Abstract
SOX5 encodes a transcription factor that is expressed in multiple tissues including heart, lung and brain. Mutations in SOX5 have been previously found in patients with amyotrophic lateral sclerosis (ALS) and developmental delay, intellectual disability and dysmorphic features. To characterize the neuronal role of SOX5, we silenced the Drosophila ortholog of SOX5, Sox102F, by RNAi in various neuronal subtypes in Drosophila. Silencing of Sox102F led to misorientated and disorganized michrochaetes, neurons with shorter dendritic arborization (DA) and reduced complexity, diminished larval peristaltic contractions, loss of neuromuscular junction bouton structures, impaired olfactory perception, and severe neurodegeneration in brain. Silencing of SOX5 in human SH-SY5Y neuroblastoma cells resulted in a significant repression of WNT signaling activity and altered expression of WNT-related genes. Genetic association and meta-analyses of the results in several large family-based and case-control late-onset familial Alzheimer’s disease (LOAD) samples of SOX5 variants revealed several variants that show significant association with AD disease status. In addition, analysis for rare and highly penetrate functional variants revealed four novel variants/mutations in SOX5, which taken together with functional prediction analysis, suggests a strong role of SOX5 causing AD in the carrier families. Collectively, these findings indicate that SOX5 is a novel candidate gene for LOAD with an important role in neuronal function. The genetic findings warrant further studies to identify and characterize SOX5 variants that confer risk for AD, ALS and intellectual disability.
Introduction
The SOX5 gene (OMIN 604975) on chromosome 12p12.1 encodes a transcription factor of the SOX family that is characterized by a DNA-binding motif HMG (high mobility group) box. Human SOX5 has two isoforms: the longer SOX5 variant encodes a 763-amino acid protein that is expressed in multiple human tissues, including heart, brain, liver, lung, kidney, spleen, and testis; and the shorter isoform of SOX5 encodes a 750-amino acids protein (N-terminally truncated by 13 amino acids compared with the longer isoform) that is specifically expressed in testis (1). SOX5 has been identified as a susceptibility candidate gene for multiple human diseases and traits by genome-wide association studies (GWAS) including higher resting heart rate (RHR) (2), abnormal electrocardiographic PR interval (3), aberrant atrial fibrillation (AF) (4) increased left ventricular mass (LVM) (5), and chronic obstructive pulmonary disease (COPD) (6). Human SOX5 is highly conserved with the Drosophila ortholog Sox102F: 71% identity and 82% amino acid similarity. We have previously shown that silencing of Sox102F in heart resulted in cardiac dysfunction in adult Drosophila (7). Sox5 deficient mice had abnormal lung development and died at birth with respiratory distress (6). Moreover, SOX5 promoted cell cycle exit of neural progenitors and its down-regulation was required for the progression of neuronal differentiation (8). Furthermore, Sox5 and Sox6 are redundant enhancers of chondroblast functions, controlling both expressions of extracellular matrix genes and cell proliferation (9). Collectively, these studies implicate that SOX5 have multi-functional roles.
Mutations in SOX5 have been found in patients with amyotrophic lateral sclerosis (ALS) (10), developmental delay or intellectual disability with prominent language delay, behavioral problems and mild dysmorphic features (11). These findings suggest that variants and mutations in SOX5 may disrupt neuronal development and/or function. However, owing to embryonic lethality in Sox5 knockout mice and chicks, to date, the neuronal function of SOX5 remains unclear.
To determine the role of SOX5 on neuronal development and function, we silenced the Drosophila ortholog of SOX5, Sox102F in type I bristle-type sensory neurons, type II dendritic arborization (DA) sensory neurons, motor neurons, mushroom bodies (MBs) and whole brain. We previously found that Sox102F regulates the expression of the WNT-related wingless (Wg) and wing development in Drosophila (7). Thus, in this study, we examined the effect of silencing of SOX5 in human SH-SY5Y neuroblastma cells on the expression of the 84 genes related to WNT-mediated signal transduction. Our current studies show that silencing of Sox102F leads to abnormal neuronal development and behavioral impairment in Drosophila. Silencing of SOX5 in human SH-SY5Y neuroblastoma cells resulted in a significant repression of WNT signaling activity and altered expression of WNT-related genes. We also tested for genetic association between SOX5 variants and risk for Alzheimer's disease (AD), the most common form of dementia in the elderly. We observed significant genetic association of SOX5 with familial late-onset AD (LOAD), and identified four novel mutations in SOX5 that segregate with disease status in LOAD families.
Results
Silencing of the Drosophila ortholog of SOX5 leads to abnormal neuronal development and behavioral impairment
We employed the UAS-Sox102F-RNAi transgenic flies and the UAS-GAL4 system as described previously (7) to silence Sox102F in various neuronal subtypes.
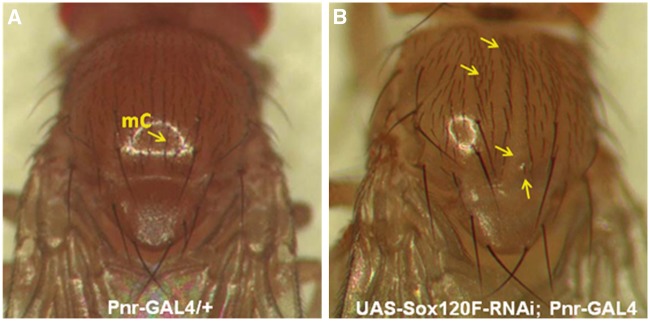
Sensory neurons . Sox102F was silenced in the type I bristle-type sensory neurons by Pnr-GAL4 in the pattern of pannier (Pnr) gene (12). Flies in which Sox102F was silenced (UAS-Sox102F-RNAi; Pnr-GAL4) exhibited misorientation and disorganization of michrochaetes (mC), accumulation of upper dorsal michrochaetes close to the head, loss of partial michrochaetes in the dorsal centre, and michrochaete morphology defects (Fig. 1B, arrows) as compared to controls (Fig. 1A).
Figure 1.
Analysis of the type I bristle-type sensory neurons. (A) Control shows normal michrochaetes (mC). (B) Silencing of Sox102F led to misorientated and disorganized mC, and accumulation of upper dorsal michrochaetes close to head, loss of partial michrochaetes in the dorsal centre and michrochaete morphology defect (arrows).
