Abstract
Background
Trichloroethylene (TCE) is a known carcinogen in humans and rodents. Previous studies of inter-strain variability in TCE metabolism were conducted in multi-strain panels of classical inbred mice with limited genetic diversity to identify gene-environment interactions associated with chemical exposure.
Objectives
To evaluate inter-strain variability in TCE metabolism and identify genetic determinants that are associated with TCE metabolism and effects using Collaborative Cross (CC), a large panel of genetically diverse strains of mice.
Methods
We administered a single oral dose of 0, 24, 80, 240, or 800 mg/kg of TCE to mice from 50 CC strains, and collected organs 24 h post-dosing. Levels of trichloroacetic acid (TCA), a major oxidative metabolite of TCE were measured in multiple tissues. Protein expression and activity levels of TCE-metabolizing enzymes were evaluated in the liver. Liver transcript levels of known genes perturbed by TCE exposure were also quantified. Genetic association mapping was performed on the acquired phenotypes.
Results
TCA levels varied in a dose- and strain-dependent manner in liver, kidney, and serum. The variability in TCA levels among strains did not correlate with expression or activity of a number of enzymes known to be involved in TCE oxidation. Peroxisome proliferator-activated receptor alpha (PPARα)—responsive genes were found to be associated with strain-specific differences in TCE metabolism.
Conclusions
This study shows that CC mouse population is a valuable tool to quantitatively evaluate inter-individual variability in chemical metabolism and to identify genes and pathways that may underpin population differences.
Keywords: population, trichloroethylene, mouse
Addressing population-level variability in susceptibility to toxic effects of environmental exposures is a persistent unmet need in human health assessments (Zeise et al., 2013). Traditional toxicity testing is conducted in a single isogenic strain of laboratory rodents (Festing, 1995), which complicates the extrapolation to the heterogeneous human population. Recent studies demonstrated the utility of the mouse population-based approach for quantitative evaluation of the variability in metabolism and toxicity (Bradford et al., 2011; Chiu et al., 2014; French et al., 2015), characterization of the susceptibility mechanisms (Harrill et al., 2009a, 2012; Koturbash et al., 2011; Yoo et al., 2015a,b), and mapping of the loci that may confer susceptibility to toxicants (Church et al., 2015; French et al., 2015; Harrill et al., 2009b).
The use of population-based mouse models in experimental biomedical research have flourished with the recent advancements in mouse genetics and the availability of the Diversity Outbred (DO) and Collaborative Cross (CC) populations (Aylor et al., 2011; Bogue et al., 2015). These mouse populations were designed to randomize genetic variation so that all components of systems can be interrogated as allele frequencies are ideal for quantitative trait locus (QTL) mapping; they are also sufficiently large to power analyses of modest interactions (Threadgill and Churchill, 2012). The genome of each CC and DO line represents a mosaic of eight parental inbred strains distributed randomly across the genome; however, because CC lines have been inbred they carry an advantage of infinitely reproducible model to support data integration and reproduction across laboratories, chemicals and study designs (Threadgill and Churchill, 2012). The CC mouse model has been highly efficient in identifying candidate genetic markers responsible for several pathophysiological conditions, genetic analysis of complex traits, identification of novel gene functions, and modeling emergent diseases (Durrant et al., 2011; Ferris et al., 2013; Kovacs et al., 2011; Rasmussen et al., 2014; Rogala et al., 2014).
Experimental toxicology is poised to make use of the CC population for investigations of the genetic and molecular determinants of inter-individual variability in toxicokinetics and toxicodynamics. For example, quantitative assessment of inter-individual variability in metabolism of several environmental contaminants of major public health relevance, such as chlorinated solvent trichloroethylene (TCE), is among the critical needs for human health assessments (Cichocki et al., 2016). TCE is a ubiquitous environmental chemical and is a carcinogen in humans (Rusyn et al., 2014). It is primarily metabolized (Figure 1) by the cytochrome P450 (CYP) system and glutathione conjugation generating qualitatively similar metabolites in both humans and rodents (Lash et al., 2014). Among the metabolites of TCE, trichloroacetic acid (TCA) is the prevalent oxidative metabolite and a known peroxisome proliferator-activated receptor alpha (PPARα) agonist that is thought to be one of the key drivers of TCE toxicity (Bull et al., 1990; Chiu et al., 2013; Corton, 2008). Previous studies have demonstrated inter-individual differences in oxidative TCE metabolism and toxicity in humans (Muller et al., 1974, 1975) and multi-strain studies in mice (Bradford et al., 2011; Yoo et al., 2015a,b). Interestingly, PPARα-mediated pathways, including the metabolism genes induced by TCE, are among the most distinct effects that are dependent on genetic background (Bradford et al., 2011).
FIG. 1.
Schematic representation of metabolism of trichloroethylene (TCE) via oxidative pathway. Names of metabolites that are chemically unstable or reactive are shown in brackets. Major enzymatic pathways are identified for cytochrome P450 enzymes (CYPs), aldehyde dehydrogenase (ALDH), alcohol dehydrogenase (ADH), UDP-glucuronosyltransferase (UGT), and GSH S-transferase zeta (GSTZ).
The objective of this study was to evaluate the extent of inter-strain variability in the oxidative metabolism of TCE, and to identify loci that may be driving these differences. We hypothesized that genetic diversity affects inter-strain differences in TCE metabolism and toxicity. We conducted a dose–response (0, 24, 80, 240, or 800 mg/kg of TCE) study in 50 CC strains. Inter-strain variability in TCA levels, as well as protein or activity levels of several TCE-metabolizing enzymes, was examined in the CC mouse population at 24 h after dosing. We identified several QTLs associated with variability in TCE metabolism that provide genetic evidence for the role of PPAR pathway in the oxidative metabolism of TCE.
MATERIALS AND METHODS
Animals and treatments
Adult male mice (8–12 weeks old) from 50 CC strains were acquired from the University of North Carolina Systems Genetics Core (Chapel Hill, NC). Animals were fed an NTP 2000 wafer diet (Zeigler Brothers, Inc., Gardners, PA) and water ad libitum. The housing room was maintained on a 12-h light–dark cycle. Animals were allowed to acclimate to the room for at least 10 days prior to beginning experimentation. The experimental design sought to maximize the number of strains relative to within-strain replications based on the power analysis for QTL mapping in mouse populations (Kaeppler, 1997); therefore, one mouse was used per strain/dose group. Male animals were used because maximal rates of TCE oxidative metabolism in rodents differ between males and females, higher concentrations of CYP-derived metabolites of TCE were found in livers of males than in females (Lash et al., 2006). Mice were orally administered a single dose of 0, 24, 80, 240, or 800 mg/kg TCE (Sigma Aldrich, St. Louis, MO) in 5% Alkamuls EL-620 vehicle (Solvay, Deptford, NJ). The highest dose was selected based on the studies of sub-chronic toxicity and carcinogenicity of TCE in mice (National Toxicology Program, 1990). Mice were sacrificed 24 h after treatment, tissues were flash frozen in liquid nitrogen and stored at −80 °C until analysis. In the highest dose group, mice from 5 CC strains died before sacrifice; therefore, all further analyses were conducted on 45 CC strains with complete dose–response data. All treatments were conducted between 8 and 11 am to limit potential diurnal variation in TCE metabolism. In life portion of the study was conducted over a 6-week time-frame to limit seasonal variations. These studies were approved by the Institutional Animal Care and Use Committees at Texas A&M University and the University of North Carolina.
