Abstract
Plasma fetuin-A is associated with type 2 diabetes, and AHSG, the gene encoding fetuin-A, has been identified as a susceptibility locus for diabetes and metabolic syndrome. Thus far, unbiased investigations of the genetic determinants of plasma fetuin-A concentrations have not been conducted. We searched for single nucleotide polymorphisms (SNPs) related to fetuin-A concentrations by a genome-wide association study in six population-based studies.
We examined the association of fetuin-A levels with ∼ 2.5 million genotyped and imputed SNPs in 9,055 participants of European descent and 2,119 African Americans. In both ethnicities, the strongest associations were centered in a region with a high degree of LD near the AHSG locus. Among 136 genome-wide significant (P < 0.05 × 10−8) SNPs near the AHSG locus, the top SNP was rs4917 (P =1.27 × 10−303), a known coding SNP in exon 6 that is associated with a 0.06 g/l (∼13%) lower fetuin-A level. This variant alone explained 14% of the variation in fetuin-A levels. Analyses conditioned on rs4917 indicated that the strong association with the AHSG locus stems from additional independent associations of multiple variants among European Americans. In conclusion, levels of fetuin-A in plasma are strongly associated with SNPs in its encoding gene, AHSG, but not elsewhere in the genome. Given the strength of the associations observed for multiple independent SNPs, the AHSG gene is an example of a candidate locus suitable for additional investigations including fine mapping to elucidate the biological basis of the findings and further functional experiments to clarify AHSG as a potential therapeutic target.
Introduction
Fetuin-A is a liver-derived protein that is involved in the metabolism of calcified minerals and the regulation of the insulin signaling (1). Fetuin-A forms complexes with circulating calcium and phosphorus and increases the solubility of these minerals (2), thereby inhibiting arterial calcium deposition. Fetuin-A also directly binds and inhibits the insulin receptor, resulting in insulin resistance (3–5). The fetuin-A knock-out mouse has improved insulin sensitivity by euglycemic clamp experiments, lower triglyceride and free fatty acid levels, resistance to weight gain, and less adiposity (6,7). High fetuin-A levels are associated with insulin resistance (8–10), interacts with circulating free fatty acids in determining insulin sensitivity and predicts incident diabetes (11–16). Thus, fetuin-A represents an emerging biomarker for improved diabetes risk assessment in clinical practice and a potential therapeutic target for primary or secondary prevention of type 2 diabetes.
The alpha-2-HS-glycoprotein (AHSG) locus on chromosome 3 (3q27) encodes the fetuin-A gene. Linkage studies have identified this region as a susceptibility locus for the metabolic syndrome and T2DM (17,18), and small case-control studies have reported strong relations between several single nucleotide polymorphisms (SNPs) identified from direct exon sequencing studies and plasma concentrations of fetuin-A (19–22). However, a comprehensive investigation of genetic determinants of fetuin-A levels has not been conducted, and it remains unknown if genetic loci distinct from the AHSG locus can be identified that regulate fetuin-A concentrations. Hence, to better understand the genetic control of fetuin-A levels we conducted a genome-wide association (GWA) analysis in six population-based studies, as part of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium.
Results
Our analyses included data from six studies of European Americans totaling 9,055 individuals and four studies comprising a total of 2,119 African Americans. The majority of the participants were female and the average age ranged from 52 years in ARIC to 75 years in CHS. Mean fetuin-A levels ranged from 0.43 ± 0.09 g/l in African Americans from CHS to 1.00 ± 0.41 g/l in European Americans in HABC. However, most studies had smaller standard deviations than HABC and mean fetuin-A levels averaged around 0.50 g/l (Table 1).
Table 1.
Descriptive characteristics of the cohorts included in the genome-wide analysis according to ancestry
| Cohort | N | Age, years | Women, % | Fetuin-A level, g/l | Prevalent diabetes, % |
|---|---|---|---|---|---|
| CHS/European | 2824 | 74.9 (5.0) | 60.6 | 0.48 (0.10) | 15.7 |
| CHS/African | 727 | 73.2 (5.6) | 63.4 | 0.43 (0.09) | 20.9 |
| ARIC/European | 485 | 53.4 (5.6) | 55.7 | 0.51 (0.07) | 0 |
| ARIC/African | 366 | 51.9 (5.4) | 55.7 | 0.47 (0.09) | 0 |
| HABC/European | 403 | 73.9 (2.9) | 56.8 | 1.00 (0.41) | 13.9 |
| HABC/African | 350 | 73.5 (3.0) | 54.9 | 0.91 (0.37) | 20.6 |
| MESA/European | 1093 | 62.7 (10.1) | 52.0 | 0.49 (0.11) | 3.9 |
| MESA/African | 678 | 62.0 (10.1) | 52.3 | 0.45 (0.10) | 10.5 |
| NHS/European | 1029 | 59.9 (6.4) | 100 | 0.46 (0.11) | 9.3 |
| FHS/European | 3592 | 40.0 (8.7) | 53.6 | 0.45 (0.18) | 2.5 |
Numbers in table are Mean (SD) or percentage. ARIC= Atherosclerosis Risk in Communities Study; CHS = Cardiovascular Health Study; FHS = Framingham Heart Study; HABC= Health, Aging, and Body Composition (Health ABC) Study; MESA= Multi-Ethnic Study of Atherosclerosis, NHS= Nurses’ Health Study.
