Abstract
Introduction:
Cigarette smoking is inversely associated with DNA methylation of the aryl hydrocarbon receptor repressor (AHRR; cg05575921). However, the association between secondhand tobacco smoke (SHS) exposure and AHRR methylation is unknown.
Methods:
DNA methylation of AHRR cg05575921 in CD14+ monocyte samples, from 495 never-smokers and 411 former smokers (having quit smoking ≥15 years) from the Multi-Ethnic Study of Atherosclerosis (MESA), was cross-sectionally compared with concomitantly ascertained self-reported SHS exposure, urine cotinine concentrations, and estimates of air pollutants at participants’ homes. Linear regression was used to test for associations, and covariates included age, sex, race, education, study site, and previous smoking exposure (smoking status, time since quitting, and pack-years).
Results:
Recent indoor SHS exposure (hours per week) was inversely associated with cg05575921 methylation (β ± SE = −0.009 ± 0.003, p = .007). The inverse effect direction was consistent (but did not reach significance) in the majority of stratified analyses (by smoking status, sex, and race). Categorical analysis revealed high levels of recent SHS exposure (≥10 hours per week) inversely associated with cg05575921 methylation (β ± SE = −0.28 ± 0.09, p = .003), which remained significant (p < .05) in the majority of stratified analyses. cg05575921 methylation did not significantly (p < .05) associate with low to moderate levels of recent SHS exposure (1–9 hours per week), urine cotinine concentrations, years spent living with people smoking, years spent indoors (not at home) with people smoking, or estimated levels of air pollutants.
Conclusions:
High levels of recent indoor SHS exposure may be inversely associated with DNA methylation of AHRR in human monocytes.
Implications:
DNA methylation is a biochemical alteration that can occur in response to cigarette smoking; however, little is known about the effect of SHS on human DNA methylation. In the present study, we evaluated the association between SHS exposure and DNA methylation in human monocytes, at a site (AHRR cg05575921) known to have methylation inversely associated with current and former cigarette smoking compared to never smoking. Results from this study suggest high levels of recent SHS exposure inversely associate with DNA methylation of AHRR cg05575921 in monocytes from nonsmokers, albeit with weaker effects than active cigarette smoking.
Introduction
DNA methylation is a biochemical modification to the DNA that occurs without changes to the underlying genetic code and is typically investigated at cytosine-[phosphate]-guanine (CpG) dinucleotides. CpG methylation can be altered by various environmental and external exposures,1 including cigarette smoking.2–8 For instance, robust and significant reductions in average methylation levels of the aryl hydrocarbon receptor repressor (AHRR) gene CpG dinucleotide cg05575921 (located on chromosome 5: 373,378, hg19) have been reported in blood and lung tissues from current and former smokers compared to never-smokers.3,6,7 Methylation levels of cg05575921 were reported to be sensitive to less than one-half pack-year of smoking exposure3 and remain lower in former smokers decades after quitting compared to never-smokers.4,7 cg05575921 methylation levels in newborn cord blood were also found to inversely associate with maternal smoking during pregnacy.2
In our recent study8 of CD14+ monocytes, a cell type known to be affected by cigarette smoke,9 we reported inverse associations between short-term (cotinine, cigarettes per day) and long-term (pack-years) estimates of tobacco smoking with cg05575921 methylation, and direct associations between time since quitting in former smokers and cg05575921 methylation, confirming previous findings from whole blood and leukocyte samples. Moreover, we observed inverse associations between cg05575921 methylation and subclinical atherosclerosis, even after adjusting for other known cardiovascular risk factors. DNA methylation may play a regulatory role in the transcription of nearby genes.10 Intriguingly, lower levels of cg05575921 methylation observed in cigarette smokers correlate with increased gene expression of AHRR.6,8,11AHRR is known to be activated by aryl hydrocarbon receptor (AhR) pathway ligands12 such as polycyclic aromatic hydrocarbons present in active and secondhand tobacco smoking13–15 as well as other forms of air pollution such as vehicle exhaust.
Secondhand tobacco smoke (SHS) exposure is a major public health concern and an established risk factor for cardiovascular diseases, respiratory disease, and lung cancer.16–19 As SHS exposure during childhood is associated with an increased risk of cardiovascular disease in adulthood,17,20 alterations in DNA methylation profiles resulting from SHS exposure represent a potential functional mechanism that could contribute to SHS-related health risks that persist long after exposure. We hypothesized that (1) exposure to SHS can reduce AHRR methylation at cg05575921 and increase transcription of AHRR in nonsmokers, and (2) cg05575921 methylation may reflect acute and long-term SHS exposure in nonsmokers. The literature, however, is currently lacking reports of the relationship between SHS exposure and methylation of cg05575921. Here, we quantify the associations between SHS exposure and methylation of cg05575921 in CD14+ monocyte samples, using self-reported SHS exposures and urine cotinine concentrations from a subset of nonsmoking participants from a population-based cohort study in several cities across the United States, the Multi-Ethnic Study of Atherosclerosis (MESA). Additionally, we incorporate estimates of ambient fine particulate matter (PM2.5) and oxides of nitrogen (NOX) concentrations predicted at participants’ residences using environmental air monitoring measures, geographic variables, and land use information to further examine the influence of air pollution on cg05575921 methylation in nonsmokers.
Methods
Study Design and Data Collection
MESA was designed to investigate the prevalence, correlates, and progression of subclinical cardiovascular disease in a population cohort of 6814 participants. MESA enrolled participants aged 45–84 years, free of cardiovascular disease at baseline, from July 2000 to July 2002 at six field centers (Baltimore, Maryland; Chicago, Illinois; Los Angeles, California; New York; St. Paul, Minnesota; and Winston-Salem, North Carolina). Since its inception, five clinic visits collected extensive clinical, sociodemographic, lifestyle, behavior, laboratory, nutrition, and medication data.21 During the MESA exam 5 (April 2010–February 2012), the MESA Epigenomic and Transcriptomic Ancillary Study collected genome-wide methylomic and transcriptomic profiles in CD14+ purified monocytes from 1264 randomly selected MESA participants (47% Caucasian, 32% Hispanic, and 21% African American) from four MESA field centers (Baltimore, Maryland; New York; St. Paul, Minnesota; and Winston-Salem, North Carolina).22 Asian American participants of MESA were not included in this ancillary study due to the limited sample size within MESA. The study protocol was approved by the Institutional Review Board at each site. All participants signed informed consent.