TCA levels in tissues
Analyses were performed by a modification of US EPA method 552.2 (Domino et al., 2003). In brief, 50 mg of kidney, 100 mg of liver, 50 mg of brain, or 100 μL of serum was homogenized in 1 mL chloroform:methanol (1:1) containing 20 µL of 2-bromobutyric acid (550 nmol/mL). Distilled deionized water (300 µL) was added to the homogenate followed by vortexing and centrifugation at 15 000 rpm for 10 min. The supernatant was transferred to a new vial containing 1.5 mL of 10% sulfuric acid in methanol and incubated in a water bath at 50 °C for 2 h. Methyl esters were extracted in 2 mL methyl tert-butyl ether followed by addition of 3 mL sodium sulfate (100 g/L). The organic layer was neutralized by addition of 3 mL saturated sodium bicarbonate solution and concentrated under a stream of nitrogen gas to a volume <50 µL. A calibration curve from 0 to 1000 nmol/g consisting of TCA spiked into tissues before extraction was run with each batch for quantitative analysis. The extracts were analyzed using a HP 6890 gas chromatography system coupled with a HP 5973 mass selective detector (Agilent Technologies, Santa Clara, CA). A 2 µL sample was introduced into gas chromatograph by splitless injection on to a 30 m × 0.25 mm, 0.25 µm HP5-ms column (Agilent Technologies). The carrier gas flow was set at a constant flow rate of 1 mL/min. The oven was held isothermally at 40 °C for 10 min and then ramped to 65 °C over the next 10 min, then to 85 °C for the next 2 min, and finally to 205 °C over 6 min. The injector and transfer line temperatures were maintained at 210 °C and 280 °C, respectively. The mass selective detector operated in the electron impact mode with the ion source set at 230 °C and electron energy at 70 eV. Methyl esters of TCA and 2-bromobutyric acid were quantified by monitoring the ion at 59 m/z. Ions at m/z 117, 119, 151, and 153 were used to identify the analytes.
Extraction of hepatic microsomes and cytosolic fractions
A published protocol was adapted for the extraction of microsomal and cytosolic fractions (Yan and Caldwell, 2004). In brief, 100 mg of liver tissue was homogenized in 1 mL ice-cold 50 mM tris base solution containing 15 mM potassium chloride and centrifuged at 9000 g for 20 min. The supernatant was transferred to a new vial and ultra-centrifuged at 100,000 g for 1 h at 4°C. The supernatant was aliquoted and the microsomal pellet was resuspended in 200 µL 50 mM tris-HCl (with 20% glycerol, pH 7.4) and stored at −80 °C. Total protein concentration was measured using a Pierce BCA Protein Assay Kit (ThermoFisher Scientific, Waltham, MA).
Alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) activity assays
ADH and ALDH activity assay kits were purchased from Sigma-Aldrich. Activity assays were performed on cytosolic extracts according to the manufacturer’s protocol.
CYP1A1 activity assay
Magnesium chloride, glucose-6-phosphate, glucose-6-phosphate dehydrogenase, resorufin, resorufin ethyl ether, and β-nicotinamide adenine dinucleotide phosphate hydrate reduced tetra sodium salt hydrate (NADPH) were purchased from Sigma-Aldrich. Microsomal incubations were setup according to a published protocol (Yan and Caldwell, 2004). In brief, 100 µL of chilled tris base buffer, 50 µL of 5 mM MgCl2, 50 µL of 5 mM glucose-6-phosphate, 50 µL of 0.5 mM NADPH, 250 µg of microsomal protein, and 10 µL of 3.3 mM resorufin ethyl ether were incubated in a 96-well plate. Formation of resorufin was monitored at excitation and emission wavelengths of 530 nm and 585 nm, respectively. A standard curve from 0 to 1 µM for resorufin was generated using heat-inactivated microsomes.
CYP2E1 protein expression
About 20–30 mg pulverized liver tissue was homogenized in 400 μL tissue protein extraction reagent (T-PER, ThermoFisher Scientific) containing HALT protease inhibitor cocktail (ThermoFisher Scientific). The homogenate was centrifuged at 10,000 g for 5 min and the proteins in the supernatant were transferred to a new vial. The isolated proteins were quantified using Pierce BCA Protein Assay Kit (ThermoFisher Scientific). Protein aliquots (20 µg) were loaded on to an SDS-PAGE containing of Mini-Protean TGX precast gels (Bio-Rad, Richmond, CA). Western blotting was performed using Trans-blot Turbo Transfer System (Bio-Rad), and Immuno-Blot PVDF membranes (Bio-Rad). Membranes were blocked using Odyssey blocking buffer (Li-Cor, Lincoln, NE) and were incubated overnight at 4 °C with 1:5000 anti-CYP2E1 antibody (Abcam, Cambridge, MA). The membranes were then washed and incubated with 1:2500 goat anti-rabbit antibody conjugated with peroxidase (Millipore, Bedford, MA). Protein bands were developed by chemiluminescent staining with WesternSure ECL Substrate visualized by C-Digit Blot-Scanner (Li-Cor). Anti-β-actin antibody (1:2500, Abcam) staining was used as a loading control.
Gene expression analysis by RT-PCR
Total RNA was isolated from 20 to 30 mg of pulverized left liver lobes in liquid nitrogen using MiRNeasy mini kit (Qiagen, Valencia, CA). RNA concentrations were measured using Nanodrop ND-1000 (Thermo Fisher Scientific). cDNA was synthesized from 2 µg RNA using High-Capacity cDNA Achieve kit (Thermo Fisher Scientific). Quantitative real-time PCR was performed using Taqman probes for genes Acox1, Acot8, Cyp4a10, Cd40, Fitm2, and Top1 (Abcam) on Light Cycler 96 (Roche, Indianapolis, IN).
Genome-wide quantitative trait locus (QTL) mapping
High-density sequences of CC genomes for strains used in this study were downloaded from the UNC Systems Genetic Core (http://csbio.unc.edu/CCstatus/index.py?run=Pseudo; last accessed March 31, 2017). Information on reference SNPs for genes were acquired from the Mouse Genome Informatics database (http://www.informatics.jax.org/; last accessed March 31, 2017). Flanking DNA sequences for each SNP in each CC strain was performed using a custom R script. QTL mapping was performed using DOQTL package (Gatti et al., 2014) using a model that considered each of the eight founder alleles separately. Only seven founder alleles were present in the mapping population at the location Acox1 QTL, resulting in overfitting of doqtl's eight-allele model. In order to better estimate the QTL effects, we simulated CC mapping populations that consisted of our actual CC lines plus an additional three simulated lines that harbored the PWK allele missing form our mapping population. We assigned each simulated line a phenotype randomly sampled the observed phenotype distribution (such that the simulated lines would not affect the QTL) and remapped in the QTL region. These simulations yielded a properly fitted model and improved estimates of the effects of the other seven alleles.
Statistical analysis
GraphPad Prism (La Jolla, CA) was used to perform statistical tests. R (v.3.1.2) was used to generate line graphs and boxplots (ggplot2). For all tests, a P < .05 was required for statistical significance. In analyses that involved multiple comparisons, a false discover-corrected q values were derived.