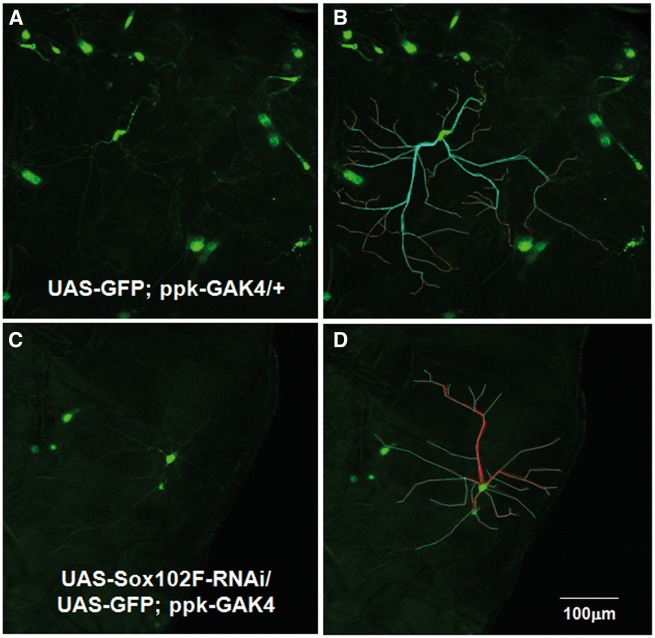
Next, Sox102F was silenced in the type II internal multiple dendritic arborization (DA) sensory neurons by ppk-GAL4, which expresses in the pattern of the pickpocket (ppk) gene in Class III and IV DA neurons (13). A GFP protein was co-overexpressed using ppk-GAL4 in the DA neurons for fluorescence imaging analysis. The DA neuron length, branching and complexity in the third instar larva were quantified (Fig. 2). The analysis parameters included: length of dendrites (µm), surface area (µm2), complexity of arborization (total number of branches), and branching complexity (%). Silencing of Sox102F in DA neurons significantly affected dendrite branching morphology and resulted in shorter DA neurons with lower complexity (Fig. 2C and D;Table 1) in third instar larva as compared to controls (Fig. 2A and B). Flies in which Sox102F was silenced exhibited an average dendrite length of 177 μm compared to 147 μm of that in controls (P = 1.2 × 10 − 11), and an average arbor surface area of 847571 μm2 compared to 652477 μm2 in controls (P = 0.04). In addition, silencing of Sox102F resulted in a reduced complexity of arbor with an average number of total branches of 21 compared to 28 in controls (P = 0.002), and that the number of either primary or secondary branches were significantly reduced (Table 1).
Figure 2.
Analysis of the DA neurons. The parameters include (A) Length of dendrites (µm)-Measure from cell body to tip of branch for each ending in arbor; (B) Surface area (µm2); (C) Complexity of arbor (total number of braches) and Branching complexity (%). (D,E) ppk-GAL4 drives expression of UAS-GFP in the DA neurons in control flies. (F,G) Silencing of Sox102F in DA neurons significantly affects dendrite branching morphology and results in shorter DA neurons with lower complexity in third instar larva in preliminary analysis compared to that in controls. Magnification: 10X; Scale bar: 100µm.
Table 1.
The effect of silencing of Sox102F on dendritic arborization neurons
| Parameters/Mean ± SD | ppk-GAL4; UAS-GFP/+ N=20 | UAS-Sox102F; ppk-GAL4; UAS-GFP N = 21 | T Test P value | |
|---|---|---|---|---|
| Average dendrite length | 176.7 ± 76 μm | 147.14 ± 58.17 μm | 1.2 × 10−11* | |
| Arbor surface area | 847571.4 ± 401269.2 μm2 | 652476.6 ± 293906.9 μm2 | 0.04 * | |
| Complexity of arbor (number of branches) | 28.35 ± 9.48 | 20.81 ± 5.67 | 0.002 * | |
| Complexity of dendrite branching (%) | Primary (%) | 8.86 ± 3.63 | 12.13 ± 4.01 | 0.005 * |
| Secondary (%) | 25.70 ± 6.45 | 31.29 ± 9.34 | 0.02 * | |
| Tertiary (%) | 34.87 ±10.94 | 37.06 ± 8.78 | NS | |
| Quaternary (%) | 17.92 ± 8.46 | 15.86 ±10.45 | NS | |
| Quinary (%) | 12.47 ± 6.35 | 7.68 ± 3.72 | 0.036 * | |
NS: not significant.
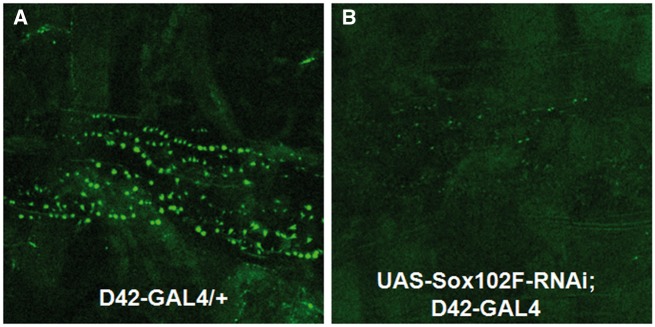
Motor neuron. To determine the effect of Sox102F on motor neurons, we silenced Sox102F using D42-GAL4. The GFP protein was co-overexpressed in motor neuron for fluorescence imaging analysis. The locomotion activities were assessed in the third instar larva (N = 110) using the larval crawling assay and in adult flies (N = 356) by climbing assays. Control larva exhibited normal locomotion function with an average of 40 peristaltic contractions per minute (40 ± 15 contractions/min, N = 55); in contrast, silencing of Sox102F significantly reduced the count of larval peristaltic contractions (33 ± 12 contractions/min, N = 55) by Student T Test (P = 0.002). Flies in which Sox102F was silenced exhibited a significant decrease in climbing height (160 ± 57 mm, N = 80) compared to controls (177 ± 44 mm, N = 176) (P = 0.002). Consistent with the reduced larval and adult locomotion abilities in the Sox102F RNAi flies, the third instar larva in which Sox102F was silenced exhibited a markedly reduced number of boutons and loss of bouton structures at the neuromuscular junction (NMJ) (Fig. 3B) compared to controls (Fig. 3A).
Figure 3.
Analysis of the NMJs. (A) Control larva showed normal NMJ boutons and structure; (B) Larva in which Sox102F was silenced exhibited a markedly reduced number of boutons and loss of bouton structures at the NMJ. Magnification: 20X; Scale bar: 100µm.
Mushroom bodies (MBs). MBs in the Drosophila brain are the center of olfactory learning and memory. We silenced Sox102F in the MBs using Tab2-GAL4, which expresses in the pattern of TAK1-associated binding protein 2 (Tab2). We then examined larval olfactory learning behavior using the fructose preference assay (examine larval preference for fructose) modified from that was described previously (14). Flies in which Sox102F was silenced in the MBs (UAS-Sox102F-RNAi; Tab2-GAL4) exhibited a significantly decreased olfactory perception behavior by fructose assay (224 fructose-preferred vs. 195 non-preferred) compared with controls (250 fructose-preferred vs. 155 non-preferred) (Fisher Exact Test P = 0.01).
