Abstract
Activating Group 1 (Gp1) metabotropic glutamate receptors (mGluRs), including mGluR1 and mGluR5, elicits translation-dependent neural plasticity mechanisms that are crucial to animal behavior and circuit development. Dysregulated Gp1 mGluR signaling has been observed in numerous neurological and psychiatric disorders. However, the molecular pathways underlying Gp1 mGluR-dependent plasticity mechanisms are complex and have been elusive. In this study, we identified a novel mechanism through which Gp1 mGluR mediates protein translation and neural plasticity. Using a multi-electrode array (MEA) recording system, we showed that activating Gp1 mGluR elevates neural network activity, as demonstrated by increased spontaneous spike frequency and burst activity. Importantly, we validated that elevating neural network activity requires protein translation and is dependent on fragile X mental retardation protein (FMRP), the protein that is deficient in the most common inherited form of mental retardation and autism, fragile X syndrome (FXS). In an effort to determine the mechanism by which FMRP mediates protein translation and neural network activity, we demonstrated that a ubiquitin E3 ligase, murine double minute-2 (Mdm2), is required for Gp1 mGluR-induced translation and neural network activity. Our data showed that Mdm2 acts as a translation suppressor, and FMRP is required for its ubiquitination and down-regulation upon Gp1 mGluR activation. These data revealed a novel mechanism by which Gp1 mGluR and FMRP mediate protein translation and neural network activity, potentially through de-repressing Mdm2. Our results also introduce an alternative way for understanding altered protein translation and brain circuit excitability associated with Gp1 mGluR in neurological diseases such as FXS.
Introduction
The ability of living organisms to adapt to constantly changing environments depends on the plasticity of their nervous systems. Neural plasticity often requires activity-dependent translation to rapidly supply specific proteins (1–4). One well-known mechanism underlying activity-mediated translation occurs through Group 1 metabotropic glutamate receptors (Gp1 mGluRs) (5,6). Activating Gp1 mGluRs, including mGluR1 and mGluR5, elicits multiple translation-dependent neural plasticity mechanisms (7–9). Dysregulated Gp1 mGluR signaling is observed in both neurological and psychiatric disorders, including fragile X syndrome (FXS) (10), autism spectrum disorders (ASDs) (11), Alzheimer’s disease (12), and epilepsy (13). However, because of the complexity of Gp1 mGluR signaling, pharmacological correction of Gp1 mGluR signaling in animal models of such diseases has been challenging. In this study, we aim to characterize novel and promising signaling pathways that underlie Gp1 mGluR-dependent protein translation and neural plasticity with the intention of revealing novel therapeutic targets.
Activating Gp1 mGluRs leads to elevated Phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) or Extracellular Signal-regulated Kinase-1/2 (ERK1/2) signaling (14–16), which in turn activates eIF4E and translation initiation (17–21). Several recent studies have also demonstrated that translation elongation is facilitated upon mGluR activation through the phosphorylation of eukaryotic elongation factor 2 (eEF2) (22–26). These mechanisms belong to general translational regulation and are often difficult to target for therapeutic intervention. To begin to search for other promising targets underlying Gp1 mGluR-mediated protein translation, we are studying murine double minute-2 (Mdm2), a ubiquitin E3 ligase which was recently shown to be dysregulated in FXS mouse model, Fmr1 knockout (KO) mice (27,28). Mdm2 directly interacts with multiple ribosomal proteins, including ribosomal protein 11 and ribosomal protein 5 (29–31). Studies indicate that interactions between Mdm2 and ribosomal proteins prevent Mdm2 from interacting with Mdm2’s substrates (29,32,33). Although Mdm2 did not appear to regulate translation in cancer cells (34), it remains unknown whether Mdm2 can participate in translational regulation under physiological conditions in normal cells. In our current study, we revealed strong association between Mdm2 and active polyribosomes. Furthermore, using a conditional Mdm2 knockdown mouse model, we uncovered that Mdm2 acts as a translational suppressor and is crucial to Gp1 mGluR-induced translational activation. These findings introduce a novel and important molecule in Gp1 mGluR signaling and open many new avenues for studying Gp1 mGluR-dependent neural plasticity.
To study Gp1 mGluR-mediated plasticity, we focus on neural network activity, a phenomenon that is less understood in regards to Gp1 mGluR signaling. Previous studies showed that activating Gp1 mGluRs triggers oscillatory network activity in the mouse cortex (35). However, the molecular mechanisms of this activation are unclear. Because altered Gp1 mGluR signaling is observed in multiple neurological disorders associated with brain circuit hyperexcitability including FXS and epilepsy, identifying the key molecules that mediate Gp1 mGluR-induced network activity is vital to developing therapies that reduce excitability. To this end, our study characterized that Gp1 mGluR-induced neural network activity requires three components: protein translation, fragile X mental retardation protein (FMRP), and Mdm2. In an effort to determine the mechanistic connection between these three components, our findings showed that FMRP mediates the ubiquitination and down-regulation of Mdm2 upon Gp1 mGluR activation. This pathway potentially leads to de-repression of translation and in turns elevates neural network activity. These data uncovered a novel mechanism by which FMRP regulates translation and a novel signaling process by which Gp1 mGluR mediates neural network activity. Our findings also introduce Mdm2 as a novel and promising molecule for studying and targeting for treatment in neurological diseases associated with hyperexcitability.
Results
Activation of Gp1 mGluR leads to translation-dependent elevation of neural network activity
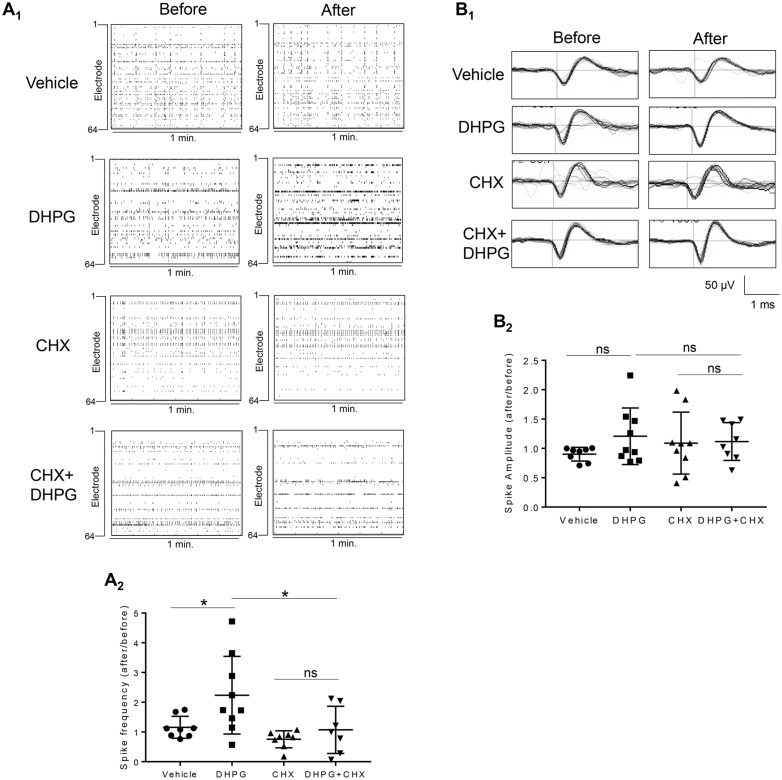
To study Gp1 mGluR-mediated neural network activity, we employed a multi-electrode array (MEA) recording system to record extracellular spontaneous spikes in primary cortical neuron cultures (36). Because this system allows recording and analysis from the same cultures before and after treatment, the variability between cultures is greatly diminished. As shown in Figure 1A, activating Gp1 mGluR using an agonist, (RS)-3,5-Dihydroxyphenylglycine (DHPG; 100 µM, 30 min), significantly elevates the frequency of spontaneous spikes (i.e. action potentials). To determine whether this elevation requires protein translation, we employed a translation inhibitor, cycloheximide (CHX, 60 µM). While CHX alone did not have an effect, pre-application of CHX for 10 min attenuates DHPG-elevated spontaneous spike frequency. The average spontaneous spike amplitude did not differ between all 4 groups (Fig. 1B). These data indicate that Gp1 mGluR-elevated spontaneous spike frequency requires protein translation.
Figure 1.
Activation of Gp1 mGluR induces protein translation-dependent elevation of spontaneous spike frequency. (A) Representative raster plots of spontaneous spikes from 1-min recordings of the same MEA dishes before and after drug treatments as labeled in the Figure (A1). Quantification of relative spontaneous spike frequency by comparing ‘after treatment’ to ‘before treatment’, and the dot plot showing the distribution of data points were on the bottom (A2) (n = 7–9). (B) Representative average traces of 1-min recording from the same single electrodes before and after drug treatments as labeled in the figure (B1). Quantification of average spontaneous spike amplitude by comparing ‘after treatment’ to ‘before treatment’, and the dot plot were on the bottom (B2) (n = 7–9). For both figures, a two-way ANOVA with Tukey test was used. Data are represented as mean ± SEM with *P < 0.05, ns: non-significant.
