Abstract
Background: The prognostic value of BRAF and KRAS mutations within microsatellite-unstable (MSI) and microsatellite-stable (MSS) subgroups of resected colon carcinoma patients remains controversial. We examined this question in prospectively collected biospecimens from stage III colon cancer with separate analysis of MSI and MSS tumors from patients receiving adjuvant FOLFOX +/− cetuximab in two adjuvant therapy trials.
Methods: Three groups were defined: BRAF Mutant, KRAS Mutant, and double wild-type. The analytic strategy involved estimation of study-specific effects, assessment of homogeneity of results, and then analysis of pooled data as no differences in patient outcome were found between treatment arms in both trials. Associations of mutations with patient outcome were analyzed, and multivariable models were adjusted for treatment and relevant factors.
Results: Four thousand four hundred eleven tumors were evaluable for BRAF and KRAS mutations and mismatch repair status; 3934 were MSS and 477 were MSI. In MSS patients, all BRAF V600E mutations (hazard ratio [HR] = 1.54, 95% confidence interval [CI] = 1.23 to 1.92, P < .001), KRAS codon 12 alterations, and p.G13D mutations (HR = 1.60, 95% CI = 1.40 to 1.83, P < .001) were associated with shorter time to recurrence (TTR) and shorter survival after relapse (SAR; HR = 3.02 , 95% CI = 2.32 to 3.93, P < .001, and HR = 1.20, 95% CI = 1.01 to 1.44, P = .04, respectively). Overall survival (OS) in MSS patients was poorer for BRAF-mutant patients (HR = 2.01, 95% CI = 1.56 to 2.57, P < .001) and KRAS-mutant patients (HR = 1.62, 95% CI = 1.38 to 1.91, P < .001) vs wild-type. No prognostic role of KRAS or BRAF mutations was seen in MSI patients. Furthermore, no interaction was found between treatment arm (with or without cetuximab) and KRAS and BRAF mutations for TTR or OS in MSS patients.
Conclusions: In a pooled analysis of resected stage III colon cancer patients receiving adjuvant FOLFOX, BRAF or KRAS mutations are independently associated with shorter TTR, SAR, and OS in patients with MSS, but not MSI, tumors. Future clinical trials in the adjuvant setting should consider these mutations as important stratification factors.
Colorectal cancer (CRC) is a common malignancy and the fourth cause of cancer-related death worldwide (1,2). Molecular testing is currently a routine part of clinical practice in these patients in the metastatic setting, with RAS and BRAF assessment recommended before any treatment in Western countries (3,4). KRAS and NRAS mutations predict resistance to epidermal growth factor receptor (EGFR) inhibitors such as cetuximab, resulting in their restricted use to patients with KRAS wild-type and more recently NRAS wild-type metastatic CRC (5–7).
BRAF mutation is a rare event found in 5% to 10% of metastatic CRCs, and while it has to date no clear predictive role to guide treatment decisions, it has a major prognostic role with particularly poor survival reported in BRAF-mutant metastatic CRCs (8). However, for stage III, nonmetastatic colon cancer patients, the prognostic role of these two mutations is still controversial, particularly among microsatellite-unstable (MSI) vs -stable tumors (MSS). We recently reported that codon 12 KRAS mutations in stage III colon cancer patients included in the PETACC8 or N0147 randomized phase III trials have a role on the time to recurrence (TTR) that was limited to distal vs proximal cancers (9,10). Regarding BRAF-mutant tumors, it has been suggested that BRAF V600E mutation is not influencing disease recurrence but only survival after relapse (SAR) (11–13). Furthermore, the role of BRAF mutations in tumors with MSI is essentially unknown. Combining a good prognostic molecular factor (MSI) and a bad one (BRAF mutation) may not allow the assessment of the true prognostic value of BRAF mutation in all subgroup populations of stage III colon cancer. Accordingly, analysis of these molecular markers in relationship to tumor location and microsatellite status (MSI/MSS) is needed.
MSI tumors represent 10% to 15% of stage III colon cancer, are enriched in BRAF mutations (12) among sporadics, and have been known to be associated with better prognosis than their MSS counterparts, although this may not be the case among FOLFOX-treated patients (10–13). Finally, the full picture of the prognostic association of these molecular markers (MSI, KRAS, BRAF) on recurrence, SAR, and then overall survival (OS) has been inconsistently reported in the current publications. We thus decided to pool data from two large recent adjuvant trials (14,15) dedicated to resected stage III colon cancer patients to assess the role of BRAF and KRAS mutations on TTR, OS, and SAR in MSS and MSI colon cancer patients after surgery and standard 5-fluorouracil (5-FU) plus oxaliplatin (FOLFOX)–based adjuvant therapy. Both trials tested the addition of cetuximab to standard FOLFOX chemotherapy, and both were negative showing no beneficial or detrimental effect of the addition of this anti-EFGR monoclonal antibody. We thus pooled the different treatment arms for our analyses. To our knowledge, this is the largest series evaluating the potential prognostic role of KRAS and BRAF mutations stratified by MSI/MSS status in stage III colon cancer patients.
Methods
This is an ancillary study of the PETACC8 trial (EUDRACT 2005-003463-23) and the N0147 trial (clinicaltrials.gov identifier NCT00079274).
Patient Characteristics
A histologically proven stage III colon adenocarcinoma was completely resected from all eligible patients for the PETACC8 and North Central Cancer Treatment Group (NCCTG) N0147 trials. Patients were randomly assigned to receive six months of either FOLFOX or FOLFOX + cetuximab with regular monitoring, as described previously (14,15). Both protocols were amended to only enroll patients with KRAS wild-type tumors, increasing the sample size. Written informed consent was required from each patient included in the planned translational program of both trials.
Microsatellite Status Determination
Mismatch repair (MMR) tumor status was determined by immunohistochemistry (IHC) or by MSI testing when IHC was indeterminate in accordance of the Bethesda criteria and as previously described for each trial (16,17). MSI phenotype tumors were defined as presenting with the loss of one or more MMR proteins’ expression by IHC or exhibiting high-level tumor DNA MSI (MSI-H) on MSI testing. Microsatellite-stable phenotype tumors were defined by normal MMR protein expression in IHC, or MSS or low-level MSI (MSI-L) status on MSI testing.
DNA Extraction and Mutation Analysis
Tumor samples were prospectively banked. Tumor DNAs were extracted from formalin-fixed, paraffin-embedded (FFPE) tissues containing more than 50% tumor cells using the QIAamp DNA Mini Kit (Qiagen). For PETACC8, molecular analysis was centralized at Georges Pompidou European Hospital, Paris, France, and was performed retrospectively for 2096 patients included before trial amendment and prospectively for the other 463 patients. For N0147, analyses were performed centrally at the Mayo Clinic, Rochester, Minnesota, retrospectively for 2110 included before trial amendment and prospectively for the other 889 patients. Detection of KRAS exon 2 hotspot mutations and BRAF V600E mutations was performed as described previously (9,10).
Statistical Analyses
The overall population and then MSS and MSI patients separately were considered for the present work. Patients were divided into three groups: group 1: wild-type for KRAS exon2 and BRAF V600E (wild-type); group 2: mutant for KRAS exon2 (KRAS-mutant); group 3: mutant for BRAF V600E (BRAF-mutant). Comparisons of patients with specific codon 12 and codon 13 KRAS mutations vs the double wild-type population were also performed. The end points for these analyses were TTR, OS, and SAR. TTR was defined as the time between the date of random assignment and the date of local or metastatic recurrence or death linked to disease recurrence, whichever occurred first. OS was defined as the time between the date of random assignment and the date of death due to all causes.
