Abstract
TDP-43 is a well known RNA binding protein involved in the pathogenesis of Amyotrophic Lateral Sclerosis (ALS) and Frontotemporal Lobar Dementia (FTLD). In physiological conditions, TDP-43 mainly localizes in the nucleus and shuttles, at least in neurons, to the cytoplasm to form TDP-43 RNA granules. In the nucleus, TDP-43 participates to the expression and splicing of RNAs, while in the cytoplasm its functions range from transport to translation of specific mRNAs. However, if loss or gain of these TDP-43 functions are affected in ALS/FTLD pathogenesis is not clear. Here, we report that TDP-43 localizes on ribosomes not only in primary neurons but also in SH-SY5Y human neuroblastoma cells. We find that binding of TDP-43 to the translational machinery is mediated by an interaction with a specific ribosomal protein, RACK1, and that an increase in cytoplasmic TDP-43 represses global protein synthesis, an effect which is rescued by overexpression of RACK1. Ribosomal loss of RACK1, which excludes TDP-43 from the translational machinery, remarkably reduces formation of TDP-43 cytoplasmic inclusions in neuroblastoma cells. Finally, we corroborate the interaction between TDP-43 and RACK1 on polyribosomes of neuroblastoma cells with mis-localization of RACK1 on TDP-43 positive cytoplasmic inclusions in motor neurons of ALS patients. In conclusions, results from this study suggest that TDP-43 represents a translational repressor not only for specific mRNAs but for overall translation and that its binding to polyribosomes through RACK1 may promote, under conditions inducing ALS pathogenesis, the formation of cytoplasmic inclusions.
Introduction
Amyotrophic lateral sclerosis (ALS) and Frontotemporal Lobar Dementia (FTLD) are devastating fatal neurodegenerative diseases, characterized by degenerating motor neurons in brain and in spinal cord (1). The causes of neurodegeneration are not yet clear, even if most of sporadic and familial ALS/FTLD cases share a pathological feature: the accumulation of ubiquitin-positive and phosphorylated cytoplasmic inclusions containing the TAR DNA-binding protein TDP-43 (2–4). The TARDBP gene encodes for a well characterized RNA binding protein, containing two RNA binding domains (RRM1 and RRM2), a glycine-rich domain in the C-terminal region for the binding with protein partners and a nuclear localization signal (NLS) (5–7). Nuclear TDP-43 regulates RNA metabolism, including RNA expression and splicing (8–10). TDP-43 also shuttles from nucleus to cytoplasm, where its function is not fully determined yet. In neurons, cytoplasmic TDP-43 forms mobile RNA granules containing mRNA Binding Proteins (mRBPs) such as FMRP, ZBP1/IMP-1 and HuD, and, at least, β-actin and CaMKII mRNAs, that suggest a role on transport, stability and translation of specific mRNAs (11–13). Interestingly, recent studies indicate that TDP-43 ALS-linked mutations might affect size and movement along axons and dendrites of these TDP-43 RNA enriched granules (14,15). TDP-43 is also recruited, with others mRBPs, to form Stress Granules (SGs), which are cytoplasmic protein:RNA complexes that modulate RNA translation during stress (16). Noteworthy, SGs, recruiting small ribosome subunits and eukaryotic initiation factors, block constitutive protein synthesis, allowing translation of protein required to stress (17–19).
Biochemical studies of ALS and FTLD cases have found that TDP-43 in cytoplasmic inclusions, besides being ubiquinated and phosphorylated, can also be cleaved in C-terminal fragments (CTF) (3,20), which accumulate in cytoplasmic inclusions of FTLD brains and in cytoplasmic aggregations of ALS spinal cords (21–23).
Another result of cytoplasmic accumulation of TDP-43 in ALS and FTLD cases is loss of nuclear TDP-43 itself (3,20). The nuclear loss of TDP-43 opens the question on whether degeneration of motor neurons depends on loss-of-nuclear or gain-of-cytoplasmic functions of TDP-43, or a combination of the two. Although transgenic murine models overexpressing wild-type and ALS-linked mutated forms of human TDP-43 show some ALS/FTLD phenotypes without cytoplasmic aggregation or nuclear loss of TDP-43 (24–26), none of these models closely recapitulate the full pathological picture of ALS and FTLD. Therefore, one cannot exclude that a combination of both events might be involved in disease development.
In recent years, classical SGs markers have been found in TDP-43 cytoplasmic inclusions and an increasing number of ALS/FTLD related proteins have been reported on SGs (21,27). These findings have led to the hypothesis that cytoplasmic inclusions may originate from SGs that failed to disassemble. However, at present, there is no data nor methodology available to determine if these inclusions are broadly composed of SG proteins or if SG proteins are themselves recruited on pre-formed inclusions.
In this report, we found that cytoplasmic TDP-43 inhibits global protein synthesis. In agreement with literature (28), we confirm that TDP-43 RNA granules contain polyribosomes, the associated translational machinery and other proteins. We also demonstrate that this occurs through a mechanism dependent on the binding to a ribosomal scaffold protein, RACK1 (Receptor Activated C Kinase 1 protein). Moreover, we determine that the localization of TDP-43 on ribosomes might promote the formation of TDP-43 cytoplasmic inclusions. Finally, we found that these results correlate with mislocalization of RACK1 on TDP-43 cytoplasmic inclusions of ALS spinal cord patients. Thus, taken together these results suggest that a cytoplasmic excess of TDP-43, following nuclear loss of TDP-43, represses global translation at the onset of motor neuron degeneration.