In addition to elevated spontaneous spike frequency, we also asked whether the pattern of spontaneous spikes is affected by Gp1 mGluR activation. If so, we further investigated whether or not it is also dependent on protein translation. We first determine the burst activity, which is a series of spontaneous spikes occurred within a short period of time. We defined a minimum of 5 spontaneous spikes with a maximum inter-spike interval of 0.1 second as a burst, as previously reported (36), and confirmed that activating Gp1 mGluR using applications of DHPG (100 µM, 15 min) elevates burst activity, as demonstrated by increased burst duration (Fig. 2A and B) and average number of spikes per burst (Fig. 2C). Remarkably, inhibiting translation by CHX attenuates DHPG-triggered burst activity, suggesting protein translation is also required for Gp1 mGluR-induced burst activity.
Figure 2.
Activation of Gp1 mGluR induces protein translation-dependent elevation of burst activity. (A) Representative traces of burst activity from cultures before and after treatment with various agents as indicated. Traces are from a designated electrode. (B, C) Quantification of relative burst duration (B) and the number of spikes per burst (C) by comparing ‘after treatment’ to ‘before treatment’, and the dot plot (n = 7–10). (D) Quantification of synchrony index from the entire MEA dishes by comparing ‘after treatment’ to ‘before treatment’, and the dot plot were on the right (n = 7–8). For the quantification above, a two-way ANOVA with Tukey test was used. Data are represented as mean ± SEM with *P < 0.05, ns: non-significant.
Our previous study demonstrated synchronization of spontaneous spikes across a culture when the neurons are hyperexcitable (36). To determine whether activating Gp1 mGluR also affects the synchrony of spontaneous spikes, we measured the synchrony index before and after DHPG treatment. As shown in Figure 2D, neither activation of Gp1 mGluR nor inhibition of translation has an effect on the synchrony of spontaneous spikes. Together, our data showed that Gp1 mGluR mediates the translation-dependent elevation of spontaneous spike frequency and burst activity.
FMRP is required for Gp1 mGluR-induced neural network activity
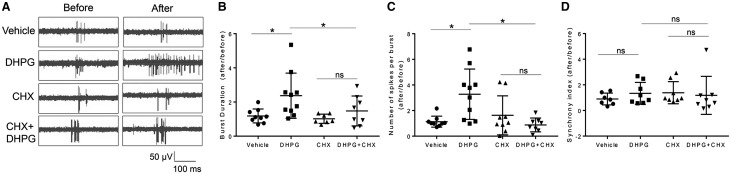
Gp1 mGluR-dependent protein translation is co-regulated by FMRP (37–39). In the absence of FMRP, Gp1 mGluR signaling is dysregulated and leads to many mGluR-associated deficits in FXS (40–45). To determine whether FMRP participates in the elevation of neural network activity upon Gp1 mGluR activation, we employed Fmr1 KO mice, in whom the gene encoding FMRP has been deleted. As shown in Figure 3A, compared to the cortical neuron cultures made from wild type (WT) mice (Fig. 3A1 and 3A3), Fmr1 KO cortical neuron cultures did not exhibit elevated spontaneous spike frequency after DHPG treatment (Fig. 3A2 and A3). The spontaneous spike amplitude did not differ between vehicle or DHPG treatment in either WT or Fmr1 KO cultures (Fig. 3B). We also asked whether elevated burst activity upon Gp1 mGluR activation requires FMRP. As shown in Figure 3C, DHPG fails to increase burst duration or the number of spikes per burst in Fmr1 KO cultures. Together, these data suggest that Gp1 mGluR-induced elevation of neural network activity, demonstrated by increased spontaneous spike frequency and burst activity, requires FMRP.
Figure 3.
Gp1 mGluR-induced elevation of neural network activity and burst activity requires FMRP. (A) Representative raster plots of spontaneous spikes from 1-min recordings of WT (A1) or Fmr1 KO (A2) cortical neuron cultures on MEA dishes before and after vehicle or DHPG treatment as labeled in the figure. Quantification of relative spontaneous spike frequency by comparing ‘after treatment’ to ‘before treatment’, and the dot plot were on the right (A3) (n = 5–7). (B) Representative average traces of 1-min recording from a designated electrode of WT or Fmr1 KO cortical neuron cultures on MEA dishes before and after vehicle or DHPG treatment as labeled in the figure (B1). Quantification of average spontaneous spike amplitude by comparing ‘after treatment’ to ‘before treatment’, and the dot plot were on the right (B2) (n = 4–8). (C1) Representative traces of burst activity from WT or Fmr1 KO cortical neuron cultures before and after vehicle or DHPG treatment as indicated. Traces are from a designated electrode. (C2, C3) Quantification of relative burst duration (C2) and the number of spikes per burst (C3) by comparing ‘after treatment’ to ‘before treatment’, and the dot plots were on the right (n = 5–9). For the quantification above, a two-way ANOVA with Tukey test was used. Data are represented as mean ± SEM with *P < 0.05, ns: non-significant.
Mdm2 is required for Gp1 mGluR-induced neural network activity
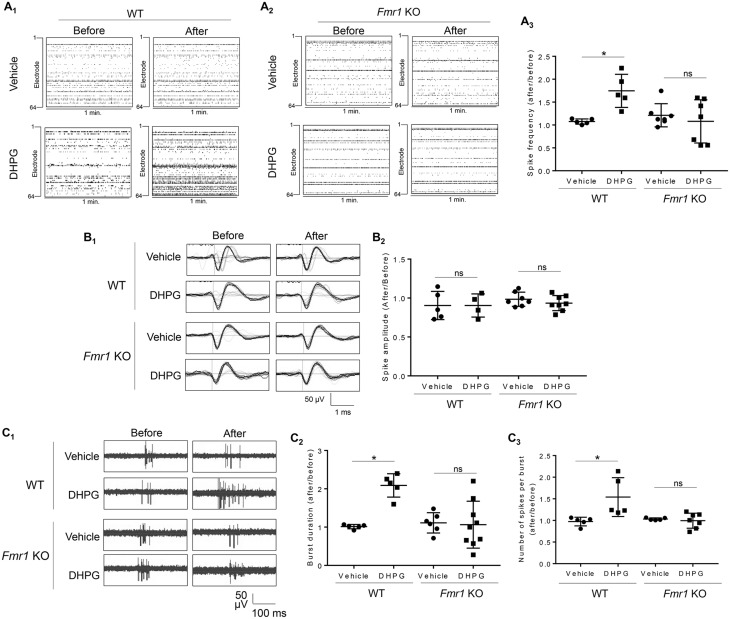
We next sought to identify the signaling pathway by which Gp1 mGluR and FMRP mediate neural network activity. Multiple recent studies have identified a connection between FMRP and Mdm2 in the neuronal and synaptic deficits observed in Fmr1 KO mice (27,28). To determine the role of Mdm2 in Gp1 mGluR-induced neural network activity, we employed a conditional Mdm2 knockdown mouse model by crossing Mdm2 floxed mice (Mdm2f/f) with Emx1-Cre mice to obtain Mdm2f/+-Emx-Cre+ and Mdm2f/+-Emx-Cre-mice. Emx1-Cre can confer Mdm2 reduction in the cortex and hippocampus beginning as early as embryonic day 10.5 (E10.5) (46,47). We used heterozygous mice (Mdm2f/+) to avoid potential apoptosis caused by complete Mdm2 knockout (48,49). As shown in Figure 4A, we observed approximately 40% knockdown of Mdm2 in the Mdm2f/+-Emx1-Cre+ cortical neuron cultures in comparison to the Mdm2f/+-Emx1-Cre- cultures at days-in-vitro (DIV 14) when the cultures were prepared on postnatal day 0.
Figure 4.
Gp1 mGluR-induced elevation of neural network activity and burst activity requires Mdm2. (A) Representative western blots of Mdm2 and Gapdh, and quantification of Mdm2 normalized to Gapdh from Mdm2f/+-Emx-Cre+ or Mdm2f/+-Emx-Cre-cortical neuron cultures at DIV 14 (n = 4). (B) Representative raster plots of spontaneous spikes from 1-min recordings of Mdm2f/+-Emx-Cre- (B1) and Mdm2f/+-Emx-Cre+(B2) cortical neuron cultures on MEA dishes before and after vehicle or DHPG treatment as labeled in the figure. Quantification of relative spontaneous spike frequency by comparing ‘after treatment’ to ‘before treatment’, and the dot plot were on the right (B3) (n = 12–16). (C) Representative average traces of 1-min recording from a designated electrode of Mdm2f/+-Emx-Cre+ or Mdm2f/+-Emx-Cre-cortical neuron cultures on MEA dishes before and after vehicle or DHPG treatment as labeled in the figure (C1). Quantification of average spontaneous spike amplitude by comparing ‘after treatment’ to ‘before treatment’, and the dot plot were on the right (C2) (n = 12–19). (D1) Representative traces of burst activity from Mdm2f/+-Emx-Cre+ or Mdm2f/+-Emx-Cre-cortical neuron cultures before and after vehicle or DHPG treatment as indicated. Traces are from a designated electrode. (D2,D3) Quantification of relative burst duration (D2) and the number of spikes per burst (D3) by comparing ‘after treatment’ to ‘before treatment’, and the dot plots were on the right (n = 11–19). For the quantification above, a Student t-test (A) or a two-way ANOVA with Tukey test (B-D) was used. Data are represented as mean ± SEM with *P < 0.05, **P < 0.01, ns: non-significant.
