation of two residues, W50 and H125, caused this phenotype. Both of these residues are conserved in Hus1 proteins from many different species (Venclovas and Thelen, 2000). The PCNA alignment does not explain how a single mutation can affect interactions with both proteins unless the mutation has a general effect on protein conformation. Although this is possible, it is not a likely explanation for our results, because the dominant negative phenotypes of our mutants suggest that the mutant proteins still participate in some normal interactions. Possibly, like Hus1N80 and Hus1N121, this class of mutants may be primarily defective in their ability to interact with Rad9 and the weakened association with Rad1 may reflect a requirement for Rad9 during the complex assembly. This explanation is most consistent with the PCNA alignment, because both mutants in this class lie in the region predicted to be involved in Rad9 interactions (Figure 7A) and residues in an equivalent region of PCNA are thought to be required for subunit–subunit interactions in S. cerevisiae and humans (Ayyagari et al., 1995; Jonsson et al., 1995).
Alternatively, these mutants may be defective in their interactions with Rad1, and this weakened interaction with Rad1 may prevent interaction with Rad9. This explanation, however, is less consistent with the PCNA alignment, because it places both Rad1- and Rad9-interacting residues in the N-terminal domain of Hus1 (Figure 7A), whereas the PCNA alignment predicts that Rad1- and Rad9-interacting residues should lie in separate domains.
In either case, the fact that a single alteration can affect both Rad1 and Rad9 interactions suggests that the association between Hus1 and Rad9 may be dependent on the Hus1-Rad1 interaction. The idea of this hierarchy is further supported by our failure to identify mutations that selectively abolished the Hus1–Rad1 interaction, even although we identified three alleles that selectively abolished the Hus1-Rad9 interaction. Further evidence supporting this idea of a hierarchy comes from S. cerevisiae where the association of Rad17 and Mec3 (homologs of Rad1 and Hus1) does not require the presence of Ddc1 (homolog of Rad9) (Kondo et al., 1999). This hierarchy of interactions is not predicted by the PCNA alignment, because in the PCNA structure, each monomer plays an equivalent role. These observations are also inconsistent with a recent study (Burtelow et al., 2001) that shows that human Hus1, Rad1, and Rad9 interact with one another in a pairwise manner and these pairwise interactions occur independently of the presence of the third subunit of the complex.
Evaluating PCNA Model for Hus1-Rad1-Rad9 Complex
We have used a number of approaches to test the validity of the PCNA model. Although we found some instances where the PCNA alignment nicely explained mutant phenotypes, we also found several instances where the model failed to predict or explain the consequences of specific mutations. The discrepancies between our results and the predictions of the PCNA model may mean that the Hus1-Rad1-Rad9 complex has some features in common with PCNA but has also unique features probably adapted to its role in checkpoint control. In this regard, it is interesting that crystallographic studies of the herpes simplex virus DNA polymerase subunit UL-42 establish that this protein consists of a PCNA fold, although like the checkpoint proteins, UL-42 does not show substantial primary sequence similarity to PCNA. Interestingly, UL-42 does not trimerize or interact with clamp-loaders, although it does act as a specific processivity factor for the viral polymerase (Zuccola et al., 2000). Thus, the presence of PCNA folds in checkpoint proteins does not necessarily imply that they form the same tertiary structure as PCNA.
It is also possible that one, two, or all three of the proteins do not actually consist of PCNA folds. So far evidence in favor of the PCNA model is indirect and derives mainly from three-dimensional structure predictions (Venclovas and Thelen, 2000; Zuccola et al., 2000). Such predictions are not definitive in the absence of direct structural analysis. Indeed, the presence of PCNA folds in Hus1, Rad1, or Rad9 may be a key test of the validity of these methods, because it represents an instance where such methods allow novel predictions about the structure and function of proteins.
Conclusively determining whether the Hus1-Rad1-Rad9 complex structurally resembles the PCNA homotrimer will require structural and biochemical analysis of the purified complex. In the meantime, PCNA model for the Hus1-Rad1-Rad9 complex is clearly valuable because it provides a number of specific biochemical and genetic hypotheses about the structure and function of this important complex that are amenable to further testing.
