Abstract
Mitochondria are thought to have originated as free-living prokaryotes. Mitochondria organelles have small circular genomes with substantial structural and genetic similarity to bacteria. Contrary to the prevailing concept of intronless mitochondria, here we present evidence that mitochondrial RNA transcripts (mtRNA) are not limited to policystronic molecules, but also processed as nuclei-like transcripts that are differentially spliced and expressed in a cell-type specific manner. The presence of canonical splice sites in the mtRNA introns and of core components of the nuclei-encoded spliceosome machinery within the mitochondrial organelle suggest that nuclei-encoded spliceosome can mediate splicing of mtRNA.
Introduction
Mitochondria are the primary energy-producing organelles in eukaryotic cells (1). Evolutionary studies of mitochondrial DNA (mtDNA) sequence composition and RNA expression, as well as the shape and size of mitochondria, suggest that these organelles were free-living prokaryotes (2–4). Both modern prokaryotes and mitochondria contain circular genomes lacking histones and introns. Mitochondria also encode their own tRNA and an RNA polymerase that displays significant sequence similarity to prokaryotes (2). In humans, the mtDNA is a closed-circular 16.5 kb double-stranded molecule encoding twenty-two tRNAs, two rRNAs and thirteen protein coding mRNAs. Mitochondrial mRNAs and rRNAs have a 3′-end poly(A) tail, similar to nucleus-encoded mRNA (5), without an intermediate pre-mRNA stage (2). Proteins regulating transcription and replication of mtDNA (2,5,6) account for a small fraction of more than 99% of molecules that are imported into mitochondrial compartments by a poorly understood system and with no clear function within the organelle (6,7).
Here, we show that the initial lack of intronic regions in human mtDNA has been overlooked, likely due to the sensitivity levels of sequencing techniques in the past and to the low abundance of spliced mitochondrial transcript variants in the cell. Taking advantage of high throughput sequencing technology, we demonstrate that transcripts from human mtDNA do have introns. Additionally, we show that spliced mitochondrial transcripts (mtRNA) can be differentially spliced and expressed in a cell-type-specific manner. We found that spliced out mtRNA introns have nuclei-like canonical splice sites, and that the majority of the ribonucleoproteins known to be part of nucleus spliceosome machinery are imported into mitochondrial compartments. Moreover, we observed that core proteins of the spliceosome complex colocalize within the mitochondria organelle and also occupy mtRNA transcripts. Together, these observations evidence the occurrence of splicing in the mtRNA and suggest that the nuclear spliceosome complex can mediate mtRNA splicing within the organelle.
Results and Discussion
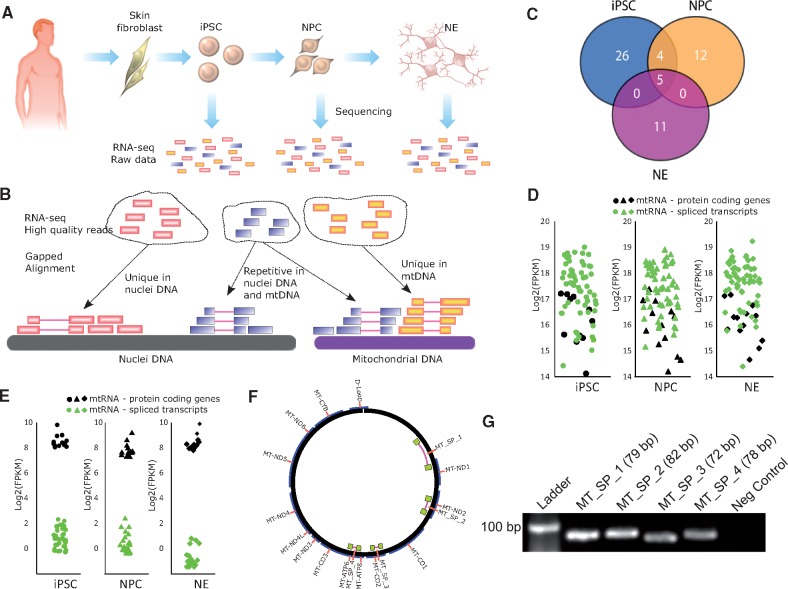
In the present study, we report evidence of a new type of mtRNA encoded by human mtDNA that is spliced in a cell-type specific manner. We used high throughput RNA sequencing data (RNA-seq) that were previously published elsewhere (7). The data report the transcriptional changes occurring in human primary skin fibroblasts (HPSF) reprogrammed into pluripotent stem cells (iPSC) that were further differentiated into neuronal progenitor cells (NPC) and postmitotic neurons (NE). The RNA-seq from these cells (Fig. 1A and 1B, Supplementary Material, Table S1) were aligned to the human reference nuclear and mitochondrial genomes to determine the expression levels of the 13 mitochondrial protein-coding genes (Supplementary Material, Table S2).