Whole brain . Sox102F was silenced in all the neurons of whole brain using Insc-GAL4. Memory function of adult flies was assessed by an adult courtship behavior assay (15). Newly emerged virgin male and female flies were collected and independently cultured for 5 days then subjected to courtship behavior analysis. Each tested fly was observed under a dissection scope for recording the time of: orientation, tapping, wing song, licking, curling, and the first copulation attempt, the final successful copulation attempt, and the total time engaged in copulation. The courtship latency, copulation latency, and the courtship index (CI) were also determined. If copulation occurred, the female was isolated in a fresh tube to determine if the copulation indeed resulted in progeny. Flies in which Sox102F was silenced in whole brain (UAS-Sox102F-RNAi; Insc-GAL4), exhibited a significant delay in the time of licking (1079 seconds (s), P = 0.02) and curling (1173s, P = 0.01) and a significantly shortened length of copulation (1082s, P = 0.02) compared to that in controls (878s, 857s, and 1324s, respectively) (Table 2). The CI was significantly decreased to 0.36 in UAS-Sox102F-RNAi; Insc-GAL4 flies compared to a CI of 0.44 in controls (P = 0.02) (Table 2). However, there were no significant differences in the time of orientation, tapping, wing song, courtship latency, copulation attempt, and copulation latency between the UAS-Sox102F-RNAi; Insc-GAL4 and control flies.
Table 2.
The effect of silencing of Sox102F in the brain on adult Drosophila courtship behavior
| Group | UAS-Sox102F-RNAi; Insc-GAL4 | Insc-GAL4/+ | T Test P value |
|---|---|---|---|
| Mean ± SD | |||
| Total number of flies (N) | 30 | 40 | |
| Courtship Index (CI) | 0.36 ± 0.06 | 0.44 ± 0.07 | 0.02* |
| Orientation (Seconds) | 858 ± 469 | 767 ± 385 | NS |
| Tapping (S) | 991 ± 623 | 863 ± 608 | NS |
| Wing Song (S) | 983 ± 440 | 865 ± 447 | NS |
| Licking (S) | 1079 ± 666 | 878 ±438 | 0.02* |
| Curling (S) | 1173 ± 362 | 857 ±415 | 0.01* |
| Courtship Latency (S) | 842 ± 485 | 731 ±419 | NS |
| Copulation Attempt (S) | 978 ±393 | 992 ± 374 | NS |
| Copulation Latency (S) | 953 ± 550 | 1122 ± 323 | NS |
| Copulation Duration (S) | 1082 ± 184 | 1324 ±217 | 0.02* |
NS: not significant.
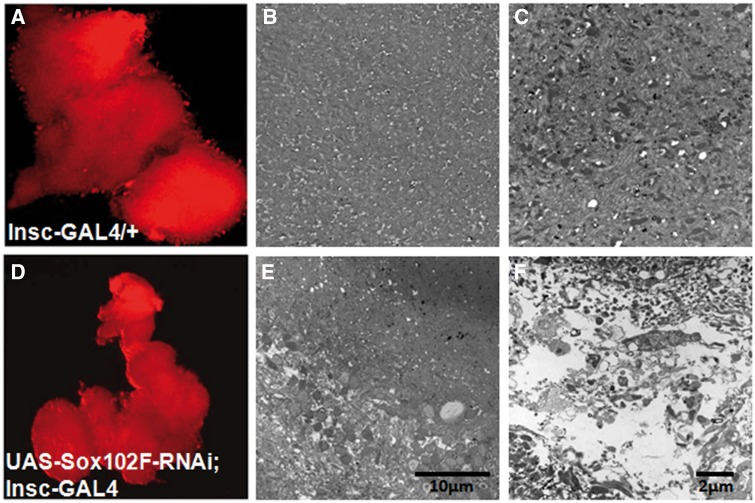
We dissected whole brain from the 30-day-old UAS-Sox102F-RNAi; Insc-GAL4 and control adult flies. Fluorescence imaging analysis by co-overeprerssing mCherry–RFP in the neuron demonstrates that control flies (Insc-GAL4-ChRFP/+) showed normal brain structure of optical lobs and the central brain; in contrast, silencing of Sox102F (UAS-Sox102F-RNAi; Insc-GAL4-ChRFP) resulted in a smaller and disorganized brain with degenerated optical lobs and central brain (Fig. 4D) compared to that of controls (Fig. 4A). Consistently, ultra-structural analysis of whole brain from 30-day-old flies by transmitted electron microscopy (TEM) demonstrated that silencing of Sox102F by Insc-GAL4 in whole brain resulted in disruption of the brain structure and neurodegeneration with the formation of vacuoles in the brain (Fig. 4E and F) compared to that in controls (Fig. 4B and C).
Figure 4.
Structure and ultra-structural analysis of central regions of brain of 30-day-old controls and UAS-Sox102F-RNAi; Insc-GAL4 flies with fluorescence imaging and TEM. (A–C) Control fly brains showed normal brain morphology. (D–F): UAS-Sox102F-RNAi; Insc-GAL4 showed a smaller and disorganized brain (D), wide-spread vacuoles and severe neurodegeneration (E,F) in the whole brain. Magnification: (A and D): 10X, scale bar: 100µm. (B,E): 2000X; scale bar 10um. (C,F): 20000X; scale bar: 2µm.
Silencing of SOX5 in human SY5Y neuronal cells affects the expression of genes involved in WNT signaling
To identify the effect of SOX5 on genes involved in WNT signaling, we silenced SOX5 in human SH-SY5Y neuroblastoma cells using SOX5-specific siRNAs in comparison to scrambled control siRNAs. Total RNA was isolated from cultured cells and reverse transcribed into cDNA. Silencing of SOX5 resulted in a 60% reduction in gene expression compared to that in controls as examined by real-time RT-PCR.
The effect of SOX5 on the expression of 84 genes involved in WNT-mediated signal transduction was examined using the Human WNT Signaling Pathway Plus RT-PCR Array analysis. Silencing of SOX5 led to a significant repression of WNT signaling with a WNT Pathway Activity Score of -0.752 (P = 1 × 10 − 7) in comparison to that of the control. A greater than 2 fold change with statistical significance was observed in the expression of 7 genes related to WNT signaling: GSK3β, WNT9A, SOX17, DAB2, FGF4, WNT11 and WNT2. Notably, silencing of SOX5 increased GSK3β expression 2.75 fold (Table 3).
Table 4.
Novel mutations in SOX5 were identified in LOAD patients
| Variants | Missense |
Splice-site | ||
|---|---|---|---|---|
| R36T | T74M | H721R | 1018-1G>A | |
| Exon | 2 | 2 | 15 | 11 |
| Code change | AGA - ACA | ACG - ATG | CAT- CGT | |
| Nucleotide Change | G107C | C221T | A2162G | Exon11-1G>A |
| CADD C-Score Predication | C-Score 24.8 | C-Score 18.64 | C-Score 11.67 | |
| INSIGHT Fitcon Scores Predication | Fitcon Score:0.447 | Fitcon Score:0.447 | Fitcon Score:0.55 | |
| PolyPhen-2 Predication | Possibly damaging (Score 0.791) | Benign (Score 0.342) | Possibly damaging (Score 0.791) | |
| Risk or Protection Allele | Risk | Protection | Risk | Risk |
| No.of carrier families | 1 | 1 | 1 | 1 |
| Number of Affected/Number of SOX5 Mutation carriers | 2/2 | 2/0 | 2/2 | 2/2 |
| Number of Unaffected/Number of SOX5 Mutation carriers | 0 | 3/3 | 2/1 | 0 |
| APOE-ε4 status | 2 affected were heterozygous | 2 unaffected were homozygous; 1 unaffected was heterozygous | All 4 were heterozygous | 1 affected was heterozygous |
Table 3.
