Abstract
Germline H255Y and K508R missense mutations in the folliculin (FLCN) gene have been identified in patients with bilateral multifocal (BMF) kidney tumours and clinical manifestations of Birt-Hogg-Dubé (BHD) syndrome, or with BMF kidney tumours as the only manifestation; however, their impact on FLCN function remains to be determined. In order to determine if FLCN H255Y and K508R missense mutations promote aberrant kidney cell proliferation leading to pathogenicity, we generated mouse models expressing these mutants using BAC recombineering technology and investigated their ability to rescue the multi-cystic phenotype of Flcn-deficient mouse kidneys. Flcn H255Y mutant transgene expression in kidney-targeted Flcn knockout mice did not rescue the multi-cystic kidney phenotype. However, expression of the Flcn K508R mutant transgene partially, but not completely, abrogated the phenotype. Notably, expression of the Flcn K508R mutant transgene in heterozygous Flcn knockout mice resulted in development of multi-cystic kidneys and cardiac hypertrophy in some mice. These results demonstrate that both FLCN H255Y and K508R missense mutations promote aberrant kidney cell proliferation, but to different degrees. Based on the phenotypes of our preclinical models, the FLCN H255Y mutant protein has lost it tumour suppressive function leading to the clinical manifestations of BHD, whereas the FLCN K508R mutant protein may have a dominant negative effect on the function of wild-type FLCN in regulating kidney cell proliferation and, therefore, act as an oncoprotein. These findings may provide mechanistic insight into the role of FLCN in regulating kidney cell proliferation and facilitate the development of novel therapeutics for FLCN-deficient kidney cancer.
Introduction
Folliculin (FLCN) is the causative gene for the inherited kidney cancer disorder Birt-Hogg-Dubé (BHD) syndrome, which predisposes patients to kidney cancer, cutaneous fibrofolliculomas and pulmonary cysts (1–3). Folliculin (FLCN) encoded by the FLCN gene, affects the regulation of a broad spectrum of metabolic processes through its interactions with the AMPK (4–6), mTORC1(7–10) and PPARGC1A (11,12) pathways, which are essential for energy homeostasis. Identification of two binding partners, folliculin-interacting proteins 1 (FNIP1) (4) and 2 (FNIP2) (5,6), which were found to be AMPK interacting molecules, has contributed to our understanding of the role of FLCN in cell metabolism. Structural analyses revealed that the C-terminus of FLCN may be distantly related to Differentially Expressed in Normal Cells and Neoplasm (DENN) domain proteins, suggesting that FLCN may play a significant role in membrane trafficking (13). These data have defined important roles for FLCN in a wide range of cellular metabolic pathways including energy sensing, membrane trafficking and mitochondrial oxidative metabolism.
Studies of several Flcn knockout mouse models have provided the basis for elucidating FLCN functions. Whole body Flcn knockout mice are embryonic lethal with vacuolation in the visceral endoderm (8), suggesting a defect in nutrient uptake and transport in the Flcn-null embryo. Kidney-targeted Flcn knockout mice developed enlarged polycystic kidneys and died at 3 weeks of age due to renal failure (7). A muscle-targeted Flcn knockout mouse model displayed red-coloured muscle with increased mitochondrial oxidative metabolism, which was Ppargc1a dependent (11). Inactivation of Flcn in murine heart resulted in cardiac hypertrophy and mortality at 3 months of age due to dilated cardiomyopathy (9). In both kidney and cardiac models, mTORC1 and Ppargc1a pathways were up-regulated and inactivation of these pathways using either drug or gene manipulations ameliorated the phenotypes of these mouse models (9,11), suggesting that increased cell proliferation and protein synthesis under Flcn-deficiency are triggered by deregulation of Ppargc1a and mTORC1. Fnip1 knockout mice exhibited B cell developmental defects (14,15), supporting a critical role for FNIP1 in the metabolism of the immune system. Although Fnip2 knockout mice did not exhibit any phenotype, a kidney-targeted Fnip1 and Fnip2 double knockout mouse model developed enlarged polycystic kidneys, which were identical to Flcn-deficient kidneys (16), suggesting that the Flcn/Fnip interaction may play a key role in kidney tumour suppression.
FLCN is a classical tumour suppressor that fits the two hit hypothesis, supported by the fact that the loss of the remaining wild-type FLCN allele leads to tumorigenesis in human and mouse kidneys (8,17). Most of the FLCN mutations in BHD syndrome are frameshift, nonsense, or splice site mutations leading to protein truncation. However, two missense mutations, FLCN H255Y(8) and FLCN K508R (18), have been identified in the germlines of patients presenting with bilateral multifocal (BMF) kidney tumours, fibrofolliculomas and pulmonary manifestations of BHD (FLCN H255Y), or with BMF kidney neoplasia as the only manifestation (FLCN K508R). A mutation in the same codon, Flcn H255R, was originally identified as causative for a BHD-like syndrome in German shepherd dogs predisposing to cutaneous dermatofibromas and kidney cancer (19). Although the identification of these two FLCN missense mutations in patients presenting with multiple kidney tumours implies that these residues are functionally important, genetic studies to validate pathogenicity in preclinical animal models have not been performed.
Using BAC transgenic technology, we have developed two mouse models harbouring the Flcn H255Y and Flcn K508R mutations. We assessed the impact of these mutations in driving aberrant kidney cell proliferation by genetically introducing the mutant transgenes into a kidney-targeted Flcn knockout mouse model and evaluating their ability to rescue the polycystic kidney phenotype. We investigated signalling molecules in pathways dysregulated under Flcn deficiency using kidney tissues harvested from the kidney-targeted Flcn knockout mouse models carrying these Flcn missense mutations. In addition, we examined the phenotypic effect of the Flcn K508R mutant expression in mice with wild-type Flcn by expressing the Flcn K508R mutant transgene in heterozygous Flcn knockout mice.
