Abstract
Phosphorylation affects ubiquitination, stability, and activity of transcriptional factors, thus regulating various cellular functions. E2F transcriptional factor 1 (E2F1) regulates paternally expressed imprinted gene 10 (Peg10) expression, thereby promoting cell proliferation. However, the effect of E2F1 stability on Peg10 expression and the molecular regulation of E2F1 stability by its phosphorylation have not been well demonstrated. Here, we describe a new pathway in which phosphorylation of E2F1 by GSK3β increases E2F1 association with the deubiquitinating enzyme, ubiquitin-specific protease 11 (USP11), which removes K63-linked ubiquitin chains thereby preventing E2F1 degradation in the nuclei. Downregulation of USP11 increases E2F1 ubiquitination and reduces E2F1 stability and protein levels, thereby decreasing Peg10 mRNA levels. Physiologically, USP11 depletion suppresses cell proliferation and wound healing in lung epithelial cells, and these effects are reversed by E2F1 and PEG10 overexpression. Thus, our study reveals a new molecular model that phosphorylation promotes substrate stability through increasing its association with a deubiquitinating enzyme. The data suggest that GSK3β and USP11 act in concert to modulate E2F1 abundance and PEG10 expression in lung epithelial cells to affect cell wound healing. This study provides new therapeutic targets to lessen lung injury by improving lung epithelial cell repair and remodeling after injury.
Keywords: deubiquitination, E2F1, phosphorylation, USP11, cell proliferation, wound healing
Introduction
Lung injury leads to the acute respiratory distress syndrome (ARDS), a disease which has an extremely high mortality rate at 30%–40% (Kahn et al., 2006), and imparts tremendous burden onto the healthcare system with ~75000 annual deaths in the United States (Rubenfeld et al., 2005). The lung epithelial lining functions as a physical barrier between inspired air and the interstitium of the lung, which is the first line of defense against particulates, noxious chemicals, and microbial pathogens. Damages in lung epithelial cells initiate inflammatory responses, increase lung microvascular permeability and the infiltration of edema fluid, with impaired oxygen exchange (Z’graggen et al., 2010; Brune et al., 2015). In the resolution of ARDS, impaired lung epithelial cell proliferation, wound healing, and epithelium repair lead affected individuals more susceptible to recurrent infection or chronic lung disease (Schneeberger and Karnovsky, 1976; Koizumi et al., 1989; Sacco et al., 2004). Lung re-epithelialization is dependent on epithelial cell proliferation and wound healing, which is vital to proper epithelial repair after injury (Freeman et al., 1993; Sugahara et al., 2006; Corvol et al., 2009). An in-depth understanding of the molecular regulation of lung epithelial cell wound healing may therefore inform new therapeutic strategies to reduce global health burden of lung injury.
The paternally expressed gene 10 (Peg10) is located in human chromosome 7q21 (Ono et al., 2001) that is genomically imprinted without expression of the maternal allele. Peg10 has been considered an ‘oncogene’ because it promotes tumor cell proliferation, migration, and invasion (Tsou et al., 2003; Ono et al., 2006; Rousseaux et al., 2013; Peng et al., 2017). Okabe et al. demonstrate that PEG10 plays an anti-apoptotic effect through association with SIAH1, an ubiquitin E3 ligase (Okabe et al., 2003). Aberrant expression of Peg10 has been described in various tumors including hepatocellular carcinoma, esophageal cancer, and lung cancer. The role of PEG10 in lung injury and repair has not been reported. In the current study, we reveal that PEG10 plays a critical role in regulation of lung epithelial cell proliferation and wound healing.
Transcriptional induction of Peg10 expression by the E2F transcription factor 1 (E2F1) has been described (Wang et al., 2008; Peng et al., 2017). Overexpression of E2F1 significantly increases Peg10 promoter activity, while knockdown of E2F1 diminishes Peg10 gene expression (Akamatsu et al., 2015). E2F1 belongs to the E2F family of DNA-binding proteins, which play a determining role in cell cycle progression and cellular proliferation via induction of various downstream target genes including the pituitary tumor transforming gene, insulin growth factor 1 receptor, and Peg10 (Stevaux and Dyson, 2002; Trimarchi and Lees, 2002; Wang et al., 2008). Peng et al. (2017) demonstrate that E2F1-induced PEG10 expression promotes pancreatic cell proliferation and migration. Recent studies have been focused on investigating the molecular regulation of E2F1 cellular abundance, especially its protein stability.
Ubiquitination is a protein post-translational modification that regulates protein stability, intracellular trafficking, and enzyme activity. The polyubiquitination of substrate proteins serves as a molecular tag that typically triggers substrate degradation. Lysine 11 (K11)- and K48-linked ubiquitin chains lead proteasomal degradation, while K63-linked chains tend to regulate substrate activity and trafficking as well as degradation (Yau and Rape, 2016). Ubiquitin E3 ligases, such as APC/C, SCF(SKP2), ROC1, have been reported to target E2F1 for its degradation in the proteasome (Marti et al., 1999; Ohta and Xiong, 2001; Budhavarapu et al., 2012). The removal of ubiquitin chains from ubiquitinated proteins is catalyzed by deubiquitinating enzymes (DUBs) which can divert substrate proteins or rescue them from degradation (Wolberger, 2014). Recent studies reveal that two proteasome-associated DUBs, UCH37 and POH1, can deubiquitinate ubiquitinated E2F1 (Ub-E2F1) (Mahanic et al., 2015; Wang et al., 2015). UCH37 removes K63-linked polyubiquitin chains from Ub-E2F1, increasing E2F1 transcriptional activation, but it has no effect on E2F1 protein abundance (Mahanic et al., 2015). POH1 removes K63- and K11-linked ubiquitin chains and increases E2F1 abundance in liver tumor cells (Wang et al., 2015).
The ubiquitin-specific protease 11 (USP11) is a member of the USP family of DUB enzymes. Catalytic study shows that USP11 preferentially depolymerizes K63- and K6-linked polyubiquitin chains, while it has moderate or lower activity for cleavage of K11- and K48-linked polyubiquitin chains (Harper et al., 2014). The study from Faesen et al. demonstrates that USP11 can cleave all types of di-Ubi, except linear di-Ubi (Faesen et al., 2011). USP11 is known to stabilize several substrates including cIAP2 (Lee et al., 2015), Mgl-1 (Lim et al., 2016), p53 (Yamaguchi et al., 2007), ALK5 (Al-Salihi et al., 2012), LPA1 (Zhao et al., 2016), TGFβ receptor II (Jacko et al., 2016). USP11 localizes to the cell membrane, the cytosol, and the nucleus (Zhao et al., 2016). The effect of USP11 on the protein stability of transcription factors has not been described. In this study, we identify that USP11 stabilizes the transcription factor E2F1 in the nuclei, which in turn upregulates Peg10 expression.
Substrate recognition by ubiquitin modifying enzymes is an area of ongoing study. Our group and others have demonstrated a clear role for the pleiotropic GSK3β kinase in directing substrate recognition by ubiquitin E3 ligases and thus facilitating protein ubiquitination (Zou et al., 2011; Zhao et al., 2012; Weathington et al., 2014). While the biochemical diversity of DUB binding specificity for ubiquitin and ubiquitin chains has been exquisitely characterized in recent years (Komander, 2010), the drivers of DUB-substrate interactions that may exist separate from the ubiquitin chain are incompletely characterized. Some DUBs are known to be enzymatically optimized by allosteric modulators and must be appropriately localized to encounter target ubiquitinated proteins (Sahtoe and Sixma, 2015), but substrate-specific modifications as determinants of DUB binding have not been well described for DUB enzymes. Phosphorylation of E2F1 by GSK3β enhances E2F1 transcriptional activity (Garcia-Alvarez et al., 2007), however, the effect of this phosphorylation on its protein stability has not been well described. Here, we show that GSK3β-mediated E2F1 phosphorylation enhances the association between E2F1 and USP11, thus decreasing E2F1 ubiquitination and enhancing E2F1 stability.
In this study, we demonstrate that suppression of USP11 DUB expression or activity diminishes lung epithelial cell proliferation and wound healing through destabilization of E2F1. Our study therefore describes a mechanism which implicates GSK3β and USP11 in the regulation of Peg10 gene expression via stabilization of transcription factor E2F1. This GSK3β−USP11−E2F1−PEG10 pathway facilitates lung epithelial cell proliferation and wound healing.