SOX5 regulates the expression of WNT genes
| Regulated Genes | Function | Fold Change |
|---|---|---|
| GSK3β | Canonical WNT pathway | 2.75 |
| WNT9A | Canonical WNT pathway | 2.40 |
| SOX17 | WNT Signaling Negative Regulation | 2.30 |
| DAB2 | Cell Proliferation/WNT Signaling Negative Regulation | 2.27 |
| FGF4 | Cell Growth & Proliferation | 2.11 |
| WNT11 | WNT/calcium pathway | 2.09 |
| WNT2 | WNT/calcium pathway | 2.00 |
Family-based association analysis and identification of functional variants in SOX5 in LOAD cohort
To test genetic association between SOX5 and AD, we analyzed the results of a GWAS using the Affymetrix Human Genome Wide SNP 6.0 arrays in three well-characterized samples of LOAD families described earlier (NIMH, NIA, NCRAD families; total 1,653 AD families, 4,006 subjects) (16–19). The diagnosis of AD was made and the age of onset for all AD cases was determined according to NINCDS/ADRDA (20) criteria. Our analyses showed that two SNPs: rs16927107 (A/G, minor allele frequency 0.1) and rs117708160 (C/G, minor allele frequency 0.15), that are located in 5’ untranslated region (UTR) in SOX5 on chromosome 12p12.1, exhibited significant association with a multivariate phenotype combining affection status and onset age (P = 5 × 10 − 6 and 3 × 10 − 5, respectively).
We then utilized recently generated whole genome sequencing (WGS) data from the NIMH AD Initiative study families (1,510 subjects; 437 AD families) to identify SOX5 functional variants that co-segregate with the disease status. Four novel variants that segregated with the disease phenotype were identified in our analysis. Three novel rare variants, including two missense mutations, (R36T, H721R) and one splice acceptor mutation 1018-1 G > A at exon 11 (rs201883079), were identified to co-segregate with AD in single families. The SOX5-R36T mutation occurred in one LOAD family with autopsy-confirmed AD and was present in both affected siblings (onset ages 77 and 78 with both heterozygous for APOE-ε4). No unaffected were available in the family and the variant has not been previously described. The SOX5-H721R mutation was present in one LOAD family two affected siblings (onset ages 70 and 75), but was also present in one of two unaffected siblings, who was 74 years old at last examination in 2008. Thus, this sib may still become affected in the future. All four siblings were heterozygous for APOE-ε4. The splice acceptor mutation occurred in one late-onset family with autopsy-confirmed AD and was present in both affected siblings (onset ages 70 and 71 with only the latter, heterozygous for APOE-ε4). No unaffected were available in the family and the variant has no listed minor allele frequency (MAF) on dbSNP. Moreover, we also found one novel missense mutation, SOX5-T74M, is possibly protective against AD as it was present in all three unaffected and neither of two affected siblings (onset ages of 67 and 70) in one late-onset AD family. Two of the unaffected were homozygous for APOE-ε4 (ages 63 and 78 when last checked) and the other was heterozygous for APOE-ε4 (age 59 at last examination).
Next we used three pathogenicity prediction algorithms to analyze the identified SOX5 missense mutations. The deleteriousness of single nucleotide variants predicted using the combined annotation dependent depletion (CADD) Score (21) showed a C-Score over 10 in all three missense mutations: 24.8 for SOX5-R36T, 18.64 for SOX5-T74M and 11.67 for SOX5- H721R, indicating the relative functionality and pathogenicity of these missense mutations. Analysis of using the INSIGHT fitness consequences of functional annotation (Fitcon) scores (22) demonstrated a Fitcon score over 0.40 in all three missense mutations, indicating these missense mutations occur in functionally conserved regions through evolution. Consistent with the clinical and genetic findings, SOX5-R36T and H721R were predicted to be “Possible damaging” mutations with a score of 0.791 while the SOX5-T74M was “Benign” change with a score of 0.342 by PolyPhen-2 (Polymorphism Phenotyping V2) analysis (23). Collectively, these data suggest that SOX5 is a novel LOAD gene with a cluster of variants in the exon 2 linked to LOAD.
Discussion
This comprehensive study of SOX5 was carried out in transgenic Drosophila models, human SH-SY5Y neuroblastoma cells and LOAD families, respectively. We have demonstrated that silencing of Sox102F in Drosophila leads to destructive neuron and abnormal behavior and that SOX5 regulates the WNT signaling pathway activity and the expression of the WNT-related genes in human neuronal cells. We have also observed that variants in SOX5 were associated with AD by family-based GWAS and identified novel mutations in AD families using whole genome sequence analyses.
Studies have shown that SOX5 modulates cell fate, controls cell proliferation, regulates cartilage formation and neuronal development (8,24,25). In mouse, Sox5 plays important roles in the specification of subcortically projecting axons in the developing cerebral cortex (26). In Sox5-null mice, the Sox5-null neocortex neurons maintain an immature differentiation state by losing and misrouting axons (27); Sox5 overexpression causes re-emergence of neurons with corticofugal features (28). Sox5 is expressed in the chick neural crest and neural progenitors and is required for the development of the glial lineage (29) and neuronal differentiation (8). Overall, previous studies indicate an important role for SOX5 in neuronal generation, differentiation and diversity.
To characterize the functional role of SOX5 in adult neuron and brain function, we silenced Sox102F, the Drosophila ortholog of SOX5, in various neuronal subtypes. Silencing of Sox102F led to misorientated and disorganized michrochaetes, shorter DA neurons with lower complexity, reduced contraction movements and loss of neuron muscular junction bouton structure, impaired olfactory perception and severe neuron degeneration in whole brain. These novel findings implicate an important functional role of SOX5 in neuronal development and brain function.