Results
Patients with FLCN H255Y and K508R mutations develop kidney neoplasia with or without cutaneous or pulmonary BHD manifestations
Case 1: Patient from NCI Family with FLCN H255Y mutation
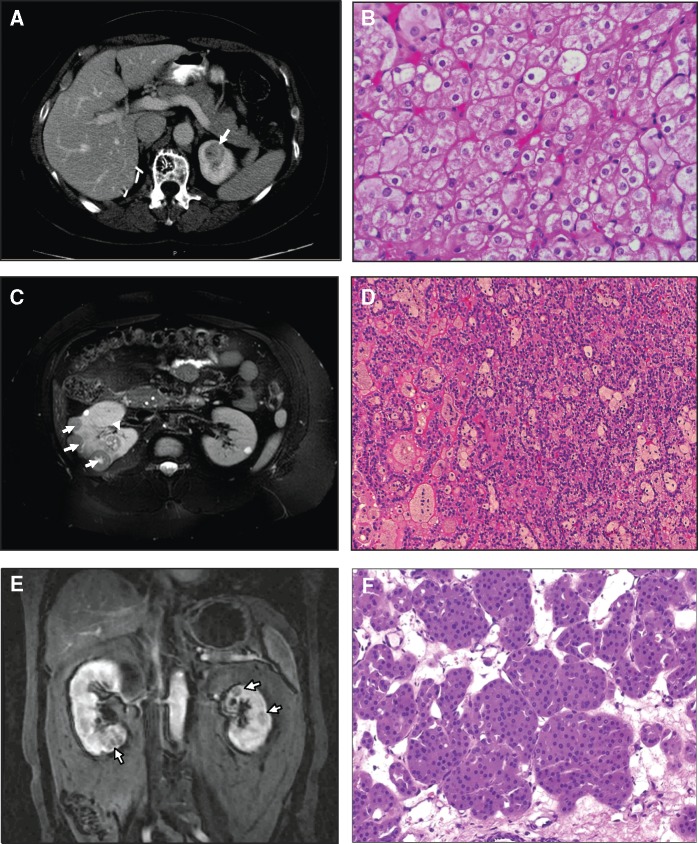
A 60 year-old female Caucasian patient presented with hematuria and was found to have bilateral renal masses on magnetic resonance imaging. A fine needle aspiration of the right kidney detected an ‘oncocytic neoplasm’ and she subsequently underwent a right partial nephrectomy at an outside institution. The tumour pathology was read as chromophobe renal carcinoma. Her history included a pneumothorax that had developed during her surgical procedure but which did not require treatment. Several family members reported history of pneumothoraces and multiple skin papules. She reported having had cutaneous papules since the age of 20, which were biopsied and found to be fibrofolliculomas. She was subsequently referred to the Urologic Oncology Branch, National Cancer Institute (NCI) for evaluation and management of the tumours in her left kidney. Dermatologic evaluation was performed at the NIH which revealed multiple cutaneous papules clinically consistent with BHD. Chest CT imaging revealed several lung cysts. Abdominal MRI imaging revealed the presence of multiple renal lesions in the left kidney. At the age of 62, she underwent a left robotic-assisted laparoscopic partial nephrectomy at the NCI for the removal of 3 hybrid tumours with features of chromophobe RCC and oncocytoma (Fig. 1A and B). Hybrid oncocytic renal tumours are characteristic of Birt-Hogg-Dubé syndrome (20). Her brother was found to have a renal oncocytoma at autopsy following his death due to an unrelated condition, and her 34 year-old daughter reported a history of spontaneous pneumothorax at age 20 and cutaneous trichodiscomas. Genetic testing for the FLCN mutation in peripheral blood DNAs from the mother and daughter revealed a germline FLCN missense mutation that exchanged a conserved histidine residue for a tyrosine (c.763 C > T, p.H255Y) (Table 1).
Figure 1.
Imaging and renal pathology of patients with FLCN H255Y and K508R mutations. Case 1: Patient from NCI Family with FLCN H255Y mutation. (A) MRI showing renal tumour in left kidney of patient with FLCN H255Y mutation. (B) Patient underwent a left partial nephrectomy in which 3 hybrid oncocytic tumours were removed. Magnification 200X. Case 2: Bilateral multifocal papillary type 1 RCC patient with FLCN K508R mutation. (C) MRI scan showing multiple renal tumours in right kidney of patient with FLCN K508R mutation. (D) This patient had a right partial nephrectomy in which 22 papillary type 1 renal tumours were removed. Magnification 150X. Case 3: Bilateral multifocal oncocytoma patient with FLCN K508R mutation. (E) MRI scan showing multiple renal tumours in right and left kidneys of patient with FLCN K508R mutation. (F) This patient had a left partial nephrectomy in which 5 renal oncocytomas were removed. Magnification 150X.
Table 1.
Clinical characteristics of BHD patients with germline FLCN H255Y and FLCN K508R mutations
| Case/Gender | Germline mutation | Age at surgery | Number of tumours removed | Histology | Somatic 2nd hit mutation | Number of tumours tested | Clinical manifestations |
|---|---|---|---|---|---|---|---|
| Case 1/female | c.763 C>T p.H255Y | 60y | 3a | Chromophobe RCC | ND | ND | Fibrofolliculomas, Lung cysts, Liver cysts |
| 62y | 3 | Hybrid oncocytic tumour | c.1432 + 1 G >T (1 tumour) | 2 | |||
| Case 2/male | c.1523A>G p.K508R | 25y | 22 | Papillary RCC type 1 | None | 4 | TAR syndrome, Mitral valve vegetations, Anti-phospholipid syndrome |
| Case 3/male | c.1523A>G p.K508R | 60y | 5 | Oncocytoma, oncocytosis, papillary adenomas | None | 2 | Hypertension, Gastroesophageal reflux, Gout, Hyperlipidemia, BPH, Irritable bowel |
| 61y | 3 | Oncocytoma with focus of papillary RCC type 1 | None | 1 |
N.D., not determined; RCC, renal cell carcinoma.
Numbering according to GenBank Accession No. NM_144997.5 with A of initiator codon designated as nucleotide1.
First surgery of Case 1 was performed at an outside institution.
Case 2: Bilateral multifocal papillary type 1 RCC patient with FLCN K508R mutation
A 25 year-old male African American patient who presented with abdominal pain to an outside institution, was imaged and was found to have bilateral multifocal tumours. Type 1 papillary renal cell carcinoma was found by core needle biopsy of the right kidney. The patient was referred to the NCI for evaluation and management. Dermatologic evaluation was not performed. Chest CT imaging revealed no pulmonary nodules. Abdominal MRI imaging revealed multiple renal tumours <3cm in size. The patient subsequently underwent a right open partial nephrectomy with removal of 22 papillary type 1 renal cell carcinomas (Fig. 1C and D). The patient is currently under active surveillance for a 1.7cm tumour in his left kidney. He reports no family history of renal tumours and no other BHD-associated manifestations; however, he has numerous medical conditions including TAR (thrombocytopenia with absent arm radius) syndrome, antiphospholipid syndrome and mitral valve vegetations. Genetic testing revealed a FLCN missense mutation in a conserved lysine mutated to arginine (c.1523A > G, p.K508R) (Table 1).
Case 3: Bilateral multifocal oncocytoma patient with FLCN K508R mutation
A 60 year-old male Caucasian patient presented with bilateral multiple renal lesions detected incidentally during workup for hematuria at an outside institution. Percutaneous renal biopsy was read as papillary renal tumour and the patient was referred to the NCI for evaluation. He subsequently underwent sequential left and right laparoscopic partial nephrectomies. Five tumours were removed from the left kidney of which three were oncocytoma and oncocytosis with papillary adenoma, and two were oncocytomas (Fig. 1E and F). Two tumours were removed from the right kidney of which one was renal oncocytoma and one was renal oncocytoma with a focus of papillary renal cell carcinoma type 1. He is currently being managed by active surveillance for additional renal tumours that are slowly increasing in size. Genetic testing revealed the same FLCN missense mutation, (c.1523A > G, p.K508R), as seen in Case 2. A dermatological evaluation and chest CT scan did not identify any other BHD-associated manifestations and he has no definitive family history suggestive of BHD (Table 1).
The diagnostic genetic testing of peripheral blood DNA from patients with bilateral multifocal kidney tumours seen at the NIH Clinical Center identified the patient in Case 1 with the germline FLCN missense mutation H255Y, who was clinically diagnosed with BHD, and the two patients in Cases 2 and 3 with the germline FLCN missense mutation K508R, who only presented with kidney neoplasia (Table 1). To evaluate the potential pathogenicity of these FLCN missense mutations, we searched for a somatic second hit mutation in the remaining FLCN allele in available kidney tumour samples from these patients by DNA sequencing. In a hybrid oncocytic tumour that developed in the patient carrying the germline FLCN H255Y mutation, we identified a somatic intron 12 splice site mutation (c.1432 +1 G > T; Table 1) predicted to result in aberrant splicing and a premature termination codon, suggesting that the loss of the wild type FLCN allele led to kidney tumorigenesis in this FLCN H255Y patient. None of the renal lesions (renal oncocytomas and papillary type 1 renal tumours) that developed in patients with the FLCN K508R mutation had second hit somatic mutations in FLCN. Contaminating normal kidney tissue can interfere with detection of a somatic mutation; alternatively, haploinsufficiency of FLCN may be sufficient to support kidney tumour development in individuals with a germline FLCN K508R mutation.