Data availability
All raw data are publically-available through the Mouse Phenome Database (http://phenome.jax.org/; ID: Rusyn8).
RESULTS
Mouse Population Variability in Metabolism of TCE to TCA
Metabolism is critical for the toxicity, mutagenicity, and carcinogenicity of TCE; TCA is the most abundant metabolite that is formed through cytochrome P450 (CYP)-dependent oxidation [Figure 1, reviewed in (Cichocki et al., 2016; Lash et al., 2014)]. Because of the focus on the dose–response effects of TCE and toxicodynamic responses among multi-tissues, a single time point of 24 h was selected. At this time point, only TCA is detectable in mouse tissues at comparable highest doses (Kim et al., 2009). We observed extensive strain-dependent variability in TCA levels in mouse tissues (liver, kidney, brain, and serum) 24 h after oral dosing with TCE (Figure 2, Supplementary Figures 1–3). At each dose (24–800 mg/kg), the capacity to metabolize TCE via the oxidative pathway varied by an order of magnitude or more among strains. Appreciable consistency exists among strains with high, intermediate, or low TCA levels across all dose groups (Supplementary Figure 1). For example, strains CC028/GeniUnc, CC059/TauUNC, and CC010/GeniUnc showed relatively high levels of TCA in liver across different dose groups (Figure 2). Similarly, strains CC042/GeniUnc, CC017/Unc, and CC055/TauUnc are low responders, and strains CC006/TauUnc, CC030/GeniUnc, and CC003/Unc show intermediate levels of TCA at 24 hrs after dosing. We observed similar trends in kidney, brain and serum (Supplementary Figures 1–3).
FIG. 2.
Liver TCA levels in CC mice 24 h after oral administration of a dose of 24 (A), 80 (B), 240 (C), or 800 (D) mg/kg of TCE. Bars colored in red, green, and blue represent strains that exhibit high, intermediate and low, respectively, levels of TCA across different treatment groups.
Next, we compared the population-level (using data from 45 CC strains) variability in TCA amounts at 24 h in liver, kidney, brain, and serum with that predicted by a population-wide physiologically based pharmacokinetic (PBPK) model (Chiu et al., 2014) that was calibrated with data from 15 classical inbred strains (Bradford et al., 2011). Figure 3 shows data from each strain (circles) compared with the PBPK model predictions (boxes). Liver TCA levels at doses of 24, 80, and 240 mg/kg for more than 85% of CC strains fall within the 95% confidence interval of the PBPK model predictions (Figure 3A). Interestingly, liver TCA levels in the 800 mg/kg dose group are higher than the predicted 95% confidence interval for more than half of CC strains (Figure 3A). Kidney TCA levels at 24 mg/kg and 80 mg/kg doses were below the limit of detection for most of the CC strains; at the 240 mg/kg dose, TCA levels fall within the 95% confidence interval for more than 85% of CC strains (Figure 3B). In 800 mg/kg dose group, about one third of the strains fall outside of the 95% confidence interval for model-predicted kidney TCA levels (Figure 3B). Similarly, while serum TCA levels at 24 and 80 mg/kg were generally within the 95% confidence limit of the model, close to half of the strains are greater than this confidence interval in 240 and 800 mg/kg dose groups (Figure 3C). The most striking feature is the difference in predicted and observed medians, nearly every CC animal is above the median value predicted by the model. Overall, these results from a much larger population-based study of TCE toxicokinetics show that the PBPK model is under-predicting the levels of TCA in different tissues, especially at the highest doses.
FIG. 3.

Inter-strain variability in formation of TCA in liver (A), kidney (B), and serum (C) 24 h after oral administration of a dose of 24, 80, 240, or 800 mg/kg of TCE. Each dot represents TCA level in a CC strain, horizontal black line indicates median value for each dose-tissue combination among CC strains. Red box and whiskers are PBPK model-derived (Chiu et al., 2014) population estimates. The boxes and the horizontal line represent 5, 95, and 50% median TCA levels. The whiskers represent upper and lower bound confidence interval for 95 and 5%, respectively, median TCA levels.
Induction of Enzyme Levels/Activity in Response to TCE Treatment
Oxidative metabolism of TCE to TCA is thought to involve a number of cytochrome P450 and other enzymes (Lash et al., 2014); therefore, we sought to examine effects of TCE exposure on several key enzymes. Specifically, because of the observed wide range of liver TCA levels among strains, we sought to examine whether basal or TCE-induced levels of major microsomal and cytosolic enzymes involved in TCE metabolism might explain the inter-strain variability in liver TCA levels. Liver Cyp2e1 protein levels (Figs. 4A–B) and liver Cyp1a1 activity (Figs. 4C–D) varied extensively among strains in both vehicle- and TCE- (800 mg/kg) treated animals. No difference of the group means was observed in response to TCE for either of these enzymes, albeit the variance in these parameters increased in TCE-treated group. This observation is concordant with a previous report that treatment with TCE was without effect on protein levels of liver Cyp2e1 across seven inbred strains (Yoo et al., 2015a).
FIG. 4.
Inter-strain differences in liver Cyp2e1 protein expression (A–B) and Cyp1a1 activity (C–D) in CC mice administered vehicle (black circles) or a single oral dose of TCE (800 mg/kg, red circles). Panels (B) and (D) are box and whiskers plots of population variability where a horizontal line represents the median, the box shows 1st and 3rd quartile ranges and the whiskers represent standard errors of the mean.
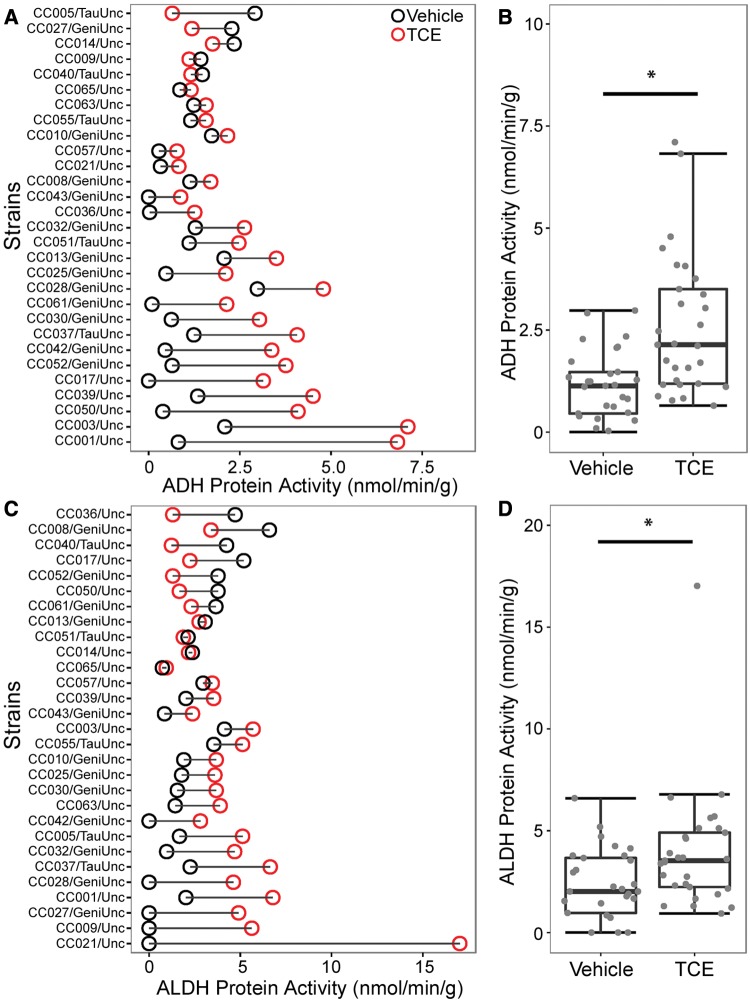
Alcohol dehydrogenases (ADH) and aldehyde dehydrogenases (ALDH) also play a key role in determining the metabolic flux of chloral hydrate, an intermediate metabolite that yields TCA (Figure 1). We examined activity of Adh and Aldh in livers of both vehicle- and TCE (800 mg/kg)-treated animals. A statistically significant increase (P < .05) in activity of both Adh and Aldh was observed with TCE exposure (Figure 5).
