Abstract
Large-scale meta-analyses of genome-wide association studies (GWAS) have identified >175 loci associated with fasting cholesterol levels, including total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and triglycerides (TG). With differences in linkage disequilibrium (LD) structure and allele frequencies between ancestry groups, studies in additional large samples may detect new associations. We conducted staged GWAS meta-analyses in up to 69,414 East Asian individuals from 24 studies with participants from Japan, the Philippines, Korea, China, Singapore, and Taiwan. These meta-analyses identified (P < 5 × 10−8) three novel loci associated with HDL-C near CD163-APOBEC1 (P = 7.4 × 10−9), NCOA2 (P = 1.6 × 10−8), and NID2-PTGDR (P = 4.2 × 10−8), and one novel locus associated with TG near WDR11-FGFR2 (P = 2.7 × 10−10). Conditional analyses identified a second signal near CD163-APOBEC1. We then combined results from the East Asian meta-analysis with association results from up to 187,365 European individuals from the Global Lipids Genetics Consortium in a trans-ancestry meta-analysis. This analysis identified (log10Bayes Factor ≥6.1) eight additional novel lipid loci. Among the twelve total loci identified, the index variants at eight loci have demonstrated at least nominal significance with other metabolic traits in prior studies, and two loci exhibited coincident eQTLs (P < 1 × 10−5) in subcutaneous adipose tissue for BPTF and PDGFC. Taken together, these analyses identified multiple novel lipid loci, providing new potential therapeutic targets.
Introduction
Cholesterol and triglyceride levels are modifiable risk factors for cardiovascular diseases (CVD), a leading cause of death worldwide (1). Genome-wide association studies (GWAS) have identified >175 loci associated with high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), total cholesterol (TC) and triglyceride levels (TG) (2–7). The largest GWAS meta-analysis published to date has been performed in populations predominantly of European ancestry (4). As lipid profiles, allele frequencies, and environmental contributions differ between populations, investigating genetic factors influencing inter-individual variation in cholesterol and triglyceride levels in non-European populations that have not been as extensively studied can discover additional loci.
Several previous studies have investigated lipid trait associations in East Asian individuals (8–13). Novel loci near MYL2 and HECTD2 were found to be associated with HDL-C in a GWAS meta-analysis of more than 26,000 Korean individuals (9). A recent meta-analysis conducted in 12,685 Chinese using an exome array identified missense variants associated with TG and LDL-C at PNPLA3 and PKD1L3, respectively (13). In addition, novel signals have been identified in East Asians at previously established loci (11,13). For example, rs2075291, encoding APOA5 Gly185Cys, is associated with TG in East Asians (MAF = 0.06) (14), but not in >6,000 African American or European individuals, likely due to a minor allele frequency (MAF) that is rare (MAF <.005) in those populations (15). Similarly, low-frequency (MAF <0.05) missense variants at CETP, LDLR and PCSK9 were significantly associated with lipid traits in Chinese, but non-polymorphic in other ancestries examined (13). To date, the largest lipid genome-wide meta-analysis conducted in East Asians was performed in 12,545 individuals with replication in up to 30,395 (9).
In addition to examining studies with similar ancestry, combining results across ancestries provides opportunities to increase power to detect novel loci. For variant associations shared across ancestry groups, the larger sample size obtained by including studies from additional groups increases statistical power compared to a single ancestry group. However, variants may interact with environmental or other genetic factors that disproportionately affect ancestry groups, generating differences in allelic effects across populations. While the fixed-effects meta-analysis method assumes allelic effects to be the same across ancestries, the Meta-ANalysis of TRans-ethnic Association studies (MANTRA) algorithm allows for heterogeneity between more diverse ancestral populations (16). This approach allows different allelic effect between populations, and has successfully discovered many novel loci (15,17–23).
To identify additional loci, we meta-analyzed genome-wide association data for HDL-C, LDL-C, TC and TG from up to 69,414 individuals of East Asian ancestry participating in the Asian Genetic Epidemiology Network (AGEN). We also performed a trans-ancestry meta-analysis combining the East Asian summary results with publicly available summary results from 187,365 individuals of European ancestry from the Global Lipids Genetic Consortium (GLGC) (4). These analyses identified plausible candidate genes to further elucidate cholesterol and triglyceride metabolism and identify possible new cholesterol management therapies.
Results
Study overview
To discover novel loci, we performed East Asian-specific meta-analyses and East Asian-European trans-ancestry meta-analyses with four lipid traits: HDL-C, LDL-C, TC, and TG. The study design is described in Supplementary Material, Fig. S1. The East Asian meta-analyses consisted of two phases. In the first phase, we conducted a genome-wide discovery stage with 25,923 individuals in eleven studies from Japan, Korea, the Philippines, China, Singapore, and Taiwan (Supplementary Material, Tables S1 and S2) followed by in silico replication in up to 26,676 individuals from five additional genome-wide studies. Prioritized variants were then selected for de novo genotyping in up to 19,936 individuals from five additional studies (see Materials and Methods). In the second phase, we conducted a second genome-wide meta-analysis by combining the original discovery stage association results (eleven GWAS from Phase 1) with 10,805 individuals from two new genome-wide studies (BES and CHNS) that were not available during the first phase discovery meta-analysis. Description of phenotype collection, genotyping, and quality control metrics for each study are shown in the Supplementary Materials and Tables S1 and S2. We defined novel loci based on P < 5×10−8 and a distance of at least 1 Mb from the reported index variant at known loci. For the trans-ancestry meta-analysis, we used MANTRA (16) to meta-analyze association results from the larger, Phase 2 East Asian meta-analysis with 187,365 individuals of European ancestry from the GLGC (4), for a total of up to 221,739 individuals, and defined new loci based on log10 Bayes factor (BF) ≥ 6.1, requiring nominal significance (fixed effects P < 0.05) in each ancestry alone. Finally, we examined association evidence for all apparently novel loci in 10,857 individuals from four additional East Asian studies (MESA, TaiChi, TaiChi-G, and TUDR).
All East Asian-specific analyses were performed with and without adjustment for body mass index (BMI). Results from both models were largely similar, with the BMI-adjusted models exhibiting stronger P-values and slightly larger beta estimates (Supplementary Material, Table S3 and S4, Figs S2 and S3). For the East Asian analyses, we present results from the BMI-adjusted models. Sensitivity analyses were performed by excluding non-fasting samples. As there were no appreciable differences, we report results including non-fasting samples for maximal sample size. In the trans-ancestry meta-analysis, we present results from the BMI-unadjusted model based on data availability from GLGC.
Four novel lipid loci identified through East Asian meta-analysis
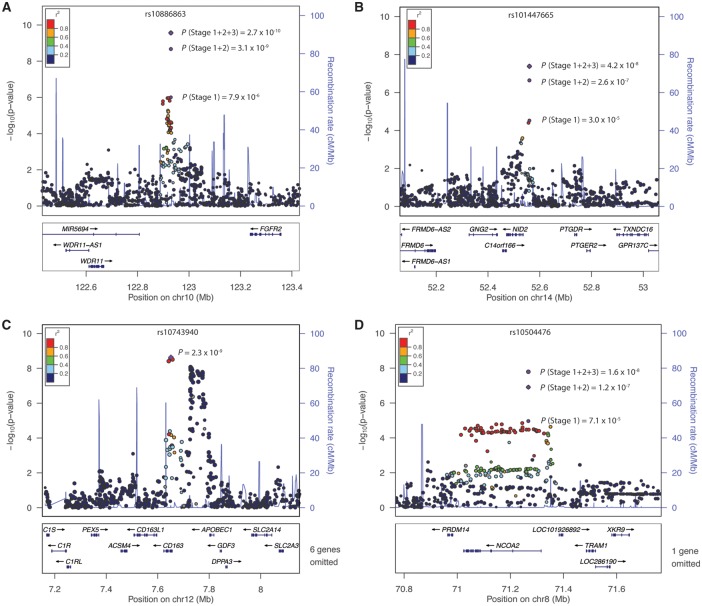
In the discovery stage from the first phase of the East Asian meta-analysis, we identified 25 previously known loci associated (P < 5×10−8) with one or more lipid traits (Supplementary Material, Tables S3 and S4, Figs S2 and S3). Based on the discovery stage, we selected 3,870 variants with P < 1×10−3 and pairwise LD r2 < 0.1 to evaluate in the in silico stage. After meta-analysis of the discovery and in silico stages, we selected 36 variants for de novo genotyping (see Materials and Methods). Meta-analyses of the three stages (totaling 21 studies, up to 69,414 subjects) identified ten additional previously known lipid loci (Supplementary Material, Table S5). We identified one novel locus associated with TG (rs10886863, near WDR11 and FGFR2, PadjBMI=2.73 × 10−10) and two novel loci associated with HDL-C (rs10504476, near NCOA2, PadjBMI=1.62 × 10−8; rs10144765, near NID2 and PTGDR, PadjBMI=4.16 × 10−8) (Table 1, Fig. 1, Supplementary Material, Table S6). In this first phase of East Asian analysis, we also observed that two loci previously reported to be associated with one lipid trait are now associated with an additional trait, including rs2792751 (near GPAM, HDL-C, PadjBMI=2.21 × 10−10), and rs1230180 (near METAP1 and ADH5, TC, PadjBMI=4.63 × 10−8) (Supplementary Material, Table S6). In the second phase of the East Asian meta-analysis, we replicated additional established loci (Supplementary Material, Table S7), and identified a third novel HDL-C association near CD163 and APOBEC1 (rs10743940, PadjBMI=2.30 × 10−9) (Table 1, Fig. 1). Forest plots for the four novel loci are presented in Supplementary Material, Fig. S4A and B. All four novel loci were validated to have the same direction of effect in up to an additional 10,857 individuals from four additional East Asian studies (Supplementary Material, Table S8).