SHS Exposure Assessment
Smoking status, age of smoking cessation, and pack-years of cigarette smoking were assessed via standard American Thoracic Society questionnaire items.23 Participants who reported smoking fewer than 100 cigarettes in their lifetimes were classified as never-smokers. Among participants who reported smoking greater than 100 cigarettes in their lifetime, those reporting not smoking during the last 30 days were initially classified as former smokers (for exclusion criteria, see Methods: Study Population). Pack-years of cigarette smoking was calculated from age of starting to quitting × (cigarettes per day/20). Self-reported SHS exposure included both recent SHS exposure, defined as the approximate number of hours per week in close contact with people smoking cigarettes indoors during the past year, and long-term SHS exposures, defined as the number of adult years with time spent indoors where there were people smoking cigarettes (not at home), and the number of adult years living with a cigarette smoker who smoked in the home. Urinary cotinine concentrations were measured via immunoassay24 (Immulite 2000 Nicotine Metabolite Assay; Diagnostic Products Corp., Los Angeles, CA) in a subsample of participants of the MESA Lung Ancillary Study (exam 5), including 688 nonsmoking participants of the MESA Epigenomic and Transcriptomic Ancillary Study (exam 5). Undetectable values of cotinine were assigned as 7.07ng/mL.
Air Pollution Assessment
PM2.5 and NOX concentrations were predicted using spatiotemporal models estimated by maximum likelihood and are available as 2-week averages from MESA Air for all participants from 1999 to 2012.25–28 Exposure models incorporated air pollution data from Environmental Protection Agency-operated Air Quality System monitors, monitors deployed by MESA Air in MESA community locations and participants’ homes, geographic data such as roadway density and land use, and dispersion model outputs.26,29 This modeling technique used the combined data to characterize seasonal and shorter-term time trends, key sources of spatial variability within the study communities, and underlying spatial and spatiotemporal correlation.26–28 For the present analyses, individual-level outdoor residential concentrations of PM2.5 and NOX were averaged over the 12 months prior to blood draw.
DNA Methylation Assessment
As described previously,22 monocytes were isolated with anti-CD14 monoclonal antibody–coated magnetic beads, using AutoMACs automated magnetic separation unit (Miltenyi Biotec, Bergisch Gladbach, Germany). Initially, flow cytometry analysis of 18 monocyte samples collected from all four MESA field centers was performed to assess the cell purification quality across the labs and technicians. The purity was >90% for all samples. DNA and RNA were isolated from samples simultaneously using the AllPrep DNA/RNA Mini Kit (Qiagen, Inc, Hilden, Germany). Illumina HumanMethylation450 BeadChips and HiScan reader were used to perform epigenome-wide methylation quantification.22 These methylation data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number (GSE56046).
Data preprocessing and quality control analyses were performed using R statistical software using Bioconductor packages. Bead-level methylation data were summarized in GenomeStudio. Because a two-channel system and both Infinium I and II assays were used, normalization was performed in several steps using the lumi package. Smooth quantile normalization was used to adjust for color bias. Next, the data were background adjusted by subtracting the median intensity value of the negative control probes. Lastly, data were normalized across all samples by standard quantile normalization applied to the bead-type intensities and combined across Infinium I and II assays and both colors. Quality control measures included checks for sex and race/ethnicity mismatches, and outlier identification by multidimensional scaling plots. To remove technical error in methylation levels associated with batch effects across the multiple chips, positional effects of the sample on the chip, we adjusted methylation M-values for chip and sample position on the chip.
RNA Sequencing
A subset of 374 samples was randomly selected from the MESA monocyte samples for RNA sequencing of mRNA as previously described.8
Study Population
To limit potential confounding effects of time since quitting with cg05575921 methylation,4 former smokers with smoking cessation dates within the past 15 years (119 former smokers) or missing (92 former smokers) were excluded from analysis. Overall, demographics were similar between former smokers included in the analysis and former smokers excluded due to missing quit dates. Other participants excluded from analysis included those with urine cotinine concentrations >100ng/mL, a threshold previously used to classify active smokers in MESA30 (five never-smokers and 18 former smokers), and participants missing covariates (two never-smokers). The resulting study population included 906 nonsmoking participants (48% Caucasian, 31% Hispanic, and 21% African American) of the MESA Epigenomic and Transcriptomic Ancillary Study with DNA methylation measured in monocytes.
Statistical Analysis
Using R statistical software, linear regression was used to examine the associations between recent and long-term SHS exposures (independent variables) and cg05575921 methylation (dependent variable). Independent variables investigated included: (1) SHS exposure status (“SHS exposed” defined as any self-reported nonzero recent or long-term SHS exposure, or having detectable cotinine), (2) self-reported hours per week of recent SHS exposure, (3) urine cotinine concentrations, (4) self-reported years of long-term SHS exposure at home, (5) self-reported years of long-term SHS exposure not at home, and (6) categories of recent exposure (no exposure: 0 hours per week, low–moderate: 1–9 hours per week, high: ≥10 hours per week). Two outliers for hours per week of SHS exposure greater than the total 168 hours in the week (two never-smokers) were excluded from the analysis. Methylation of cg05575921 was measured using the “M-value” (the log ratio of the methylated to the unmethylated intensity in monocyte samples,31 adjusted for chip and sample position on chip). Although associations were performed using cg05575921 M-values, M-values were transformed into the beta-value, an estimate of the percent methylation of an individual CpG site within a sample, that ranges from 0 to 1 [M is logit(beta-value)]31 in figures to aid interpretability of cg05575921 methylation levels. Model covariates included age, sex, ethnicity/race, smoking status, pack-years and time since quitting (former smokers only), education (three levels: no high school degree/diploma, high school degree/diploma, degree or certification in addition to a high school degree), study site, and estimated residual sample contamination with nonmonocyte cell types (neutrophils, B cells, T cells, and natural killer cells). Although the CD14+ purified monocyte samples were >90% pure, as described previously,22 we estimated residual sample contamination by leveraging transcriptomic measurements from the same monocyte samples to generate separate enrichment scores for neutrophils, B cells, T cells, monocytes, and natural killer cells. We implemented a gene set enrichment analysis32 to calculate the enrichment scores, using the gene signature of each blood cell type in the ranked list of expression values for each CD14+ sample. The cell type–specific signature genes were selected from previously defined lists33 and passed the following additional quality control filters: at least fourfold more highly expressed in the targeted cell type than in any other cell populations and low expression levels in monocytes.