Results
TDP-43 RNA granules include the translational machinery
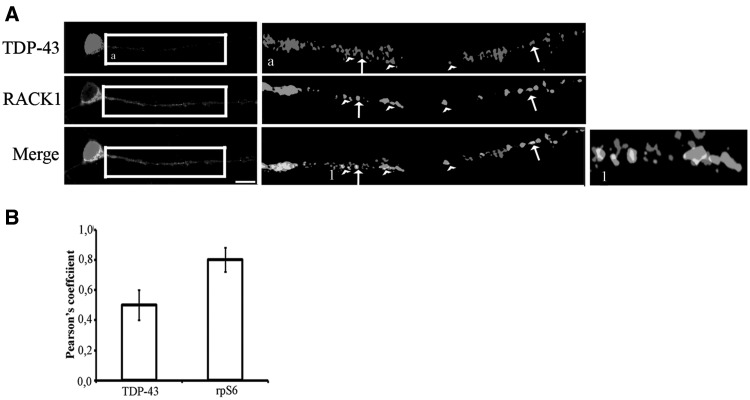
To test whether cytoplasmic TDP-43 might regulate global protein synthesis, we initially examined the association of TDP-43 to the translational machinery. To this end, mouse embryonic hippocampal cells, which express TDP-43 in cytoplasm (11,14,15), were double immunostained for TDP-43 and ribosomal protein RACK1, a specific ribosomal docking site for several proteins and kinases such as ZBP1/IMP-1, AGO-1, PKC and Src (29–31). Labeling of TDP-43 showed a strong nuclear localization and a granular pattern in soma and along neurites (Fig. 1A), as previously described. RACK1 strongly appeared in the soma and showed a punctated pattern similar to that of TDP-43 along neurites (Fig. 1A). Co-localization analysis showed a significant overlapping between TDP-43 and RACK1 in the soma and in granules of neurites (0.5 Pearson’s coefficient in bar graph, Fig. 1B). However, a part of RACK1 granules was adjacent to those of TDP-43 (Fig. 1A, enlarged images). A double labeling of neurons with RACK1 and 40S ribosomal protein S6, rpS6 demonstrated strong co-localization, indicating that most part of granules labeled by RACK1 corresponded to ribosome/polyribosomes (Supplementary Material Fig. S1 and 0.8 coefficient Pearson’s Fig. 1B). These results suggest that a pool of cytoplasmic TDP-43 granules interact with the translational machinery under physiological conditions.
Figure 1.
Part of TDP-43 enriched granules may contain translational machinery. (A) Embryonic hippocampal mouse cells immunolabeled by TDP-43 and RACK1. RACK1 has been used to indicate ribosomes and polyribosomes. Magnification of boxed regions is reported in a. The heads of arrows indicate the partial or adjacent co-localization of TDP-43 and RACK1, while the arrows indicate the full co-localization of TDP-43/RACK1. 1 is magnification of regions indicated by the heads and the arrows. Scale bar=10 µm. (B) Graph reporting the correlation measured by Pearson coefficient of the puncta signal between TDP-43 and RACK1 (TDP-43 graph bar) and that between rpS6 and RACK1 (rpS6 graph bar). All bar represent mean ± SD; n=70 puncta from three independent experiments. Blue = DAPI.
TDP-43 is recruited on the translational machinery by RACK1
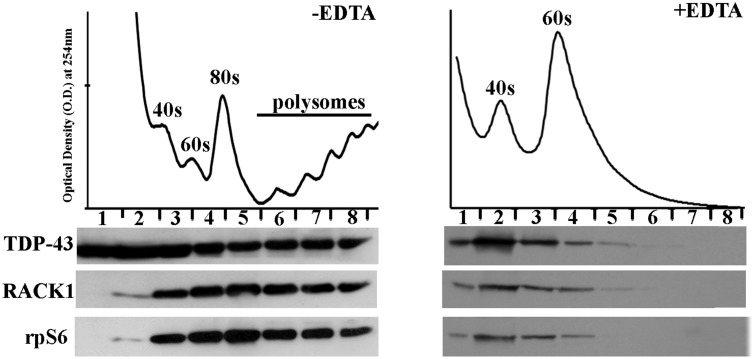
Since it has previously been reported that RACK1 functions as a ribosomal scaffold for several other proteins, we next investigated whether the association of TDP-43 with ribosomes requires RACK1. Performing a polysome profile purification in SH-SY5Y human neuroblastoma cells, which express TDP-43 in nucleus and cytoplasm (Supplementary MaterialFig. S2), we initially examined whether RACK1 and TDP-43 co-sedimented with ribosomal subunits. A total lysate of human neuroblastoma SH-SY5Y cells was loaded on 10–50% sucrose density gradient, and free ribosomes and polysomes were separated through ultracentrifugation. Next, polysome profile was obtained by adsorbance at 254 nm and free ribosomes and polyribosomes were collected in fractions. Immunoblotting experiments on collected fractions revealed that TDP-43, RACK1 and rpS6 co-sedimented in the same ribosome and polyribosomes fractions (Fig. 2A, left). Moreover, when polysomes profiles were performed with EDTA treatment, which dissociates polyribosomes to 40S and 60S free ribosome subunits, TDP-43 distribution shifted in the ribosome-enriched fractions, marked by RACK1 and rpS6 localization (Fig. 2A, right), indicating that TDP-43 associated to ribosome subunits.