The cognitive hallmark of AD is an extraordinary inability to form new memories. Precise and rapid synaptic vesicle recycling is crucial for synaptic transmission and plays an important role in synaptic plasticity. At the NMJ, pre-synaptic motor neurons depolarize the post-synaptic muscle (30). Silencing of Sox102F resulted in loss of NMJ bouton structure. In addition, MBs are the learning and memory center of Drosophila, silencing of Sox102F led to an impaired olfactory perception. Furthermore, courtship behavior has been widely used to examine the activity and coordination, as well as learning and memory, as it involves the exchange of various sensory stimuli including visual, auditory, and chemosensory signals between males and females, which lead to a complex series of well characterized motor behaviors culminating in successful copulation (31). Silencing of Sox102F significantly affected adult courtship behavior and exhibited a significantly delay in the mating steps (licking and curling), a statistically significant shortened length of copulation and a reduced CI. Moreover, silencing of Sox102F in whole brain led to severe neurodegeneration. Histological analysis in adult brain showed the degenerated brain structure, which was consistent with behavioral changes.
The WNT family of secreted growth factors regulates the developmental processes of cell fate and polarity and cell maintenance processes. WNT signaling consists of canonical WNT signaling (WNT/β-catenin pathway) that is dependent on the β-catenin pathway, whereas noncanonical WNT signaling involves β-catenin independent pathways, which comprise different types of WNT ligands and receptors (32). Aβ binds to the extracellular cysteine-rich domain of the Frizzled receptor (Fz) inhibiting WNT/β-catenin signaling (33). Activation of WNT signaling by Huperzine A enhances the nonamyloidogenic pathway in an AD transgenic mouse model, suggesting a sustained loss of WNT signaling function may be involved in the Aβ-dependent neurodegeneration observed in AD brain (33). Sox5 regulates the timing of cell cycle exit by triggering the expression of the negative regulator of the pathway axin2, which opposes WNT-beta-catenin activity on cell cycle progression in chick (8). In addition, we previously found that silencing of Sox102F increased the WNT-related Wg expression in the wing disc of Drosophila (7). These previous studies supported a functional role for SOX5 in WNT signaling. We have now demonstrated that silencing of SOX5 resulted in a statistically significant repression of WNT signaling pathway activity. SOX5 regulated the expression of 7 of the 84 genes related to WNT signaling pathway including GSK3β. GSK3β is a constitutively active, proline-directed serine/threonine kinase that directly controls the level of β-catenin phosphorylation, which leads to its consequent degradation by the proteasome pathway (33). Studies have shown that GSK3β plays a pivotal and central role in the pathogenesis of both sporadic and familial forms of AD. GSK3β is involved in the formation of paired helical filament (PHF)-tau, which is an integral component of the neurofibrillary tangle (NFT) deposits that disrupt neuronal function (34,35). It is suggested that over-activity of GSK3β accounts for memory impairment, tau hyper-phosphorylation, increased β-amyloid production and local plaque-associated microglial-mediated inflammatory responses; all of which are hallmark characteristics of AD (34). Silencing of SOX5 increased GSK3β expression, which indicates a functional link between SOX5 and GSK3β in AD pathogenesis. Silencing of SOX5 also led to an increased expression of two known negative regulators of WNT signaling: Disabled-2 (DAB2) and SOX17. The adaptor molecule DAB2 stabilizes the β-catenin degradation complex (36). SOX17, a homolog of SOX5, changes β-catenin, SFRP1 and WNT/Frizzled expression (37). FGF4 is a direct transcriptional target for LEF1 and WNT signaling (38). It has been shown that FGF infusion or gene transfer restores neurogenesis in the subventricular zone and hippocampal functions in aged mice and mouse models of AD and has therapeutic implications for neurocognitive disorders (39). WNT11 and WNT2 are components of noncanonical WNT signaling, while WNT9A is the components of canonical WNT signaling [26] (40).
Moreover, bone morphogenic proteins (BMPs) as members of the transforming growth factor-β (TGF-β) superfamily are important regulators of neurogenesis and neuronal cell fate determination during development. The hippocampus from either AD patients or APP transgenic mice exhibited significantly increased BMP6 expression accompanied by defects in hippocampal neurogenesis compared to controls (41). The WNT and the BMP signaling pathways are functionally integrated in many biological processes including stem cell maintenance and cell fate specifications. WNT and BMP ligands are expressed in overlapping or complementary manners. For example, the enhancer of the even-skipped gene (eve) has a BMP response element (Smad1/5/8 and Smad4 binding sites) next to a WNT response element which integrates synergy between WNT and BMP signaling (42). In mice Sox5 is essential for activation of BMP directed target gene expression in embryos and explants, that it physically interacts with BMP Smad1/5/8, and this interaction is essential for recruitment of Smad1/4 to BMP regulatory elements (43). Functional studies are warranted to further investigate the regulating effects of SOX5 on genes related to WNT-signaling as well as BMP or TGF-β signaling.
SOX5 variants have been previously associated with two neurological disorders: ALS and developmental delay or intellectual disability. Two missense mutations in SOX5, Q362P and E367Q, five amino acids apart, were identified in two of the 190 ALS patients and not detected in 190 normal controls (10). Several mutations including a reciprocal translocation breakpoint within SOX5 and variable deletions in SOX5 that range in size from 72 to 466 kb have been identified in patients with developmental delay or intellectual disability, which are associated with prominent language delay, behavior problems and mild dysmorphic features (11). In line with the above findings in ALS, we demonstrate here the SOX5 association with LOAD, another neurodegenerative disease attributed to SOX5. Furthermore, four novel functional variants in SOX5 that modify susceptibility to LOAD predicted to have functionality and pathogenicity by computer algorithms annotations. Three of the four novel mutations in the SOX5 gene, two missense mutations SOX5-R36T and H721R and one splice-site variant, segregated with the affection status, suggesting that these three SOX5 variants have an important role in the onset of AD, probably by loss of SOX5 function. In contrast to the deleterious SOX5-R36T and H721R mutations, the AD-linked SOX5-T74M mutation may be acting in a more complex gain-of-function fashion to confer protection from AD risk. Notably, missense mutations in SOX5 identified in LOAD families are clustered in the N and C-termini of SOX5 (R36T, T74M, and H721R); in contrast, the distribution of two missense mutations in ALS patients is in the middle part of SOX5 (Q362P and E367Q) (10). Further studies are warranted to determine the relationship between the specific type and location of SOX5 missense mutations and neuronal development, function and AD neuropathogenesis.
Structurally SOX5 consists of an HMG domain (542-612 amino-acid) and a pattern of the TonB-dependent receptor protein signature 1 (PS00430, TONB_DEPENDENT –REC_1_) at the N-terminal of the SOX5 protein (1–82 amino-acid) (http://prosite.expasy.org/). Two of the four identified SOX5 missense mutations (R36T and T74M) are localized in the exon 2 encoding the N-terminal TonB-dependent receptor like region. These data suggest that the TonB-dependent receptor like region in SOX5 may be important in the functional role of SOX5, disruption of which may lead to AD pathogenesis. It is known that the short form of SOX5 differs from the long form in length with a 13 amino acid truncation in N-terminal (1). Both SOX5-R36T and T74M are located in the N-terminal, which may cause alternative splicing of the SOX5 exon 2 to alter the expression of SOX5 long and/or short isolforms. Notably, SOX5-R36T is associated with an increased AD risk, while SOX5-T74M confers protection from AD risk in three subjects of an AD family who had known increased risk for AD from either homozygous or heterozygous for APOE-ε4. However, the protective mechanism of SOX5-T74M on LOAD, distinct from SOX5-R36T and H721R mutations which increase AD risk, is unclear. One possibility would be if the T74M variant stabilizes SOX5 similar to the PMP22 mutation, T118M, that causes Charcot Marie Tooth, type 1A (CMT1A), by an overall increase in the stability of the mutant PMP22 protein (44).
