Abstract
Multiple techniques for quantification of hippocampal subfields from in vivo MRI have been proposed. Linking in vivo MRI to the underlying histology can help validate and improve these techniques. High-resolution ex vivo MRI can provide an intermediate modality to map information between these very different imaging modalities. This article evaluates the ability to match information between in vivo and ex vivo MRI in the same subjects. We perform rigid and deformable registration on 10 pairs of in vivo (3 T, 0.4 × 0.4 × 2.6 mm3) and ex vivo (9.4 T, 0.2 × 0.2 × 0.2 mm3) scans, and describe differences in MRI appearance between these modalities qualitatively and quantitatively. The feasibility of using this dataset to validate in vivo segmentation is evaluated by applying an automatic hippocampal subfield segmentation technique (ASHS) to in vivo scans and comparing SRLM (stratum/radiatum/lacunosum/moleculare) surface to manual tracing on corresponding ex vivo scans (and in 2 cases, histology). Regional increases in thickness are detected in ex vivo scans adjacent to the ventricles and were not related to scanner, resolution differences, or susceptibility artefacts. Satisfactory in vivo/ex vivo registration and subvoxel accuracy of ASHS segmentation of hippocampal SRLM demonstrate the feasibility of using this dataset for validation, and potentially, improvement of in vivo segmentation methods.
Keywords: hippocampus, in vivo, MRI, postmortem, subfields
Accurate delineation of anatomical structures is critical for quantifying human brain morphometry derived from neuroimaging data. However, almost invariably, delineation of brain structures on in vivo MRI relies on heuristic rules that are based on macroscopic features and landmarks. By contrast, neuroanatomists define brain regions (e.g., Brodmann areas) in terms of cytoarchitectonic features such as the size, shape, and density of neuronal cell bodies. The validity of heuristic rules, used widely for manual and automated in vivo MRI delineation, vis-à-vis the “true” cytoarchitectonic boundaries is questionable, especially given the significant intersubject variability in both macroscopic and microscopic aspects of the human brain (Thompson et al. 1996; Xie et al. 2014). Dense 3D histological imaging (e.g., BigBrain (Amunts et al. 2013)) and ex vivo MRI microscopy (e.g. (Fatterpekar et al. 2002)) can allow for more precise definition of anatomical boundaries. However, without a way to translate this information into the in vivo MRI domain, these high-resolution ex vivo modalities have little to offer in support of quantitative in vivo MRI analysis. Although a few animal studies used ex vivo imaging to help define in vivo image quantification (Li et al. 2009; Stille et al. 2013), there has been relatively little work in the MRI literature on mapping anatomical definitions from the ex vivo domain to the in vivo domain, particularly in humans.
One strategy, exemplified by the work of Amunts et al. (2005) and Augustinack et al. (2013), is to combine information from multiple ex vivo specimens into a probabilistic map and to embed this map onto a brain MRI template used for in vivo analysis. For example, Augustinack et al. (2013) labeled the perirhinal cortex in a set of ex vivo MRI scans, mapped each of these scans onto the Freesurfer T1-MRI whole brain template, and computed the spatial density map for these anatomical regions. By registering new in vivo scans to the template, one can estimate the location of the perirhinal cortex in the in vivo MRI scans. Such an approach is objective and data-driven. However, since information from multiple specimens is combined into a single average, the anatomical information mapped to a given subject's in vivo MRI space does not account for that subject's individual anatomy. Hence, the use of this strategy for informing and validating in vivo MRI segmentation protocols is limited.
Another more direct way to map information derived from histology into the space of in vivo MRI is to obtain in vivo MRI and ex vivo histology in the same set of subjects and use intermodality image registration to superimpose histology data onto the in vivo MRI. Such an approach poses 2 challenges. First, obtaining in vivo and ex vivo imaging in human subjects requires either long-term studies in large research cohorts or studies in populations with low life expectancy (Clark et al. 2012). Second, registration of in vivo MRI and histology data is difficult given the differences in the resolution, dimensionality (2D vs. 3D), and contrast mechanisms of these 2 modalities. Ultrahigh resolution ex vivo MRI (Fatterpekar et al. 2002; Yushkevich et al. 2008) can serve as an intermediate modality to map information from histological sections to in vivo MRI.
An example of a brain region for which such an approach would be of interest is the medial temporal lobe (MTL). Segmentation of MTL subregions, such as hippocampal subfields, from in vivo MRI scans has received increasing attention (Yushkevich et al. 2015b) because of their involvement in declarative memory (Milner 2005) and neuropsychiatric diseases (Small et al. 2011). Adler et al. (2014) already demonstrated the feasibility of mapping information from dense serial histology of the human MTL to high-resolution ex vivo MRI. However, to our knowledge there has been no prior work quantitatively comparing in vivo MRI and ex vivo MRI of the MTL, or of other brain regions, in the same subjects. It is unclear to what extent physiological changes during end-of-life events, brain extraction protocols, and fixation methods affect the structure of the human MTL; and whether it is possible to overcome these potential effects to map information from the ex vivo to the in vivo domain.
To address this gap in knowledge, the current study aims to characterize the differences and similarities between in vivo and ex vivo imaging of the MTL in the same subjects. This study was carried out using imaging data obtained from ten autopsies in which the subject had had research-quality, high-resolution in vivo brain MRI within the last ~1–3 years of life. Autopsy specimens were fixed in formalin and imaged at 0.2 × 0.2 × 0.2 mm3 resolution at 9.4 T. The in vivo and ex vivo MRI scans were registered based on manually outlined outer hippocampal boundaries. To evaluate the quality of in vivo/ex vivo registration, we labeled the stratum radiatum lacunosum moleculare (SRLM), which defines most of the border between dentate gyrus (DG) and cornu ammonis (CA), in both sets of scans and report distance statistics. We summarized, qualitatively, the visual comparison of aligned in vivo and ex vivo scan pairs. We then performed thickness measurements in corresponding locations of in vivo and ex vivo scans to quantify the differences between in vivo and ex vivo scans. We analyzed additional scans of ex vivo specimens at lower field strength, at lower resolution and in water to determine whether the differences between in vivo and ex vivo scans could be explained by interscanner effects, partial volume effects or susceptibility artefacts due to the hyperintense signal of the water, mimicking the properties of cerebrospinal fluid (CSF). Additionally, to demonstrate how the in vivo/ex vivo dataset could be used to validate in vivo segmentation, we applied the Automated Segmentation of Hippocampal Subfields (ASHS) (Yushkevich et al. 2015a) algorithm to in vivo scans and compare the resulting SRLM surface to those traced in the ex vivo images.
Material and Methods
Subjects
Subjects participating in neuroimaging research studies at the Penn Frontotemporal Degeneration Center and the Penn Memory Center were included in the current study if a high-resolution T2-weighted scan, optimized for the segmentation of MTL structures, was obtained during life and the subjects then underwent an autopsy in the Center for Neurodegenerative Disease Research (Toledo et al. 2014). As part of their involvement in ongoing research at these centers, subjects consented to autopsy and use of their clinical and imaging data for research studies. The studies were in accordance with the declaration of Helsinki and were approved by the IRB of the University of Pennsylvania. Of the 12 subjects who met these criteria, one was excluded because of tissue damage during autopsy and one because the time between the in vivo scan and autopsy exceeded 3 years, leaving ten subjects for the current study. Their mean age at death was 67.3 ± 8.8 years and 6 were male. All subjects received either a Mild Cognitive Impairment (MCI) or dementia diagnosis during life (Table 1).
Table 1.