Figure 2.
TDP-43 co-sediments with translational components on polysomal profiling from SH-SY5Y cells. Ribosome and polyribosome collected fractions were immunoblotted for TDP-43 to examine the localization of TDP-43 on free ribosome subunits and polyribosomes indicated by RACK1 and rpS6 proteins (left). EDTA treatment, which disassembles polyribosomes in free ribosomal subunits, enriched TDP-43 on free dissociated ribosome subunits as indicated by RACK1 and rpS6 (right).
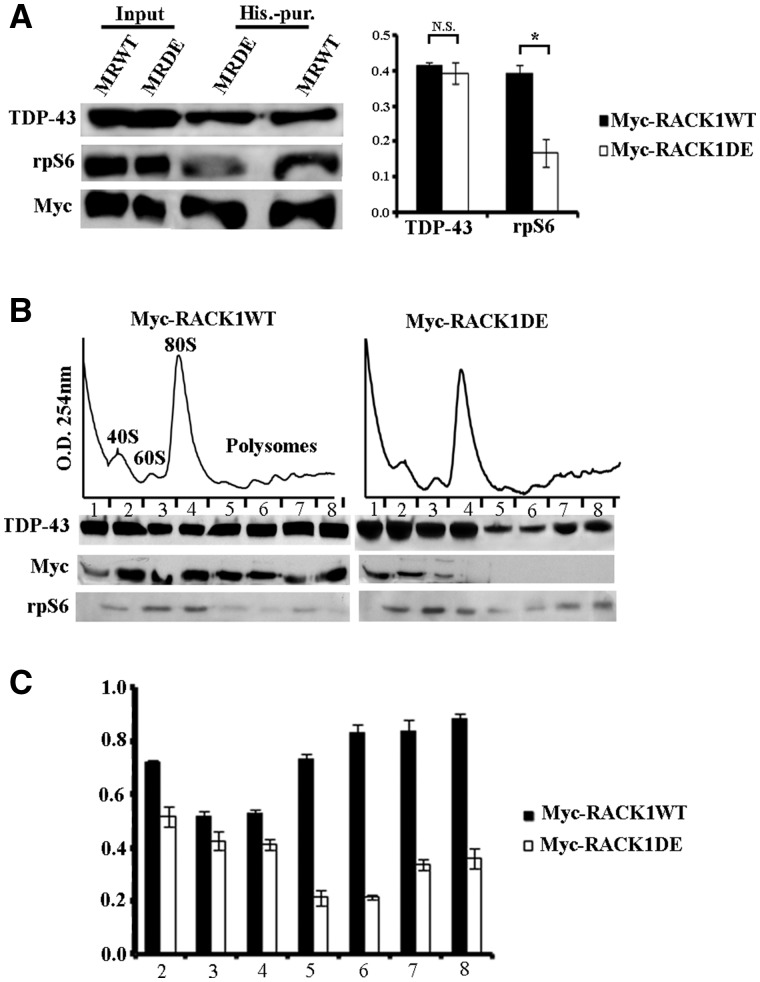
Next, we determined whether RACK1 was needed for binding of TDP-43 to ribosome/polyribosomes. For this purpose, we generated SH-SY5Y cells stably overexpressing His-Myc tagged RACK1 carrying mutations in R38D/K40E (RACK1DE), which is unable to bind to ribosomes (32,33), and isolated RACK1 (WT and DE) by a histidine purification assay. We initially verified whether TDP-43 bound this mutated form of RACK1 protein. Immunoblotting experiments on imidazole eluted proteins indicated that TDP-43 co-purified with RACK1DE as well as RACK1WT. A decreased amount rpS6 was co-purified with RACK1DE pull downs, which confirmed the inability of RACK1DE to bind to translational machinery and validated the specificity of the co-purification (Fig. 3A). Moreover, we also investigated if the interaction of TDP-43 with RACK1WT was dependent on RNA. Co-purification by histidine pull down assay in presence of RNase, showed a not significant decrease of the binding between TDP-43 and RACK1WT (Supplementary MaterialFig. S3). Next, given that TDP-43 bound RACK1DE as well as RACK1WT, and that RACK1DE mutation inhibited binding to ribosomes, it was reasonable to hypothesize that RACK1DE might exclude TDP-43 from ribosome/polyribosomes. Thus, we measured the amount of TDP-43 bound to the translational machinery in the polysome profile performed on RACK1DE overexpressing cells. Immunoblots on collected fractions showed that TDP-43 was less associated to the translational machinery in RACK1DE expressing cells compared to RACK1WT cells (Fig. 3B and bar graph). In particular, the decreasing of TDP-43 was more marked in polyribosome fractions than in free ribosome subunits, suggesting that TDP-43 preferentially associated with actively translating ribosomes. Moreover, these results indicated that binding of TDP-43 to translational machinery is mediated by the scaffold ribosomal protein RACK1.