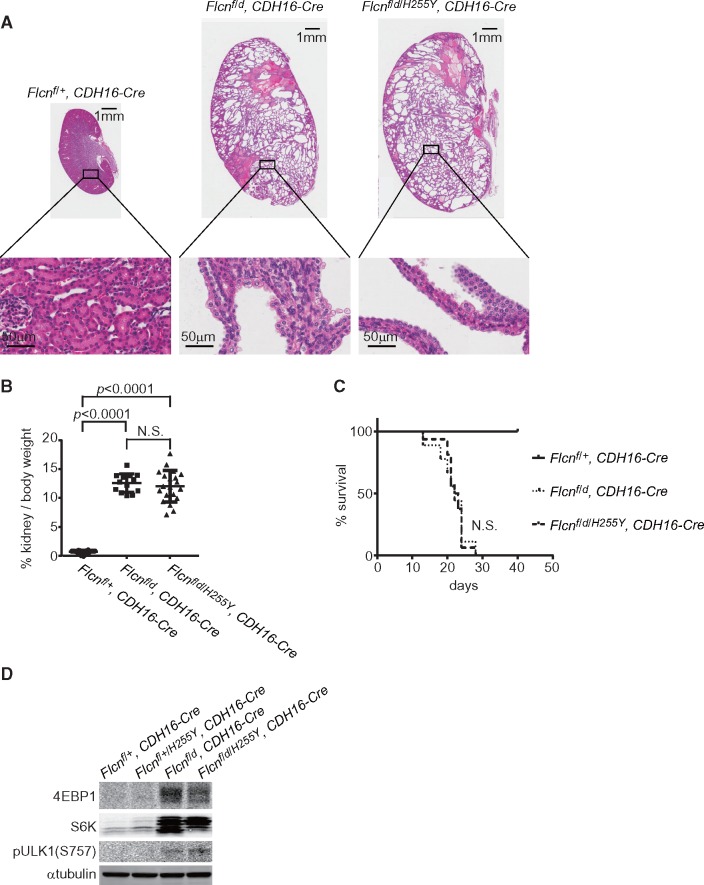
Expression of a BAC transgene carrying the Flcn H255Y mutation in kidney-targeted Flcn knockout mice did not rescue the polycystic kidney phenotype
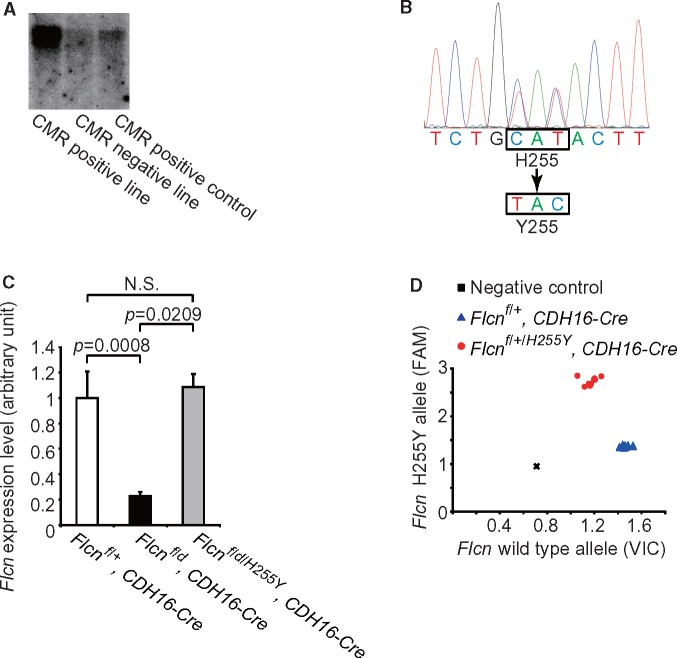
To evaluate the contribution of the FLCN missense mutants to aberrant kidney cell proliferation in these patients, we established genetically engineered mouse models carrying each of these missense mutations as transgenes randomly inserted into the mouse genome using BAC transgenic technology (Figs 2 and 3) (21). We genetically introduced the Flcn mutant transgenes into our kidney-targeted Flcn knockout mouse model, which develops enlarged polycystic kidneys and dies at 3 weeks of age due to renal failure (7), in order to determine if expression of the Flcn missense mutant protein would rescue or fail to rescue the polycystic kidney phenotype. Expression of the Flcn H255Y mutant in the kidney-targeted Flcn knockout mouse model did not alter the histology (Fig. 4A), the kidney/body weight ratio (Fig. 4B) or affect survival of the kidney-targeted Flcn knockout mice (Fig. 4C). Kidney-targeted Flcn knockout mice expressing the Flcn H255Y allele developed multi-cystic kidneys that were 10-fold heavier than control kidneys, and died due to renal failure by 3 weeks of age. Histologically, hyperplastic kidney cells were seen protruding into the cystic lumen. Western blot results showed that expression of Flcn H255Y had no additional effect on the activated mTORC1 pathway signalling observed in Flcn deficient kidneys (Fig. 4D). These data indicate that the Flcn H255Y mutant protein has lost its tumour suppressive properties and cannot rescue the aberrant kidney cell proliferation seen in Flcn-deficient kidneys, supporting the idea that the FLCN H255Y missense mutation inherited in the germline of the Case 1 BHD patient was responsible for BHD-associated phenotypes in this individual.
Figure 2.
Establishment of a Flcn H255Y mutant transgenic mouse model using BAC recombineering technology. (A) Screening for BAC transgene integration using Southern blotting. The probe detecting the chloramphenicol resistant (CMR) cassette was used for identifying BAC transgenic founder mice. (B) Sequencing result of mouse tail DNA confirming the correct mutant sequence encoding a histidine to tyrosine exchange. (C) Real-time PCR showing Flcn expression levels in kidneys of Flcnf/+, CDH16-Cre control mice, Flcn f/d, CDH16-Cre kidney-targeted knockout mice, and Flcn f/d/H255Y, CDH16-Cre kidney-targeted knockout mice expressing the Flcn H255Y mutation. (D) Representative genotyping results for Flcnf/+, CDH16-Cre mice and Flcn f/+/H255Y, CDH16-Cre mice using SNP real-time PCR and mouse tail DNA. N.S., no significance.
Figure 3.
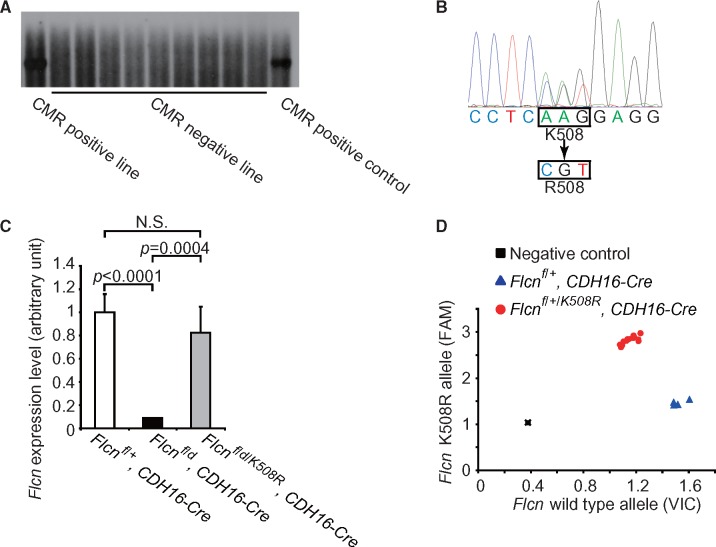
Establishment of a Flcn K508R mutant transgenic mouse model using BAC recombineering technology. (A) Screening for BAC transgene integration using Southern blotting. The probe detecting the chloramphenicol resistant (CMR) cassette was used for identifying BAC transgenic founder mice. (B) Sequencing result of mouse tail DNA confirming the correct mutant sequence encoding a lysine to arginine exchange. (C) Real-time PCR showing Flcn expression levels in kidneys of Flcnf/+, CDH16-Cre control mice, Flcn f/d, CDH16-Cre kidney-targeted knockout mice, and Flcn f/d/K508R, CDH16-Cre kidney-targeted knockout mice expressing the Flcn K508R mutation. (D) Representative genotyping results for Flcnf/+, CDH16-Cre mice and Flcn f/+/K508R, CDH16-Cre mice using SNP real-time PCR. N.S., no significance.