For comparisons of baseline characteristics, categorical factors were analyzed with χ2 tests and continuous factors were compared with standard parametric or nonparametric tests. Continuous variables are presented as the mean (SD) and median (interquartile range [IQR]).
TTR and OS curves were estimated with the Kaplan-Meier method. Differences between groups of patients were stratified by log-rank tests and the Cox model. Kaplan-Meier curves and forest plots were used for all of these analyses. Factors included in the multivariable analyses were treatment group and baseline prognostic factors identified in univariate analyses or clinically relevant. The assumption of proportionality was checked graphically using Schoenfeld's residuals. In case of doubt, an interaction time factor has been included in the model. Analyses were carried out according with a two-sided statistical significance level of 5%. Results were not adjusted for multiple comparisons. All statistical analyses were performed by Fédération Francophone de Cancérologie Digestive (FFCD) statisticians with the SAS statistical software package (version 9.4). The database was locked in June 2015.
Results
Study population
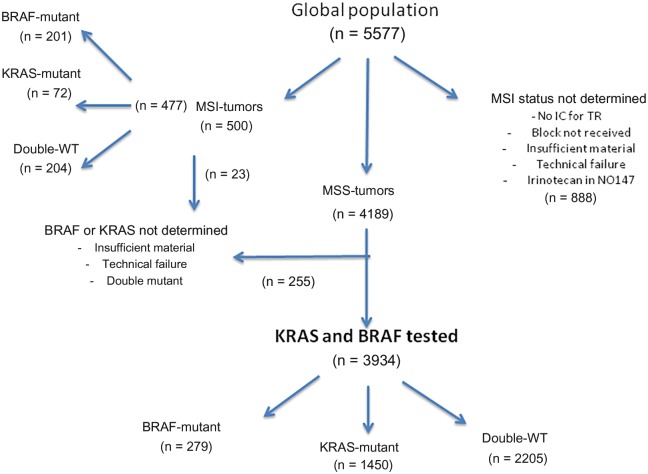
Among the 5577 patients included in the PETACC8 and N0147 phase III studies, 4189 were MSS, of which 3934 met all criteria for molecular analysis (sufficient material and no technical failure). Among 500 MSI tumors, 477 had complete data for KRAS and BRAF status (Figure 1). Demographic and clinical characteristics of the patients in the molecular study (n = 4411) were not statistically significantly different from those of the randomly assigned population (n = 5577) (Supplementary Table 1, available online).
Figure 1.
Flow chart of molecular analysis of PETACC8-N0147 trials molecular study evaluating the association of BRAF and KRAS mutations with time to recurrence and overall survival in microsatellite-stable and microsatellite-unstable populations. IC = informed consent; MSI = microsatellite-unstable; MSS = microsatellite-stable; TR = translational research; WT = wild-type.
KRAS and BRAF Results
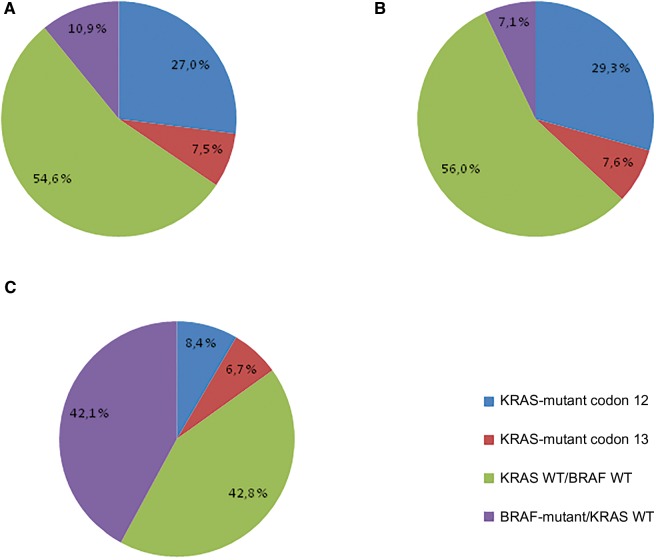
Out of the 4411 patients in the molecular study, 2409 (54.6%) were double wild-type, 1522 (34.5%) were KRAS mutant (exon 2), and 480 (10.9%) were BRAF mutant (Figure 2A). Altogether, KRAS mutations were located on codon 12 in 27.0% of the cases and on codon 13 in 7.5% of the cases. KRAS mutations were more frequent in men in proximal and N1 stage colon cancer. BRAF mutations were more frequent in women in T3 stage and proximal colon cancer (Table 1). Out of the 3934 MSS tumors, 2205 (56.0%) were double wild-type, 1450 (36.9%) were KRAS mutant, and 279 (7.1%) were BRAF mutant (Figure 2B). Out of the 477 MSI tumors, 204 (42.8%) were double wild-type, 72 (15.1%) were KRAS mutant, and 201 (42.1%) were BRAF mutant (Figure 2C). In the whole data set, as all patients didn’t provide consent for the current translational research project, KRAS-mutated tumors were more frequently found in the FOLFOX group and BRAF-mutated tumors were more frequently found in the FOLFOX + cetuximab arm. Moreover, in N0147, as after the amendment to restrict random assignment to only patients with KRAS wild-type tumors (patients with KRAS-mutant tumors were all treated with FOLFOX), KRAS-mutated tumors were more frequently found in the FOLFOX group.
Figure 2.
KRAS and BRAF mutation frequency in overall (A), microsatellite-stable (B), and microsatellite-unstable (C) tumors. MSI = microsatellite-unstable; MSS = microsatellite-stable; WT = wild-type.
Table 1.