Figure 1.
Analysis of spliced mitochondrial RNA transcripts. (A) Reprogramming of HPSF into human induced pluripotent stem cells (iPSC). The iPSC were then differentiated into neuronal progenitor cells (NPC) and postmitotic neurons (NE). RNA-seq data from these three cell types were analyzed. (B) High quality reads from RNA-seq were aligned against the human mitochondrial DNA and to the human genomic DNA to identify non-redundant mtRNA spliced reads. (C) Venn diagram showing the number of common or exclusive mitochondrial spliced transcripts in iPSC, NPC and NE. (D) Expression levels (including repetitive reads) in FPKM of the 13 known protein-coding genes from mitochondria and the respective spliced transcripts found in iPSC, NPC and NE. (E) Expression levels (excluding repetitive and non-spliced reads) in FPKM of the 13 known protein-coding genes from mitochondria and the respective spliced transcripts found in iPSC, NPC and NE. (F) Mitochondrial DNA locus of the four spliced transcripts identified and tested by PCR. (G) PCR amplification products of the four spliced mtRNA transcripts observed in HPSF.
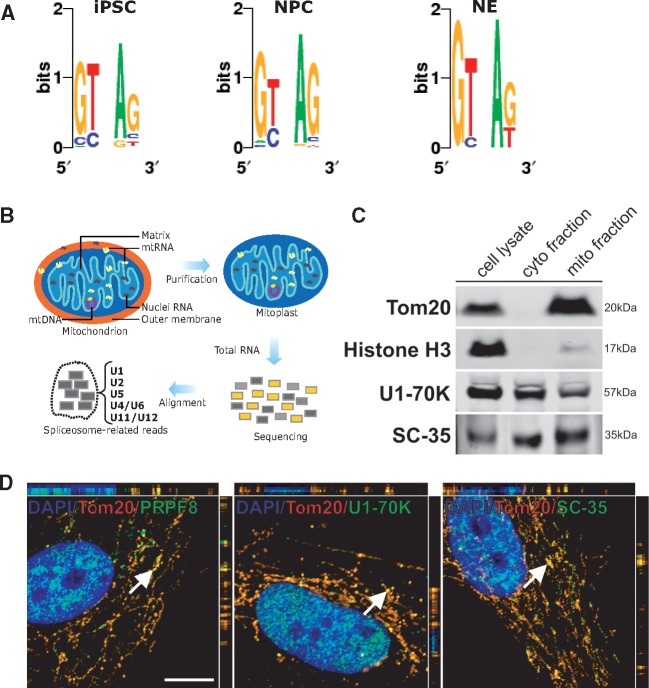
Several alignments were found to contain gaps when uniquely aligned to the reference mtDNA (unaligned to nuclear DNA), likely representing splicing mtRNA events, with introns removed out after processing of primary mtRNA (Fig. 1B). By requiring that bona fide spliced mtRNA reach a certain read threshold (section Material and Methods), we found that iPSC, NPC and NE encode 35, 21 and 16 spliced mtRNA, respectively (Fig. 1C, Supplementary Material, Table S3). Spliced mtRNA were also found using a more restrictive cut-off criteria (Supplementary Material, Fig. S1A–B). Five spliced transcripts were common among the three cell types, and four were common between iPSC and NPC. Spliced out introns from mtRNA range from 55 bp to 13,846 bp in length. To calculate the expression of mtDNA transcripts, we did not remove the PCR replicates to avoid biased expression values (8). Compared to the 13 protein coding genes, the expression level of the detected spliced mtRNA was similar (Fig. 1D). As the detected spliced mtRNA overlap with some of the already known mitochondrial genes, we measured the expression profile based on only spliced and non-repetitive reads (Fig. 1E, Supplementary Material, Table S4). Additionally, spliced mtRNA were found throughout the mtDNA except in the small 1,122 bp D-loop region (Fig. 1F) (9). We confirmed that spliced mtRNAs were also present in primary cells. In HPSF, the spliced mtRNAs MT_SP_1, MT_SP_2, MT_SP_3 and MT_SP_4 (Fig. 1F) could be detected by PCR analysis (Fig. 1G, Supplementary Material, Table S5), matching the expected sizes for the observed fragments. Furthermore, using the same primers, the identity (nucleotide content) of these fragments was confirmed by SANGER sequencing (Supplementary Material, Table S5). The sequenced fragments were also aligned to the human reference genome (hg19) to confirm that they mapped uniquely to mitochondrial DNA. Our PCR experiment suggests that even low expressed spliced mtRNA (Fragments Per Kilobase of transcript per Million mapped reads; FPKM < 1) could be real as we confirmed by Sanger and bioinformatics analysis. Next, we analyzed the intronic sequences from all spliced mtRNA found in iPSC, NPC and NE. The dinucleotides GT and AG were frequently found at the 5' and 3' intron boundaries, respectively (Fig. 2A). This observation indicates that major spliceosome components are likely mediating splicing within the mitochondria.