Figure 3.
TDP-43 associates the translational machinery by the binding to RACK1. (A) Left, immunoblotting for TDP-43, rpS6 and Myc antibodies on proteins eluted by histidine purification from SH-SY5Y cells overxpressing Myc-RACK1WT (MRWT) and Myc-RACK1DE (MRDE) proteins. The amount of TDP-43 purified from MRWT and MRDE imidazole eluted proteins was similar, while the level of rpS6 was reduced in purified MRDE. Right, quantification of TDP-43 and rpS6 co-purified with MRWT or MRDE measured by densitometry on bands related to immunoblottings of three independent experiments. All bar graphs represent the mean and Standard Deviation (S.D.) of TDP-43 or rpS6 level normalized to amount of purified Myc-fusion proteins. Student’s t-test was used to calculate P-values *<0.01. (B) Amount of TDP-43, examined by immunoblotting, on ribosomal fractions collected by polysome profiling conducted on SH-SY5Y cells overexpressing MRWT or MRDE proteins. The localization of TDP-43 on translational machinery was reduced by the overexpression of MRDE. The graphic in C quantifies amount of TDP-43 normalized to level of rpS6 on ribosome and polyribosome fractions (from 2 to 8 fractions) measured by densitometry on bands related to immunoblottings of three independent experiments. All bar graphs represent the mean and S.D.
RACK1 regulates the localization of TDP-43 on Stress Granules (SGs)
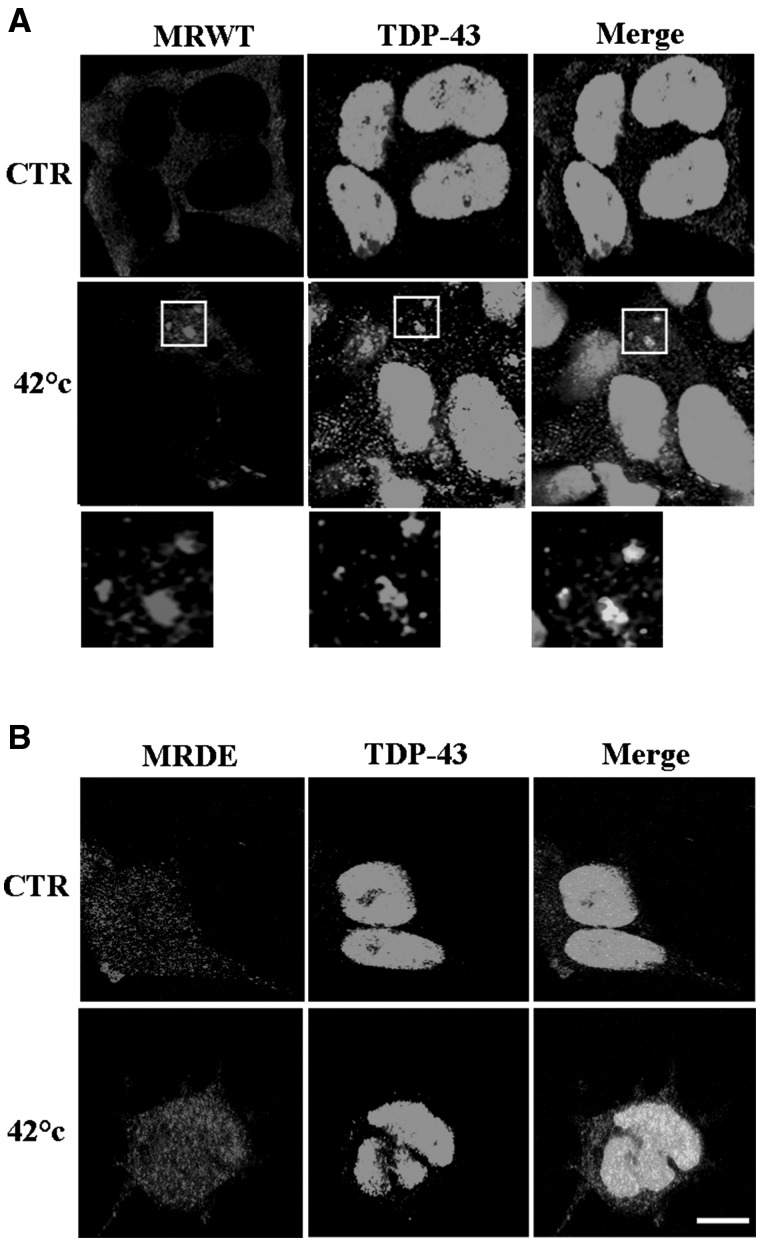
Considering that ribosome subunits are included in SGs (16,34), we next investigated if RACK1 might also control the recruitment of TDP-43 on these cytoplasmic structures. We first verified whether TDP-43 and RACK1 co-localized on the same SGs. Immunofluorescence experiments conducted on SH-SY5Y cells stressed by heat shock revealed a percentage of co-localization between RACK1 and TDP-43 higher than that measured for TDP-43 and TIAR, the classical marker used for SGs (Supplementary MaterialFig. S4A and B and bar graph in C). Moreover, the number of SGs labeled by TDP-43 was lower than those visualized by RACK1, suggesting that TDP-43 probably localized on SGs later than RACK1 (Supplementary MaterialFig. S4A and bar graph in D), which would be consistent with a mechanism in which RACK1 promotes the recruitment of TDP-43 on SGs. To confirm this possibility, localization of TDP-43 on SGs was investigated in RACK1WT or RACK1DE SH-SY5Y overexpressing cells stressed by heat shock Immunofluorescence studies, using TDP-43 and Myc antibodies to label endogenous TDP-43 and exogenous RACK1WT or RACK1DE, revealed a co-localization between TDP-43 and RACK1WT on most of same SGs (Fig. 4A), while neither TDP-43 nor RACK1DE displayed SG labelling (Fig. 4B), even if SGs were induced by heat stress as revealed by TIAR immunofluorescences (Supplementary MaterialFig. S5). Thus, these results indicate that RACK1 not only mediates association of TDP-43 to the translational machinery, but also to SGs.