Demographic and clinical characteristics—overall population patients*
| Characteristics | N0147 |
PETACC8 |
||||||
|---|---|---|---|---|---|---|---|---|
| Kras-mutant/ Braf WT | Kras WT/ Braf WT | Braf-mutant/ Kras WT | Total | Kras-mutant/ Braf WT | Kras WT/ Braf WT | Braf-mutant/ Kras WT | Total | |
| MMR status, No. (%) | 990 | 1454 | 337 | 2781 | 532 | 955 | 143 | 1630 |
| pMMR | 945 (95.5) | 1331 (91.5) | 189 (56.1) | 2465 (88.6) | 505 (94.9) | 874 (91.5) | 90 (62.9) | 1469 (90.1) |
| dMMR | 45 (4.5) | 123 (8.5) | 148 (43.9) | 316 (11.4) | 27 (5.1) | 81 (8.5) | 53 (37.1) | 161 (9.9) |
| Treatment arm, No. (%) | 990 | 1454 | 337 | 2781 | 532 | 955 | 143 | 1630 |
| FOLFOX | 660 (66.7) | 723 (49.7) | 154 (45.7) | 1537 (55.3) | 280 (52.6) | 478 (50.1) | 67 (46.9) | 825 (50.6) |
| FOLFOX+ cetuximab | 330 (33.3) | 731 (50.3) | 183 (54.3) | 1244 (44.7) | 252 (47.4) | 477 (49.9) | 76 (53.1) | 805 (49.4) |
| Sex, No. (%) | 990 | 1454 | 337 | 2781 | 532 | 955 | 143 | 1630 |
| Male | 510 (51.5) | 838 (57.6) | 119 (35.3) | 1467 (52.8) | 295 (55.5) | 570 (59.7) | 65 (45.5) | 930 (57.1) |
| Female | 480 (48.5) | 616 (42.4) | 218 (64.7) | 1314 (47.2) | 237 (44.5) | 385 (40.3) | 78 (54.5) | 700 (42.9) |
| Age, No. | 990 | 1454 | 337 | 2781 | 532 | 955 | 143 | 1630 |
| Mean (SD), y | 57.43 (10.79) | 55.88 (11.00) | 64.37 (8.91) | 57.46 (11.01) | 59.90 (9.31) | 58.83 (9.70) | 59.94 (9.28) | 59.28 (9.54) |
| Median, y | 58.00 | 56.00 | 65.00 | 58.00 | 61.00 | 60.00 | 61.00 | 60.00 |
| Q1, Q3, y | 50.00, 65.00 | 49.00, 64.00 | 58.00, 70.00 | 50.00, 66.00 | 54.00, 67.00 | 53.00, 66.00 | 54.00, 67.00 | 54.00, 67.00 |
| Min, Max, y | 22.00, 85.00 | 19.00, 84.00 | 31.00, 86.00 | 19.00, 86.00 | 23.00, 74.00 | 19.00, 75.00 | 27.00, 74.00 | 19.00, 75.00 |
| ECOG PS, No. (%) | 988 | 1452 | 336 | 2776 | 509 | 926 | 136 | 1571 |
| 0 | 766 (77.5) | 1122 (77.3) | 246 (73.2) | 2134 (76.9) | 410 (80.6) | 758 (81.9) | 102 (75.0) | 1270 (80.8) |
| 1 | 213 (21.6) | 322 (22.2) | 87 (25.9) | 622 (22.4) | 97 (19.1) | 164 (17.7) | 34 (25.0) | 295 (18.8) |
| 2 | 9 (0.9) | 8 (0.6) | 3 (0.9) | 20 (0.7) | 2 (0.4) | 3 (0.3) | 0 (0.0) | 5 (0.3) |
| 3 | – | – | – | – | 0 (0.0) | 1 (0.1) | 0 (0.0) | 1 (0.1) |
| Tumor-sidedness, No. (%) | 987 | 1452 | 337 | 2776 | 527 | 948 | 143 | 1618 |
| Left | 400 (40.5) | 899 (61.9) | 52 (15.4) | 1351 (48.7) | 289 (54.8) | 644 (67.9) | 36 (25.2) | 969 (59.9) |
| Right | 573 (58.1) | 536 (36.9) | 280 (83.1) | 1389 (50.0) | 225 (42.7) | 298 (31.4) | 106 (74.1) | 629 (38.9) |
| Both | 14 (1.4) | 17 (1.2) | 5 (1.5) | 36 (1.3) | 13 (2.5) | 6 (0.6) | 1 (0.7) | 20 (1.2) |
| N stage, No. (%) | 990 | 1454 | 337 | 2781 | 532 | 955 | 143 | 1630 |
| N1 | 607 (61.3) | 858 (59.0) | 163 (48.4) | 1628 (58.5) | 340 (63.9) | 603 (63.1) | 76 (53.1) | 1019 (62.5) |
| N2 | 383 (38.7) | 596 (41.0) | 174 (51.6) | 1153 (41.5) | 192 (36.1) | 352 (36.9) | 67 (46.9) | 611 (37.5) |
| Grouped T stage, No. (%) | 989 | 1454 | 337 | 2780 | 532 | 955 | 142 | 1629 |
| T1/T2 | 150 (15.2) | 236 (16.2) | 32 (9.5) | 418 (15.0) | 43 (8.1) | 104 (10.9) | 7 (4.9) | 154 (9.5) |
| T3 | 699 (70.7) | 1072 (73.7) | 250 (74.2) | 2021 (72.7) | 365 (68.6) | 663 (69.4) | 104 (73.2) | 1132 (69.5) |
| T4 | 140 (14.2) | 146 (10.0) | 55 (16.3) | 341 (12.3) | 124 (23.3) | 188 (19.7) | 31 (21.8) | 343 (21.1) |
| Grouped histological grade, No. (%) | 990 | 1454 | 337 | 2781 | 525 | 944 | 141 | 1610 |
| G3/G4 | 204 (20.6) | 325 (22.4) | 164 (48.7) | 693 (24.9) | 91 (17.3) | 156 (16.5) | 53 (37.6) | 300 (18.6) |
| G1/G2 | 786 (79.4) | 1129 (77.6) | 173 (51.3) | 2088 (75.1) | 434 (82.7) | 788 (83.5) | 88 (62.4) | 1310 (81.4) |
ECOG = Eastern Oncology Cooperative Group; MMR = mismatch repair; PS = performance status; WT = wild-type.
Outcome in the Whole Population
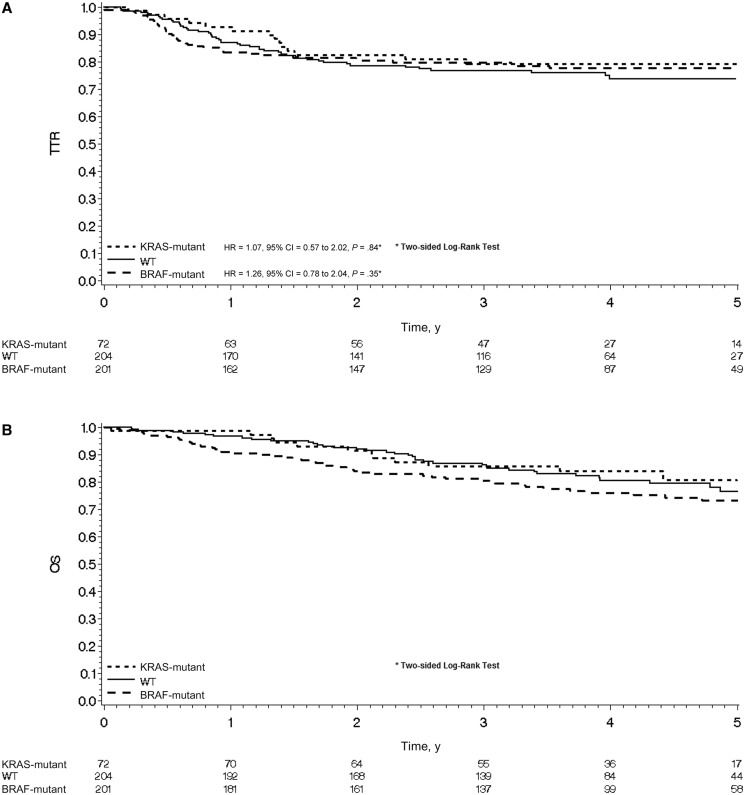
Median follow-up durations were 4.1 years (95% CI = 4.0 to 4.2 years), 4.2 years (95% CI = 4.1 to 4.3 years), and 4.5 years (95% CI = 4.3 to 4.8 years) for patients with wild-type tumors, KRAS-mutated tumors, and BRAF-mutated tumors, respectively. Both mutations were associated with shorter TTR and OS compared with wild-type patients (Supplementary Figure 1, A and B, available online).