Table 1.
New associations with lipid traits identified in the East Asian meta-analysis (P < 5 × 10−8)
| Trait | Variant | Chr | Position | Locus | N | EA/NEA | EAF | Adjusted for age, sex |
Adjusted for age, sex, BMI |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P-value | Phet | Beta | SE | P-value | Phet | ||||||||
| TG | rs10886863 | 10 | 122,929,493 | WDR11-FGFR2 | 45,245 | C/T | 0.64 | 0.041 | 0.007 | 2.71 × 10−8 | 0.275 | 0.041 | 0.006 | 2.73 × 10−10 | 0.175 |
| HDL-C | rs10743940 | 12 | 7,651,138 | CD163-APOBEC1 | 34,243 | A/T | 0.30 | 0.050 | 0.009 | 7.00 × 10−9 | 0.105 | 0.050 | 0.008 | 2.30 × 10−9 | 0.163 |
| HDL-C | rs10504476 | 8 | 71,267,629 | NCOA2 | 42,731 | G/A | 0.60 | 0.040 | 0.007 | 2.11 × 10−7 | 0.993 | 0.042 | 0.007 | 1.62 × 10−8 | 0.812 |
| HDL-C | rs10144765 | 14 | 52,559,930 | NID2-PTDGR | 54,176 | G/C | 0.27 | 0.031 | 0.007 | 1.63 × 10−6 | 0.626 | 0.035 | 0.007 | 4.16 × 10−8 | 0.380 |
East Asian meta-analysis results from Phases 1 and 2 (without and with adjustment for BMI). Genome-wide significant association is defined as P < 5 × 10−8. Physical positions based on hg19. Effect alleles are associated with higher triglyceride and HDL-C trait values. Beta estimates reflect per allele effects of variants on inverse normal transformed traits. rs10743940 at CD163-APOBEC1 attained genome-wide significance at Phase 2 (N up to 34,421). The other three loci attained genome-wide significance from combined analysis of Phase 1 discovery, in silico, and de novo stages (N up to 61,607 for HDL-C, and N up to 45,839 for TG).
Chr, chromosome; EA, effect allele; NEA, non-effect allele; EAF, effect allele frequency; SE, standard error; TG, triglycerides; HDL-C, high-density lipoprotein cholesterol.
Figure 1.
Regional association plots of the four loci identified from East Asian meta-analysis. (A) WDR11-FGFR2, (B) NID2-PTGDR, (C) CD163-APOBEC1, (D) NCOA2. –log10 (P-values) are plotted against the hg19 genomic coordinates at each locus. The purple circle represents the lead variant, which exhibited the strongest evidence of association at the locus among HapMap-imputed variants. Variants are colored based on 1000 Genomes Project Phase 3 East Asian LD with the lead variant. Results shown in A,B, and D are from Phase 1 of the East Asian meta-analysis, while C is from Phase 2 of the East Asian meta-analysis. Stage 1 indicates the Phase 1 discovery stage results, 1 + 2 indicates the results after the in silico stage, and 1 + 2 + 3 indicates the results after the de novo genotyping stage.
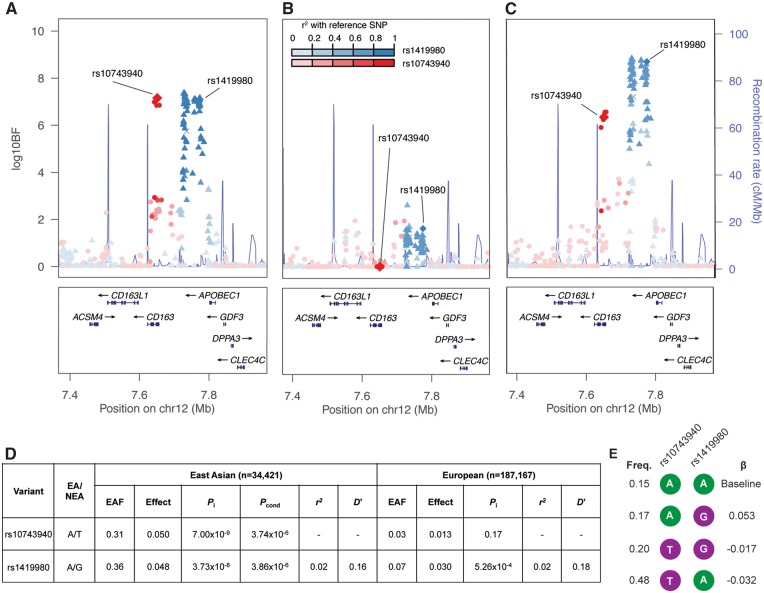
To explore the presence of multiple signals at novel lipid-associated loci, we performed approximate conditional analyses at the WDR11-FGFR2, CD163-APOBEC1, NCOA2, and NID2-PTGDR loci using Genome-wide Complex Trait Analysis (GCTA) (24) within a 500 kb window upstream and downstream of the index variant. The only evidence of a second signal was identified at the CD163-APOBEC1 locus (rs1419980; Pinitial=3.7 × 10−8; Pjoint=3.9 × 10−6), approximately 123 kb downstream of the first signal (rs10743940) (Fig. 2A). The LD between the two variants computed using 1000 Genomes Phase 3 East Asians is r2=0.02 and D’=0.16, suggesting the two signals are essentially independent from one another.
Figure 2.
HDL-C locus CD163-APOBEC1 exhibits two signals. (A) The two signals, rs10743940 and rs1419980, identified in Phase 2 of the East Asian meta-analysis, shown with the log10BF on the y-axis. (B) log10BF results from the GLGC European data showing the variant associations at the locus. (C) Trans-ancestry meta-analysis. Variants are colored based on LD with the lead variants, rs10743940 (red) and rs1419980 (blue) based on 1000 Genomes Project Phase 3 East Asian (A) and European (B and C) LD. (D) Fixed effect association results at two lead variants (rs10743940 and rs1419980) in East Asians and Europeans. Pinitial is the P value result from the unconditioned analysis. Pconditional is the joint conditional P value for each variant after conditioning on the other variant. (E) Haplotypes of rs10743940 and rs1419980 in the China Health and Nutrition Survey data. Alleles associated with lower HDL-C are shown in purple while alleles associated with higher HDL-C are shown in green. Haplotype association was performed with HDL-C inverse normalized residuals after adjusting for age, age2, sex, and BMI from the China Health and Nutrition Survey study using the most frequent haplotype as the reference.
Trans-ancestry meta-analysis
In the trans-ancestry analysis of up to 222,739 East Asian and European samples, we validated 151 established lipid loci at log10BF ≥ 6.1 (Supplementary Material, Table S9). We further replicated two loci (LOC100996634 and CTC1-PFAS) recently reported in similar meta-analysis of >4,700 Mexican individuals with the GLGC European samples (23) (Supplementary Material, Table S10). We also observed 10 novel loci associated with one or more of the four lipid traits (five with HDL-C, three with TG, one with LDL-C, and one with TC) and one locus associated with both LDL-C and TC (Table 2). All of the lead variants showed moderate evidence of association in the Europeans (fixed effect P-values 5.30 × 10−4 to 6.42 × 10−8) and demonstrated at least nominal association (fixed effect P-values 0.048 to 2.06 × 10−8) in the East Asians. The lead variants at all novel loci are common (MAF > 0.05 in East Asians and Europeans). The loci with the strongest evidence of an association were observed with variants near CD163-APOBEC1 (log10BF = 8.14, HDL-C), SNTB1 (log10BF = 7.48, HDL-C), BPTF (log10BF = 7.88, TG), and GPR180 (log10BF = 7.36, TG). We present fixed-effect meta-analysis results in each ancestry for comparison. The fixed-effect meta-analysis P-values were significant at only six of the ten loci, with comparable allelic sizes across the East Asians and Europeans.
Table 2.