For associations reaching a Bonferroni significance threshold (α = 0.05, Bonferroni corrected for multiple testing of six independent variables), secondary analyses were performed to investigate potential interactions between SHS exposure and cg05575921 methylation by smoking status, sex, race/ethnicity, and education levels. Also, to investigate the potential for population stratification, the first 10 principal components (derived from genotypic data as previously described34 and estimated using Eigenstrat35) were included as covariates in place of self-reported race/ethnicity. Analyses of PM2.5 and NOX (independent variables) and cg05575921 methylation (dependent variable) included the following covariates: age, sex, ethnicity/race, education (three levels), smoking status, hours of SHS exposure, pack-years and time since quitting (former smokers only), study site, estimated residual sample contamination with non-monocyte cell types, and chip and position on chip of Illumina 450K microarray.
We estimate 80% power to detect at least 1.4% variance in cg05575921 methylation explained by the number of hours per week in close contact with people smoking indoors (n = 889, SD = 5.1), 1.4% of methylation variance explained by the number of years living or around people smoking indoors (n = 890, SD = 11.3), and 1.8% variance in cg05575921 methylation explained by urine cotinine (n = 688, SD = 5.3), based on a power calculation using a simple linear regression (α = 0.05, Bonferroni corrected for multiple testing of six independent variables), performed using Quanto v1.2.
Results
The overall population characteristics of the 906 nonsmokers, aged 54–93 years, representing three race/ethnicities (48% Caucasian, 31% Hispanic, and 21% African American) are shown in Table 1. Approximately 60% of population (SHS exposed group) reported either recent or long-term SHS exposure, or had detectable urine cotinine suggestive of SHS exposure. Compared to the unexposed group, the SHS exposed group had more former smokers and Caucasian participants, and fewer participants without a high school degree or of Hispanic race/ethnicity. DNA methylation of cg05575921 in the SHS exposed group (n = 544, mean beta-value ± SD = 0.81 ± 0.07) was not significantly different than cg05575921 methylation in the SHS unexposed group (n = 362, mean beta-value = 0.82 ± 0.06) after adjusting for covariates including age, sex, race, smoking status, and pack-years of active smoking exposure (β ± SE = −0.015 ± 0.037, p = .68).
Table 1.
Characteristics of the Study Population
| Overall (N = 906) | SHS exposure | Smoking status | |||
|---|---|---|---|---|---|
| Unexposed (n = 362) | Exposed (n = 544) | Never (n = 495) | Former (≥15 y, n = 411) | ||
| Mean ± SD or % | Mean ± SD or % | Mean ± SD or % | Mean ± SD or % | Mean ± SD or % | |
| Demographic | |||||
| Mean age (y) | 70 ± 9 | 70 ± 9 | 71 ± 9 | 70 ± 10 | 71 ± 9 |
| Gender (% female) | 54 | 52 | 56 | 62 | 44*** |
| Former smokers (%) | 45 | 35 | 52*** | 0a | 100 |
| Education | |||||
| No HS degree (%) | 14 | 20 | 10*** | 17 | 11** |
| HS degree/diploma (%) | 37 | 29 | 42*** | 34 | 39 |
| >HS degree (%) | 49 | 51 | 48 | 49 | 50 |
| Race | |||||
| Caucasian (%) | 48 | 40 | 53*** | 44 | 53** |
| Hispanic (%) | 31 | 41 | 25*** | 34 | 28* |
| African American (%) | 21 | 19 | 22 | 22 | 19 |
| SHS exposure | |||||
| Recent (≥1h/wk, %) | 21 | 0a | 36 | 20 | 24 |
| Urine cotinine (>7.1ng/mL, %)b | 5.4 | 0a | 9.2 | 4.4 | 6.8 |
| Long-term in home (≥1 y, %) | 36 | 0a | 59 | 31 | 43*** |
| Long-term not at home (≥1 y, %) | 37 | 0a | 60 | 29 | 47*** |
HS = high school; SHS = secondhand tobacco smoke.
aCharacteristic used to define group; unexposed is defined as none of the four SHS exposures, the exposed group is defined as at least one of the four SHS exposures.
bAvailable in a subset of the population (n = 688).
***p < .001; **.001 ≤ p < .01; *.01 ≤ p < .05; SHS exposed group compared with unexposed group, former smokers compared with never-smoker group; t test to compare mean, and chi-square test to compare proportions.
Comparisons of the 495 never-smokers and 411 former smokers (all having quit smoking for at least 15 years) revealed never-smokers had higher frequencies of women and participants without a high school degree, and a lower frequency of long-term SHS exposure, compared to the former smokers. As previously reported,8 former smokers had significantly lower cg05575921 methylation compared to never-smokers (β ± SE = −0.20 ± 0.04, p = 3.8×10−6), even after adjusting for demographics and pack-years of active smoking history.