Figure 4.
Expression of BAC transgene carrying Flcn H255Y mutant allele does not rescue kidney-targeted Flcn knockout mouse phenotype. (A) Histology of Flcn-deficient kidney with Flcn H255Y mutant expression (Flcn f/d/H255Y, CDH16-Cre) at 3 weeks of age compared to Flcn-deficient kidney (Flcn f/d, CDH16-Cre) and control kidney (Flcnf/+, CDH16-Cre). Multi-cystic kidneys are shown with hyperplastic kidney cells protruding into the cystic lumen in both Flcn f/d/H255Y, CDH16-Cre mice and Flcn f/d, CDH16-Cre mice. (B) Kidney to body weight ratio of kidney-targeted Flcn knockout mice with Flcn H255Y mutant expression (Flcn f/d/H255Y, CDH16-Cre) at 3 weeks of age compared to kidney-targeted Flcn knockout mice (Flcn f/d, CDH16-Cre) and control mice (Flcnf/+, CDH16-Cre). No significant difference was seen between Flcn f/d/H255Y, CDH16-Cre and Flcn f/d, CDH16-Cre mice. (C) Kaplan-Meier survival plot of kidney-targeted Flcn knockout mice with Flcn H255Y mutant expression compared to kidney-targeted Flcn knockout and control mice; n= 8 mice for each genotype. No significant difference was seen between Flcn f/d/H255Y, CDH16-Cre and Flcn f/d, CDH16-Cre mice. (D) Western blot analysis of mTORC1 downstream effectors in Flcn-deficient kidneys with Flcn H255Y mutant expression, Flcn-deficient kidneys and control kidneys. N.S., no significance.
Expression of a BAC transgene carrying the Flcn K508R mutation in kidney-targeted Flcn knockout mice partially rescued the polycystic kidney phenotype
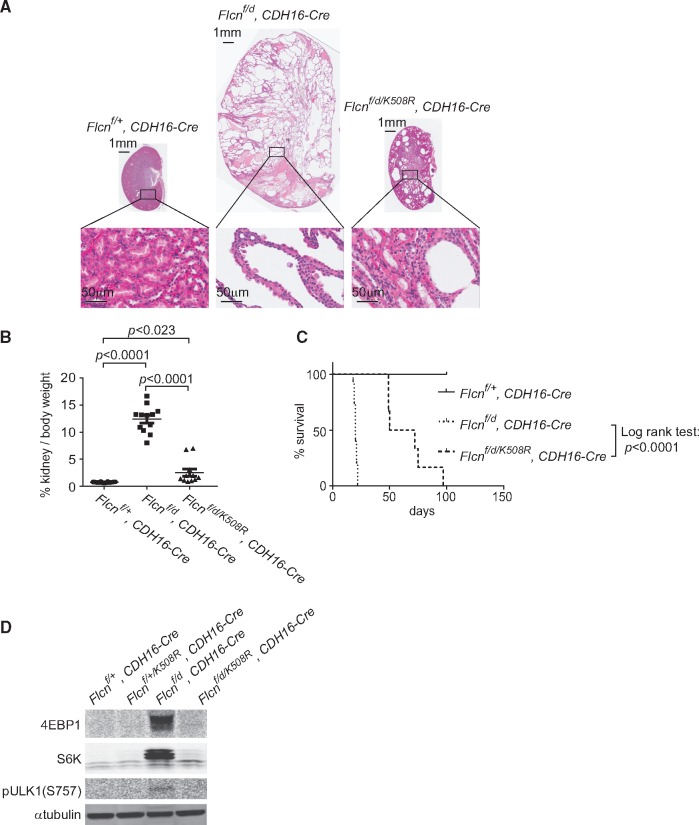
On the other hand, expression of the Flcn K508R mutant protein in the kidney-targeted Flcn knockout mouse model partially reversed the polycystic kidney phenotype (Fig. 5A) and significantly decreased the kidney/body weight ratio at 3 weeks of age (12.46 vs 2.58%; P < 0.0001) (Fig. 5B). Median survival of kidney-targeted Flcn knockout mice was increased 3-fold (20 d vs 61d, P < 0.0001; Fig. 5C) by the expression of Flcn K508R mutant protein. Furthermore, mTORC1 pathway signalling was repressed in the kidney-targeted Flcn knockout mouse model by expression of the Flcn K508R mutant protein (Fig. 5D). However, although the expression of the Flcn K508R mutant partially reversed the phenotype of the kidney-targeted Flcn knockout mouse model, none of these mice survived beyond 4 months of age. The Flcn K508R mutant protein was unable to sustain suppression of the aberrant kidney cell proliferation eventually leading to the development of multiple cysts, renal failure and premature death of the mice. These results support the idea that the FLCN K508R missense mutation is responsible for the kidney tumour phenotype in patients who inherit this mutation, but should be classified as a ‘weak’ mutation relative to the FLCN H255Y mutation.
Figure 5.
Expression of BAC transgene carrying Flcn K508R mutant allele partially, but not completely rescues the kidney-targeted Flcn knockout mouse phenotype. (A) Histology of Flcn-deficient kidney with Flcn K508R mutant expression (Flcn f/d/K508R, CDH16-Cre) at 3 weeks of age compared to Flcn-deficient kidney (Flcn f/d, CDH16-Cre) and control kidney (Flcnf/+, CDH16-Cre). Fewer cysts develop in Flcn f/d/K508R, CDH16-Cre mice compared to Flcn f/d, CDH16-Cre mice with no protruding hyperplastic cells. (B) Kidney to body weight ratio of kidney-targeted Flcn knockout mice with Flcn K508R mutant expression (Flcn f/d/K508R, CDH16-Cre) at 3 weeks of age showed significantly decreased % kidney/body weight relative to kidney-targeted Flcn knockout mice (Flcn f/d, CDH16-Cre). (C) Kaplan-Meier survival plot of kidney-targeted Flcn knockout mice with Flcn K508R mutant expression demonstrates 3-fold increase in survival (61 vs 20 days) compared to kidney-targeted Flcn knockout mice; n= 8 mice for each genotype. (D) Western blot analysis of mTORC1 downstream effectors in Flcn-deficient kidneys with Flcn K508R mutant expression, Flcn-deficient kidneys and control kidneys at 3 weeks of age showing suppression of the mTOR pathway activation in the Flcn f/d/K508R, CDH16-Cre mice compared with Flcn f/d, CDH16-Cre mice. N.S., no significance.