In multivariable analysis including all clinically or statistically significant variables, both mutations remained associated with shorter TTR and OS (Table 2). Interestingly, OS was shorter in this pooled analysis, mixing patients with KRAS mutations, BRAF mutation, and double wild-type in patients treated with FOLFOX + cetuximab compared with FOLFOX alone.
Table 2.
Multivariable Cox proportional hazards regression models for TTR and OS—overall population
| Factors | TTR |
OS |
||
|---|---|---|---|---|
| HR (95% CI) | P* | HR (95% CI) | P* | |
| Mutational status | ||||
| Double WT | 1.00 (referent) | 1.00 (referent) | ||
| KRAS-mutant | 1.59 (1.40 to 1.81) | <.001 | 1.51 (1.29 to 1.77) | <.001 |
| BRAF-mutant | 1.27 (1.04 to 1.56) | .02 | 1.49 (1.20 to 1.86) | <.001 |
| Sex | ||||
| Male | 1.00 (referent) | 1.00 (referent) | ||
| Female | 0.88 (0.78 to 0.99) | .04 | 0.76 (0.66 to 0.88) | <.001 |
| Age, y | ||||
| ≤70 | 1.00 (referent) | 1.00 (referent) | ||
| >70 | 0.97 (0.80 to 1.17) | .76 | 1.50 (1.24 to 1.82) | <.001 |
| Histopathology grade | ||||
| G1/G2 | 1.00 (referent) | 1.00 (referent) | ||
| G3/G4 | 1.19 (1.04 to 1.36) | .01 | 1.40 (1.20 to 1.63) | <.001 |
| Site | ||||
| Left | 1.00 (referent) | 1.00 (referent) | ||
| Right | 1.02 (0.90 to 1.16) | .73 | 1.43 (1.22 to 1.66) | <.001 |
| N stage | ||||
| N1 | 1.00 (referent) | 1.00 (referent) | ||
| N2 | 2.36 (2.09 to 2.66) | <.001 | 2.29 (1.98 to 2.64) | <.001 |
| T stage | ||||
| T1/T2 | 1.00 (referent) | 1.00 (referent) | ||
| T3 | 2.55 (1.93 to 3.37) | <.001 | 2.20 (1.59 to 3.05) | <.001 |
| T4 | 4.83 (3.58 to 6.51) | <.001 | 4.12 (2.91 to 5.84) | <.001 |
| Treatment | ||||
| Folfox + cetuximab | 1.00 (referent) | 1.00 (referent) | ||
| Folfox | 0.93 (0.83 to 1.05) | .24 | 0.82 (0.71 to- 0.94) | <.001 |
| ECOG performance status | ||||
| 0 | 1.00 (referent) | 1.00 (referent) | ||
| 1/2 | 1.12 (0.98 to 1.29) | .10 | 1.46 (1.25 to 1.71) | <.001 |
Two-sided P values were calculated using Cox proportional hazards models.
CI = confidence Interval; ECOG = Eastern Oncology Cooperative Group; HR = hazard ratio; OS = overall survival; TTR = time to recurrence; WT = wild-type.
Outcome in MSS Patients
In the MSS cohort, KRAS-mutated tumors were equally numerous in both treatment arms, as were BRAF-mutated tumors. An interaction test between mutational status (wild-type, BRAF and KRAS) and treatment was not statistically significant (TTR P = .38, OS P = .16), leading to the conclusion that the association of KRAS and BRAF mutations and TTR and OS could be analyzed independently of the treatment received.
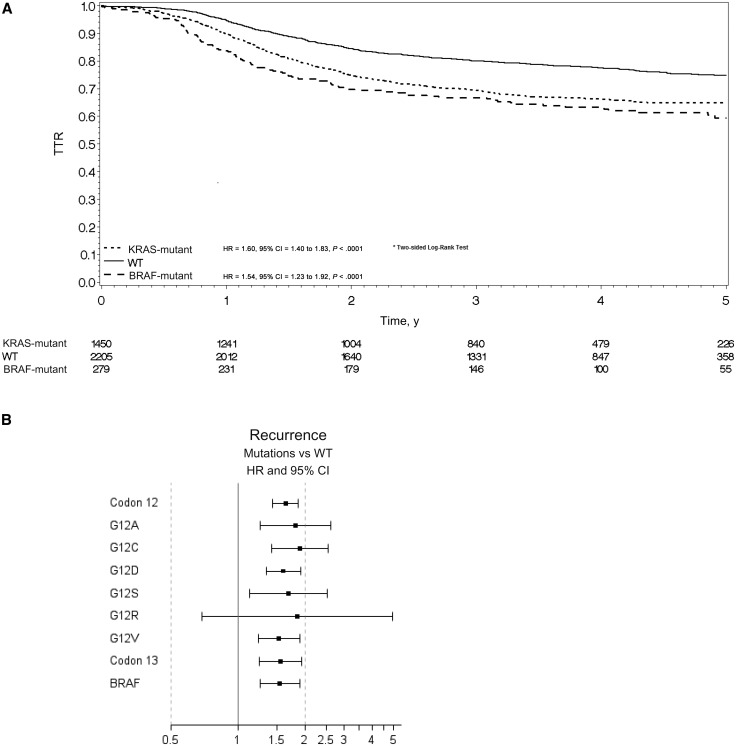
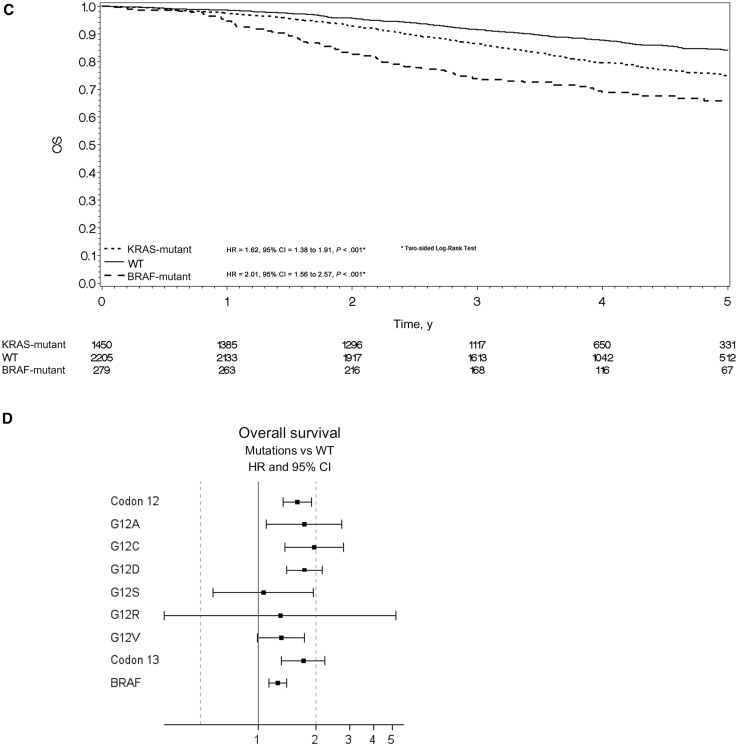
Compared with double wild-type patients, TTR was statistically significantly shorter in KRAS-mutated patients (HR = 1.60, 95% CI = 1.40 to 1.83, P < .001) and in BRAF-mutated patients (HR = 1.54, 95% CI = 1.23 to 1.92, P < .001) (Figure 3A). The rates of patients alive and without disease recurrence at three years were 66.8%, 69.5%, and 80.0% in the BRAF-mutant, KRAS-mutant, and double wild-type groups, respectively. All codon 12 alterations, p.G13D mutation, and BRAF V600E mutation were associated with shorter TTR. Figure 3B summarizes the role of individual mutation on TTR.