Demographics, fixation time and time between 2 scans for each subject
| Subject | Age at death (years) | Sex | Diagnosis during life | Neuropathological diagnosis | Fixation time (days) | Time between scans (years) |
|---|---|---|---|---|---|---|
| 1 | 60 | M | Logopenic variant primary progressive aphasia (PPA) | Alzheimer's disease (AD), high probability | 82 | 2.2 |
| 2 | 75 | M | Dementia with Lewy bodies (DLB) | DLB | 31 | 3.0 |
| 3 | 61 | F | Semantic variant PPA | Frontotemporal lobar degeneration (FTLD) with TDP-43 inclusions | 43 | 2.8 |
| 4 | 74 | F | Amnestic MCI | AD, high probability | 24 | 3.1 |
| 5 | 54 | M | AD | AD, high probability | 47 | 2.8 |
| 6 | 67 | F | Nonfluent/agrammatic variant PPA | Corticobasal degeneration | 34 | 2.0 |
| 7 | 83 | M | Behavioral variant frontotemporal dementia (FTD) | Progressive supranuclear palsy (PSP) and AD, low probability | 43 | 2.5 |
| 8 | 73 | M | Behavioral variant FTD | PSP | 22 | 2.2 |
| 9 | 61 | F | Behavioral variant FTD | FTLD TDP-43 | 27 | 0.9 |
| 10 | 65 | M | Behavioral variant FTD | FTLD TDP-43 | 27 | 1.3 |
| Average | 67.3 | 60% M | 38.0 | 2.3 |
AD, Alzheimer's disease; DLB, dementia with Lewy bodies; FTD, frontotemporal dementia; FTLD, frontotemporal lobar degeneration; MCI, mild cognitive impairment; PPA, primary progressive aphasia; PSP, progressive supranuclear palsy.
MR Imaging, Sampling of Postmortem Tissue and Alignment of MRI Scans
In vivo: A high-resolution T2-weighted sequence was acquired on a 3 T Siemens Trio scanner with an 8-channel array coil, angulated perpendicular to the long axis of the hippocampus, with: an in-plane resolution of 0.4 × 0.4 mm2, a slice thickness of 2 mm (30 interleaved slices), a gap of 0.6 mm and acquisition time: 7:12 min. A T1-weighted gradient echo MRI was obtained with a 1.0 × 1.0 × 1.0 mm3 resolution and acquisition time: 5:13 min.
Ex vivo: Brain specimens were fixed in 10% neutral buffered formalin (for an average of 38.0±17.7 days, range: 22–82 days). Brain specimens were immersed in a cylinder of Fomblin, an inert oil without MR signal, for scanning. A standard spin echo multislice sequence was acquired oriented roughly perpendicular to the long axis of the hippocampus on a 9.4 T Varian scanner with a custom M2M quadrature transmit/receive radiofrequency coil, with a 0.2 × 0.2 × 0.2 mm3 resolution. The average time between the 2 scans was 2.3 years. See Supplementary Material 1 for more details on the scanning protocol. The dataset will be made available upon publication via https://www.nitrc.org/.
To investigate whether scanner type and resolution played a role in any measured differences between in vivo and ex vivo MRI, 4 ex vivo samples were rescanned at the 3 T scanner using the aforementioned in vivo protocol and at 9.4 T at a lower resolution of 0.4 × 0.4 × 2.6 mm3 matching the in vivo T2-weighted MRI and at 9.4 T in water to mimic the effects of CSF in the in vivo scans (resolution: 0.4 × 0.4 × 2.6 mm3; see Supplementary Table 1.1 for more details on the scanning protocols).
Registration
We 3D-printed a cylindrical phantom with a diameter of 30.5 mm and a length of 80 mm, fitting in the cylinder used for the ex vivo tissue samples, and holes with a diameter of 2 mm, 4 mm apart, along the longitudinal axis and orthogonal to the longitudinal axis of the phantom, see Figure 1.1 in Supplementary Material 1. We scanned the phantom with both the 3 T and 9.4 T scanner. The 3 T scanner showed no discernable distortion, however, the 9.4 T scanner showed a linear scaling effect of 6% horizontal, 3% vertical, and 11% longitudinal. Additional nonlinear distortions were found at the ends of the phantom. Using this information, we linearly rescaled all the ex vivo images obtained at the 9.4 T scanner and to address the nonlinear distortion we performed deformable registration, described below.
We first roughly manually aligned the in vivo T2-weighted and ex vivo images using HistoloZee, interactive software (http://picsl.upenn.edu/software/histolozee/). After this initial alignment, the outer boundary of the hippocampus proper was manually traced on both scans by author LEMW on every in vivo slice and on every 13th 0.2 mm ex vivo slice, which were subsequently interpolated (see Supplementary Material 1 page 4 for more information on Segmentation interpolation). Next, rigid registration was performed (Avants et al. 2008; Avants et al. 2011) between the binary segmentation images. This registration was used for the qualitative comparison and thickness measurements. Lastly, we performed deformable diffeomorphic registration (Avants et al. 2008, 2011) using these same segmentations. Deformable registration parameters were selected to ensure a very smooth deformation field to account for physical distortions due to tissue handling and 9.4 T MRI scanner nonlinear distortions (see for more information Supplementary Material 1 page 2). We also attempted deformable registration between ex vivo and in vivo scans based on MRI intensity, using both mutual information and cross-correlation metrics, but found this registration to be less reliable than registering hippocampal outlines, due to substantial differences in MRI appearance and presence of artefacts due to air bubbles and residual water in ex vivo scans (Figure 2i).
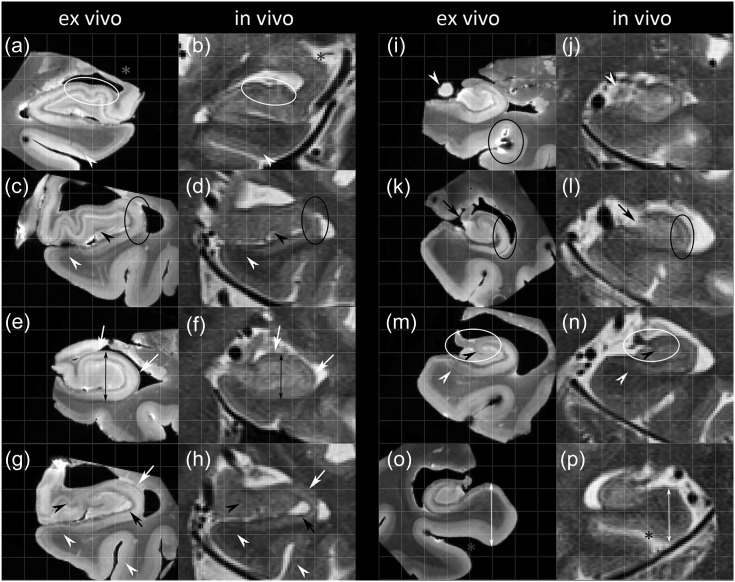
Figure 2.
Comparison of in vivo and ex vivo imaging throughout the length of the hippocampus in 5 subjects. The white circles (a,b and m,n) show examples of locations where the registration was successful and black circles are examples where the alignment was slightly off. Similarities are: multiple layers in the para-, pre-, or subiculum (white open arrowheads in c,d, g,h, and m,n) and in the perirhinal cortex (white open arrowheads in a,b, and g,h), the ‘x’-like strip of CA3 in the head (black open arrowheads in g,h), the endfolial pathway (black open arrowheads in m,n) and a small dark strip, potentially part of SRLM, in the head separating the DG and CA3 (open black arrowheads in c,d). Compared with ex vivo images, on in vivo images the hippocampus (see, e.g., black arrows in e,f), the extrahippocampal regions (white arrows in o,p) and CA appears smaller (white arrows in e,f and g,h), cysts appear larger (black arrows in g,h), and there are partial volume voxels obscuring the medial boundary of the DG (black arrows in k,l) and the outer boundaries of the uncus (white open arrowheads in i,j). In addition, the location of the uncus (grey stars in a,b) is different and there is a widening of the collateral sulcus ex vivo (grey stars in o,p). In (i) a localized MR artefact due to an air bubble can be observed. CA, cornu ammonis; DG, dentate gyrus; SRLM, stratum radiatum lacunosum moleculare.