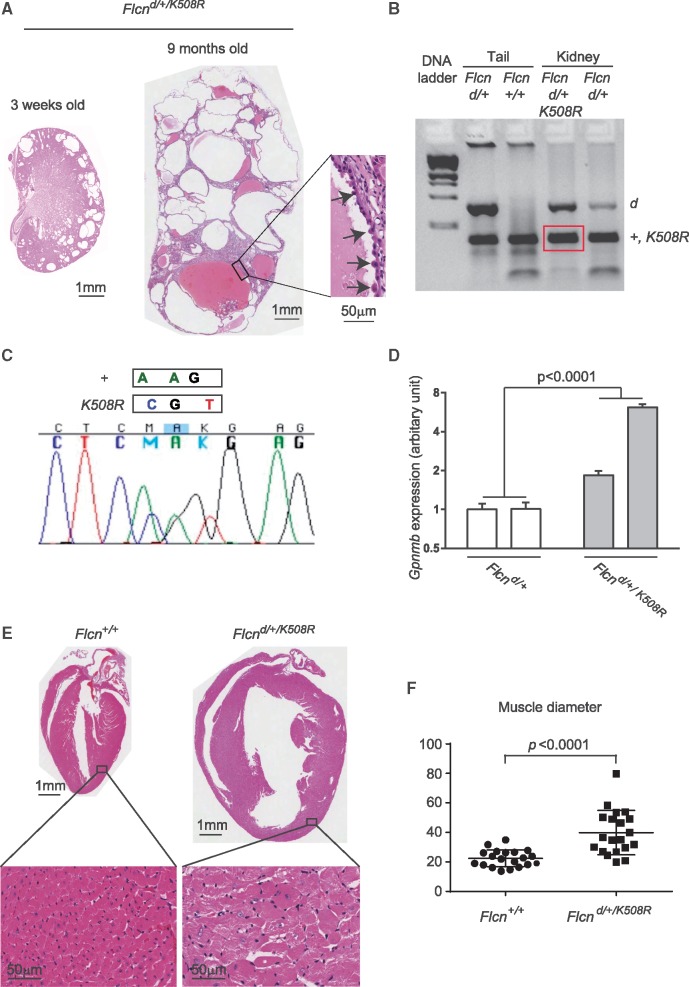
Expression of the Flcn K508R transgene in heterozygous Flcn knockout mice promotes polycystic kidneys and cardiac hypertrophy in a portion of mice
Heterozygous Flcn knockout mice (Flcn d/+) begin to develop renal cysts and tumours after 10 months of age, with a median lesion-free survival of 23 months and an average 3.5 lesions per animal; however, Flcn d/+ mice do not display any phenotype in other organs including heart (8). Flcn deficiency targeted to the murine heart leads to cardiac hypertrophy and mortality after 12 weeks of age (9). Notably, expression of the Flcn K508R mutant transgene in heterozygous Flcn knockout mice that harbour one wild-type Flcn allele and one deleted Flcn allele in all tissues produced enlarged kidneys filled with multiple large cysts (Fig. 6A, average >40 cysts per histologic cross-section) and also cardiac hypertrophy (Fig. 6E and F) by 9 months of age in a portion of the mice. PCR-based genotyping of macrodissected formalin fixed paraffin embedded (FFPE) cystic kidney epithelial cells from Flcn d/+/K508R mice produced a PCR product that indicated the presence of the wild-type Flcn allele and/or Flcn K508R transgene (both are the same size; Fig. 6B). Sequencing of the PCR product verified the presence of both wild-type Flcn and the Flcn K508R transgene in the kidney cysts (Fig. 6C). Although penetrance was reduced (multi-cystic kidneys in 11/42 mice, 26.2%; cardiac hypertrophy in 3/42 mice, 7.1%), the development of these phenotypes in mice that express both the Flcn K508R mutant and Flcn wild-type proteins suggests that the Flcn K508R mutant protein may have a dominant negative effect on wild-type Flcn in these tissues, which could explain the absence of a somatic second hit mutation in kidney tumour tissue from patients who carry the germline FLCN K508R mutation (Table 1).
Figure 6.
Expression of BAC transgene carrying Flcn K508R mutant alleles in heterozygous Flcn knockout mice drives a late onset Flcn-deficient phenotype. (A) Histology of polycystic kidneys that developed in a portion of heterozygous Flcn knockout mice with Flcn K508R mutant expression (Flcn d/+/K508R) at 3 weeks and 9 months of age, respectively. Insert shows hyperplastic cells protruding into the cystic lumen.(B) PCR-based genotyping of DNA from macrodissected FFPE Flcn d/+/K508R cystic kidney tissue generated PCR products of the correct size for the Flcn deleted allele and for the Flcn wild-type allele and/or Flcn K508R transgene (both are the same size). DNA from macrodissected FFPE Flcn d/+ kidneys was genotyped and included as a control along with PCR-based genotyping of mouse tail DNA for comparison. +, wild-type allele; d, delete allele; Kid, kidney tissue. (C) Sequence verification of both wild-type Flcn and Flcn K508R transgene in PCR product from (B) amplified from DNA extracted from macrodissected Flcn d/+/K508R cystic kidneys. +, wild-type allele. (D) Gpnmb expression (read-out of Flcn deficiency) measured by qRT-PCR was statistically significantly higher in Flcn d/+/K508R kidneys compared to Flcn d/+ mouse kidneys. Statistical significance measured by two-way ANOVA. Two representative mouse kidneys for each genotype are shown. P <0.05 for significance. (E) Histology of cardiac hypertrophy that developed in a portion of heterozygous Flcn knockout mice with Flcn K508R mutant expression at 9 months of age. (F) Muscle diameter analysis of (E).
We investigated whether the Flcn K508R mutant protein had an effect on wild-type Flcn function by evaluating levels of Gpnmb, a transcriptional target of TFE3 whose activity is negatively regulated by FLCN (22). In support of a dominant negative phenotype, Gpnmb expression was statistically significantly higher in Flcn d/+/K508R kidneys compared to Flcn d/+ control kidneys (Fig. 6D).
Flcn H255Y mutant protein is unstable with a reduced half-life compared to Flcn K508R mutant protein or Flcn WT protein
In order to determine whether the H255Y and K508R missense mutations had an effect on FLCN protein stability, we stably transduced HA-tagged FLCN H255Y, FLCN K508R or wild-type (WT) FLCN constructs into UOK257, a FLCN-deficient kidney tumour cell line established from a BHD patient (4), using a lentiviral doxycycline-inducible system. We performed quantitative RT-PCR after doxycycline induction, and found equivalent expression of FLCN WT, FLCN H255Y and FLCN K508R mRNAs. However, western analysis revealed reduced FLCN H255Y protein relative to FLCN WT and FLCN K508R proteins (Supplementary Material, Fig. S1). Furthermore, when we performed western analysis of UOK257 cells expressing WT FLCN and FLCN H255Y in the presence of cycloheximide, an inhibitor of protein translation, we found that FLCN H255Y was degraded more rapidly suggesting a shorter protein half-life (Supplementary Material, Fig. S2).
Molecular modeling predictions suggest that a conformational change may result from the K508 to R508 replacement leading to protein instability
Using NACCESS software developed at the University of Manchester, UK (23), we calculated the solvent accessible surface area on the human FLCN C-terminal crystal structure (PDBID: 3V42). The predictions suggest that K508 in FLCN may be exposed on the protein surface in a relatively small area [total accessible surface area of K508 = 39.17Å (relative accessibility = 19.5%), accessible surface area of K508 side chain = 38.96Å (relative accessibility = 23.9%) and accessible surface area of K508 main chain = 0.21Å (relative accessibility = 0.6%)]. Free energy changes predicted using FoldX software (24) suggest that the R508 exchange is destabilizing with a ΔG of 1.0 kcal mol − 1. Therefore, the functional consequence of the K508R mutation might be due to a conformational change caused by the replacement of lysine with arginine rather than loss of a posttranslational modification of K508 upon R508 replacement. This conformational change might affect protein-protein interactions or the posttranslational modification of surrounding amino acid residues, which may weaken the tumour suppressor function of FLCN resulting in only partial rescue of the kidney-targeted Flcn-knockout mouse model, and the aberrant kidney cell proliferation and cardiac hypertrophy observed in the Flcnd/+/K508R mouse model.