Results
Downregulation or inhibition of USP11 suppresses Peg10 expression
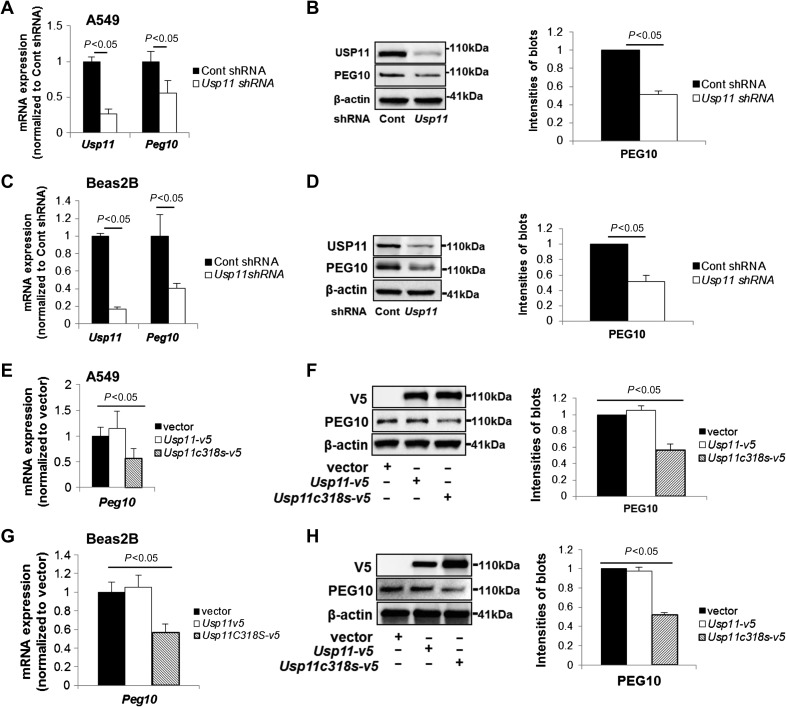
When we evaluated the effects of USP11 knockdown on transcript abundance in a microarray experiment, we identified that Peg10 gene expression was reduced in USP11 downregulated cells (Supplementary Figure S1). We pursued further study of PEG10 because it is known to stimulate cell proliferation and migration (Xiong et al., 2012; Deng et al., 2014; Li et al., 2016), however, upstream molecular regulation of Peg10 transcription by the ubiquitin system has not been well studied. To confirm this relationship, A549 cells and Beas2B cells were each transfected with Usp11 shRNA for 72 h. Downregulation of USP11 significantly reduced both Usp11 and Peg10 mRNA levels in both cell types (Figure 1A–D). Immunoblots for USP11 and PEG10 proteins demonstrated correlation to the low mRNA levels in Usp11 shRNA-transfected cells.
Figure 1.
Knockdown or inhibition of USP11 reduces PEG10 mRNA and protein levels. (A–D) A549 cells (A, B) or Beas2B cells (C, D) were transfected with control shRNA or Usp11 shRNA for 72 h. (A and C) RNA was extracted and then analyzed by qRT-PCR with Usp11 and Peg10 primers. The relative expressions of Usp11 and Peg10 were normalized to Cont shRNA. (B and D) Cell lysates were analyzed by immunoblotting with USP11, PEG10, and β-actin antibodies. PEG10 protein levels were quantified by ImageJ software. (E–H) A549 cells (E, F) or Beas2B cells (G, H) were transfected with empty vector plasmid, V5-tagged USP11, or V5-tagged USP11 mutant C318S (Usp11c318s-v5) plasmid for 48 h. Peg10 mRNA expression (E, G) and PEG10 protein levels (F, H) were determined. Shown are representative blots from at least three independent experiments.
We next tested if USP11 DUB activity is required for this effect on Peg10 expression, and a catalytic dominant negative mutant of USP11 (USP11C318S) (Schoenfeld et al., 2004) was overexpressed in A549 cells and Beas2B cells. Inhibition of USP11 by USP11C318S overexpression likewise diminished PEG10 protein and mRNA levels in both A549 and Beas2B cells compared to empty vector or USP11 wild type-overexpressed cells (Figure 1E–H). These data suggest that the deubiquitinase activity of USP11 is essential for Peg10 mRNA expression.
Downregulation of USP11 diminishes Peg10 expression by reducing E2F1 levels
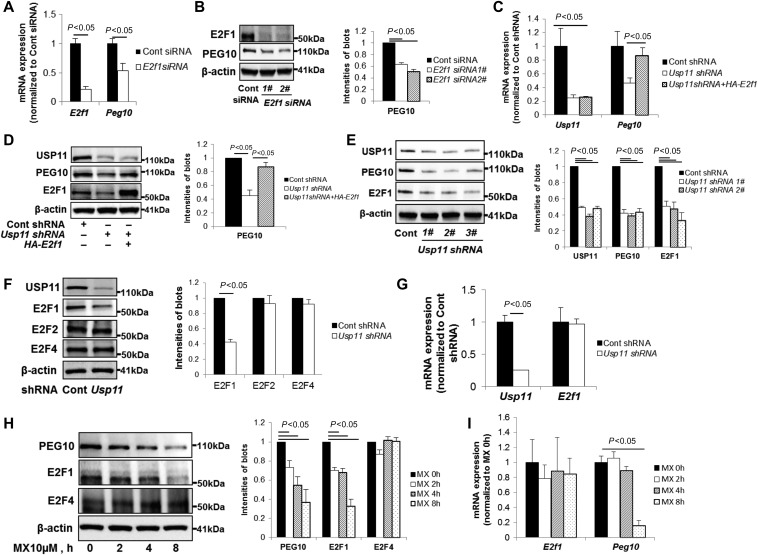
Transcriptional factor, E2F1 has been shown to bind to Peg10 promoter region and upregulate Peg10 expression (Wang et al., 2008; Peng et al., 2017). To evaluate this relationship in lung epithelial cells, we transfected E2f1 siRNA in A549 cells. E2f1 siRNA reduced both E2f1 and Peg10 mRNA levels and protein expression (Figure 2A and B), suggesting that E2F1 regulates Peg10 gene expression in lung epithelial cells. To examine whether the effect of USP11 on Peg10 gene expression is dependent on E2F1, we overexpressed E2F1 in USP11 depleted cells. Suppression of USP11 caused a reduction in Peg10 mRNA and protein levels, with rescue of this effect by overexpression of E2F1 (Figure 2C and D), indicating that E2F1 is an intermediate in USP11 regulation of Peg10 expression. We also observed that E2F1 protein levels were decreased in these USP11 depleted cells (Figure 2D and E), suggesting that USP11 modulates E2F1 levels. To determine whether USP11 specifically reduces E2F1 levels, we examined levels of other two E2F1 family members E2F2 and E2F4. Usp11 shRNA exerted suppression of E2F1 protein levels without altering E2F2 and E2F4 abundance (Figure 2F). We next tested if USP11 modulation of E2F1 is transcriptional, and observed that knockdown of USP11 did not alter E2f1 mRNA by qRT-PCR (Figure 2G). To evaluate if USP11 DUB activity mediates E2F1 expression, we used mitoxantrone (MX), a small molecule recently reported to inhibit USP11 (Burkhart et al., 2013). MX treatment reduced the protein mass of endogenous PEG10 and E2F1, not E2F4 (Figure 2H). Reduction of Peg10 mRNA levels, but not E2f1 mRNA levels, were observed in MX treated cells (Figure 2I). These results demonstrate that USP11 regulates Peg10 expression via modulation of E2F1 protein stability, not changes of mRNA.
Figure 2.
Knockdown or inhibition of USP11 diminishes Peg10 through reduction of E2F1 protein levels. (A) A549 cells were transfected with control siRNA or E2f1 siRNA for 72 h. RNA was extracted and analyzed by qRT-PCR with E2f1 and Peg10 primers. The relative expression of E2f1 and Peg10 were normalized to Cont siRNA. (B) A549 cells were transfected with control siRNA or two different E2f1 siRNAs for 72 h. Cell lysates were analyzed by immunoblotting with E2F1, PEG10, and β-actin antibodies. PEG10 protein levels were quantified by ImageJ software. (C and D) A549 cells were transfected with control shRNA or Usp11 shRNA for 24 h, and then transfected with or without HA-tagged E2F1 for additional 48 h. Usp11 and Peg10 mRNA expressions (C) and PEG10 protein levels (D) were determined. (E) A549 cells were transfected with control shRNA or three different Usp11 shRNAs for 72 h. USP11, PEG10, and E2F1 protein levels were determined. (F and G) A549 cells were transfected with control shRNA or Usp11 shRNA for 72 h. E2F1, E2F2, and E2F4 protein levels (F) and Usp11 and E2f1 mRNA expressions (G) were determined. (H and I) A549 cells were treated with 10 μM mitoxantrone (MX) for 0, 2, 4, 8 h. PEG10, E2F1, and E2F4 protein levels (H) and E2f1 and Peg10 mRNA expressions (I) were determined.