Figure 2.
Presence of nuclei-encoded spliceosome proteins within mitochondria. (A) Analysis of the dinucleotide motifs on the 5′ and 3′ ends of introns in spliced mtRNA from iPSC, NPC and NE. (B) Schematic of the detection of nucleus-encoded spliceosome in mitoplast RNA-seq data. (C) Western blot analysis for Tom20, Histone H3, U1-70K and SC-35 in total cell lysate compared with equal protein amounts of cytosolic- and mithochondrial-enriched fractions from HPSF. (D) Co-localization of spliceosomal proteins and mitochondria in HPSF. Cells were fixed and stained for the mitochondrial translocase Tom20, and three spliceosomal proteins PRPF8, U1-70k and SC-35. White arrows indicate random sites chosen for three-dimensional-z-stack analysis. Scale bar, 10 μm (images correspond to merged panels showed on Supplementary Material Figure S1C).
Based on these findings, we hypothesize that mtRNA is being spliced by the nuclei-encoded spliceosome machinery, which is imported into the mitochondria. Analyzing RNA-seq from human mitoplasts (5) (Fig. 2B, Supplementary Material, Table S6), we found evidence that 60 out of 66 spliceosome-associated RNAs are located within mitochondria. These include components of both the major (U2-type, formed by U1, U2, U4, U5, and U6) and minor (U12-type, formed by U5, U6atac, U4atac, U11 and U12) spliceosomes (10). Such core spliceosome-related proteins are overrepresented in different cell compartments including nuclei, cytosol, and are also adhered to mitochondria. In order to confirm that these molecules could be found within the mitochondria, we investigated if the U1-70K and SC-35 proteins were present in mitochondria-enriched fractions isolated from HPSF (Fig. 2C). A comparable protein expression level of these proteins was observed in total cell lysates and in cytosolic fractions.
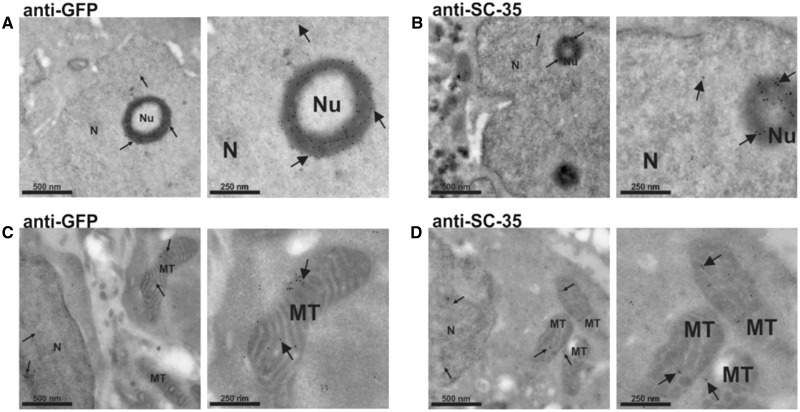
To show that core spliceosome proteins were located within mitochondria, we performed immunofluorescence experiments using HPSF. We observed that PRPF8, U1-70K and SC-35 proteins co-localize with a specific protein from the organelle, Tom20 (Fig. 2D, Supplementary Material, Fig. S1C). Next, we performed an electron microscopy (EM) assay with gold particles using labeled antibodies against SC-35 protein in a different human cell line (HEK293) (Fig. 3A–D). Gold particles were associated with the cell nucleus and also with internal mitochondrial compartments, supporting the hypothesis that nuclei-encoded spliceosome proteins, such as SC-35 expression, are not restricted to nuclei but also present within the mitochondria.
Figure 3.