A statistically significant association was observed between the average number of hours per week of recent SHS exposure and cg05575921 methylation (n = 889, β ± SE = −0.009 ± 0.003, p = 7.05×10−3), as shown in Figure 1. No significant associations were observed between other continuous measures of SHS exposure and cg05575921 methylation, including urine cotinine concentrations as a biomarker of recent exposure (n = 688, β ± SE = −0.001 ± 0.004, p = .70) or long-term SHS exposure estimates: years of SHS exposure in home (n = 890, β ± SE = −0.0009 ± 0.0016, p = .56), and years of SHS exposure not at home (n = 875, β ± SE = −0.003 ± 0.002, p = .15). Estimated concentrations of PM2.5 and NOX were also investigated for association with cg05575921 methylation in nonsmokers; however, neither fine particulate matter (PM2.5: n = 646, β ± SE = −0.03 ± 0.04, p = .45) nor oxides of nitrogen (NOX: n = 646, β ± SE = −0.08 ± 0.08, p = .33) concentration estimates significantly (p < .05) associated with cg05575921 methylation, as shown in Supplementary Figure 1.
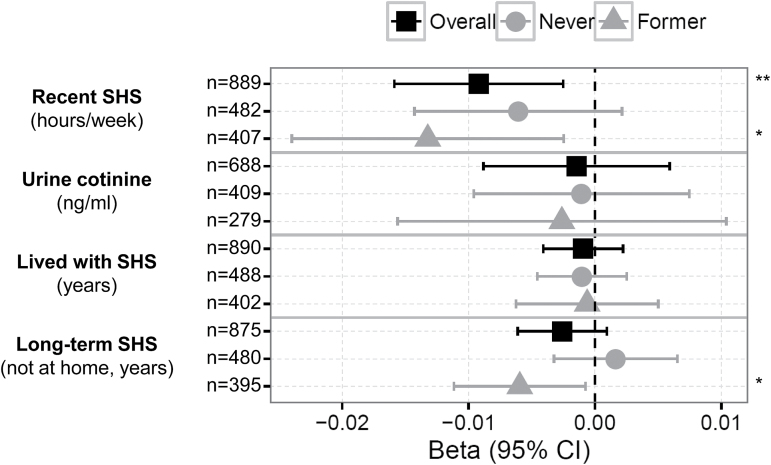
Figure 1.
Estimated effects of recent and long-term secondhand tobacco smoke (SHS) exposure on cg05575921 methylation in nonsmokers. Forest plots show SHS-related exposure effect sizes on cg05575921 methylation (x-axis, β and 95% confidence interval [CI]) measured in CD14+ monocyte samples, including recent SHS exposure (hours per week of close contact with indoor SHS, urine cotinine concentrations), and long-term SHS exposure (years exposed to SHS at home, and not at home). SHS-related effect sizes on methylation are shown overall and stratified by smoking status (never-smokers and former smokers having quit for at least 15 years). Covariates included age, sex, race/ethnicity, education level, smoking status, pack-years, time since quitting smoking (former smokers only), site of data collection, and estimates of residual sample contamination with non-monocyte cell types. Significance indicated as follows: **p < .01; *.01 ≤ p < .05.
The number of hours per week of recent SHS exposure uniquely explained 0.6% of the variance of cg05575921 methylation. Other than hours per week of recent SHS exposure, other covariates in the model also uniquely explained variation in cg05575921 methylation including: pack-years of active smoking (3.2%), smoking status (2.0%), sex (0.9%), age (0.7%), race (0.5%), and residual monocyte sample contamination with non-monocyte cell types (0.6%). Lower cg05575921 methylation significantly associated with: the number of pack-years of active smoking (β ± SE = −0.009 ± 0.001, p = 4.2×10−9), former smoker for at least 15 years versus never-smoker status (β ± SE = −0.20 ± 0.04, p = 3.3×10−6), male versus female (β ± SE = −0.12 ± 0.04, p = 1.4×10−3), older age (β ± SE = −0.006 ± 0.002, p = 2.6×10−3), African American race/ ethnicity compared to Caucasian (β ± SE = −0.10 ± 0.05, p = .04) and Hispanic (β ± SE = −0.15 ± 0.06, p = .01) race/ethnicity, and residual monocyte sample contamination with B cells (β ± SE = −0.6 ± 0.3, p = .02). Further analysis (1) utilizing principal components derived from genotypic data in place of self-reported ethnicity, and (2) excluding estimates of monocyte sample contamination did not alter the significant association between hours of SHS exposure and cg05575921 methylation.
The inverse relationship between the number of hours of SHS exposure and cg05575921 methylation had a statistically significant (p < .05) interaction by participant education level, but not by smoking status, sex, or race/ethnicity. In stratified analyses by education level, a significant (p < .05) inverse association was observed in subsets of participants without high school degrees/diplomas and with high school degrees/diplomas, but not in the subset of participants with additional degrees beyond a high school degree, as shown in Supplementary Figure 2A. The inverse relationship between hours per week of recent SHS exposure and cg05575921 methylation was consistent in stratified analyses by smoking status, sex, and race/ethnicity, although did not reach significance (p < .05) in many of the strata. For instance, Figure 1 illustrates the nominally significant effect sizes for the association between hours of recent SHS exposure and cg05575921 methylation among the former smokers (β ± SE = −0.013 ± 0.005, p = .015) but not among never-smokers (β ± SE = −0.006 ± 0.004, p = .15).