Discussion
In the absence of a validated assay for FLCN function, the functional consequence of FLCN missense mutations may be investigated through evaluating their effect on kidney phenotype in an in vivo model. Here, we have evaluated two FLCN missense mutations, H255Y and K508R, in genetically engineered mice, and demonstrated that they both are pathogenic mutations that promote aberrant kidney cell proliferation, but to different degrees, with the potential for malignancy. Expression of the Flcn H255Y mutant transgene could not reverse the enlarged kidney size, polycystic histology or 3-week lifespan of mice with Flcn-deficient kidneys, mirroring the phenotype of kidney-targeted homozygous Flcn deletion. The Flcn K508R expression only partially abrogated the Flcn-deficient phenotype leading eventually to highly cystic kidneys, renal failure and shortened lifespan in the Flcn f/d/K508R, CDH-16 Cre mice, suggesting that both Flcn H255Y and K508R mutant proteins were functionally impaired resulting in aberrant kidney cell proliferation. Expression of the Flcn K508R mutant in heterozygous Flcn knockout mice caused the development of enlarged multi-cystic kidneys and cardiac hypertrophy in 26 and 7% of mice, respectively, by 9 months of age, indicating that the Flcn K508R mutant protein may have a potential dominant negative effect on the function of wild-type Flcn to suppress cell growth of kidney cells and cardiomyocytes. Of note, somatic second hit mutations were not detected in renal tumour samples from human BHD patients with the germline FLCN K508R mutation, further supporting the potential of a dominant negative effect conferred by the FLCN K508R mutant on wild-type FLCN. The altered secondary structure of FLCN with K508R mutation may potentially disrupt the formation of a protein complex with wild type FLCN and/or other interacting partners (i.e., FNIP1, FNIP2, AMPK) and therefore lead to altered FLCN signalling resulting in the kidney neoplasia that develops in the FLCN K508R patients.
Although the Flcn pathway is essential to murine cardiac homeostasis, cardiac manifestations have not been reported in human BHD syndrome, suggesting that FLCN haploinsufficiency does not by itself cause cardiac hypertrophy in humans. Mitral valve vegetation was observed in one of the FLCN K508R patients (Case 2); however, this patient was diagnosed with TAR syndrome, a rare genetic disorder in which heart defects were frequently reported (25). In our mouse model, we observed cardiac hypertrophy in heterozygous Flcn knockout mice with Flcn K508R transgene expression, indicating that the Flcn K508R mutant protein may have a dominant negative effect on wild-type Flcn in the heart, leading to cardiac hypertrophy. These observations suggest that increased copy number of FLCN K508R mRNA (and presumably elevated FLCN mutant protein levels) above a certain threshold may predispose to cardiac hypertrophy. Therefore, careful cardiac follow up might be considered in the management of FLCN K508R patients.
Expression of the Flcn H255Y mutant transgene in Flcn-deficient kidneys did not affect kidney size or histology, indicating that Flcn H255Y mutant protein could not suppress the aberrant kidney cell proliferation. In vitro experiments have shown that the expression levels of the FLCN H255R (26,27) and H255P (28) mutant proteins in mammalian cells are consistently low, suggesting that the FLCN H255Y mutant protein might also be unstable. Our western blot data and cycloheximide chase experiment support the idea that the Flcn H255Y protein is unstable in agreement with these published data. Psi-blast based secondary structure prediction software PSI-PRED (29) predicts that amino acids 249-265 of the FLCN protein, wherein residue H255 resides, may form an α-helix with W251, A252, F258 and A259. Based on a conserved domain database (CDD) search (30), the residue H255 is well conserved across the species and residues 251 and 258 tend to be aromatic amino acids, raising the possibility that residue H255 might form pi stacking interactions together with residues W251 and F258. The replacement of Histidine 255 with Tyrosine or Arginine might cause a change in the electrostatic or aromatic property of the helix, respectively, leading to protein instability.
Gene variants of unknown significance (VUS) are identified much more frequently as more and more whole genome/exome sequencing data are being analysed, and this presents a challenge to physicians and genetic counsellors who manage and counsel these patients. Missense variants in genes that are not in known functional domains, splice sites or regulatory regions present a particular challenge for predicting pathogenicity, and the available predictive programs do not always accurately identify damaging/pathogenic alterations. There is no functional test for FLCN protein beyond evaluating its interaction with binding partners FNIP1 and FNIP2. Here, we have introduced two variants of unknown significance, which were identified in patients in our clinic, into mouse models and observed a clinical phenotype, which represents the ultimate test of pathogenicity. Currently, mouse model development using BAC transgenic technology has been largely replaced by gene editing using CRISPR/Cas9 technology thereby enabling rapid and efficient in vivo modeling of gene mutations as a mechanism for confirming the pathogenicity of VUS alterations.
In summary, germline missense mutations H255Y and K508R in the FLCN gene were associated with kidney tumours in patients with BHD clinical manifestations but also with kidney neoplasia as the sole phenotypic feature. Based on phenotypes resulting from their expression in kidney-targeted Flcn-deficient mouse models, FLCN H255Y and K508R are predicted to be pathogenic mutations. Based on the preclinical models presented here, the FLCN K508R mutant protein may have a dominant negative effect on wild-type FLCN in kidney and heart. These findings provide further mechanistic insight into the role of FLCN in the regulation of kidney cell proliferation and will aid in the development of novel therapeutics for kidney cancer.
Materials and Methods
Patients
Patients known or suspected to be affected with Birt-Hogg-Dubé syndrome were evaluated in the Urologic Oncology Branch, Center for Cancer Research, National Cancer Institute under protocol # 02-0159 approved by the NIH Institutional Review Board and each provided written informed consent. Each patient received genetics counseling and was evaluated for clinical manifestations of Birt-Hogg-Dubé syndrome with a dermatologic examination, pulmonary computed tomography (CT) scan and abdominal magnetic resonance imaging (MRI).