Electron microscopic distribution of SC-35 in HEK293 cells transfected with a SC35-GFP-flag. Cells were stained with a primary antibody against GFP (A and C) or SC-35 (B and D) protein, and a secondary antibody conjugated to 10 nm colloidal gold. Proteins are diffusely distributed throughout the nucleoplasm. A cluster of splicing factors is also concentrated in a ring shape structure around the nucleolus (A and B). Regions of SC-35 immunostaining extend out the nucleus and the signal is clearly found within the mitochondria (C and D), suggesting that the protein is being imported into this organelle. Images on the right panels are higher magnifications from those on the left panels. Arrows highlight the location of some gold particles. N: nucleus; Nu: nucleolus; MT: mitochondria. Scale bars are indicated.
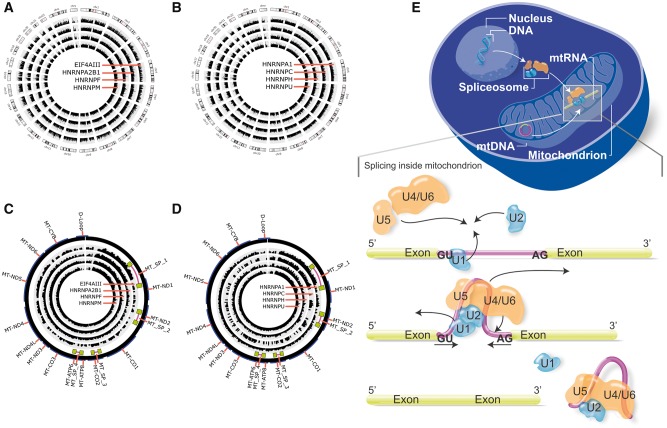
We also verified whether the core spliceosomal proteins were occupying the mtRNA by analyzing cross-linking immunoprecipitated RNA-seq data (Clip-seq) from human and mouse cells (Supplementary Material, Table S7). In Clip-seq data from human HeLa and HEK 293T cells, we found that the spliceosome-related proteins EIF4AIII and the heterogeneous nuclear ribonucleoparticles (HNRNP) proteins HNRNPC, HNRNPH, HNRNPU, HNRNPM, HNRNPF, HNRNPA2B1 and HNRNPA1 are not exclusively occupying nuclei-encoded RNA (Fig. 4A and B, respectively), but also the mtDNA loci of those PCR-confirmed spliced mtRNA (Fig. 4C and D, Supplementary Material, Table S8). These HNRNP regulate alternative splicing by interacting with splice sites of pre-mRNA (10–12). Additionally, ELF4AIII participates on assembling near exon–exon junctions of mRNAs as a result of splicing (11), but its occupancy is reported to rarely occur on exon-exon junctions (13). A similar observation was found when we analyzed the ELF4AIII protein occupancy. Analysis of Clip-seq data shows ELF4AIII protein occupying exon-intron junctions, exon or introns (Supplementary Material, Fig. S2A) compared to exon-exons regions (Supplementary Material, Fig. S2B). In Clip-seq from mouse embryonic fibroblast and the brain, we found that the core spliceosome-related proteins SRSF1, SC-35 (also known as SRSF2) and TDP43 also occupy mouse mtRNA (Supplementary Material, Table S7). These proteins, including PRPF8 and U1-70K, interact with pre-mRNA and some small nuclear ribonucleoproteins (12) influencing alternative splicing (14), exon inclusion (15), and definition of cleavage sites (11). Next, we screened in silico for spliced mtRNA and found that MT_SP_1 encodes for a bacterial protein YP_001313728 (Supplementary Material, Fig. S3A), a class of acyltransferase enzyme involved in storage lipid synthesis (16). We also found that MT_SP_2 mtRNA encodes for a bacterial protein WP_015855296 (Supplementary Material, Fig. S3B), a type of serine endoprotease enzyme that cleaves peptide bonds to link amino acid residues in proteins (17).
Figure 4.
Representation of core spliceosome proteins in human somatic and non-somatic chromosomes, and an overview of the mtRNA splicing mediated nuclei-encoded spliceosome proteins. (A and C) Clip-seq data of the core spliceosome proteins ELF4AIII, HNRNPA2B1, HNRNPF, HNRNPM occupying mitochondrial RNA transcripts and nuclei RNA transcripts, respectively; (B and D) Clip-seq data of the core spliceosome proteins HNRNPA1, HNRNPC, HNRNPH, HNRNPU occupying mitochondrial RNA transcripts and nuclei RNA transcripts, respectively. Data coverage (internal histograms) were normalized according to sample size and a log base 2 scale was applied over read coverage, ranging from 0 to 100 for both mitochondrial DNA (circular genome) and nuclei DNA (linear chromosomes 1-22, X and Y). (E) Schematic model of nuclei-encoded spliceosome moving from nuclei to cytosol, and then imported into mitochondrial compartments to occupy and splice mitochondrial RNA transcripts.