For the thickness measurements on the in vivo T1-weighted scan, the original high-resolution ex vivo 9.4 T image was aligned with the T1-weighted in vivo MRI scan using only rigid registration. The rigid registration was done likewise for the 0.4 × 0.4 × 2.6 mm3 9.4 T ex vivo scans, the 9.4 T scans in water and the 3 T scans of the 4 rescanned ex vivo samples.
Assessment of Hippocampal Substructure Alignment
To evaluate the ability of rigid and deformable registrations based on hippocampal outlines to align hippocampal sub-structures, we manually labeled the inner boundary of SRLM in all 10 pairs of in vivo and ex vivo scans. SRLM was labeled on every in vivo MRI slice and every 0.2 13th ex vivo MRI slice, and interpolated and trimmed in 3D to obtain a surface mesh (as described in Supplementary Material 1). Point-to-point surface distance statistics (root mean squared distance and quantiles) between in vivo and ex vivo SRLM surface meshes were computed for the rigid and deformable registration.
Hippocampal Subfield Thickness Measurements
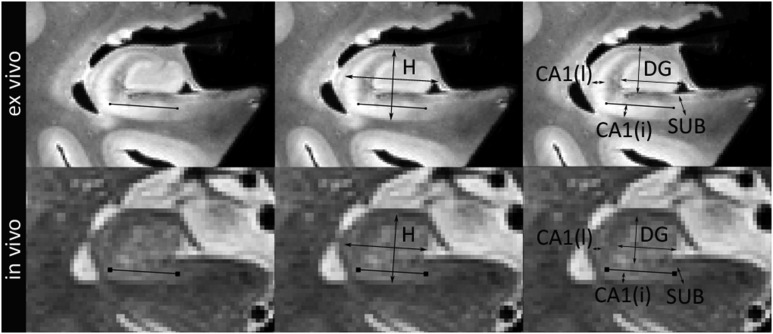
Thickness measurements were performed by one rater (LEMW) using the annotation tool in ITK-SNAP version 2.4 (Yushkevich et al. 2006). The measurements were performed on a single coronal slice in the hippocampal body, one slice posterior to the last slice on which the uncus could be visualized on the original in vivo T2-weighted image and on the corresponding ex vivo slice. Structures such as the SRLM, alveus and fimbria were not included in the measurements as we aimed to measure differences in grey matter. The hippocampal body shows intersubject variability in shape and rotation relative to surrounding structures. To account for this in our measurements, we defined the long axis of the subiculum/CA1 area inferior to the DG in the hippocampus by placing markers in the middle of the subiculum inferior to the most medial point of the DG and one inferior to the most lateral point of the SRLM (Figure 1). A straight line between these 2 points defined the long axis of the subiculum/CA1. This line, defined separately for the in vivo and ex vivo images, was used as a reference to perform height and width measurements of the hippocampus (defined as CA and DG, only when referring to the thickness measurements) and the combined CA3 and DG. For both subregions, width was measured as the longest line parallel to the reference line and height as the longest line perpendicular to the reference line. Thickness of the subiculum was measured at the most medial location of the DG. A straight line was drawn perpendicular to the long axis of the subiculum at this point. The thickness of CA1 was measured on the most lateral point of the hippocampus (Adachi et al. 2003) from which a straight line was drawn perpendicular to the long axis of CA1 at this point. Thickness of CA1 is also assessed at another location; measuring a straight line perpendicular to the long axis of CA1 at the middle point of the width of the hippocampus. It should be noted that CA1 might cross over in subiculum at this point (Duvernoy et al. 2005), but we chose this location because it is easily identifiable. All measurements were performed on the in vivo T2- and ex vivo proton density weighted images.
Figure 1.
Scheme for thickness measurements in hippocampal regions ex vivo (top row) and in vivo (bottom row). The black line shown in the first column defines the axis of the hippocampus. Parallel and perpendicular to this line, width and height of the hippocampus (second column) and DG and CA3 (third column, referred to as DG) were assessed. Thickness of the SUB and CA1 are measured perpendicular to the long axis of these structures (third column). Thickness of the SUB is measured at the most medial point of the DG, thickness of CA1 is measured at the most lateral point of CA and at the middle point of the width of the hippocampus.
H, Hippocampus; SUB, subiculum; CA, cornu ammonis; DG, dentate gyrus. Note that for the measurements hippocampus is defined as CA and DG.
On the T1-weighted in vivo images and the lower resolution 0.4 × 0.4 × 2.6 mm3 3 and 9.4 T ex vivo images and the 9.4 T images in water, the fimbria and alveus could not be consistently distinguished from the grey matter and were therefore included in the measurements of these image pairs since it was not feasible to measure grey matter size separately from these white matter structures. On the T1-weighted in vivo image, we only measured height and width of the hippocampus, because the SRLM necessary for the measurements of the subfields, could not be visualized consistently. On the low resolution 0.4 × 0.4 × 2.6 mm3 3 and 9.4 T ex vivo images and the 9.4 T images in water, subfield thickness was measured unless the SRLM could not be sufficiently visualized in that specific location. Measurements of all subfields could be performed in at least 3 out of 4 specimens.
All measurements were performed 3 times with a one-week interval and averaged to reduce the effect of measurement errors.
Statistical analysis: The consistency of the thickness measurements was assessed using the intraclass correlation coefficient (ICC) using SPSS version 20 (SPSS, Inc., Chicago, IL, USA). The ICC variant that measured absolute agreement under a two-way random analysis of variance model was used. The thickness measurements on in vivo and ex vivo MRI were compared using Wilcoxon Signed Rank Tests (two-sided). The P-value was set at 0.05.
Assessment of Hippocampal Substructure Segmentation by ASHS
We also processed all 10 in vivo scans with ASHS (Yushkevich et al. 2015a) and inferred the SRLM boundary from the ASHS segmentations as the boundary between the DG label and SUB, CA1, and CA2 labels. We manually segmented the ex vivo images using the same set of labels as in ASHS, and similarly inferred the SRLM boundary from manual ex vivo segmentations. Note that this second set of in vivo and ex vivo segmentations picks the mid surface of SRLM, rather than the inner boundary. As above, we computed distance statistics between ASHS-derived SRLM and ex vivo SRLM (based on the deformable in vivo to ex vivo alignment).
Results
Section 1: Alignment and Registration of the In Vivo and the Ex Vivo Images
The performance of the registration was analyzed by calculating distance statistics between SRLM boundaries in both images. Table 2 shows a median distance of 0.29 mm for the rigid registration, and 0.25 mm for the deformable registration, suggesting that the smooth deformation recovers some of the nonlinear scanner and physical manipulation effects. In both cases, the median distance is below the in vivo voxel size (0.4 × 0.4 mm2 in-plane). Supplementary Figure 2.1 visualizes the manually traced hippocampal outer surfaces (which are used for rigid/deformable alignment) and SRLM surfaces for an in vivo/ex vivo pair after rigid and deformable alignment.
Table 2.
RMS and distances between the SRLM in the in vivo images compared with the ex vivo images
| Distance | |||||
|---|---|---|---|---|---|
| RMS (mm) | 50th perc. (mm) | 75th perc. (mm) | 90th perc. (mm) | 95th perc. (mm) | |
| IV–SRLM to rigid EV–SRLM | 0.45 | 0.29 | 0.50 | 0.72 | 0.93 |
| IV–SRLM to deformed EV–SRLM | 0.40 | 0.25 | 0.43 | 0.65 | 0.82 |
| ASHS to deformed EV segmentation | 0.56 | 0.35 | 0.59 | 0.93 | 1.18 |
ASHS, automated segmentation of hippocampal subfields; EV, ex vivo; IV, in vivo; RMS, root mean square; SRLM, stratum radiatum lacunosum moleculare.