We next employed primary cortical neuron cultures made from Mdm2f/+-Emx1-Cre+ or Mdm2f/+-Emx1-Cre- mice to determine whether and how Mdm2 is involved in regulating neural network activity. As shown in Figure 4B, DHPG elevated spontaneous spike frequency in the Mdm2f/+-Emx1-Cre- cultures (Fig. 4B1 and B3), similar to what we observed in WT cultures (Figs 1A and 3A). However, this elevation was not detected in the Mdm2f/+-Emx1-Cre+ cultures (Fig. 4B2 and 4B3). The spontaneous spike amplitude before and after treatment was not different in both genotypes (Fig. 4C). Likewise, when we analyzed burst activity, we found that the DHPG-induced elevation of burst duration and the number of spikes per burst were only observed in the Mdm2f/+-Emx1-Cre- cultures but not in the Mdm2f/+-Emx1-Cre+ cultures (Fig. 4D). These data introduced a novel function of Mdm2 in neuroplasticity and demonstrated the necessity of Mdm2 in Gp1 mGluR-mediated elevation of neural network activity.
Knocking down Mdm2 does not affect basal synaptic transmission or Gp1 mGluR signaling
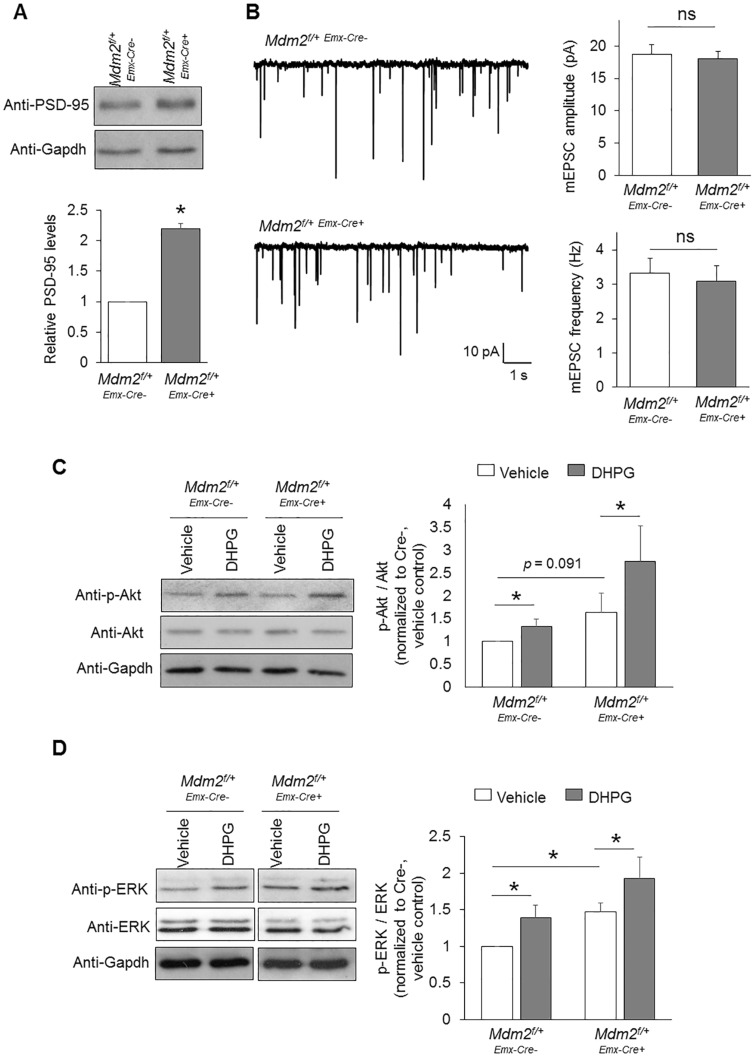
Despite the connection between FMRP and Mdm2 that was identified in the previous studies (27,28), it remains unknown whether and how Mdm2 participates in Gp1 mGluR signaling. To answer this question, we first determined whether knocking down Mdm2 affects basal synaptic activity. Because Mdm2 mediates the ubiquitination of post-synaptic density protein 95 (PSD-95) (28,50), we tested whether knocking down Mdm2 affects the levels of PSD-95. As shown in Figure 5A, the levels of PSD-95 were indeed elevated in Mdm2f/+-Emx1-Cre+ cultures when compared to Mdm2f/+-Emx1-Cre- cultures, confirming the role of Mdm2 in mediating PSD-95 half-life. However, when we performed whole-cell patch-clamp recording to obtain miniature excitatory post-synaptic current (mEPSC) from Mdm2f/+-Emx1-Cre- and Mdm2f/+-Emx1-Cre+ cortical neurons, we did not find any significant difference in either mEPSC amplitude or mEPSC frequency (Fig. 5B) despite the changes in basal PSD-95 levels. We reason that other substrates of Mdm2 might counter the effect of PSD-95 and maintain synaptic transmission. Together, these data suggest that increased basal level of PSD-95 in Mdm2f/+-Emx1-Cre+ cultures is unlikely the mechanism behind DHPG-induced changes in network activity.
Figure 5.
Knocking down Mdm2 does not alter synaptic function or Gp1 mGluR signaling. (A) Representative western blots of PSD-95 and Gapdh, and quantification from Mdm2f/+-Emx-Cre+ or Mdm2f/+-Emx-Cre-cortical neuron cultures (n = 8). (B) Patch-clamp recording from Mdm2f/+-Emx-Cre+ or Mdm2f/+-Emx-Cre-cortical neurons at DIV 14. Representative mEPSC traces, and quantification of mEPSC amplitude and frequency are shown (n = 21 for both Mdm2f/+-Emx-Cre+ and Mdm2f/+-Emx-Cre-neurons). (C, D) Representative western blots of Akt, p-Akt, ERK, p-ERK and Gapdh, and quantification from Mdm2f/+-Emx-Cre+ or Mdm2f/+-Emx-Cre-cortical neuron cultures after vehicle or DHPG treatment for 30 min (n = 7 and 6 for (C) and (D), respectively). For the quantification above, a one-sample t-test (A), Student t-test (B) or a two-way ANOVA with Tukey test (C, D) was used. Data are represented as mean ± SEM with *P < 0.05, **P < 0.01, ns: non-significant.
We next asked whether knocking down Mdm2 disrupts the activation of downstream signaling pathways of Gp1 mGluR, such as MEK-ERK and PI3K-Akt pathways (51). To this end, we assessed the activity of ERK and Akt by quantifying the phosphorylation of ERK and Akt upon treatments of vehicle or DHPG in Mdm2f/+-Emx1-Cre- and Mdm2f/+-Emx1-Cre+ cultures. As shown in Figure 5C, although Akt was slightly activated basally in Mdm2f/+-Emx1-Cre+ cultures, Mdm2f/+-Emx1-Cre- and Mdm2f/+-Emx1-Cre+ cultures showed similar activation of Akt upon the treatment of DHPG. When we studied ERK, we first observed basally elevated phosphorylation of ERK in Mdm2f/+-Emx1-Cre+ cultures (Fig. 5D). This was likely due to a compensatory effect when Mdm2 is knocked down because Mdm2 is a downstream molecule of ERK signaling (52). However, upon DHPG treatments, Mdm2f/+-Emx1-Cre- and Mdm2f/+-Emx1-Cre+ cultures showed similar activation of ERK (Fig. 5D). These data conclude that, although knocking down Mdm2 affects basal activity of MEK-ERK and PI3K-Akt pathways, it does not disrupt Gp1 mGluR-induced activation of either pathway.