Diabetes was defined as fasting blood glucose >125 mg/dL, a random blood glucose of >200 mg/dL, or use of insulin or oral hypoglycemic agents.
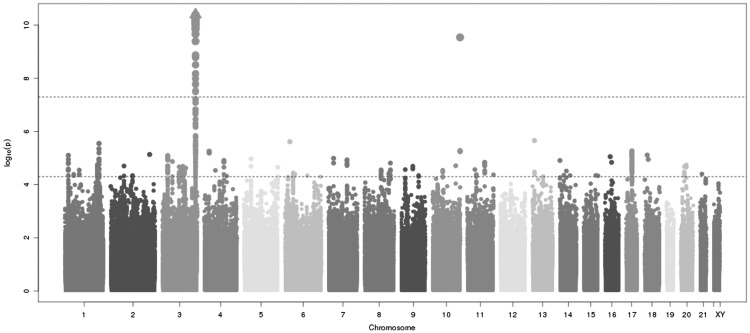
Separate meta-analyses of 2.5 million SNPs for European and African American participants who contributed to the study-specific genome-wide association analyses of fetuin-A levels, identified a very strong signal on chromosome 3q27 (Fig. 1, Supplementary Material, Fig. S1). In total, 136 SNPs at the AHSG locus achieved genome-wide significance among European Americans. The top SNP, rs4917 (P-value =1.27×10−303), is located in the AHSG gene which encodes the fetuin-A protein. The genetic region and the linkage disequilibrium (LD) of SNPs with rs4917 according to r2 in CEU are displayed in Supplementary Material, Figure S2A and B. The triangles represent the top two non-synonymous SNPs (rs4917 in purple and rs4918 in red). Detailed results on the 136 SNPs are available in the online Supplementary Material, Table S3. Though the list of genes in closest proximity to the SNPs includes FETUB, CRYGS, DNAJB11, KGN1, and TBCCD1, these genes are all within close distance to AHSG, as shown in Supplementary Material, Figure S2A. Each additional copy of the rs4917 minor allele (T variant allele, minor allele frequency =0.32) was associated with approximately 0.066 ±0.002 g/l lower fetuin-A level in each of the cohorts (Table 2). rs4917 alone explained 14% of the variation in fetuin-A levels among European Americans.
Figure 1.
Meta-analysis of six genome-wide association analyses of fetuin-A levels in a total of 9,055 European Americans. Figure displays p-value for association for each SNP on a log10 scale
Table 2.
Association of the top SNPs (rs4917 in European Americans; rs1900618 in African Americans) on chromosome 3 with fetuin-A levels in European and African Americans
| rs4917 |
||||||||
|---|---|---|---|---|---|---|---|---|
| European Americans (MAF: 0.32) |
African Americans (MAF: 0.26) |
|||||||
| Cohort | N | β | SE | p | N | β | SE | P |
| CHS | 2742 | −0.0632 | 0.0023 | 2.47E-163 | 725 | −0.0447 | 0.0046 | 3.58E-22 |
| ARIC | 485 | −0.0546 | 0.0046 | 1.03E-32 | 366 | −0.0302 | 0.0082 | 2.48E-04 |
| HABC | 403 | −0.11 | 0.031 | 4.44E-04 | 350 | 0.001 | 0.029 | 0.98 |
| MESA | NA | NA | NA | NA | 678 | −0.0433 | 0.0061 | 1.13E-12 |
| NHS | 741 | −0.069 | 0.053 | 6.71E-50 | NA | NA | NA | NA |
| FHS | 3592 | −0.080 | 0.0032 | 3.40E-80 | NA | NA | NA | NA |
| Combined | 7963 | −0.0657 | 0.0018 | 1.27E-303 | 2119 | −0.0413 | 0.0034 | 1.20E-34 |
|
rs1900618 |
||||||||
|
European Americans (MAF: 0.33) |
African Americans (MAF: 0.33) |
|||||||
| Cohort | N | β | SE | p | N | β | SE | p |
| CHS | 2742 | −0.0635 | 0.0023 | 2.27E-162 | 725 | −0.0505 | 0.0040 | 2.58E-36 |
| ARIC | 485 | −0.0542 | 0.0046 | 2.10E-32 | 366 | −0.0345 | 0.0074 | 3.02E-06 |
| HABC | 403 | 0.11 | 0.031 | 4.44E-04 | 350 | −0.015 | 0.027 | 0.59 |
| MESA | NA | NA | NA | NA | 678 | −0.0514 | 0.0057 | 3.35E-19 |
| NHS | 741 | −0.0691 | 0.0053 | 6.57E-50 | NA | NA | NA | NA |
| FHS | 3592 | −0.0805 | 0.0042 | 1.03E-80 | NA | NA | NA | NA |
| Combined | 7963 | −0.0659 | 0.0018 | 6.44E-303 | 2119 | −0.0477 | 0.003 | 1.58E-56 |
The rs4917 and rs1900618 were identified as the top SNPs among European Americans and African Americans, respectively. The SNPs are in almost perfect LD in HapMap CEU population (Caucasians): D| =1 and r2 = 0.96. In HapMap YRI population= D| =1 and r2 = 0.57.