Figure 2 illustrates the quality of the rigid registration between the in vivo and ex vivo images (we focus on rigid registration because it is used for the qualitative and quantitative comparisons in Sections 2–4), showing that the registration is generally better in the body sections and slightly worse in the head and tail sections. The white circles in (a,b) and (m,n) illustrate examples where the alignment is good whereas the black circles in (c,d) and (k,l) demonstrate locations where the alignment is slightly off. In addition, the grey stars in figure show that certain parts of the specimen ex vivo appear dislocated relative to the rest of the specimen. Specifically, Figure 2a,b show compression of the temporal horn and Figure 2o,p show widening of the collateral sulcus, compared with the in vivo image. This is likely partly due to mechanical deformation of the tissue during extraction and placing it in a cylinder for the postmortem scan (see Figure 2.2 in Supplementary Material 2).
Section 2: Qualitative Comparison of In Vivo and Ex Vivo Images
When comparing the in vivo with the ex vivo images we noted several similarities and differences. Figure 2 shows that similarities can be observed both in general shape, for example, outer borders, digitations and dark band, and in smaller features. Figure 2c,d and 2g,h (white open arrowheads) show, for example, that multiple layers can be distinguished in the parasubiculum, presubiculum and/or subiculum and similarly in (a,b and g,h) (white open arrowheads) in the perirhinal cortex. Other features that can be appreciated in both images are: the ‘x’-like strip of CA3 in the head (black open arrowheads in g,h), the endfolial pathway (Lim et al. 1997) (black open arrowheads in m,n) and a small dark strip in the head separating the DG and CA3 (black open arrowheads in c,d). Interestingly, loss of definition of the uncus due to partial volume effects in the transition from head to hippocampal body (white open arrowhead in j), which is often observed in vivo, actually translates to the last small tip of the uncus on the ex vivo image (white open arrowhead in i). This comparison of in vivo clinical images with ex vivo MRI confirms that certain macrostructural and microstructural features of the hippocampus can be observed in vivo.
Several differences can also be observed. Global differences can be observed with the in vivo tissue appearing slightly smaller than the ex vivo tissue, both for the hippocampus (see, e.g., black arrows e,f) and the extrahippocampal regions (white arrows in o,p). As noted in the previous section, some local differences can be observed, such as the displacement of the uncus (grey stars in a,b) and a widening of the collateral sulcus (grey stars in o,p) likely due to the placement of the ex vivo specimen in a cylinder or handling of the specimen in general. Other local differences are that on in vivo MRI compared with ex vivo, CA in general appears smaller (white arrows in e,f and g,h), cysts appear larger (black arrows in g,h), and there are partial volume voxels obscuring the medial boundary of the DG (black arrows in k,l). These differences need to be accounted for when translating information from the ex vivo to the in vivo domain. To further characterize and quantify these differences, we performed thickness measurements in a single slice in the body (Section 3) and we will describe how these similarities observed on in vivo and ex vivo MRI can help guide in vivo segmentation (Section 4).
Section 3: Comparison of Thickness Measurements In Vivo and Ex Vivo
Intrarater ICC for average measures was 0.925–0.996 for almost all of the measurements, except for the in vivo measurement of CA1 thickness adjacent to the parahippocampal white matter (ICC = 0.72). See Supplementary Table 2.1 for the ICCs per region.
Table 3 shows that all subfields, except for the width of DG and CA3, had on average smaller size on the T2-weighted in vivo images as compared with the ex vivo images, though it should be noted that there was intersubject variability. Differences were between 0.83% and 10.27% for most regions except for CA1 thickness at the most lateral point which was 39.48% smaller on in vivo images as compared with ex vivo. As assessed by Wilcoxon Rank tests, size differences reached significance for the height of the hippocampus and for the thickness of CA1 at the most lateral point. The size difference for DG and CA3 width reached a trend level with larger in vivo thickness compared with the ex vivo thickness.
Table 3.
Comparison of thickness of the hippocampus and hippocampal subfields between in vivo/ex vivo image pairs, ex vivo 3 and 9.4 T image pairs, low resolution/high-resolution ex vivo 9.4 T image pairs and water/Fomblin ex vivo 9.4 T image pairs
| In vivo | Ex vivo | In vivo–Ex vivo | 3 T Ex vivo–9.4 T Ex vivo | LR Ex vivo–HR Ex vivo | H2O Ex vivo–Fomblin Ex vivo | |||
|---|---|---|---|---|---|---|---|---|
| Mean (SD) in mm | Mean (SD) in mm | Diff. in mm | Diff. in % | Neg diff. in | Diff. in % | Diff. in % | Diff. in % | |
| Number | 10 | 10 | 10 | 10 | 10 | 4 | 4 | 4 |
| Height H | 6.24 (0.54) | 6.54 (0.56) | −0.30** | −4.75 | 8 | 0.63 | 1.95 | 1.11 |
| Width H | 8.05 (0.91) | 8.29 (0.75) | −0.24 | −3.10 | 7 | −1.54 | 1.46 | −0.68 |
| Thickness SUB | 1.82 (0.27) | 2.01 (0.27) | −0.19 | −10.27 | 6 | −2.31 | −5.14 | −2.61 |
| Thickness CA1 lat. | 0.69 (0.10) | 1.04 (0.14) | −0.34** | −39.48 | 9 | 3.33 | −3.53 | −2.64 |
| Thickness CA1 inf. | 1.31 (0.17) | 1.45 (0.28) | −0.14 | −8.99 | 6 | 8.67 | 7.95** | 2.46 |
| Height DG and CA3 | 4.19 (0.58) | 4.22 (0.57) | −0.03 | −0.83 | 6 | −2.98 | 2.84 | 2.56 |
| Width DG and CA3 | 6.54 (0.88) | 6.22 (0.73) | 0.32* | 4.81 | 2 | −1.41 | 1.67 | 2.19 |
Mean ± SD are displayed. **P < 0.05; *P < 0.10. LR, low resolution; HR, high resolution; H, hippocampus; SUB, subiculum; CA, cornu ammonis; DG, dentate gyrus. Note that for the measurements hippocampus is defined as CA and DG.
A comparison of the ratios of hippocampal subfields to the hippocampus in vivo and ex vivo (Figure 2.3 in Supplementary Material 2), showed that most ratios were similar in vivo and ex vivo, except for the ratios of CA1 thickness at the most lateral point and CA3&DG width to the width of the hippocampus. This further supports the above findings that there is a general effect of regions appearing smaller in vivo than ex vivo, but that there are also disproportionate effects in certain areas, that is, CA1 thickness at the most lateral point and DG and CA3 width.
When measuring height and width of the hippocampus on T1-weighted images, we found that both are also smaller on T1-weighted in vivo images, as compared with ex vivo images (height: IV = 7.26 (0.74) mm; EV = 7.60 (0.92) mm; difference = −4.37%; P = 0.09; width: IV = 8.34 (0.69) mm; EV = 8.68 (0.86) mm; difference = −3.86%; P = 0.24). These results are similar to those of the measurements in the T2-weighted in vivo images and indicate that the observed differences are not dependent on specific contrast characteristics of the T2-weighted images.
