Abstract
Although many genes that specify neocortical projection neuron subtypes have been identified, the downstream effectors that control differentiation of those subtypes remain largely unknown. Here, we demonstrate that the LIM domain-binding proteins Ldb1 and Ldb2 exhibit dynamic and inversely correlated expression patterns during cerebral cortical development. Ldb1-deficient brains display severe defects in proliferation and changes in regionalization, phenotypes resembling those of Lhx mutants. Ldb2-deficient brains, on the other hand, exhibit striking phenotypes affecting layer 5 pyramidal neurons: Immature neurons have an impaired capacity to segregate into mature callosal and subcerebral projection neurons. The analysis of Ldb2 single-mutant mice reveals a compensatory role of Ldb1 for Ldb2 during corticospinal motor neuron (CSMN) differentiation. Animals lacking both Ldb1 and Ldb2 uncover the requirement for Ldb2 during CSMN differentiation, manifested as incomplete CSMN differentiation, and ultimately leading to a failure of the corticospinal tract.
Keywords: brain development, corticospinal motor neurons, differentiation, Ldb
Introduction
The identification of key factors that specify neocortical pyramidal neuron subtypes, such as Satb2 (callosal; Alcamo et al. 2008; Britanova et al. 2008; Leone et al. 2015), Tbr1 (layer 6 corticothalamic; Hevner et al. 2001; Han et al. 2011; McKenna et al. 2011), and Fezf2 (layer 5 subcerebral; Chen B et al. 2005; Chen JG et al. 2005; Molyneaux et al. 2005; Shim et al. 2012), has dramatically increased our understanding of initial cell fate specification in the neocortex. However, it has become clear that initial specification is only the first step in a cascade of events that culminates in the manifestation of appropriate subtype-specific identity. The downstream regulatory mechanisms that control the differentiation of neuronal subtypes remain poorly understood. Here, we focus on corticospinal motor neurons (CSMNs) in layer 5B of sensorimotor areas, which control fine and precise voluntary movements. They provide a valuable model to study differentiation due to their thorough molecular characterization (Arlotta et al. 2005; Chen B et al. 2005; Chen JG et al. 2005; Molyneaux et al. 2005; Shim et al. 2012) and distinct axonal trajectory to the spinal cord via the corticospinal tract (CST; Jones et al. 1982). While genes such as Sox5 (Kwan et al. 2008), Ctip2 (Arlotta et al. 2005), CoupTF1 (Tomassy et al. 2010), and Bhlhb5 (Joshi et al. 2008) have been implicated in some aspects of CSMN differentiation, the genetic program that governs their maturation remains poorly understood. CSMNs are susceptible to damage and death in neurological conditions such as stroke, spinal cord injury, and neurodegenerative disorders such as amyotrophic lateral sclerosis. A comprehensive elucidation of the genetic cascade that drives CSMN specification and differentiation will thus support efforts to design optimal cell replacement therapies for the treatment of these debilitating conditions.
In this study, we describe the roles of Ldb adaptor proteins (LIM domain-binding proteins; also called Clim and NLI), which bind to LIM domains of LIM homeodomain (LIM-HD) and LIM-only (Lmo) proteins (Agulnick et al. 1996; Jurata et al. 1996; Bach et al. 1997). Ldb proteins have an intrinsic capability for dimerization, which allows LIM-HD proteins to interact with other LIM-HD proteins and/or regulatory proteins such as Otx, GATA, and bHLH (Wadman et al. 1994, 1997; Bach et al. 1997; Jurata and Gill 1997; Visvader et al. 1997; Breen et al. 1998; Meier et al. 2006). This capacity for dimerization enables LIM-HD proteins to form both homomeric and heteromeric complexes [reviewed in Matthews and Visvader (2003)].
There are 2 Ldb family members in vertebrates, 4 in zebrafish, and 1 each in Caenorhabditiselegans and Drosophila melanogaster [reviewed in Matthews and Visvader (2003)]. During cortical development, Ldb proteins may interact with the Lmo proteins Lmo3 and Lmo4, which are expressed in the developing cortex as early as E9.5 (Kenny et al. 1998; Sugihara et al. 1998; Bulchand et al. 2003). A germline deletion of Lmo4 leads to defects in neural tube closure (Tse et al. 2004; Lee et al. 2005) and perinatal lethality, while a neocortex-specific knockout of Lmo4 alters the formation of somatosensory barrel fields (Huang et al. 2009). A more recent study has shown that Lmo4 forms a complex with Ngn2 and Ldb1 that co-activates Ngn2-dependent transcription (Asprer et al. 2011). A second major group of interaction partners for Ldb proteins is the LIM-HD family of Lhx proteins. Lhx5-deficient animals show disruptions of hippocampal development (Zhao et al. 1999), and Lhx1/Lhx5 compound mutants show defects in Purkinje cell differentiation (Zhao et al. 2007). Lhx2 plays several important roles during corticogenesis: The deletion of Lhx2 results in neocortex hypoplasia and aplasia of the hippocampal anlagen (Porter et al. 1997), while Lhx2 also functions as a cortical selector gene that specifies the neuroepithelium to adopt a cortical fate (Mangale et al. 2008). In addition, Lhx2 specifies the regional fate of telencephalic neurons (Chou et al. 2009). Collectively, these studies suggest that Lhx and Lmo family members play varied and distinct roles during brain development.
To explore the role of Ldb proteins in cortical development, we took advantage of a conditional allele of Ldb1 (Zhao et al. 2007), which we combined with Emx1-Cre to limit recombination to the dorsal neocortex (Gorski et al. 2002). Using Emx1-Cre allowed us to bypass both the lethality of the Ldb1 germline null allele (Mukhopadhyay et al. 2003) and the early selector function of Lhx2 (Mangale et al. 2008). To address the possibility that Ldb1 and Ldb2 play redundant roles during cortical development, we also analyzed Ldb2 mutants and mice lacking both Ldb1 and Ldb2.
Our studies reveal that Ldb2 is upregulated specifically in layer 5 during development of the cerebral cortex. Ldb1 exhibits an inversely correlated expression pattern: As cortical development proceeds, its early widespread expression in the cortical plate is progressively excluded from layer 5, concomitant with the acquisition of Ldb2 expression. Ldb1-deficient mutants show a complex phenotype that includes proliferation defects and a fate change of lateral neocortex into piriform cortex, reminiscent of Lhx2 null mutants (Chou et al. 2009). Ldb2-deficient mice display a normal CST, but layer 5 neurons show a dramatic compensatory upregulation of Ldb1 and fail to properly segregate into callosal projection neurons (CPNs) and subcerebral projection neurons (SCPNs). The analysis of compound nulls reveals incomplete molecular differentiation of CSMNs and a failure of the CST at the pyramidal decussation.
Materials and Methods
Generation of Ldb2 Germline Null Allele
An FRT-neo cassette was cloned into the AspI sites of an 8.1-kb genomic Ldb2 fragment (see Supplementary Fig. 1B). The Ldb2-Neo fragment was excised using ClaI and SmaI, and then cloned into a pDTA vector using ClaI and EcoRV sites. PCR-positive embryonic stem cell clones were screened using Southern blots, a 5′ probe (3.3 kb) was used on AspI-cut DNA, and a 3′ probe (1.4 kb) was used on XbaI-cut DNA.
Animals
Mice hemizygous for either Nestin-Cre or Emx-Cre, carrying Ldb1lox/wt and Ldb20/wt, were crossed with Ldb1lox/lox;Ldb20/0 mice to obtain mutant and control littermates. For some experiments, animals were also hemizygous for either Z/EG or Golli-τ-EGFP. See Supplementary Methods for genotyping. The morning of the vaginal plug observed was considered as E0.5. For proliferation assays, pregnant females were injected with 1 mg BrdU (in PBS) 2 h prior to analysis. In utero electroporations were performed as previously described (Ohtsuka et al. 2001). Briefly, a plasmid-expressing Cre recombinase, driven by a CAG promoter, was in utero electroporated into the ventricles of E12.5 embryos.
Histology, Immunocytochemistry, Antibodies, and In Situ Hybridization
Standard methods of immunohistochemistry were used for immunocytochemical stainings. Animals were perfused with 4% paraformaldehyde in PBS, cryoprotected in 30% sucrose in PBS, and either embedded in OCT tissue-tek for cryosectioning or directly cut on a sliding microtome at 70–100 µm. For BrdU stainings, sections were immersed in 10 mM citric acid (pH 6.0) at 94 °C for 25 min before proceeding with standard immunohistochemistry. In situ hybridization was carried out as previously described (Frantz et al. 1994); see Supplementary Methods for the complete list of probes.
We used a rabbit-anti-Ldb1 antibody (gift of Dr Paul Love, National Institutes of Health), rat-anti-Ctip2 (Abcam), mouse-anti-TuJ1 (Covance), both mouse- and rabbit-anti-Satb2 (Abcam), rabbit-anti-Tbr2 (Abcam), rat-anti-BrdU (Accurate Chemical), rabbit-anti-Tbr1 (Abcam), goat-anti-Lhx2 (Santa Cruz Biotech), goat-anti-Lmo4 (Santa Cruz Biotech), both rabbit- and chicken-anti-GFP (Abcam), rat-ant-L1 (Millipore), and rabbit-anti-PKCγ (Santa Cruz Biotech). Alexa Fluor-coupled secondary antibodies were used to detect primary antibodies.
Confocal images were acquired on a Zeiss LSM 510 meta confocal microscope, and epifluorescent images were acquired on a Nikon 80i microscope with a Hamamatsu Orca ER camera. Images were postprocessed using ImageJ and Adobe Photoshop CS3.
Retrograde Tracings
For retrograde tracings from the cervical spinal cord, P28 animals were anesthetized with isoflurane and placed on a stereotactic set-up for surgery. Cervical spinal cord was surgically exposed and 0.2–0.3 µL of fluorescent-labeled latex beads (Lumafluor, Inc.) were injected into the ventral dorsal funiculus using a stereotactic injector (Stoelting). Animals were sacrificed 48–60 h after surgery to allow for transport of the beads.