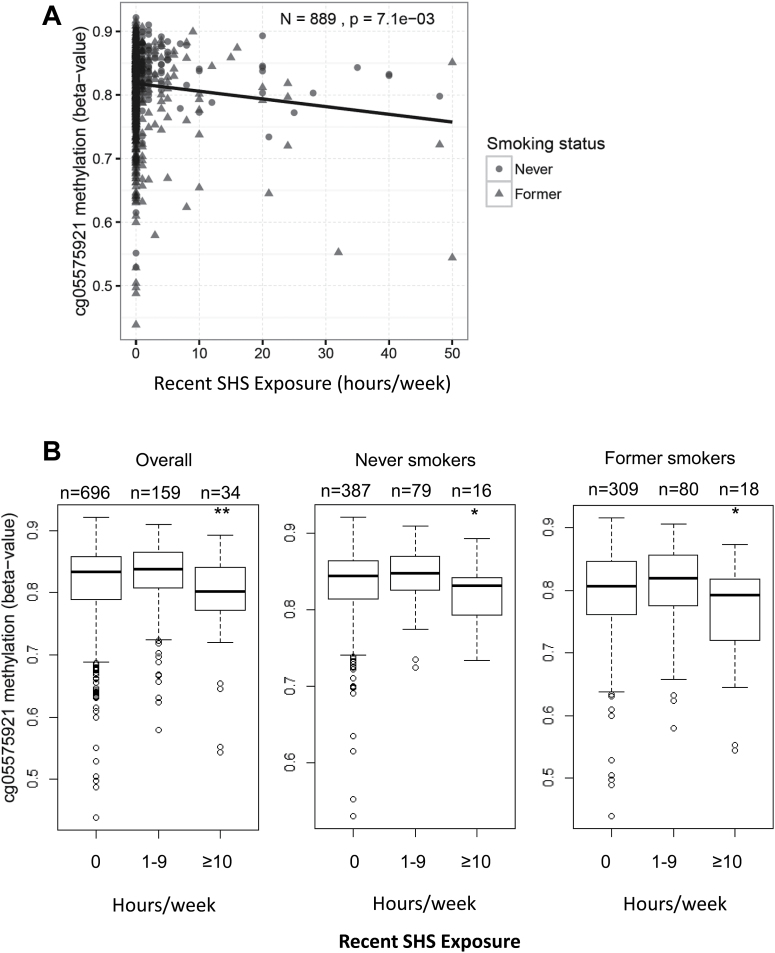
The number of hours per week of recent SHS exposure was weakly correlated with cg05575921 methylation (partial correlation = −0.09; 95% confidence interval: −0.16, −0.03), as shown in Figure 2A. Categorical analysis of recent SHS exposure (0 hours per week, 1–9 hours per week, and ≥10 hours per week), as shown in Figure 2B, revealed that participants reporting at high levels of recent SHS exposure (≥10 hours per week) had significantly lower cg05575921 methylation compared to participants reporting no recent SHS exposure (β ± SE = −0.28 ± 0.09, p = .0025). However, an inverse association was not detected between low to moderate levels of recent SHS exposure (1–9 hours per week) compared to no recent SHS exposure (β ± SE = 0.08 ± 0.04, p = .08). High levels of recent exposure to SHS (≥10 hours per week) were reported in 3.8% of the overall population, including 4.4% of former and 3.3% of never-smokers. High levels of recent SHS exposure remained significantly associated with lower cg05575921 methylation levels in both subsets of never-smokers (β ± SE = −0.24 ± 0.11, p = .033) and former smokers (β ± SE = −0.34 ± 0.15, p = .018) compared to those reporting no recent SHS exposure. The inverse relationship between high levels of recent SHS exposure and cg05575921 methylation was consistent across the majority of sex and race/ethnicity specific strata, although not among participants with additional education beyond high school, as shown in Supplementary Figure 2B. High levels of recent exposure were reported by 4.0% of participants without a high school degree/diploma, 6.2% of participants with a high school degree/diploma, and 2.0% of participants with degrees in addition to high school degree.
Figure 2.
Hours per week of recent indoor secondhand tobacco smoke (SHS) exposure and cg05575921 methylation in nonsmokers. (A) Scatterplot of cg05575921 methylation in CD14+ monocyte samples (y-axis: converted to beta-values) versus hours per week of recent indoor SHS exposure (x-axis). (B) Boxplot of cg05575921 methylation (y-axis, beta-value) by categories of recent SHS exposure per week (none: 0 hours, low–moderate: 1–9 hours, heavy: ≥10 hours), in all nonsmokers (overall), and stratified by smoking status (never-smokers and former smokers having quit for at least 15 years). Covariates included age, sex, race/ethnicity, education level, smoking status, pack-years, time since quitting smoking (former smokers only), site of data collection, and estimates of residual sample contamination with non-monocyte cell types; significance indicated as follows: **p < .01; *.01 ≤ p < .05.
Given the previous relationships reported between DNA methylation and transcription of nearby genes,10,22 we further examined the association between cg05575921 methylation and nearby gene expression (transcription start site within 1 MB of cg05575921) within the subset (n = 271) of the nonsmokers with both DNA methylation and RNA sequencing results. A nominally significant relationship was observed between cg05575921 methylation and AHRR mRNA expression (β ± SE = −0.31 ± 0.12, p = .013); however, no significant (p < .05) effect on AHRR expression was detectable with either hours per week of SHS exposure (β ± SE = −0.006 ± 0.013, p = .65) or high levels of SHS exposure (≥10 hours per week, β ± SE = −0.22 ± 0.34, p = .52) compared to participants reporting no recent SHS exposure.
Discussion
Results from this population-based cohort study of nonsmokers provide evidence for an inverse association between the number of hours recently in close contact with people smoking cigarettes indoors and DNA methylation of AHRR, particularly with high levels (≥10 hours per week) of recent SHS exposure. Suggestive inverse associations were observed across the majority of sex and race/ethnicity specific strata, but not in participants with degrees and certifications beyond high school. We also investigated associations between cg05575921 methylation and urine cotinine concentrations, low to moderate recent SHS exposure (1–9 hours per week), years spent with SHS exposure in home, years spent with SHS exposure not at home, and estimated concentrations of air pollutants (PM2.5 and NOX), although none of these associations reached statistical significance (p < .05).