Mdm2 mediates Gp1 mGluR-induced protein translation
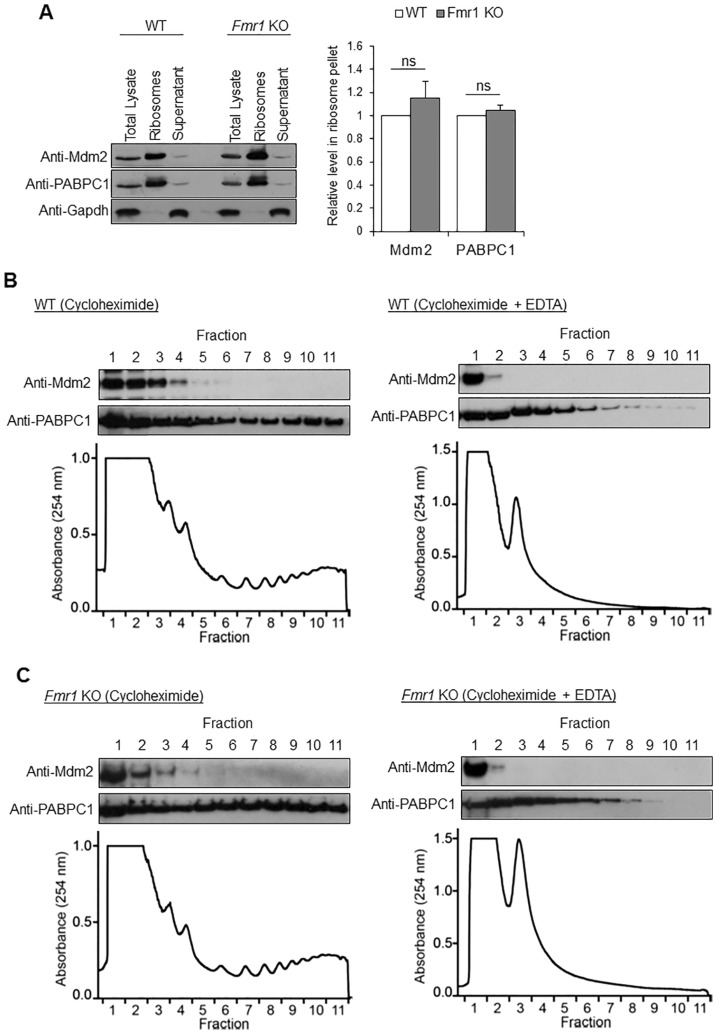
Because Mdm2 directly interacts with multiple ribosomal proteins (29–31), we next asked whether Mdm2 functions to mediate Gp1 mGluR-induced protein translation. We first determined whether Mdm2 associates with intact ribosomes. Using sucrose density ultracentrifugation to isolate ribosomes from the brains of 3-week-old mice, we found that Mdm2 is highly enriched in the ribosome pellet (Fig. 6A), suggesting Mdm2 associates with ribosomes. Interestingly, the distribution of Mdm2 in ribosomes did not differ between WT and Fmr1 KO brains, suggesting the association between Mdm2 and ribosomes is independent of FMRP. To determine whether Mdm2 associates more with monosomes or polysomes, we utilized polysome profiling by sucrose gradient sedimentation using mouse brain homogenates. As shown in Figure 6B and C, Mdm2 is detected in the lighter fractions, likely with monosomes or individual ribosomal subunits, as compared to PABPC1 which is heavily associated with polyribosomes. This phenomenon is consistent between WT and Fmr1 KO brains. Furthermore, EDTA, a ribosome-disrupting agent, successfully collapsed the polysome gradient and shifted both Mdm2 and PABPC1 to lighter fractions. These data indicate that Mdm2 might be involved in Gp1 mGluR-mediated protein translation by interacting with ribosomes and ribosomal components.
Figure 6.
Mdm2 associates with ribosomes. (A) Representative western blots of Mdm2, poly(A) binding protein cytoplasmic 1 (PABPC1) and Gapdh after sucrose density ultracentrifugation of WT or Fmr1 KO whole brains. PABPC1 and Gapdh serve as positive and negative controls, respectively, for ribosomes. Quantification is shown on the right (n = 4). (B, C) Representative western blots of Mdm2 and PABPC1, and polysome analysis from WT (B) or Fmr1 KO (C) brains. EDTA was used as a control to validate the fractionation by disrupting ribosomes.
We next asked whether and how Mdm2 affects protein translation. We employed rabbit reticulocyte lysate system to perform in vitro translation with total RNA extracted from WT cortical neuron cultures. As shown in Figure 7A, the global translation efficiency in the presence of recombinant Mdm2 (lane 2), as measured by puromycin incorporation, was significantly lower than in reaction mixtures without Mdm2 (lane 1) or with control Glutathione S-Transferase (GST) protein (lane 3). These results suggest that Mdm2 functions as a translational suppressor. To determine whether such suppressive activity requires Mdm2’s association with ribosomal proteins, we utilized two mutant Mdm2s, D294A-Mdm2 and T306A-S307A-Mdm2, which exert reduced affinity to interact with ribosomal proteins (53). When we expressed WT or either of the mutant Mdm2s in HEK 293 cells and used puromycin to label newly synthesized proteins, we found that while WT-Mdm2 reduces global translation, both of the mutants fail to exhibit significant activity toward translational suppression (Fig. 7B). These data suggest that Mdm2 represses translation likely through its association with ribosomal proteins.
Figure 7.
Mdm2 mediates Gp1 mGluR-induced protein translation. (A) Representative western blots of puromycin after in vitro translation using rabbit reticulocyte lysate and purified total RNA from WT cortical neuron cultures. Quantification (n = 5) and the purity of recombinant Mdm2 and GST were shown on the right. (B) Representative western blots of puromycin, Mdm2 and Gapdh after 30-min labeling of puromycin in HEK 293 cells transfected with HA-WT-Mdm2, HA-D294A-Mdm2 or HA-T306A-S307A-Mdm2. Quantification is shown on the right (n = 12). (C) Representative western blots of puromycin and Gapdh, and quantification after 30-min labeling of puromycin in Mdm2f/+-Emx-Cre+ or Mdm2f/+-Emx-Cre-cortical neuron cultures at DIV 14 (n = 6). (D) Representative western blots of Pcdh7 and Gapdh, and quantification after vehicle or DHPG treatment for 30 min in Mdm2f/+-Emx-Cre+ or Mdm2f/+-Emx-Cre-cortical neuron cultures at DIV 14 (n = 12). For the quantification above, Student t-test (A, B) or a two-way ANOVA with Tukey test (C, D) was used. Data are represented as mean ± SEM with *P < 0.05, ns: non-significant.
To further validate the role of Mdm2 in Gp1 mGluR-mediated protein translation, we employed primary cortical neuron cultures prepared from Mdm2f/+-Emx1-Cre+ or Mdm2f/+-Emx1-Cre- mice. Using puromycin to label newly synthesized proteins in cultures, we found that the Mdm2f/+-Emx1-Cre+ cultures showed basally elevated protein translation in comparison to the Mdm2f/+-Emx1-Cre- cultures (Fig. 7C, comparing lanes 1 and 3). When we applied DHPG to activate Gp1 mGluR signaling and translation, we found that elevated protein translation is occluded in Mdm2f/+-Emx1-Cre+ cultures (Fig. 7C, comparing lanes 3 and 4). Because Gp1 mGluR-induced translation often occurs to FMRP target mRNAs (54), we tested the expression of one of the known FMRP target, protocadherin-7 (55), upon DHPG treatment for 30 min. As shown in Figure 7D, the levels of Pcdh7 protein were moderately elevated, and further elevation upon DHPG treatment was occluded in Mdm2f/+-Emx1-Cre+ cultures. Altogether, our data suggest that Mdm2 functions as a novel translational suppressor and is required for Gp1 mGluR-induced translation activation.
FMRP mediates Gp1 mGluR-induced Mdm2 ubiquitination and down-regulation
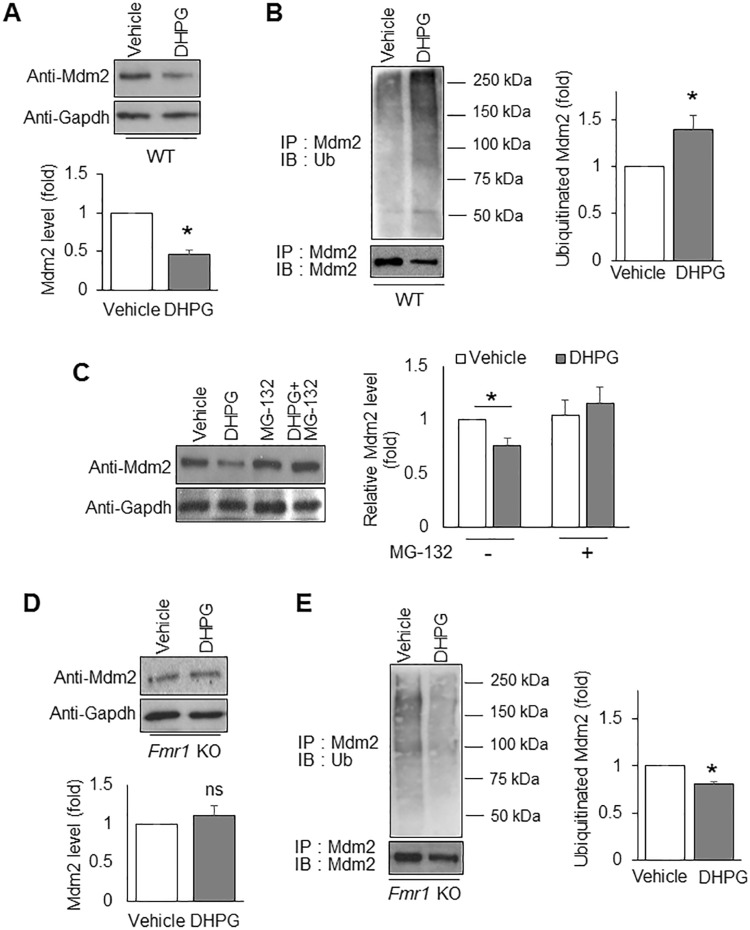
Because Mdm2 inhibits protein translation and is required for Gp1 mGluR-induced translation activation (Fig. 7), we hypothesized that activating Gp1 mGluR will de-repress Mdm2’s function so as to permit protein translation. To test this idea, we evaluated the protein level of Mdm2 before and after DHPG treatment. As shown in Figure 8A, DHPG treatment significantly reduced Mdm2 levels. Because this reduction was observed within 30 min, we hypothesized that the process involves protein degradation. To this end, we measured Mdm2 ubiquitination by performing immunoprecipitation of Mdm2 followed by western blotting with the anti-ubiquitin antibody. We found that DHPG treatment elevates Mdm2 ubiquitination (Fig. 8B), which is consistent with the reduction of total Mdm2 levels (Fig. 8A). Furthermore, the downregulation of Mdm2 upon DHPG treatment was inhibited by a proteasome inhibitor MG132 (10 µM) (Fig. 8C), conforming that the downregulation is resulted from proteasome-mediated degradation. These data suggest that activating Gp1 mGluR-induced protein translation likely occurs in part through de-repression of Mdm2’s function in translational suppression.