New associations with lipid traits identified in trans-ancestry analysis of East Asian and Europeans (log10BF ≥ 6.1)
| Trait | Variant | Chr | Position | Locus | EA/NEA | East Asian Meta-Analysis |
European Meta-Analysis |
Trans-ancestry Meta-Analysis |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | EAF | Beta | SE | P-value | N | EAF | Beta | SE | P-value | N | Phet | P-value | log10(BF) | ||||||
| HDL-C | rs6822892 | 4 | 157,734,675 | PDGFC | G/A | 34,308 | 0.31 | 0.018 | 0.008 | 3.00 × 10−2 | 186,999 | 0.35 | 0.019 | 0.004 | 1.93 × 10−7 | 221,307 | 0.890 | 3.65 × 10−8 | 6.55 |
| HDL-C | rs4714556 | 6 | 41,993,229 | CCND3-TAF8 | G/A | 25,618 | 0.61 | 0.029 | 0.009 | 1.85 × 10−3 | 187,015 | 0.37 | 0.018 | 0.004 | 5.74 × 10−6 | 212,633 | 0.176 | 1.52 × 10−7 | 6.80 |
| HDL-C | rs6982451 | 8 | 71,014,079 | NCOA2 | G/A | 34,254 | 0.64 | 0.037 | 0.008 | 8.34 × 10−6 | 187,090 | 0.79 | 0.017 | 0.004 | 8.43 × 10−6 | 221,344 | 0.009 | 1.05 × 10−7 | 6.57 |
| HDL-C | rs4871137 | 8 | 121,868,551 | SNTB1 | G/T | 21,783 | 0.30 | 0.041 | 0.011 | 1.85 × 10−4 | 187,072 | 0.35 | 0.021 | 0.004 | 1.93 × 10−7 | 208,855 | 0.072 | 1.80 × 10−9 | 7.48 |
| HDL-C | rs1419980 | 12 | 7,774,892 | CD163-APOBEC1 | G/A | 32,933 | 0.32 | 0.048 | 0.008 | 2.06 × 10−8 | 94,311 | 0.07 | 0.030 | 0.010 | 5.27 × 10−4 | 127,244 | 0.003 | 1.45 × 10−8 | 8.14 |
| LDL-C | rs2239620 | 6 | 52,452,585 | TRAM2 | G/A | 31,737 | 0.73 | 0.023 | 0.009 | 1.46 × 10−2 | 89,888 | 0.63 | 0.027 | 0.005 | 5.59 × 10−7 | 121,625 | 0.644 | 4.02 × 10−8 | 6.52 |
| TC | rs2239619 | 6 | 52,453,220 | TRAM2 | A/C | 31,978 | 0.72 | 0.021 | 0.009 | 2.50 × 10−2 | 187,297 | 0.61 | 0.019 | 0.004 | 5.63 × 10−7 | 219,275 | 0.851 | 5.79 × 10−8 | 6.32 |
| TC | rs941408 | 19 | 2,814,181 | THOP1 | T/C | 19,042 | 0.46 | 0.035 | 0.013 | 5.67 × 10−3 | 180,459 | 0.29 | 0.020 | 0.004 | 1.71 × 10−6 | 199,501 | 0.229 | 8.39 × 10−8 | 6.52 |
| TG | rs1341267 | 13 | 95,284,980 | GPR180 | C/A | 27,672 | 0.68 | 0.026 | 0.010 | 9.80 × 10−3 | 175,109 | 0.42 | 0.018 | 0.003 | 8.30 × 10−7 | 202,781 | 0.445 | 2.75 × 10−8 | 7.36 |
| TG | rs8030477 | 15 | 73,085,815 | ADPGK | T/C | 27,664 | 0.25 | 0.031 | 0.010 | 2.52 × 10−3 | 174,898 | 0.69 | 0.018 | 0.004 | 5.76 × 10−6 | 202,562 | 0.251 | 1.22 × 10−7 | 6.52 |
| TG | rs12602912 | 17 | 65,870,073 | BPTF | T/C | 26,379 | 0.63 | 0.018 | 0.009 | 4.83 × 10−2 | 170,841 | 0.22 | 0.024 | 0.004 | 1.29 × 10−7 | 197,220 | 0.967 | 2.08 × 10−8 | 7.88 |
Trans-ancestry meta-analysis (MANTRA) combining fixed-effects meta-analysis summary statistics of up to 187,365 Europeans from the Global Lipids Genetics Consortium and up to 34,374 East Asians from Phase 2 of East Asian genome-wide meta-analysis. A log10(BF) ≥ 6.1 is defined as genome-wide significant. Results are unadjusted for BMI. Physical positions based on hg19. Effect alleles are associated with higher trait values. Beta estimates reflect per allele effects of variants on inverse normal transformed traits. P-values are obtained from fixed-effect meta-analysis within each ancestries, or combined across both ancestries. Phet represents the P-value for the Cochran's Q test for heterogeneity from fixed-effect meta-analysis.
Chr, chromosome; EA, effect allele; NEA, non-effect allele; EAF, effect allele frequency; SE, standard error; BF, Bayes factor; HDL-C, high-density lipoprotein cholesterol; TG, triglycerides.
Two of the novel loci identified in the East Asian HDL-C meta-analyses, CD163-APOBEC1 and NCOA2, were also identified in the trans-ancestry analyses with different lead variants (Table 2). The LD between the lead variants of NCOA2 (rs10504476 and rs6982451, East Asian LD r2=0.79) suggests the East Asian and trans-ancestry-identified variants are both part of the same signal. However, at CD163-APOBEC1, the lead East Asian variant is not in LD with the lead trans-ancestry-identified variant (rs10743940 and rs1419980, East Asian LD r2=0.02). Instead, the lead trans-ancestry variant is the lead variant for the second signal at this locus in the East Asian analysis after conditional analysis on the first signal. The locus association plots in East Asians, Europeans, and the trans-ancestry analyses (Fig. 2A–C) suggest that the first signal at CD163-APOBEC1 (lead variant: rs10743940) may be East-Asian-specific (MAF = 0.30), and the second signal (lead variant: rs1419980) is shared between the Europeans and East Asians. The allele frequency of rs1419980 is higher in the East Asians (MAF = 0.32) compared to Europeans (MAF = 0.07) and reached significance in East Asians (PadjBMI=1.9×10−8; PunadjBMI=5.4×10−8). No evidence of association was observed at rs10743940 in 94,169 Europeans (MAF = 0.03; P = 0.17), and European LD between the two lead variants was 0.02. The strongest association at this locus in Europeans was at rs7132326 near the second signal (MAF = 0.05; log10BF = 2.62; P = 9.92 × 10−5; European LD r2=0.03 and 0.66 with rs10743940 and rs1419980 respectively) (Fig. 2B). Among the ten loci and using a Bonferroni threshold (P < 0.05/10 = 0.005), only CD163-APOBEC1 exhibited evidence of heterogeneity (Phet=0.003), possibly due to the presence of the second signal in East Asians.
The remaining two novel signals identified in the East Asian meta-analysis near WDR11-FGFR2 (rs10886863: log10BF = 2.17; fixed-effect P-value = 2.23 × 10−3) and NID2-PTDGR (rs10144765: log10BF = 2.77; fixed-effect P-value = 1.60 × 10−3) were not significant in the trans-ancestry meta-analysis. Allele frequencies of the East Asian lead variants were notably lower in Europeans, and no evidence of association was observed in GLGC at either variant despite being well-powered to detect the association (rs10886863: MAF = 0.03, P = 0.13; rs10144765: MAF = 0.03, P = 0.21) (Supplementary Material, Table S11).
In addition to the trans-ancestry analysis of lipid traits unadjusted for BMI, we performed a sensitivity analysis by meta-analyzing the BMI-adjusted results from the East Asian meta-analysis with the BMI-unadjusted European results (Supplementary Material, Table S12). We identified nine loci that were also observed in the BMI-unadjusted analysis, although the index variant was different at one of the loci. Two additional loci were identified in the BMI-adjusted analysis. However, two loci identified in the original analysis were not found in the BMI-adjusted analysis.
Biological annotations
We investigated multiple resources to identify candidate genes at each of the novel loci from the East Asian and trans-ancestry meta-analyses (Table 3). Nonsynonymous variants in high LD (European and East Asian r2 > 0.80) with an index variant exist at two loci: the first (East Asian) signal at CD163-APOBEC1 (CD163, I324V) and the locus near ADPGK (BBS4, I182T). We examined eQTLs in blood, liver, and adipose and found two loci coincident (r2 > 0.80 in both East Asians and Europeans) with eQTLs in subcutaneous adipose (Table 3, Supplementary Material, Table S13). At eight loci, the index variants associated with a lipid trait showed at least nominal evidence of association in GWAS for other metabolic traits (Supplementary Material, Table S13). For example, the index variant at PDGFC, associated with HDL-C, is also nominally associated with type 2 diabetes and four glycemic traits, and the variant at WDR11-FGFR2, associated with TG, is also associated with adiponectin levels (P = 4.4 × 10−9) in East Asians. Seven traits across five genes met a multiple testing p-value threshold of 0.0033 (15 variants). In a manual literature search, we identified at least 1 potential candidate gene at nine loci (Table 3, Supplementary Material).
Table 3.