Figure 4.
RACK1 mediates recruitment on SGs of TDP-43. (A, B), TDP-43 positive SGs were analysed on SH-SY5Y cells overexpressing RACK1WT (A) and RACK1DE (B) proteins stressed by heat shock at 42 °C for 1 h. MRDE overexpression reduced the accumulation of TDP-43 on SGs, indicated by arrows in MRWT overexpressing cells. Scale bar = 10 µm.
Figure 5.
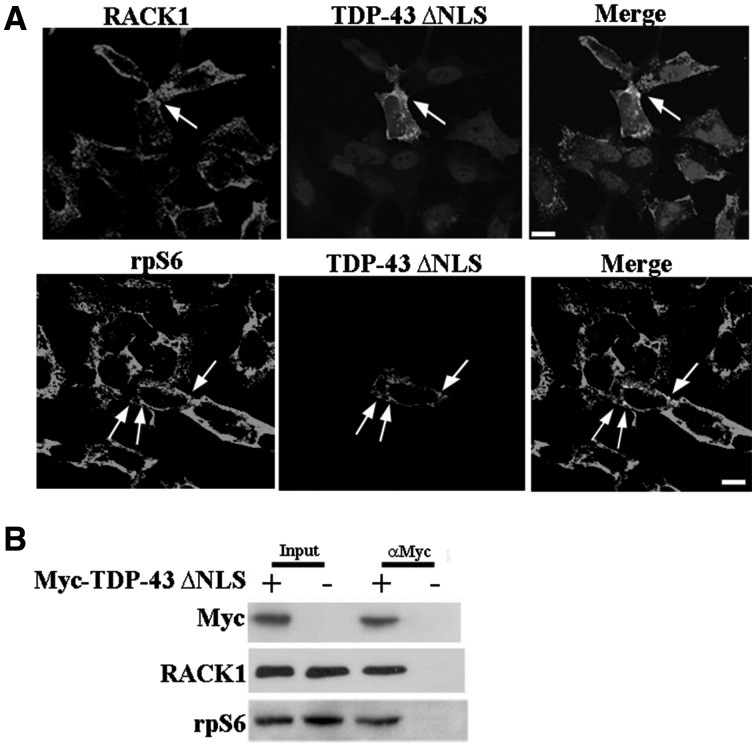
ΔNLS-TDP-43 associates translational machinery. (A) The SH-SY5Y cells overexpressing Myc-ΔNLS-TDP-43 showed co-localization in cytoplasm with RACK1 and rpS6, which was more pronounced in ΔNLS-TDP-43 cytoplasmic inclusions indicated by arrows. Scale bar = 10 µm. (B) RACK1 and rpS6 proteins were eluted with ΔNLS-TDP-43 purified by Myc immunoprecipitation assay from SH-SY5Y cells overexpressing Myc-ΔNLS-TDP-43 protein.
Ribosomal TDP-43 reduces protein synthesis
Although the localization of TDP-43 on ribosomes/polyribosomes has been found by several studies (17,35,36), which role TDP-43 performs on global translation is still not fully determined. One possible reason might be the low amount of TDP-43 in the cytoplasm. Thus, a mutant in the Nuclear Localization Signal of TDP-43 (ΔNLS-TDP-43) fused with a Myc tag, (25,27,37,38), has been used to increase the amount of cytoplasmic TDP-43. Immunofluorescence studies in transiently transfected SH-SY5Y cells revealed that ΔNLS-TDP-43 diffused in the cytoplasm and, partially, also in the nucleus (Fig. 5A). Moreover, a low number of cells exhibited cytoplasmic inclusions, in which TIAR was also included (Supplementary MaterialFig. S6). To examine whether ΔNLS-TDP-43 was part of the translational machinery, we co-labeled ΔNLS-TDP43 with RACK1 or rpS6 antibodies. Double labeling experiments showed a diffuse co-localization between ΔNLS-TDP43 and RACK1 or rpS6 in the cytoplasm, which was more evident in cells containing ΔNLS-TDP43 inclusions (Fig. 5A). To further investigate whether ΔNLS-TDP43 bound to RACK1 and rpS6, we performed a co-immunoprecipitation assay, using Myc antibody to immunoprecipitate ΔNLS-TDP43. As negative control, we used SH-SY5Y transfected with Myc alone. By immunoblotting subsequent to Myc-immunoprecipitation, RACK1 and rpS6 only appeared in extracts co-purified with ΔNLS-TDP43 and not with those purified in the negative control (Fig. 5B). These results indicated that ΔNLS-TDP43 associates to the translational machinery as well as endogenous TDP-43 and thus it could be used to examine the activity of TDP-43 on protein synthesis.