Together, our findings suggest that a number of different spliced human mtRNA are located into several loci in mtDNA (Supplementary Material, Table S3). Variations in the expression levels of spliced mtRNA suggest that splicing in mtDNA is cell-type dependent. Since the mtRNA introns exhibit canonical splice sites, splicing of mtRNA seems to be mediated by the imported spliceosome into the mitochondria (Fig. 4E), but further analyses will be needed to confirm this hypothesis.
Materials and Methods
RNA-seq from reprogrammed human primary skin fibroblasts
Human primary skin fibroblasts (HPSF) were reprogrammed into a pluripotent stage (iPSC) and used to generate neural progenitor cells (NPC) and post-mitotic neurons (NE), as previously described (18). Dermal biopsies were obtained from healthy individuals following informed written consent under protocols approved by the University of California San Diego. One sample of each cell type was subjected to Illumina high throughput sequencing for RNA-seq of mRNA and are available at public databases (7) under GEO accession number: GSE47626. All samples used in this study were negative for mycoplasma contamination.
Mitochondrial transcriptome expression analysis and splicing detection
To detect spliced mitochondrial RNA (mtRNA) in human cells and whether they vary in a cell-type specific manner, a bionformatics pipeline was applied to prepare and align the RNA-seq sequences from iPSC, NPC and NE. To prepare the data, we used the software NGS QC Toolkit (19)(using default parameters and PHRED score Q20) over the RNA-seq for high-quality reads recovery and to remove possible sequence contaminants (amplicons, adapters). Next, repetitive sequences were removed since PCR-based experiments used during cDNA library preparation and sequencing-by-synthesis step are prone to RNA template switching on generating false alternative transcripts (20,21). The high quality reads were then gapped-aligned with the software Tophat2 (22) (minimum intron length: 50 nt, non-canonical splice sites allowed) to the human reference genome (Hg19), including mitochondrial DNA (mtDNA) and chromosome-unmapped scaffolds. Reads uniquely aligned to mtDNA were used to calculate the expression profile, in FPKM, of 13 known mitochondrial genes. After excluding redundant reads that could align to both nuclei genome and mtDNA (nuclei genome has copies of some mtDNA fragments, known as Numts (23)), reads that were uniquely spliced to mtDNA were analyzed. Using the coordinates of the gap formed by the spliced read alignment, we extracted the introns and only those with 3 read coverage and a minimum of 10 bp exon length were preserved. Additionally, the up- and downstream portions of each spliced fragment, corresponding to exons, were limited to a minimum length of 10 nt long. Each set of reads sharing the same intron was then subjected to a de novo transcriptome assembly with the software Edena (24) (minimum overlapping length: 20 nt) into a consensus sequence, representing a spliced mitochondrial RNA (spliced mtRNA). The FPKM for each known mtDNA gene and spliced mtRNA was calculated.
Spliceosome machinery component detection
To detect the spliceosome complex within the mitochondria, it was used a set of 66 nucleotide sequences (Supplementary Material, Table S6) from SpliceosomeDB (25), subdivided into major (U2-type, formed by U1, U2, U4, U5 and U6 stable snRNP) and minor (U12-type, formed by U11, U12, U4atac, and U6atac, together with U5) spliceosomes (10,25–27). Next, we downloaded RNA-seq data from small RNA (SRA under accession number SRR314795) from mitoplasts (mitochondria without outer membrane) extracted from human osteosarcoma 143B cells (5). This database excludes the possibility to consider molecules (including RNA and proteins) that could be attached to the mitochondria outer membrane. The small RNA-seq data was then aligned to the spliceosome sequences. The alignment was based on a non-gapped approach using the software Bowtie2 (28) (0 mismatch allowed, seed length 22, other parameters were default). Based on the number of aligned fragments per sequence, we calculated the FPKM (Supplementary Material, Table S6).
RNA isolation and PCR
Total RNA was isolated from HPSF using the RNeasy Plus MiniKit (Qiagen, USA) according to manufacturer’s instructions. One microgram of total RNA was used for cDNA preparation using the QuantiTect Reverse Transcription Kit (Qiagen, USA), as suggested by manufacturer, and PCR reaction mixtures were prepared in a total volume of 25 μl containing 2 μl of cDNA, 5 pmol of each primer (Supplementary Material, Table S5), 2.5 mM MgCl2, 0.5 mM dNTPs, 1X PCR buffer, 1 unit of Expand High Fidelity PCR System (Roche, USA) and dH2O up to 25 μl. PCR program was started at 94°C for 4 min, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 30 s and completed with a final extension of 72°C for 10 min. PCR products were separated in 4% low melting agarose gel electrophoresis, allowing the identification of the fragments by size (Supplementary Material, Table S4). PCR products were cloned into the pCR 2.1 TOPO vector from TOPO TA Cloning Kit (Invitrogen, USA), according to manufacturer's instructions. About 5 clones from each library were subjected to SANGER sequencing including standard vector-specific M13 forward and reverse primers by Eton Biosciences (San Diego, USA). Sequenced products were then compared to the in silico spliced mtRNA predictions.