Biological candidate genes at novel loci
| Locus | Trait | Lead SNP | Chr | Position | Nearest Gene | Nearest Gene (kb) | Number of Genes within 100 kb | Literature Candidate | Nonsynonymous Variants (r2>0.8) | eQTL Gene (P<5×10−8) | Other Trait Associations of Similar Effect Direction(P < 0.05) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PDGFC | HDL-C | rs6822892 | 4 | 157,734,675 | PDGFC | 0.0 | 1 | PDGFC | FI*, HOMA-IR, 2hr G, 2hr I, T2D | ||
| CCND3-TAF8 | HDL-C | rs4714556 | 6 | 41,993,229 | CCND3 | 0.0 | 4 | 2hr I | |||
| NCOA2 | HDL-C | rs10504476 | 8 | 71,267,629 | NCOA2 | 0.0 | 1 | NCOA2 | |||
| SNTB1 | HDL-C | rs4871137 | 8 | 121,868,551 | SNTB1 | 44.2 | 1 | SNTB1 | FG, 2hr G, WHR* | ||
| CD163-APOBEC1 | HDL-C | rs10743940 | 12 | 7,651,138 | CD163 | 0.0 | 3 | APOBEC1 | CD163 (I342V) | ||
| CD163-APOBEC1 | HDL-C | rs1419980 | 12 | 7,774,892 | APOBEC1 | 27.1 | 3 | APOBEC1 | |||
| NID2-PTGDR | HDL-C | rs10144765 | 14 | 52,559,930 | NID2 | 24.1 | 2 | PTGDR, PTGER2 | |||
| TRAM2 | LDL-C, TC | rs2239620 | 6 | 52,452,585 | TRAM2-AS1 | 3.8 | 4 | ||||
| THOP1 | TC | rs941408 | 19 | 2,814,181 | THOP1 | 0.5 | 8 | S1PR4 | HOMA-B, SCC, UACR | ||
| WDR11-FGFR2 | TG | rs10886863 | 10 | 122,929,493 | WDR11 | 263.1 | 0 | FGFR2 | Adiponectin*, CAD | ||
| GPR180 | TG | rs1341267 | 13 | 95,284,980 | GPR180 | 1.9 | 5 | GPR180 | SBP | ||
| ADPGK | TG | rs8027181 | 15 | 73,088,869 | ADPGK-AS1 | 0.0 | 3 | BBS4 | BBS4 (I182T) | BMI*, FI*, HOMA-B, WHR | |
| BPTF | TG | rs12602912 | 17 | 65,870,073 | BPTF | 0.0 | 1 | PITPNC1 | BPTF | BMI*, HbA1c, T2D |
Biological candidate genes at each of the novel lipid loci identified in the East Asian and trans-ancestry meta-analyses. For loci associated with more than one trait, the lead variant was selected based on the trait with the smallest P-value. For loci detected in both East Asian and trans-ancestry analyses, the lead variant was selected from the East Asian meta-analysis results. Annotation of nearest gene and number of nearby genes were obtained from UCSC Genome Browser. Literature candidates were selected after manual investigation of genes within 100 kb of the index variant for biological relevance to lipids. Detailed decription of literature candidates and references can be found in Supplementary Text. eQTLs were identified in METSIM subcutaneous adipose (see methods). Detailed information about the METSIM eQTL results can be found in Supplementary Table 10. Detailed information about the other trait associations can be found in Supplementary Table 13. Physical positions based on hg19. For "Other Trait Associations", asterisks (*) indicate other traits that met a multiple testing threshold of 0.0033 (15 variants).
CAD, coronary artery disease; DBP, distolic blood pressure; FG, fasting glucose; FI, fasting insulin; G, glucose; I, insulin; MA, microalbuminuria; SBP, systolic blood pressure; SC, serum creatinine; SCC, serum cystatin C T2D, type 2 diabetes; UACR, urinary albumin-creatinine ratio; WHR, waist-hip ratio.
Discussion
This study represents the largest GWAS meta-analysis performed for cholesterol and triglyceride associations with individuals of East Asian ancestry to date. The two-phase East Asian meta-analysis identified three novel loci associated with HDL-C and one novel locus associated with TG. At one of the novel HDL-C loci, CD163-APOBEC1, we identified two signals, one of which is shared with Europeans. We also conducted a trans-ancestry meta-analysis of East Asians and Europeans identifying 10 novel loci associated with HDL-C, LDL-C, TC, and TG. Several of the identified loci harbor candidate genes based on eQTLs and literature review.
Despite prior studies with sample sizes of nearly 200,000 European individuals, four loci were identified in the East Asian meta-analysis that have not been described previously. In the European data, these lead variants were not nominally significant (all P > 0.13; Supplementary Material, Table S14). The loci at WDR11-FGFR2, NID2-PTDGR, NCOA2, and CD163-APOBEC1 may have been identified in East Asians because their allele frequencies are higher (MAF 0.27–0.40) compared to Europeans (MAF 0.03–0.13). The higher frequency among East Asians allowed us to detect these associations in a smaller sample size.
As expected, the trans-ancestry meta-analysis replicated most (151 of the 157) loci reported in the European data alone. Of the loci that did not replicate, most exhibited opposite direction of effects between East Asians and Europeans (ASAP3, ADH5, AMPD3, and FTO) (Supplementary Material, Table S9). Meta-analysis association results from multiple ancestries provide an opportunity to increase the overall power to detect novel loci (15,17–22). For all ten loci identified here, the P-values in the European samples were suggestive (P < 1×10−6), and the increase in sample size from the addition of the East Asians allowed these loci to be considered significant.
Our data suggested two essentially independent signals are present at the novel HDL-C locus, CD163-APOBEC1. The lead variant of the first signal, rs10743940, is located within an intron of CD163. The lead variant of the second signal, rs1419980, is located 124 kb downstream of the first signal, 71 kb from the 5’ end of CD163 and 27 kb from the 3’ end of APOBEC1. Both variants were significant in the East Asian meta-analysis, and the joint conditional P-value for both variants reached locus-wide significance (P < 1×10−5) at Pjoint = 4 × 10−6. The lead variant at the second signal is also the lead variant that arose from the trans-ancestry meta-analysis, suggesting the second signal is shared between Europeans and East Asians. Of note, the European lead variant in the GLGC dataset, rs7132326, is in moderate LD (European LD r2=0.66) with the lead variant of the second (shared) signal, rs1419980. While the first signal lies within CD163, a gene that encodes the CD163 hemoglobin scavenger receptor protein, the nearby gene, APOBEC1 (Apolipoprotein B mRNA Editing Enzyme Catalytic Subunit 1), is an excellent candidate gene for both signals. APOBEC1 is known to play a role in cholesterol control and metabolism, as knockout of the Apobec1 gene in mice resulted in a significant decrease in HDL-C levels (25,26). While we were able to detect two separate signals at this locus, these variants may still by partially tagged by a different variant not present in the HapMap reference panel. Larger sample sizes and dense imputation reference panels are needed to further characterize this locus.
At the novel TG-associated locus WDR11-FGFR2, nearby variants have previously been associated with adiponectin levels in East Asians (27). The lead variant for the adiponectin signal (rs3943077; P = 3.0×10−14) is located 15 kb from the lead TG-associated variant rs10886863, and the two variants exhibit weak LD (East Asian r2=0.23, D’=0.85). The lead variant alleles associated with lower TG levels are associated with higher adiponectin levels. The variants are located within a predicted enhancer region based on chromatin marks (28) 260 kb from the 3’ end of WDR11 and 308 kb from the 3’ end of FGFR2. FGFR2 encodes the fibroblast growth factor receptor 2 and is known to play a role in lipogenesis (29). FGFR2 is also highly expressed in adipose tissue where triglycerides are stored (30), suggesting FGFR2 as a better candidate gene at this locus. Another TG-associated locus, ADPGK, is located approximately 50kb downstream of BBS4, which has also been shown to play a role in the TG accumulation in adipocytes (31).
At least three additional loci discovered in our analysis harbor genes known to play roles in lipid metabolism. NCOA2 (nuclear receptor coactivator 2) is a member of the nuclear hormone receptor coactivator family (32) and is known to play an important role in both adipogenesis and lipid metabolism (33–36). Mice with a complete knockout of Ncoa2 demonstrated increased energy expenditure, an inability to store fat in the adipocytes, and an inability to absorb dietary fat from the gut (33). The mice also have significantly decreased levels of key regulatory enzymes required for fatty acid and cholesterol biosynthesis (35). Two variants within NCOA2 were recently reported to be nominally associated (P < 0.05) with obesity and dyslipidemia in 529 Chinese individuals (37). At another locus, SNTB1 encodes syntrophin beta 1, which interacts with ABCA1 (ATP-binding cassette transporter A1), a major regulator of cellular HDL-C (38). Additionally, rs4871137 was recently reported as a novel pleiotropic locus for lipids and inflammation (39). Finally, four hundred kb upstream of the BPTF locus, PITPNC1 encodes a phosphatidylinositol transfer protein and is known to have play several roles in lipid metabolism including the binding and transferring of phosphatidic acid (40), regulating the synthesis of phosphinositides (41), cellular distribution and metabolism of lipids (42).
G-protein coupled receptors (GPCR) are commonly used as therapeutic targets, particularly in the treatment of dyslipidemia (43). In our meta-analyses, we identified three loci at or near four genes known to encode GPCRs (GPR180, S1PR4, PTGDR, and PTDGER2). At the TG-associated locus, GPR180 encodes a GPCR in the rhodopsin family with a suggested role in vascular remodeling (44). S1PR4, an HDL-C-associated locus, encodes a sphingosine-1-phosphate receptor that binds lysophospholipids or lysphingolipids and is involved in endothelial differentiation (45). Finally, PTGD and PTGER are located approximately 174 kb downstream and 221 kb downstream, respectively, of HDL-associated variant rs10144765. PTGDR and PTGER2 encode the prostaglandin D2 and E2 receptors that are members of the guanine nucleotide-binding GPCR superfamily and mediate allergic and airway inflammation (46). Previously, this family of GPCRs has been used as a therapeutic target for coronary artery disease and atherosclerosis (47).
In conclusion, our East Asian GWAS meta-analysis for HDL-C, LDL-C, TC, and TG provides evidence for novel HDL-C loci near CD163-APOBEC1, PTGDR, and NCOA2, and a new TG locus near WDR11-FGFR2. The trans-ancestry meta-analysis also provides evidence for eight additional novel loci, including several with previously undiscovered roles in lipid biology. Additional loci will likely be discovered with the use of more dense reference panels, larger sample sizes, and larger trans-ancestry meta-analyses, further expanding our understanding of the genetic basis of cholesterol and triglyceride levels.