Results
Ldb1 and Ldb2 Show Inversely Correlated Expression Patterns During Cortical Development
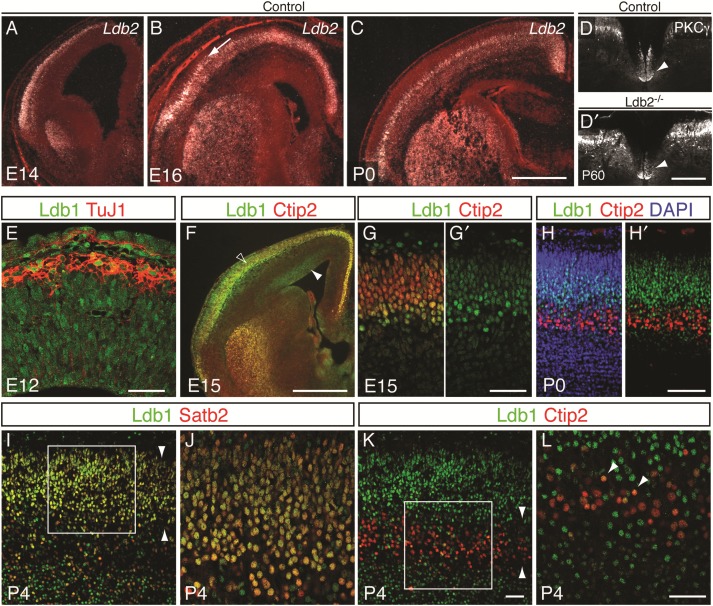
The Lim domain-binding protein Ldb2 (also known as Clim1 or NLI) is an excellent candidate for the study of CSMN differentiation. First, Ldb2 is expressed in the neocortex (Bulchand et al. 2003) in a pattern that overlaps with Fezf2, which is required for CSMN fate specification (Chen B et al. 2005; Chen JG et al. 2005; Molyneaux et al. 2005). Second, Ldb2 expression is abolished specifically in layer 5B of Fezf2−/− mice (Chen B et al. 2005; Molyneaux et al. 2005), implicating Ldb2 as a potential downstream effector of Fezf2 in the genesis of the CST. Third, there is a delay in the onset of Ldb2 expression in layer 5 neurons, suggesting a role in differentiation but not in initial specification of CSMN identity: in a microarray screen for genes expressed differentially in newly postmitotic neurons between E12.5 and E16.5, Ldb2 showed a dramatic increase in expression by E14.5, a time that correlates with the birth and migration of layer 5 neurons, with even more elevated expression by E16.5 (data not shown). These results were corroborated by in situ hybridization: at E12.5, Ldb2 expression is barely detectable in the neocortex (see Supplementary Fig. 1A and Bulchand et al. 2003). However, by E14.5, we detect a dramatic upregulation in layer 5B of the cortical plate (Fig. 1A) with robust expression at E16.5 (Fig. 1B) and P0 (Fig. 1C). Due to its restricted and delayed expression pattern in layer 5B, we hypothesized that Ldb2 might be involved in maturation but not in initial specification of CSMNs. To investigate this possibility, we examined the status of the CST in P60 mice lacking Ldb2 (see Supplementary Fig. 1B,C) by immunolabeling for protein kinase C γ (PKCγ), a specific marker for the CST (Mori et al. 1990). The analysis of cervical spinal cord cross-sections reveals an intact CST in Ldb2 mutants (Fig. 1D′) that is indistinguishable from controls (Fig. 1D).
Figure 1.
Inverse correlation of expression of Ldb1 and Ldb2 in layer 5 neurons. (A–C) In situ hybridization for Ldb2. High Ldb2 expression is first detected in the cortical plate at E14 (A) and robust expression is seen in layer 5B at E16 (B; arrow) and P0 (C). The corticospinal tract, labeled by PKCγ expression in the ventral dorsal funiculus (arrowheads in D and D′), is unaffected by loss of Ldb2 (D′) compared with controls (D). (E–L) Immunocytochemistry on coronal brain sections shows progressive exclusion of Ldb1 protein (green in E–L) from layer 5 neurons during cortical development. (E) At E12, Ldb1 is expressed by neurons of the cortical plate, labeled with TuJ1, and by progenitors lining the ventricular surface (arrowhead). (F) Low-power magnification shows Ldb1 expression at E15 in the VZ (arrowhead) and partial overlap with Ctip2 in the cortical plate (open arrowhead). (G and G′) High-power confocal imaging reveals partial exclusion of Ldb1 from Ctip2+ neurons while some Ctip2+ neurons still coexpress Ldb1. (H and H′) At P0, Ldb1 and Ctip2 expression have segregated to a large extent. (I) By P4, Ldb1 largely overlaps with Satb2. Arrowheads delineate upper layers 2–4. (J) High-power confocal imaging of boxed region in I shows virtually all Satb2+ neurons coexpressing Ldb1 (appearing yellow). (K) Doublestaining of Ldb1 and Ctip2 at P4 shows almost complete exclusion of Ldb1 from Ctip2+ layer 5 neurons (layer 5 delineated by arrowheads). (L) High-power confocal picture of boxed area in K confirms the absence of Ldb1 in Ctip2+ neurons, although a small fraction shows low levels of Ldb1 (arrowheads). Scale bars: 500 µm in (C) for (A–C); 300 µm in (D′) for (D,D′); 100 µm in (E), 500 µm in (F); 50 µm in (G′) for (G,G′); 100 µm in (H′) for (H,H′); 100 µm in (K) for (I,K); 100 µm in (L) for (J,L).
The lack of CST aberrations in Ldb2-null mice prompted us to examine potential functional redundancy and compensation of Ldb2 by its close family member Ldb1 during CSMN differentiation. Strikingly, Ldb1 is widely expressed across the cortical wall at E12.5 (Fig. 1E). At this age, newly generated neurons of layer 6 are migrating outward to populate the cortical plate. Immunocytochemical analyses reveal that Ldb1 is expressed both by neural progenitors that line the apical surface and by differentiated neurons in the cortical plate, as revealed by counterstaining with TuJ1 (Fig. 1E). However, during mid-corticogenesis, at E15.5, Ldb1 begins to become progressively excluded from layer 5B SCPNs: At E15.5, Ldb1 shows partial coexpression with Ctip2 (Fig. 1F), a transcription factor expressed by layer 5 SCPN (Arlotta et al. 2005, 2008), and hints of differential expression levels within layer 5 become obvious. While some Ctip2+ layer 5 neurons strongly express Ldb1, the majority shows a reduced expression level (Fig. 1G,G′), suggesting a downregulation of Ldb1 in Ctip2+ SCPNs. Indeed, by P0, expressions of Ctip2 and Ldb1 are mostly exclusive: Ldb1 is expressed by upper layer neurons and cells in layer 6 (Fig. 1H,H′). By postnatal day 4 (P4), when upper layer neurons have reached their final destination, Ldb1 is coexpressed with Satb2 (Fig. 1I,J), a marker for CPNs (Alcamo et al. 2008; Britanova et al. 2008; Leone et al. 2015), but is excluded from Ctip2+ neurons in layer 5 (Fig. 1K,L). These data reveal that Ldb2 and Ldb1 display inversely correlated expression patterns that suggest a specific requirement for Ldb2 in layer 5 SCPNs during differentiation.
Loss of Ldb1 Leads to Cortical Proliferation Defects
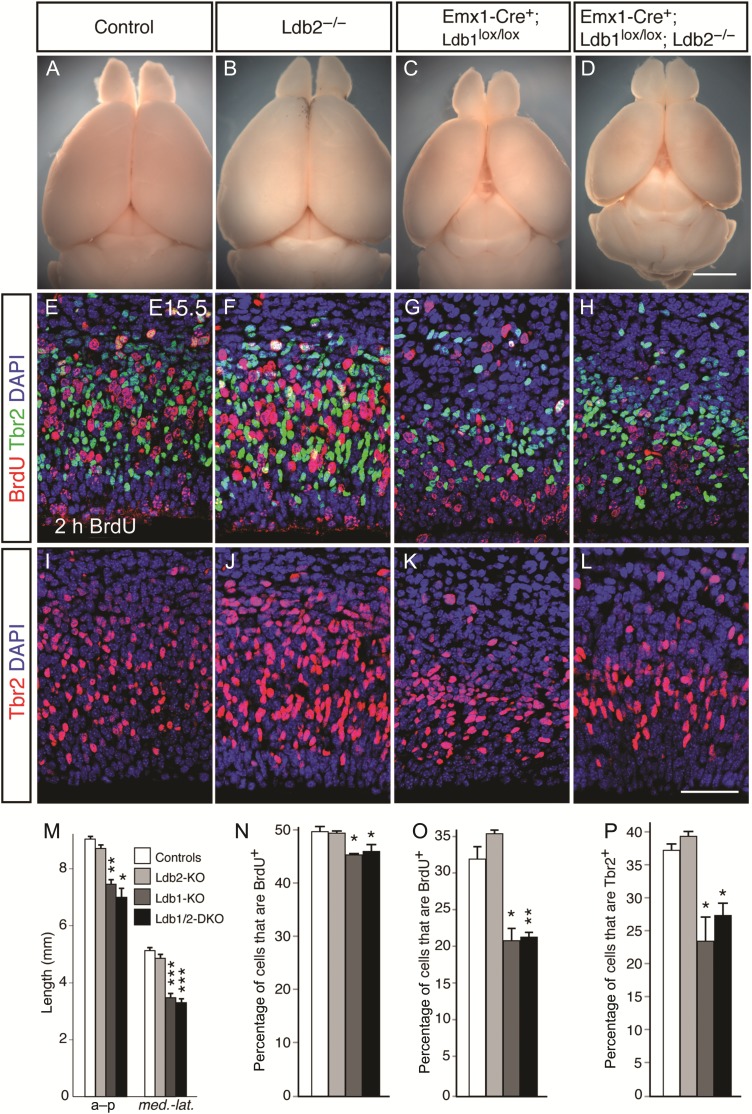
We generated a germline null allele of Ldb2 by replacing its third exon, which encodes part of the essential Lim-binding domain, with a neomycin cassette, thus leading to a truncated, nonfunctional Ldb2 allele (see Supplementary Fig. 1B,C and Methods). Ldb2-deficient animals (Ldb2-KO) do not display any obvious defects in the gross morphology of the brain (Fig. 2B). However, the cerebral hemispheres of Emx1-Cre+;Ldb1lox/lox mutants (Ldb1-KO) and Emx1-Cre+;Ldb1lox/lox;Ldb2−/− double knockouts (Ldb1/2-DKO) are strikingly smaller (Fig. 2C,D) than littermate controls (Fig. 2A), even though the mutants survive into adulthood, showing a statistically significant reduction in both anterior–posterior (Fig. 2E; a–p) and medio-lateral (Fig. 2E; med.-lat.) lengths. We reasoned that the size reduction in the cortical hemispheres could be due to defects in proliferation, differentiation, or cell survival.