Cross-linking immunoprecipitation of spliceosomal proteins
RNA-seq data from experiments based on cross-linking immunoprecipitation-high-throughput sequencing (Clip-seq) using spliceosomal proteins were obtained from published studies (Supplementary Material, Table S7 and S8). In mouse, Clip-seq was performed in embryonic fibroblasts using the proteins SRSF1 and SRSF2 (15), and in the whole brain tissue using the protein TDP43 (29). In humans, Clip-seq was performed in HeLa cells using the proteins HNRNPC (30) and ELF4AIII (13); and in HEK 293T cells using HNRNPH (31) proteins. Moreover, HEK 293T cells were also used to perform Clip-seq using the proteins HNRNPU, HNRNPM, HNRNPF, HNRNPA2B1 and HNRNPA1 (32). These RNA-seq datasets were gapped-aligned with TopHat2 (22) (v2.0.10) against human (build hg19) and mouse (build mm10) genomes and mitochondrial DNA (mtDNA), according to the sample specie. Non-redundant reads aligned exclusively to mtDNA were isolated and used to calculate the FPKM expression for those mtDNA loci of the four PCR-tested spliced mtRNA. The expression was normalized using a geometric-based approach available in Cuffdiff software (v2.1.1) (33). Alignments were also used to show read coverage of Clip-seq data over the mtDNA. Absolute numbers of reads were normalized by the sequence library size, and a logarithm base 2 scale was plotted for mtDNA (Fig. 4A and B) and nuclei chromosomes (Fig. 4C and D). Graphical representations were created using Circos (34) software package.
Immunofluorescence analysis of human mitochondrial and spliceosomal proteins
HPSF were fixed in 4% paraformaldehyde, permeabilized and blocked with 0.1% Triton X-100 and 2.5% BSA. Immunofluorescence was performed by incubating cells with PRPF8 (1:250, Abcam), SC-35, clone 1SC-4F11 (1:500, Millipore), U1-70K (1:500, Millipore) and Tom20 (1:1000, FL-145 or FL-10, Santa Cruz) antibodies, followed by incubation with secondary antibodies (Alexa488-conjugated donkey anti-rabbit/mouse IgG and Alexa555-conjugated donkey anti-rabbit or Alexa555-conjugated goat anti-mouse, 1:1000, Life Technologies). Nuclei were stained with DAPI (Life Technologies) and slides were mounted using ProLong Gold antifade reagent (Invitrogen). Fluorescent signals were detected using a Zeiss Apotome 2 inverted microscope and images were processed with CorelDRAW Graphics Suite X5 (Corel). The co-localization of splicing factor proteins with mitochondrial preprotein translocases was evaluated from individual projections of z-stacks optical sections and scanned at 0.7-µm increments that correlated with the resolution valued at z-plane.
Western blot
A mitochondria-enriched fraction from HPSF was obtained using the Mitochondria Isolation Kit for Cultured Cells (Thermo Scientific), according to manufacturer’s protocol. Twenty micrograms of protein from the whole cell lysate were compared with equal amounts of protein from the cytosolic and mitochondrial fractions. Proteins were separated on Bolt® 4–12% Bis-Tris Plus Gels (Life Technologies), transferred to an Immuno-Blot® PVDF membrane (Bio-Rad), blocked and probed with primary antibodies against the 20-kDa mitochondrial pre-protein translocases of the outer membrane (Tom20; 1:1500; Santa Cruz), Histone H3 (17kDa; 1:1000; Cell Signaling), U1 small nuclear ribonucleoprotein 70 kDa (U1-70K; 1:500; Millipore) and splicing factor SC-35 (1:1000; Millipore). Next, membranes were incubated with IRDye secondary antibodies (800CW or 680RD; 1:4000; Li-Cor) and proteins were visualized using the ODYSSEY® CLx Infrared Imaging System (Li-Cor).