NA = SNP not available.
The plots in Supplementary Material, Figure S2 show that the strongest associations are centered in a region with a high degree of LD. However, as shown in Supplementary Material, Figure S2B, five of the SNPs with extremely low P-values (< 5×10−235) were not in strong LD with rs4917/rs4918 (light blue shaded dots, Supplementary Material, Fig. S2B). These five SNPs (rs13098866, rs13080283, rs2077119, rs1029353, and rs2070635) were instead in tight LD with each other (r2 < 0.6). Further, when we repeated the genome-wide association analyses conditioning on rs4917 to uncover any additional, independently associated SNPs, 34 SNPs at the AHSG locus were associated with fetuin-A concentrations (P< 5×10−8). The five SNPs from Supplementary Material, Figure S2B were among the SNPs that remained strongly statistically significantly associated with fetuin-A levels in this analysis (all P < 5×10−28 in the conditional analysis). Among the 34 SNPs, ten SNPs were new hits (P <5×10−8 in the conditional analysis only). The 10 SNPs were not in LD with rs4917, indicating that the strong association with fetuin-A levels observed for the AHSG locus stems from more than a single hit (Data for the 10 SNPs shown in Table 3). The Manhattan plot for European Americans also shows that other associations on chromosome 3 were elevated, though still not statistically significant upon adjustment for rs4917 (Supplementary Material, Fig. S5).
Table 3.
Additional SNPs that were statistically significantly associated with fetuin-A levels in European Americans only in the genome-wide analysis conditioned on rs4917
| Association results from meta-analyses conditioned on rs4917 |
Prior association results (unconditional analysis) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP/coded allele | Position (chr 3) | MAF | β | SE | P | Closest Gene* | Distanceϒ | β | SE | P |
| rs6787344/c | 187822535 | 0.15 | −0.0277 | 0.0023 | 3.82E-32 | AHSG | 733 | 0.000 | 0.003 | 0.89 |
| rs4831/c | 187813663 | 0.84 | 0.0259 | 0.0024 | 2.92E-28 | AHSG | 120 | −0.003 | 0.003 | 0.23 |
| rs4686432/t | 187800551 | 0.15 | −0.0246 | 0.0023 | 3.70E-27 | AHSG | 12992 | 0.002 | 0.003 | 0.48 |
| rs9842063/a | 187804207 | 0.89 | 0.0287 | 0.0028 | 4.13E-25 | AHSG | 9336 | 0.001 | 0.003 | 0.71 |
| rs9873987/t | 187802726 | 0.09 | −0.0323 | 0.0032 | 2.00E-23 | AHSG | 10817 | −0.002 | 0.004 | 0.58 |
| rs9872086/t | 187828594 | 0.79 | 0.0175 | 0.0026 | 1.13E-11 | AHSG | 6792 | −0.002 | 0.003 | 0.46 |
| rs6444146/t | 187794361 | 0.91 | 0.0177 | 0.0028 | 1.79E-10 | DNAJB11 | 8079 | −0.003 | 0.003 | 0.30 |
| rs9841006/t | 187775829 | 0.09 | −0.0172 | 0.0028 | 5.42E-10 | DNAJB11 | 4669 | 0.003 | 0.003 | 0.31 |
| rs13317898/t | 187740514 | 0.93 | 0.0216 | 0.0035 | 9.94E-10 | CRYGS | 1588 | −0.002 | 0.004 | 0.62 |
| rs12330837/a | 187768400 | 0.08 | −0.0191 | 0.0033 | 6.98E-09 | TBCCD1 | 594 | 0.009 | 0.004 | 0.03 |
Closest gene: shows the gene-encoding region most closely situated to the given SNP.