USP11 deubiquitinates and stabilizes E2F1
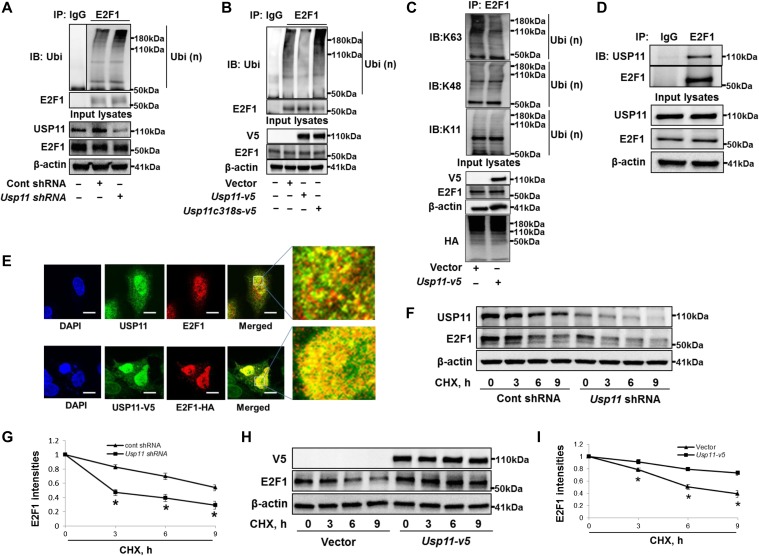
E2F1 degradation is known to be mediated by the ubiquitin-proteasome system (Marti et al., 1999; Ohta and Xiong, 2001; Wang et al., 2015). We showed that E2F1 degradation was inhibited by proteasome inhibitor (MG132), but not by lysosome/autophagy inhibitor (Bafilomycin A1) (Supplementary Figure S2), suggesting that E2F1 degradation is in the proteasome in lung epithelial cells. We hypothesized that USP11 DUB activity regulates E2F1 protein stability by removing polyubiquitin chains from Ub-E2F1, thereby preventing its degradation. E2F1 ubiquitination was examined in USP11-overexpressed or downregulated cells by in vivo ubiquitination assay. Knockdown of USP11 or overexpression of USP11C318S increased detectable amounts of Ub-E2F1 (Figure 3A and B), while overexpression of USP11 significantly reduced Ub-E2F1 in lysates from A549 cells (Figure 3B and C) and murine lung epithelial cells (Supplementary Figure S3). We evaluated several Ub chain linkages for Ub-E2F1, and observed that USP11 specifically reduced K63-linked polyubiquitination (Figure 3C and Supplementary Figure S4), consistent with the known USP11 biochemistry (Harper et al., 2014). Co-immunoprecipitation (Co-IP) studies revealed that E2F1 was associated with USP11 protein in A549 cells (Figure 3D) as well as HEK293 cells (Supplementary Figure S5A). Co-immunostaining showed that both endogenous and overexpressed USP11 and E2F1 were co-localized in the nuclei (Figure 3E). In the presence of the protein synthesis inhibitor cycloheximide (CHX), native cellular E2F1 protein was degraded more rapidly in USP11-knockdown cells compared with control cells (Figure 3F and G), while overexpression of USP11 significantly stabilized the E2F1 protein (Figure 3H and I). Likewise, V5-tagged USP11 could stabilize overexpressed E2F1 (Supplementary Figure S5B). These data collectively suggest that USP11 interacts with and deubiquitinates Ub-E2F1 to prevent its degradation and stabilize E2F1. Another DUB, POH1, has been reported to stabilize E2F1 in liver cells (Wang et al., 2015). We found that knockdown of POH1 dramatically eliminated both E2F1 protein as well as mRNA levels (Supplementary Figure S6), suggesting that POH1 regulates E2F1 in part through modulation of E2f1 transcription, not protein stability in lung epithelial cells. These differences may be due to differences between the liver cells used in that study and our lung epithelial cell-based system.
Figure 3.
USP11 deubiquitinates and stabilizes E2F1. (A) A549 cells were transfected with control shRNA or Usp11 shRNA for 72 h. In vivo ubiquitination assay was performed with a modified IP. Denatured cell lysates were immunoprecipitated with control IgG or an E2F1 antibody, and the immunoprecipitated complexes were immunoblotted with antibodies against ubiquitin and E2F1. Input cell lysates were immunoblotted with USP11, E2F1, and β-actin antibodies. (B) A549 cells were transfected with empty vector, Usp11-v5, or Usp11c318s-v5 plasmid for 48 h and analyzed by in vivo ubiquitination assay with a modified IP. (C) A549 cells were transfected with empty vector or V5-tagged USP11 plasmid for 48 h and analyzed by in vivo ubiquitination assay with a modified IP. (D) Endogenous E2F1 in A549 cells was immunoprecipitated with IgG or an E2F1 antibody, and then analyzed by immunoblotting with USP11 and E2F1 antibodies. Input cell lysates were immunoblotted with USP11, E2F1, and β-actin antibodies. (E) A549 cells were immunostained with USP11 and E2F1 antibodies (upper). USP11-V5 and HA-E2F1 co-overexpressed A549 cells were immuostained with V5 and HA antibodies (lower). Scale bar, 5 μm. Yellow indicates co-localization. (F) A549 cells were transfected with control shRNA or Usp11 shRNA for 72 h, and then treated with CHX (300 μg/ml) for 0, 3, 6, and 9 h. Cell lysates were immunoblotted with USP11, E2F1, and β-actin antibodies. (G) E2F1 intensities were measured by ImageJ software. *P < 0.01, compared to cont shRNA-transfected cells. (H) A549 cells were transfected with empty vector or Usp11-v5 plasmid for 48 h, and then treated with CHX (300 μg/ml) for 0, 3, 6, and 9 h. Cell lysates were immunoblotted with V5, E2F1, and β-actin antibodies. (I) E2F1 intensities were measured by ImageJ software. *P < 0.01, compared to empty vector-transfected cells. Shown are representative blots from at least three independent experiments.
Nuclear USP11 is essential for regulating E2F1
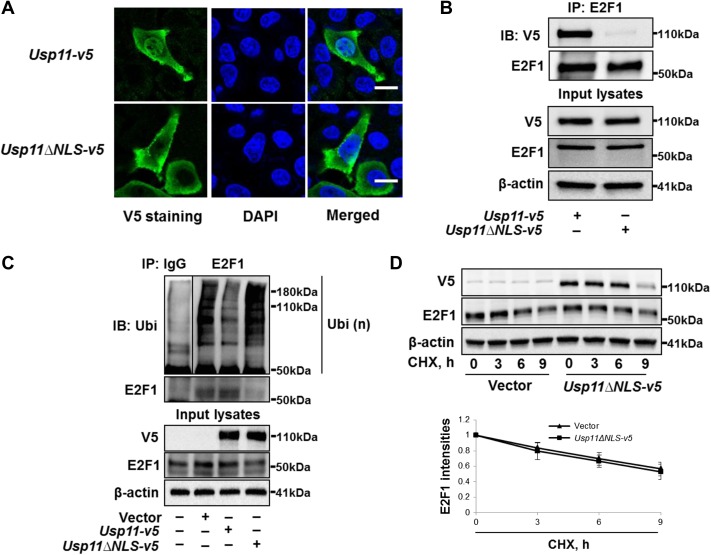
Nuclear translocation is necessary for E2F transcription factors’ activity (Muller et al., 1997); we next examined whether subcellular localization impacts USP11 stabilization of E2F1. We showed that both E2F1 and USP11 were co-localized in the nuclei (Figure 3C). Bioinformatic analysis (by nls-maper.iab.keio.ac.jp/) shows that USP11 contains a potential nuclear localization signal (NLS) at amino acids 445–452. Thus, we generated a V5-tagged NLS deletion mutant of USP11 (USP11ΔNLS-V5). USP11 wild type was found in the plasma membrane, cytoplasm, and nuclei, while USP11ΔNLS was localized on the plasma membrane and in the cytoplasm, but not in nuclei (Figure 4A). Immunoprecipitation (IP) of whole cell lysates revealed that E2F1 was associated with wild type USP11 but not USP11ΔNLS (Figure 4B), indicating that E2F1 is associated with nuclear USP11 through docking to amino acids in the NLS of USP11. Furthermore, USP11ΔNLS overexpression failed to reduce Ub-E2F1 levels (Figure 4C) or stabilize E2F1 protein (Figure 4D). From these data, we conclude that USP11 protein primarily compartmentalized within the nucleus exerts DUB activity on Ub-E2F1 to modulate E2F1 stability.
Figure 4.
USP11 deubiquitinates and stabilizes E2F1 in the nuclei. (A) A549 cells grown on glass-bottom dishes were transfected with Usp11-v5 plasmid or V5-tagged USP11 mutant with deletion of aa445–452 (Usp11ΔNLS-v5) for 48 h. Immunostaining was performed to examine the localization of V5-tagged proteins (green). Nuclei were stained by DAPI (blue). Scale bar, 5 μm. (B) A549 cells were transfected with Usp11-v5 plasmid or Usp11ΔNLS-v5 plasmid for 48 h, and cell lysates were analyzed by IP. (C) A549 cells were transfected with vector, Usp11-v5 plasmid, or Usp11c318s-v5 plasmid for 48 h and analyzed by in vivo ubiquitination assay with a modified IP. (D) A549 cells were transfected with empty vector or Usp11ΔNLS-v5 for 48 h, and then were treated with CHX (300 μg/ml) for 0, 3, 6, and 9 h. Cell lysates were immunoblotted with V5, E2F1, and β-actin antibodies. E2F1 intensities were measured by ImageJ software. Shown are representative blots from at least three independent experiments.