Figure 3.
Time to recurrence (TTR) and overall survival (OS) for microsatellite-stable (MSS) population. A) Kaplan-Meier curve for time to recurrence (TTR) in the MSS population by mutational status. Hazard ratios and 95% confidence intervals were calculated using the adjusted Cox proportional hazard model. B) Univariate analysis was performed using the Cox proportional hazard model for TTR in the MSS population by specific mutations. Solid circles represent hazard ratio, and open-ended horizontal lines represent the 95% confidence intervals. C) Kaplan-Meier curve for overall survival (OS) in the MSS population by mutational status. Hazard ratios and 95% confidence intervals were calculated using the adjusted Cox proportional hazard model. D) Univariate analysis was performed using the Cox proportional hazard model for death in the MSS population by specific mutations. Solid circles represent hazard ratio, and open-ended horizontal lines represent the 95% confidence intervals. CI = confidence interval; HR = hazard ratio; OS = overall survival; TTR = time to recurrence; WT = wild-type.
OS was also statistically significantly shorter in KRAS-mutated patients (HR = 1.62, 95% CI = 1.38 to 1.91, P < .001) and in BRAF-mutated patients (HR = 2.01, 95% CI = 1.56 to 2.57, P < .001) (Table 3 and Figure 3C). The rates of patients alive at three years were 73.9%, 86.3%, and 91.4% in the BRAF-mutant, KRAS-mutant, and double wild-type groups, respectively. All KRAS codon 12 alterations, p.G13D mutation, and BRAF V600E mutation were associated with shorter OS. Figure 3D summarizes the role of individual mutation on OS.
Table 3.
Multivariate Cox proportional Hazards Regression models for TTR and OS in MSS and MSI Patients
| TTR |
OS |
|||
|---|---|---|---|---|
| Population and Factors | HR (95%CI) | P* | HR (95%CI) | P* |
| MSS Patients | ||||
| Mutational Status | ||||
| Double WT | 1.00 (referent) | 1.00 (referent) | ||
| KRAS Mutant | 1.60 (1.40 to 1.82) | <.001 | 1.62 (1.38 to 1.91) | <.001 |
| BRAF Mutant | 1.54 (1.23 to 1.92) | <.001 | 2.01 (1.56 to 2.57) | <.001 |
| Sex | ||||
| Male | 1.00 (referent) | 1.00 (referent) | ||
| Female | 0.89 (0.79 to 1.01) | .07 | 0.80 (0.69 to 0.94) | .005 |
| Age | ||||
| ≤ 70 years | 1.00 (referent) | 1.00 (referent) | ||
| > 70 years | 0.99 (0.81 to 1.22) | .97 | 1.56 (1.27 to 1.92) | <.001 |
| Histopathology Grade | ||||
| G1/G2 | 1.00 (referent) | 1.00 (referent) | ||
| G3/G4 | 1.25 (1.08 to 1.44) | .003 | 1.48 (1.25 to 1.75) | <.001 |
| N Stage | ||||
| N1 | 1.00 (referent) | 1.00 (referent) | ||
| N2 | 2.22 (1.95 to 2.52) | <.001 | 2.15 (1.84 to 2.51) | <.001 |
| T Stage | ||||
| T1/T2 | 1.00 (referent) | 1.00 (referent) | ||
| T3 | 2.50 (1.88 to 3.31) | <.001 | 2.10 (1.50 to 2.93) | <.001 |
| T4 | 4.70 (3.47 to 6.37) | <.001 | 3.97 (2.78 to 5.68) | <.001 |
| ECOG PS | ||||
| 0 | 1.00 (referent) | 1.00 (referent) | ||
| 1/2 | 1.11 (0.96 to 1.28) | .18 | 1.44 (1.21 to 1.70) | <.001 |
| MSI patients | ||||
| Mutational Status | ||||
| Double WT | 1.00 (referent) | 1.00 (referent) | ||
| KRAS Mutant | 1.04 (0.57 to 1.90) | .91 | 1.07 (0.57 to 2.02) | .84 |
| BRAF Mutant | 0.94 (0.58 to 1.51) | .80 | 1.26 (0.78 to 2.04) | .35 |
| Sex | ||||
| Male | 1.00 (referent) | 1.00 (referent) | ||
| Female | 0.89 (0.57 to 1.37) | .59 | 0.52 (0.34 to 0.81) | .004 |
| Age | ||||
| ≤ 70 years | 1.00 (referent) | 1.00 (referent) | ||
| > 70 years | 0.95 (0.52 to 1.75) | .87 | 1.88 (1.13 to 3.13) | .02 |
| Histopathology Grade | ||||
| G1/G2 | 1.00 (referent) | 1.00 (referent) | ||
| G3/G4 | 0.93 (0.62 to 1.38) | .70 | 1.19 (0.78 to 1.81) | .41 |
| N Stage | ||||
| N1 | 1.00 (referent) | 1.00 (referent) | ||
| N2 | 3.56 (2.33 to 5.45) | <.001 | 3.05 (1.99 to 4.66) | <.001 |
| T Stage | ||||
| T1/T2 | 1.00 (referent) | 1.00 (referent) | ||
| T3 | 7.59 (1.05 to 54.98) | .05 | 7.16 (0.99 to 51.97) | .05 |
| T4 | 13.91 (1.87 to 103.47) | .01 | 11.52 (1.54 to 86.26) | .02 |
| ECOG PS | ||||
| 0 | 1.00 (referent) | 1.00 (referent) | ||
| 1/2 | 1.25 (0.80 to 1.95) | .32 | 1.76 (1.14 to 2.70) | .01 |
Two-sided p-values were calculated using Cox proportional hazards models.
WT=Wild-type; TTR=Time To Recurrence; OS =Overall Survival; HR=Hazard Ratio; CI = Confidence Interval; MSS= Microsatellite stable; MSI = Microsatellite unstable.
These results were also true when looking at FOLFOX arms or FOLFOX + cetuximab arms separately (Supplementary Figure 2, available online).
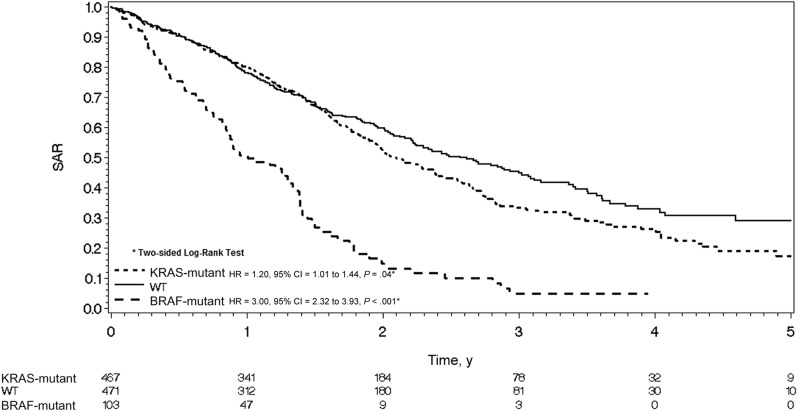
SAR was also shorter in KRAS- and BRAF-mutated patients. Median SARs were 2.57, 2.09, and 1.0 year in double wild-type, KRAS-mutated (HR = 1.20, 95% CI = 1.01 to 1.44, P = .04), and BRAF-mutated (HR = 3.02, 95% CI = 2.32 to 3.93, P < .001) populations, respectively (Figure 4).