In summary, the findings from our study indicate that SOX5 is a novel LOAD candidate gene with an important role in neuronal function. Furthermore, SOX5 holds significant potential for the presence of additional variants that could confer risk for AD, ALS and intellectual disability.
Materials and Methods
Drosophila studies
Transgenic flies and fly culture . Fly culture and crosses were carried out according to standard procedures. The following fly strains were used: UAS-Sox102F-RNAi from the TRiP Project at Harvard Medical School (http://www.flyrnai.org/TRiP-HOME.html), Pnr-GAL4, Ppk-GAL4, D42-GAL4, Tab2-GAL4, and Insc-GAL4-ChRFP from the Bloomington Drosophila Stock Center at Indiana University (http://flystocks.bio.indiana.edu/).
Analysis of the DA neurons . The DA neurons were dissected from the third instar larvae, and analyzed by fluorescence imaging and quantified using NIH Image J version 1.47 software (http://rsbweb.nih.gov/ij/). The analysis parameters included 1) Length of dendrites (µm)-Measure from cell body to tip of branch for each ending in arbor; 2) Surface area (µm2); 3) Complexity of arbor (total number of branches) and Branching complexity (%).
Larva crawling assay, the drosophila larval NMJ dissection and imaging and adult climbing assay. The third instar larvae from each experimental group were gently placed in a 150x25mm tissue culture dish containing 2% agarose gel. Each larva was observed to determine the number of full peristaltic contractions that occurred in 1 min (Peristaltic Contractions/min; Full anterior to posterior movement = 1 contraction) as described previously (31). The Drosophila larval NMJs were dissected from the third instar larva by removing all internal organs while leaving the body wall intact and imaged using an Olympus confocal scope as described previously (45).
Behavior assays
Larva learning fructose assay. The third instar larva was transferred back and forth 3 times each between 1% agarose plates that either did or did not contain appetitive fructose. The memory of larva was examined by the selective preference for fructose in an agorase plate. In brief, 5 larvae were placed on a 1% agarose plate that had half of the plate contained fructose and the other half did not. The number of larva that was attracted to the fructose or control side was recorded.
Adult courtship assay. Male and female flies were collected within 6 h of hatching so as to ensure all flies remained virgins until time of experiment. Flies were cultured for 5 days prior to mating assays. Courtship behavior was examined as described previously (31). In brief, one male and one female were placed into a mating ring made of a 35×10 mm Petri dish containing a layer of 2% agarose gel. Flies were observed under a dissection microscope for the following behaviors: orientation, tapping, wing song, licking, curling, copulation attempt for 30 min or until successful copulation occurred. The time at which each of these actions occurred for each set of flies was recorded as well as the time of the first copulation attempt, the beginning of the final successful copulation, and the total time engaged in copulation. The courtship latency was defined as the total amount of time elapsed before any type of courtship action was observed in the male. The copulation latency was defined as the total amount of time elapsed before successful copulation began. Courtship index (CI) was calculated by dividing the time spent in courtship divided by the total time until copulation. If copulation occurred, the female was isolated in a fresh tube to determine if the copulation indeed resulted in progeny.
Whole brain structural analysis by fluorescence and transmission electron microscopy
Whole brains from the 30-day-old control or the Sox102F RNAi flies were dissected for whole brain structure and ultrastructure analyses. Adult brain imaging was done as described previously (46). In brief, the Insc-GAL4-ChRFP and UAS-Sox102F-RNAi; Insc-GAL4-ChRFP brains were fixed with 4% formaldehyde and imaged with an Olympus 1X70 confocal microscope. For TEM, 15 brains from the Insc-GAL4-ChRFP and UAS-Sox102F-RNAi; Insc-GAL4-ChRFP flies were fixed, embedded, sectioned, examined and imaged as described previously (7).
Human SH-SY5Y neuroblastma cell studies
Human SH-SY5Y neuroblastoma cells were cultured as described previously (47). SOX5 was silenced by RNAi using pools of three to five SOX5-specific 19-25 nt siRNAs (Santa Cruz Biotech, CA) designed to silence SOX5 expression. Real-time RT-PCR was performed to examine the expression of SOX5 in SH-SY5Y cells. Total RNA was isolated from the cultured cells using RNeasy Mini Kit (Qiagen, CA). The cDNA was synthesized using SUPERSCRIPT Preamplification System for First-Strand cDNA Synthesis (Life Technologies, CA). Real-time RT-PCR quantification with human SOX5 and GAPDH sense and antisense primers was done on an iCycler (BIO-RAD, CA) using SYBR Green PCR Core Reagents (Qiagen, MD) according to the manufacturer’s instructions. The sequences of primers are: SOX5 5’ AGGGACT CCCGAGAGCTTAG, 3’ CTGTTGCTGGAGCAAATTGA; RT-PCR product size 238 bp; hGAPDH 5’ CCACCC AGAAGACTGTGGAT, 3’ TTCTAGA CGGCA GGTCAGGT; RT-PCR product size 203 bp. The house-keeping gene hGAPDH was used as an internal control and was co-amplified under the same PCR conditions. All standards and unknown samples were run in triplicates per reaction and repeated 3 times. The fluorescence intensity was calculated using iCycler software version 3.1. The expression of SOX5 was given as relative number of copies (%) of mRNA molecules, as calibrated by co-amplification of hGAPDH. The expressions of 84 genes related to WNT-mediated signal transduction were examined using the Human WNT Signaling Pathway RT2 Profiler PCR Plus Array and analyzed using RT2 Profiler PCR Plus Array Data Analysis software according to the manufacturer’s instructions (Qiagen, CA). The experiments were repeated 3 times and over-or down-regulated genes with the average expression levels over 2-fold with a statistically significant P value <0.05 were shown.
Statistical analysis
The Student t-test and Fisher’s Exact test were performed to statistically compare the difference between the experimental groups and p < 0.05 was defined as statistically significant. Mean ± Standard Deviation (SD) of each parameter was reported for each experimental group.
Human studies
Subjects. AD Families: Three well-characterized samples from several large collections of AD families (NIMH, NIA, NCRAD; total 1,653 AD families, 4,006 subjects) (16–19) were utilized in the GWAS analysis as described previously (16). Of the above study families, the NIMH AD Initiative study cohort of 1510 subjects belonging to 437 families, were used in the whole-genome analysis for rare functional variants in SOX5.