Figure 2.
Proliferation defects in Ldb1-deficient mutants. (A–D) Whole mount brain preparations of 2-month-old animals show dramatic size reduction of cortical hemispheres in Ldb1-KO (C) and Ldb1/2-DKO (D) compared with controls (A) and Ldb2-KO (B). (E–L) Reduction in proliferative cells in Ldb1-KO and Ldb1/2-DKO. Two-hour BrdU pulse labeling at E15.5 reveals a dramatic reduction in the fraction of BrdU-labeled proliferating cells (red in E–H) in Ldb1-KO (G) and Ldb1/2-DKO (H) compared with control (E). No obvious change in BrdU incorporation was found in Ldb2-KO (F) compared with controls. (I–L) Immunocytochemical analysis for SVZ marker Tbr2 (red in I–L) shows striking reduction in the number of Tbr2+ progenitors in Ldb1-KO (K) and Ldb1/2-DKO (L) compared with control (I). (M) Quantification of cortical size reductions shows a statistically significant reduction in both anterior–posterior (a–p) and medio-lateral dimensions (med.-lat.) in Ldb1-KO and Ldb1/2-DKO compared with controls and Ldb2-KO. (N) Quantification of BrdU incorporation after a 2-h pulse at E12.5 shows a significant reduction in BrdU incorporation at E12.5 in Ldb1-KO and Ldb1/2-DKO compared with controls. (O) Reduction in BrdU+;Tbr2− progenitor fraction in VZ at E15.5 shows a reduction in the percentage of BrdU+ progenitor cells in both Ldb1-KO and Ldb1/2-DKO compared with controls. (P) Reduction in the percentage of Tbr2+ intermediate progenitors at E15.5 in Ldb1-KO and Ldb1/2-DKO compared with controls. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. Scale bars: 2.5 mm in (D) for (A–D); 50 µm in (L) for (E–L).
To measure progenitor proliferation, we injected pregnant females with BrdU at E12.5, sacrificed embryos 2 h later, and processed brain sections for BrdU immunohistochemistry. Quantification revealed a small but statistically significant reduction in BrdU incorporation at E12.5 in Ldb1-KO compared with controls (Fig. 1N; 45.3 ± 0.3% vs. 49.8 ± 1.0%; P = 0.02) and in Ldb1/2-DKO compared with controls (Fig. 1N; 46.2 ± 1.2% vs. 49.8 ± 1.0%; P = 0.02). To investigate whether radial glia or intermediate progenitor proliferation is affected by the loss of Ldb1/2, we analyzed brains at E15.5 for BrdU incorporation after a 2-h pulse. Colabeling with antibodies against Tbr2 allowed us to distinguish between radial glia progenitors (Tbr2−) and subventricular zone (SVZ) intermediate progenitors (Tbr2+; Englund et al. 2005). To determine the fraction of proliferating radial glia progenitors, we counted the number of BrdU+;Tbr2− cells between the apical ventricular surface and the dorsal border of the Tbr2+ domain (Fig. 2E–H). In control mice, 31.9 ± 1.4% of Tbr2− cells were BrdU+ (Fig. 2O; n = 3 animals per genotype). In Ldb2-KO animals, this fraction was 35.5 ± 0.1%, not significantly different from controls (P = 0.064). However, in Ldb1-KO and Ldb1/2 DKO mice, the fraction of BrdU+;Tbr2− cells was reduced significantly to 20.5 ± 2.7% (P = 0.017) and 21.6 ± 0.5% (P = 0.005), respectively. These data demonstrate that Ldb1 is an essential cofactor for proliferation in radial glia progenitors. No statistically significant difference was found between Ldb1-KO and Ldb1/2-DKO, suggesting that the loss of Ldb2 in an Ldb1-KO background does not lead to a further defect in proliferation (P = 0.373).
The findings that both Ldb1-KO and Ldb1/2-DKO mice show significant deficits in proliferating radial glia imply that their proliferative output may be affected by the loss of Ldb1. To determine whether production of radial glia-derived SVZ progenitors is compromised, we examined the number of Tbr2+ cells across the 4 genotypes at E15.5 (Fig. 2I–L). Both Ldb1-KO and Ldb1/2 DKO mice showed obvious deficits in the number of Tbr2+ cells (Fig. 2K,L, respectively). In controls, 36.9 ± 0.7% of cells between the ventricular surface and the dorsal border of the Tbr2+ domain expressed Tbr2 (Fig. 2P; n = 3 animals). In Ldb2-KO mice, 38.9 ± 0.4% of cells were Tbr2+ (P = 0.035; n = 3 animals), a small increase. Strikingly, in Ldb1-KO and Ldb1/2-DKO mice, these fractions were reduced significantly to 23.6 ± 4.2% (P = 0.043) and 27.4 ± 2.2% (P = 0.020), respectively.
These findings suggest that a reduced proliferative output by ventricular cells in the absence of Ldb1 does indeed compromise the generation of SVZ progenitors. To assess whether subsequent SVZ proliferation is affected, we counted the fraction of BrdU+ cells among the Tbr2+ progenitors following a 2-h pulse of BrdU. However, we did not find a statistically significant change in Ldb1-KO or Ldb1/2-DKO compared with controls (data not shown), suggesting that although the initial production of the SVZ pool is compromised in both mutants, the remaining population of SVZ progenitors proliferates normally.
Ldb1 can bind to Lmo4 and Neurogenin2 (Ngn2) in a protein complex that enhances the transcription of Ngn2-dependent targets such as Tbr2 (Asprer et al. 2011). Lmo4, in turn, is expressed by SVZ progenitors at E15.5 (Huang et al. 2009). It is conceivable that the absence of Ldb1 protein in Ldb1-KO and Ldb1/2-DKO mice disables the Lmo4/Ngn2 complex and leads to a reduction in downstream gene activation, including that of Tbr2. However, our observations suggest that a requirement for Ldb1 for Tbr2 expression does not seem to be absolute, given that Tbr2+ progenitors are not lost entirely in Ldb1-KO and Ldb1/2-DKO brains.
Lack of Hippocampus and Defects in Regionalization in Ldb1-Deficient Brains
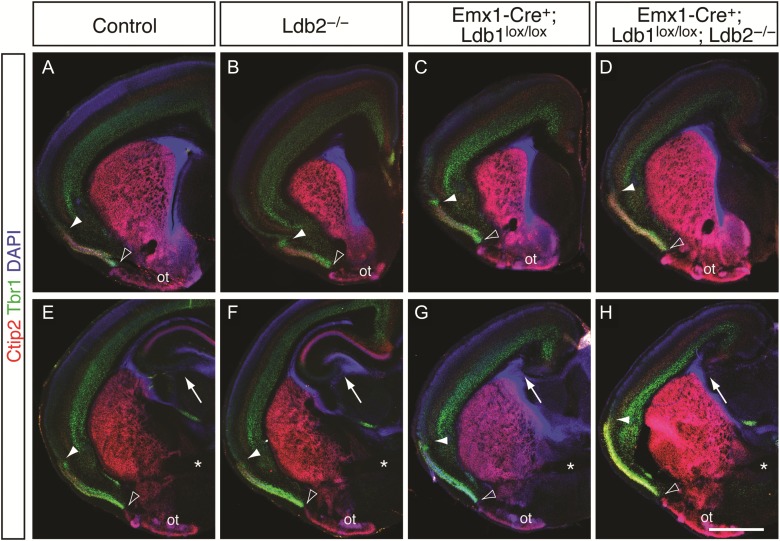
We next ascertained whether the production of specific cortical cell types in postnatal brains is affected by the loss of Ldb1 and/or Ldb2. Immunocytochemical analysis at P4 for Ctip2, which is expressed by layer 5 neurons, and Tbr1, a T-box transcription factor expressed by layer 6 cells (Hevner et al. 2001), revealed no obvious gross defects in the formation of layers 5 and 6 in Ldb2-KO mice (Fig. 3B,F) compared with controls (Fig. 3A,E). Despite their smaller sizes, Ldb1-KO and Ldb1/2-DKO brains showed a correct overall organization of layers 5 and 6 (Fig. 3C,D, respectively). However, these lines exhibited a loss of the hippocampal formation (Fig. 3G,H, respectively), a result anticipated by previous reports that the Ldb-interacting proteins Lhx5 (Zhao et al. 1999) and Lhx2 (Porter et al. 1997) are required for hippocampal development. It is plausible that the absence of Ldb1, which normally functions as an adaptor protein for Lhx transcription factors, renders both of these Lim homeobox proteins nonfunctional, leading to a failure of hippocampal specification in Ldb1-deficient brains. In accordance with this hypothesis, we observed reduced expression of signaling molecules such as Wnt5a and Bmp7, which are required by hippocampal precursor cells, in Ldb1-KO brains at E12.5 (data not shown). Lhx2 itself, which is expressed predominantly by neurons of layers 2–4 at P4, did not show any obvious changes in any of the mutants compared with controls (see Supplementary Fig. 3M–P), aside from a slight, qualitative reduction in expression which is likely due to the lower neuronal numbers in Ldb1-KO and Ldb1/2-DKO.