Figure 8.
FMRP mediates Gp1 mGluR-induced ubiquitination and down-regulation of Mdm2. (A) Representative western blots of Mdm2 and Gapdh from WT cortical neuron cultures after vehicle or DHPG treatment for 30 min (n = 4). (B) Representative western blots of Ubiquitin and Mdm2 from WT cortical neuron cultures after immunoprecipitation with anti-Mdm2 antibody (n = 4). (C) Representative western blots of Mdm2 and Gapdh from WT cortical neuron cultures after vehicle, MG132, DHPG or DHPG + MG132 treatment for 30 min. The cells in all four conditions were pre-treated with translation inhibitor cycloheximide (60 µM, 10 mins) to avoid potential interference from translation after Gp1 mGluR activation (n = 8). (D) Representative western blots of Mdm2 and Gapdh from Fmr1 KO cortical neuron cultures after vehicle or DHPG treatment for 30 min (n = 4). (E) Representative western blots of Ubiquitin and Mdm2 from Fmr1 KO cortical neuron cultures after immunoprecipitation with anti-Mdm2 antibody (n = 4). For the quantification above, a one-sample t-test was used (A, B and D, E) to compare DHPG-treated groups to vehicle-treated groups while a two-way ANOVA with Tukey test was used for (C). Data are represented as mean ± SEM with *P < 0.05, ns: non-significant.
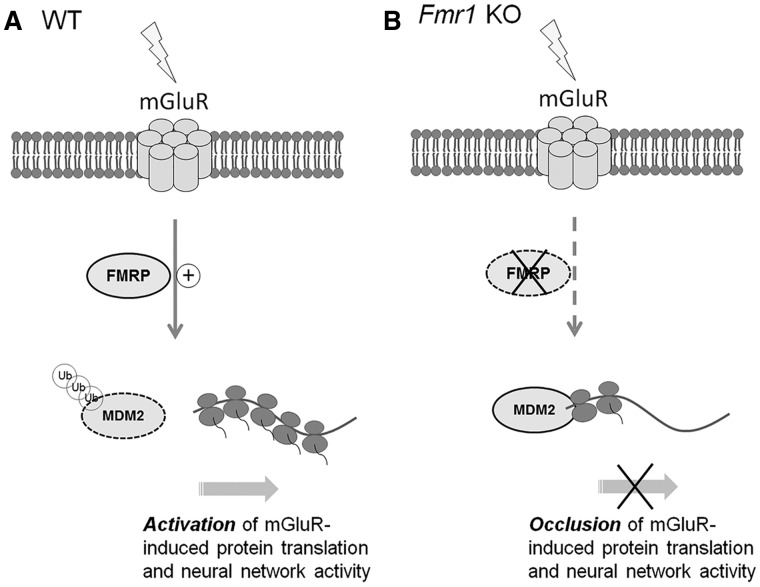
Because Gp1 mGluR-induced protein translation (37–39) and neural network activity (Fig. 3) requires FMRP, we next asked whether Gp1 mGluR-induced Mdm2 ubiquitination and down-regulation requires FMRP. As shown in Figure 8D and E, Fmr1 KO cortical neuron cultures exhibited neither ubiquitination nor down-regulation of Mdm2 upon DHPG treatment. In fact, Mdm2 ubiquitination was significantly reduced (Fig. 8E). These results indicate that FMRP is required for Gp1 mGluR-induced Mdm2 ubiquitination and down-regulation. They also suggested a novel signaling process by which FMRP and Mdm2 mediate protein translation and neural network activity upon Gp1 mGluR activation (Fig. 9).
Figure 9.
A working model for Mdm2 in Gp1-mGluR- and FMRP-dependent elevation of protein translation and neural network activity. (A) In WT neurons, activation of Gp1 mGluR triggers FMRP-dependent ubiquitination and down-regulation of Mdm2, subsequently leading to activation of protein translation and elevation of neural network activity. (B) In Fmr1 KO neurons, Mdm2 does not undergo increased ubiquitination or down-regulation upon Gp1 mGluR activation. Both protein translation and neural network activity induced by Gp1 mGluR activation are occluded in Fmr1 KO or Mdm2 conditional knockdown neurons.
Discussion
A novel mechanism by which FMRP regulates translation
FMRP is an RNA-binding protein with nearly 1,000 target genes identified through high throughput screening (55). It is well accepted that FMRP usually acts as a translational suppressor by hindering ribosomes from moving along its target mRNAs (55). However, because of the numerous targets regulated by FMRP, the mechanisms by which FMRP affects protein translation are complex and involve many indirect signaling pathways, leading to a global effect on translation. Our current study suggests a novel signaling pathway by which FMRP mediates protein ubiquitination and down-regulation to affect protein translation through de-repression of Mdm2 (Fig. 9). It is currently unknown what molecular pathway mediates Mdm2 degradation upon Gp1 mGluR activation and how FMRP participates in this mechanism. Our previous study showed that FMRP functions to limit phosphorylation of Mdm2. It is therefore possible that the phosphorylation status of Mdm2 is crucial to permitting its degradation upon Gp1 mGluR activation. Future work is necessary to test this possibility.
Although both Mdm2 and FMRP associate with ribosomal components, due to the lack of known RNA-binding motifs, Mdm2 likely provides a more general translation suppressive activity than FMRP. Because our data showed that Mdm2 is involved in FMRP-associated translation potentially through its association with ribosomal proteins, and also because FMRP regulates the expression of Mdm2 through direct binding to Mdm2 mRNA (27), it is possible that FMRP might exert other functions to mediate global translation. Further dissecting these functions would offer new molecular targets for designing new therapies to ameliorate translation-associated neuronal deficits in FXS.
Gp1 mGluR-mediated neural network activity
Activating Gp1 mGluR has multiple effects on neuroplasticity. We focus on neural network activity because it is relevant to brain circuit excitability, and hyperexcitability is seen in multiple mGluR-associated neurological disorders including epilepsy, FXS, and autism spectrum disorders (56–58). Using a MEA system, our study found that activation of Gp1 mGluR elevates spontaneous spike frequency and burst activity, two indicators of hyperexcitability. Although Gp1 mGluR signaling was basally activated in FXS, and elevated burst activity was observed in Fmr1 KO mice (35), we were unable to appropriately compare burst activity basally between WT and Fmr1 KO cultures in MEA recordings due to high variability between cultures and the necessity to compare activity internally within the same culture. Instead, we focused on the neural network and burst activity upon Gp1 mGluR activation, which was clearly dysregulated in Fmr1 KO cortical neuron cultures. Importantly, we revealed that this phenomenon is mediated by Mdm2. A previous study demonstrated that Gp1 mGluR activation elevates neuronal excitability through downregulation of dendritic h-channels (59), while another study showed that such elevation of neuronal excitability depends on protein translation (13). Because we found that Gp1 mGluR activation leads to the elevation of translation by down-regulating Mdm2 in this study, and because FMRP also undergoes degradation upon Gp1 mGluR activation (60), it would be important to determine whether Mdm2, FMRP and h-channels undergo the same degradation pathway, and whether downregulation of h-channels contributes to the elevation of network activity that we observed. Furthermore, our previous studies showed that chronic neuronal activity stimulation triggers Mdm2-mediated degradation of p53, and this degradation homeostatically down-regulates the synchronization of spontaneous network activity without affecting spontaneous spike frequency or burst activity (35). This study, together with our current study, suggests that Mdm2 is a multifaceted regulator that mediates different properties of neural network activity, depending on the stimulation paradigms. It again shows the complexity of mGluR signaling and emphasizes the necessity of elucidating downstream mechanisms to identify better and more specific therapeutic targets for mGluR-associated diseases.