FIG. 5.
Inter-strain differences in liver ADH (A–B) and ALDH (C–D) activity in CC mice administered vehicle (black circles) or a single oral dose of TCE (800 mg/kg, red circles). Panels (B) and (D) are box and whiskers plots of population variability where a horizontal line represents the median, the box shows 1st and 3rd quartile ranges and the whiskers represent standard errors of the mean. The asterisks (*) denote statistical significant differences (P < .05) between TCE and vehicle treated groups (Mann–Whitney test).
Because we observed large differences in liver levels of TCA among strains, we also examined whether TCE-induced peroxisomal proliferation varied. While TCE and TCA are PPARα ligands (Maloney and Waxman, 1999; Zhou and Waxman, 1998), it is well known that expression and protein levels of PPARα are not affected by TCE (Fang et al., 2013; Ramdhan et al., 2008) and other peroxisome proliferators (Ito et al., 2007) but its transcription factor activity is affected. Therefore, we examined transcript levels of two PPARα-responsive genes, cytochrome P450 subfamily 4 polypeptide 10 (Cyp4a10) and acyl-CoA oxidase 1 (Acox1) in livers of vehicle and TCE (800 mg/kg) treated animals. Both Acox1 and Cyp4a10 transcript levels were induced (P < .05) by TCE (Figure 6). Similar to our observation in a smaller population of inbred strains (Bradford et al., 2011) strain-specific effects were prominent. The expression of both transcripts was either not affected or increased in TCE-treated animals in all strains (with the exception of Cyp4a10 levels in strain CC037/TauUnc). The magnitude of the effects on PPARα-mediated gene expression was much more robust than effects on Cyp2e1, Cyp1a1, Adh, or Aldh (Figs. 4 and 5).
FIG. 6.
Inter-strain differences in liver Acox1 (A–B) and Cyp4a10 (C–D) transcript levels in CC mice administered vehicle (black circles) or a single oral dose of TCE (800 mg/kg, red circles). Panels (B) and (D) are box and whiskers plots of population variability where a horizontal line represents the median, the box shows 1st and 3rd quartile ranges and the whiskers represent standard errors of the mean. The asterisks (*) denote statistical significant differences (P < .05) between TCE and vehicle treated groups (Mann–Whitney test).
Correlation of Enzyme Levels/Activity With TCA Liver Concentrations
To further examine the relationships between TCA levels in different tissues and other phenotypes evaluated in this population-based mouse study, we performed correlation (Spearman rank) analysis using data on CC mice from the 800 mg/kg dose group (Supplementary Table 1). We observed that TCA levels correlate positively among all tissues examined, with the highest correlation among liver, brain and kidney (Figure 7A). In addition, we found that liver TCA levels positively correlated with induction of Acox1 transcript levels in the liver, and the transcriptional effects of TCE on Acox1 and Cyp4a10, both markers of activation of transcription factor PPARα, were also significantly correlated (Figure 7B). These data suggest that inter-strain differences in TCE metabolism may be associated with TCE-mediated responses in CC mice. Notably, liver levels of TCA did not correlate with either basal or TCE-affected protein levels or activity of Cyp2e1, Cyp1a1, Adh, or Aldh, an observation that is similar to the findings of Yoo et al. (2015a).
FIG. 7.
Correlation analysis (Spearman) among toxicokinetic and toxicodynamic phenotypes across the population of CC strains. A, Scatter plots of TCA levels at 24 h after dosing with 800 mg/kg of TCE in multiple tissues of 45 CC strains. Trendlines and corresponding r and P-values are shown. B, Correlation between expression of Acox1, Cyp4a10, and liver TCA levels in responses to TCE treatment. Full correlation matrix is included as Supplementary Table 1.
QTL Mapping
To identify genomic regions associated with inter-strain differences in TCE metabolism and transcriptional changes, we mapped QTL using a model that considered each of the eight founder alleles separately using liver TCA and Acox1 transcript levels in the highest treatment group. We identified quantitative trait loci (QTL) on chromosome 2 for each of the phenotypes (Figs. 8A and C). Further, we observed that NOD/ShiLTJ and WSB/EiJ founder alleles have most pronounced effects on QTLs associated with TCA formation and Acox1 induction (Figs. 8B and D).
FIG. 8.
QTL mapping of TCE toxicokinetic and toxicodynamic phenotypes. Log transformed liver TCA (A–B) and Acox1 (C–D) transcript levels measured at 24-h time point in CC mice administered with a single oral dose of TCE (800 mg/kg) were mapped to the mouse genome polymorphisms among CC strains. Panels A and C show logarithmic odds ratio (LOD) scores across the whole mouse genome (chromosomes 1 through X). The red line represents a permutation-based significance threshold (n = 1000 permutations). Plots B and D show the effect of the CC founder strain alleles on chromosome 2 (top) and a zoom-in on the significant regions with corresponding LOD scores.
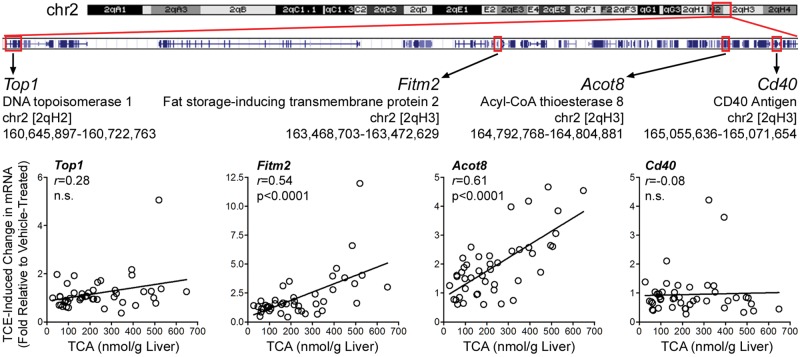
There are many protein-coding genes in the locus associated with the variability in liver TCA levels across the population (Table 1). From this list, candidate genes were selected based on the available information for their function that could be linked to the effects of TCE. We chose acyl-coenzyme A thioesterase 8 (Acot8) and Fat Storage Inducing Transmembrane protein 2 (Fitm2) as candidate genes for further analysis because of their role in PPAR signaling, as well as several neighboring genes not involved in PPAR signaling or metabolism, Cluster of Differentiation 40 (Cd40) and Topoisomerase 1 (Top1). We performed RT-PCR to further examine whether transcript levels of the candidate and neighboring genes are associated with the phenotype, level of TCA in liver, used for mapping. Induction of Acot8 and Fitm2 expression was significantly positively correlated with liver TCA; Cd40 and Top1 transcript levels showed no correlation with this phenotype (Figure 9).