Materials and Methods
Study populations
The East Asian meta-analyses were performed with studies participating in the Asian Genetic Epidemiology Network (AGEN), a consortium of genetic epidemiology studies of type 2 diabetes and related traits conducted in individuals of East Asian ancestry (see URLs, Supplementary Methods). The first phase of the East Asian meta-analysis involved a three-stage design: i) meta-analysis of genome-wide association results of up to 25,164 Chinese, Japanese, Filipino, Korean, and Malay individuals from eleven GWAS (Phase 1 discovery); ii) in silico replication in five independent studies of up to 26,676 Japanese, Chinese, and Korean individuals (Phase 1 in silico), and iii) further de novo genotyping in up to 19,936 Japanese, Chinese, and Korean individuals from five additional studies (Phase 1 de novo) (Supplementary Material, Fig. S1 and Table S1). In Phase 2 of the East Asian analysis, we meta-analyzed the Phase 1 discovery data with newly available GWAS data from two additional studies with 10,805 Chinese individuals (BES and CHNS; combined n ≤ 34,421). Both phases of East Asian meta-analyses included 21 population-based and five case-control studies with a total of 69,414 individuals. In the trans-ancestry meta-analysis, we meta-analyzed the results from the larger, Phase 2 of the East Asian meta-analysis with the publicly available summary results for up to 187,365 individuals of European ancestry from the Global Lipids Genetic Consortium (GLGC) (4), for a combined total of up to 221,585 individuals. The lead variant at each novel locus identified from the East Asian and trans-ancestry meta-analyses was then evaluated in four independent studies of up to 10,886 East Asian individuals (MESA, TaiChi, TaiChi-G, and TUDR). Institutional review boards approved all study protocols at their respective sites, and written informed consent was obtained from all participants.
Genotyping, imputation, and quality control
Studies included in East Asian genome-wide discovery, and in silico replication were genotyped using commercially available Affymetrix or Illumina genome-wide genotyping arrays. Quality control criteria implemented in each study, including variant call rate and Hardy-Weinberg equilibrium (HWE), were summarized in Supplementary Material, Table S2. We present principal component plots with either HapMap or 1000 Genomes Project for a subset of the studies, representing each of the major East Asian populations from China, Japan Korea, Philippines, and Singapore (Supplementary Material, Fig. S6). All studies performed genotype imputation to HapMap Project Phase II reference panel (JPT + CHB for all studies except CLHNS, which used JPT + CHB + CEU, and SiMES, which used JPT + CHB + CEU + YRI) of ∼2.4 million variants using Beagle (48), IMPUTE (49), or MACH (50). The two newly available GWAS, CHNS and BES, were imputed to the 1000 Genomes Project Phase 3 reference panel (51) using the University of Michigan imputation server (52). Only variants included in the HapMap reference panel were included in the meta-analyses. Variants with poor imputation quality (IMPUTE or BEAGLE quality score < 0.50 or MACH r2 < 0.30) or with a minor allele frequency < 5% were excluded from the association analyses. De novo genotyping for variants that showed compelling evidence of association from the Phase 1 discovery and in silico meta-analysis was performed in five additional studies using the Sequenom MassARRAY iPLEX system or TaqMan assays (Supplementary Material, Table S1). All reported European and East Asian LD information was calculated based on the 1000 Genomes Phase 3 reference panel using LDLink. East Asian and European minor allele frequencies were drawn from the East Asian Phase 2 and GLGC respectively.
Phenotype measurements
Plasma blood lipid levels were measured by standard biochemical methods in mg/dl. Studies with fasting blood lipids contributed data to all four lipid analyses, while non-fasting studies contributed to the HDL-C and TC analyses only. If both fasting and non-fasting samples were available, analyses were performed separately for fasting and non-fasting samples. Directly measured LDL-C levels were used when available; otherwise, we estimated LDL-C using Friedewald formula only on fasting samples: if TG ≤400 mg/dl, LDL-C = TC – HDL-C – (TG/5); if TG > 400, LDL-C values were set to missing (53). Individuals on lipid lowering medications were excluded from all analyses.
Statistical analysis and variant prioritization
Association analyses within each cohort . In each individual study, residuals for each lipid trait were computed by including age, age2, sex, and other study-specific covariates (e.g. principal components, sample recruitment sites) in a linear regression model (Supplementary Material, Table S2). In a second model, BMI was included as an additional covariate. The residuals were then inverse-normalized to approximate a normal distribution with a mean value of 0 and standard deviation of 1. For studies ascertained based on case or control status, trait transformation was computed in cases and controls separately. Linear regression models assuming an additive mode of inheritance were applied to test for genotype-phenotype association with normalized residuals. EastAsianphase 1 and 2 GWAS discovery meta-analyses. The meta-analyses for associations between the four lipid traits with ∼2.4 million variants were performed by two independent analysts, each using Stouffer sample-size weighted fixed effects meta-analysis implemented in METAL (54). We used a fixed effects meta-analysis as this approach assumes the same underlying allelic effects on disease susceptibility and is more appropriate among homogeneous studies from the same ancestry (16). Study-specific summary statistics were corrected using genomic-control (λGCranges 0.989 – 1.102) prior to meta-analysis; overall meta-analysis results were also corrected for genomic control (Supplementary Material, Table S1). Cochran’s Q-test was used to assess for the presence of heterogeneity. We obtained effect sizes by performing an inverse-variance weighted meta-analysis on study-specific inverse normal transformed trait values. The same protocol was used in the second phase of the East Asian meta-analysis where the eleven Phase 1 discovery studies were combined with two newly available cohorts (BES and CHNS). East Asian Phase 1 in silico replication. We selected 3,870 variants with P-value < 1 × 10−3 in either model (with and without BMI adjustment) from the Phase 1 discovery meta-analysis to follow-up in 26,676 individuals from five GWAS studies. The P-value threshold was determined from visually observing the quantile-quantile (Q-Q) plots for each trait after excluding known loci (Supplementary Material, Fig. S2). For uniformity, we applied the same threshold to all four lipid traits. To prioritize variants for in silico replication, we used the ‘-clump’ command in PLINK (55) using the HapMap CHB + JPT as reference, and setting an East Asian LD threshold r2 < 0.10 and a physical distance of 1 Mb. We then meta-analyzed study summary statistics from the in silico replication with the data from the eleven studies in Phase 1. EastAsianphase 1 de novo genotyping. For the 79 lead variants that showed suggestive evidence of association (P < 5 × 10−6) in the East Asian Phase 1 discovery and in silico meta-analysis, we compared the physical locations to known index variants collected from literature (2,4,9) and considered variants to be at an established locus if they were located within 1 Mb and/or in LD (r2 ≥ 0.1 using HapMap JPT + CHB) of a reported index variant. To prioritize variants for de novo genotyping, we grouped variants into six tiers: (1) variants at loci not previously reported to be associated with any of the four lipid traits; (2) variants at loci previously reported to be associated with a different lipid trait; (3) variants at previously known loci, but in low LD (r2 < 0.10) with the published index variant(s); (4) variants at previously known loci and in LD (r2 ≥ 0.10) with an index variant identified in a study of European participants; (5) variants at loci discovered (P < 5×10−8) after meta-analysis of the discovery and in silico stages; (6) or variants in the Major Histocompatibility Complex (MHC) region. When multiple variants at a locus met these tiers, variants with larger discovery sample sizes or known variant consequences were selected. Constrained by resources and the capacity of de novo genotyping, the top 36 variants (27 from Tier 1, 5 from Tier 2, and 4 from Tier 3) were selected for de novo genotyping in 19,936 individuals from five studies. We combined study-level summary statistics from the Phase 1 discovery, in silico, and de novo stages in a joint meta-analysis.
Trans-ancestry meta-analysis
For the trans-ancestry meta-analysis, we meta-analyzed the larger East Asian Phase 2 genome-wide summary statistics (n ≤ 34,421) with European summary statistics from GLGC using the Meta-ANalysis of Trans-ethnic Association Studies (MANTRA) software (16). We defined log10BF ≥ 6.1 as genome-wide significant, approximately comparable to P < 5 × 10−8 (56).
Conditional analysis
To test for multiple signals at the four novel loci identified in the East Asian meta-analysis, we carried out approximate conditional analysis using summary level association statistics as implemented in Genome-Wide Complex Trait (GCTA) program (24). We used allele frequency and LD estimates from the largest Chinese study, CHNS, to approximate the reference variance-covariance matrix of genotypes. Analyses were also conducted using the SCES study to approximate the reference variance-covariance genotype matrix; results were similar (data not shown).
Further evaluation in four additional East Asian studies
Four additional East Asian cohorts became available for further examination of the novel loci: MESA, TaiChi, TaiChi-G, and TUDR (Supplementary Material, Table S2). Each study provided BMI-adjusted or BMI-unadjusted results for the index variant at each novel locus that exhibited significant results in either the East Asian or trans-ancestry meta-analyses. Results for up to 10,857 individuals were available after meta-analysis (Supplementary Material, Table S8).
Associations with other metabolic traits and outcomes
We used the Type 2 Diabetes Knowledge Portal (57) to explore associations of the newly identified loci with other metabolic traits and outcomes. Association summary statistics from the following consortia: coronary artery disease from CARDIoGRAM (58), kidney-related traits from CKDGen (59), type 2 diabetes from DIAGRAM (60), BMI and waist-to-hip ratio from GIANT (61,62), and glycemic traits from MAGIC (63–66) were available for query on the portal (last assessed August 8, 2016). Additionally, we used available data from AGEN East Asian adiponectin (27) and ICBP-GWAS (systolic and diastolic blood pressure) (67). Effect sizes for each data set were obtained from publicly available summary statistic files.