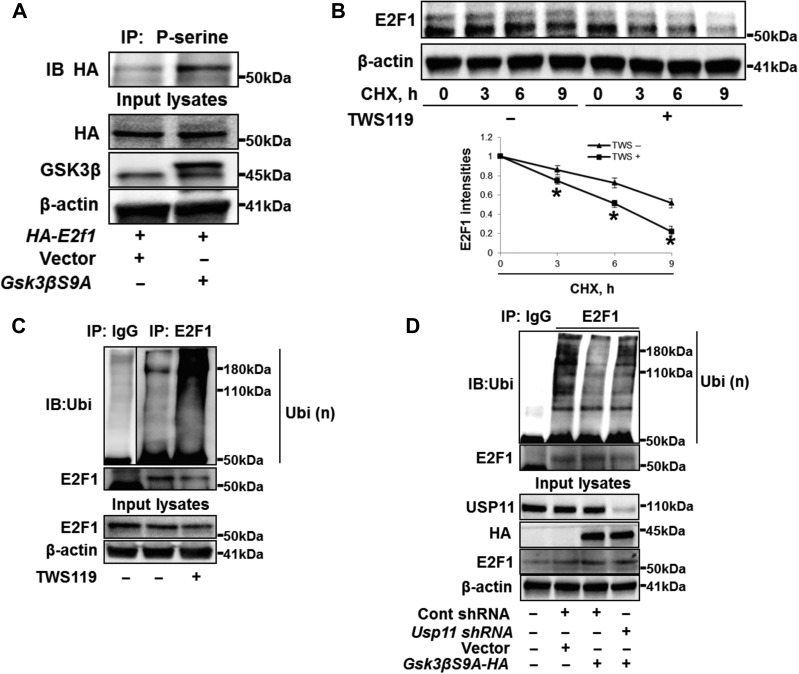
GSK3β-mediated phosphorylation of E2F1 enhances E2F1 deubiquitination and stability
Nuclear E2F1 has been characterized as a GSK3β substrate, with downstream regulation of E2F1 transcriptional activity (Garcia-Alvarez et al., 2007; Zhou et al., 2008). Overexpression of a hyperactive form of GSK3β (GSK3βS9A) (Li et al., 2004; Jang et al., 2011) caused an increase in serine phosphorylation of E2F1 (Figure 5A), not USP11 (Supplementary Figure S7), suggesting that E2F1 is phosphorylated by GSK3β in lung epithelial cells. To test whether GSK3β modulates E2F1 stability, cells were treated with the GSK3β inhibitor TWS119 prior to CHX exposure. TWS119 enhanced E2F1 degradation (Figure 5B), suggesting that GSK3β-mediated phosphorylation may promote E2F1 stability. When we examined GSK3β antagonism as a regulator of E2F1 ubiquitination, we observed that TWS119 treatment increased the amount of Ub-E2F1 in cells (Figure 5C), while overexpression of GSK3βS9A eliminated the Ub-E2F1 (Figure 5D). Furthermore, depletion of USP11 attenuated the effect of GSK3βS9A (Figure 5D), suggesting that USP11 activity is essential for GSK3β-mediated reduction of Ub-E2F1. Taken together, GSK3β affects E2F1 stability with increased phosphorylation and deubiquitination of E2F1.
Figure 5.
GSK3β phosphorylates E2F1 and facilities E2F1 deubiquitination and stability. (A) A549 cells were transfected with vector or GSK3β mutant with S9A (Gsk3βS9A) for 24 h, and then transfected with HA-tagged E2F1 (HA-E2f1) plasmid for 24 h. Cell lysates were analyzed by IP. (B) A549 cells were treated with CHX (300 μg/ml) with or without TWS119 (10 μM) for 0, 3, 6, and 9 h. Cell lysates were immunoblotted with E2F1 and β-actin antibodies. E2F1 intensities were measured by ImageJ software. *P < 0.01, compared to TWS(−) treated cells. (C) A549 cells were treated with TWS119 (10 μM) for 2 h and analyzed by in vivo ubiquitination assay with a modified IP. (D) A549 cells were transfected with Cont shRNA, Usp11 shRNA, or Gsk3βS9A-HA for 48 h as indicated and analyzed by in vivo ubiquitination assay with a modified IP.
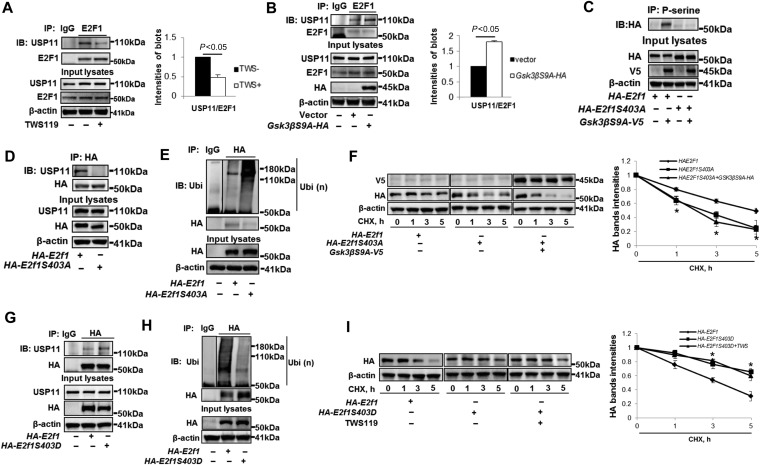
GSK3β promotes association between E2F1 and USP11
We next tested if E2F1 phosphorylation by GSK3β modulated the association between E2F1 and USP11. TWS119 weakened the binding between E2F1 and USP11 (Figure 6A), while overexpression of the active form of GSK3β enhanced their association (Figure 6B). These data indicate that E2F1 phosphorylation by GSK3β enhances its association with USP11. Inhibition of other protein kinases, such as Erk pathway and PKCδ, had no effect on the association between E2F1 and USP11 (Supplementary Figure S8). To further evaluate the hypothesis that phosphorylation stabilizes protein stability via increasing its association with a DUB, we examined the effect of E2F1 phosphorylation site mutation on its interaction with USP11. It has been shown that GSK3β phosphorylates E2F1 on serine residue 403 (S403) (Garcia-Alvarez et al., 2007), which was confirmed by use of E2F1S403A mutant in the current study. As shown in Figure 6C, GSK3β failed to phosphorylate E2F1S403A mutant. Interestingly, this mutant was not able to associate with USP11 (Figure 6D). Further, we found that E2F1S403A had higher level of ubiquitination (Figure 6E) and exhibited unstable (Figure 6F). Overexpression of GSK3βS9A had no effect on E2F1S403A stability (Figure 6F). In addition to use of phosphorylation site mutant, we generated a plasmid coding for a mutation that mimic phosphorylation of E2F1 (E2F1S403D). Consistent with our conclusion, E2F1S403D increased the association with USP11 (Figure 6G), resulting in a reduced ubiquitination (Figure 6H) and an increased stability (Figure 6I). Further, inhibition of GSK3β by TWS119 had no effect on E2F1S403D stability (Figure 6I). This study reveals that phosphorylation of E2F1 by GSK3β promotes its interaction with USP11, ultimately reducing its ubiquitination and increasing its stability.
Figure 6.
GSK3β-mediated phosphorylation facilities the association between E2F1 and USP11. (A) A549 cells were treated with or without TWS119 (10 μM) for 2 h. Cell lysates were analyzed by IP. Immunoprecipitated USP11 and E2F1 were quantified, and ratio of USP11/E2F1 was shown. (B) A549 cells were transfected with empty vector or Gsk3βS9A-HA for 48 h. Cell lysates were analyzed by IP. Immunoprecipitated USP11 and E2F1 were quantified, and ratio of USP11/E2F1 was shown. (C) A549 cells were transfected with HA-E2f1 or HA-E2f1S403A with or without Gsk3βS9A-v5 plasmid for 48 h. Cell lysates were analyzed by IP. (D) A549 cells were transfected with HA-E2f1 or HA-E2f1S403A plasmid for 48 h. Cell lysates were analyzed by IP. (E) A549 cells were transfected with HA-E2f1 or HA-E2f1S403A plasmid for 48 h and analyzed by in vivo ubiquitination assay with a modified IP. (F) A549 cells were transfected with HA-E2f1, HA-E2f1S403A, or Gsk3βS9A-v5 plasmid for 48 h as indicated, and then treated with CHX (300 μg/ml) for 1–5 h. Cell lysates were immunoblotted with V5, HA, and β-actin antibodies. HA band intensities were measured by ImageJ software. *P < 0.01, compared to HA-E2F1-overexpressed cells. (G) A549 cells were transfected with HA-E2f1 or HA-E2f1S403D for 48 h. Cell lysates were analyzed by IP. (H) A549 cells were transfected with HA-E2f1 or HA-E2f1S403D for 48 h and analyzed by in vivo ubiquitination assay with a modified IP. (I) A549 cells were transfected with HA-E2f1 or HA-E2f1S403D for 48 h, and then treated with CHX (300 μg/ml) with or without TWS119 (10 μM) for 1–5 h. Cell lysates were immunoblotted with HA and β-actin antibodies. HA band intensities were measured by ImageJ software. *P < 0.01, compared to HA-E2F1-overexpressed cells. Shown are representative blots from at least three independent experiments.