GWAS. Genotypes were generated using the Affymetrix Human Genome Wide SNP 6.0 arrays and following stringent quality control. 906,600 single nucleotide polymorphisms (SNPs) were utilized for family-based association analysis using FBAT v2.0.1 (48). The p-values are reported based on the Liptak test statistic (49). The Liptak method may attain higher power levels, than the traditional FBAT approach, by combining the Z-statistics that correspond to the p-values of the family-based test (the within family information) with the rank-based p-values for population-based analysis (the between family information). To optimize statistical power, age of onset and AD affection status were tested together, as a combined phenotype, by use of the multivariate extension of the FBAT method, FBAT-GEE (Generalized Estimating Equation) (50).
WGS. We performed WGS in the NIMH AD families using Illumina's HiSeq 2500 ultra-high-throughput sequencing system and pair-end sequencing method with a minimum 48X coverage. The variants were re-called with FreeBayes software (https://github.com/ekg/freebayes) and fully annotated GEMINI database (http://gemini.readthedocs.org/en/latest/) was created for downstream analyses. The variants were analyzed for the presence of rare variants that co-segregate with the disease status in the harboring families (manuscript in preparation). The possible impact of amino acid substitutions in SOX5 on the structure and function of SOX5 was analyzed using a combination of functional prediction tools, including Combined Annotation Dependent Depletion (CADD) C-score (21) and INSIGHT Fitcon scores (22), and PolyPhen-2 (Polymorphism Phenotyping v2) software (23) (http://genetics.bwh.harvard.edu/pph2/index.shtml).
Acknowledgments
Conflict of Interest statement. None declared.
Funding
This work was supported by the Cure Alzheimer's Fund [to R.E.T], the National Institute of Health [R01AG014713 and R01MH60009 to R.E.T; R03AR063271 and R15EB019704 to A.L.], the National Science Foundation [NSF1455613 to A.L.] and a Massachusetts General Hospital ECOR Award [to A.L.].
References
- 1. Ikeda T., Zhang J., Chano T., Mabuchi A., Fukuda A., Kawaguchi H., Nakamura K., Ikegawa S. (2002) Identification and characterization of the human long form of Sox5 (L-SOX5) gene. Gene, 298, 59–68. [DOI] [PubMed] [Google Scholar]
- 2. Eijgelsheim M., Newton-Cheh C., Sotoodehnia N., de Bakker P.I., Muller M., Morrison A.C., Smith A.V., Isaacs A., Sanna S., Dorr M., et al. (2010) Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum. Mol. Genet., 19, 3885–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pfeufer A., van Noord C., Marciante K.D., Arking D.E., Larson M.G., Smith A.V., Tarasov K.V., Muller M., Sotoodehnia N., Sinner M.F., et al. (2010) Genome-wide association study of PR interval. Nat. Genet., 42, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olesen M.S., Holst A.G., Jabbari J., Nielsen J.B., Christophersen I.E., Sajadieh A., Haunso S., Svendsen J.H. (2012) Genetic loci on chromosomes 4q25, 7p31, and 12p12 are associated with onset of lone atrial fibrillation before the age of 40 years. Can. J. Cardiol., 28, 191–195. [DOI] [PubMed] [Google Scholar]
- 5. Della-Morte D., Beecham A., Rundek T., Wang L., McClendon M.S., Slifer S., Blanton S.H., Di Tullio M.R., Sacco R.L. (2011) A follow-up study for left ventricular mass on chromosome 12p11 identifies potential candidate genes. BMC Med. Genet., 12, 100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hersh C.P., Silverman E.K., Gascon J., Bhattacharya S., Klanderman B.J., Litonjua A.A., Lefebvre V., Sparrow D., Reilly J.J., Anderson W.H., et al. (2011) SOX5 is a candidate gene for chronic obstructive pulmonary disease susceptibility and is necessary for lung development. Am. J. Respir. Crit. Care. Med., 183, 1482–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li A., Ahsen O.O., Liu J.J., Du C., McKee M.L., Yang Y., Wasco W., Newton-Cheh C.H., O'Donnell C.J., Fujimoto J.G., et al. (2013) Silencing of the Drosophila ortholog of SOX5 in heart leads to cardiac dysfunction as detected by optical coherence tomography. Hum. Mol. Genet., 22, 3798–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martinez-Morales P.L., Quiroga A.C., Barbas J.A., Morales A.V. (2010) SOX5 controls cell cycle progression in neural progenitors by interfering with the WNT-beta-catenin pathway. EMBO Rep., 11, 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lefebvre V., Li P., de Crombrugghe B. (1998) A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J., 17, 5718–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daoud H., Valdmanis P.N., Gros-Louis F., Belzil V., Spiegelman D., Henrion E., Diallo O., Desjarlais A., Gauthier J., Camu W., et al. (2011) Resequencing of 29 candidate genes in patients with familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol., 68, 587–593. [DOI] [PubMed] [Google Scholar]
- 11. Lamb A.N., Rosenfeld J.A., Neill N.J., Talkowski M.E., Blumenthal I., Girirajan S., Keelean-Fuller D., Fan Z., Pouncey J., Stevens C., et al. (2012) Haploinsufficiency of SOX5 at 12p12.1 is associated with developmental delays with prominent language delay, behavior problems, and mild dysmorphic features. Hum. Mutat., 33, 728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramain P., Heitzler P., Haenlin M., Simpson P. (1993) pannier, a negative regulator of achaete and scute in Drosophila, encodes a zinc finger protein with homology to the vertebrate transcription factor GATA-1. Development, 119, 1277–1291. [DOI] [PubMed] [Google Scholar]
- 13. Kuo C.T., Jan L.Y., Jan Y.N. (2005) Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc. Natl. Acad. Sci. U S A, 102, 15230–15235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Apostolopoulou A.A., Widmann A., Rohwedder A., Pfitzenmaier J.E., Thum A.S. (2013) Appetitive associative olfactory learning in Drosophila larvae. J. Vis. Exp., 18, pii: 4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chu Y., Yang E., Schinaman J.M., Chahda J.S., Sousa-Neves R. (2013) Genetic analysis of mate discrimination in Drosophila simulans. Evolution, 67, 2335–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bertram L., Lange C., Mullin K., Parkinson M., Hsiao M., Hogan M.F., Schjeide B.M., Hooli B., Divito J., Ionita I., et al. (2008) Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am. J. Hum. Genet., 83, 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herold C., Hooli B.V., Mullin K., Liu T., Roehr J.T., Mattheisen M., Parrado A.R., Bertram L., Lange C., Tanzi R.E. (2016) Family-based association analyses of imputed genotypes reveal genome-wide significant association of Alzheimer's disease with OSBPL6, PTPRG, and PDCL3. Mol. Psychiatry, 21,1608–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wijsman E.M., Pankratz N.D., Choi Y., Rothstein J.H., Faber K.M., Cheng R., Lee J.H., Bird T.D., Bennett D.A., Diaz-Arrastia R., et al. (2011) Genome-wide association of familial late-onset Alzheimer's disease replicates BIN1 and CLU and nominates CUGBP2 in interaction with APOE. PLoS Genet., 7, e1001308.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blacker D., Bertram L., Saunders A.J., Moscarillo T.J., Albert M.S., Wiener H., Perry R.T., Collins J.S., Harrell L.E., Go R.C., et al. (2003) Results of a high-resolution genome screen of 437 Alzheimer's disease families. Hum. Mol. Genet., 12, 23–32. [DOI] [PubMed] [Google Scholar]