ACKNOWLEDGMENTS
We are indebted to Harmon Zuccola for help with the modeling of PCNA folds in Hus1, Rad1, and Rad9. We thank Suresh Subramani for the gift of strains, Musetta Leung for help in increasing antibodies against Rad9, and Bob Weiss and Sarah Evans for valuable comments on this manuscript. This study was supported by National Institutes of Health grant GM-50015 and by an Howard Hughes Medical Institute Predoctoral Fellowship to C.F.K.
REFERENCES
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo MP, Downey KM, So AG, Wang TS. Schizosaccharomyces pombe proliferating cell nuclear antigen mutations affect DNA polymerase delta processivity. J Biol Chem. 1996;271:15971–15980. doi: 10.1074/jbc.271.27.15971. [DOI] [PubMed] [Google Scholar]
- Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, Hoekstra M, Carr AM. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Bluyssen HA, van Os RI, Naus NC, Jaspers I, Hoeijmakers JH, de Klein A. A human and mouse homolog of the Schizosaccharomyces pombe rad1+ cell cycle checkpoint control gene. Genomics. 1998;54:331–337. doi: 10.1006/geno.1998.5582. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Russell P. DNA replication checkpoint control. Front Biosci. 1999;1:D841–D848. doi: 10.2741/boddy. [DOI] [PubMed] [Google Scholar]
- Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage-responsive checkpoint complex. J Biol Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- Cai RL, Yan-Neale Y, Cueto MA, Xu H, Cohen D. HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J Biol Chem. 2000;275:27909–27916. doi: 10.1074/jbc.M000168200. [DOI] [PubMed] [Google Scholar]
- Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr AM. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;20:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- Hartwell L, Weinert T. Checkpoints: Controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Henderson DS, Wiegand UK, Norman DG, Glover DM. Mutual correction of faulty PCNA subunits in temperature-sensitive lethal mus209 mutants of Drosophila melanogaster. Genetics. 2000;154:1721–1733. doi: 10.1093/genetics/154.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Humphrey T. DNA damage and cell cycle control in Schizosaccharomyces pombe. Mutat Res. 2000;451:211–226. doi: 10.1016/s0027-5107(00)00051-8. [DOI] [PubMed] [Google Scholar]
- Jonsson ZO, Podust VN, Podust LM, Hubscher U. Tyrosine 114 is essential for the trimeric structure and the functional activities of human proliferating cell nuclear antigen. EMBO J. 1995;14:5745–5751. doi: 10.1002/j.1460-2075.1995.tb00261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanter-Smoler G, Knudsen KE, Jimenez G, Sunnerhagen P, Subramani S. Separation of phenotypes in mutant alleles of the Schizosaccharomyces pombe cell-cycle checkpoint gene. rad1+ Mol Biol Cell. 1995;6:1793–1805. doi: 10.1091/mbc.6.12.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z. PCNA: structure, functions and interactions. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- Kelman Z, Hurwitz J. Protein-PCNA interactions: a DNA-scanning mechanism? Trends Biochem Sci. 1998;23:236–238. doi: 10.1016/s0968-0004(98)01223-7. [DOI] [PubMed] [Google Scholar]
- Kelman Z, Zuo S, Arroyo MP, Wang TS, Hurwitz J. The C-terminal region of Schizosaccaromyces pombe proliferating cell nuclear antigen is essential for DNA polymerase activity. Proc Natl Acad Sci USA. 1999;96:9515–9520. doi: 10.1073/pnas.96.17.9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Matsumoto K, Sugimoto K. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol Cell Biol. 1999;19:1136–1143. doi: 10.1128/mcb.19.2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrub CF, al-Khodairy F, Ghazizadeh H, Carr AM, Enoch T. Molecular analysis of hus1+, a fission yeast gene required for S-M and DNA damage checkpoints. Mol Gen Genet. 1997;254:389–399. doi: 10.1007/pl00008606. [DOI] [PubMed] [Google Scholar]