Figure 6.
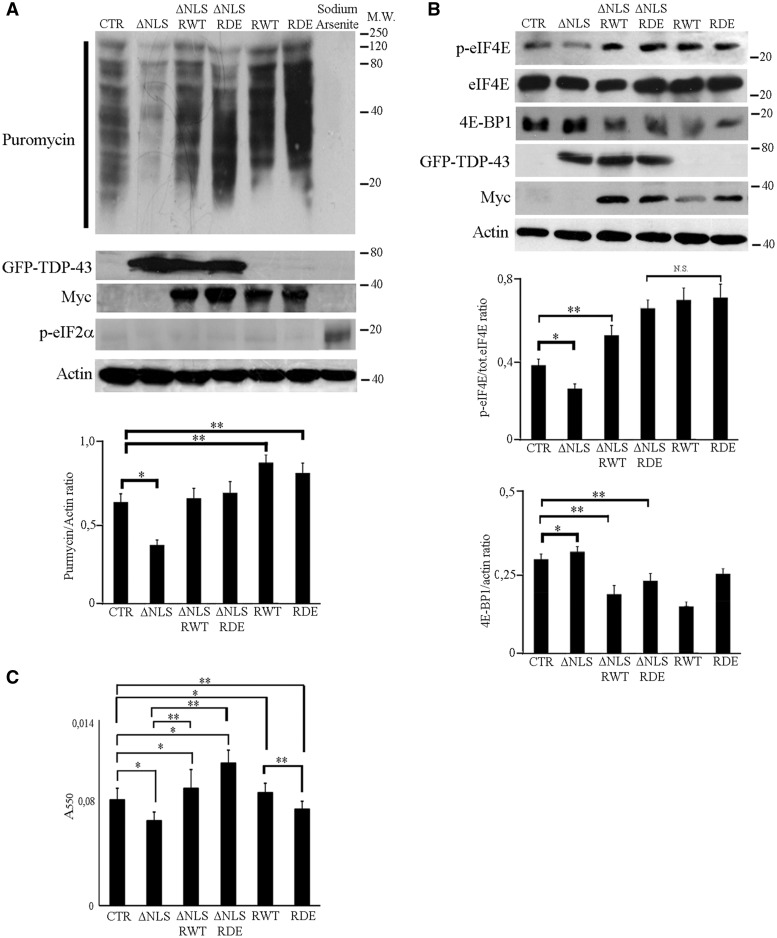
ΔNLS-TDP-43 overexpression reduces protein synthesis and cell viability. (A) SUnSET assay revealed a decrease of puromycin incorporation by expression of GFP-ΔNLS-TDP-43 compared to control (SH-SY5Y transfected with empty vectors, CTR) which was rescued by co-expression with RACK1WT or RACK1DE. Puromycin incorporation was increased in cells overxpressing RACK1WT or RACK1DE. Sodium Arsenite stress blocked protein synthesis rate by eIF2a phosphorylation and inhibited puromycin incorporation. To avoid interaction with myc tag fused to RACK1WT or RACK1DE, the cells were transfected with GFP-ΔNLS-TDP-43. The immunoblottings were performed with TDP-43 antibody to visualize the ΔNLS-TDP-43. The bar graph summarizes the level of total puromycilated proteins normalized to amount of actin measured by densitometry on immunoblottings of three independent experiments. (B) Immunoblottings for p-eIF4E, total eIF4E and 4E-BP1 showed a decrease of eIF4E phosphorylation and increase of 4E-BP1 induced by ΔNLS-TDP-43 overxpression which were rescued by the overexpression of RACK1WT or RACK1DE. The bar graph for p-eIF4E summarizes the level of eIF4E phosphorylated normalide to total eIF4E, while total 4E-BP1 was normalized to amount of actin measured by densitometry on immunoblottings of three independent experiments All bar graphs represent the mean and S.D. Student’s t-test was used to calculate P-values. *P<0.05; **P< 0.01. (C) MTT assay shows a reduction of cell viability in cells transfected with GFP-ΔNLS-TDP-43 cDNA, which is rescued by the overexpression of RACK1WT or RACK1DE. All bar graphs represent the mean and S.D. Student’s t-test was used to calculate P-values. *P<0.01 and **P<0.05.