Figure 4.
Kaplan-Meier curve for survival after relapse in the microsatellite-stable population by mutational status. Hazard ratios and 95% confidence intervals were calculated using the Cox proportional hazard model. CI = confidence interval; HR = hazard ratio; SAR = survival after relapse; WT = wild-type.
In multivariable analysis, all tested variables were statistically significantly associated with shorter TTR. BRAF mutations (P < .001), KRAS mutations (P < .001), high-grade tumors (P = .003), pT3 and pT4 (P < .001), and pN2 (P < .001) remained associated with shorter TTR (Table 3). Similar results were obtained for OS as shown in Table 3.
Outcome for MSI Patients
In the MSI cohort, there was no difference for TTR in patients with KRAS-mutated tumors (HR = 1.04, 95% CI = 0.57 to 1.90, P = .91) or BRAF-mutated tumors (HR = 0.94, 95% CI = 0.58 to 1.51, P = .80) as compared with wild-type patients (Figure 5A).
Figure 5.
A) Kaplan-Meier curve for time to recurrence in the microsatellite-unstable (MSI) population by mutational status. Hazard ratio and 95% confidence intervals were calculated using the adjusted Cox proportional hazard model or (B) Kaplan-Meier curve for overall survival in the MSI population by mutational status. Hazard ratios and 95% confidence intervals were calculated using the adjusted Cox proportional hazard model. CI = confidence interval; HR = hazard ratio; OS = overall survival; TTR = time to recurrence; WT = wild-type.
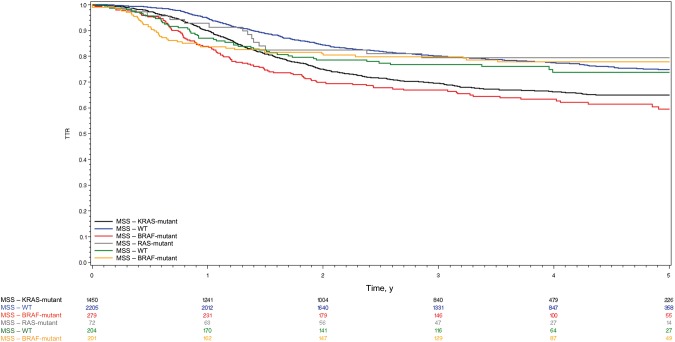
Considering OS, as shown in Figure 5B, no statistically significant differences in patient outcome were seen for KRAS- or BRAF-mutated as compared with wild-type patients (HR = 1.07, 95% CI = 0.57 to 2.02, P = .84; HR = 1.26, 95% CI = 0.78 to 2.04, P = .35). In multivariable analysis among patients with MSI tumors, only pT4 stage (P = .01) and pN2 stage (P < .001) were associated with a statistically significantly shorter TTR (Table 3). Similar results were obtained for OS with pT4 stage (P = .02) and pN2 stage (P < .001) but also age over 70 years (P = .02), male sex (P = .004), and Eastern Cooperative Oncology Group performance status (ECOG-PS) of 1–2 (P = .01), which were all associated with shorter OS (Table 3). No association between KRAS and BRAF mutational status and TTR or OS was observed in multivariable analysis for MSI patients (Table 3). Because of the low number of events to date in the MSI population, SAR for each subgroup was not analyzed in this study. Figure 6 summarizes TTR in all subgroups (ie, MSI and MSS, mutated or not for KRAS and BRAF).
Figure 6.
Kaplan-Meier curve for time to recurrence in the microsatellite-stable and the microsatellite-unstable population according to mutational status. MSI = microsatellite-unstable; MSS = microsatellite-stable; TTR = time to recurrence; WT = wild-type.
Discussion
The aim of this study was to determine the independent prognostic value of KRAS and BRAF mutations in MSS and MSI subgroups using prospectively collected stage III colon cancer specimens from patients receiving FOLFOX ± cetuximab from two large adjuvant randomized trials. We found that BRAF and KRAS mutations are independently associated with shorter TTR, SAR, and OS in patients with MSS, but not MSI, tumors. The prognostic role of KRAS and BRAF mutations in nonmetastatic colon cancer has been inconsistent. While in some recent publications, including those of PETACC8 and N0147, their poor prognostic value on tumor recurrence has been suggested in MSS patients, contradictory results have been published (9–11,13,16,18–26). Moreover, very few analyses have examined patients treated with standard FOLFOX, reported OS results, analyzed the prognostic role of each individual mutation, and specifically assessed the effect of KRAS and BRAF mutations in MSI patients because of the low incidence of MSI tumors. A recent paper by Cuba et al. suggested that KRAS and BRAF mutations may be poor prognostic factors in MSI early-stage colon cancer (27). This paper included a relatively small number of patients (n = 143) and mixed stage II and stage III tumors. Furthermore, patient receipt of adjuvant chemotherapy was not controlled, which limits the validity of the reported results. To our knowledge, our study is the largest series evaluating the potential prognostic role of KRAS and BRAF mutations stratified by MSI/MSS status in stage III colon cancer patients treated with a homogeneous and standard adjuvant chemotherapy regimen.
Discrepancies regarding the prognostic value of KRAS mutations in the current literature (9,10,13,18–20,23,24) may be due to the heterogeneity of the study populations, the influence of primary tumor site, tumor stage, and adjuvant treatment received. No association between KRAS mutations and relapse or survival was found in the PETACC-3 trial among 1404 colon cancer patients treated with 5-FU +/− irinotecan (10). In contrast, the QUASAR study (20), which mainly included stage II patients, showed an increased risk of recurrence among patients with KRAS-mutated tumors, which was not affected by adjuvant chemotherapy. The RASCAL population-based studies (18,26) showed that only one specific KRAS mutation, p.G12V, was associated with poorer outcome, suggesting differences between mutations. Similarly, Imamura et al. showed that KRAS mutations at codon 12 but not at codon 13 (HR = 1.25, 95% CI = 0.85 to 1.84) negatively affected cancer-specific survival (22). In contrast, data from the N0147 trial showed that KRAS mutations at codons 12 (HR = 1.52, 95% CI = 1.28 to 1.8, P < .0001) and 13 (HR = 1.36, 95% CI = 1.04 to 1.72, P = .025) both had prognostic value (22). Interestingly, in most studies MSI status was not included in the analyses. In the present work, we have for the first time evaluated a clinical trial cohort with a sufficient number of patients to analyze the individual prognostic role of codons 12 and 13 on TTR and OS in MSI (n = 477) and MSS (n = 3934) patients, respectively.