Figure 3.
Lack of hippocampus and expansion of piriform cortex into lateral neocortex in Ldb1-deficient mutants. (A–H) Neocortical markers Ctip2 (red in A–H) and Tbr1 (green in A–H), which are expressed in layers 5 and 6, respectively, are also expressed in layer 2 of piriform cortex (demarcated by open and filled arrowheads), and Ctip2 is robustly detected in the olfactory tubercle (ot). (A–D) Neocortical organization appears largely intact in Ldb mutants (B–D), but piriform cortex is expanded into lateral neocortex in Ldb1-KO (C) and Ldb1/2-DKO (D) compared with controls (A) and Ldb2-KO (B). Open arrowheads demarcate the ventral boundary of the piriform cortex, bordering the olfactory tubercle. Filled arrowheads indicate the dorsal/lateral boundary of the piriform cortex. (E–H) Sections at more posterior level show a similar expansion of piriform cortex, where the piriform cortex in Ldb1-KO (G) and Ldb1/2-DKO (H) dramatically extends into the lateral neocortex. The hippocampus in both Ldb1-KO and Ldb1/2-DKO (arrows in G,H) is absent compared with controls and Ldb2-KO (arrows in E,F). Posterior sections in E–H were matched using the anterior commissure (asterisks in E–H) as a landmark. Scale bars: 1.3 mm in (H) for (A–H).
Prior studies showed that the neocortex-specific deletion of Lhx2 leads to a fate conversion of neocortex into three-layered piriform cortex (Chou et al. 2009). The interaction of Ldb and Lhx proteins in this process generates the prediction that Ldb1-deficient brains should phenocopy this transformation. Indeed, the piriform cortex of mice lacking Ldb1 expands into lateral neocortex, reminiscent of Lhx2 knockout mice. In controls, piriform cortex, which borders the lateral neocortex at the rhinal fissure, is marked by strong immunoreactivity against Ctip2 and Tbr1 in layer 2, and the olfactory tubercle (Fig. 3A,E; Chou et al. 2009). While the piriform cortex of Ldb2-KO mice appears similar to controls (Fig. 3B,F), the markers expand into the lateral neocortex in Ldb1-KO mice (Fig. 3C,G) and in Ldb1/2-DKO brains (Fig. 3D,H). Fluorescent in situ hybridization for the piriform cortex-specific markers Liprin β1 (Ppfibp1) and Slc6a7 confirmed these findings: Both markers, which are normally not detected in the dorsal neocortex, are ectopically upregulated in the Ldb1-KO neocortex. High-power confocal microscopic analysis of motor cortex allowed us to perform colocalization with Satb2, revealing that control brains express no Liprin β1 (see Supplementary Fig. 2A,A′) in motor cortex, but Ldb1-KO mice show a striking expression of Liprin β1 (see Supplementary Fig. 2B,B′). Slc6a7 is detected at low levels in control motor cortex (see Supplementary Fig. 2C,C′), but is robustly upregulated in Ldb1-KO brains (see Supplementary Fig. 2D,D′). The vast majority of Liprin β1+ cells do not coexpress Satb2, suggesting that the Ldb1-deficient motor cortex houses 2 discrete cell populations—one piriform, the other neocortical—that are intermingled and coexist within a regionally hybrid area.
Compensatory Upregulation of Ldb1 in Ldb2−/− Ctip2+ Layer 5 Neurons
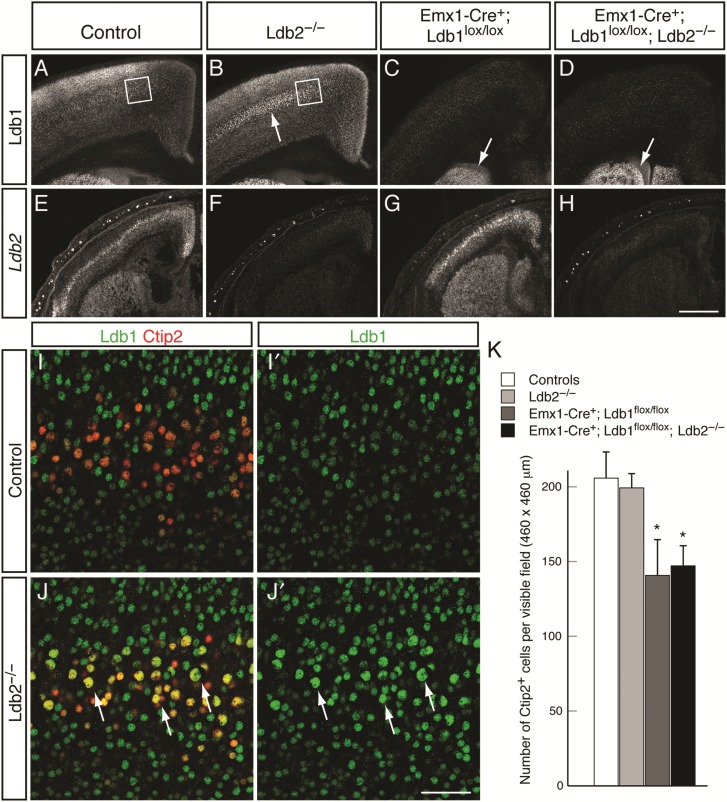
The progressive exclusion of Ldb1 from Ctip2+ layer 5 neurons is concomitant with the emerging expression of Ldb2 and is consistent with the idea that Ldb2 might repress Ldb1 expression in CSMNs. We thus hypothesized that the loss of Ldb2 results in the ectopic expression of Ldb1 in CSMNs, which can then compensate for Ldb2 function in CSMN differentiation. To test this hypothesis, we first assessed P4 coronal sections for Ldb1 expression. Low-power magnification reveals a striking upregulation of Ldb1 in layer 5 of Ldb2-KO (Fig. 4B) compared with controls (Fig. 4A). Both Ldb1-KO and Ldb1/2-DKO (Fig. 4C,D) show Emx1-Cre-dependent loss of Ldb1 in the neocortex, while striatal expression is maintained. Ldb2 expression, on the other hand, is abolished in both Ldb2-KO and Ldb1/2-DKO (Fig. 4F,H) compared with controls (Fig. 4E), but appears unchanged in Ldb1-KO (Fig. 4G), suggesting that Ldb2 cannot compensate for the loss of Ldb1. To further investigate the compensatory upregulation of Ldb1 in Ldb2 mutants, we performed high-power confocal microscopy for Ldb1 in Ctip2+ layer 5B SCPN at P4. In controls, the vast majority of Ctip2+ SCPN do not express Ldb1 (Fig. 4I,I′). However, in Ldb2 mutants, an overwhelming majority of Ctip2+ neurons were also Ldb1+ (Fig. 4J,J′), suggesting that the loss of Ldb2 leads to an ectopic, compensatory expression of Ldb1 in layer 5B SCPN.
Figure 4.
Compensatory upregulation of Ldb1 in Ldb2-deficient layer 5 neurons. (A–D) Immunocytochemical analysis of Ldb1 expression at P4 reveals striking upregulation of Ldb1 in layer 5 of Ldb2-KO neocortex (arrow in B) compared with control (A). Neocortex-specific loss of Ldb1 protein in pyramidal neurons of Ldb1-KO (C) and Ldb1/2-DKO (D). Remaining striatal expression of Ldb1 (arrows in C,D) confirms the specificity of the Emx1-Cre-dependent recombination. (E–H) In situ hybridization for Ldb2 confirms loss of Ldb2 mRNA in Ldb2-KO (F) and Ldb1/2-DKO (H), but no change in Ldb2 expression in Ldb1-KO (G) compared with control (E). (I–J′) High-power confocal analysis of layer 5 (boxed areas in A,B) confirms compensatory upregulation of Ldb1 (green in I–J′) in Ldb2-deficient Ctip2+ layer 5 neurons (red in J): In controls, Ctip2+ layer 5 neurons do not express Ldb1 (I,I′). Strikingly, in Ldb2-KO, almost all Ctip2+ neurons are coexpressing Ldb1 (yellow cells in J), including the large nucleated CSMN neurons (arrows in J,J′). A few Ctip2+ neurons are detected that do not express Ldb1 (arrows in J; appearing red in J); however, all of them appear to have small nuclei. (K) Quantification of the absolute number of Ctip2+ neurons across genotypes at P4. A statistically significant decrease in the number of Ctip2+ neurons was found in Ldb1-KO and Ldb1/2-DKO compared with controls; however, Ldb1/2-DKO do not show any further reduction from Ldb1-KO. *P ≤ 0.05. Scale bars: 500 µm in (H) for (A–H); 50 µm in (J′) for (I–J′).
Having shown that the loss of Ldb1 leads to proliferation defects, we wished to ascertain whether the absolute number of Ctip2+ SCPNs was affected. Quantification of the number of Ctip2+ neurons in the M1 motor area of 3 animals per genotype revealed no statistically significant changes in the number of Ctip2+ neurons in Ldb2-KO mice compared with controls [Fig. 4K; 206 ± 17 cells per visible field (controls) vs. 199 ± 10 (Ldb2-KO; P = 0.38)]. In agreement with the proliferation defects observed (Fig. 2), both Ldb1-KO and Ldb1/2-DKO brains showed statistically significant decreases in the number of Ctip2+ neurons [141 ± 24 (Ldb1-KO; P = 0.047) and 147 ± 13 (Ldb1/2-DKO; P = 0.02)]. Importantly, we did not detect a further reduction in the numbers of Ctip2+ neurons in Ldb1/2-DKO compared with Ldb1-KO mice (P = 0.42), suggesting that the additional loss of Ldb2 in an Ldb1-KO background does not adversely affect survival of Ctip2+ neurons.