In addition, we investigated whether the described difference in thickness between the in vivo and ex vivo images can be explained by a difference in scanner or resolution. Since the low resolution, 0.4 × 0.4 × 2.6 mm3, ex vivo images obtained at 3 and 9.4 T cannot be directly compared with the in vivo image because of slight differences in angulation, we therefore compare these images indirectly by calculating a thickness difference with the high-resolution ex vivo images (for each of these comparisons the high-resolution image is sliced parallel to the low resolution image after rigid registration). Table 3 shows that thickness differences between the 3 and 9.4 T ex vivo images and between the low resolution, 0.4 × 0.4 × 2.6 mm3, and high-resolution 9.4 T ex vivo images were much smaller than the observed differences for the in vivo/ex vivo comparisons for CA1 (3 vs. 9.4 T: 3.33%; LR vs. HR: −3.53%; IV vs. EV: −39.48% including alveus and −19.89 without) and hippocampal height (3 T vs. 9.4 T: 0.63%; LR vs. HR: 1.95%; IV vs. EV: −.75% including alveus and fimbria and −4.51 without). The observed differences between in vivo and ex vivo for hippocampal height and CA1 thickness are therefore most likely not due to scanner or resolution differences. (It should be noted that the thickness measurements in the in vivo/ex vivo pairs excluded alveus and fimbria as we aimed to measure differences in grey matter, while they were included in the ex vivo 3 T/9.4 T and LR/HR measurements. To verify the observed differences for hippocampal height and CA1 thickness, we repeated those for the in vivo/ex vivo pairs including the WM and found similar results, though smaller [Hippocampal height: −4.51%, P = 0.01; CA1 thickness lat.: −19.89%, P = 0.01]. This may be due to partial voluming of these white matter structures with CSF in vivo.) Additionally, we investigated the effect of the hyperintense signal of water, as a proxy for CSF in the in vivo scans, and again found differences much smaller, especially for the CA1 region (H2O vs. fomblin EV: −2.64; IV vs. EV: −39.48% including alveus and −19.89 without; and for hippocampal height: H2O vs. fomblin EV: 1.11%; IV vs. EV: −4.75% including alveus and fimbria and −4.51 without; right column of Table 3), than for the in vivo/ex vivo comparisons which indicates that the hyperintense signal of water did not notably affect the thickness measurements. In Supplementary Figure 2.4 the image pairs are depicted for 2 of the samples for each of the experiments.
Finally, the effect of fixation time with the difference in thickness between the in vivo and ex vivo scans was investigated. We found variable and nonsignificant associations (P > 0.18) for most of the subfields, except for CA3 and DG height (Spearman's Rho = 0.67; P = 0.04) suggesting that a larger fixation time is associated with smaller thickness ex vivo relative to in vivo (see Table 2.2 in Supplementary Material 2 for correlations with each region).
Section 4: Ad Hoc Observations Relevant to In Vivo Segmentation
This dataset can also help guide in vivo segmentation by validating a number of features that are currently used in in vivo segmentation protocols. The outer shape and white matter band are used in all subfield protocols for T2-weighted images and indeed appear similar in the in and ex vivo images. Another question that arises when performing in vivo segmentations is whether or not the most anterior slice of the hippocampus and the most anterior slice of the DG can be identified correctly, of which the latter is often used as a landmark guiding segmentation of other subfields. An experienced manual rater (J.B.P.) identified the most anterior slice of the hippocampus and DG on in vivo MRI, blind to the ex vivo images. The most anterior slice of the hippocampus and DG were either correctly identified or only one 2.6 mm slice off, indicating that the in vivo identification of these regions is close to what can be observed on ex vivo MRI. Observed discrepancies are most likely at least partly attributable to small errors in alignment, partial volume effects and motion artefacts, which may have obscured the visibility of these regions on the in vivo images. We also investigated further how consistently the endfolial pathway can be observed on in vivo images in body and tail slices as this can help guide the segmentation of CA3. Unfortunately, this white matter band could only be identified 47% (intersubject range: 0–83%) of the inspected slices, indicating that it may not be a reliable landmark for in vivo segmentations on T2-weighted 3 T images with similar resolution. An important landmark frequently used for determining the most anterior slice of the body, is the uncus. Loss of definition of the uncus due to partial volume effects in the transition from head to body, actually translates to the last small tip of the uncus on the ex vivo image, as described in Section 2. Dependent on the slice planning of the MRI, partial volume voxels of the uncus can only be observed in some subjects. In other subjects the uncus is clearly defined in the most posterior slice of the head and is no longer visible on the consecutive first slice of the body. A similar in vivo slice with partial volume voxels could be identified in 5 of the 10 subjects and in all these subjects these partial volume voxels translated to a clearly defined uncus on the ex vivo images, extending for 1–4 subsequent posterior slices (0.2–0.8 mm). On the other hand, partial volume voxels causing a loss of definition of the medial boundary of the DG did not clearly correspond to tissue on ex vivo MRI. It is not clear how these voxels should be handled, especially since in some subjects DG width in vivo was smaller, and in some cases larger, than ex vivo scans. A final observation with regard to manual segmentation is that the inferior boundary of the subiculum and adjacent presubiculum and parasubiculum cannot be discerned in all slices. Because the inferior layer is relatively hypointense, the boundary with the parahippocampal white matter can be obscured on the in vivo images, and was difficult to identify on ~60% of the slices. In some slices it was even difficult to observe this boundary on the ultrahigh-resolution ex vivo images (see, e.g., 2a). This suggests that current segmentation protocols may undersegment subiculum and underestimate subiculum thickness and volume. These observations are summarized in Table 4.
Table 4.
Summary of observed features on in vivo MRI, compared with ex vivo MRI, and their relevance for in vivo segmentation on 0.4 × 0.4 × 2.6 mm3 3 T MRI
| Observed feature on in vivo MRI | Relevance for in vivo segmentation |
| Outer shape and white matter band | Similar to ex vivo MRI and supports the usefulness of these boundaries for in vivo segmentations. |
| Most anterior slice hippocampus and most anterior slice of the DG | Detection of this border closely approximates the border on the ex vivo images, but not perfectly (max. 1 slice mismatch). This difference is likely due to alignment errors, which occurred especially in the head. |
| Loss of definition of the medial boundary of the DG | In some cases the width of the DG&CA3 is larger in vivo than ex vivo. Although this may reflect a loss of definition due to partial volume voxels, this was not observed consistently. |
| Loss of definition of the uncus at the transition from head to body | These partial volume voxels translate to actual tissue on ex vivo MRI in all 5 cases in which a similar slice could be identified and extended for 1–4 slices posterior to this slice in the ex vivo image. This indicates these partial volume voxels should perhaps be segmented as uncal tissue. |
| Endfolial pathway | Could not be consistently observed, only on 47% (intersubject range 0–83%) of the slices, and may not be a reliable marker for segmentation. |
| Inferior boundary of the SUB and adjacent pre- and para-SUB | The boundary with the white matter of the PHG is difficult to discern ~60% of the slices. This may lead to an underestimation of thickness/volume. |
CA, cornu ammonis; DG, dentate gyrus; PHG, parahippocampal gyrus; SUB, subiculum.
Section 5. Evaluation of In Vivo Automatic Labeling of the SRLM Border in ASHS
When comparing the SRLM surface inferred from the ASHS segmentation (based on the location of the border of the DG with CA1/CA2/subiculum) with the SRLM surface computed from the manual segmentation of the ex vivo images using an analogous protocol, the median surface-to-surface distance was 0.35 mm, which is only slightly higher than the median distance between the SRLM traced manually in the in vivo scans and the ex vivo SRLM surface. Further, it should be noted that in the in vivo images, SRLM is sometimes one voxel thick and is then counted by ASHS towards DG, whereas in the ex vivo segmentations we were able to split SRLM evenly between the DG and surrounding CA1/CA2/subiculum. This may account for the slightly larger median distance in this comparison.
Discussion
We were able to register in vivo and ex vivo images of the hippocampus obtained in the same subjects satisfactorily, with median distance between the SRLM within one 0.4 × 0.4 mm2 voxel for both rigid and deformable registration. When comparing the in vivo and ex vivo images, we observed several similarities in overall shape, but also in microstructural features. There also were differences between the 2 image modalities, with the most salient one being a difference in size, especially in the CA area. We further characterized this with thickness measurements and found that most regions were larger on ex vivo images compared with in vivo images, with a difference between 1% and 10% for most regions, but 39% for CA1 thickness at the most lateral point. Finally, when comparing the ASHS segmentation of SRLM, that is, the boundary between SUB/CA1/CA2 and DG, with an ex vivo manual segmentation, a median distance of less than one 0.4 × 0.4 mm2 voxel was found.