Distance: is the distance in base pairs from the closest known gene.
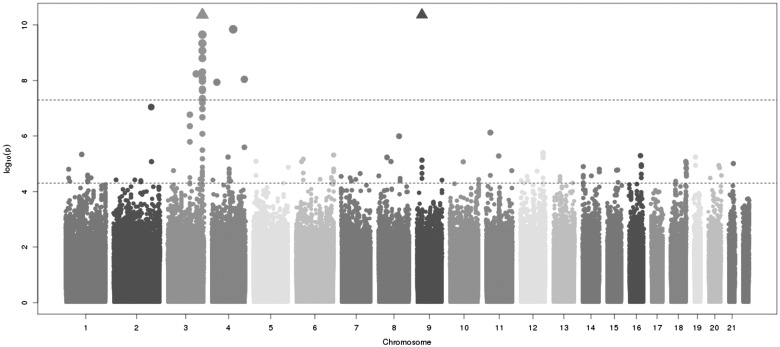
The AHSG locus also had the strongest association in the meta-analysis of four genome-wide studies that contributed data from 2,119 African American participants (Fig. 2, Supplementary Material, Fig. S3). In total, 42 SNPs at the AHSG locus were associated with fetuin-A levels with p values < 5×10−8 among the African Americans (Supplementary Material, Fig. S4 and Table S4). Each copy of the rs4917 minor allele was associated with 0.041 ±0.003 g/l lower fetuin-A concentration (P-value = 1.20×10−34) and this variant alone explained 13% of the variation in fetuin-A levels, but another AHSG variant (rs1900618) was the top SNP in the genome-wide analysis of the African American samples. rs4917 and rs1900618 are in complete LD according to D' from HapMap YRI population, but the two SNPs were only moderately correlated (r2=0.57) and had slightly different MAFs in African American participants only (0.26 versus 0.33). In contrast, both D| and r2 were nearly 1 in HapMap CEU. Overall, Supplementary Material, Figure S4 shows the extent of LD was less pronounced among African American participants. Of note the five variants that were in tight LD with each other but not with rs4917/rs4918 were also among the genome-wide significant SNPs in African Americans (Supplementary Material, Table S4). Because of the modest correlation of rs4917 with just a handful of the 42 fetuin-A associated SNPs, the SNPs that were significant in the conditional analyses (conditioning on rs4917) mostly overlapped with the significant SNPs in the unconditional analyses (Supplementary Material, Fig. S6). Two exceptions were rs1486336 and rs958629 that reached significance only in the conditional analysis (and had borderline p values in the unconditional analysis (P ∼1×10−7).
Figure 2.
Meta-analysis of six genome-wide association analyses of fetuin-A levels in a total of 2,119 African Americans. Figure displays p-value for association for each SNP on a log10 scale
Discussion
Very little is known about the genetic determination of circulating fetuin-A levels, a marker of diabetes and the metabolic syndrome. To our knowledge, the present study represents the first unbiased genome-wide search for genetic variants associated with fetuin-A concentrations. In the present paper, all of the major genetic determinants of fetuin-A levels were located in the fetuin-A encoding gene; AHSG. We further provide evidence that the genetic variation of importance to fetuin-A levels co-localize to the AHSG locus in both European and African Americans.
In both ethnicities, variants in and near the AHSG locus showed the strongest associations with fetuin-A levels. The most highly associated SNPs in the two ethnicities were not identical (rs4917 in European and rs1900618 in African-Americans), but showed remarkably similar associations that were in close proximity (within 1 kb distance) and were in very high linkage disequilibrium, suggesting that they may mark the same causal variant. Though the top association was different (rs1900618) among African American participants, the high degree of LD in this region does not allow for conclusions on the potential causal variant marked by these SNPs. Interestingly, genetic variation in AHSG has not been examined in African American samples before.
We found that rs4917 alone explained 14% of the variation in fetuin-A in European and 13% in African Americans individuals. In comparison, genome-wide association studies have identified several genetic loci that contribute to the levels of other biomarkers such as fibrinogen, CRP or adiponectin. Multigenetic risk scores that include all the independent genome-wide significant loci for each of these traits (e.g. up to 23 independent loci for fibrinogen) have been found to explain < 4% of the variation in fibrinogen and up to 5% for CRP and adiponectin (23–25).