Exploration of expression quantitative trait loci (eQTL) at novel loci
We searched publicly available eQTL databases, GTEx (68,69), the Blood eQTL Browser (70), and the University of Chicago eQTL browser (71) to identify cis-eQTLs at the observed loci in blood, liver, and adipose tissue. We also searched for cis-eQTLs in subcutaneous adipose tissue data from the METSIM study (72). Coincident eQTLs were identified in the METSIM data if the lead eSNP and the GWAS index variant were in high LD (r2>0.80) in both Europeans and East Asians to accommodate the eQTL source and the European ancestry composition of the trans-ancestry data. Reciprocal conditional analyses were also performed using the METSIM data to determine if the GWAS index variant and the lead eSNP were part of the same eQTL signal.
Determining the most biologically relevant gene(s) at each novel locus
We used SNIPPER (see URLs) to identify nearby genes 500 kb upstream or downstream of the index variant at each of the novel loci. SNIPPER returns information about each gene from NCBI Entrez Gene, Gene Ontology terms, KEGG pathways, and PubMed. We included the following keywords as search terms for PubMed: lipid, cholesterol, triglyceride, and metabolic. After analyzing results from SNIPPER, we then conducted an additional traditional literature review through PubMed and OMIM to identify candidate genes. A gene was considered a potential candidate if there was an apparent link to lipid biology, based on prior studies of that particular gene.
Web Resources
Summary statistics for the East Asian meta-analyses are available at: https://blog.nus.edu.sg/agen/summary-statistics/
AGEN, http://blog.nus.edu.sg/agen/
IMPUTE, http://mathgen.stats.ox.ac.uk/impute/impute_v2.html
MACH, http://www.sph.umich.edu/csg/abecasis/MACH/inde x. html
Beagle, http://faculty.washington.edu/browning/beagle/beagle. html
University of Michigan Imputation Server, https://imputationserver.sph.umich.edu/index.html
SNPTEST, https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html
PLINK, https://www.cog-genomics.org/plink2
ProbABEL, http://www.genabel.org/
GCTA, http://cnsgenomics.com/software/gcta/
SAS, http://www.sas.com/software/sas9/
LDLink, http://analysistools.nci.nih.gov/LDlink/
Type 2 Diabetes Knowledge Portal, http://www.type2diabetesgenetics.org/
ICBP-GWAS data, http://locuszoom.sph.umich.edu/locus zoom/
GTEx, http://www.gtexportal.org/home/
Blood eQTL Browser, http://genenetwork.nl/bloodeqtl brow ser/
University of Chicago eQTL Browser, http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/
HaploReg, http://www.broadinstitute.org/mammals/haploreg/haploreg.php
SNIPPER, http://csg.sph.umich.edu/boehnke/snipper
NCBI Entrez Gene, http://www.ncbi.nlm.nih.gov/gene
Gene Ontology, http://geneontology.org/
KEGG Pathways, http://www.genome.jp/kegg/pathway.html
PubMed, http://www.ncbi.nlm.nih.gov/pubmed
OMIM, http://www.ncbi.nlm.nih.gov/omim All URLs were last accessed on March 1, 2017.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors thank all investigators, staff members, and study participants for their contributions to all the participating studies. We are grateful to the NUH Tissue Repository and the Genome Institute of Singapore, Agency for Science, Technology and Research, Singapore which provided services for tissue archival and genotyping (SCES, SiMES, SP2, SCHS-MI), Siew-Hong Low (National University of Singapore) for supervising fieldwork and Renwei Wang for maintenance SCHS database, and SCHS founding Principal Investigator, Mimi C. Yu. We also thank Andrew P. Morris for his advice with MANTRA analyses.
Conflict of Interest statement. None declared.
Funding
The Beijing Eye Study (BES) was funded by National Natural Science Foundation of China (NSFC, No. 81170890 and 81570835). The CAGE Network Studies were supported by grants for the Core Research for Evolutional Science and Technology (CREST) from the Japan Science Technology Agency; the Program for Promotion of Fundamental Studies in Health Sciences, National Institute of Biomedical Innovation Organization (NIBIO); and the Grant of National Center for Global Health and Medicine (NCGM). The China Health and Nutrition Survey (CHNS) was supported by the China National Institute of Nutrition and Food Safety; the China Center for Disease Control; the National Institutes of Health (R01HD30880, R01HL108427, and R01DK104371); the Fogarty International Center of the National Institutes of Health; the China-Japan Friendship Hospital; the Chinese Ministry of Health; and the Carolina Population Center (R24 HD050924). The Cebu Longitudinal Health and Nutrition Survey (CLHNS) was supported by National Institutes of Health grants DK078150, TW005596 and HL085144; pilot funds from RR020649, ES010126, and DK056350; and the Office of Population Studies Foundation. The Genetic Epidemiology Network of Salt-Sensitivity (GenSalt) study was supported by research grants (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung, and Blood Institute, National Institutes of Health, and partially supported by the Collins C. Diboll Private Foundation, New Orleans, LA, USA. The Korean Association Resource (KARE) study was supported by grants from Korea Centers for Disease Control and Prevention (4845-301, 4851-302, 4851-307) and an intramural grant from the Korea National Institute of Health (2016-NI73001-00). The Korean-Cancer Prevention Study-II (KCPSII) was funded by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (HI14C2686). The Shanghai Breast Cancer Study (SBCS) was supported by grants from the US National Institutes of Health (R01CA124558, R01CA064277, and R01CA090899). The Singapore Chinese Eye Study (SCES), and Singapore Malay Eye Study (SiMES) are supported by the National Medical Research Council (NMRC), Singapore (grants 0796/2003, 1176/2008, 1149/2008, STaR/0003/2008, 1249/2010, CG/SERI/2010, CIRG/1371/2013, and CIRG/1417/2015), and Biomedical Research Council (BMRC), Singapore (08/1/35/19/550 and 09/1/35/19/616). The Singapore Prospective Study Program (SP2) was supported by the individual research grant and clinician scientist award schemes from the National Medical Research Council (NMRC) and the Biomedical Research Council (BMRC) of Singapore. The Shanghai Women's Health Study and Shanghai Men's Health Studies (SWMHS) were supported by grants from the US National Institutes of Health (R37CA070867, R01CA082729, UM1CA182910, UM1CA173640). GWAS were supported by R37CA070867, R01CA082729, R01CA124558, R01CA148667, R01CA122364, R01CA122756, and R01CA137013, as well as Ingram Professorship and Research Reward funds from the Vanderbilt University School of Medicine. Sample preparation and genotyping assays at Vanderbilt were conducted at the Survey and Biospecimen Shared Resources and Vanderbilt Microarray Shared Resource, which were supported in part by Vanderbilt-Ingram Cancer Center (P30CA068485). The Taiwan Super Control Study (TWSC) was supported by a grant from Academia Sinica, Taipei, Taiwan (IBMS 40-05-GMM). The Dongfeng-Tongji cohort study (DFTJ-cohort) was supported by the grant from the National Natural Science Foundation (grant NSFC-81230069 to T. Wu, grant NSFC-81473051 and 81522040 to M. He). The Health Examinee (HEXA) was supported by grants from Korea Centers for Disease Control and Prevention (4845-301, 4851-302, 4851-307) and an intramural grant from the Korea National Institute of Health (2016-NI73001-00). The CAGE Amagasaki Study was supported in part by grants from SENSHIN Medical Research Foundation. Health2 was supported by grants from Korea Centers for Disease Control and Prevention (4845-301, 4851-302, 4851-307) and an intramural grant from the Korea National Institute of Health (2016-NI73001-00). The Nutrition and Health of Aging Population in China (NHAPC) study was supported by the Major Project of the Ministry of Science and Technology of China (2016YFC1304903), the National Natural Science Foundation of China (81471013, 30930081, 81321062, 81170734 and 81471013), and the Chinese Academy of Sciences (KJZD-EW-L14). The Singapore Chinese Health Study (SCHS) was supported by the National Institutes of Health, USA (RO1CA144034 and UM1CA182876), the nested case-control study of myocardial infarction by the Singapore National Medical Research Council (NMRC 1270/2010) and genotyping by the HUJ-CREATE Programme of the National Research Foundation, Singapore (Project Number 370062002). The Shanghai Diabetes Inpatient Database (SDIID) was supported by National Science Foundation of China (81322010). The work at Shanghai Jiao Tong University Affiliated Sixth People’s Hospital for Shanghai Diabetes Inpatient Database (SDIID) and Shanghai Diabetes Study (SHDS) was supported by grants from National 973 Program (2011CB504001), NSFC (81322010 and 81170735), Shanghai Rising Star Program (12QH1401700), Shanghai Talent Development Grant (2012041), Excellent Young Medical Expert of Shanghai (XYQ2011041) and National Young Top Talent Support Program. The Suzhou Metabolic Syndrome Study (SMSS) was supported by The National Natural Science Foundation of China (81320108026), and is a Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions, China. The Multi-Ethnic Study of Atherosclerosis study (MESA) was supported by the Multi-Ethnic Study of Atherosclerosis (MESA) contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169 and by grants UL1-TR-000040 and UL1-RR-025005 from NCRR. Funding for MESA SHARe genotyping was provided by NHLBI Contract N02-HL-6-4278. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The TaiChi consortium (TaiChi) study was supported by the National Health Research Institutes, Taiwan (PH-100-SP-01, BS-094-PP-01, PH-100-PP-03), the National Science Council, Taiwan (Grant Nos NSC 98-2314-B-075A-002-MY3, NSC 96-2314-B-002-151, NSC 96-2314-B-002-152, NSC 98-2314-B-002-122-MY2, NSC 100-2314-B-002-115, NSC 101-2325-002-078, 101-2314-B-075A-006-MY3), the National Taiwan University Hospital, Taiwan (NTUH 98-N1266, NTUH 100-N1775, NTUH 101-N2010, NTUH 101-N, VN101-04, NTUH 101-S1784). The TaiChi-G study was supported by the National Health Research Institutes, Taiwan (PH-100-SP-01, BS-094-PP-01, PH-100-PP-03), the National Science Council, Taiwan (Grant Nos NSC 98-2314-B-075A-002-MY3, NSC 96-2314-B-002-151, NSC 96-2314-B-002-152, NSC 98-2314-B-002-122-MY2, NSC 100-2314-B-002-115, NSC 101-2325-002-078, 101-2314-B-075A-006-MY3), the National Taiwan University Hospital, Taiwan (NTUH 98-N1266, NTUH 100-N1775, NTUH 101-N2010, NTUH 101-N, VN101-04, NTUH 101-S1784). The Taiwan-US Diabetic Retinopathy (TUDR) study was supported by the National Eye Institute of the National Institutes of Health (EY014684 to J.I.R. and Y.-D.I.C.) and ARRA Supplement (EY014684-03S1, -04S1), the Eye Birth Defects Foundation Inc., the National Science Council, Taiwan (MOST 104-2314-B-075A-006 -MY3 to W.H.S., MOST 104-2314-B-075A-007 to I.T.L.) and the Taichung Veterans General Hospital, Taichung, Taiwan (TCVGH-HK1028001, TCVGH-HK1048002, TCVGH-HK1058001 to W.J.L ., TCVGH-VHCY1058605 to K.H.L., TCVGH-VTA105T4-3 to I.T.L.). DNA handling and genotyping were supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881 and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. Cassandra N. Spracklen was supported by the American Heart Association Postdoctoral Fellowship (15POST24470131). Peng Chen was supported by Changbai Mountain Scholar Award Program of Jilin Province China, and National Recruitment Program for Young Professionals of the Thousand Talents Plan China. Shengxu Li was partly supported by grant 13SDG14650068 from American Heart Association. Maren E. Cannon was supported by National Institutes of Health F31 HL127984. Tamara S. Roman was supported by National Institutes of Health R01 DK072193. Ching-Yu Cheng was supported by an award from NMRC (CSA/033/2012). Yoon Shin Cho acknowledged support from the National Research Foundation of Korea (NRF) grant (NRF-2012M3A9D1054534).
References
- 1. Wannamethee G., Shaper A.G. (1994) The association between heart rate and blood pressure, blood lipids and other cardiovascular risk factors. J. Cardiovasc. Risk, 1, 223–230. [DOI] [PubMed] [Google Scholar]
- 2. Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature, 466, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asselbergs F.W., Guo Y., van Iperen E.P., Sivapalaratnam S., Tragante V., Lanktree M.B., Lange L.A., Almoguera B., Appelman Y.E., Barnard J., et al. (2012) Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am. J. Hum. Genet., 91, 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S., et al. (2013) Discovery and refinement of loci associated with lipid levels. Nat. Genet., 45, 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peloso G.M., Auer P.L., Bis J.C., Voorman A., Morrison A.C., Stitziel N.O., Brody J.A., Khetarpal S.A., Crosby J.R., Fornage M., et al. (2014) Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am. J. Hum. Genet., 94, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Surakka I., Horikoshi M., Magi R., Sarin A.P., Mahajan A., Lagou V., Marullo L., Ferreira T., Miraglio B., Timonen S., et al. (2015) The impact of low-frequency and rare variants on lipid levels. Nat. Genet., 47, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Leeuwen E.M., Karssen L.C., Deelen J., Isaacs A., Medina-Gomez C., Mbarek H., Kanterakis A., Trompet S., Postmus I., Verweij N., et al. (2015) Genome of The Netherlands population-specific imputations identify an ABCA6 variant associated with cholesterol levels. Nat. Commun., 6, 6065.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kamatani Y., Matsuda K., Okada Y., Kubo M., Hosono N., Daigo Y., Nakamura Y., Kamatani N. (2010) Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet., 42, 210–215. [DOI] [PubMed] [Google Scholar]
- 9. Kim Y.J., Go M.J., Hu C., Hong C.B., Kim Y.K., Lee J.Y., Hwang J.Y., Oh J.H., Kim D.J., Kim N.H., et al. (2011) Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat. Genet., 43, 990–995. [DOI] [PubMed] [Google Scholar]
- 10. Tan A., Sun J., Xia N., Qin X., Hu Y., Zhang S., Tao S., Gao Y., Yang X., Zhang H., et al. (2012) A genome-wide association and gene-environment interaction study for serum triglycerides levels in a healthy Chinese male population. Hum. Mol. Genet., 21, 1658–1664. [DOI] [PubMed] [Google Scholar]
- 11. Wu Y., Marvelle A.F., Li J., Croteau-Chonka D.C., Feranil A.B., Kuzawa C.W., Li Y., Adair L.S., Mohlke K.L. (2013) Genetic association with lipids in Filipinos: waist circumference modifies an APOA5 effect on triglyceride levels. J. Lipid Res., 54, 3198–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou L., He M., Mo Z., Wu C., Yang H., Yu D., Yang X., Zhang X., Wang Y., Sun J., et al. (2013) A genome wide association study identifies common variants associated with lipid levels in the Chinese population. PLoS One, 8, e82420.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang C.S., Zhang H., Cheung C.Y., Xu M., Ho J.C., Zhou W., Cherny S.S., Zhang Y., Holmen O., Au K.W., et al. (2015) Exome-wide association analysis reveals novel coding sequence variants associated with lipid traits in Chinese. Nat. Commun., 6, 10206.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kao J.T., Wen H.C., Chien K.L., Hsu H.C., Lin S.W. (2003) A novel genetic variant in the apolipoprotein A5 gene is associated with hypertriglyceridemia. Hum. Mol. Genet., 12, 2533–2539. [DOI] [PubMed] [Google Scholar]
- 15. Wu Y., Waite L.L., Jackson A.U., Sheu W.H., Buyske S., Absher D., Arnett D.K., Boerwinkle E., Bonnycastle L.L., Carty C.L., et al. (2013) Trans-ethnic fine-mapping of lipid loci identifies population-specific signals and allelic heterogeneity that increases the trait variance explained. PLoS Genet., 9, e1003379.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morris A.P. (2011) Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol., 35, 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keller M.F., Reiner A.P., Okada Y., van Rooij F.J., Johnson A.D., Chen M.H., Smith A.V., Morris A.P., Tanaka T., Ferrucci L., et al. (2014) Trans-ethnic meta-analysis of white blood cell phenotypes. Hum. Mol. Genet., 23, 6944–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu C.T., Buchkovich M.L., Winkler T.W., Heid I.M. African Ancestry Anthropometry Genetics Consortium, GIANT Consortium Borecki I.B., Fox C.S., Mohlke K.L., North K.E., et al. (2014) Multi-ethnic fine-mapping of 14 central adiposity loci. Hum. Mol. Genet., 23, 4738–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mahajan A., Go M.J., Zhang W., Below J.E., Gaulton K.J., Ferreira T., Horikoshi M., Johnson A.D., Ng M.C., Prokopenko I., et al. (2014) Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet., 46, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu Y., Li H., Lu L., Manichaikul A., Zhu J., Chen Y.D., Sun L., Liang S., Siscovick D.S., Steffen L.M., et al. (2016) Genome-wide meta-analyses identify novel loci associated with n-3 and n-6 polyunsaturated fatty acid levels in Chinese and European-ancestry populations. Hum. Mol. Genet., 25, 1215–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fuchsberger C., Flannick J., Teslovich T.M., Mahajan A., Agarwala V., Gaulton K.J., Ma C., Fontanillas P., Moutsianas L., McCarthy D.J., et al. (2016) The genetic architecture of type 2 diabetes. Nature, 536, 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu C.T., Raghavan S., Maruthur N., Kabagambe E.K., Hong J., Ng M.C., Hivert M.F., Lu Y., An P., Bentley A.R., et al. (2016) Trans-ethnic Meta-analysis and Functional Annotation Illuminates the Genetic Architecture of Fasting Glucose and Insulin. Am. J. Hum. Genet., 99, 56–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Below J.E., Parra E.J., Gamazon E.R., Torres J., Krithika S., Candille S., Lu Y., Manichakul A., Peralta-Romero J., Duan Q., et al. (2016) Meta-analysis of lipid-traits in Hispanics identifies novel loci, population-specific effects, and tissue-specific enrichment of eQTLs. Sci. Rep., 6, 19429.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang J., Ferreira T., Morris A.P., Medland S.E. GIANT Consortium, DIAGRAM Consortium Madden P.A., Heath A.C., Martin N.G., et al. (2012) Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet., 44, 369–375. S361-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakamuta M., Chang B.H., Zsigmond E., Kobayashi K., Lei H., Ishida B.Y., Oka K., Li E., Chan L. (1996) Complete phenotypic characterization of apobec-1 knockout mice with a wild-type genetic background and a human apolipoprotein B transgenic background, and restoration of apolipoprotein B mRNA editing by somatic gene transfer of Apobec-1. J. Biol. Chem., 271, 25981–25988. [DOI] [PubMed] [Google Scholar]