The observed differences in size were unexpected given the well-known effects of aging and neurodegenerative diseases on the hippocampus (Barnes et al. 2009; Neumann et al. 2006) and shrinkage effects due to formalin fixation (Mouritzen Dam 1979). However, to the best of our knowledge, no research has been done on the comparison of the same human subjects studied by in vivo and ex vivo MR images. One explanation for aforementioned differences in hippocampal size is that they were due to MRI-related variables. Factors to consider that may influence these discrepancies could be the differences in resolution or in the scanners. We examined these possibilities by rescanning ex vivo brain tissue at a lower resolution at 9.4 T, as well as at 3 T. The observed differences for these comparisons were much smaller than the in vivo/ex vivo differences and it is therefore likely that other factors than resolution and scanner have led to these differences. Although the nonlinear distortions of the 9.4 T, as measured using the phantom, could affect measured thickness of the specimen, these distortions were smooth, occurred at the end of the phantom, and therefore probably did not affect the thickness measurements performed in the middle of the hippocampus. Another example is that in the in vivo scans, CA1 width is measured adjacent to CSF in the temporal horn, which is hyperintense in the T2-weighted image, whereas in the ex vivo scans the CSF is replaced by fomblin, which lacks MRI signal and appears hypointense. This difference in the contrast between the temporal horn and CA1 may affect the apparent size of CA1 by locally distorting the reconstructed MRI image due to slight susceptibility artefacts (Schenck 1996). To further investigate this, we therefore rescanned tissue samples in water to mimic the effect of CSF and found minimal differences with the same tissue scanned in fomblin. This suggests that susceptibility artefacts in the in vivo scans likely did not affect the thickness measurements.
Another possible explanation is that part of the difference in size between the in vivo and ex vivo hippocampus is due to an actual difference in size. It has been proposed that tissue changes may occur during or after death since the agonal state is thought to cause hypoxia and ischemia which is assumed to result in brain swelling (Shen et al. 1993; Maxeiner and Behnke 2008). This would indeed affect the hippocampus (Nikonenko et al. 2009), and especially the CA1 area (de la Torre et al. 1992). This would support our finding of the largest difference in CA1 thickness in our dataset. Formalin fixation could also be hypothesized to underlie the differences reported here. However, even though brain swelling has been reported in the initial period of fixation, 10% neutral buffered formalin is generally found to cause shrinkage after several weeks (Mouritzen Dam 1979). Another hypothesis is that an increase in size could result from brain extraction, for example by a relief of intracranial pressure after autopsy. Finally, our dataset consisted of subjects with a diagnosis of MCI or dementia. Pathological changes in the hippocampus due to MCI or dementia could potentially affect the thickness measurements 1) by obscuring the visualization of the inner structure and 2) by local neurodegeneration. However, the dark band and the anchor points needed to perform the thickness measurements could be identified on all images. Additionally, we observed larger hippocampal size in the ex vivo images compared with the in vivo images, which is opposite to what would be expected if the pathologies played a role. As the effect of scanner, resolution and CSF are at most minor, it is likely that in this study physical changes in the tissue (e.g., due to end-of-life events or brain extraction) play an important role in the observed differences in hippocampal size between the in vivo and ex vivo images.
Although, to the best of our knowledge, there are no human studies comparing same-subject in vivo and ex vivo MRI, several animal studies of this kind have been performed. However, no clear picture emerges with some studies reporting no differences (Scheenstra et al. 2009) and others a significant decrease in hippocampal volume, or volume of another brain region, on ex vivo images compared with in vivo images (Wehrl et al. 2015). In addition, it is unclear how well these animal studies compare to our study with human brain tissue given a number of methodological differences, such as the divergent circumstances under which humans and animals are scanned in vivo (awake vs. anesthetized) resulting in differences in resolution and image quality as well as in fixatives used and fixation times. For example, it is unclear how the fixation time varying from 24 h to 7 days in the animal studies compares to the average of 39 days of fixation in the current study.
It would be interesting to know what caused the size difference between the ex vivo and in vivo hippocampus, however, this information is not necessary for translating cytoarchitectonic information from the ex vivo to the in vivo domain. In this study, we indeed showed that, by performing deformable registration we were able to register the ex vivo images onto the in vivo images, which can be used to map information from ex vivo to in vivo. An evaluation of this method showed that alignment of the SRLM in the 2 image groups was well within a 0.4 × 0.4 mm2 voxel. Additionally, we were able to partially validate ASHS. While histology is necessary to definitively identify the boundaries between individual hippocampal subfields (e.g., CA2/CA3), the high-resolution ex vivo MRI provides excellent contrast between the SRLM, the so-called “dark band”, and the hippocampal gray matter. The SRLM is a major feature in the definition of subfield boundaries—for example, the CA1/DG boundary is almost entirely defined by the SRLM. When comparing the SRLM surface inferred from the in vivo ASHS segmentation with that inferred from manual ex vivo segmentation, median surface distance was again within a voxel, and only 0.1 mm (0.35 vs. 0.25) larger than the median distance between manually traced in vivo and ex vivo SRLM surfaces. This confirms that ASHS can accurately pick up this anatomical boundary on the in vivo images.
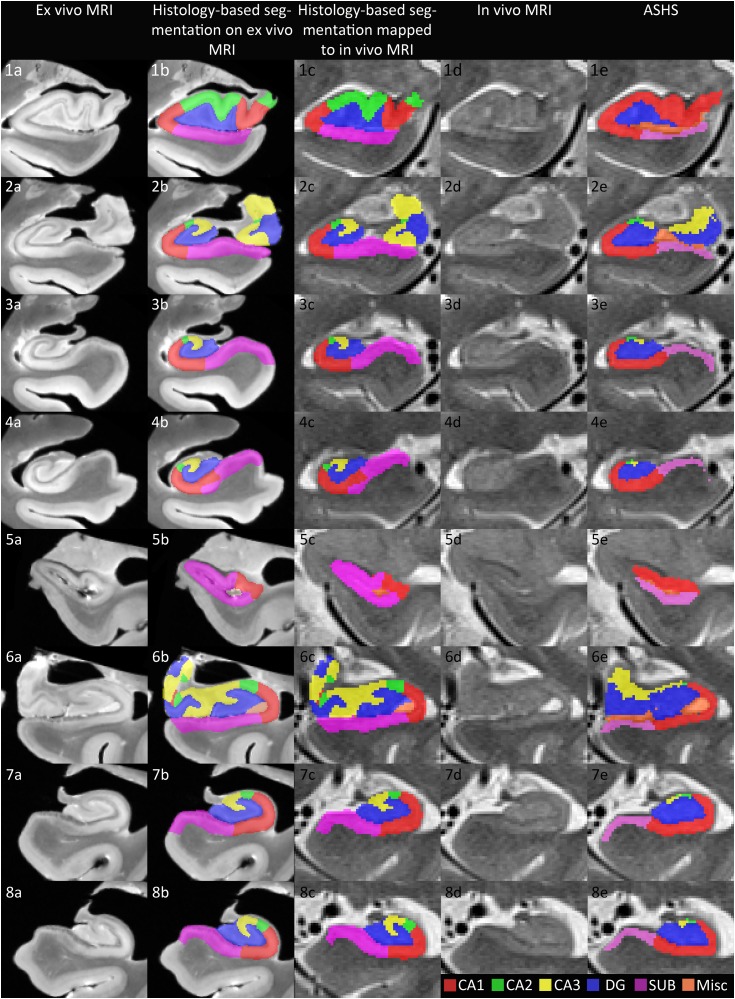
Taking this one step further, we compared a segmentation on the ex vivo MR images, based on histological annotations, with ASHS (Yushkevich et al. 2015a) performed on the in vivo images of 2 subjects for whom this data is available (more information on the histology data and segmentations can be found in Supplementary Material 1). Figure 3 illustrates that the segmentations show similarities in general. At the same time, there are a number of areas in which the histology annotations can clearly improve the in vivo segmentation. For example, the boundary between CA1 and the subiculum is placed more medial in ASHS compared with the histological annotation (Figure 3.3, 3.4, 3.7 and 3.8), the location of CA2 and CA3 follows a more complex pattern than generated by the in vivo segmentation of ASHS (Figure 3.1, 3.2 and 3.6) and the segmentation of CA3 in the body sections ex vivo (Figure 3.3, 3.4, 3.7 and 3.8) is more extended than in the in vivo segmentation protocol of ASHS. These differences between ASHS and the histological annotations on ex vivo MRI result partly from the limited visualization of certain features, such as the endfolial pathway, on in vivo MRI, and partly because limited information is available on the complex pattern of subfields in the head and the between-subject variability of certain boundaries, such as the CA1/subiculum boundary and it is therefore difficult to incorporate this in vivo segmentation protocols.