- 20. McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. (1984) Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology, 34, 939–944. [DOI] [PubMed] [Google Scholar]
- 21. Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet., 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arbiza L., Gronau I., Aksoy B.A., Hubisz M.J., Gulko B., Keinan A., Siepel A. (2013) Genome-wide inference of natural selection on human transcription factor binding sites. Nat. Genet., 45, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smits P., Li P., Mandel J., Zhang Z., Deng J.M., Behringer R.R., de Crombrugghe B., Lefebvre V. (2001) The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell, 1, 277–290. [DOI] [PubMed] [Google Scholar]
- 25. Shim S., Kwan K.Y., Li M., Lefebvre V., Sestan N. (2012) Cis-regulatory control of corticospinal system development and evolution. Nature, 486, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leone D.P., Srinivasan K., Chen B., Alcamo E., McConnell S.K. (2008) The determination of projection neuron identity in the developing cerebral cortex. Curr. Opin. Neurobiol., 18, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kwan K.Y., Lam M.M., Krsnik Z., Kawasawa Y.I., Lefebvre V., Sestan N. (2008) SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc. Natl. Acad. Sci. U S A, 105, 16021–16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lai T., Jabaudon D., Molyneaux B.J., Azim E., Arlotta P., Menezes J.R., Macklis J.D. (2008) SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron, 57, 232–247. [DOI] [PubMed] [Google Scholar]
- 29. Perez-Alcala S., Nieto M.A., Barbas J.A. (2004) LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development, 131, 4455–4465. [DOI] [PubMed] [Google Scholar]
- 30. Menon K.P., Carrillo R.A., Zinn K. (2013) Development and plasticity of the Drosophila larval neuromuscular junction. Wiley Interdiscip. Rev. Dev. Biol., 2, 647–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nichols C.D., Becnel J., Pandey U.B. (2012) Methods to assay Drosophila behavior. J. Vis. Exp., 7, pii: 3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wan W., Xia S., Kalionis B., Liu L., Li Y. (2014) The role of Wnt signaling in the development of Alzheimer's disease: a potential therapeutic target?. Biomed. Res. Int., 2014, 301575.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Inestrosa N.C., Toledo E.M. (2008) The role of Wnt signaling in neuronal dysfunction in Alzheimer's Disease. Mol. Neurodegener., 3, 9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Engel T., Hernandez F., Avila J., Lucas J.J. (2006) Full reversal of Alzheimer's disease-like phenotype in a mouse model with conditional overexpression of glycogen synthase kinase-3. J. Neurosci., 26, 5083–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crespo-Biel N., Theunis C., Borghgraef P., Lechat B., Devijver H., Maurin H., Van Leuven F. (2014) Phosphorylation of protein Tau by GSK3beta prolongs survival of bigenic Tau.P301LxGSK3beta mice by delaying brainstem tauopathy. Neurobiol. Dis., 67, 119–132. [DOI] [PubMed] [Google Scholar]
- 36. Prunier C., Hocevar B.A., Howe P.H. (2004) Wnt signaling: physiology and pathology. Growth Factors., 22, 141–150. [DOI] [PubMed] [Google Scholar]
- 37. Chen H.L., Chew L.J., Packer R.J., Gallo V. (2013) Modulation of the Wnt/beta-catenin pathway in human oligodendroglioma cells by Sox17 regulates proliferation and differentiation. Cancer Lett., 335, 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kratochwil K., Galceran J., Tontsch S., Roth W., Grosschedl R. (2002) FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1(-/-) mice. Genes Dev., 16, 3173–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li J.S., Yao Z.X. (2013) Modulation of FGF receptor signaling as an intervention and potential therapy for myelin breakdown in Alzheimer's disease. Med. Hypotheses, 80, 341–344. [DOI] [PubMed] [Google Scholar]
- 40. Katoh M. (2007) Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem Cell Rev., 3, 30–38. [DOI] [PubMed] [Google Scholar]
- 41. Crews L., Adame A., Patrick C., Delaney A., Pham E., Rockenstein E., Hansen L., Masliah E. (2010) Increased BMP6 levels in the brains of Alzheimer's disease patients and APP transgenic mice are accompanied by impaired neurogenesis. J. Neurosci., 30, 12252–12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Itasaki N., Hoppler S. (2009) Crosstalk between Wnt and bone morphogenic protein signaling: A turbulent relationship. Dev. Dyn., 239, 16–23. [DOI] [PubMed] [Google Scholar]
- 43. Nordin K., LaBonne C. (2014) Sox5 Is a DNA-binding cofactor for BMP R-Smads that directs target specificity during patterning of the early ectoderm. Dev. Cell, 31, 374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kumar C.V., Swetha R.G., Anbarasu A., Ramaiah S. (2014) Computational Analysis Reveals the Association of Threonine 118 Methionine Mutation in PMP22 Resulting in CMT-1A. Adv. Bioinformatics, 2014, 502618.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brent J.R., Werner K.M., McCabe B.D. (2009) Drosophila larval NMJ dissection. J. Vis. Exp., 24, 1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alayari N.N., Vogler G., Taghli-Lamallem O., Ocorr K., Bodmer R., Cammarato A. (2009) Fluorescent labeling of Drosophila heart structures. J. Vis. Exp., 72, 4334.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xie Z., Romano D.M., Tanzi R.E. (2005) RNA interference-mediated silencing of X11alpha and X11beta attenuates amyloid beta-protein levels via differential effects on beta-amyloid precursor protein processing. J. Biol. Chem., 280, 15413–15421. [DOI] [PubMed] [Google Scholar]
- 48. Horvath S., Xu X., Laird N.M. (2001) The family based association test method: strategies for studying general genotype–phenotype associations. Eur. J. Hum. Genet., 9, 301–306. [DOI] [PubMed] [Google Scholar]
- 49. Laoutidis Z.G., Luckhaus C. (2014) The liptak-stouffer test for meta-analyses. Biol. Psychiatry, 77, e1–e2. [DOI] [PubMed] [Google Scholar]
- 50. Lange C., Silverman E.K., Xu X., Weiss S.T., Laird N.M. (2003) A multivariate family-based association test using generalized estimating equations: FBAT-GEE. Biostatistics, 4, 195–206. [DOI] [PubMed] [Google Scholar]