- Kostrub CF, Knudsen K, Subramani S, Enoch T. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 1998a;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrub CF, Lei EP, Enoch T. Use of gap repair in fission yeast to obtain novel alleles of specific genes. Nucleic Acids Res. 1998b;26:4783–4784. doi: 10.1093/nar/26.20.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Lieberman HB, Hopkins KM, Nass M, Demetrick D, Davey S. A human homolog of the Schizosaccharomyces pombe rad9+ checkpoint control gene. Proc Natl Acad Sci USA. 1996;93:13890–13895. doi: 10.1073/pnas.93.24.13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. In: Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink GR, editors. Vol. 194. San Diego: Academic Press; 1991. pp. 795–823. [DOI] [PubMed] [Google Scholar]
- Mossi R, Hubscher U. Clamping down on clamps and clamp loaders–the eukaryotic replication factor C. Eur J Biochem. 1998;254:209–216. [PubMed] [Google Scholar]
- Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Rfc5, in cooperation with Rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5888–5896. doi: 10.1128/mcb.20.16.5888-5896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima H. Cell cycle checkpoints, chromosome stability and the progression of cancer. Hum Cell. 1997;10:221–230. [PubMed] [Google Scholar]
- Onel K, Koff A, Bennett RL, Unrau P, Holloman WK. The REC1 gene of Ustilago maydis, which encodes a 3′->5′ exonuclease, couples DNA repair and completion of DNA synthesis to a mitotic checkpoint. Genetics. 1996;143:165–174. doi: 10.1093/genetics/143.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker AE, Vande Weyer I, Laus MC, Ostoveen I, Yon J, Verhasselt P, Luyten WH. A human homologue of the Schizosaccharomyces pombe rad1+ checkpoint gene encodes an exonuclease. J Biol Chem. 1998;273:183332–183339. doi: 10.1074/jbc.273.29.18332. [DOI] [PubMed] [Google Scholar]
- Rauen M, Burtelow MA, Dufault VM, Karnitz LM. The human checkpoint protein hRad17 interacts with the PCNA-like proteins hRad1, hHus1, and hRad9. J Biol Chem. 2000;275:29767–29771. doi: 10.1074/jbc.M005782200. [DOI] [PubMed] [Google Scholar]
- Sheldrick KS, Carr AM. Feedback controls and G2 checkpoints: fission yeast as a model system. Bioessays. 1993;15:775–781. doi: 10.1002/bies.950151202. [DOI] [PubMed] [Google Scholar]
- Shimada M, Okuzaki D, Tanaka S, Tougan T, Tamai KK, Shimoda C, Nojima H. Replication factor C3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, plays a role in both replication and damage checkpoints. Mol Biol Cell. 1999;10:3991–4003. doi: 10.1091/mbc.10.12.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge RP, Udell CM, Casselman R, Davey S. The human G2 checkpoint control protein hRAD9 is a nuclear phosphoprotein that forms complexes with hRAD1 and hHUS1. Mol Biol Cell. 1999;10:1985–1995. doi: 10.1091/mbc.10.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E, Enoch T. S-phase and DNA-damage checkpoints: a tale of two yeasts. Curr Opin Cell Biol. 1996;8:781–787. doi: 10.1016/s0955-0674(96)80078-0. [DOI] [PubMed] [Google Scholar]
- Thelen MP, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T. PCNA, a multifunctional ring on DNA. Biochim Biophys Acta. 1998;1443:23–39. doi: 10.1016/s0167-4781(98)00204-8. [DOI] [PubMed] [Google Scholar]
- Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmer E, Karnitz LM. Human homologs of Schizosaccharomyces pombe Rad1, Hus1, and Rad9 form a DNA damage-responsive protein complex. J Biol Chem. 1999;274:567–570. doi: 10.1074/jbc.274.2.567. [DOI] [PubMed] [Google Scholar]
- Weinert T. DNA damage checkpoints update: getting molecular. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- Weiss RS, Kostrub CF, Enoch T, Leder P. Mouse Hus1, a homolog of the Schizosaccharomyces pombe hus1+ cell cycle checkpoint gene. Genomics. 1999;59:32–39. doi: 10.1006/geno.1999.5865. [DOI] [PubMed] [Google Scholar]
- Zhang G, Gibbs E, Kelman Z, O'Donnell M, Hurwitz J. Protein-PCNA interactions: a DNA-scanning mechanism? Proc Natl Acad Sci USA. 1999;96:1869–1874. doi: 10.1073/pnas.96.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccola HJ, Filman DJ, Coen DM, Hogle JM. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol Cell. 2000;5:267–278. doi: 10.1016/s1097-2765(00)80422-0. [DOI] [PubMed] [Google Scholar]