To probe TDP-43 role in translation, we performed SUrface SEnsing of Translation (SUnSET) technique. SUnSET consists in a non-radioactive method, in which protein synthesis is evaluated through puromycin incorporation, measured by immunoblotting using an anti-puromycin antibody. We performed SUnSET to measure overall translation in SH-SY5Y cells overexpressing GFP-ΔNLS-TDP-43 cDNA and, considering that RACK1 might regulate the binding of endogenous TDP-43 to the translational machinery, in SH-SY5Y co-expressing GFP-ΔNLS-TDP-43 and RACK1WT or RACK1DE proteins. We used GFP-ΔNLS-TDP-43 instead of Myc-ΔNLS-TDP43 to avoid possible interferences between the Myc tag fused to ΔNLS-TDP43 and the one fused to RACK1WT or RACK1DE protein. Nevertheless, ΔNLS-TDP43 features were not affected (data not shown). Immunoblots for puromycin indicated that overexpression of ΔNLS-TDP-43 significantly reduced translation compared to controls (cells transfected with an empty vector), while the overexpression of RACK1WT or RACK1DE increased puromycin incorporation (Fig. 6A). Interestingly, when ΔNLS-TDP-43 was co-expressed with RACK1WT or RACK1DE, the puromycin incorporation was comparable to controls, which suggests a rescue of translation by either RACK1WT or RACK1DE. These results indicate that TDP-43 can inhibit protein synthesis, and that RACK1 can suppress the translational repressor activity of TDP-43
To understand how ΔNLS-TDP43 reduce translation, we initially determined the level of eukaryotic initiation 2 alpha (eIF2alpha) phosphorylation, that is reported to precede SG formation and to induce a prolonged translational repression in Drosophila overexpressing TDP-43 (16,39). By immunoblotting, we found that the overexpression of ΔNLS-TDP-43 in SH-SY5Y cells did not stimulate the phosphorylation of eIF2α (Fig. 6A), indicating that ΔNLS-TDP-43 impaired protein synthesis through an eIF2α phosphorylation-independent mechanism.
To further find which mechanism use ΔNLS-TDP43 to affected protein synthesis, the level of eukaryotic initiation 4E phosphorylation (eIF4E), which promotes translation (40), and the amount of 4E eukaryotic binding protein (4E-BP1), a translational repressor (41), were measured by immunoblotting. The phosphorylation of eIF4E was slightly reduced by the overexpression of ΔNLS-TDP-43 compared to control. The co-expression with RACK1WT or RACK1DE in ΔNLS-TDP-43 overexpressing cells increased phosphorylated eIF4E, more in ΔNLS-TDP43/RACK1DE co-expressing cells than in those co-expressing ΔNLS-TDP43/RACK1wt. This increase of eIF4E phosphorylation was dependent on the RACK1 overexpression because SH-SY5Y cells overexpressing RACK1WT or RACK1DE showed more phosphorylated eIF4E than control cells (Fig. 6B). In parallel, the overexpression of ΔNLS-TDP-43 increased the amount of 4E-BP1 with respect to control; in contrast the overexpression of RACK1WT or RACK1DE rescued it to control level. As seen for eIF4E phosphorylation, the recovery of 4E-BP1 amount was related to RACK1 overexpression. In fact, the 4E-BP1 expression was reduced in both RACK1WT or RACK1DE overexpressing cells.
Moreover, we also us wondered whether TDP-43 may block protein synthesis by including components of 60S ribosomal subunits in cytoplasmic aggregates. To this end, an immunolabeling using an antibody for eIF6/p27BBP,a protein associated to 60S ribosomal subunits (30), was conducted on GFP-ΔNLS-TDP-43 overexpressing cells. eIF6/p27BBP appeared in the nucleus, in particular in the nucleolus, and in cytoplasm as expected, but no co-localization was visible in GFP-ΔNLS-TDP-43 cytoplasmic (Supplementary MaterialFig. S7). This result suggests that TDP-43 cytoplasmic inclusions only contain element of 40S ribosomal subunits.
Figure 7.
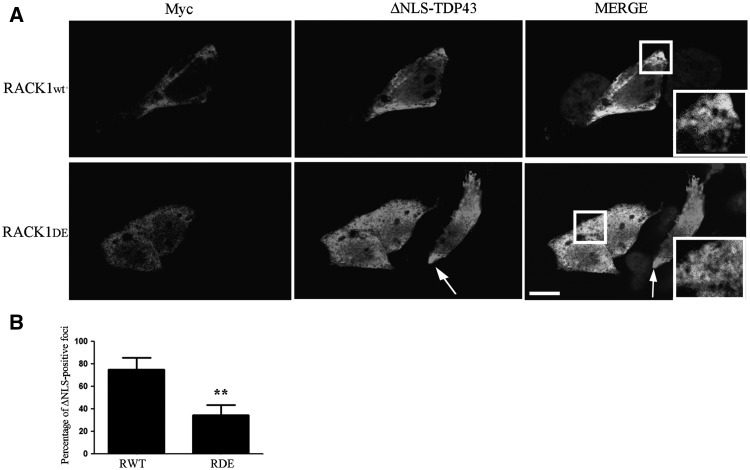
Myc-RACK1DE overexpression inhibits the formation of ΔNLS-TDP-43 cytoplasmic inclusions. (A) SH-SY5Y co-expressing RACK1WT or RACK1DE and GFP-ΔNLS-TDP-43 cDNAs were labeled with myc and TDP-43 antibodies and showed that TDP-43 positive cytoplasmic inclusions induced by the overexpression of ΔNLS-TDP-43 were reduced by the RACK1DE expression. The arrows indicate the ΔNLS-TDP-43 cytoplasmic aggregates in RACK1WT overexpressing cells and in cells overexpressing alone GFP-ΔNLS-TDP-43. Scale bar = 10 µm. (B) The graph shows the quantification of cells positive for DNLS-TDP-43 that present cytoplasmatic foci of TDP-43 itself. For each genotype, at least 60 cells were analysed. Statistic applied was t-test, paired, two tails (**P-value≤0.005%).