USP11 promotes cell proliferation and wound healing through stabilization of E2F1
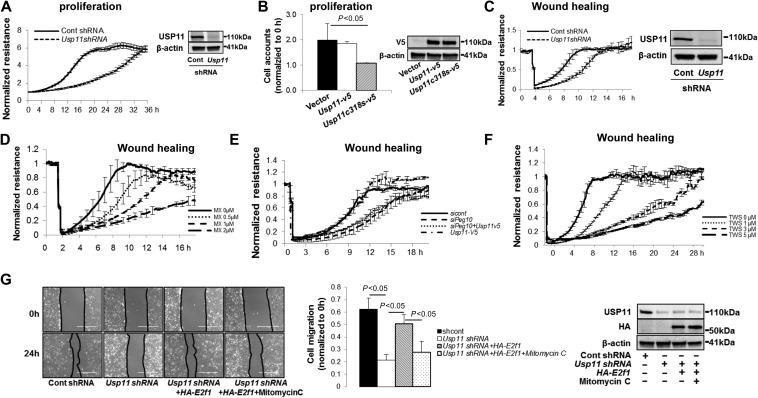
To investigate USP11 activity in the context of cell proliferation and wound healing in lung epithelial cells, we used electric cell-substrate impedance sensing (ECIS) system. Increases in resistance indicate higher levels of cell confluency. We observed that downregulation of USP11 by Usp11 shRNA markedly suppressed A549 cell proliferation (Figure 7A). Inhibition via USP11C318S overexpression reduces cell growth (Figure 7B). Usp11 shRNA or MX treatment significantly reduced wound healing (Figure 7C and D). This cell behavior correlates directly the changes in PEG10 levels described above. Pharmacologically, MX is also a topoisomerase II inhibitor, which is used clinically to induce apoposis in cancer cells, therefore the effects of MX may be not totally dependent on inhibition of USP11 (Bellosillo et al., 1998). Accordingly, downregulation of PEG10 by Peg10 siRNA transfection in A549 cells significantly reduced wound healing, an effect that was not reversed in USP11-overexpressed cells (Figure 7E). This implies that the effect of USP11 on cell wound healing is due to downstream Peg10 expression. Further, we demonstrated that inhibition of GSK3β by TWS119 significantly reduced wound healing in a dose-dependent manner (Figure 7F). In an orthogonal assessment of A549 wound healing, scratch assays were performed and wound healing determined by measuring the area of wound closure. USP11 knockdown significantly decreased wound healing in this assay, with restoration by overexpression of E2F1. To further evaluate the effect of cell proliferation in wound healing mediated by E2F1 stability, we treated cells with mitomycin C, an inhibitor of cell proliferation. As shown in Figure 7G, mitomycin C significantly suppressed E2F1-mediated wound healing, suggesting that cell proliferation plays a critical role in the wound healing. Taken together, we conclude that USP11 promotes cell proliferation and wound healing via stabilization of nuclear E2F1, which leads to increases in PEG10 expression in lung epithelial cells. These results implicate that USP11 plays an important role in lung epithelial cell transcriptional control, proliferation, and repair after injury.
Figure 7.
USP11 regulates cell proliferation and wound healing through modulating E2F1 stability and PEG10 expression. (A) A549 cells cultured in D60 dishes were transfected with control shRNA or Usp11 shRNA for 72 h, and then transferred to 8-well gold microelectrode plates (50000 cells/well) for proliferation assay. Cell proliferation was traced for up to 40 h with an ECIS system. The rest of the cells were analyzed by USP11 and β-actin immunoblotting. (B) A549 cells were transfected with empty vector plasmid, Usp11-v5, or Usp11c318s-v5 plasmid for 24 h, and then cultured on 6-well plates for proliferation assay. Cell numbers at 20 h were accounted. Cell lysates were analyzed by V5 and β-actin immunoblotting. (C) A549 cells cultured in D60 dishes were transfected with control shRNA or Usp11 shRNA for 72 h, and then transferred to 8-well gold microelectrode plates (200000 cells/well) for wound healing assay. Confluent cells were wounded after at least 4 h of growth. Wounding conditions were 20 sec, 1400 μA, 60000 Hz. Wound healing was immediately traced following wounding via an ECIS system up to 24 h. The rest of the cells were analyzed by USP11 and β-actin immunoblotting. (D) A549 cells were cultured on 8-well gold microelectrode plates (200000 cells/well) treated with 0, 0.5, 1, and 2 μM MX for wound healing assay. (E) A549 cells cultured in D60 dishes were transfected with control siRNA or Peg10 siRNA for 24 h, and then transfected with Usp11-v5 plasmid for additional 24 h. Cells were then transferred to 8-well gold microelectrode plates (200000 cells/well) for wound healing assay. (F) A549 cells cultured on 8-well gold microelectrode plates (200000 cells/well) were treated with 0, 1, 3, and 5 μM TWS119 for wound healing assay. (G) A549 cells cultured in 6-well plates were transfected with control shRNA or Usp11 shRNA for 24 h, and then transfected with empty vector or HA-E2f1 plasmid for additional 48 h. Monolayers were scratched using a sterile 10 μl pipette tip, and then cells were treated with or without mitomycin C (10 μM) and digitally photographed at 0 h and 24 h. The extent of wound healing was quantified using ImageJ software and the percentage of wound closure was calculated. Cells were collected for immunoblotting with USP11, HA, and β-actin antibodies.
Discussion
Recent studies on molecular mechanisms of lung repair and remodeling after injury indicate that epithelial cell proliferation and wound healing are integral to lung tissue repair (Gonzalez-Lopez and Albaiceta, 2012; Chen and Fine, 2016). As a universal modulator of cell physiology, ubiquitination directs protein signaling activity, stability, and localization (Yau and Rape, 2016). In the current study, we describe the deubiquitinase activity of USP11 as a driver of lung epithelial cell proliferation through stabilization of the transcription factor E2F1, which in turn increases expression of its target gene, Peg10. Phosphorylation of E2F1 by GSK3β enhances Ub-E2F1 binding to USP11, which facilitates deubiquitination and stabilization of E2F1 in the nuclei.
There is emerging evidence that USP11 DUB activity is integral to a wide range of cellular responses, including DNA repair (Wiltshire et al., 2010), viral RNA replication (Li et al., 2014), apoptosis (Lee and Song, 2016), and cytokine release (Zhao et al., 2016). USP11 has therefore been implicated in the physiology of multiple diseases (Bayraktar et al., 2013; Wu et al., 2014). The role of USP11 in lung repair and remodeling has not been investigated prior to this report. PEG10 promotes the migration of human Burkitt’s lymphoma cells (Xiong et al., 2012), trophoblast cells (H. Chen et al., 2015), and A549 lung epithelial cells (Deng et al., 2014) with upregulated expression of matrix metalloproteinase (MMP)-2 and MMP-9. Both MMP-2 (Gonzalez-Lopez et al., 2011) and MMP-9 (O’Kane et al., 2009) have been shown to be involved in alveolar epithelial repair in experimental models of wound healing. The current study links PEG10 expression and USP11 activity in lung epithelial cell proliferation and wound healing. Al-Salihi et al. (2012) has demonstrated that USP11 stabilizes TβRI, which enhances TGFβ-1 signaling, while PEG10 has been shown to inhibit TGFβ-1 signaling through interaction with TβRI (Akamatsu et al., 2015). While the interplay between USP11 and PEG10 in TGFβ-1 signaling has not been closely examined, our data may suggest that reduction of PEG10 expression by USP11 depletion may exert opposing, perhaps homeostatic inputs into TGFβ-1 signaling, which is also involved in lung repair and remodeling.
E2F transcription factors play a critical role in the control of cellular proliferation and apoptosis (Koizumi et al., 1989; DeGregori et al., 1997; Dyson, 1998; Lissy et al., 2000; Wang et al., 2015). It has been reported that Peg10 expression was directly regulated by E2F1 (Wang et al., 2008; Akamatsu et al., 2015). We confirmed in A549 cells that knockdown of E2F1 resulted in decreased Peg10 mRNA, with concomitant decrease in PEG10 protein levels. E2F1 ubiquitination and stability has been investigated, and E2F1 degradation has been known to be mediated by the ubiquitin-proteasome system (Hofmann et al., 1996; Campanero and Flemington, 1997). Consistent with their finding, our data show that proteasome inhibitor prolonged E2F1 half-life. We show that E2F1 is modified by K63-linked ubiquitin chains. K63-linkages are often non-proteolytic, however accumulating evidence show that K63 Ubi linkage can facilitate substrate degradation in the proteasome system (Babu et al., 2005; Kim et al., 2007). Our data suggest that, at least in lung epithelial cells, K63-linked polyubiquitination of E2F1 shuttles E2F1 for degradation in the proteasome. Wang et al. (2015) have shown that the deubiquitinating enzyme, POH1 stabilizes E2F1 protein in liver cells. Here, we reveal that a non-proteasome-associated DUB, USP11, deubiquitinates and stabilizes E2F1. This phenomenon is specific for E2F1, and not E2F2 or E2F4. Wang et al. (2015) show that USP11 has no effect on E2F1 stability, while we show that depletion of POH1 eliminates E2F1 levels through reduction of E2f1 mRNA levels. The differences between the two studies may in part be due to cell type differences; they investigated E2F1 stability in human liver tissues, while we used human lung epithelial cells. It is also possibly caused by the nature of two different DUBs. Since POH1 is a proteasome-associated DUB, the effects observed in the Wang’s study may relate to Ub-E2F1 that has localized to the nuclear proteasome, while the effects of USP11 we observed may occur in nucleoplasmic foci. Our study corroborates observations from Wang et al. (2015) that E2F1 stability is regulated by removal of K63-linked ubiquitin chains in the nucleus.