Immuno electron microscopy
Immuno-gold electron microscopy (EM) was performed at the CMM Electron Microscopy Facility at University of California San Diego. To confirm the presence of splicing factors within the mitochondria, assays were performed using HEK293 cells transfected with a SC35-GFP-flag. After transfection, cells were kept in suspension for 3 days to form spheres, and fixed using 4% paraformaldehyde in 0.1M phosphate buffer (pH 7.4). Fixed cells were pelleted and washed with 0.15 M glycine/phosphate buffer, embedded in 10% gelatin/phosphate buffer and infused with 2.3 M sucrose/phosphate buffer. Blocks of cells with 1 mm3 were mounted onto specimen holders and snap frozen in liquid nitrogen. Ultracryomicrotomy was carried out at –100 °C on a Leica Ultracut UCT with EM FCS cryoattachment (Leica, Bannockburn, IL) using a Diatome diamond knife (Diatome US, Hatfield, PA). 80 to 90 nm frozen sections were picked up with a 1:1 mixture of 2.3 M sucrose and 2% methyl cellulose (15cp) as described by Liou (35), and transferred onto Formvar and carbon-coated copper grids. Immunolabeling was performed according to the Tokuyasu technique (36). Briefly; grids were placed on 2% gelatin at 37 °C for 20 min, rinsed with 0.15 M glycine/PBS and the sections were blocked using 1% cold water fish-skin gelatin. Previously titrated, primary antibodies against GFP (Living Colors A.V. (JL-8), Clontech Laboratories) or SC-35 (clone 1SC-4F11; EMD-Millipore) protein were diluted 1:10 in 1% BSA/PBS. Incubation with primary antibodies for 2 h at room temperature was followed by 10-nm gold-conjugated goat anti-mouse IgG & IgM (Jackson ImmunoResearch, West Grove, PA) diluted 1:25 in 1% BSA/PBS, for 30 min at room temperature. Grids were analyzed using a Tecnai G2 Spirit BioTWIN transmission electron microscope equipped with an Eagle 4k HS digital camera (FEI, Hilsboro, OR).
Accession numbers
Human RNA-seq samples from induced pluripotent stem cells (iPC), neuronal progenitor cells (NPC) and neurons (NE) are available under NCBI - BioProject accession PRJNA126459. Human RNA-seq samples from mitoplasts (mitochondria without outer membrane) are available under NCBI – BioProject accession PRJNA144321. Cross-linking immunoprecipitation (Clip-seq) libraries used to analyze mtRNA occupied by spliceosome-related proteins are available under NCBI – BioProject accession code PRJNA190888 (protein SRSF1, SRSF2, organism: Mus musculus), PRJNA142049 (protein TDP43, organism: Mus musculus), PRJEB2253 (protein hNRNPC, organism: Homo sapiens), PRJNA130865 (protein HNRNPH, organism: Homo sapiens), PRJNA156301 (protein HNRNPU, HNRNPM, HNRNPF, HNRNPA2B1, HNRNPA1 organism: Homo sapiens), PRJNA174879 (protein ELF4AIII, organism: Homo sapiens).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
A.R.M. was supported by the National Alliance for Research on Schizophrenia and Depression (NARSAD) Independent Investigator Grant. R.H.H. was supported by an International Rett Syndrome Foundation (IRSF) fellow (grant #2925). We thank Dr. T. Bos for assistance with SC35-GFP-flag vector construction, and Mr. T. Meerloo for his expert assistance with electron microscopy.
Conflict of Interest statement. None declared.
Funding
California Institute for Regenerative Medicine (CIRM) DISC1-08825, TR2-01814 and TR4-06747, National Institutes of Health through the R01MH108528, R01MH109885, R01MH100175, R21MH107771, R56MH109587, R01MH094753, R01MH103134, U19MH107367, and P01NICHD033113, National Alliance for Research on Schizophrenia and Depression (NARSAD) Independent Investigator Grant, International Rett Syndrome Foundation (IRSF) (grant #2925).