Impaired Segregation of Layer 5 Neurons in Ldb2−/− Mice
The upregulation of Ldb1 in Ldb2-KO layer 5B SCPNs motivated us to further examine this population of neurons. Recent work by Macklis and colleagues suggests that Ldb2 and Lmo4 expression can be used as markers to progressively demarcate layer 5 neurons as they differentiate toward either a SCPN or CPN fate, respectively (Azim et al. 2009). Although Lmo4 and Ldb2 are coexpressed by many layer 5 neurons at mid-corticogenesis, the 2 markers segregate into 2 distinct populations with little overlap by P6. We hypothesized that if Ldb2 is required for immature layer 5 neurons to acquire a SCPN identity, then loss of Ldb2 might interfere with this process.
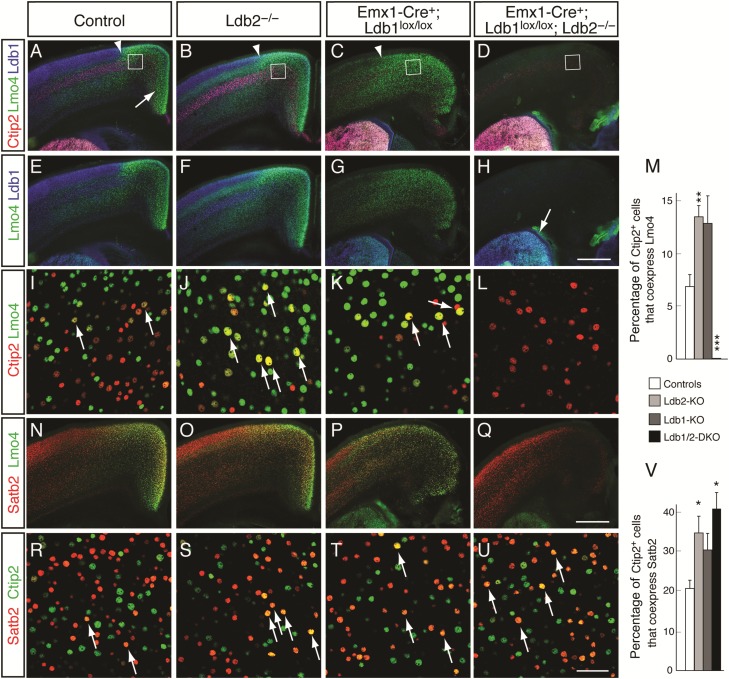
At P4, Lmo4 is highly expressed in motor cortex of wild-type controls (Fig. 5A,E) within layer 5, delineated using Ctip2 (in red, Fig. 5A–D), and in layer 6. In agreement with previous reports, Lmo4 is also detected in the upper layers of somatosensory cortex (Bulchand et al. 2003; Huang et al. 2009). Ldb1 is most strongly expressed by upper layer neurons of somatosensory cortex, and more weakly in motor and cingulate cortex (Figs 4A and 5A,E). In Ldb2-KO mice, we detected a subtle upregulation in Lmo4 expression in layers 4 and 5 at low-power magnification (Fig. 5B,F); however, high-power confocal pictures from layer 5 of motor cortex revealed a dramatic increase in the coexpression of Lmo4 with Ctip2 (Fig. 5J) compared with controls (Fig. 5I). Quantification revealed a statistically significant increase from 6.9 ± 1.1% of Ctip2+ cells coexpressing Lmo4 (controls) to 13.5 ± 1.1% (Ldb2-KO; P = 0.002; n = 4; Fig. 5M). Unexpectedly, we find striking changes in Lmo4 expression in Ldb1-KO (Fig. 5C,G,K) and Ldb1/2-DKO mice (Fig. 5D,H,L): In Ldb1-KO brains, the motor/somatosensory boundary appears shifted laterally, and Lmo4 expression is elevated diffusely over the cortical wall (Fig. 5C). In Ldb1/2-DKO mice, Lmo4 expression is completely abolished across the neocortex (Fig. 5D,H,L; 0% Lmo4/Ctip2 coexpression; P = 0.0001; n = 4), while striatal expression remains unaffected (Fig. 5H), due to the Emx1-Cre specificity to the neocortex.
Figure 5.
Impaired segregation of immature layer 5 neurons in the absence of Ldb2. (A–L) Immunocytochemical analysis shows striking Ldb1/2-dependent changes in Lmo4 expression. Lmo4 expression (green in A–L) is high in the cingulate cortex of controls (arrow in A), extends into motor cortex, and shows lower expression levels in layers 2/3 lateral to the motor/somatosensory boundary (arrowheads in A–C). Lmo4 is also expressed by subsets of neurons of layers 4–6. Ctip2 (red in A–D) was used to mark layer 5 neurons and Ldb1 (blue in A–H) as a reference marker. Low-power magnification reveals a subtle upregulation of Lmo4 in layers 4 and 5 of Ldb2-KO (B,F,J; arrows in J) compared with controls (A,E,I), while Ldb1-KO displays a striking ectopic upregulation of Lmo4 across the cortical wall (C,G,K). Lmo4 is completely lost in the neocortex of Ldb1/2-DKO (D,H,L), but striatal expression of both Ldb1 and Lmo4 (arrow in H) is maintained due to Emx1-Cre's neocortex specificity. (I–L) High-power confocal pictures of M1/M2 motor area of layer 5 (boxed in A–D) show colocalization between Ctip2 (red in I–L) and Lmo4 (green in I–L). The number of Ctip2+;Lmo4+ double-positive neurons (yellow in I–L) is increased in Ldb2-KO (J) and Ldb1-KO (K) compared with controls (I). Cortical Lmo4 immunoreactivity is abolished in layer 5 of Ldb1/2-DKO (L). (M) Quantification of Ctip2/Lmo4 coexpression reveals a statistically significant increase in coexpression in layer 5 neurons of Ldb2-KO compared with controls, and a dramatic loss in Ldb1/2-DKO. (N–U) Colabeling with a CPN marker Satb2 (red in N–U) reveals partial overlap between Lmo4 (green in N–Q) and Satb2 in controls (N) and Ldb2-KO brains (O). Satb2 expression is qualitatively reduced in Ldb1-KO (P) and Ldb1/2-DKO (Q), but Satb2+ CPNs are present in Ldb1/2-DKO (Q), suggesting that these neurons have downregulated Lmo4. (R–U) Colabeling of Satb2 and Ctip2 shows very few Ctip2+;Satb2+ (yellow in R) double-positive neurons in layer 5 motor cortex of controls (arrows in R). An increased fraction of Ctip2+ neurons coexpress Satb2 in Ldb2-KO (arrows in S) and Ldb1-KO (T). A further increase in Ctip2+;Satb2+ double-positive layer 5 neurons (arrows in U) is found in motor cortex of Ldb1/2-DKO (U). (V) Quantification of Ctip2/Satb2 coexpression shows statistically significant increases in the fraction of double-positive layer 5 neurons in Ldb2-KO and Ldb1/2-DKO compared with controls. *P ≤ 0.05; **P ≤ 0.01; ***P ≤0.001. Scale bars: 500 µm in (H) for (A–H) and in (Q) for (N–Q); 50 µm in (U) for (I–L) and (R–U).
The striking loss of cortical Lmo4 expression in Ldb1/2-DKO mice could be the result of a downregulation of Lmo4 (with the neurons themselves still present), or a loss of the population of Lmo4+ neurons. To distinguish between these possibilities, we performed immunocytochemistry for the CPN marker Satb2. The analysis of control (Fig. 5N) and Ldb2-KO mice (Fig. 5O) revealed partial overlap between Satb2 and Lmo4, and a qualitative reduction in Satb2 immunoreactivity in Ldb1-KO brains (Fig. 5P). Given the proliferation defects seen in Ldb1 mutants, the latter is likely due to a global reduction in the neuronal numbers (Fig. 2). Ldb1/2-DKO mice showed no further reduction in Satb2 expression (Fig. 5Q), suggesting that Satb2+ neurons are still present—albeit at reduced numbers—but have indeed downregulated Lmo4.
To further investigate the failure in segregation of layer 5 neuronal subtypes, we analyzed high-power confocal pictures for colocalization of Satb2 and Ctip2. We hypothesized that the impaired segregation should also manifest itself in an increase in Satb2/Ctip2 colocalization in layer 5 motor cortex neurons. Quantification indeed revealed that in motor cortex of Ldb2-KO mice, 34.5 ± 4.6% of Ctip2+ cells coexpressed Satb2, compared with 20.4 ± 1.7% in controls (Fig. 5R,V; n = 3, P = 0.045). Ldb1-KO mice showed a nonsignificant increase in coexpression to 30.0 ± 5.3% (Fig. 5T,V; n = 3, P = 0.192). We found that 41.1 ± 5.1% of Ctip2+ cells coexpress Satb2 in Ldb1/2-DKO animals (Fig. 5U,V; n = 3, P = 0.018), a statistically significant increase compared with controls. The fact that we did not observe a further increase in Satb2/Ctip2 coexpression in Ldb1/2-DKO compared with Ldb2-KO mice (P = 0.393) implies that the loss of Ldb1 does not compound the segregation defect observed in Ldb2-KO mutants. Taken together, the data suggest that Ldb2 is required by immature layer 5 neurons to segregate into CPNs and SCPNs, and that the upregulation of Ldb1 in Ldb2-KO Ctip2+ neurons fails to functionally compensate for the loss of Ldb2.
Incomplete Differentiation of CSMNs in Ldb1/2-DKO Mutants
The results above show that loss of Ldb2 impairs the early differentiation of layer 5 neurons, and suggests that it might affect CSMN differentiation and ultimately their ability to form cortical efferents. To further investigate the potential compensatory actions of ectopic Ldb1 expression in Ldb2−/− mutants, we evaluated CSMN differentiation. Significantly, none of the single or compound mutants show changes in the expression of layer 5B SCPN-enriched transcription factors Fezf2 (see Supplementary Fig. 3A–D) or Ctip2 (see Supplementary Fig. 3E–H), confirming that Ldb proteins do not regulate initial specification or migration of layer 5B SCPN.