When placed in the context of previous studies, several of the over 100 variants with genome-wide significant associations in our analysis have been identified previously. In particular, rs4917 and rs4918 are both missense variants located in the last two exons of the AHSG gene (exons 6 and 7, respectively) (26–28). Both SNPs have been associated with a similar magnitude of difference in plasma fetuin-A as observed in our study (0.06 ± 0.002 g/l lower levels in variant carriers) (22). While rs4917 and rs4918 do not appear to be related to metabolic markers including insulin levels, lipids, BMI, and fasting glucose in other studies (20,28–30) associations with risk of cardiovascular disease have been reported for both general (22,31) and renal patient populations (32).
AHSG expression is controlled by a number of transcriptional factors (TFs) such as C/EBP-[beta], NF-1, HNF-3[beta], AP-1 and ER[alpha] (33–36). The promoter SNP rs2248690, which has also been strongly associated with fetuin-A levels (26,29), modifies AHSG transcription by altering the affinity of AP-1 (26). Though this variant is located in the opposite region of AHSG, it is in LD with the exonic rs4917 and rs4918 SNPs (r2>0.80 CEU), that potentially alter the DNA binding of several TFs including AP-1 and ER[alpha] (37). A transcriptional complex comprising ERα and AP-1 may contribute to estrogen-induced transcription of AHSG (36). A potential regulatory function stemming from the coding regions harboring rs4917 and rs4918 thus warrants further study.
In genome-wide analysis conditioning on rs4917, we identified 34 SNPs that were genome-wide significant in European Americans. Five SNPs were also significant in the main analysis (rs13098866, rs13080283, rs2077119, rs1029353, and rs2070635) because they were not in LD with rs4917, but in high LD with each other. Among these, rs2077119 in the 5′ untranslated region has previously been found to be associated with markers of insulin resistance in adipocytes (28), and with the risk of diabetes (20). A borderline association with risk of CHD has been reported for the intron variant rs2070635 (22). Another ten SNPs were only significantly associated with plasma fetuin-A levels in the analysis conditioned on rs4917. These 10 SNPs were in LD with rs2077119 (D′ > 0.7 in HapMap CEU).
In conclusion, genetic variation in AHSG is strongly associated with fetuin-A levels. While we cannot exclude the possibility that other genetic loci play a role in fetuin-A concentrations, our sample size of over 9,000 European and 2,000 African Americans identified SNPs in the AHSG with P-values for their association lower than 1×10−300 indicating a very low chance that this is a false positive finding and highlighting the power inherent in analyses of this trait. Further work is needed to identify the functional variants and to understand the underlying biology of these associations. Our finding of a strong link between AHSG SNPs and fetuin-A levels provide a useful framework for the continued investigation of the causal variants in the AHSG gene.
Materials and Methods
We used data on genetic variation and fetuin-A levels from six population-based studies from the U.S. The following studies from the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium were included: Atherosclerosis Risk in Communities Study (ARIC), the Cardiovascular Health Study (CHS), the Multi-Ethnic Study of Atherosclerosis (MESA), and the Framingham Heart Study. In addition, investigators from the Health, Aging, and Body Composition (Health ABC) and Nurses’ Health Study contributed data. Local ethical committees at each institution approved the individual study protocols. Detailed methods of each study population are included in the online supplement. Various genotyping platforms were used by the different studies, however, all studies imputed their genotyped data to HapMap release 22 CEU and YRI reference panels. Details on genotyping platform, quality control and imputation specifics are available in online in Supplementary Materials, Tables S1 and S2.
Fetuin-A measures
Fetuin-A was measured in plasma using enzyme linked immunosorbent assays (ELISA) from Epitope Diagnostics, San Diego, CA (CHS, MESA, HABC, and ARIC), Biovendor, Candler, NC (FHS), or from R&D Systems, Minneapolis, MN (NHS). Each study reported CV’s < 14%.
Statistical analysis
The associations between genotypes and fetuin-A level were analyzed within each cohort using linear regression in an additive model. All analyses were adjusted for age, sex, eigenvectors for population stratification, and if applicable, field center. Analyses were conducted separately for European and African Americans. See details for study-specific methods in Supplementary Material, Tables S1 and S2.
To combine results across cohorts, we performed an inverse variance–weighted meta-analysis using the software package METAL (38). Cohort-specific standard errors were adjusted using genomic control. In GWAS, we chose P = 5 × 10−8 as the threshold for significance (39). To investigate whether the multiple SNPs associated in the respective region were due to linkage disequilibrium (LD) with the top SNP or if multiple independent signals existed, we performed a meta-analysis based on models conditioned on the SNP with the smallest P-value.
To assess the variation in fetuin-A levels explained by our top SNPs, we compared the r2 values from a model with just basic covariates versus a model that also included rs4917.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors thank the staff and participants of the individual studies for their important contributions, and acknowledge the essential role of the Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) Consortium in the development and support of this manuscript.