Other mGluR-dependent neuronal plasticity mechanisms
Another neuroplasticity mechanism that is well-known to be mediated by mGluR is long-term synaptic depression (LTD) (61). LTD describes a long-lasting attenuation of excitatory synaptic strength (62). LTD is critical to properly establishing activity- or experience-dependent synaptic connections during circuit development (63) and to learning and memory consolidation in the mature brain (64). Inducing mGluR-LTD, as with mGluR-regulated neural network activity (Figs 1 and 2), requires de novo protein translation. Because Mdm2 mediates Gp1 mGluR-mediated translation, it is anticipated that Mdm2 also participates in mGluR-LTD and will be an important future direction. mGluR-LTD is often associated with local protein translation. Local protein translation is thought to efficiently provide specific proteins, such as activity-regulated cytoskeleton-associated protein (Arc; also known as Arg3.1) (54,65), at close proximity to the synapses, thereby allowing for rapid weakening of the synapses. Mdm2 can be found in dendrites and post-synaptic terminals (28) and therefore can potentially contribute to local protein translation. A broader effect of Mdm2 is waiting to be discovered.
Mdm2 in FXS and in cancer biology
Mdm2 is a ubiquitin E3 ligase that has been well-characterized in the field of cancer biology, and the tumor suppressor p53 is one of its major substrates (66). Mdm2-mediated p53 ubiquitination leads to inactivation of p53-dependent transcriptional regulation and ultimately alters the cell cycle (67–69). Unbiased screening of direct p53 target mRNAs has revealed many brain-enriched genes (70). However, the function and regulation of Mdm2-p53 signaling in neuronal cells is largely unclear. Recently, several studies have begun to uncover the functions of Mdm2-p53 signaling in neurodevelopment and neuroplasticity (27,36,71). However, it remains unknown whether or not p53 is also involved in Mdm2-mediated protein translation. If this is the case, it would open another entirely new avenue for studying mGluR-dependent plasticity. Given that a number of small molecules related to Mdm2-p53 signaling are currently available, and some of them are currently on clinical trials, exploring if these small molecules could affect Mdm2-Gp1 mGluR signaling could open the possibility of developing new potential therapies for treating neurological diseases associated with dysregulated Gp1 mGluR signaling.
Materials and Methods
All experimental protocols involving mice were performed in accordance with the guidelines and regulations set forth by the Institutional Animal Care and Use Committee at the University of Illinois at Urbana-Champaign.
Reagents
Cycloheximide (CHX) and picrotoxin was from Santa Cruz Biotechnology. Dimethyl sulfoxide (DMSO) was from Fisher Scientific. Tetrodotoxin was from Cayman Chemical. DHPG was from Abcam. MG132 was from Selleck Chemical. Puromycin was from MP Biomedicals. DMSO was used as vehicles in this study. The antibodies used in this study were purchased from Santa Cruz Biotechnology (anti-Mdm2), GenScript Corporation (anti-Gapdh), Millipore (anti-puromycin), Enzo Life Sciences (anti-Akt), Bioss (anti-Pcdh7), Abcam (anti-PABPC1) and Cell Signaling (anti-ubiquitin, anti-p-Akt, anti-ERK and anti-p-ERK). Recombinant Mdm2 protein was from Abcam and GST was from Sigma. HRP-conjugated secondary antibodies were from Santa Cruz Biotechnology.
In vitro translation, transfection, immunoprecipitation and western blotting
In vitro translation was conducted using rabbit reticulocyte lysate (RRL, Promega). In brief, 15 µl of RRL was mixed with 100 ng of total RNA extracted from WT cortical neuron cultures at DIV 14. After initial reaction for 5 min, puromycin (10 µg/ml) was added for another 10 min to label actively translating proteins. At the end of the experiment, SDS buffer was added to stop the reaction followed by standard western blotting.
Transfection in HEK cells was performed using Lipofectamine 3000 (72). For immunoprecipitation, 80 µg of total protein lysate was mixed with 1 µg of primary antibody for 1 h at 4 °C with constant rotation. Then 20 µl of Protein A/G beads were added for another 1 h. At the end of incubation, beads were washed with buffer (50 mM Tris, pH 7.4, 120 mM NaCl, 0.5% Nonidet P-40) for 3 times before subjecting to western blotting.
For western blotting after SDS-PAGE, the gel was transferred onto a polyvinylidene fluoride (PVDF) membrane. After blocking with 1% Bovine Serum Albumins in TBST buffer (20 mM Tris pH 7.5, 150 mM NaCl, 0.1% Tween-20), the membrane was incubated with the a primary antibody for overnight at 4 °C, followed by a 30-min wash with TBS. The membrane was then incubated with an HRP-conjugated secondary antibody for 1 h, followed by another 30-min wash. The membrane was then developed with an ECL (enhanced chemiluminescence) solution as described (65).
MEA recording
All the multi-electrode array recordings were done using an Axion Muse 64-channel system in single well MEAs (M64-GL1-30Pt200, Axion Biosystems) inside a 5% CO2, 37 °C incubator. Field potentials (voltage) at each electrode relative to the ground electrode were recorded with a sampling rate of 25 kHz. After 30 min of recording the baseline (before), drug(s) indicated in each experiment was added, and the MEA dish was immediately put back into the recording device for another 30 min of recording (after). Due to changes in network activity caused by physical movement of the MEA, only the last 15 min of each recording were used in data analyses as performed previously (36). AxIS software (Axion Biosystems) was used for the extraction of spikes (i.e. action potentials) from the raw electrical signal obtained from the Axion Muse system. After filtering, a threshold of ±6 standard deviations was independently set for each channel; activity exceeding this threshold was counted as a spike. The settings for burst detection were a minimum of 5 spikes with a maximum inter-spike interval of 0.1 s. Synchrony index was computed through AxIS software, based on a previously published algorithm (73), by taking the cross-correlation between two spike trains, removing the portions of the cross-correlogram that are contributed by the auto-correlations of each spike train, and reducing the distribution to a single metric. A value of 0 corresponds to no synchrony and a value of 1 corresponds to perfect synchrony.
Patch-clamp electrophysiology
Whole-cell patch-clamp recordings were made at room temperature (23–25 °C) in a submersion chamber continuously perfused with artificial cerebrospinal fluid (aCSF) containing (in mM): 119 NaCl, 2.5 KCl, 4 CaCl2, 4 MgCl2, 1 NaH2PO4, 26 NaHCO3 and 11 D-Glucose, saturated with 95% O2/5% CO2 (pH 7.4, 310 mOsm). aCSF was supplemented with 1 μM TTX and 100 μM picrotoxin for mEPSC measurements to block action potential-dependent EPSCs and GABAA receptors, respectively. Whole cell recording pipettes (∼4–6 MΩ) were filled with intracellular solution containing (in mM): 130 K-gluconate, 6 KCl, 3 NaCl, 10 HEPES, 0.2 EGTA, 4 Mg-ATP, 0.4 Na-GTP, 14 Tris-phosphocreatine, 2 QX-314 (pH 7.25, 285 mOsm). Neurons at DIV 11–14 were used for electrophysiological analyses. Membrane potential was clamped at –60 mV. All recordings were performed with Clampex 10.6 and Multiclamp 700B amplifier interfaced with Digidata 1440A data acquisition system (Molecular Devices). Recordings were filtered at 1 kHz and digitized at 10 kHz. Miniature excitatory postsynaptic currents (mEPSCs) were analyzed with Mini Analysis Program (Synaptosoft) with a 5-pA threshold level.
Isolation of total ribosomes
Whole brains were lysed using a glass dounce glass homogenizer in 1 ml polysome buffer [(20 mM Tris-HCl pH 7.5, 15 mM MgCl2, 200 mM KCl, 1% TritonX-100) with cycloheximide (100 µg/ml), DTT (2 mM), and heparin (1 mg/ml) added fresh]. Homogenates were spun in a microfuge at 13,000g, 4 °C for 20 min. The supernatant was carefully removed into a new tube, taking care not to disturb the cell debris pellet. Fresh polysome buffer was added to the lysate to a final volume of 1.1 ml. 1 ml of diluted supernatant was carefully layered onto 2.5 ml of sucrose cushion [(20 mM Tris-HCl pH 7.5, 15 mM MgCl2, 200 mM KCl, 1% TritonX-100, 25% w/v Sucrose) with cycloheximide (100 µg/ml), DTT (2 mM), and heparin (1 mg/ml) added fresh] in an ultracentrifuge tube (Beckman Coulter, #349622); the remaining 100 µl of supernatant was saved as input (total lysate). Samples were subjected to ultracentrifugation at 149,000g, 4° for 2 h, in a Beckman TLA-100.3 rotor. Following ultracentrifugation, supernatant was recovered and transferred to a fresh ultracentrifuge tube taking care to maintain the solution distribution. The resulting polysome pellet was resuspended in 100 µl of polysome buffer. The supernatant was subjected to further ultracentrifugation at 100,000g, 4 °C for 24 h, in a Beckman TLA-100.3 rotor. Following the second ultracentrifugation, the supernatant was recovered and saved as the soluble protein fraction. All resulting samples (total lysate, polysome fraction, and ultracentrifugation supernatant were subjected to western blotting.