Figure 3.
Comparison of a segmentation based on histological annotations (second column ex vivo 9.4 T MRI; mapped to in vivo 3 T T2-weighted MRI in the third column) with automated segmentation of hippocampal subfields (ASHS) on in vivo 3 T T2-weighted MRI (fifth column) in the same subjects. This figure illustrates that the segmentations show similarities in general. At the same time, there are a number of areas in which the histology annotations can clearly improve the in vivo segmentation. For example, the boundary between CA1 and the subiculum is placed more medial in ASHS compared with the histological annotation (Figure 3.3, 3.4, 3.7, and 3.8), the location of CA2 and CA3 follows a more complex pattern than generated by the in vivo segmentation of ASHS (Figure 3.1, 3.2, and 3.6) and the segmentation of CA3 in the body sections ex vivo (Figure 3.3, 3.4, 3.7, and 3.8) is more extended than in the in vivo segmentation protocol of ASHS. These differences between ASHS and the histological annotations on ex vivo MRI result partly from the limited visualization of certain features, such as the endfolial pathway, on in vivo MRI, and partly because limited information is available on the complex pattern of subfields in the head and the between-subject variability of certain boundaries, such as the CA1/subiculum boundary and how to translate this to heuristic rules for an in vivo segmentation protocol. ASHS, automated segmentation of hippocampal subfields; CA, cornu ammonis; DG, dentate gyrus; Mics, miscellaneous; SUB, subiculum.
Datasets such as this one containing a set of subjects for whom in vivo MRI, ex vivo MRI and histology is available, can benefit in vivo imaging research in 2 ways: firstly by enabling validation of in vivo segmentation protocols and automatic techniques, allowing objective comparison of heuristic-based protocols taking into account intersubject variability. Secondly, such a dataset could aid the development of more anatomically correct automatic segmentation techniques. For example, methods like ASHS use multiatlas label fusion (Wang et al. 2012) in which a set of annotated example in vivo MRI scans are used jointly to label new in vivo MRI scans. The current ASHS atlas, which is based on heuristic rule-based manual segmentations of in vivo MRI, could in the future be replaced by an atlas consisting of in vivo images for which ex vivo MRI and histology are available, and in which subfields boundaries are labeled based on histological ground truth rather than heuristics (as in Fig. 3 third column). When this histology-based atlas is used to label new in vivo MRI scans, it is likely that the anatomical accuracy of the segmentation will be higher than using the current atlas. Another way in which the current dataset can advance the field is by using the ex vivo domain to interpret image features in in vivo scans with the goal of informing in vivo segmentation. In general, it was confirmed that the outer boundaries and the appearance of the dark band were very similar on the in vivo and ex vivo images. Several additional similarities were observed. For example, loss of definition of the uncus in the transition from head to body due to partial volume voxels in vivo consistently reflected actual uncus tissue ex vivo. Another issue for in vivo segmentations is determining the most anterior border of the hippocampus and of the DG. These anterior boundaries could be approximated closely, but not perfectly which may in part be due to small errors in the alignment, partial volume and motion artefacts in the in vivo images. Another example is that features, such as the endfolial pathway could be observed in most cases but not consistently on each slice and might therefore be less useful for the segmentation of CA3 in vivo. Of note, the comparison of the ex vivo segmentations with corresponding in vivo MR images revealed that some of the features that guided the more complex CA3 segmentation in the head of the hippocampus can be observed on in vivo MRI (see Figure 2.5 in Supplementary Material 2 for an illustration). These examples illustrate of the added value of obtaining ex vivo MRI besides histology. As this dataset is growing, these and future questions could be addressed and will help with improving in vivo segmentation protocols.
In conclusion, this is the first study to register and compare in vivo and ex vivo images of the hippocampus in the same human subjects. We report several similarities as well as some differences between the 2 image modalities, with the most salient difference in regional thickness, which is likely due to physical changes of the tissue. The satisfactory in vivo/ex vivo registration and subvoxel accuracy of ASHS segmentation of hippocampal SRLM demonstrate the feasibility of using this dataset for validation, and potentially, improvement of in vivo segmentation methods. Although this study focused on the MTL, several aspects of the methods, such as the software used for registration, can also be useful for other brain regions. In addition, certain results, such as the overall larger hippocampal size on ex vivo compared with in vivo MRI, may also be generalizable to other brain areas and the reported results may therefore serve as a knowledge base for future investigations.
Abbreviations
AD = Alzheimer's disease; ASHS = automated segmentation of hippocampal subfields; CA = cornu ammonis; CSF = cerebrospinal fluid; DG = dentate gyrus; DLB = dementia with Lewy bodies; EV = Ex vivo; FTD = frontotemporal dementia; FTLD = frontotemporal lobar degeneration; H = Hippocampus; HR = high resolution; ICC = intraclass correlation coefficient; IV = In vivo; MCI = mild cognitive impairment; Mics = miscellaneous; MTL = medial temporal lobe; PHG = parahippocampal gyrus; PPA = primary progressive aphasia; PSP = progressive supranuclear palsy; RMS = root mean square; SRLM = stratum radiatum lacunosum moleculare; SUB = subiculum; T = Tesla.
Supplementary Material
Notes
We thank the subjects and their families. We also acknowledge the physicians at the neuropathology department for performing the autopsies and the research staff at the Penn Memory Center and the Penn FTD Center for recruiting subjects. Conflict of Interest: Dr. Trojanowski serves as an Associate Editor of Alzheimer's & Dementia. He may accrue revenue on patents submitted by the University of Pennsylvania wherein he is an inventor including Modified avidin-biotin technique, Method of stabilizing microtubules to treat Alzheimer's disease, Method of detecting abnormally phosphorylated tau, Method of screening for Alzheimer's disease or disease associated with the accumulation of paired helical filaments, Compositions and methods for producing and using homogeneous neuronal cell transplants, Rat comprising straight filaments in its brain, Compositions and methods for producing and using homogeneous neuronal cell transplants to treat neurodegenerative disorders and brain and spinal cord injuries, Diagnostic methods for Alzheimer's disease by detection of multiple MRNAs, Methods and compositions for determining lipid peroxidation levels in oxidant stress syndromes and diseases, Compositions and methods for producing and using homogenous neuronal cell transplants, Method of identifying, diagnosing and treating alpha-synuclein positive neurodegenerative disorders, Mutation-specific functional impairments in distinct tau isoforms of hereditary frontotemporal dementia and parkinsonism linked to chromosome-17: genotype predicts phenotype, Microtubule stabilizing therapies for neurodegenerative disorders, and Treatment of Alzheimer's and related diseases with an antibody. He is a co-inventor on patents submitted the University of Pennsylvania wherein he is an inventor that have generated income he has received from the sale of Avid to Eli Lily including Amyloid plaque aggregation inhibitors and diagnostic imaging agents. None of the other authors has anything to disclose.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/
Funding
This work was supported by the National Institute of Health grants (Grant numbers R01 AG 037376, R01 AG040271, P01 AG017586, R01 NS044266, R01 AG038490, P30 AG010124, P30 NS045839, P41 EB015893; and P01 AG032953, P01 AG017586 and P50 NS053488 to J.B.T.).