Conflict of Interest statement. None declared.
Funding
The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN 268201100006C, HHSN268201100007C, HHSN268201100008C, HH SN2 68201100009C, HHSN268201100010C, HHSN2 68201100011C, and HHSN268201100012C). National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HSN268200625226C. Infrastructure was partly supported by grant number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. Funding for fetuin-A measurements was provided by R01-DK56918. The authors thank the staff and participants of the ARIC study for their important contributions.
NHLBI contracts HHSN268201200036C, HHSN268200800007C, HHSN268200960009C,N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086 supported CHS research; and NHLBI grants HL080295, HL087652, HL105756, R01HL094555 & HL085251 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG023629 from the National Institute on Aging (NIA). A full list of CHS investigators and institutions can be found at http://chs-nhlbi.org/. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
National Heart, Lung, and Blood Institute's Framingham Heart Study (N01-HC-25195, HHSN268201500001 & HL25195) supported the Framingham Heart Study, and its contract with Affymetrix, Inc., for genotyping services (contract number N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center.
NIA contracts N01AG62101, N01AG62103, and N01AG62106 supported the Health, Aging, and Body Composition (Health ABC) Study. Fetuin-A was measured with support from American Diabetes Association: 1-08-IG-01. The genome-wide association study was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. Mike Nalls' participation was supported in part by the Intramural Research Program of the NIH, National Institute on Aging NIA contract Z01-AG000932-03 and utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Md. (http://biowulf.nih.gov).
National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators supported the Multi-Ethnic Study of Atherosclerosis (MESA) and the MESA SHARe project. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079, UL1-TR-000040, and DK063491. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, California, USA) and the Broad Institute of Harvard and MIT (Boston, Massachusetts, USA) using the Affymetrix Genome-Wide Human SNP Array 6.0. Fetuin-A was measured with support from NHLBI R21HL091217.
UM1 CA186107, HL34594, CA87969, CA49449, HL35464, CA55075, R01 HL088521 and CA167552 from the National Institutes of Health, Bethesda, MD, with additional support for genotyping from Merck Research Laboratories, North Wales, PA supported the Nurses’ Health Study (NHS).
References
- 1. Denecke B., Graber S., Schafer C., Heiss A., Woltje M., Jahnen-Dechent W. (2003) Tissue distribution and activity testing suggest a similar but not identical function of fetuin-B and fetuin-A. Biochem. J., 376, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schafer C., Heiss A., Schwarz A., Westenfeld R., Ketteler M., Floege J., Muller-Esterl W., Schinke T., Jahnen-Dechent W. (2003) The serum protein alpha 2-Heremans-Schmid glycoprotein/fetuin-A is a systemically acting inhibitor of ectopic calcification. J. Clin. Invest., 112, 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Auberger P., Falquerho L., Contreres J.O., Pages G., Le Cam G., Rossi B., Le Cam A. (1989) Characterization of a natural inhibitor of the insulin receptor tyrosine kinase: cDNA cloning, purification, and anti-mitogenic activity. Cell, 58, 631–640. [DOI] [PubMed] [Google Scholar]
- 4. Srinivas P.R., Wagner A.S., Reddy L.V., Deutsch D.D., Leon M.A., Goustin A.S., Grunberger G. (1993) Serum alpha 2-HS-glycoprotein is an inhibitor of the human insulin receptor at the tyrosine kinase level. Mol. Endocrinol., 7, 1445–1455. [DOI] [PubMed] [Google Scholar]
- 5. Cintron V.J., Ko M.S., Chi K.D., Gross J.P., Srinivas P.R., Goustin A.S., Grunberger G. (2001) Genetic mapping and functional studies of a natural inhibitor of the insulin receptor tyrosine kinase: the mouse ortholog of human alpha2-HS glycoprotein. Int. J. Exp. Diabetes Res,. 1, 249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mathews S.T., Rakhade S., Zhou X., Parker G.C., Coscina D.V., Grunberger G. (2006) Fetuin-null mice are protected against obesity and insulin resistance associated with aging. Biochem. Biophys. Res. Commun., 350, 437–443. [DOI] [PubMed] [Google Scholar]