While several studies have shown that overexpression of a DUB does not change steady-state levels of the substrate proteins (Berthouze et al., 2009; Yun et al., 2015; Savio et al., 2016), most of the time DUB overexpression does lead to a prolonged cellular half-life of the substrate. For example, USP2a (Liu et al., 2013) and Cezanne-1 (Pareja et al., 2012) deubiquitinate and stabilize EGFR, while overexpression of these DUBs fail to increase basal levels of EGFR. We presume that the steady-state protein levels are regulated by homeostatic transcriptional mechanisms. Here, we similarly show that overexpression of USP11 significantly reduced E2F1 ubiquitination and prolonged its half-life, but USP11 overexpression did not increase state-steady levels of E2F1 (Figure 3H) nor Peg10 expression (Figure 1E–H).
While substrate phosphorylation can generate ‘phosphodegron’ signals that recruit ubiquitin ligases to interact with substrate (Alessandrini et al., 1997; Zhao et al., 2012), the effect of substrate phosphorylation on the association between the substrate and DUB has not been described to our knowledge. GSK3β is an intracellular protein kinase that has been implicated in regulation of metabolism (Cross et al., 1995), protein synthesis (Ferkey and Kimelman, 2000), cytokine release (Suber et al., 2017), and cell cycle (Diehl et al., 1998). Here, we demonstrate that phosphorylation of E2F1 by GSK3β enhances the association between E2F1 and USP11, thereby promoting E2F1 deubiquitination and stability. A prior report demonstrates that GSK3β binds to E2F1 and regulates its transcriptional activity (Garcia-Alvarez et al., 2007), and our study reveals a new relationship between these molecules, whereby GSK3β facilitates stabilization of E2F1. The cellular context of GSK3β activation, access to E2F1, and specific phosphorylation sites are important topics for future studies.
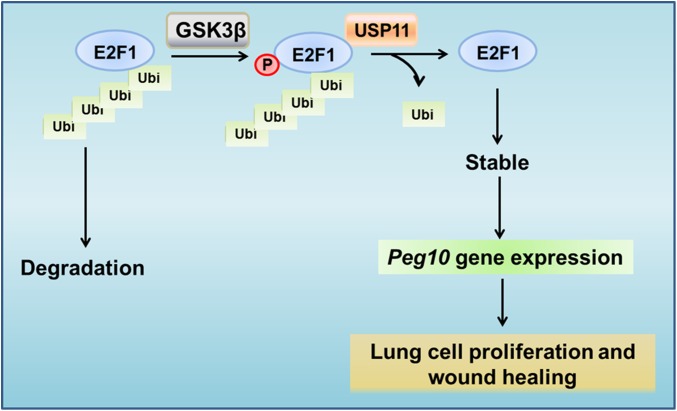
In conclusion, we propose an undescribed signal cascade for protein stabilization. In our model, a protein kinase (GSK3β) phosphorylates a substrate (E2F1), thereby enhancing the association between that substrate and a deubiquitinating enzyme (USP11). The DUB removes polyubiquitin chains and stabilizes substrate to increase its biological activity (Peg10 expression) to facilitate cell behavior (lung epithelial cell proliferation and wound healing) (Figure 8). This GSK3β/E2F1/USP11/PEG10 pathway may play an important role in the repair and remodeling after lung injury, implying that better understanding of USP11 biology might someday inform ARDS therapy.
Figure 8.
GSK3β-mediated phosphorylation of E2F1 promotes its stability through increasing its association with USP11. GSK3β phosphorylates E2F1, which increases E2F1 interaction with USP11. USP11 binds to E2F1, leading to deubiquitination and stabilization of E2F1. E2F1 activates Peg10 promoter and increases Peg10 mRNA expression. Elevation of PEG10 levels promotes lung cell proliferation and wound healing.
Materials and methods
Cells and reagents
A549 cells (American Type Culture Collection) were cultured with RPMI 1640 medium containing 2 mM glutamine with 10% fetal bovine serum (FBS). The murine lung epithelial cell line (MLE12) and human airway epithelial cell line (Beas2B, American Type Culture Collection) were cultured in HITES medium complemented with 10% FBS. Cells were maintained in a 37°C incubator in the presence of 5% CO2. V5 antibody, the mammalian expression plasmid pcDNA3.1/V5-His TOPO, Escherichia coli Top 10 competent cells, lipofectamine 2000 transfection reagent were purchased from Invitrogen (Lifetechnologies). USP11 antibody and PEG10 antibody were obtained from Abcam. Cycloheximide (CHX), mitoxantrone (MX), Bafilomycin A1, mitomycin C, and β-actin antibody were from Sigma Aldrich. E2F1 antibody, E2F2 antibody, E2F4 antibody, human E2f1 siRNA, human Peg10 siRNA, immunobilized protein A/G beads were from Santa Cruz Biotechnology. Proteasome inhibitor MG132 was from EMD Millipore. HA tag, GSK3β antibodies, ubiquitin antibody were from Cell Signaling Technologies. Horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse secondary antibodies were obtained from Bio-Rad Laboratories, Inc. TWS119 was from Cayman Chemical. All materials used in the experiments are commercially available.
Construction of plasmid
Human Usp11 cDNA was inserted into pcDNA3.1D/His-V5 TOPO vector. Intracellular domain 445–452 deletion mutants of USP11 were generated by PCR with specific primers designed to target USP11 cDNA sequence. Site directed mutagenesis was performed to generate USP11 or E2F1 mutants according to the manufacturer’s instructions (Agilent Technologies). Plasmid pRcCMV-HA-E2F1 was a gift from William Kaelin (Addgene plasmid #21667).
Plasmid transfection
A549 or Beas2B cells were subcultured on 6-well plates, 35-mm plates, or 100-mm dishes to 70%–90% confluence. Lipofectamine 2000 transfection reagent was added to the mixture containing varying amounts of plasmid and 200 μl of Opti-medium, and then incubated for 10 min to allow transfection reagent/DNA complexes to form. The mixture was added directly to the cells with complete medium. MLE12 cells grown on 100-mm plates (70%–90% confluence) were transfected with plasmids using Lonza electroporation transfection according to the manufacturer’s protocol, and then cultured for 48 h. Protein transient expression were confirmed by western blotting.
Cell lysis and immunoblotting
Following the aforementioned cellular treatments, cells were washed with cold PBS and collected in cell lysis buffer containing 20 mM Tris HCl (pH 7.4), 150 mM NaCl, 2 mM EGTA, 5 mM β-glycerophosphate, 1 mM MgCl2, 1% Triton X-100, 1 mM sodium orthovanadate, 10 μg/ml protease inhibitors, 1 μg/ml leupeptin, and 1 μg/ml pepstatin. Protein concentrations of the samples were then determined with use of a Bio-Rad protein assay kit (Bio-Rad Laboratories, Inc.) using BSA as a standard. An equal amount of cell lysates (20 μg) was subjected to SDS-PAGE gel, and then transferred to membranes. Blots were washed with 25 mM Tris HCl (pH 7.4), 137 mM NaCl, and 0.1% Tween20 (TBST) and incubated with primary antibodies in 5% BSA in TBST for overnight. The membranes were then washed three times at 10 min intervals with TBST prior to addition of secondary antibodies for an additional 1 h. Blots were developed with an enhanced chemiluminescence detection kit according to manufacturer’s instruction.
Co-immunoprecipitation
Equal amounts of cell lysates (1 mg) were incubated with specific primary antibodies overnight at 4°C, followed by the addition of 40 μl protein A/G agarose beads and incubation for an additional 2 h at 4°C. The immunoprecipitated complex was then washed 3 times with ice cold 1% Triton in PBS. The 2× dye with β-ME was added to the beads and heated at 100°C for 10 min and analyzed by immunoblotting with indicated antibodies.
In vivo ubiquitination assay
In vivo ubiquitination assay were performed with a modified IP. Cells were treated with MG132 and leupeptin for 3 h, and then cells were washed and collected with cold PBS. The cells in PBS were then centrifuged; the supernatant was removed, followed by adding 1 μl of ubiquitin aldehyde and 1 μl of NEM to the cell pellet. Based on the size of the pellet, 50–80 μl of 2% SDS lysis buffer was added. The cells were then boiled at 100°C for 10 min following sonication. And then, the samples were diluted with 500–800 μl of 1× TBS. Regular IP procedure was then followed out.
Immunofluorescence staining
A549 cells were grown to 60% confluence on 35-mm glass-bottom culture dishes and were transfected with plasmids for 48 h. Cells were washed with PBS, and fixed with 3.7% formaldehyde for 20 min. After blocking in 5% BSA in TBST for 1 h, cells were incubated with 1:200 dilution of primary antibody for 1 h, followed by a 1:200 dilution of fluorescence-conjugated secondary antibodies sequentially for immunostaining. Images were captured by a Nikon ECLIPSE TE 300 inverted confocal microscope.