While previous studies in humans have identified significant relationships between active smoking habits and DNA methylation measured in blood and lung tissues,5,6 as well as associations between maternal smoking habits during pregnancy and DNA methylation patterns in new born cord blood,2 no previous studies have reported associations between SHS exposure and DNA methylation in humans. In mice, one recent study found prenatal SHS exposure was associated with lower global methylation, as well as significant DNA methylation alterations to inflammatory mediators compared to controls.36 However, two other studies in mice reported no significant changes in DNA methylation or in gene expression in lung tissue of key epigenetic modifiers after SHS exposure.37,38 These studies were performed on a relatively small number of mice and may have been underpowered to detect effects of SHS exposure on DNA methylation, and also may have missed significant associations with DNA methylation profiles not examined by the microarrays used to measure DNA methylation.37
Our results in a human population support an inverse relationship between high levels (≥10 hours per week) of recent SHS exposure and DNA methylation of AHRR (cg05575921), but not between low to moderate (1–9 hours per week) levels of SHS exposure and cg05575921 methylation. The relationship between high levels of SHS exposure and cg05575921 methylation did not appear to be modified by smoking status, sex, or race/ethnicity; however, educational attainment was identified as a potential modifier. In analyses stratified by educational attainment, high levels of recent SHS exposure only significantly associated with lower cg05575921 methylation in the subsets of participants without degrees beyond a high school degree/diploma. One explanation for this could be that our analysis lacked the power necessary to detect the effect of SHS exposure on cg05575921 methylation in the subset of participants with degrees beyond a high school degree/diploma, as participants without a degree beyond a high school were more than twice as likely to report high levels of SHS exposure than participants with degrees beyond a high school degree/diploma.
Although our sample sizes were relatively small in stratified analyses, inverse associations between high levels of SHS exposure and AHRR methylation were detected in both subsets of never-smokers and former smokers having quit for at least 15 years. However, the estimated effect size of high levels of SHS exposure on cg05575921 methylation was larger in former smokers than in never-smokers, which could have resulted from an increased power to detect an effect in former smokers because of an increased prevalence of high levels of SHS exposure. Results from continuous analysis of the number of hours per week of recent SHS exposure and AHRR methylation revealed the correlation between hours of recent SHS exposure and cg05575921 methylation (r = −0.09) was weaker than the correlations we previously observed with cg05575921 methylation in current smokers with the number of cigarettes per day (r = −0.39) and pack-years of active smoking (r = −0.34).8 The correlation between recent SHS exposure and cg05575921 methylation was also weaker than the correlations in former smokers with pack-years active smoking (r = −0.35), and the number of years since quitting smoking (r = 0.35).8 In this study of nonsmokers, including never-smokers and former smokers having quit for at least 15 years, we found a larger proportion of variance of cg05575921 methylation uniquely explained by pack-years of active smoking and by former versus never smoking status than explained by recent SHS exposure. Combined, these results suggest that while high levels of SHS exposure may have similar effects on cg05575921 methylation in never-smokers and former smokers, previous active smoking exposure, even 15 years prior to the methylation measurement, may be a stronger influence on cg05575921 methylation levels than recent SHS exposure in former smokers.
Our results also indicate that cg05575921 methylation may not accurately reflect self-reported cumulative or long-term levels of SHS exposure. However, we cannot rule out the potential for cg05575921 methylation to reflect long-term SHS exposure, as we may have been underpowered in our analyses. Future longitudinal studies will be necessary to better understand the relationship between recent and long-term SHS exposure and cg05575921 methylation, as well as the potential reversibility of these modifications during periods of time without SHS exposure, which may be similar to observations of increasing cg05575921 methylation levels with time since quitting among former smokers.7,8
As DNA methylation is associated with alterations in transcription of nearby genes,22 and cg05575921 lies within a predicted poised enhancer in the AHRR gene,8 the biological implications of reduced cg05575921 methylation include alterations in AHRR expression. As expected based on our previous findings in current smokers,8 here we report significant associations between reduced cg05575921 methylation and increased AHRR expression in nonsmokers. Although we did not identify a significant association directly between recent SHS exposure and AHRR gene expression, a previous transcriptomic study of mice did report upregulated Ahrr gene expression in mice exposed to SHS compared to controls,39 supporting a potential role for AHRR in the response to SHS. AHRR is a transcription factor which is known to form a complex with the AhR nuclear translocator (ARNT) to repress the AhR signaling pathway and hypoxia-inducible factor (HIF)-dependent signaling.40 Given that AHRR can be upregulated by the AhR signaling pathway,12,41 a pathway known to regulate transcription of cytochrome P450 genes in response to environmental AhR ligands,41 we hypothesize that reduced methylation of AHRR associated with recent SHS exposure could reflect components of SHS activating the AhR pathway, such as polycyclic aromatic hydrocarbons or other harmful or potentially harmful constituents of SHS.14 Future studies of the effects of the individual components of SHS on AHRR cg05575921 methylation will be required to determine if a specific component of cigarette smoke and SHS may be functionally linked to methylation or gene expression alterations in humans.
It is known that differences in nicotine and carcinogen metabolism exist between race/ethnic groups.42,43 Intriguingly, in addition to the AhR signaling pathway being responsible for activation of phase I and II carcinogen metabolism genes,44 genetic variation within AHRR (rs2292596) has been associated with differences in induction of a carcinogen metabolism gene (CYP1A2).45 However, we did not observe a significant statistical interaction between SHS exposure and cg05575921 methylation by race, suggesting that either SHS exposure–related alterations in cg05575921 methylation may not differ by race or that there was insufficient power to detect such an interaction.
Limitations of this study include the cross-sectional nature of the SHS exposure and methylation measurements, and the potential for unaccounted variables which may also influence DNA methylation patterns. For instance, cg05575921 methylation may be influenced by other exposures (ie, wood burning exposure, previous active smoking in former smokers, air pollution), although no significant associations were detected between air pollution levels estimated at participants’ homes and cg05575921 methylation. Methylation is also known to vary by cell type46; therefore, DNA methylation measured in monocyte samples purified from peripheral blood may not be applicable to all cell types. Previous studies have found inverse associations between cg05575921 methylation and active smoking habits in multiple tissues, including whole blood and lung macrophages11,47 supporting the potential for these results to be valid in other cell types. In this study, the results did not appreciably change with and without the adjustment for residual sample contamination with non-monocyte cell types; however, estimates of B-cell contamination significantly associated with cg05575921 methylation, suggesting that cell type may be important to consider in future studies of SHS exposure and DNA methylation.