TABLE 1.
Significant Quantitative Trait Loci and Genes in These Loci That Were Associated With Inter-Strain Differences in Liver TCA and TCE-Induced Induction in Acox1 Expression in Liver
| Phenotype | Genomic Position (Mb) | Protein Coding Genes in the Locus |
|---|---|---|
| TCA | Chr 2: 157.685– 180.142 Mb | CTNNBL1, VSTM2L, TTI1, RPRD1B, TGM2, BPI, LBP, SNHG11, RALGAPB, ADIG, ARHGAP40, SLC32A1, ACTR5, PPP1R16B, FAM83D, DHX35, MAFB, TOP1,PLCG1, ZHX3, LPIN3, EMILIN3, CHD6, PTPRT, CISD3B, SRSF6, L3MBTL1, SGK2, IFT52, MYBL2, GTSF1L, TOX2, JPH2, OSER1, GDAP1L1, FITM2,R3HDML, HNF4A, TTPAL, SERINC3, PKIG, ADA, WISP2, KCNK15, RIMS4, YWHAB, PABPC1L, TOMM34, STK4, KCNS1, WFDC5, WFDC12, WFDC15A, WFDC15B, SVS2, SVS3B, SVS4, SVS3A, SVS6, SVS5, SLPI, MATN4, RBPJL, SDC4, SYS1, TRP53TG5, DBNDD2, PIGT, WFDC2, SPINT3, WFDC6A, EPPIN, WFDC8, WFDC6B, WFDC16, WFDC9, WFDC10, WFDC11, WFDC13, SPINT4, SPINT5, WFDC3, DNTTIP1, UBE2C, TNNC2, SNX21,ACOT8, ZSWIM3, ZSWIM1, SPATA25, NEURL2, CTSA, PLTP, PCIF1, ZFP335, MMP9, SLC12A5, NCOA5, CD40,CDH22, SLC35C2, ELMO2, ZFP663, ZFP334, OCSTAMP, SLC13A3, TRP53RKA, SLC2A10, EYA2, ZMYND8, NCOA3, SULF2, PREX1, TRP53RKB, ARFGEF2, CSE1L, STAU1, DDX27, ZNFX1, KCNB1, PTGIS, B4GALT5, SLC9A8, SPATA2, RNF114, SNAI1, UBE2V1, TMEM189, CEBPB, PTPN1, FAM65C, PARD6B, ADNP, DPM1, MOCS3, KCNG1, NFATC2, ATP9A, SALL4, ZFP64, TSHZ2, ZFP217, BCAS1, CYP24A1, PFDN4, DOK5, CBLN4, MC3R, FAM210B, AURKA, CSTF1, CASS4, RTFDC1, GCNT7, FAM209, TFAP2C, BMP7, SPO11, RAE1, RBM38, CTCFL, PCK1, ZBP1, PMEPA1, ANKRD60, RAB22A, VAPB, STX16, NPEPL1, NELFCD, CTSZ, TUBB1, ATP5E, SLMO2, ZFP831, EDN3, ETOHI1, ZFP931, PHACTR3, SYCP2, PPP1R3D, FAM217B, CDH26, CDH4, TAF4, LSM14B, PSMA7, SS18L1, MTG2, HRH3, OSBPL2 |
| Acox1 induction by TCE in liver | Chr 2: 41.836– 43.374 Mb | LRP1B |
Genes in bold font were selected for RT-PCR analysis (Figure 9).
FIG. 9.
QTL region on distal chromosome 2 associated with liver TCA levels. RefSeq genes that are located on chr2 [qH2-qH3] in the region between 160,640,000 and 165,200,000 (a sub-set of the entire QTL, see Table 1) are shown with a position marked for the candidate genes (abbreviation, full name and physical genomic location are shown) that were examined for the relationship between TCE-induced expression and TCA level in livers of CC mice treated with a single oral dose of 800 mg/kg of TCE. Scatter plots show correlation among gene expression for Top1, Fitm2, Acot8, and Cd40 and liver TCA levels. Best fit linear regression lines are plotted and corresponding r and P values are depicted on each plot.
The QTL associated with strain-specific variability in TCE-induced liver expression of Acox1 (Table 1) contained only one protein coding gene, Low Density Lipoprotein Receptor-Related-Protein 1B (Lrp1b). Even though, Lrp1b is a tumor suppressor gene regulated via the PPARγ signaling pathways (Liu et al., 2000), it was neither expressed in livers of the mice included in this study, nor was it induced by TCE.
To further explore the role of PPARα signaling in the variability associated with TCE metabolism, we considered the potential contribution from the genetic variants of PPARα. There are two non-synonymous coding SNPs (rs16820391 and rs16820392) in PPARα in the CC founder strains. The frequency of distribution of A/G missense substitution for both SNPs was around 50% across CC strains included in this study. No statistically significant difference was observed in liver TCA levels among strains with PPARα genotypes (A/G) for either SNP (data not shown). It is well established that retinoid X receptor alpha (RXRα), a type II nuclear receptor is capable of interacting with PPARα and PPARγ by forming heterodimers thereby mediating fatty acid and lipid metabolism (Berger and Moller, 2002; Chandra et al., 2008). A non-synonymous SNP (rs16817900) generating a C/T missense substitution was identified among the CC founder strains and was also present in 4 out of 45 CC strains examined. Interestingly, we found that in strains with RXRα C genotype, liver TCA levels were about 2-fold lower (in both mean and median) than in T allele strains (Supplementary Figure 4). Although, no statistical significant difference was observed between C/T genotypes and liver TCA levels because only 4 strains harbor the C allele this finding suggests PPAR-RXR signaling as contributing to differences in oxidative metabolism of TCE among CC strains.
DISCUSSION
The focus of this study was on characterizing the extent of population-level variability in chemically mediated responses in a diverse population, as well as on exploring the mechanisms of such variability using genetic mapping. Although several previous studies used classical inbred strains to assess inter-individual variability in TCE metabolism and toxicity, they were limited in the degree of genetic diversity. In the CC mouse population, genetic variants are randomly distributed across the genome, creating a mosaic genome with potentially significant differences in cellular functions as compared with those in the founder strains. Consequently, the CC lines permit high-resolution QTL mapping, enabling the identification of the genes that may be associated with the phenotypic variability at the genome level.