We show here that in MSS patients, all codon 12 alterations, and the p.G13D mutation are associated with shorter TTR and OS, even if rare mutations such as G12S and G12R do not reach statistical significance because of broad confidence intervals. KRAS mutations are also associated with a shorter SAR as compared with double wild-type patients. In contrast, KRAS mutations do not seem to be associated with shorter TTR or OS in MSI patients. This seems in accordance with our prior data from PETACC8 and N0147 populations showing that the association of KRAS mutations with prognosis was stronger in distal compared with proximal tumors (9,10), which is the predominant location of MSI cancers.
When looking at the prognostic value of BRAF mutations, it has been mainly suggested that BRAF was associated with shorter OS because of a poor SAR (11,13,19,20,24), but its role on tumor recurrence has been less clear. Moreover, the low incidence of this mutation (5%–10%) precluded most reports from definitive conclusions. We show here, in 480 BRAF-mutated stage III colon cancer patients, that BRAF mutation is associated with shorter TTR and OS as compared with wild-type patients in MSS but not in MSI patients. BRAF mutation is also associated with a shorter SAR as compared with double wild-type patients, as previously reported (10–12) and in accordance with the particularly poor prognosis of BRAF-mutated colon cancer in the metastatic setting (8). Interestingly, SARs reported here for our three study groups are in perfect agreement with the recently published data from the TRIBE trial (8) in a twofold larger population.
The BRAF pathway leads to both MSI and MSS tumors (28), with MLH1 hypermethylation being the event that confers MSI status to these tumors of known better prognosis (29). Mixing a good prognostic factor and a bad one in these MSI/BRAF-mutated tumors may have impacted previous results, suggesting that BRAF mutational status was not associated with increased disease recurrence in colon cancer. In fact, the present analysis shows that MSI/BRAF-mutated tumors have an excellent outcome. At the opposite end of the spectrum, the poor prognostic value of BRAF mutation in MSS patients on disease recurrence, clearly demonstrated here, may be explained by the upregulation of genes regulating epithelial mesenchymal transition and matrix remodeling that may facilitate the metastatic process as described previously (30). This group of MSS BRAF-mutant colon cancer is still poorly studied, but recent studies have already shown that they often display altered immunophenotype with reduced CDX2 and increased CK7 expression and share molecular and clinical features of both the BRAF-related serrated and KRAS-related traditional pathways of colorectal tumorigenesis with more TP53 mutations and less CIMP phenotype than their MSI counterparts (31,32).
Three retrospective analyses of randomized adjuvant trials suggested that BRAF mutation was independently associated with shorter OS but not with disease-free or recurrence-free survival. Recently, data from the NCCTG N0147 trial showed that BRAF mutation was statistically significantly associated with shorter DFS in multivariable analysis (HR = 1.37, 95% CI = 1.08 to 1.70, P = .009) (23). However, the adverse prognostic value of BRAF mutation was limited to MSS patients after stratification on MMR status. Similar results were reported in PETACC8 for DFS and confirmed its adverse role on OS (16). In this last report, longer DFS and a trend toward longer OS in BRAF-mutant MSI patients were also suggested. This result is not confirmed in this pooled analysis, with KRAS-mutant, BRAF-mutant, or double wild-type tumors having very similar outcomes in MSI patients.
Altogether we have identified six patients groups defined by tumor MSI, BRAF and KRAS status, with different outcomes. Four of them, that is, MSS double wild-type, MSI double wild-type, MSI KRAS-mutated, and MSI BRAF-mutated, seem to have a relatively favorable outcome, with long-term (five-year) survival without recurrence ranging from 74% to 80%. Together, these four groups account for 61% of the overall study population. At the opposite end of the spectrum, MSS/KRAS-mutated and MSS/BRAF-mutated tumors, representing 37% and 7% of our study population, respectively, have a less favorable outcome, with long-term (five-year) survival without relapse ranging from 59% to 65%. The same results are observed when looking at SAR and thus OS, which is the resultant of the two previous parameters. Beyond MSI status and KRAS and BRAF, other molecular factors may also influence patient outcome and add value to this molecular classification. Expanded RAS, including KRAS and NRAS assessment, would likely be helpful in the next steps to improving our classification, but also PI3KCA, HER2 (33), MET (34–36), and other molecular alterations that may play a prognostic role in the prognosis of resected colon cancer and will be studied in the future in this pooled study population.
Limitations include the retrospective design of these pooled analyses, the absence of expanded RAS mutation assessment as recommended by European Society for Medical Oncology and National Comprehensive Cancer Network guidelines in the metastatic setting, the relative underrepresentation of KRAS-mutant tumors because of the amendments restricting recruitment to KRAS wild-type patients in both trials in 2008, and the relatively low frequency of stage III MSI tumors.
In patients undergoing surgical resection of CRC, prognosis and management are currently based entirely on TNM classification. Though T and n stages remain major prognostic factors in the present analyses, stage-independent prognostic factors may influence patient outcome. We show here that in MSS stage III colon cancer patients, who represent 90% of the overall stage III population, KRAS and BRAF mutations are biologically significant and clinically relevant prognostic molecular markers for TTR, SAR, and OS. This does not seem to be the case for the MSI counterpart population. MSI status assessment and KRAS and BRAF mutational status determination are thus important stratification factors for future clinical trials dedicated to this population. These molecular assessments in daily practice may also be discussed because they provide potentially important prognostic information for our patients and will help to tailor treatment in case of disease recurrence.
Funding
The PETACC8 study was sponsored by the Fédération Francophone de Cancérologie Digestive (FFCD), which was responsible for study management. Merck KGaA and Sanofi-Aventis supported the study: Merck provided the study cetuximab and financial support for study management; Sanofi-Aventis provided financial support for the provision of oxaliplatin to Belgian sites when necessary.
The N0147 trial was conducted as a collaborative trial of the North Central Cancer Treatment Group (NCCTG), Mayo Clinic, which is now part of the Alliance for Clinical Trials in Oncology. The study and was supported in part by grants from the US National Cancer Institute to the NCCTG (grant numbers CA-25224, CA-32102, CA-14028, CA-49957, CA-21115 CA-12027, CA-37377. CA-3704, CA-35103, CA-35113, CA-35272), the NCCTG Biospecimen Resource (grant number CA-114740), the Alliance for Clinical Trials in Oncology (grant numbers U10CA1808821 and U10CA180882). Bristol-Myers Squibb, ImClone Systems Inc., Sanofi Aventis, and Pfizer provided unrestricted support for the trial. Bristol-Myers Squibb provided cetuximab to the NCCTG for conduct of the study.
F. Sinicrope was supported, in part, by a US National Cancer Institute Senior Scientist Award (K05CA-142885).
Notes
The study was sponsored by “Fédération Francophone de Cancérologie Digestive” (FFCD), which was responsible for study management; for design and conduct of the study; for collection, management, analysis, and interpretation of the data; for preparation, review, and approval of the manuscript; and for the decision to submit the manuscript for publication.
Merck KGaA and Sanofi-Aventis were not involved in the design; the collection, analysis, or interpretation of the data; the writing or submission. The National Cancer Institute participated in the design and conduct of the trial, but not in the data collection, analysis, and interpretation, nor in the preparation, review, or approval of the manuscript. Bristol-Myers Squibb, ImClone, sanofi-aventis, and Pfizer had no role in the design or conduct of the trial; in the collection, management, analysis, or interpretation of the data; or in the preparation, review, or approval of the manuscript.
Julien Taieb and Frank Sinicrope had full access to the data and had final responsibility for the decision to submit for publication.