Generation of BAC Transgenic Mice and Genotyping
BAC transgenic mice carrying either Flcn H255Y or Flcn K508R mutant alleles were generated using BAC recombineering technology (21). A clone encompassing the mouse Flcn gene was isolated from a CITB 129/Svj BAC library (Invitrogen). Galactokinase gene (galK) expression cassettes were generated by PCR with 50 bp 5’ and 3’ homology arms homologous to sequences flanking the H255 and K508 codons, respectively. The PCR products were digested with DpnI, and galK targeting cassettes were gel purified. The galK targeting cassette was transformed into a galK deficient SW102 E. coli strain with the λ prophage recombineering system, which contained the BAC covering the Flcn genomic region. Correctly recombined clones were selected on galactose-containing minimal media. Then galK cassettes were replaced by 103bp double-stranded oligos containing the Y255 and R508 mutations, respectively, by recombineering technology in SW102. The recombined galK negative clones were selected on 2-deoxy-galactose minimal plates. The introduced mutations were verified by sequencing. After sequence verification, the Flcn mutant BAC clones were used to generate BAC transgenic mice by pronuclear microinjection of fertilized C57BL/6 eggs. Tail snip DNAs from the offspring were screened for single or low copy number integration of the transgene by Southern blotting. A probe detecting the chloramphenicol resistant (CMR) cassette was used for identifying founder mice. Mice carrying Flcn H255Y or K508R mutant alleles were mated with mice carrying Flcn deleted (d) alleles or floxed (f) alleles, and/or the cadherin 16 (CDH16)-Cre transgene that targets Cre recombinase expression in the distal nephron (31). Three different founder BAC transgenic mouse strains for each of the two mutations were expanded and evaluated in the Flcn f/d, CDH16 Cre mouse genetic rescue experiments; similar phenotypic results were obtained for the three strains carrying each mutation. SNP genotyping of mouse tail DNA for the H255Y mutant alleles was carried out with Taqman probes 5′- cgtgtctgcacacttcctt-3′-VIC for wild type allele and 5′-cgtgtctgtatacttcctt-3′-FAM for mutant allele, which detect the amplicon from the following primers: forward, 5′- gctctctgacctccttgaccag-3′; reverse, 5′- tgggttgttctaatggctcacag-3′. SNP genotyping of mouse tail DNA for K508R mutant alleles was carried out with Taqman probes 5′-tctgcctcaaggaggaatg-3′-VIC for wild type allele and 5′-tctgcctccgtgaggaatg-3′-FAM for mutant allele, which detect the amplicon from the following primers: forward, 5′-ccagaacctgtctgtggatgtg-3′; reverse, 5′-ctggagaagctgtcagagcaag-3′. Genotyping was also performed by targeted sequencing of mouse tail DNA using these primers for K508R: forward, 5’aggcctctgccttatccagt-3’; reverse, 5’-aaggctggggcttacttcat-3’; and for H255Y: forward, 5’gtgcccagaggatgaacact-3’; reverse, 5’-agatgggtgactttgggttg-3’. Mice were housed in National Cancer Institute animal facilities and euthanized by CO2 asphyxiation for analyses according to the National Cancer Institute Frederick Animal Care and Use Committee guidelines. Animal care procedures followed the National Cancer Institute Animal Care and Use Committee guidelines.
Tissue macrodissection, DNA extraction and PCR-based allelotyping
Cystic regions of mouse kidneys were hand macrodissected from formalin fixed paraffin embedded (FFPE) tissue slides and placed into 1.5 ml microcentrifuge tubes. One drop of mineral oil was added to each tube, and heated to 65 °C for 2 min. 175 μl of 0.2 mg/ml Proteinase K (Ambion), TD-S0 (Autogen) was added to each sample. Samples were placed into a Thermomixer R (Eppendorf, North America) for 5 min at 65 °C, and then at 55 °C for 68 h with 5 min. of shaking at 1400 rpm every 30 min; adding 10 μl of 20 mg/ml Proteinase K (Ambion) to each sample every 24 h. Lysates were incubated at 90 °C for 2 h, centrifuged for 5 min at 14,000g at 4 °C, and transferred to fresh tubes leaving the paraffin behind. 10 μg RNase A (Sigma) was added to the lysate, and incubate for 30 min. at 37 °C. DNA was isolated using the phenol-based AutoGenprep 245T Animal Tissue DNA Extraction Kit (Autogen) according to the manufacturer’s method. DNA was suspended in 10 mM Tris, pH 8.0. Yield and purity were determined by NanoDrop 1000 spectrophotometer (NanoDrop technologies). DNA was stored at 4°C until subsequent assay/analyses. PCR protocol was previously reported (4). Primers used for PCR and sequencing are listed above under genotyping.
Real-time PCR
Total RNA was isolated from mouse kidney specimens using TRIzol reagent (Invitrogen), and total RNA was reverse transcribed to cDNA using a Superscript III reverse transcriptase kit (Invitrogen). Quantitative real-time polymerase chain reaction (PCR) was performed with the 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA) using SYBR Green PCR Master Mix (Fermentas, Glen Burnie, CA). Signal intensity obtained from the Real-Time PCR System was described in relative units; each value was normalized using β-actin. Primer sequences are as follows: mouse Flcn-forward, 5′-gctcctgaaggcatgcggtagca-3′; mouse Flcn-reverse, 5′-cagcaagcttctccatctggaccag- 3′; mouse Gpnmb-forward, 5’-gctacttcagagccaccatcacaa-3’; mouse Gpnmb-reverse, 5’-ggagatgatcgtacaggcttcca-3’; mouse β-actin forward, 5′-gacatggagaagatctggca-3′; and mouse β-actin reverse, 5′-ggtctcaaacatgatctgggt-3′.
Western Blotting
Frozen kidney samples were homogenized in RIPA buffer (20mMTris·HCl at pH 7.5, 150mM NaCl, 1 mM EDTA, 1.0% Triton X-100, 0.5% deoxycholate, 0.1% SDS) supplemented with Complete protease inhibitor mixture and the PhosStop phosphatase inhibitor mixture (Roche). For immunoblotting, 20 μg protein was loaded in each well. Antibodies used for western blotting included total S6K, 4EBP1, phospho-Ulk1(Ser757), HA(6E2) and α−tubulin (all from Cell Signaling), and FLCN monoclonal antibody (4) at 1:1,000 dilution. Antibody–protein complexes were detected using an Odyssey imager (LI-COR Biotechnology).
Statistical Analysis
Experimental data are summarized as the mean values with SD. Statistical analyses were performed using a two-tailed Student t test (SPSS Statistics version 20), and two-way ANOVA, and differences were considered to be statistically significant at a value of P < 0.05. Survival curves were obtained using GraphPad Prism version 6.01.
Animal Care
National Cancer Institute–Frederick is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals. Animal care was provided in accordance with the procedures outlined in the National Research Council’s ‘Guide for Care and Use of Laboratory Animals’.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Dr. Peter Igarashi for CDH16-Cre transgenic mice, and Louise Cromwell and Nicole Morris for excellent technical support with the mouse studies. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Conflict of Interest Statement. None Declared.
Funding
This project has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under Contract HHSN261200800001 E. (LSS) M.B. was supported by JSPS KAKENHI Grant Number 15604475 and 15551829. M.L. was supported by an annual outbound fellowship from Fondazione Italiana Ricerca sul Cancro, ‘Fellowship For Abroad 2011’.