Using a CC mouse population, we aimed to increase precision of the estimates for the extent of inter-individual variability in TCE metabolism. Data from this study was compared with a previous study (Bradford et al., 2011) where a multi-strain panel was used to assess variability in TCE metabolism. In the Bradford et al. (2011) study, inter-strain differences in the TCA levels in serum at 24 h post-dosing at 2,100 mg/kg TCE were around 4- to 6-fold across strains. In CC population, we observed more than an order of magnitude differences in the level of TCA at 24 h in tissues (using 800 mg/kg TCE dose data), further supporting the importance of genetic variability in TCE metabolism. We also found that select strains are low, intermediate or high “metabolizers” of TCE to TCA. To determine the concordance in population-level variability among two population-based studies, median estimates of TCA levels were derived using a PBPK model (Chiu et al., 2014) and compared with the data in this study. We found that inter-strain varaibility in TCA levels in the CC mice was concordant with PBPK model predictions at lower doses across tissues. However, at the 800 mg/kg dose, nearly half of all strains were above the predicted ranges across different tissues. Metabolism of TCE in the rat is a saturable process at doses around 1,000 mg/kg, but in the mouse TCE metabolism is linear up to a dose of 2,000 mg/kg (Prout et al., 1985). Thus, our results are unlikely to be due to metabolic saturation and suggest that genetic diversity among CC mice may play a role in inter-strain differences in TCE toxicokinetics. The fact that model-derived median predictions were about one half to an order of magnitude below the observed values across all tissues examined is an indication that our data may be useful for updating the model with a larger population and data from additional tissues. In addition, further exploration of strain-dependent variability in concentration–time responses is needed. One additional test for the hypothesis that genetic diversity plays a role in inter-strain differences in toxicokinetics would be selection of strains representing population-level variability in TCA levels for a toxicokinetic study that will evaluate TCE metabolite levels at concentration–time across various tissues.
Several enzymes are involved in the oxidative metabolism of TCE to TCA (Lash et al., 2000). Genes associated with CYP, ADH, and ALDH families are known to be polymorphic and to result in inter-individual differences in their activity among individuals (Pastino et al., 2000). Little evidence for the inter-individual differences in TCE metabolism through oxidation exist in human populations or in experimental animal studies. Most studies examined the role of polymorphisms in CYP2E1 and ADH/ALDH pathway, genes that are known not only to be polymorphic, but also contain polymorphisms that have known linkages to human cancer susceptibility (Li et al., 2016). CYP2E1 is the principal enzyme responsible for metabolism of TCE in humans and rodents (Pastino et al., 2000). Other CYP isoforms including CYP1A1 have also been associated with TCE metabolism. We observed little difference in CYP2E1 and CYP1A1 activity levels in both TCE- and vehicle-treated mice. Yoo et al. (2015a) reported suggestive, but only marginally significant (P = .06) correlation between liver TCA and basal CYP2E1 protein levels in a panel of 7 mouse inbred strains that are parental strains for CC. In the present study, we did not find a correlation between TCA and basal CYP2E1 protein levels in liver suggesting that a more complex milieu of oxidative enzymes may be responsible for TCE metabolism to TCA.
Interestingly, 10-fold or greater inter-strain differences were observed in ADH and ALDH activities in both vehicle- and TCE-treated animals. We also observed significant increases in ALDH and ADH activity with TCE exposure. Further, the extent of variability in these enzymes across mouse strains was consistent with previously reported variability in ALDH and ADH activity in human cryopreserved hepatocytes (Bronley-DeLancey et al., 2006). However, no association was observed between TCA levels and activity of ADH or ALDH, again suggesting that multiple factors may be responsible for the observed differences in TCA levels. We also observed no correlations between TCE-metabolizing enzyme levels or activity, indicating lack of potential additive interactions among these enzymes in certain strains. Thus, while mouse population-derived data demonstrate the utility of the CC mouse population in capturing the population-level variability observed in humans, they also show the complexities of the oxidative metabolism of TCE whereby multiple enzymes play a role in TCE toxicokinetics.
TCA and other products of oxidative metabolism of TCE are well known to be PPARα agonists and peroxisome proliferation is one mechanism of liver toxicity and carcinogenesis of TCE (Bull et al., 1990; Rusyn et al., 2014). In our study, we hypothesized that strains with higher TCA levels would exhibit greater effects on PPARα-mediated signaling. Indeed, we observed significant induction and positive correlation in the transcription of PPARα-responsive Acox1 and Cyp4a10 genes with TCE treatment, with levels of Acox1 transcripts and TCA levels in liver also positively correlated. This is an important finding as it demonstrates the association between TCE metabolism and toxicodynamics. It also confirms the findings reported in Bradford et al. (2011) that peroxisome proliferator activated receptor-mediated molecular networks, consisting of the metabolism genes known to be induced by TCE, represent some of the most pronounced molecular effects of TCE treatment in mouse liver that are dependent on genetic background. These results are also consistent with findings that Acox1 and Cyp4a10 transcript levels are induced with repeated exposure to TCE and are positively correlated with each other and with liver TCA levels (Yoo et al., 2015a).
While a strong and reproducible concordance between liver TCA levels and induction of peroxisome proliferation at a population level confirms the traditional sequence of events: metabolism of TCE to TCA which then acts as a PPARa ligand and induces peroxisomal proliferation, the outcome of the haplotype-associated mapping afforded by CC population suggests that a feedback loop also exists. A novel outcome of this study is the identification of candidate genes at QTLs associated with TCE metabolism to TCA. Among several protein-coding genes in the locus significantly associated with liver TCA levels among strains, Acot8 and Fitm2 were identified as candidate genes and were significantly correlated with TCA levels. These results are intriguing as Acot8 and Fitm2 are PPAR-responsive genes, suggesting the role of PPAR signaling in TCE metabolism, in addition to the role of TCE metabolites in peroxisome proliferation. A recent study showed significantly lower levels of TCA in the liver and kidney in PPARα-null and -humanized mice after either single or repeated exposures to TCE, suggesting a direct role of PPARα signaling in TCE toxicokinetics (Yoo et al., 2015c).
Another indication that genetic variants associated with PPARα signaling are associated with TCE metabolism to TCA is the observation that strains carrying a non-synonymous SNP variant (C genotype) in RXRα are associated with relatively low levels of TCA. PPARs are nuclear receptors that require dimerization with the nuclear receptor RXRα to form a heterodimer. The C genotype of RXRα is carried in the CAST/Eij strain among the CC founders. Although, no statistically significant differences in TCA levels were observed between C and T genotypes of RXRα, strains with C genotype demonstrated 2-fold lower mean and median TCA levels compared with strains carrying the T genotype. Results of previous studies also suggest that the CAST-specific alleles may be a significant contributor to the variability associated with TCE metabolism. Yoo et al. (2015a,b) found that TCA levels in serum, liver and kidneys after exposure to TCE were the lowest in CAST/Eij strain as compared with other CC parental strains. A similar observation was reported by Bradford et al. (2011) in a comparison of 15 mouse inbred strains. Despite differences in dose and study designs, consistently low TCA levels in CAST/Eij strains across studies are highly reproducible, supporting the suggestion that genetic variants in RXRα and other genes may contribute to the observed inter-strain variability in TCE metabolism.
This study, while informative with respect to quantitation and discovery of the genetic drivers of inter-individual variability, has a number of limitations. First, while we surveyed a dose–response in a large population of CC strains, we did not examine intra-strain reproducibility. In order to maximize the power of the study design with respect to mapping, we maximized the number of strains, not replicates (Kaeppler, 1997). Consistency of dose–response curves within each strain and concordance of major effects and relationships among the phenotypes with other multi-strain studies suggests that strain-specific effects are reproducible; however, this needs to be tested in subsequent studies. Studies also need to examine toxicokinetics of TCE in a concentration–time study design. Second, this study examined only a single time point and a more complete understanding of the effects that inter-strain differences in TCE metabolism may have on the apical toxicity phenotypes is needed though sub-chronic and chronic studies. Our data on strain-specific metabolism of TCE will be crucial for selection of strains for follow up longer term studies because it is unlikely that such studies can be done in a comparably large mouse population. While highly informative, population-based studies are challenging in terms of cost and logistics; thus, acute treatment-based surveys in large populations will likely be a very useful approach for narrowing the number of strains while preserving the extend of population variability through data-drive study designs and strain selection.