The power to detect a dose–response relationship between SHS exposure and cg05575921 methylation was limited by the small number of participants reporting recent SHS exposure greater than 10 hours per week, or with detectable urine cotinine. As serum cotinine is currently the preferred SHS biomarker, and chromatography-based analytical methods may be more sensitive and specific to cotinine compared to immunoassay,48 the use of urine cotinine concentrations ascertained by immunoassay was not ideal to quantify recent SHS exposure. Additionally, cotinine concentrations are subject to considerable individual variability in the rates of nicotine metabolism and elimination,49 and do not reflect long-term or cumulative SHS exposure, while self-reported SHS exposures are limited by recall biases and the potential lack of knowledge of SHS exposures by participants.50 Finally, this study was limited to methylation of one CpG dinucleotide within AHRR (cg05575921) that was previously found to be strongly modified by active smoking.2,5,6 Methylation profiles of hundreds of other CpG dinucleotides have been associated with active smoking47 and also have the potential to be associated with SHS exposure. Larger studies will be necessary to quantify SHS exposure associations with DNA methylation on the genome-wide scale.
In conclusion, in combination with previous findings of cg05575921 methylation as a potential dose-dependent biomarker of cigarette smoke exposure in current and former smokers, these findings support the ability of cg05575921 methylation to reflect high levels of recent SHS exposure. However, the effect of recent SHS exposure on cg05575921 methylation appears to be much weaker than the effects of active smoking. Larger studies are necessary to validate findings across different race/ethnic backgrounds and education levels, and to determine if there is a significant inverse association between long-term SHS exposure and DNA methylation.
Supplementary Material
Supplementary Figures 1 and 2 can be found online at http://www.ntr.oxfordjournals.org
Funding
This work was supported by the National Heart, Lung, and Blood Institute grant 1R01HL101250 to Wake Forest University Health Sciences, for the MESA Epigenomic and Transcriptomic Ancillary Study. Urinary cotinine measurements are available to the MESA study courtesy of MESA Lung contract R01-HL077612. The research described in this publication was funded in part by the US Environmental Protection Agency through RD831697 to the University of Washington (MESA Air); it has not been subjected to the Agency’s required peer and policy review and therefore does not necessarily reflect the views of the Agency and no official endorsement should be inferred. ANA was supported by National Institute of Environmental Health Sciences grant 1R01ES025216. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01- HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC- 95169, UL1-TR-001079, UL1-TR-000040, and DK063491.
Declaration of Interests
None declared.
Supplementary Material
Acknowledgments
The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions.
References
- 1. Christensen BC, Houseman EA, Marsit CJ, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Joubert BR, Håberg SE, Nilsen RM, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120(10):1425–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Philibert RA, Beach SR, Brody GH. Demethylation of the aryl hydrocarbon receptor repressor as a biomarker for nascent smokers. Epigenetics. 2012;7(11):1331–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zeilinger S, Kühnel B, Klopp N, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8(5):e63812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Philibert RA, Beach SR, Lei MK, Brody GH. Changes in DNA methylation at the aryl hydrocarbon receptor repressor may be a new biomarker for smoking. Clin Epigenetics. 2013;5(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shenker NS, Polidoro S, van Veldhoven K, et al. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22(5):843–851. [DOI] [PubMed] [Google Scholar]
- 7. Guida F, Sandanger TM, Castagné R, et al. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum Mol Genet. 2015;24(8):2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reynolds LM, Wan M, Ding J, et al. DNA methylation of the aryl hydrocarbon receptor repressor associations with cigarette smoking and subclinical atherosclerosis. Circ Cardiovasc Genet. 2015;8(5):707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bergmann S, Siekmeier R, Mix C, Jaross W. Even moderate cigarette smoking influences the pattern of circulating monocytes and the concentration of sICAM-1. Respir Physiol. 1998;114(3):269–275. [DOI] [PubMed] [Google Scholar]
- 10. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. [DOI] [PubMed] [Google Scholar]
- 11. Monick MM, Beach SR, Plume J, et al. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(2):141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haarmann-Stemmann T, Abel J. The arylhydrocarbon receptor repressor (AhRR): structure, expression, and function. Biol Chem. 2006;387(9):1195–1199. [DOI] [PubMed] [Google Scholar]
- 13. Lodovici M, Akpan V, Evangelisti C, Dolara P. Sidestream tobacco smoke as the main predictor of exposure to polycyclic aromatic hydrocarbons. J Appl Toxicol. 2004;24(4):277–281. [DOI] [PubMed] [Google Scholar]
- 14. Lee HL, Hsieh DP, Li LA. Polycyclic aromatic hydrocarbons in cigarette sidestream smoke particulates from a Taiwanese brand and their carcinogenic relevance. Chemosphere. 2011;82(3):477–482. [DOI] [PubMed] [Google Scholar]
- 15. Suwan-ampai P, Navas-Acien A, Strickland PT, Agnew J. Involuntary tobacco smoke exposure and urinary levels of polycyclic aromatic hydrocarbons in the United States, 1999 to 2002. Cancer Epidemiol Biomarkers Prev. 2009;18(3):884–893. [DOI] [PubMed] [Google Scholar]
- 16. Teo KK, Ounpuu S, Hawken S, et al. ; INTERHEART Study Investigators Tobacco use and risk of myocardial infarction in 52 countries in the INTERHEART study: a case-control study. Lancet. 2006;368(9536):647–658. [DOI] [PubMed] [Google Scholar]
- 17. West HW, Juonala M, Gall SL, et al. Exposure to parental smoking in childhood is associated with increased risk of carotid atherosclerotic plaque in adulthood: the Cardiovascular Risk in Young Finns Study. Circulation. 2015;131(14):1239–1246. [DOI] [PubMed] [Google Scholar]