References
- 1. Gray M.W., Burger G., Lang B.F. (2001) The origin and early evolution of mitochondria. Genome Biol., 2, REVIEWS1018.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asin-Cayuela J., Gustafsson C.M. (2007) Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci., 32, 111–117. [DOI] [PubMed] [Google Scholar]
- 3. Timmis J.N., Ayliffe M.A., Huang C.Y., Martin W. (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet., 5, 123–135. [DOI] [PubMed] [Google Scholar]
- 4. Gray M.W. (1999) Mitochondrial Evolution. Science, 283, 1476–1481. [DOI] [PubMed] [Google Scholar]
- 5. Mercer T.R., Neph S., Dinger M.E., Crawford J., Smith M.A., Shearwood A.-M.J., Haugen E., Bracken C.P., Rackham O., Stamatoyannopoulos J.A.. et al. (2011) The human mitochondrial transcriptome. Cell, 146, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gaspari M., Larsson N.-G., Gustafsson C.M. (2004) The transcription machinery in mammalian mitochondria. Biochim. Biophys. Acta – Bioenerg., 1659, 148–152. [DOI] [PubMed] [Google Scholar]
- 7. Marchetto M.C.N., Narvaiza I., Denli A.M., Benner C., Lazzarini T.A., Nathanson J.L., Paquola A.C.M., Desai K.N., Herai R.H., Weitzman M.D.. et al. (2013) Differential L1 regulation in pluripotent stem cells of humans and apes. Nature, 503, 525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parekh S., Ziegenhain C., Vieth B., Enard W., Hellmann I. (2016) The impact of amplification on differential expression analyses by RNA-seq. Sci. Rep., 6, 25533.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang R., Zhang F., Wang C., Wang S., Shiao Y.-H., Guo Z. (2010) Identification of sequence polymorphism in the D-Loop region of mitochondrial DNA as a risk factor for hepatocellular carcinoma with distinct etiology. J. Exp. Clin. Cancer Res., 29, 130.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Korneta I., Magnus M., Bujnicki J.M. (2012) Structural bioinformatics of the human spliceosomal proteome. Nucleic Acids Res., 40, 7046–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan C.C. (2004) eIF4A3 is a novel component of the exon junction complex. RNA, 10, 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature, 456, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saulière J., Murigneux V., Wang Z., Marquenet E., Barbosa I., Le Tonquèze O., Audic Y., Paillard L., Roest Crollius H., Le Hir H. (2012) CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat. Struct. Mol. Biol., 19, 1124–1131. [DOI] [PubMed] [Google Scholar]
- 14. Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., König J., Hortobágyi T., Nishimura A.L., Zupunski V.. et al. (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci., 14, 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pandit S., Zhou Y., Shiue L., Coutinho-Mansfield G., Li H., Qiu J., Huang J., Yeo G.W., Ares M., Fu X.-D. (2013) Genome-wide Analysis Reveals SR Protein Cooperation and Competition in Regulated Splicing. Mol. Cell, 50, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Röttig A., Steinbüchel A. (2013) Acyltransferases in bacteria. Microbiol. Mol. Biol. Rev., 77, 277–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Di Cera E. (2009) Serine proteases. IUBMB Life, 61, 510–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marchetto M.C.N., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. (2010) A Model for Neural Development and Treatment of Rett Syndrome Using Human Induced Pluripotent Stem Cells. Cell, 143, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patel R.K., Jain M. (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One, 7, e30619.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cocquet J., Chong A., Zhang G., Veitia R.A. (2006) Reverse transcriptase template switching and false alternative transcripts. Genomics, 88, 127–31. [DOI] [PubMed] [Google Scholar]
- 21. Houseley J., Tollervey D. (2010) Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS One, 5, e12271.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol., 14, R36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hazkani-Covo E., Zeller R.M., Martin W. (2010) Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet., 6, e1000834.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hernandez D., François P., Farinelli L., Osterås M., Schrenzel J. (2008) De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res., 18, 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cvitkovic I., Jurica M.S. (2013) Spliceosome database: a tool for tracking components of the spliceosome. Nucleic Acids Res., 41, D132–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rymond B. (2007) Targeting the spliceosome. Nat. Chem. Biol., 3, 533–535. [DOI] [PubMed] [Google Scholar]
- 27. Matlin A.J., Clark F., Smith C.W.J. (2005) Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol., 6, 386–398. [DOI] [PubMed] [Google Scholar]
- 28. Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.-C., Sun E., Wancewicz E., Mazur C.. et al. (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci., 14, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. König J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J. (2010) iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol., 17, 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katz Y., Wang E.T., Airoldi E.M., Burge C.B. (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods, 7, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huelga S.C., Vu A.Q., Arnold J.D., Liang T.Y., Liu P.P., Yan B.Y., Donohue J.P., Shiue L., Hoon S., Brenner S.. et al. (2012) Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep., 1, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol., 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. (2009) Circos: an information aesthetic for comparative genomics. Genome Res., 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liou W., Geuze H.J., Slot J.W. (1996) Improving structural integrity of cryosections for immunogold labeling. Histochem. Cell Biol., 106, 41–58. [DOI] [PubMed] [Google Scholar]
- 36. Tokuyasu K.T. (1980) Immunochemistry on ultrathin frozen sections. Histochem. J., 12, 381–403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.