In summary, our results show that the CC mouse population is a valuable tool for assessing inter-individual variability and for investigating the genetic basis of the phenotypic differences. The mosaic genomes of CC enabled identification of the candidate genes underlying differences in TCE metabolism and support a role for PPAR signaling not only in TCE toxicity, but also in TCE metabolism. This observation further supports the need for caution suggested by (Yoo et al., 2015c) in interpreting apparent species differences in PPAR signaling, because they may result from not only receptor-mediated toxicodynamic effects, but also modulation of toxicokinetics. Consequently, these results suggest the need to investigate interplay between varying genetic backgrounds and interspecies differences, and thereby improve the ability of experiment studies to predict risks to human populations.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Dr Joseph A. Cichocki for sharing R-based scripts for graphical representation of the data in the study. We also appreciate technical assistance provided by Drs Stephen Sweet, Terry Wade, and Andrew Hillhouse of Texas A&M University. The authors do not have any conflict of interests.
FUNDING
U.S. Environmental Protection Agency (STAR RD83561202); National Institute of Environmental Health Sciences (P42 ES005948).
REFERENCES
- Aylor D. L., Valdar W., Foulds-Mathes W., Buus R. J., Verdugo R. A., Baric R. S., Ferris M. T., Frelinger J. A., Heise M., Frieman M. B., et al. (2011). Genetic analysis of complex traits in the emerging collaborative cross. Genome Res. 21, 1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J., Moller D. E. (2002). The mechanisms of action of PPARs. Annu. Rev. Med. 53, 409–435. [DOI] [PubMed] [Google Scholar]
- Bogue M. A., Churchill G. A., Chesler E. J. (2015). Collaborative Cross and Diversity Outbred data resources in the Mouse Phenome Database. Mamm. Genome 26, 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford B. U., Lock E. F., Kosyk O., Kim S., Uehara T., Harbourt D., DeSimone M., Threadgill D. W., Tryndyak V., Pogribny I. P., et al. (2011). Interstrain differences in the liver effects of trichloroethylene in a multistrain panel of inbred mice. Toxicol. Sci. 120, 206–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronley-DeLancey A., McMillan D. C., McMillan J. M., Jollow D. J., Mohr L. C., Hoel D. G. (2006). Application of cryopreserved human hepatocytes in trichloroethylene risk assessment: relative disposition of chloral hydrate to trichloroacetate and trichloroethanol. Environ. Health Perspect. 114, 1237–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull R. J., Sanchez I. M., Nelson M. A., Larson J. L., Lansing A. J. (1990). Liver tumor induction in B6C3F1 mice by dichloroacetate and trichloroacetate. Toxicology 63, 341–359. [DOI] [PubMed] [Google Scholar]
- Chandra V., Huang P. X., Hamuro Y., Raghuram S., Wang Y. J., Burris T. P., Rastinejad F. (2008). Structure of the intact PPAR-gamma-RXR-alpha nuclear receptor complex on DNA. Nature 456, 350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W. A., Campbell J. L. Jr, Clewell H. J. 3rd, Zhou Y. H., Wright F. A., Guyton K. Z., Rusyn I. (2014). Physiologically based pharmacokinetic (PBPK) modeling of interstrain variability in trichloroethylene metabolism in the mouse. Environ. Health Perspect. 122, 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W. A., Jinot J., Scott C. S., Makris S. L., Cooper G. S., Dzubow R. C., Bale A. S., Evans M. V., Guyton K. Z., Keshava N., et al. (2013). Human health effects of trichloroethylene: key findings and scientific issues. Environ. Health Perspect. 121, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church R. J., Gatti D. M., Urban T. J., Long N., Yang X., Shi Q., Eaddy J. S., Mosedale M., Ballard S., Churchill G. A., et al. (2015). Sensitivity to hepatotoxicity due to epigallocatechin gallate is affected by genetic background in diversity outbred mice. Food Chem. Toxicol. 76, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocki J. A., Guyton K. Z., Guha N., Chiu W. A., Rusyn I., Lash L. H. (2016). Target organ metabolism, toxicity, and mechanisms of trichloroethylene and perchloroethylene: Key similarities, differences, and data gaps. J. Pharmacol. Exp. Ther. 359, 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corton J. C. (2008). Evaluation of the role of peroxisome proliferator-activated receptor alpha (PPARalpha) in mouse liver tumor induction by trichloroethylene and metabolites. Crit. Rev. Toxicol. 38, 857–875. [DOI] [PubMed] [Google Scholar]
- Domino M. M., Pepich B. V., Munch D. J., Fair P. S., Xie Y. (2003). Method 552.3: Determination of haloacetic acids and dalapon in drinking water by liquid-liquid microextraction, derivatization, and gas chromatography with electron capture detection. US EPA. DIANE Publishing, Collingdale, PA.
- Durrant C., Tayem H., Yalcin B., Cleak J., Goodstadt L., de Villena F. P. M., Mott R., Iraqi F. A. (2011). Collaborative cross mice and their power to map host susceptibility to Aspergillus fumigatus infection. Genome Res. 21, 1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z. Z., Krausz K. W., Tanaka N., Li F., Qu A., Idle J. R., Gonzalez F. J. (2013). Metabolomics reveals trichloroacetate as a major contributor to trichloroethylene-induced metabolic alterations in mouse urine and serum. Arch. Toxicol. 87, 1975–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris M. T., Aylor D. L., Bottomly D., Whitmore A. C., Aicher L. D., Bell T. A., Bradel-Tretheway B., Bryan J. T., Buus R. J., Gralinski L. E., et al. (2013). Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathog. 9, e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festing M. F. W. (1995). Use of a multistrain assay could improve the Ntp carcinogenesis bioassay. Environ. Health Perspect. 103, 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French J. E., Gatti D. M., Morgan D. L., Kissling G. E., Shockley K. R., Knudsen G. A., Shepard K. G., Price H. C., King D., Witt K. L., et al. (2015). Diversity outbred mice identify population-based exposure thresholds and genetic factors that influence benzene-induced genotoxicity. Environ. Health Perspect. 123, 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D. M., Svenson K. L., Shabalin A., Wu L. Y., Valdar W., Simecek P., Goodwin N., Cheng R., Pomp D., Palmer A., et al. (2014). Quantitative trait locus mapping methods for diversity outbred mice. G3 (Bethesda) 4, 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill A. H., Desmet K. D., Wolf K. K., Bridges A. S., Eaddy J. S., Kurtz C. L., Hall J. E., Paine M. F., Tidwell R. R., Watkins P. B. (2012). A mouse diversity panel approach reveals the potential for clinical kidney injury due to DB289 not predicted by classical rodent models. Toxicol. Sci. 130, 416–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill A. H., Ross P. K., Gatti D. M., Threadgill D. W., Rusyn I. (2009a). Population-based discovery of toxicogenomics biomarkers for hepatotoxicity using a laboratory strain diversity panel. Toxicol. Sci. 110, 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill A. H., Watkins P. B., Su S., Ross P. K., Harbourt D. E., Stylianou I. M., Boorman G. A., Russo M. W., Sackler R. S., Harris S. C., et al. (2009b). Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res. 19, 1507–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Yamanoshita O., Kurata Y., Kamijima M., Aoyama T., Nakajima T. (2007). Induction of peroxisome proliferator-activated receptor alpha (PPARalpha)-related enzymes by di(2-ethylhexyl) phthalate (DEHP) treatment in mice and rats, but not marmosets. Arch. Toxicol. 81, 219–226. [DOI] [PubMed] [Google Scholar]