- 18. Zhong L, Goldberg MS, Gao YT, Jin F. A case-control study of lung cancer and environmental tobacco smoke among nonsmoking women living in Shanghai, China. Cancer Causes Control. 1999;10(6):607–616. [DOI] [PubMed] [Google Scholar]
- 19. Jaakkola JJ, Jaakkola MS. Effects of environmental tobacco smoke on the respiratory health of children. Scand J Work Environ Health. 2002;28(suppl 2):71–83. [PubMed] [Google Scholar]
- 20. Mendelson MM, de Ferranti SD. Childhood environmental tobacco smoke exposure: a smoking gun for atherosclerosis in adulthood. Circulation. 2015;131(14):1231–1233. [DOI] [PubMed] [Google Scholar]
- 21. Bild DE, Bluemke DA, Burke GL, et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol. 2002;156(9):871–881. [DOI] [PubMed] [Google Scholar]
- 22. Liu Y, Ding J, Reynolds LM, et al. Methylomics of gene expression in human monocytes. Hum Mol Genet. 2013;22(24):5065–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ferris BG. Epidemiology Standardization Project (American Thoracic Society). Am Rev Respir Dis. 1978;118(6 pt 2):1–120. [PubMed] [Google Scholar]
- 24. Rodriguez J, Jiang R, Johnson WC, MacKenzie BA, Smith LJ, Barr RG. The association of pipe and cigar use with cotinine levels, lung function, and airflow obstruction: a cross-sectional study. Ann Intern Med. 2010;152(4):201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaufman JD, Adar SD, Allen RW, et al. Prospective study of particulate air pollution exposures, subclinical atherosclerosis, and clinical cardiovascular disease: the Multi-Ethnic Study of Atherosclerosis and Air Pollution (MESA Air). Am J Epidemiol. 2012;176(9):825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keller JP, Olives C, Kim SY, et al. A unified spatiotemporal modeling approach for predicting concentrations of multiple air pollutants in the Multi-Ethnic Study of Atherosclerosis and air pollution. Environ Health Perspect. 2015;123(4):301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sampson PD, Szpiro AA, Sheppard L, Lindstr+¦m J, Kaufman JD. Pragmatic estimation of a spatio-temporal air quality model with irregular monitoring data. Atmos Environ. 2011;45(36):6593–6606. [Google Scholar]
- 28. Szpiro AA, Sampson PD, Sheppard L, Lumley T, Adar SD, Kaufman J. Predicting intra-urban variation in air pollution concentrations with complex spatio-temporal dependencies. Environmetrics. 2009;21(6):606–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cohen MA, Adar SD, Allen RW, et al. Approach to estimating participant pollutant exposures in the Multi-Ethnic Study of Atherosclerosis and Air Pollution (MESA Air). Environ Sci Technol. 2009;43(13):4687–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lovasi GS, Diez Roux AV, Hoffman EA, Kawut SM, Jacobs DR, Jr, Barr RG. Association of environmental tobacco smoke exposure in childhood with early emphysema in adulthood among nonsmokers: the MESA-lung study. Am J Epidemiol. 2010;171(1):54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Du P, Zhang X, Huang CC, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abbas AR, Baldwin D, Ma Y, et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6(4):319–331. [DOI] [PubMed] [Google Scholar]
- 34. Fox CS, White CC, Lohman K, et al. ; CARDIoGRAM Consortium Genome-wide association of pericardial fat identifies a unique locus for ectopic fat. PLoS Genet. 2012;8(5):e1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. [DOI] [PubMed] [Google Scholar]
- 36. Lee JW, Jaffar Z, Pinkerton KE, et al. Alterations in DNA methylation and airway hyperreactivity in response to in utero exposure to environmental tobacco smoke. Inhal Toxicol. 2015;27(13):724–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tommasi S, Zheng A, Yoon JI, Li AX, Wu X, Besaratinia A. Whole DNA methylome profiling in mice exposed to secondhand smoke. Epigenetics. 2012;7(11):1302–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tommasi S, Zheng A, Besaratinia A. Expression of epigenetic modifiers is not significantly altered by exposure to secondhand smoke. Lung Cancer. 2015;90(3):598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tommasi S, Zheng A, Besaratinia A. Exposure of mice to secondhand smoke elicits both transient and long-lasting transcriptional changes in cancer-related functional networks. Int J Cancer. 2015;136(10):2253–2263. [DOI] [PubMed] [Google Scholar]
- 40. Karchner SI, Jenny MJ, Tarrant AM, et al. The active form of human aryl hydrocarbon receptor (AHR) repressor lacks exon 8, and its Pro 185 and Ala 185 variants repress both AHR and hypoxia-inducible factor. Mol Cell Biol. 2009;29(13):3465–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hahn ME, Allan LL, Sherr DH. Regulation of constitutive and inducible AHR signaling: complex interactions involving the AHR repressor. Biochem Pharmacol. 2009;77(4):485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pérez-Stable EJ, Herrera B, Jacob P, III, Benowitz NL. Nicotine metabolism and intake in black and white smokers. JAMA. 1998;280(2):152–156. [DOI] [PubMed] [Google Scholar]
- 43. Wang H, Park SL, Stram DO, et al. Associations between genetic ancestries and nicotine metabolism biomarkers in the Multiethnic Cohort Study. Am J Epidemiol. 2015;182(11):945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moorthy B, Chu C, Carlin DJ. Polycyclic aromatic hydrocarbons: from metabolism to lung cancer. Toxicol Sci. 2015;145(1):5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hung WT, Lambert GH, Huang PW, Patterson DG, Jr, Guo YL. Genetic susceptibility to dioxin-like chemicals’ induction of cytochrome P4501A2 in the human adult linked to specific AhRR polymorphism. Chemosphere. 2013;90(9):2358–2364. [DOI] [PubMed] [Google Scholar]
- 46. Kundaje A, Meuleman W, Ernst J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin Epigenetics. 2015;7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Avila-Tang E, Al-Delaimy WK, Ashley DL, et al. Assessing secondhand smoke using biological markers. Tob Control. 2013;22(3):164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Swan GE, Benowitz NL, Lessov CN, Jacob P, III, Tyndale RF, Wilhelmsen K. Nicotine metabolism: the impact of CYP2A6 on estimates of additive genetic influence. Pharmacogenet Genomics. 2005;15(2):115–125. [DOI] [PubMed] [Google Scholar]
- 50. Galán I, Mayo E, López MJ, et al. Validity of self-reported exposure to second-hand smoke in hospitality venues. Environ Res. 2014;133:1–3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.