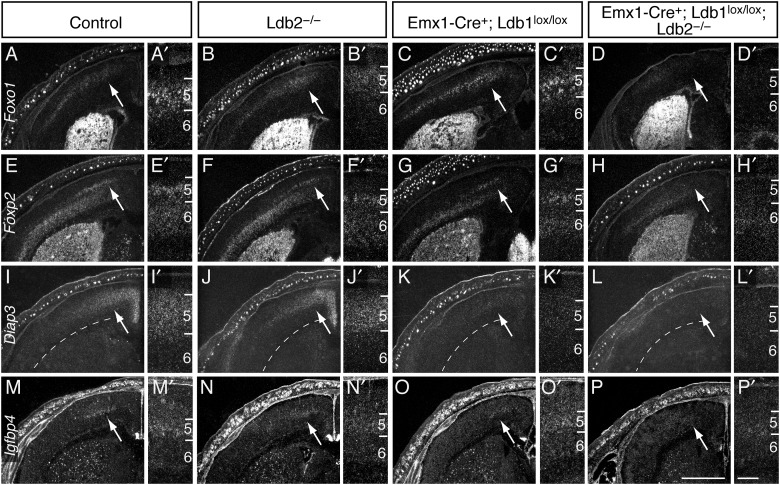
Despite appropriate initial specification and survival, CSMNs exhibit incomplete molecular differentiation in compound mutants. In situ hybridization on coronal P1 sections reveals a loss of the CSMN-enriched genes Foxo1 (Fig. 6A–D′), Foxp2 (Fig. 6E–H′), and Diap3 (Fig. 6I–L′) in Ldb1/2-DKO mice. Notably, Foxp2, which is also expressed by layer 6 neurons, is abolished in layer 5B motor cortex of Ldb1/2-DKO brains, while layer 6 expression is unaffected, in support with the idea that loss of Ldb1/2 specifically affects layer 5 CSMN differentiation. Expression of Igfbp4, which is normally upregulated during terminal CSMN differentiation, is also abrogated in Ldb1/2-DKO mice (Fig. 6M–P′). Similarly, Lmo3, one of the potential binding partners for Ldb proteins, shows robust expression in layer 5 cells of controls and Ldb2-KO mice (see Supplementary Fig. 3I,J′) at E15.5; however, Lmo3 expression is completely abolished in the neocortex of Ldb1/2-DKO brains (see Supplementary Fig. 3L,L′), similar to Lmo4. The loss of differentiation markers specifically in Ldb1/2-DKO mutants suggests that the compensatory upregulation of Ldb1 in layer 5B SCPN masks a phenotype in Ldb2 single mutants.
Figure 6.
Incomplete molecular differentiation of CSMNs in Ldb2-deficient mutants. In situ hybridization on coronal P1 sections reveals loss of the differentiation markers Foxo1 (A–D′), Foxp2 (E–H′), Diap3 (I–L′), and Igfbp4 (M–P′) in layer 5 of motor cortex (arrows) in Ldb1/2-DKO mice. Note that layer 6 expression of Foxp2 is not affected by the loss of Ldb genes, confirming the specific roles of Ldb1/2 in layer 5 neurons. Scale bars: 1 mm in (P, main panels), 0.2 mm in (P′, inset panels).
CSMNs in Ldb1/2-DKO Mice Fail to Extend Their Axons Past the Pyramidal Decussation
The impairment in CSMN differentiation in Ldb1/2-DKO mutants prompted us to further investigate CSMN maturation. In particular, we wished to examine whether the differentiation defect would manifest itself at the level of axonal targeting to the spinal cord. We therefore analyzed CST formation in all 4 lines. To investigate subcortical projection pathways, we took advantage of the Z/EG Cre-dependent EGFP reporter line, which expresses EGFP upon Cre-mediated recombination (Novak et al. 2000). This system, combined with the Emx1-Cre allele, allowed us to label cortical efferent tracts such as the CST using EGFP.
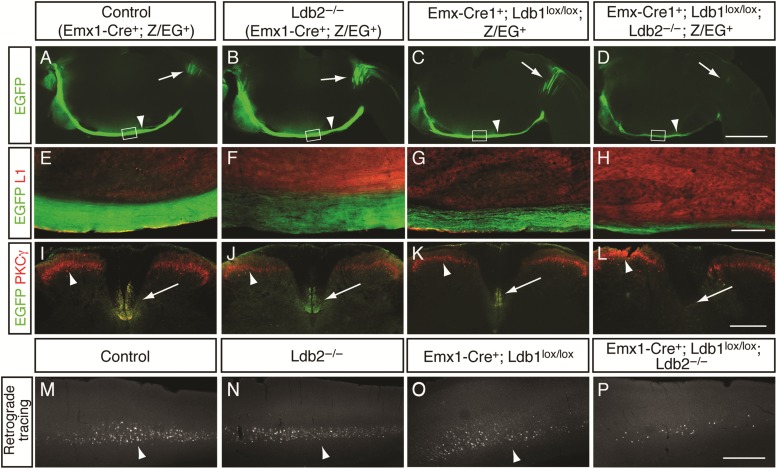
At P4, EGFP+ CST axons extend along the brainstem and cross into the dorsal funiculus of the spinal cord at the pyramidal decussation in control mice (Fig. 7A). The tract appears intact in both Ldb2-KO and Ldb1-KO brains (Fig. 7B,C, respectively); however, there is a striking failure of the CST at the pyramidal decussation in Ldb1/2-DKO mice (Fig. 7D). Among 8 Ldb1/2-DKO mutants examined, no EGFP+ fibers were detected distal to the pyramidal decussation. High-power magnifications of the brainstem (boxed areas in Fig. 7A–D) revealed robust EGFP staining in the CST of controls (Fig. 7E) and Ldb2-KO mice (Fig. 7F). Ldb1-KO brains show a modest decrease in tract size and the tract appears defasciculated at this level (Fig. 7G), while Ldb1/2-DKO animals show a further reduction in tract size (Fig. 7H).
Figure 7.
Failure of corticospinal tract in Ldb1/2-DKO mutants at the pyramidal decussation. (A–L) Emx1-Cre-dependent EGFP expression of the Z/EG reporter was used to specifically label neocortical pyramidal neurons and their efferent projections. (A–D) Sagittal brain sections of P4 animals reveal EGFP labeling in the CST along the brainstem (arrowheads in A–D), and the pyramidal decussation (arrows in A–D) of controls (A), Ldb2-KO (B), and Ldb1-KO (C). (D) In Ldb1/2-DKO, however, the tract fails at the decussation, and only very few crossing axons are detected (arrow in D). (E–H) High-power magnification of sagittal sections through the CST along the brainstem (boxed in A–D), counterstained with L1 (red in E–H), and reveals robust Z/EG-EGFP labeling in the CST of controls (E) and Ldb2-KO (F). A reduction in size of the tract is observed in Ldb1-KO (G), in agreement with the reduced number of Ctip2+ neurons in Ldb1-KO (Fig. 4). A further reduction in tract size is observed in Ldb1/2-DKO (H). (I–L) Cross-sections through the cervical spinal cord of 4-week-old animals show the absence of CST in Ldb1/2-DKO. Immunocytochemical analysis for EGFP (green in I–L) and PKCγ (red in I–L) reveals the CST in ventral dorsal funiculus of controls (arrow in I), Ldb2-KO (J), and Ldb1-KO (K), but the absence of the tract in Ldb1/2-DKO (arrow in L). PKCγ also labels spinal cord interneurons (arrowheads in I–L). (M–P) Microinjection of fluorescent microspheres into the C2 level of the cervical spinal cord of 4-week-old animals shows retrogradely labeled CSMN in layer 5. Robust back-labeling in layer 5 is seen in controls (arrowhead in M) and Ldb2-KO (arrowhead in N). Fewer cells are found in layer 5 of Ldb1-KO (arrowhead in O), and only very few labeled neurons are seen in Ldb1/2-DKO (P). Scale bars: 1 mm in (D) for (A–D); 250 µm in (H) for (E–H); 300 µm in (L) for (I–L); 500 µm in (P) for (M–P).
To exclude the possibility of a developmental delay, we analyzed cross-sections through the cervical spinal cord at P60 (Fig. 7I–L). In controls and single mutants, the CST is readily visualized by either PKCγ or EGFP (Fig. 7I–K), but the tract is absent in Ldb1/2-DKO mice (Fig. 7L), consistent with the earlier failure of axons to progress beyond the pyramidal decussation in Ldb1/2-DKO mutants. Given the reduced number of Ctip2+ layer 5 neurons in Ldb1-KO and Ldb1/2-DKO mice (Fig. 4K), a reduction in Ldb1/2-DKO tract size was expected compared with controls. However, since the number of Ctip2+ neurons in Ldb1/2-DKO cortices is comparable with that in Ldb1-KO mice, neuronal numbers are unlikely to account for the further reduction in tract size. The absence of axons that extend past the pyramidal decussation in the Ldb1/2-DKO mice suggests severe defects at the pyramidal decussation.
To determine conclusively whether any CST axons in Ldb1/2-DKO mutants reach the spinal cord, we performed retrograde axonal tracing by injecting red fluorescent latex microspheres into the cervical ventral dorsal funiculus of 4-week-old mice. Animals were sacrificed 48 h after surgery to allow for transport of the tracer. Controls and single mutants revealed robust back-labeled CSMNs in layer 5B of the cortex (Fig. 7M–O), but only very few cortical neurons were back-labeled in Ldb1/2-DKO brains (Fig. 7P), confirming that the vast majority of CSMN axons fail to project beyond the pyramidal decussation into the spinal cord.