- 7. Mathews S.T., Singh G.P., Ranalletta M., Cintron V.J., Qiang X., Goustin A.S., Jen K.L., Charron M.J., Jahnen-Dechent W., Grunberger G. (2002) Improved insulin sensitivity and resistance to weight gain in mice null for the AHSG gene. Diabetes, 51, 2450–2458. [DOI] [PubMed] [Google Scholar]
- 8. Ix J.H., Shlipak M.G., Brandenburg V.M., Ali S., Ketteler M., Whooley M.A. (2006) Association between human fetuin-A and the metabolic syndrome: data from the Heart and Soul Study. Circulation, 113, 1760–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mori K., Emoto M., Yokoyama H., Araki T., Teramura M., Koyama H., Shoji T., Inaba M., Nishizawa Y. (2006) Association of serum fetuin-A with insulin resistance in type 2 diabetic and nondiabetic subjects. Diabetes Care, 29, 468.. [DOI] [PubMed] [Google Scholar]
- 10. Stefan N., Hennige A.M., Staiger H., Machann J., Schick F., Krober S.M., Machicao F., Fritsche A., Haring H.U. (2006) Alpha2-Heremans-Schmid glycoprotein/fetuin-A is associated with insulin resistance and fat accumulation in the liver in humans. Diabetes Care, 29, 853–857. [DOI] [PubMed] [Google Scholar]
- 11. Stefan N., Fritsche A., Weikert C., Boeing H., Joost H.G., Haring H.U., Schulze M.B. (2008) Plasma fetuin-A levels and the risk of type 2 diabetes. Diabetes, 57, 2762–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stefan N., Haring H.U., Schulze M.B. (2008) Association of fetuin-A level and diabetes risk. JAMA, 300, 2247. author reply 2247-2248. [DOI] [PubMed] [Google Scholar]
- 13. Ix J.H., Biggs M.L., Mukamal K.J., Kizer J.R., Zieman S.J., Siscovick D.S., Mozzaffarian D., Jensen M.K., Nelson L., Ruderman N., et al. (2012) Association of fetuin-a with incident diabetes mellitus in community-living older adults: the cardiovascular health study. Circulation, 125, 2316–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun Q., Cornelis M.C., Manson J.E., Hu F.B. (2013) Plasma Levels of Fetuin-A and Hepatic Enzymes and Risk of Type 2 Diabetes in Women in the U.S. Diabetes, 62, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stefan N., Haring H.U. (2013) Circulating fetuin-A and free fatty acids interact to predict insulin resistance in humans. Nat. Med., 19, 394–395. [DOI] [PubMed] [Google Scholar]
- 16. Pal D., Dasgupta S., Kundu R., Maitra S., Das G., Mukhopadhyay S., Ray S., Majumdar S.S., Bhattacharya S. (2012) Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat. Med., 18, 1279–1285. [DOI] [PubMed] [Google Scholar]
- 17. Vionnet N., Hani E.H., Dupont S., Gallina S., Francke S., Dotte S., De Matos F., Durand E., Lepretre F., Lecoeur C., et al. (2000) Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21-q24. Am. J. Hum. Genet., 67, 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kissebah A.H., Sonnenberg G.E., Myklebust J., Goldstein M., Broman K., James R.G., Marks J.A., Krakower G.R., Jacob H.J., Weber J., et al. (2000) Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc. Natl. Acad. Sci. USA, 97, 14478–14483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siddiq A., Lepretre F., Hercberg S., Froguel P., Gibson F. (2005) A synonymous coding polymorphism in the alpha2-Heremans-schmid glycoprotein gene is associated with type 2 diabetes in French Caucasians. Diabetes, 54, 2477–2481. [DOI] [PubMed] [Google Scholar]
- 20. Andersen G., Burgdorf K.S., Sparso T., Borch-Johnsen K., Jorgensen T., Hansen T., Pedersen O. (2008) AHSG tag single nucleotide polymorphisms associate with type 2 diabetes and dyslipidemia: studies of metabolic traits in 7,683 white Danish subjects. Diabetes, 57, 1427–1432. [DOI] [PubMed] [Google Scholar]
- 21.(2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature, 447, 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fisher E., Stefan N., Saar K., Drogan D., Schulze M.B., Fritsche A., Joost H.G., Haring H.U., Hubner N., Boeing H., et al. (2009) Association of AHSG gene polymorphisms with fetuin-A plasma levels and cardiovascular diseases in the EPIC-Potsdam study. Circ. Cardiovasc. Genet., 2, 607–613. [DOI] [PubMed] [Google Scholar]