Cell scratch assay
A549 cells were cultured on 6-well plates until 100% confluence. Monolayers were scratched using a sterile 10 μl pipette tip. Cells were digitally photographed at 0 h and 24 h after stimulation. The extent of cell migration was quantified using ImageJ software. The percentage of wound closure was calculated as follows: [(wound area at 0 h – wound area at 24 h)/wound area at 0 h] × 100%.
ECIS-based cell proliferation and wound healing assays
ECIS Zθ device and 96W1E arrays were used (Applied Biophysics Inc.). Cell proliferation was recorded immediately following addition of the cells at multiple frequencies, for up to 40 h. For the wound healing assay, confluent cells were wounded after at least 24 h of growth. Wounding conditions were 20 sec, 1400 μA, 60000 Hz. The wound healing was immediately traced following wounding for up to 28 h. Recording and Analysis of the data were performed using the ECIS software (V1.2.215.OPC, 28 Nov 2015). Increases in resistance indicate higher levels of cell confluency.
Real-time qRT-PCR and microarray
Total RNA was isolated from cells using the NucleoSpin RNA extraction kit (Clontech Laboratories, Inc.) following manufacturer’s instructions. The isolated RNA was quantified using a spectrophotometry. cDNA preparation was preformed according to standard procedures using the iScript cDNA Synthesis kit (Bio-Rad) and T100 Thermal Cyclers system (Bio-Rad). Real-time PCR was performed by iQ SYBR Green Supermix (Applied Biosystems, Life Technologies) using the CFX96 real-time PCR detection system (Bio-Rad). The primers used for detecting mRNA levels were: human Usp11-F: 5′-GAAGAGAACGGACGGCGAT-3′; human Usp11-R: 5′-CGTGCTGTGGCTCTCTATCC-3′; human Gapdh-F: 5′-CTCTGCTCCTCCTGTTCGAC-3′; human Gapdh-R: 5′-GACTCCGACCTTCACCTTCC-3′; human Peg10-F: 5′-GTCCTCGCGTGAAATAAGCG-3′; human Peg10-R: 5′-CCCAGCTGTAGCTTCACTTCT-3′; human E2f1-F: 5′-CTTGGAGGGACCAGGGTTTC-3′; human E2f1-R: 5′-TTTCCAAACAGGCTGGGAGG-3′. Levels of transcripts were normalized to Gapdh, and then normalized to the mean of controls as indicated in the figures. For microarray, the total RNAs from control cells and Usp11 shRNA-transfected cells with three separate experiments were submitted to Genomic Research co-facility at the University of Pittsburgh. Microarray was performed using Affymetrix RNA microarray technology and transcriptome analysis was performed using Tuxedo, Tophat, and Cufflinks software.
Statistics
All results were subjected to statistical analysis using two-way analysis of variance, and, wherever appropriate, student t-test. Data are expressed as mean ± SD of triplicate samples from at least three independent experiments.
Supplementary Material
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This study was supported by the US National Institutes of Health (HL131665 to Y.Z. and R01GM115389 to J.Z.), American Lung Association Biomedical Research Grant RG350146 (J.Z.), and American Heart Association 16GRNT30660001 (Y.Z.).
Conflict of interest: none declared.
References
- Akamatsu S., Wyatt A.W., Lin D., et al. (2015). The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep. 12, 922–936. [DOI] [PubMed] [Google Scholar]
- Al-Salihi M.A., Herhaus L., Macartney T., et al. (2012). USP11 augments TGFβ signalling by deubiquitylating ALK5. Open Biol. 2, 120063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessandrini A., Chiaur D.S., and Pagano M. (1997). Regulation of the cyclin-dependent kinase inhibitor p27 by degradation and phosphorylation. Leukemia 11, 342–345. [DOI] [PubMed] [Google Scholar]
- Babu J.R., Geetha T., and Wooten M.W. (2005). Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J. Neurochem. 94, 192–203. [DOI] [PubMed] [Google Scholar]
- Bayraktar S., Gutierrez Barrera A.M., Liu D., et al. (2013). USP-11 as a predictive and prognostic factor following neoadjuvant therapy in women with breast cancer. Cancer J. 19, 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellosillo B., Colomer D., Pons G., et al. (1998). Mitoxantrone, a topoisomerase II inhibitor, induces apoptosis of B-chronic lymphocytic leukaemia cells. Br. J. Haematol. 100, 142–146. [DOI] [PubMed] [Google Scholar]
- Berthouze M., Venkataramanan V., Li Y., et al. (2009). The deubiquitinases USP33 and USP20 coordinate β2 adrenergic receptor recycling and resensitization. EMBO J. 28, 1684–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune K., Frank J., Schwingshackl A., et al. (2015). Pulmonary epithelial barrier function: some new players and mechanisms. Am. J. Physiol. Lung Cell. Mol. Physiol. 308, L731–L745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhavarapu V.N., White E.D., Mahanic C.S., et al. (2012). Regulation of E2F1 by APC/C Cdh1 via K11 linkage-specific ubiquitin chain formation. Cell Cycle 11, 2030–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart R.A., Peng Y., Norris Z.A., et al. (2013). Mitoxantrone targets human ubiquitin-specific peptidase 11 (USP11) and is a potent inhibitor of pancreatic cancer cell survival. Mol. Cancer Res. 11, 901–911. [DOI] [PubMed] [Google Scholar]
- Campanero M.R., and Flemington E.K. (1997). Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc. Natl Acad. Sci. USA 94, 2221–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., and Fine A. (2016). Stem cells in lung injury and repair. Am. J. Pathol. 186, 2544–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Sun M., Liu J., et al. (2015). Silencing of paternally expressed gene 10 inhibits trophoblast proliferation and invasion. PLoS One 10, e0144845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvol H., Flamein F., Epaud R., et al. (2009). Lung alveolar epithelium and interstitial lung disease. Int. J. Biochem. Cell Biol. 41, 1643–1651. [DOI] [PubMed] [Google Scholar]
- Cross D.A., Alessi D.R., Cohen P., et al. (1995). Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789. [DOI] [PubMed] [Google Scholar]
- DeGregori J., Leone G., Miron A., et al. (1997). Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl Acad. Sci. USA 94, 7245–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Hu Y., Ding Q., et al. (2014). PEG10 plays a crucial role in human lung cancer proliferation, progression, prognosis and metastasis. Oncol. Rep. 32, 2159–2167. [DOI] [PubMed] [Google Scholar]
- Diehl J.A., Cheng M., Roussel M.F., et al. (1998). Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12, 3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. (1998). The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Faesen A.C., Luna-Vargas M.P., Geurink P.P., et al. (2011). The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 18, 1550–1561. [DOI] [PubMed] [Google Scholar]
- Ferkey D.M., and Kimelman D. (2000). GSK-3: new thoughts on an old enzyme. Dev. Biol. 225, 471–479. [DOI] [PubMed] [Google Scholar]
- Freeman B.A., Panus P.C., Matalon S., et al. (1993). Oxidant injury to the alveolar epithelium: biochemical and pharmacologic studies. Res. Rep. Health Eff. Inst. 54, 1–30; discussion 31–39. [PubMed] [Google Scholar]
- Garcia-Alvarez G., Ventura V., Ros O., et al. (2007). Glycogen synthase kinase-3β binds to E2F1 and regulates its transcriptional activity. Biochim. Biophys. Acta 1773, 375–382. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Lopez A., and Albaiceta G.M. (2012). Repair after acute lung injury: molecular mechanisms and therapeutic opportunities. Crit. Care 16, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Lopez A., Astudillo A., Garcia-Prieto E., et al. (2011). Inflammation and matrix remodeling during repair of ventilator-induced lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 301, L500–L509. [DOI] [PubMed] [Google Scholar]
- Harper S., Gratton H.E., Cornaciu I., et al. (2014). Structure and catalytic regulatory function of ubiquitin specific protease 11 N-terminal and ubiquitin-like domains. Biochemistry 53, 2966–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann F., Martelli F., Livingston D.M., et al. (1996). The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 10, 2949–2959. [DOI] [PubMed] [Google Scholar]
- Jacko A.M., Nan L., Li S., et al. (2016). De-ubiquitinating enzyme, USP11, promotes transforming growth factor β-1 signaling through stabilization of transforming growth factor β receptor II. Cell Death Dis. 7, e2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H.D., Shin J.H., Park D.R., et al. (2011). Inactivation of glycogen synthase kinase-3β is required for osteoclast differentiation. J. Biol. Chem. 286, 39043–39050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn J.M., Caldwell E.C., Deem S., et al. (2006). Acute lung injury in patients with subarachnoid hemorrhage: incidence, risk factors, and outcome. Crit. Care Med. 34, 196–202. [DOI] [PubMed] [Google Scholar]
- Kim H.T., Kim K.P., Lledias F., et al. (2007). Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386. [DOI] [PubMed] [Google Scholar]
- Koizumi A., Sageshima M., Wada Y., et al. (1989). Immature alveolar/blood barrier and low disaturated phosphatidylcholine in fetal lung after intrauterine exposure to O,O,S-trimethylphosphorothioate. Arch. Toxicol. 63, 331–335. [DOI] [PubMed] [Google Scholar]