Previous reports have shown a role for Lhx2 as a cortical selector gene (Mangale et al. 2008) and also as a fate determinant in that loss of Lhx2 can lead to a respecification of cortical pyramidal neurons to piriform cortex identity (Chou et al. 2009). To investigate whether the observed failure of the CST in Ldb1/2-DKO mutants could be due to an early fate respecification, we deleted Ldb1 specifically in the cortical plate by electroporation of a CAG-Cre plasmid at E12.5, bypassing the early specification period during which Lhx2 is required, thereby ensuring Lhx2 function for proper neocortical specification. Ai9 reporter-positive embryos carrying either Ldb1lox/lox or Ldb1lox/wt and either Ldb2+/− or Ldb2−/− were electroporated in utero with a Cre plasmid at E12.5 and analyzed at postnatal day 5 (P5) for CST formation, similar to the method described earlier. Cortical Cre electroporation leads to robust recombination of the Ai9 reporter allele, in turn labeling the CST in controls (see Supplementary Fig. 4A), Ldb2-KO (see Supplementary Fig. 4B), and Ldb1-KO mice (see Supplementary Fig. 4C). In Cre-electroporated Ldb1/2 double mutants, however, the CST reaches the pyramidal decussation, but fails to project further caudally (see Supplementary Fig. 4D). The defect in the CST following both Emx1-Cre-induced deletion and Cre electroporation suggests that the failure of CST at the pyramidal decussation is not due to fate misspecification, which the late ablation using the Cre electroporation would bypass, but instead reveals an intrinsic defect of the tract to project further than the pyramidal decussation in the absence of both Ldb proteins.
The defects in the axonal projections of Ldb1/2-DKO layer 5 CSMNs led us to investigate whether other subcortical tracts might also be affected by the loss of Ldb1/2. To visualize layer 6 corticothalamic projections, we took advantage of the golli-τ-EGFP transgene (golli-EGFP; Jacobs et al. 2007), in which a τ-EGFP cassette, under transcriptional control of 1.3 kb of the golli promoter of the myelin basic protein gene, directs EGFP expression to deep layer pyramidal neurons, including layer 6 corticothalamic neurons. In sagittal sections of P4 animals, golli-EGFP labeled subcortical efferents, including corticothalamic projections, in both controls and Ldb2-KO mice (see Supplementary Fig. 5A,B). Both Ldb1-KO (see Supplementary Fig. 5C) and Ldb1/2-DKO brains (see Supplementary Fig. 5D) show an obvious reduction in neocortical EGFP expression and reduced corticothalamic innervation (likely due to the reduced number of pyramidal neurons in the mutants), but overall thalamic innervation appears normal in both mutants, suggesting that the formation of corticothalamic projections requires neither Ldb1 nor Ldb2.
Taken together, Ldb2 is required by layer 5B CSMNs for differentiation and axonal pathfinding into the spinal cord, but the compensatory upregulation of Ldb1 in Ldb2−/− mice masks the defects, revealing the requirement for Ldb2 in the compound Ldb1/2-DKO mutants.
Altered Expression of Axon Guidance Receptors in Ldb Mutants
The failure of Ldb1/2-DKO CSMN axons to reach the spinal cord and loss of CSMN differentiation markers prompted us to investigate whether axon guidance receptors involved in pathfinding also show altered expression. Neuropilin 1 (Nrp1) is a membrane-bound coreceptor for Semaphorins and Plexins, and both Nrp1 and Semaphorins play roles in axon guidance [reviewed in Huber et al. (2003)]. Nrp1 expression is detected in layer 5 neurons at P1 in controls and Ldb2-KO mice (see Supplementary Fig. 6A,B). In Ldb1-KO layer 5 neurons, Nrp1 expression appears reduced (see Supplementary Fig. 6C,C′) and is absent in Ldb1/2-DKO animals (although low, diffuse expression over the cortical wall remains; Supplementary Fig. 6D,D′). No changes in the expression of Semaphorins 3A, 3B, and 3C were found in any of the 4 genotypes for (data not shown).
We then asked whether Eph receptors [reviewed in Flanagan (2006)] showed changes in expression across the 4 genotypes. No changes were found for EphA3, A5, A7, B1, and B2 (data not shown). EphA4 is expressed by layer 5 neurons and a subset of upper layer neurons at P1; its expression is maintained in Ldb1-KO and Ldb2-KO mice, but qualitatively reduced in layer 5 of Ldb1/2-DKO animals (data not shown). EphA6 is strongly expressed by layer 5 neurons at P1 (see Supplementary Fig. 6E,E′). Expression is maintained in single mutants (see Supplementary Fig. 6F,G′), but abolished in layer 5 motor cortex of Ldb1/2-DKO animals (see Supplementary Fig. 6H,H′). EphA6 knockout animals suffer from learning and memory impairments (Savelieva et al. 2008), but changes in CST formation have not been reported.
These data indicate that Ldb1/2 regulates the expression of Nrp1, EphA6, and EphA4. Since several Eph receptors and their ligands have been implicated in defects in CST formation [reviewed in Canty and Murphy (2008)], it is conceivable that EphA6 and/or EphA4 are involved in the defects at the pyramidal decussation of Ldb1/2-DKO animals.
Discussion
We describe a novel involvement of the Lim domain-binding proteins Ldb1 and Ldb2 during the development of CSMNs. We find that Ldb1 and Ldb2 show inversely correlated expression patterns during cortical development, with a striking localization of Ldb2 to layer 5 and a concomitant exclusion of Ldb1 from that layer. Loss of Ldb1 leads to defects in progenitor proliferation and regionalization. Ldb2 is required during 2 discrete phases of layer 5 neuron differentiation. It first plays a role during segregation of immature layer 5 neurons into CPNs and SCPNs, resulting in increased fractions of Ctip2+/Lmo4+ and Ctip2+/Satb2+ double-positive neurons in Ldb2-KO layer 5. Second, Ldb2 is required for differentiation and pathfinding of CSMNs. Ldb1 is able to compensate for the loss of Ldb2 for both CSMN differentiation and pathfinding, thus permitting axons to reach the spinal cord in the Ldb2 single knockout, ultimately revealing the phenotype in compound Ldb1/2 double knockouts, in which CSMNs fail to extend their axons past the pyramidal decussation due to incomplete molecular differentiation.
Complex Roles of Ldb Proteins Due to Interactions with Multiple Binding Partners
Ldb proteins can interact with Lim-HD and Lmo proteins through their Lim-interacting domain. Specific roles for Ldb proteins in the brain have not been described yet, but indirect evidence suggests their involvement. For example, the proliferation defects in Lhx2-deficient brains and the associated hypoplasia of the neocortex (Porter et al. 1997) correspond well with the proliferation defects we observe in Ldb1-KO brains. Given that Ldb1 strongly binds to Lhx2 (Agulnick et al. 1996), it is plausible that Ldb1 functions as an essential cofactor for Lhx2 function during early stages of cortical development. In support of this model, we observe a regionalization fate change in Ldb1-deficient brains, which leads to a shift of piriform cortex into lateral neocortex, concomitant with the appearance of piriform cortex markers in dorsal neocortex. These findings are reminiscent of the changes in regionalization described in a conditional Lhx2 knockout (Chou et al. 2009), further supporting the notion that Lhx2 function requires Ldb1. Intriguingly, the changes in regionalization observed in the Ldb1-KO and Ldb1/2-DKO mice are less pronounced than those observed in the Lhx2 knockout, suggesting that Lhx2 signaling could be mediated, at least in part, independently of Ldb1.
Inversely Correlated Expression Patterns and Compensatory Role of Ldb1 in Ldb2−/− Mutants
Our analysis reveals a novel, progressive exclusion of Ldb1 from layer 5 neurons as they begin to express Ldb2 during differentiation. The inverse correlation between the expression patterns of the 2 family members suggests a regulatory feedback loop that governs the expression of the 2 proteins. It is tempting to speculate that the upregulation of Ldb2 suppresses the expression of Ldb1 in layer 5 neurons. Indeed, our data confirm this hypothesis: In Ldb2-deficient mutants, we find a striking upregulation of Ldb1 in layer 5 neurons, suggesting a compensatory role of Ldb1 in Ldb2-KO animals. The expression of Ldb2 in layer 5 neurons and the concomitant suppression of Ldb1 suggest that the selective presence of Ldb2 could act as a switch for differentiation of CSMNs, similar to the combinatorial code involving Ldb1, Lhx3, and Isl1 in the spinal cord, where Ldb1–Lhx3 interaction triggers V2 interneuron differentiation, while motor neurons are generated upon Ldb1-Isl1 binding (displacing Lhx3; Thaler et al. 2002). The major binding partners for Ldb proteins in the developing cortex include Lmo3, Lmo4, and Lhx2, all of which are expressed in the cortical plate between E15.5 and P3. It is conceivable that the switch from Ldb1 to Ldb2 rearranges the interacting protein complexes, in a cell type-specific manner, and consequently changes the differentiation path.
Impaired Segregation of Layer 5 CPNs and Subcerebral PNs in Ldb2-Deficient Brains
The failure in segregation of early layer 5 neurons in Ldb2-KO mutants supports the model of a rearrangement of protein interactions by changes in Ldb expression patterns: Lmo4 is widely coexpressed with Ctip2 and Ldb2 in immature layer 5 neurons at E15.5 (Azim et al. 2009). As cortical development proceeds, these layer 5 neurons begin to differentiate into either CPNs or SCPNs, and eventually 2 distinct populations emerge. CPNs downregulate Ctip2 and Ldb2, but maintain expression of Ldb1, Lmo4, and Satb2. SCPNs instead maintain Ctip2 and Ldb2, while downregulating Ldb1, Lmo4, and Satb2. It is tempting to speculate that the suppression of Ldb1 in layer 5 neurons thus initiates a switch in these cells to acquire a subcerebral (SCPN) fate. In agreement with this model, in Ldb2-KO animals, increased fractions of layer 5 neurons coexpressing Lmo4/Ctip2 and Satb2/Ctip2 are found, suggesting that these neurons fail to differentiate properly into either CPNs or SCPNs in the absence of Ldb2.