Polysome profiling
Whole brains were extracted, immediately frozen in liquid nitrogen, and stored at −80 °C. Brains were pulverized under liquid nitrogen using a previously cooled, RNAse free mortar and pestle. The powder obtained was transferred to a 10-cm plate on dry ice while subsequent brains were pulverized. Afterward, 1 ml of lysis buffer (10 mM Tris-HCl at pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mg/ml heparin, 1% Nonidet-P40, .5% deoxycholate, 40 mM dithiothreitol, 1 U/ml SUPERaseIn RNAse inhibitor [Thermo Fisher], and 150 µg/ml cycloheximide) was added to the tissue powder. Next, re-suspended powder was scraped from the plate and transferred to a microcentrifuge tube with pipetting 10 times to lyse the cells. The cell nuclei and cell debris were removed by centrifugation (2,000g, 10 min, at 4 °C). The supernatant was transferred to a fresh tube and then centrifuged again at (16,000g, 7.5 min, at 4 °C). 400 µl of supernatant was layered onto a 10 ml linear sucrose gradient (15–45% sucrose [w/v] made using a Biocomp Gradient Master) and centrifuged in a SW41Ti rotor (Beckman) for 125 min at 38,000 rpm and 4 °C, with the brake off. Polysome profiles were recorded using a UA-6 absorbance (ISCO) detector at 254 nm and fractions were collected along the gradient. Fractions were used for subsequent western blot analysis.
Statistical analysis
For multiple comparisons, a two-way ANOVA and Tukey post-hoc test were performed. Student t-test was used in paired condition (Figs 5B, 6A and 7A). One sample t-tests were used when experimental group is normalized to control groups (Fig. 8). In all figures, error bars represent SEM and *P < 0.05, **P < 0.01.
Acknowledgements
We thank Dr. Kathryn Jewett, Steven Saletta and Shelya Zeng for their technical help.
Conflict of Interest statement. None declared.
Funding
This work was supported by University of Illinois at Urbana-Champaign (N-P.T.), Brain and Behavior Research Foundation (NARSAD Young Investigator Award to N-P.T.) and National Institute of Health grants (R01HL126845 to A.K.; 5T32-GM070421 to J.S.; R01NS083402 to H.J.C.; and R01CA095441, R01CA172468, R01CA127724, R21CA190775, and R21CA201889 to H.L.).
References
- 1. Bramham C.R., Wells D.G. (2007) Dendritic mRNA: transport, translation and function. Nat. Rev. Neurosc., 8, 776–789. [DOI] [PubMed] [Google Scholar]
- 2. Heise C., Gardoni F., Culotta L., di Luca M., Verpelli C., Sala C. (2014) Elongation factor-2 phosphorylation in dendrites and the regulation of dendritic mRNA translation in neurons. Front. Cell. Neurosci., 8, 35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ruiz C.R., Shi J., Meffert M.K. (2014) Transcript specificity in BDNF-regulated protein synthesis. Neuropharmacology, 76 Pt C, 657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang D.O., Martin K.C., Zukin R.S. (2010) Spatially restricting gene expression by local translation at synapses. Trends Neurosci., 33, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Esseltine J.L., Willard M.D., Wulur I.H., Lajiness M.E., Barber T.D., Ferguson S.S. (2013) Somatic mutations in GRM1 in cancer alter metabotropic glutamate receptor 1 intracellular localization and signaling. Mol. Pharmacol., 83, 770–780. [DOI] [PubMed] [Google Scholar]
- 6. Luscher C., Huber K.M. (2010) Group 1 mGluR-dependent synaptic long-term depression: mechanisms and implications for circuitry and disease. Neuron, 65, 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gladding C.M., Fitzjohn S.M., Molnar E. (2009) Metabotropic glutamate receptor-mediated long-term depression: molecular mechanisms. Pharmacol. Rev., 61, 395–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loweth J.A., Tseng K.Y., Wolf M.E. (2013) Using metabotropic glutamate receptors to modulate cocaine's synaptic and behavioral effects: mGluR1 finds a niche. Curr. Opin. Neurobiol., 23, 500–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rondard P., Pin J.P. (2015) Dynamics and modulation of metabotropic glutamate receptors. Curr. Opin. Pharmacol., 20, 95–101. [DOI] [PubMed] [Google Scholar]
- 10. Bear M.F., Huber K.M., Warren S.T. (2004) The mGluR theory of fragile X mental retardation. Trends Neurosci., 27, 370–377. [DOI] [PubMed] [Google Scholar]
- 11. Kleijer K.T., Schmeisser M.J., Krueger D.D., Boeckers T.M., Scheiffele P., Bourgeron T., Brose N., Burbach J.P. (2014) Neurobiology of autism gene products: towards pathogenesis and drug targets. Psychopharmacology, 231, 1037–1062. [DOI] [PubMed] [Google Scholar]
- 12. Kumar A., Dhull D.K., Mishra P.S. (2015) Therapeutic potential of mGluR5 targeting in Alzheimer's disease. Front. Neurosci., 9, 215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao W., Chuang S.C., Young S.R., Bianchi R., Wong R.K. (2015) Extracellular glutamate exposure facilitates group I mGluR-mediated epileptogenesis in the hippocampus. J. Neurosci., 35, 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kinkl N., Sahel J., Hicks D. (2001) Alternate FGF2-ERK1/2 signaling pathways in retinal photoreceptor and glial cells in vitro. J. Biol. Chem., 276, 43871–43878. [DOI] [PubMed] [Google Scholar]
- 15. Liu S., Wierod L., Skarpen E., Grosvik H., Duan G., Huitfeldt H.S. (2013) EGF activates autocrine TGFalpha to induce prolonged egf receptor signaling and hepatocyte proliferation. Cell. Physiol. Biochem., 32, 511–522. [DOI] [PubMed] [Google Scholar]
- 16. Wang H., Cheng H., Shao Q., Dong Z., Xie Q., Zhao L., Wang Q., Kong B., Qu X. (2014) Leptin-promoted human extravillous trophoblast invasion is MMP14 dependent and requires the cross talk between Notch1 and PI3K/Akt signaling. Biol. Reprod., 90, 78.. [DOI] [PubMed] [Google Scholar]
- 17. Bhattacharya A., Kaphzan H., Alvarez-Dieppa A.C., Murphy J.P., Pierre P., Klann E. (2012) Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron, 76, 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Di Prisco G.V., Huang W., Buffington S.A. (2014) Translational control of mGluR-dependent long-term depression and object-place learning by eIF2alpha. Nat. Neurosci., 17, 1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsu C.C., Bonnen P.E., Placzek A.N., Sidrauski C., Krnjevic K., Kaufman R.J., Walter P., Costa-Mattioli M., Trinh M.A., Ma T.. et al. (2014) The eIF2alpha kinase PERK limits the expression of hippocampal metabotropic glutamate receptor-dependent long-term depression. Nat. Neurosci., 21, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aguilar-Valles A., Matta-Camacho E., Khoutorsky A. (2015) Inhibition of Group I metabotropic glutamate receptors reverses autistic-like phenotypes caused by deficiency of the translation repressor eIF4E binding protein 2. J. Neurosci., 35, 11125–11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sonenberg N., Banko J.L., Hou L., Poulin F., Sonenberg N., Klann E. (2006) Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J. Neurosci., 26, 2167–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heise C., Gardoni F., Culotta L., di Luca M., Verpelli C., Sala C. (2014) Elongation factor-2 phosphorylation in dendrites and the regulation of dendritic mRNA translation in neurons. J. Neurosci., 8, 35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kenney J.W., Sorokina O., Genheden M., Sorokin A., Armstrong J.D., Proud C.G. (2015) Dynamics of elongation factor 2 kinase regulation in cortical neurons in response to synaptic activity. J. Neurosci., 35, 3034–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCamphill P.K., Farah C.A., Anadolu M.N., Hoque S., Sossin W.S. (2015) Bidirectional regulation of eEF2 phosphorylation controls synaptic plasticity by decoding neuronal activity patterns. J. Neurosci., 35, 4403–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Park S., Park J.M., Kim S., Kim J.A., Shepherd J.D., Smith-Hicks C.L., Chowdhury S., Kaufmann W., Kuhl D., Ryazanov A.G.. et al. (2008) Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron, 59, 70–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Verpelli C., Piccoli G., Zibetti C., Zanchi A., Gardoni F., Huang K., Brambilla D., Di Luca M., Battaglioli E., Sala C. (2010) Synaptic activity controls dendritic spine morphology by modulating eEF2-dependent BDNF synthesis. J. Neurosci., 30, 5830–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Y., Stockton M.E., Bhuiyan I., Eisinger B.E., Gao Y., Miller J.L., Bhattacharyya A., Zhao X. (2016) MDM2 inhibition rescues neurogenic and cognitive deficits in a mouse model of fragile X syndrome. Sci. Transl. Med., 8, 336ra361.