N0147 represents a collaborative effort between the North Central Cancer Treatment Group and the Cancer and Leukemia Group B (now collectively named the Alliance for Clinical Trials in Oncology); Eastern Cooperative Oncology Group; National Cancer Institute of Canada Clinical Trials Group; National Surgical Adjuvant Breast and Bowel Project; and Southwest Oncology Group.
J. Taieb received honoraria from Sanofi and Merck KGaA. Jean-François Emile received honoraria from Merck-Serono. J. Tabernero received honoraria from Amgen, Merck KGaA, and Sanofi. G. Folprecht declared research funding from Merck KGaA and honoraria from Merck KGaA, Roche, Lilly, BMS, and Amgen; P. Laurent-Puig declared providing advisory roles and lectures for Sanofi, Merck Serono, Amgen, Roche, Genomic Health, Myriad Genetics, and Pfizer. All remaining authors have declared no conflicts of interest.
Supplementary Material
References
- 1. Boyle P, Langman JS.. ABC of colorectal cancer: Epidemiology. BMJ. 2000;321(7264):805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. [DOI] [PubMed] [Google Scholar]
- 3. Benson AB 3rd, Venook AP, Bekaii-Saab T, et al. Colon cancer, version 3.2014. J Natl Compr Canc Netw. 2014;12(7):1028–1059. [DOI] [PubMed] [Google Scholar]
- 4. Van Cutsem E, Cervantes A, Nordlinger B, et al. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(suppl 3):iii1–iii9. [DOI] [PubMed] [Google Scholar]
- 5. Bokemeyer C, Bondarenko I, Hartmann JT, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: The OPUS study. Ann Oncol. 2011;22(7):1535–1546. [DOI] [PubMed] [Google Scholar]
- 6. Lievre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26(3):374–379. [DOI] [PubMed] [Google Scholar]
- 7. Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319(9):525–532. [DOI] [PubMed] [Google Scholar]
- 8. Loupakis F, Cremolini C, Masi G, et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med. 2014;371(17):1609–1618. [DOI] [PubMed] [Google Scholar]
- 9. Blons H, Emile JF, Le Malicot K, et al. Prognostic value of KRAS mutations in stage III colon cancer: Post hoc analysis of the PETACC8 phase III trial data set. Ann Oncol. 2014;25(12):2378–2385. [DOI] [PubMed] [Google Scholar]
- 10. Sinicrope FA, Mahoney MR, Yoon HH, et al. Analysis of molecular markers by anatomic tumor site in stage III colon carcinomas from Adjuvant Chemotherapy Trial NCCTG N0147 (Alliance). Clin Cancer Res. 2015;21(23):5294–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gavin PG, Colangelo LH, Fumagalli D, et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: An assessment of their prognostic and oxaliplatin predictive value. Clin Cancer Res. 2012;18(23):6531–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ogino S, Shima K, Meyerhardt JA, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: Results from intergroup trial CALGB 89803. Clin Cancer Res. 2012;18(3):890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: Results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28(3):466–474. [DOI] [PubMed] [Google Scholar]
- 14. Alberts SR, Sargent DJ, Nair S, et al. Effect of oxaliplatin, fluorouracil, and leucovorin with or without cetuximab on survival among patients with resected stage III colon cancer: A randomized trial. JAMA. 2012;307(13):1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Taieb J, Tabernero J, Mini E, et al. Oxaliplatin, fluorouracil, and leucovorin with or without cetuximab in patients with resected stage III colon cancer (PETACC-8): An open-label, randomised phase 3 trial. Lancet Oncol. 2014;15(8):862–873. [DOI] [PubMed] [Google Scholar]
- 16. Taieb J, Zaanan A, Le Malicot K, et al. Prognostic effect of BRAF and KRAS mutations in patients with stage III colon cancer treated with leucovorin, fluorouracil, and oxaliplatin with or without cetuximab: A post hoc analysis of the PETACC-8 Trial. JAMA Oncol. 2016; in press. [DOI] [PubMed] [Google Scholar]
- 17. Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96(4):261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: The ′RASCAL II′ study. Br J Cancer. 2001;85(5):692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farina-Sarasqueta A, van Lijnschoten G, Moerland E, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21(12):2396–2402. [DOI] [PubMed] [Google Scholar]
- 20. Hutchins G, Southward K, Handley K, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29(10):1261–1270. [DOI] [PubMed] [Google Scholar]
- 21. Imamura Y, Morikawa T, Liao X, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer Res. 2012;18(17):4753–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morikawa T, Kuchiba A, Qian ZR, et al. Prognostic significance and molecular associations of tumor growth pattern in colorectal cancer. Ann Surg Oncol. 2012;19(6):1944–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sinicrope FA, Shi Q, Smyrk TC, et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology. 2015;148(1):88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yokota T. Are KRAS/BRAF mutations potent prognostic and/or predictive biomarkers in colorectal cancers? Anticancer Agents Med Chem. 2011;12(2):163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoon HH, Tougeron D, Shi Q, et al. KRAS codon 12 and 13 mutations in relation to disease-free survival in BRAF-wild-type stage III colon cancers from an adjuvant chemotherapy trial (N0147 alliance). Clin Cancer Res. 2014;20(11):3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: The multicenter "RASCAL" study. J Natl Cancer Inst. 1998;90(9):675–684. [DOI] [PubMed] [Google Scholar]
- 27. Benedix F, Meyer F, Kube R, et al. Influence of anatomical subsite on the incidence of microsatellite instability, and KRAS and BRAF mutation rates in patients with colon carcinoma. Pathol Res Pract. 2012;208(10):592–597. [DOI] [PubMed] [Google Scholar]
- 28. de Cuba EM, Snaebjornsson P, Heideman DA, et al. Prognostic value of BRAF and KRAS mutation status in stage II and III microsatellite instable colon cancers. Int J Cancer. 2016;138(5):1139–1145. [DOI] [PubMed] [Google Scholar]
- 29. Leggett B, Whitehall V.. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138(6):2088–2100. [DOI] [PubMed] [Google Scholar]
- 30. Kim YH, Kakar S, Cun L, et al. Distinct CpG island methylation profiles and BRAF mutation status in serrated and adenomatous colorectal polyps. Int J Cancer. 2008;123(11):2587–2593. [DOI] [PubMed] [Google Scholar]
- 31. De Sousa EMF, Wang X, Jansen M, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–618. [DOI] [PubMed] [Google Scholar]
- 32. Popat S, Hubner R, Houlston RS.. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23(3):609–618. [DOI] [PubMed] [Google Scholar]
- 33. Bond CE, Umapathy A, Ramsnes I, et al. p53 mutation is common in microsatellite stable, BRAF mutant colorectal cancers. Int J Cancer. 2012;130(7):1567–1576. [DOI] [PubMed] [Google Scholar]
- 34. Ogino S, Liao X, Imamura Y, et al. Predictive and prognostic analysis of PIK3CA mutation in stage III colon cancer intergroup trial. J Natl Cancer Inst. 2013;105(23):1789–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seo AN, Park KU, Choe G, et al. Clinical and prognostic value of MET gene copy number gain and chromosome 7 polysomy in primary colorectal cancer patients. Tumour Biol. 2015;36(12):9813–9821. [DOI] [PubMed] [Google Scholar]
- 36. Kavuri SM, Jain N, Galimi F, et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov. 2015;5(8):832–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.