Incomplete Differentiation of SCPNs in Ldb Mutant Mice
Recent evidence suggests that specification and differentiation are comprised of a series of functionally distinct steps: The transcription factor Fezf2 is required by layer 5 CSMNs for specification, and Fezf2 knockout animals lack a CST and the majority of CSMN differentiation factors (Chen B et al. 2005; Molyneaux et al. 2005). In contrast, although mice lacking the transcription factor BhlhB5 also display defects in the CST, their layer 5 CSMNs appear largely correctly specified, but fail to properly differentiate (Joshi et al. 2008). The expression of both Ctip2 and Fezf2 in Ldb1/2-DKO layer 5 neurons indicates that CSMNs are correctly specified in the absence of Ldb1 and Ldb2. However, numerous markers of CSMN differentiation, including Diap3, Foxo1, Foxp2, and Igfbp4, are absent in Ldb1/2-DKO brains, representing a significant fraction of the markers lost in Fezf2 mutants. These data suggest that while initial specification of CSMNs appears unaffected by the loss of Ldb1 and Ldb2, differentiation of CSMNs is incomplete.
The defect in the CST of Ldb1/2-DKO animals is surprisingly similar to the defect described in Bhlhb5−/− mice; axons in both mutant strains fail to extend past the pyramidal decussation, and both strains show loss of some differentiation factors. The pyramidal decussation is a major crossing point for the CST, where the majority of axons cross from the ventral hindbrain to the ventral dorsal funiculus in the spinal cord. It is conceivable that this crossing represents a vulnerable point along the axons' trajectories, and thus is highly susceptible to changes in axon guidance receptor expression. Indeed, our data reveal alterations in the expression of several axon guidance receptors, including EphA6, EphA4, and Nrp1. It is conceivable that the loss of several axon guidance receptors could lead to a failure of axon crossing and extension at the pyramidal decussation.
Taken together, our data suggest a hierarchical relationship between the different stages of pyramidal neuron specification and differentiation: While Fezf2 is required early for initial CSMN specification, Ldb2 plays an essential role during late differentiation, perhaps through the rearrangement of protein complexes, when CSMNs extend their axons and refine their identity. Further unraveling of the genetic program that governs differentiation of CSMNs will have pivotal clinical implications for the development of cell replacement therapies for spinal cord injuries and neurodegenerative disorders.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
This work was supported by the National Institutes of Health (MH051864) to S.K.M.
Supplementary Material
Notes
We are grateful to Dr Paul Love for providing anti-Ldb1 antibody. We thank Dr James Weimann for technical help with retrograde injections, Dr Simon Hippenmeyer for helpful suggestions on the manuscript, Elizabeth Alcamo and Alan Agulnick for previous experiments, and Dr Chris Kaznowski for technical help. Conflict of Interest: None declared.
References
- Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. 1996. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature. 384:270–272. [DOI] [PubMed] [Google Scholar]
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Fariñas I, Grosschedl R, McConnell SK. 2008. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 57:364–377. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. 2005. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 45:207–221. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. 2008. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 28:622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asprer JS, Lee B, Wu CS, Vadakkan T, Dickinson ME, Lu HC, Lee SK. 2011. LMO4 functions as a co-activator of neurogenin 2 in the developing cortex. Development. 138:2823–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim E, Shnider SJ, Cederquist GY, Sohur US, Macklis JD. 2009. Lmo4 and Clim1 progressively delineate cortical projection neuron subtypes during development. Cereb Cortex. 19(Suppl 1):i62–i69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach I, Carrière C, Ostendorff HP, Andersen B, Rosenfeld MG. 1997. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 11:1370–1380. [DOI] [PubMed] [Google Scholar]
- Breen JJ, Agulnick AD, Westphal H, Dawid IB. 1998. Interactions between LIM domains and the LIM domain-binding protein Ldb1. J Biol Chem. 273:4712–4717. [DOI] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D et al. . 2008. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 57:378–392. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Subramanian L, Tole S. 2003. Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev Dyn. 226:460–469. [DOI] [PubMed] [Google Scholar]
- Canty AJ, Murphy M. 2008. Molecular mechanisms of axon guidance in the developing corticospinal tract. Prog Neurobiol. 85:214–235. [DOI] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. 2005. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 102:17184–17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N. 2005. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci USA. 102:17792–17797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SJ, Perez-Garcia CG, Kroll TT, O'Leary DD. 2009. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat Neurosci. 12:1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. 2005. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 25:247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JG. 2006. Neural map specification by gradients. Curr Opin Neurobiol. 16:59–66. [DOI] [PubMed] [Google Scholar]
- Frantz GD, Weimann JM, Levin ME, McConnell SK. 1994. Otx1 and Otx2 define layers and regions in developing cerebral cortex and cerebellum. J Neurosci. 14:5725–5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. 2002. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 22:6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Kwan KY, Shim S, Lam MM, Shin Y, Xu X, Zhu Y, Li M, Sestan N. 2011. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci USA. 108:3041–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. 2001. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 29:353–366. [DOI] [PubMed] [Google Scholar]
- Huang Z, Kawase-Koga Y, Zhang S, Visvader J, Toth M, Walsh CA, Sun T. 2009. Transcription factor Lmo4 defines the shape of functional areas in developing cortices and regulates sensorimotor control. Dev Biol. 327:132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AB, Kolodkin AL, Ginty DD, Cloutier JF. 2003. Signaling at the growth cone: ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci. 26:509–563. [DOI] [PubMed] [Google Scholar]
- Jacobs EC, Campagnoni C, Kampf K, Reyes SD, Kalra V, Handley V, Xie YY, Hong-Hu Y, Spreur V, Fisher RS et al. . 2007. Visualization of corticofugal projections during early cortical development in a tau-GFP-transgenic mouse. Eur J Neurosci. 25:17–30. [DOI] [PubMed] [Google Scholar]
- Jones EG, Schreyer DJ, Wise SP. 1982. Growth and maturation of the rat corticospinal tract. Prog Brain Res. 57:361–379. [DOI] [PubMed] [Google Scholar]
- Joshi PS, Molyneaux BJ, Feng L, Xie X, Macklis JD, Gan L. 2008. Bhlhb5 regulates the postmitotic acquisition of area identities in layers II-V of the developing neocortex. Neuron. 60:258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Gill GN. 1997. Functional analysis of the nuclear LIM domain interactor NLI. Mol Cell Biol. 17:5688–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Kenny DA, Gill GN. 1996. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci USA. 93:11693–11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny DA, Jurata LW, Saga Y, Gill GN. 1998. Identification and characterization of LMO4, an LMO gene with a novel pattern of expression during embryogenesis. Proc Natl Acad Sci USA. 95:11257–11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. 2008. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci USA. 105:16021–16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Jurata LW, Nowak R, Lettieri K, Kenny DA, Pfaff SL, Gill GN. 2005. The LIM domain-only protein LMO4 is required for neural tube closure. Mol Cell Neurosci. 28:205–214. [DOI] [PubMed] [Google Scholar]
- Leone DP, Heavner WE, Ferenczi EA, Dobreva G, Huguenard JR, Grosschedl R, McConnell SK. 2015. Satb2 regulates the differentiation of both callosal and subcerebral projection neurons in the developing cerebral cortex. Cereb Cortex. 25:3406–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV et al. . 2008. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 319:304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JM, Visvader JE. 2003. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 4:1132–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B. 2011. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 31:549–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier N, Krpic S, Rodriguez P, Strouboulis J, Monti M, Krijgsveld J, Gering M, Patient R, Hostert A, Grosveld F. 2006. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 133:4913–4923. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. 2005. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 47:817–831. [DOI] [PubMed] [Google Scholar]
- Mori M, Kose A, Tsujino T, Tanaka C. 1990. Immunocytochemical localization of protein kinase C subspecies in the rat spinal cord: light and electron microscopic study. J Comp Neurol. 299:167–177. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M, Teufel A, Yamashita T, Agulnick AD, Chen L, Downs KM, Schindler A, Grinberg A, Huang SP, Dorward D et al. . 2003. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development. 130:495–505. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. 2000. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 28:147–155. [PubMed] [Google Scholar]
- Ohtsuka T, Sakamoto M, Guillemot F, Kageyama R. 2001. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J Biol Chem. 276:30467–30474. [DOI] [PubMed] [Google Scholar]
- Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D et al. . 1997. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 124:2935–2944. [DOI] [PubMed] [Google Scholar]
- Savelieva KV, Rajan I, Baker KB, Vogel P, Jarman W, Allen M, Lanthorn TH. 2008. Learning and memory impairment in Eph receptor A6 knockout mice. Neurosci Lett. 438:205–209. [DOI] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, Sestan N. 2012. Cis-regulatory control of corticospinal system development and evolution. Nature. 486:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara TM, Bach I, Kioussi C, Rosenfeld MG, Andersen B. 1998. Mouse deformed epidermal autoregulatory factor 1 recruits a LIM domain factor, LMO-4, and CLIM coregulators. Proc Natl Acad Sci USA. 95:15418–15423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. 2002. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 110:237–249. [DOI] [PubMed] [Google Scholar]
- Tomassy GS, De Leonibus E, Jabaudon D, Lodato S, Alfano C, Mele A, Macklis JD, Studer M. 2010. Area-specific temporal control of corticospinal motor neuron differentiation by COUP-TFI. Proc Natl Acad Sci USA. 107:3576–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse E, Smith AJ, Hunt S, Lavenir I, Forster A, Warren AJ, Grutz G, Foroni L, Carlton MB, Colledge WH et al. . 2004. Null mutation of the Lmo4 gene or a combined null mutation of the Lmo1/Lmo3 genes causes perinatal lethality, and Lmo4 controls neural tube development in mice. Mol Cell Biol. 24:2063–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Mao X, Fujiwara Y, Hahm K, Orkin SH. 1997. The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc Natl Acad Sci USA. 94:13707–13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman I, Li J, Bash RO, Forster A, Osada H, Rabbitts TH, Baer R. 1994. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. EMBO J. 13:4831–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grütz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Kwan KM, Mailloux CM, Lee WK, Grinberg A, Wurst W, Behringer RR, Westphal H. 2007. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc Natl Acad Sci USA. 104:13182–13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sheng HZ, Amini R, Grinberg A, Lee E, Huang S, Taira M, Westphal H. 1999. Control of hippocampal morphogenesis and neuronal differentiation by the LIM homeobox gene Lhx5. Science. 284:1155–1158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.