- Komander D. (2010). Mechanism, specificity and structure of the deubiquitinases. Subcell. Biochem. 54, 69–87. [DOI] [PubMed] [Google Scholar]
- Lee E.W., Seong D., Seo J., et al. (2015). USP11-dependent selective cIAP2 deubiquitylation and stabilization determine sensitivity to Smac mimetics. Cell Death Differ. 22, 1463–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.W., and Song J. (2016). USP11: a key regulator of cIAP2 stability and sensitivity to SMAC mimetics. Mol. Cell. Oncol. 3, e1029829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Ryder J., Su Y., et al. (2004). Overexpression of GSK3βS9A resulted in tau hyperphosphorylation and morphology reminiscent of pretangle-like neurons in the brain of PDGSK3β transgenic mice. Transgenic Res. 13, 385–396. [DOI] [PubMed] [Google Scholar]
- Li Q., Zhang Y.Y., Chiu S., et al. (2014). Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle. PLoS Pathog. 10, e1004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Xiao R., Tembo K., et al. (2016). PEG10 promotes human breast cancer cell proliferation, migration and invasion. Int. J. Oncol. 48, 1933–1942. [DOI] [PubMed] [Google Scholar]
- Lim K.H., Suresh B., Park J.H., et al. (2016). Ubiquitin-specific protease 11 functions as a tumor suppressor by modulating Mgl-1 protein to regulate cancer cell growth. Oncotarget 7, 14441–14457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissy N.A., Davis P.K., Irwin M., et al. (2000). A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature 407, 642–645. [DOI] [PubMed] [Google Scholar]
- Liu Z., Zanata S.M., Kim J., et al. (2013). The ubiquitin-specific protease USP2a prevents endocytosis-mediated EGFR degradation. Oncogene 32, 1660–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanic C.S., Budhavarapu V., Graves J.D., et al. (2015). Regulation of E2 promoter binding factor 1 (E2F1) transcriptional activity through a deubiquitinating enzyme, UCH37. J. Biol. Chem. 290, 26508–26522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti A., Wirbelauer C., Scheffner M., et al. (1999). Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1, 14–19. [DOI] [PubMed] [Google Scholar]
- Muller H., Moroni M.C., Vigo E., et al. (1997). Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol. Cell. Biol. 17, 5508–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kane C.M., McKeown S.W., Perkins G.D., et al. (2009). Salbutamol up-regulates matrix metalloproteinase-9 in the alveolar space in the acute respiratory distress syndrome. Crit. Care Med. 37, 2242–2249. [DOI] [PubMed] [Google Scholar]
- Ohta T., and Xiong Y. (2001). Phosphorylation- and Skp1-independent in vitro ubiquitination of E2F1 by multiple ROC-cullin ligases. Cancer Res. 61, 1347–1353. [PubMed] [Google Scholar]
- Okabe H., Satoh S., Furukawa Y., et al. (2003). Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 63, 3043–3048. [PubMed] [Google Scholar]
- Ono R., Kobayashi S., Wagatsuma H., et al. (2001). A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73, 232–237. [DOI] [PubMed] [Google Scholar]
- Ono R., Nakamura K., Inoue K., et al. (2006). Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38, 101–106. [DOI] [PubMed] [Google Scholar]
- Pareja F., Ferraro D.A., Rubin C., et al. (2012). Deubiquitination of EGFR by Cezanne-1 contributes to cancer progression. Oncogene 31, 4599–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y.P., Zhu Y., Yin L.D., et al. (2017). PEG10 overexpression induced by E2F-1 promotes cell proliferation, migration, and invasion in pancreatic cancer. J. Exp. Clin. Cancer Res. 36, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux S., Debernardi A., Jacquiau B., et al. (2013). Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci. Transl. Med. 5, 186ra166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenfeld G.D., Caldwell E., Peabody E., et al. (2005). Incidence and outcomes of acute lung injury. N. Engl. J. Med. 353, 1685–1693. [DOI] [PubMed] [Google Scholar]
- Sacco O., Silvestri M., Sabatini F., et al. (2004). Epithelial cells and fibroblasts: structural repair and remodelling in the airways. Paediatr. Respir. Rev. 5(Suppl A), S35–S40. [DOI] [PubMed] [Google Scholar]
- Sahtoe D.D., and Sixma T.K. (2015). Layers of DUB regulation. Trends Biochem. Sci. 40, 456–467. [DOI] [PubMed] [Google Scholar]
- Savio M.G., Wollscheid N., Cavallaro E., et al. (2016). USP9X controls EGFR fate by deubiquitinating the endocytic adaptor Eps15. Curr. Biol. 26, 173–183. [DOI] [PubMed] [Google Scholar]
- Schneeberger E.E., and Karnovsky M.J. (1976). Substructure of intercellular junctions in freeze-fractured alveolar-capillary membranes of mouse lung. Circ. Res. 38, 404–411. [DOI] [PubMed] [Google Scholar]
- Schoenfeld A.R., Apgar S., Dolios G., et al. (2004). BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol. Cell. Biol. 24, 7444–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux O., and Dyson N.J. (2002). A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14, 684–691. [DOI] [PubMed] [Google Scholar]
- Suber T., Wei J., Jacko A.M., et al. (2017). SCFFBXO17 E3 ligase modulates inflammation by regulating proteasomal degradation of glycogen synthase kinase-3β in lung epithelia. J. Biol. Chem. 292, 7452–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara K., Tokumine J., Teruya K., et al. (2006). Alveolar epithelial cells: differentiation and lung injury. Respirology 11(Suppl), S28–S31. [DOI] [PubMed] [Google Scholar]
- Trimarchi J.M., and Lees J.A. (2002). Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3, 11–20. [DOI] [PubMed] [Google Scholar]
- Tsou A.P., Chuang Y.C., Su J.Y., et al. (2003). Overexpression of a novel imprinted gene, PEG10, in human hepatocellular carcinoma and in regenerating mouse livers. J. Biomed. Sci. 10, 625–635. [DOI] [PubMed] [Google Scholar]
- Wang B., Ma A., Zhang L., et al. (2015). POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat. Commun. 6, 8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Xiao Y., Hu Z., et al. (2008). PEG10 directly regulated by E2Fs might have a role in the development of hepatocellular carcinoma. FEBS Lett. 582, 2793–2798. [DOI] [PubMed] [Google Scholar]
- Weathington N.M., Snavely C.A., Chen B.B., et al. (2014). Glycogen synthase kinase-3β stabilizes the interleukin (IL)-22 receptor from proteasomal degradation in murine lung epithelia. J. Biol. Chem. 289, 17610–17619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire T.D., Lovejoy C.A., Wang T., et al. (2010). Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J. Biol. Chem. 285, 14565–14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C. (2014). Mechanisms for regulating deubiquitinating enzymes. Protein Sci. 23, 344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.C., Lin Y.C., Liu C.H., et al. (2014). USP11 regulates PML stability to control Notch-induced malignancy in brain tumours. Nat. Commun. 5, 3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Qin J., Zheng Y., et al. (2012). PEG10 promotes the migration of human Burkitt’s lymphoma cells by up-regulating the expression of matrix metalloproteinase-2 and -9. Clin. Invest. Med. 35, E117–E125. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Kimura J., Miki Y., et al. (2007). The deubiquitinating enzyme USP11 controls an IκB kinase α (IKKα)-p53 signaling pathway in response to tumor necrosis factor α (TNFα). J. Biol. Chem. 282, 33943–33948. [DOI] [PubMed] [Google Scholar]
- Yau R., and Rape M. (2016). The increasing complexity of the ubiquitin code. Nat. Cell Biol. 18, 579–586. [DOI] [PubMed] [Google Scholar]
- Yun S.I., Kim H.H., Yoon J.H., et al. (2015). Ubiquitin specific protease 4 positively regulates the WNT/β-catenin signaling in colorectal cancer. Mol. Oncol. 9, 1834–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Z’graggen B.R., Tornic J., Muller-Edenborn B., et al. (2010). Acute lung injury: apoptosis in effector and target cells of the upper and lower airway compartment. Clin. Exp. Immunol. 161, 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wei J., Dong S., et al. (2016). Destabilization of lysophosphatidic acid receptor 1 reduces cytokine release and protects against lung injury. EBioMedicine 10, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wei J., Mialki R.K., et al. (2012). F-box protein FBXL19-mediated ubiquitination and degradation of the receptor for IL-33 limits pulmonary inflammation. Nat. Immunol. 13, 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Zhang L., Wang A., et al. (2008). The association of GSK3β with E2F1 facilitates nerve growth factor-induced neural cell differentiation. J. Biol. Chem. 283, 14506–14515. [DOI] [PubMed] [Google Scholar]
- Zou C., Butler P.L., Coon T.A., et al. (2011). LPS impairs phospholipid synthesis by triggering β-transducin repeat-containing protein (β-TrCP)-mediated polyubiquitination and degradation of the surfactant enzyme acyl-CoA:lysophosphatidylcholine acyltransferase I (LPCAT1). J. Biol. Chem. 286, 2719–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.