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsai N.P., Wilkerson J.R., Guo W., Maksimova M.A., DeMartino G.N., Cowan C.W., Huber K.M. (2012) Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell, 151, 1581–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Y., He Y., Jin A., Tikunov A.P., Zhou L., Tollini L.A., Leslie P., Kim T.H., Li L.O., Coleman R.A.. et al. (2014) Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc. Natl Acad. Sci. U S A., 111, E2414–E2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Macias E., Jin A., Deisenroth C., Bhat K., Mao H., Lindstrom M.S., Zhang Y. (2010) An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell, 18, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou X., Hao Q., Zhang Q., Liao J.M., Ke J.W., Liao P., Cao B., Lu H. (2015) Ribosomal proteins L11 and L5 activate TAp73 by overcoming MDM2 inhibition. Cell Death Differ., 22, 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu S., Tackmann N.R., Yang J., Zhang Y. (2017) Disruption of the RP-MDM2-p53 pathway accelerates APC loss-induced colorectal tumorigenesis. Oncogene, 36,1374–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miliani de Marval P.L., Zhang Y. (2011) The RP-Mdm2-p53 pathway and tumorigenesis. Oncotarget, 2, 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dai M.S., Zeng S.X., Jin Y., Sun X.X., David L., Lu H. (2004) Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol, 24, 7654–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hays S.A., Huber K.M., Gibson J.R. (2011) Altered neocortical rhythmic activity states in Fmr1 KO mice are due to enhanced mGluR5 signaling and involve changes in excitatory circuitry. J. Neurosci., 31, 14223–14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jewett K.A., Christian C.A., Bacos J.T., Lee K.Y., Zhu J., Tsai N.P. (2016) Feedback modulation of neural network synchrony and seizure susceptibility by Mdm2-p53-Nedd4-2 signaling. Molecular Brain, 9, 32.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ronesi J.A., Huber K.M. (2008) Metabotropic glutamate receptors and fragile x mental retardation protein: partners in translational regulation at the synapse. Sci. Signal., 1, pe6.. [DOI] [PubMed] [Google Scholar]
- 38. Santoro M.R., Bray S.M., Warren S.T. (2012) Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Ann. Rev. Pathol., 7, 219–245. [DOI] [PubMed] [Google Scholar]
- 39. Waung M.W., Huber K.M. (2009) Protein translation in synaptic plasticity: mGluR-LTD, Fragile X. Curr. Opin. Neurobiol., 19, 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Auerbach B.D., Bear M.F. (2010) Loss of the fragile X mental retardation protein decouples metabotropic glutamate receptor dependent priming of long-term potentiation from protein synthesis. J. Neurophysiol., 104, 1047–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gross C., Nakamoto M., Yao X., Chan C.B., Yim S.Y., Ye K., Warren S.T., Bassell G.J. (2010) Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J. Neurosci., 30, 10624–10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. He C.X., Portera-Cailliau C. (2013) The trouble with spines in fragile X syndrome: density, maturity and plasticity. Neuroscience, 251, 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pop A.S., Gomez-Mancilla B., Neri G., Willemsen R., Gasparini F. (2014) Fragile X syndrome: a preclinical review on metabotropic glutamate receptor 5 (mGluR5) antagonists and drug development. Psychopharmacology, 231, 1217–1226. [DOI] [PubMed] [Google Scholar]
- 44. Scharf S.H., Jaeschke G., Wettstein J.G., Lindemann L. (2015) Metabotropic glutamate receptor 5 as drug target for Fragile X syndrome. Curr. Opin. Pharmacol., 20, 124–134. [DOI] [PubMed] [Google Scholar]
- 45. Thomas A.M., Bui N., Graham D., Perkins J.R., Yuva-Paylor L.A., Paylor R. (2011) Genetic reduction of group 1 metabotropic glutamate receptors alters select behaviors in a mouse model for fragile X syndrome. Behav. Brain Res., 223, 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gorski J.A., Talley T., Qiu M., Puelles L., Rubenstein J.L., Jones K.R. (2002) Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci., 22, 6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Young K.M., Fogarty M., Kessaris N., Richardson W.D. (2007) Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J. Neurosci., 27, 8286–8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jones S.N., Roe A.E., Donehower L.A., Bradley A. (1995) Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature, 378, 206–208. [DOI] [PubMed] [Google Scholar]
- 49. Montes de Oca Luna R., Wagner D.S., Lozano G. (1995) Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature, 378, 203–206. [DOI] [PubMed] [Google Scholar]
- 50. Tsai N.P., Wilkerson J.R., Guo W., Huber K.M. (2017) FMRP-Dependent Mdm2 Dephosphorylation is required for MEF2-Induced Synapse Elimination. Hum. Mol. Genet., 26, 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ronesi J.A., Huber K.M. (2008) Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. J. Neurosci., 28, 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang J.Y., Zong C.S., Xia W., Yamaguchi H., Ding Q., Xie X., Lang J.Y., Lai C.C., Chang C.J., Huang W.C.. et al. (2008) ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat. Cell Biol., 10, 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Q., Xiao H., Chai S.C., Hoang Q.Q., Lu H. (2011) Hydrophilic residues are crucial for ribosomal protein L11 (RPL11) interaction with zinc finger domain of MDM2 and p53 protein activation. J. Biol. Chem., 286, 38264–38274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Niere F., Wilkerson J.R., Huber K.M. (2012) Evidence for a fragile X mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered Arc translation and long-term depression. J. Neurosci., 32, 5924–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W.. et al. (2011) FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell, 146, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Amaral D.G., Schumann C.M., Nordahl C.W. (2008) Neuroanatomy of autism. Trends Neurosci., 31, 137–145. [DOI] [PubMed] [Google Scholar]
- 57. Fassio A., Raimondi A., Lignani G., Benfenati F., Baldelli P. (2011) Synapsins: from synapse to network hyperexcitability and epilepsy. Semin. Cell Dev. Biol., 22, 408–415. [DOI] [PubMed] [Google Scholar]
- 58. Levitt P., Eagleson K.L., Powell E.M. (2004) Regulation of neocortical interneuron development and the implications for neurodevelopmental disorders. Trends Neurosci., 27, 400–406. [DOI] [PubMed] [Google Scholar]
- 59. Brager D.H., Johnston D. (2007) Plasticity of intrinsic excitability during long-term depression is mediated through mGluR-dependent changes in I(h) in hippocampal CA1 pyramidal neurons. J. Neurosci., 27, 13926–13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nalavadi V.C., Muddashetty R.S., Gross C., Bassell G.J. (2012) Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J. Neurosci., 32, 2582–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Klein M.E., Castillo P.E., Jordan B.A. (2015) Coordination between Translation and Degradation Regulates Inducibility of mGluR-LTD. Cell Reports, doi: 10.1016/j.celrep.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stanton P.K., Sejnowski T.J. (1989) Associative long-term depression in the hippocampus induced by hebbian covariance. Nature, 339, 215–218. [DOI] [PubMed] [Google Scholar]
- 63. Hua J.Y., Smith S.J. (2004) Neural activity and the dynamics of central nervous system development. Nat. Neurosci., 7, 327–332. [DOI] [PubMed] [Google Scholar]
- 64. Lai C.S., Franke T.F., Gan W.B. (2012) Opposite effects of fear conditioning and extinction on dendritic spine remodelling. Nature, 483, 87–91. [DOI] [PubMed] [Google Scholar]
- 65. Waung M.W., Pfeiffer B.E., Nosyreva E.D., Ronesi J.A., Huber K.M. (2008) Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron, 59, 84–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Devine T., Dai M.S. (2013) Targeting the ubiquitin-mediated proteasome degradation of p53 for cancer therapy. Curr. Pharma. Des., 19, 3248–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Goldberg Z., Vogt Sionov R., Berger M., Zwang Y., Perets R., Van Etten R.A., Oren M., Taya Y., Haupt Y. (2002) Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation. EMBO J., 21, 3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang Z., Zhang R. (2008) Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation. EMBO J., 27, 852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vazquez A., Bond E.E., Levine A.J., Bond G.L. (2008) The genetics of the p53 pathway, apoptosis and cancer therapy. Nat. Rev. Drug Discov., 7, 979–987. [DOI] [PubMed] [Google Scholar]
- 70. Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z.. et al. (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell, 124, 207–219. [DOI] [PubMed] [Google Scholar]
- 71. Jewett K.A., Zhu J., Tsai N.P. (2015) The tumor suppressor p53 guides glua1 homeostasis through Nedd4-2 during chrnoic elevation of neuronal activity. J. Neurochem., 135, 226–233. [DOI] [PubMed] [Google Scholar]
- 72. Zhu J., Lee K.Y., Jewett K.A., Man H.Y., Chung H.J., Tsai N.P. (2017) Epilepsy-associated gene Nedd4-2 mediates neuronal activity and seizure susceptibility through AMPA receptors. PLoS Genet., 13, e1006634.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Eggermont J.J. (2006) Properties of correlated neural activity clusters in cat auditory cortex resemble those of neural assemblies. J. Neurophysiol., 96, 746–764. [DOI] [PubMed] [Google Scholar]