- 23. Dehghan A., Dupuis J., Barbalic M., Bis J.C., Eiriksdottir G., Lu C., Pellikka N., Wallaschofski H., Kettunen J., Henneman P., et al. (2011) Meta-Analysis of Genome-Wide Association Studies in > 80 000 Subjects Identifies Multiple Loci for C-Reactive Protein Levels. Circulation, 123, 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sabater-Lleal M., Huang J., Chasman D., Naitza S., Dehghan A., Johnson A.D., Teumer A., Reiner A.P., Folkersen L., Basu S., et al. (2013) Multiethnic meta-analysis of genome-wide association studies in > 100 000 subjects identifies 23 fibrinogen-associated Loci but no strong evidence of a causal association between circulating fibrinogen and cardiovascular disease. Circulation, 128, 1310–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dastani Z., Hivert M.-F., Timpson N., Perry J.R.B., Yuan X., Scott R.A., Henneman P., Heid I.M., Kizer J.R., Lyytikäinen L.-P., et al. (2012) Novel Loci for Adiponectin Levels and Their Influence on Type 2 Diabetes and Metabolic Traits: A Multi-Ethnic Meta-Analysis of 45,891 Individuals. PLoS. Genet., 8, e1002607.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inoue M., Takata H., Ikeda Y., Suehiro T., Inada S., Osaki F., Arii K., Kumon Y., Hashimoto K. (2008) A promoter polymorphism of the alpha2-HS glycoprotein gene is associated with its transcriptional activity. Diabetes Res. Clin. Pract., 79, 164–170. [DOI] [PubMed] [Google Scholar]
- 27. Osawa M., Tian W., Horiuchi H., Kaneko M., Umetsu K. (2005) Association of alpha2-HS glycoprotein (AHSG, fetuin-A) polymorphism with AHSG and phosphate serum levels. Hum. Genet, 116, 146–151. [DOI] [PubMed] [Google Scholar]
- 28. Dahlman I., Eriksson P., Kaaman M., Jiao H., Lindgren C.M., Kere J., Arner P. (2004) alpha2-Heremans-Schmid glycoprotein gene polymorphisms are associated with adipocyte insulin action. Diabetologia, 47, 1974–1979. [DOI] [PubMed] [Google Scholar]
- 29. Mussig K., Staiger H., Machicao F., Machann J., Hennige A.M., Schick F., Claussen C.D., Fritsche A., Haring H.U., Stefan N. (2009) AHSG gene variation is not associated with regional body fat distribution–a magnetic resonance study. Exp Clin Endocrinol. Diabetes, 117, 432–437. [DOI] [PubMed] [Google Scholar]
- 30. Jensen M.K., Bartz T.M., Djoussé L., Kizer J.R., Zieman S.J., Rimm E.B., Siscovick D.S., Psaty B.M., Ix J.H., Mukamal K.J. (2013) Genetically Elevated Fetuin-A Levels, Fasting Glucose Levels, and Risk of Type 2 Diabetes: The Cardiovascular Health Study. Diabetes Care, 36, 3121–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laugsand L.E., Ix J.H., Bartz T.M., Djousse L., Kizer J.R., Tracy R.P., Dehghan A., Rexrode K., Lopez O.L., Rimm E.B., et al. (2015) Fetuin-A and risk of coronary heart disease: A Mendelian randomization analysis and a pooled analysis of AHSG genetic variants in 7 prospective studies. Atherosclerosis, 243, 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Verduijn M., Prein R.A., Stenvinkel P., Carrero J.J., le Cessie S., Witasp A., Nordfors L., Krediet R.T., Boeschoten E.W., Dekker F.W. (2011) Is fetuin-A a mortality risk factor in dialysis patients or a mere risk marker? A Mendelian randomization approach. Nephrol. Dial. Transplant., 26, 239–245. [DOI] [PubMed] [Google Scholar]
- 33. Falquerho L., Paquereau L., Vilarem M.J., Galas S., Patey G., Le Cam A. (1992) Functional characterization of the promoter of pp63, a gene encoding a natural inhibitor of the insulin receptor tyrosine kinase. Nucleic. Acids Res., 20, 1983–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Banine F., Gangneux C., Mercier L., Le Cam A., Salier J.P. (2000) Positive and negative elements modulate the promoter of the human liver-specific alpha2-HS-glycoprotein gene. Eur. J. Biochem., 267, 1214–1222. [DOI] [PubMed] [Google Scholar]
- 35. Woltje M., Tschoke B., von Bulow V., Westenfeld R., Denecke B., Graber S., Jahnen-Dechent W. (2006) CCAAT enhancer binding protein beta and hepatocyte nuclear factor 3beta are necessary and sufficient to mediate dexamethasone-induced up-regulation of alpha2HS-glycoprotein/fetuin-A gene expression. J. Mol. Endocrinol., 36, 261–277. [DOI] [PubMed] [Google Scholar]
- 36. Qiu C., Liu X., Wang J., Zhao Y., Fu Q. (2014) Estrogen increases the transcription of human alpha2-Heremans-Schmid-glycoprotein by an interplay of estrogen receptor alpha and activator protein-1. Osteoporos Int., 7, 7.. [DOI] [PubMed] [Google Scholar]
- 37. Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M., et al. (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature, 473, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol., 34, 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.