- Kaeppler S. M. (1997). Quantitative trait locus mapping using sets of near-isogenic lines: Relative power comparisons and technical considerations. Theor. Appl. Genet. 95, 384–392. [Google Scholar]
- Kim S., Kim D., Pollack G. M., Collins L. B., Rusyn I. (2009). Pharmacokinetic analysis of trichloroethylene metabolism in male B6C3F1 mice: Formation and disposition of trichloroacetic acid, dichloroacetic acid, S-(1,2-dichlorovinyl)glutathione and S-(1,2-dichlorovinyl)-L-cysteine. Toxicol. Appl. Pharmacol. 238, 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I., Scherhag A., Sorrentino J., Sexton K., Bodnar W., Swenberg J. A., Beland F. A., Pardo-Manuel Devillena F., Rusyn I., Pogribny I. P. (2011). Epigenetic mechanisms of mouse interstrain variability in genotoxicity of the environmental toxicant 1,3-butadiene. Toxicol. Sci. 122, 448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs A., Ben-Jacob N., Tayem H., Halperin E., Iraqi F. A., Gophna U. (2011). Genotype is a stronger determinant than sex of the mouse gut microbiota. Microb. Ecol. 61, 423–428. [DOI] [PubMed] [Google Scholar]
- Lash L. H., Chiu W. A., Guyton K. Z., Rusyn I. (2014). Trichloroethylene biotransformation and its role in mutagenicity, carcinogenicity and target organ toxicity. Mutat. Res. Rev. Mutat. Res. 762, 22–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash L. H., Fisher J. W., Lipscomb J. C., Parker J. C. (2000). Metabolism of trichloroethylene. Environ. Health Perspect. 108 Suppl 2, 177–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash L. H., Putt D. A., Parker J. C. (2006). Metabolism and tissue distribution of orally administered trichloroethylene in male and female rats: Identification of glutathione- and cytochrome P-450-derived metabolites in liver, kidney, blood, and urine. J. Toxicol. Environ. Health A 69, 1285–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Zhao Z., Sun M., Luo J., Xiao Y. (2016). ALDH2 gene polymorphism in different types of cancers and its clinical significance. Life Sci. 147, 59–66. [DOI] [PubMed] [Google Scholar]
- Liu C. X., Musco S., Lisitsina N. M., Yaklichkin S. Y., Lisitsyn N. A. (2000). Genomic organization of a new candidate tumor suppressor gene, LRP1B. Genomics 69, 271–274. [DOI] [PubMed] [Google Scholar]
- Maloney E. K., Waxman D. J. (1999). Trans-activation of PPARalpha and PPARgamma by structurally diverse environmental chemicals. Toxicol. Appl. Pharmacol. 161, 209–218. [DOI] [PubMed] [Google Scholar]
- Muller G., Spassovski M., Henschler D. (1974). Metabolism of trichloroethylene in man. II. Pharmacokinetics of metabolites. Arch. Toxicol. 32, 283–295. [DOI] [PubMed] [Google Scholar]
- Muller G., Spassowski M., Henschler D. (1975). Metabolism of trichloroethylene in man. III. Interaction of trichloroethylene and ethanol. Arch. Toxicol. 33, 173–189. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program (1990). Carcinogenesis studies of trichloroethylene (Without Epichlorohydrin) (CAS No. 79-01-6) in F344/N rats and B6C3F1 mice (Gavage Studies). Natl. Toxicol. Program Tech. Rep. Ser. 243, 1–174. [PubMed] [Google Scholar]
- Pastino G. M., Yap W. Y., Carroquino M. (2000). Human variability and susceptibility to trichloroethylene. Environ. Health Perspect. 108 Suppl 2, 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prout M. S., Provan W. M., Green T. (1985). Species differences in response to trichloroethylene. I. Pharmacokinetics in rats and mice. Toxicol. Appl. Pharmacol. 79, 389–400. [DOI] [PubMed] [Google Scholar]
- Ramdhan D. H., Kamijima M., Yamada N., Ito Y., Yanagiba Y., Nakamura D., Okamura A., Ichihara G., Aoyama T., Gonzalez F. J., et al. (2008). Molecular mechanism of trichloroethylene-induced hepatotoxicity mediated by CYP2E1. Toxicol. Appl. Pharmacol. 231, 300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A. L., Okumura A., Ferris M. T., Green R., Feldmann F., Kelly S. M., Scott D. P., Safronetz D., Haddock E., LaCasse R., et al. (2014). Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science 346, 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogala A. R., Morgan A. P., Christensen A. M., Gooch T. J., Bell T. A., Miller D. R., Godfrey V. L., de Villena F. P. M. (2014). The collaborative cross as a resource for modeling human disease: CC011/Unc, a new mouse model for spontaneous colitis. Mamm. Genome 25, 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusyn I., Chiu W. A., Lash L. H., Kromhout H., Hansen J., Guyton K. Z. (2014). Trichloroethylene: Mechanistic, epidemiologic and other supporting evidence of carcinogenic hazard. Pharmacol. Ther. 141, 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D. W., Churchill G. A. (2012). Ten years of the collaborative cross. Genetics 190, 291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Caldwell G. (2004). Optimization in Drug Discovery: In Vitro Methods. Humana Press, Totowa, NJ. [Google Scholar]
- Yoo H. S., Bradford B. U., Kosyk O., Shymonyak S., Uehara T., Collins L. B., Bodnar W. M., Ball L. M., Gold A., Rusyn I. (2015a). Comparative analysis of the relationship between trichloroethylene metabolism and tissue-specific toxicity among inbred mouse strains: Liver effects. J. Toxicol. Environ. Health A 78, 15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. S., Bradford B. U., Kosyk O., Uehara T., Shymonyak S., Collins L. B., Bodnar W. M., Ball L. M., Gold A., Rusyn I. (2015b). Comparative analysis of the relationship between trichloroethylene metabolism and tissue-specific toxicity among inbred mouse strains: Kidney effects. J. Toxicol. Environ. Health A 78, 32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. S., Cichocki J. A., Kim S., Venkatratnam A., Iwata Y., Kosyk O., Bodnar W., Sweet S., Knap A., Wade T., et al. (2015c). The contribution of peroxisome proliferator-activated receptor alpha to the relationship between toxicokinetics and toxicodynamics of trichloroethylene. Toxicol. Sci. 147, 339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeise L., Bois F. Y., Chiu W. A., Hattis D., Rusyn I., Guyton K. Z. (2013). Addressing human variability in next-generation human health risk assessments of environmental chemicals. Environ. Health Perspect. 121, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. C., Waxman D. J. (1998). Activation of peroxisome proliferator-activated receptors by chlorinated hydrocarbons and endogenous steroids. Environ Health Perspect 106 Suppl 4, 983–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data are publically-available through the Mouse Phenome Database (http://phenome.jax.org/; ID: Rusyn8).