References
- Adachi M, Kawakatsu S, Hosoya T, Otani K, Honma T, Shibata A, Sugai Y. 2003. Morphology of the inner structure of the hippocampal formation in Alzheimer disease. AJNR Am J Neuroradiol. 24(0195-6108; 0195-6108; 8):1575–1581. [PMC free article] [PubMed] [Google Scholar]
- Adler DH, Pluta J, Kadivar S, Craige C, Gee JC, Avants BB, Yushkevich PA. 2014. Histology-derived volumetric annotation of the human hippocampal subfields in postmortem MRI. Neuroimage. 84:505–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts K, Kedo O, Kindler M, Pieperhoff P, Mohlberg H, Shah NJ, Habel U, Schneider F, Zilles K. 2005. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anat Embryol (Berl). 210(0340-2061; 0340-2061; 5-6):343–352. [DOI] [PubMed] [Google Scholar]
- Amunts K, Lepage C, Borgeat L, Mohlberg H, Dickscheid T, Rousseau ME, Bludau S, Bazin PL, Lewis LB, Oros-Peusquens AM, et al. 2013. BigBrain: an ultrahigh-resolution 3D human brain model. Science. 340(6139):1472–1475. [DOI] [PubMed] [Google Scholar]
- Augustinack JC, Huber KE, Stevens AA, Roy M, Frosch MP, van der Kouwe AJ, Wald LL, Van Leemput K, McKee AC, Fischl B, et al. 2013. Predicting the location of human perirhinal cortex, Brodmann's area 35, from MRI. Neuroimage. 64:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 12(1):26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. 2011. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 54(3):2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J, Bartlett JW, van de Pol LA, Loy CT, Scahill RI, Frost C, Thompson P, Fox NC. 2009. A meta-analysis of hippocampal atrophy rates in Alzheimer's disease. Neurobiol Aging 30(1558-1497; 0197-4580; 11:1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CM, Pontecorvo MJ, Beach TG, Bedell BJ, Coleman RE, Doraiswamy PM, Fleisher AS, Reiman EM, Sabbagh MN, Sadowsky CH, et al. 2012. Cerebral PET with florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid-beta plaques: a prospective cohort study. Lancet Neurol. 11(8):669–678. [DOI] [PubMed] [Google Scholar]
- de la Torre JC, Fortin T, Park GA, Butler KS, Kozlowski P, Pappas BA, de Socarraz H, Saunders JK, Richard MT. 1992. Chronic cerebrovascular insufficiency induces dementia-like deficits in aged rats. Brain Res. 582(2):186–195. [DOI] [PubMed] [Google Scholar]
- Duvernoy HM, Cattin E, Naidich T, Fatterpekar GM, Raybaud C, Risold PY, Sakvolini U, Scarabino T. 2005. The human hippocampus Berlin, Heidelberg, Germany: Springer Verlag. [Google Scholar]
- Fatterpekar GM, Naidich TP, Delman BN, Aguinaldo JG, Gultekin SH, Sherwood CC, Hof PR, Drayer BP, Fayad ZA. 2002. Cytoarchitecture of the human cerebral cortex: MR microscopy of excised specimens at 9.4 tesla. AJNR Am J Neuroradiol. 23(0195-6108; 0195-6108; 8):1313–1321. [PMC free article] [PubMed] [Google Scholar]
- Li X, Yankeelov TE, Rosen GD, Gore JC, Dawant BM. 2009. Enhancement of histological volumes through averaging and their use for the analysis of magnetic resonance images. Magn Reson Imaging. 27(3):401–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Mufson EJ, Kordower JH, Blume HW, Madsen JR, Saper CB. 1997. Connections of the hippocampal formation in humans: II. the endfolial fiber pathway. J Comp Neurol. 385(3):352–371. [PubMed] [Google Scholar]
- Maxeiner H, Behnke M. 2008. Intracranial volume, brain volume, reserve volume and morphological signs of increased intracranial pressure—a post-mortem analysis. Leg Med (Tokyo). 10(6):293–300. [DOI] [PubMed] [Google Scholar]
- Milner B. 2005. The medial temporal-lobe amnesic syndrome. Psychiatr Clin North Am. 28(0193-953; 0193-953; 3):599–611. [DOI] [PubMed] [Google Scholar]
- Mouritzen Dam A. 1979. Shrinkage of the brain during histological procedures with fixation in formaldehyde solutions of different concentrations. J Hirnforsch. 20(2):115–119. [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 314(5796):130–133. [DOI] [PubMed] [Google Scholar]
- Nikonenko AG, Radenovic L, Andjus PR, Skibo GG. 2009. Structural features of ischemic damage in the hippocampus. Anat Rec (Hoboken). 292(12):1914–1921. [DOI] [PubMed] [Google Scholar]
- Scheenstra AE, van de Ven RC, van der Weerd L, van den Maagdenberg AM, Dijkstra J, Reiber JH. 2009. Automated segmentation of in vivo and ex vivo mouse brain magnetic resonance images. Mol Imaging. 8(1):35–44. [PubMed] [Google Scholar]
- Schenck JF. 1996. The role of magnetic susceptibility in magnetic resonance imaging: MRI magnetic compatibility of the first and second kinds. Med Phys. 23(6):815–850. [DOI] [PubMed] [Google Scholar]
- Shen WC, Shieh TT, Shih TP, Chang CY, Su MC, Lee SK, Ho WL. 1993. MRI of postmortem brains. Gaoxiong Yi Xue Ke Xue Za Zhi. 9(12):690–697. [PubMed] [Google Scholar]
- Small SA, Schobel SA, Buxton RB, Witter MP, Barnes CA. 2011. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat Rev Neurosci. 12(1471-0048; 1471-003; 10):585–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stille M, Smith EJ, Crum WR, Modo M. 2013. 3D reconstruction of 2D fluorescence histology images and registration with in vivo MR images: application in a rodent stroke model. J Neurosci Methods. 219(1):27–40. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Schwartz C, Lin RT, Khan AA, Toga AW. 1996. Three-dimensional statistical analysis of sulcal variability in the human brain. J Neurosci. 16(13):4261–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo JB, Van Deerlin VM, Lee EB, Suh E, Baek Y, Robinson JL, Xie SX, McBride J, Wood EM, Schuck T, et al. 2014. A platform for discovery: the University of Pennsylvania integrated neurodegenerative disease biobank. Alzheimers Dement. 10(4):477,84.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Suh JW, Das SR, Pluta J, Craige C, Yushkevich PA. 2012. Multi-atlas segmentation with joint label fusion. IEEE Trans Pattern Anal Mach Intell. 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrl HF, Bezrukov I, Wiehr S, Lehnhoff M, Fuchs K, Mannheim JG, Quintanilla-Martinez L, Kohlhofer U, Kneilling M, Pichler BJ, et al. 2015. Assessment of murine brain tissue shrinkage caused by different histological fixatives using magnetic resonance and computed tomography imaging. Histol Histopathol. 30(5):601–613. [DOI] [PubMed] [Google Scholar]
- Xie L, Pluta J, Wang H, Das SR, Mancuso L, Kliot D, Avants BB, Ding SL, Wolk DA, Yushkevich PA. 2014. Automatic clustering and thickness measurement of anatomical variants of the human perirhinal cortex. Med Image Comput Comput Assist Interv. 17(Pt 3):81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Avants BB, Pluta J, Minkoff D, Detre JA, Grossman M, Gee JC. 2008. Shape-based alignment of hippocampal subfields: evaluation in postmortem MRI. Med Image Comput Comput Assist Interv. 11:510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. 2006. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 31(3):1116–1128. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Pluta JB, Wang H, Xie L, Ding SL, Gertje EC, Mancuso L, Kliot D, Das SR, Wolk DA. 2015. a. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum Brain Mapp. 36(1):258–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Amaral RS, Augustinack JC, Bender AR, Bernstein JD, Boccardi M, Bocchetta M, Burggren AC, Carr VA, Chakravarty MM, et al. 2015. b. Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: towards a harmonized segmentation protocol. Neuroimage. 1(111):526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.