- 26. Rosenberg B.R., Hamilton C.E., Mwangi M.M., Dewell S., Papavasiliou F.N. (2011) Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3' UTRs. Nat. Struct. Mol. Biol., 18, 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu Y., Gao H., Li H., Tabara Y., Nakatochi M., Chiu Y.F., Park E.J., Wen W., Adair L.S., Borja J.B., et al. (2014) A meta-analysis of genome-wide association studies for adiponectin levels in East Asians identifies a novel locus near WDR11-FGFR2. Hum. Mol. Genet., 23, 1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature, 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Konishi M., Nakamura H., Miwa H., Chambon P., Ornitz D.M., Itoh N. (2008) Role of Fgf receptor 2c in adipocyte hypertrophy in mesenteric white adipose tissue. Mol. Cell Endocrinol., 287, 13–19. [DOI] [PubMed] [Google Scholar]
- 30. Yang C., Wang C., Ye M., Jin C., He W., Wang F., McKeehan W.L., Luo Y. (2012) Control of lipid metabolism by adipocyte FGFR1-mediated adipohepatic communication during hepatic stress. Nutr. Metab. (Lond), 9, 94.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aksanov O., Green P., Birk R.Z. (2014) BBS4 directly affects proliferation and differentiation of adipocytes. Cell Mol. Life Sci., 71, 3381–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mangelsdorf D.J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., et al. (1995) The nuclear receptor superfamily: the second decade. Cell, 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chopra A.R., Kommagani R., Saha P., Louet J.F., Salazar C., Song J., Jeong J., Finegold M., Viollet B., DeMayo F., et al. (2011) Cellular energy depletion resets whole-body energy by promoting coactivator-mediated dietary fuel absorption. Cell Metab., 13, 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Duteil D., Chambon C., Ali F., Malivindi R., Zoll J., Kato S., Geny B., Chambon P., Metzger D. (2010) The transcriptional coregulators TIF2 and SRC-1 regulate energy homeostasis by modulating mitochondrial respiration in skeletal muscles. Cell Metab., 12, 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jeong J.W., Kwak I., Lee K.Y., White L.D., Wang X.P., Brunicardi F.C., O'Malley B.W., DeMayo F.J. (2006) The genomic analysis of the impact of steroid receptor coactivators ablation on hepatic metabolism. Mol. Endocrinol., 20, 1138–1152. [DOI] [PubMed] [Google Scholar]
- 36. Wang Z., Qi C., Krones A., Woodring P., Zhu X., Reddy J.K., Evans R.M., Rosenfeld M.G., Hunter T. (2006) Critical roles of the p160 transcriptional coactivators p/CIP and SRC-1 in energy balance. Cell Metab., 3, 111–122. [DOI] [PubMed] [Google Scholar]
- 37. Lu Y., Habtetsion T.G., Li Y., Zhang H., Qiao Y., Yu M., Tang Y., Zhen Q., Cheng Y., Liu Y. (2015) Association of NCOA2 gene polymorphisms with obesity and dyslipidemia in the Chinese Han population. Int. J. Clin. Exp. Pathol., 8, 7341–7349. [PMC free article] [PubMed] [Google Scholar]
- 38. Okuhira K., Fitzgerald M.L., Sarracino D.A., Manning J.J., Bell S.A., Goss J.L., Freeman M.W. (2005) Purification of ATP-binding cassette transporter A1 and associated binding proteins reveals the importance of beta1-syntrophin in cholesterol efflux. J. Biol. Chem., 280, 39653–39664. [DOI] [PubMed] [Google Scholar]
- 39. Ligthart S., Vaez A., Hsu Y.H. Inflammation Working Group of the, C.C. Pmi Wg X.C.P., LifeLines Cohort S., Stolk R., Uitterlinden A.G., Hofman A., Alizadeh B.Z., et al. (2016) Bivariate genome-wide association study identifies novel pleiotropic loci for lipids and inflammation. BMC Genomics, 17, 443.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garner K., Hunt A.N., Koster G., Somerharju P., Groves E., Li M., Raghu P., Holic R., Cockcroft S. (2012) Phosphatidylinositol transfer protein, cytoplasmic 1 (PITPNC1) binds and transfers phosphatidic acid. J. Biol. Chem., 287, 32263–32276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cockcroft S. (2012) The diverse functions of phosphatidylinositol transfer proteins. Curr. Top Microbiol. Immunol., 362, 185–208. [DOI] [PubMed] [Google Scholar]
- 42. Balla T. (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev., 93, 1019–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Overington J.P., Al-Lazikani B., Hopkins A.L. (2006) How many drug targets are there?. Nat. Rev. Drug Discov., 5, 993–996. [DOI] [PubMed] [Google Scholar]
- 44. Tsukada S., Iwai M., Nishiu J., Itoh M., Tomoike H., Horiuchi M., Nakamura Y., Tanaka T. (2003) Inhibition of experimental intimal thickening in mice lacking a novel G-protein-coupled receptor. Circulation, 107, 313–319. [DOI] [PubMed] [Google Scholar]
- 45. Van Brocklyn J.R., Graler M.H., Bernhardt G., Hobson J.P., Lipp M., Spiegel S. (2000) Sphingosine-1-phosphate is a ligand for the G protein-coupled receptor EDG-6. Blood, 95, 2624–2629. [PubMed] [Google Scholar]
- 46. Shiraishi Y., Asano K., Nakajima T., Oguma T., Suzuki Y., Shiomi T., Sayama K., Niimi K., Wakaki M., Kagyo J., et al. (2005) Prostaglandin D2-induced eosinophilic airway inflammation is mediated by CRTH2 receptor. J. Pharmacol. Exp. Ther., 312, 954–960. [DOI] [PubMed] [Google Scholar]
- 47. Ishizuka T., Matsui T., Okamoto Y., Ohta A., Shichijo M. (2004) Ramatroban (BAY u 3405): a novel dual antagonist of TXA2 receptor and CRTh2, a newly identified prostaglandin D2 receptor. Cardiovasc. Drug Rev., 22, 71–90. [DOI] [PubMed] [Google Scholar]
- 48. Browning B.L., Browning S.R. (2009) A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet., 84, 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marchini J., Howie B., Myers S., McVean G., Donnelly P. (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet., 39, 906–913. [DOI] [PubMed] [Google Scholar]
- 50. Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol., 34, 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fuchsberger C., Abecasis G.R., Hinds D.A. (2015) minimac2: faster genotype imputation. Bioinformatics, 31, 782–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Friedewald W.T., Levy R.I., Fredrickson D.S. (1972) Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem., 18, 499–502. [PubMed] [Google Scholar]
- 54. Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang X., Chua H.X., Chen P., Ong R.T., Sim X., Zhang W., Takeuchi F., Liu X., Khor C.C., Tay W.T., et al. (2013) Comparing methods for performing trans-ethnic meta-analysis of genome-wide association studies. Hum. Mol. Genet., 22, 2303–2311. [DOI] [PubMed] [Google Scholar]
- 57.T2D-GENES Consortium, GoT2D Consortium, DIAGRAM Consortium. Assessed 2016 December 1. http://www.type2diabetesgenetics.org/home/portalHome
- 58. Schunkert H., Konig I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C., et al. (2011) Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet., 43, 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kottgen A., Pattaro C., Boger C.A., Fuchsberger C., Olden M., Glazer N.L., Parsa A., Gao X., Yang Q., Smith A.V., et al. (2010) New loci associated with kidney function and chronic kidney disease. Nat. Genet., 42, 376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segre A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., et al. (2012) Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet., 44, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., et al. (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature, 518, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Magi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E., et al. (2015) New genetic loci link adipose and insulin biology to body fat distribution. Nature, 518, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., et al. (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet., 42, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saxena R., Hivert M.F., Langenberg C., Tanaka T., Pankow J.S., Vollenweider P., Lyssenko V., Bouatia-Naji N., Dupuis J., Jackson A.U., et al. (2010) Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet., 42, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Soranzo N., Sanna S., Wheeler E., Gieger C., Radke D., Dupuis J., Bouatia-Naji N., Langenberg C., Prokopenko I., Stolerman E., et al. (2010) Common variants at 10 genomic loci influence hemoglobin A(1)(C) levels via glycemic and nonglycemic pathways. Diabetes, 59, 3229–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Strawbridge R.J., Dupuis J., Prokopenko I., Barker A., Ahlqvist E., Rybin D., Petrie J.R., Travers M.E., Bouatia-Naji N., Dimas A.S., et al. (2011) Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes, 60, 2624–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Huang S.J., et al. (2011) Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature, 478, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. GTEx Consortium (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mele M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J., et al. (2015) Human genomics. The human transcriptome across tissues and individuals. Science, 348, 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet., 45, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. (2008) Mapping the genetic architecture of gene expression in human liver. PLoS Biol., 6, e107.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Civelek M., Wu Y., Pan C., Raulerson C.K., Ko A., He A., Tilford C., Saleem N.K., Stancakova A., Scott L.J., et al. Genetic regulation of adipose gene expression and integration with GWAS loci and cardio-metabolic traits. Am. J. Hum. Genet, 100, 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.