References
- 1. Birt A. R., Hogg G. R., Dube W. J. (1977) Hereditary multiple fibrofolliculomas with trichodiscomas and acrochordons. Arch. Dermatol., 113, 1674–1677. [PubMed] [Google Scholar]
- 2. Zbar B., Alvord W.G., Glenn G., Turner M., Pavlovich C.P., Schmidt L., McClellan W., Choyke P., Weirich G., Hewitt S. M., et al. (2002) Risk of renal and colonic neoplasms and spontaneous pneumothorax in the Birt-Hogg-Dube syndrome. Cancer Epidemiol. Biomarkers Prev ., 11, 393–400. [PubMed] [Google Scholar]
- 3. Nickerson M. L., Warren M.B., Toro J.R., Matrosova V., Glenn G., Turner M.L., Duray P., Merino M., Choyke P., Pavlovich C.P., et al. (2002) Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell, 2, 157–164. [DOI] [PubMed] [Google Scholar]
- 4. Baba M., Hong S.B., Sharma N., Warren M.B., Nickerson M.L., Iwamatsu A., Esposito D., Gillette W.K., Hopkins R.F. III, Hartley J.L., et al. (2006) Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc. Natl. Acad. Sci. USA, 103, 15552–15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hasumi H., Baba M., Hong S.B., Hasumi Y., Huang Y., Yao M., Valera V.A., Linehan W.M., Schmidt L.S. (2008) Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene, 415, 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takagi Y., Kobayashi T., Shiono M., Wang L., Piao X., Sun G., Zhang D., Abe M., Hagiwara Y., Takahashi K., et al. (2008) Interaction of folliculin (Birt-Hogg-Dube gene product) with a novel Fnip1-like (FnipL/Fnip2) protein. Oncogene ,27, 5339–5347. [DOI] [PubMed] [Google Scholar]
- 7. Baba M., Furihata M., Hong S.B., Tessarollo L., Haines D.C., Southon E., Patel V., Igarashi P., Alvord W.G., Leighty R., et al. (2008) Kidney-targeted Birt-Hogg-Dube gene inactivation in a mouse model: Erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys. J. Natl. Cancer Inst., 100, 140–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hasumi Y., Baba M., Ajima R., Hasumi H., Valera V.A., Klein M.E., Haines D.C., Merino M.J., Hong S.B., Yamaguchi T.P., et al. (2009) Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc. Natl. Acad. Sci. USA, 106, 18722–18727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hasumi Y., Baba M., Hasumi H., Huang Y., Lang M., Reindorf R., Oh H., Sciarretta S., Nagashima K., Haines D.C., et al. (2014) Folliculin (Flcn) inactivation leads to murine cardiac hypertrophy through mTORC1 deregulation. Hum. Mol. Genet., 23, 5706–5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hartman T. R., Nicolas E., Klein-Szanto A., Al-Saleem T., Cash T.P., Simon M.C., Henske E.P. (2009) The role of the Birt-Hogg-Dube protein in mTOR activation and renal tumorigenesis. Oncogene, 28, 1594–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hasumi H., Baba M., Hasumi Y., Huang Y., Oh H., Hughes R.M., Klein M.E., Takikita S., Nagashima K., Schmidt L.S., et al. (2012) Regulation of mitochondrial oxidative metabolism by tumor suppressor FLCN. J. Natl. Cancer Inst., 104, 1750–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yan M., Gingras M.C., Dunlop E.A., Nouët Y., Dupuy F., Jalali Z., Possik E., Coull B.J., Kharitidi D., Dydensborg A.B., et al. (2014) The tumor suppressor folliculin regulates AMPK-dependent metabolic transformation. J. Clin. Invest., 124, 2640–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nookala R. K., Langemeyer L., Pacitto A., Ochoa-Montaño B., Donaldson J.C., Blaszczyk B.K., Chirgadze D.Y., Barr F.A., Bazan J.F., Blundell T.L. (2012) Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol., 2, 120071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baba M., Keller J.R., Sun H.W., Resch W., Kuchen S., Suh H.C., Hasumi H., Hasumi Y., Kieffer-Kwon K.R., Gonzalez C.G., et al. (2012) The folliculin-FNIP1 pathway deleted in human Birt-Hogg-Dube syndrome is required for murine B-cell development. Blood, 120, 1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park H., Staehling K., Tsang M., Appleby M.W., Brunkow M.E., Margineantu D., Hockenbery D.M., Habib T., Liggitt H.D., Carlson G., et al. (2012) Disruption of Fnip1 reveals a metabolic checkpoint controlling B lymphocyte development. Immunity, 36, 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hasumi H., Baba M., Hasumi Y., Lang M., Huang Y., Oh H.F., Matsuo M., Merino M.J., Yao M., Ito Y., et al. (2015) Folliculin-interacting proteins Fnip1 and Fnip2 play critical roles in kidney tumor suppression in cooperation with Flcn. Proc. Natl. Acad. Sci. USA, 112, E1624–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vocke C. D., Yang Y., Pavlovich C.P., Schmidt L.S., Nickerson M.L., Torres-Cabala C.A., Merino M.J., Walther M.M., Zbar B., Linehan W.M. (2005) High frequency of somatic frameshift BHD gene mutations in Birt-Hogg-Dube-associated renal tumors. J. Natl. Cancer Inst., 97, 931–935. [DOI] [PubMed] [Google Scholar]
- 18. Toro J. R., Wei M.H., Glenn G.M., Weirich M., Toure O., Vocke C., Turner M., Choyke P., Merino M.J., Pinto P.A., et al. (2008) BHD mutations, clinical and molecular genetic investigations of Birt-Hogg-Dube syndrome: a new series of 50 families and a review of published reports. J. Med. Genet., 45, 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lingaas F., Comstock K.E., Kirkness E.F., Sørensen A., Aarskaug T., Hitte C., Nickerson M.L., Moe L., Schmidt L.S., Thomas R., et al. (2003) A mutation in the canine BHD gene is associated with hereditary multifocal renal cystadenocarcinoma and nodular dermatofibrosis in the German Shepherd dog. Hum. Mol. Genet., 12, 3043–3053. [DOI] [PubMed] [Google Scholar]
- 20. Pavlovich C.P., Walther M.M., Eyler R.A., Hewitt S.M., Zbar B., Linehan W.M., Merino M.J. (2002) Renal tumors in the Birt-Hogg-Dubésyndrome. Am. J. Surg. Pathol., 26,1542–1552. [DOI] [PubMed] [Google Scholar]
- 21. Warming S., Costantino N., Court D.L., Jenkins N.A., Copeland N.G. (2005) Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res., 33,e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hong S.B., Oh H., Valera V.A., Baba M., Schmidt L.S., Linehan W.M. (2010) Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS One., 5, e15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hubbard S.J., Thornton J.M. (1993) ′NACCESS′, Computer Program, Department of Biochemistry and Molecular Biology, University College London.
- 24. Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. (2005) The FoldX web server: an online force field. Nucleic Acids Res., 33(Webserver issue),W382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Albers C. A., Newbury-Ecob R., Ouwehand W. H., Ghevaert C. (2013) New insights into the genetic basis of TAR (thrombocytopenia-absent radii) syndrome. Curr Opin. Genet. Dev ., 23, 316–323. [DOI] [PubMed] [Google Scholar]
- 26. Hong S. B., Oh H., Valera V.A., Stull J., Ngo D.T., Baba M., Merino M.J., Linehan W.M., Schmidt L.S. (2010) Tumor suppressor FLCN inhibits tumorigenesis of a FLCN-null renal cancer cell line and regulates expression of key molecules in TGF-beta signaling. Mol. Cancer, 9, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Slegtenhorst M., Khabibullin D., Hartman T.R., Nicolas E., Kruger W.D., Henske E.P. (2007) The Birt-Hogg-Dube and tuberous sclerosis complex homologs have opposing roles in amino acid homeostasis in Schizosaccharomyces pombe. J. Biol. Chem., 282, 24583–24590. [DOI] [PubMed] [Google Scholar]
- 28. Nahorski M.S., Reiman A., Lim D.H., Nookala R.K., Seabra L., Lu X., Fenton J., Boora U., Nordenskjöld M., Latif F., et al. (2011) Birt-Hogg-Dubésyndrome-associated FLCN mutations disrupt protein stability. Hum. Mutat., 32, 921–929. [DOI] [PubMed] [Google Scholar]
- 29. Buchan D.W.A., Minneci F., Nugent T.C.O., Bryson K., Jones D.T. (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res., 41 (Webserver issue), W340–W348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., et al. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res., 43 (Database issue), D222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shao X., Somlo S., Igarashi P. (2002) Epithelial-specific Cre/lox recombination in the developing kidney and genitourinary tract. J. Am. Soc. Nephrol ., 13, 1837–1846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.