Next, as it has been reported that cytoplasmic mislocalization of TDP-43 is toxic to neurons (42), we also measured the cell viability in SH-SY5Y cells overexpressing GFP-ΔNLS-TDP-43 protein and in those co-expressing GFP-ΔNLS-TDP-43 and RACK1WT or RACK1DE proteins. By MTT assay, we observed that the overexpression of GFP-ΔNLS-TDP-43 protein was significantly more toxic compared to controls, but the co-expression with RACK1WT or RACK1DE rescued this toxicity (Fig. 6C). Interestingly, while the overexpression of RACK1WT significantly increased the cell viability, in contrast, RACK1DE overexpression slightly appeared more toxic than control and RACK1WT overexpression. These results indicate that the cytoplasmic mislocalization of TDP-43 reduces the cell viability, but RACK1 can mitigate this effect.
Binding of ΔNLS-TDP-43 to polyribosomes favors formation of ΔNLS-TDP-43 cytoplasmic inclusions
Even if the origin of TDP-43 cytoplasmic inclusions has not been fully determined (16,43,44), we wondered whether the association of TDP-43 to the translation machinery might be involved on its metabolism. For this purpose, we used confocal analysis to examine ΔNLS-TDP-43 cytoplasmic inclusions in SH-SY5Y cells co-expressing RACK1WT or RACK1DE. We observed that GFP-ΔNLS-TDP-43 formed cytoplasmic inclusions in RACK1WT cells, but its localization appeared diffused in cells overexpressing RACK1DE. (Fig. 7A and B, bar graph). These results indicated that although an excess of ΔNLS-TDP-43 induced cytoplasmic inclusions, expression of RACK1DE, probably excluding it from polysomes, inhibited formation of granules positive for ΔNLS-TDP-43.
RACK1 is included in TDP-43 positive aggregates of spinal cord ALS patients
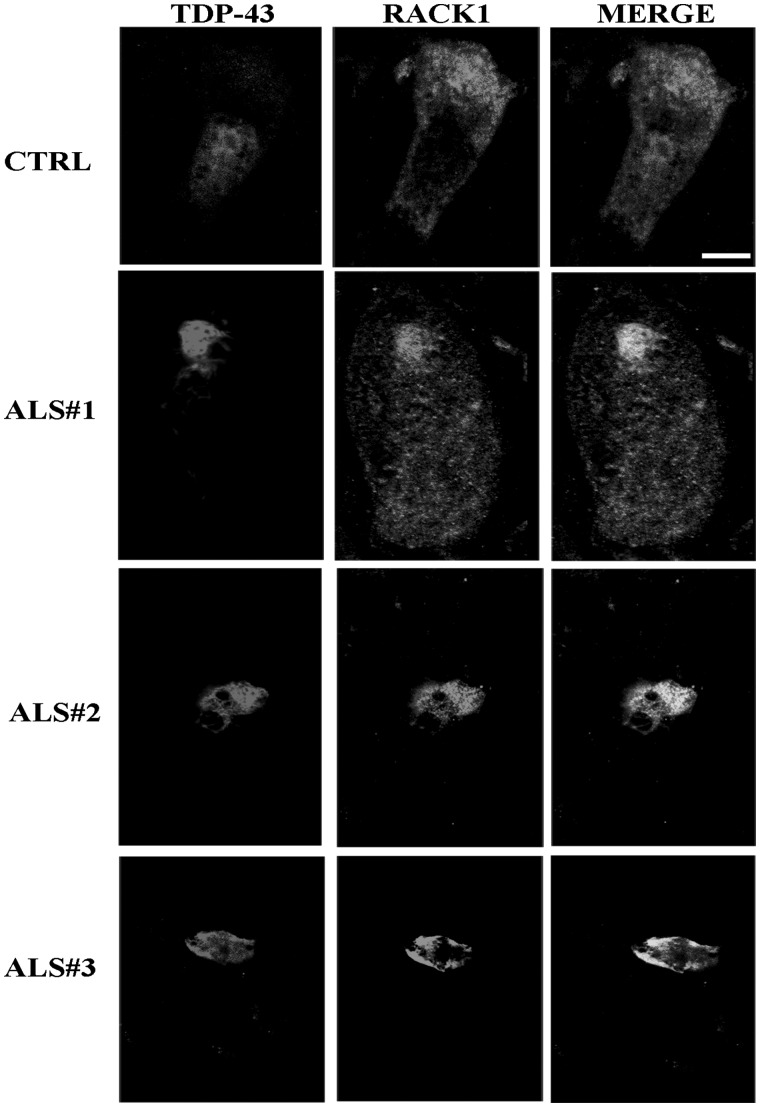
Finally, to further corroborate the interaction between TDP-43 and RACK1, we examined whether positive TDP-43 cytoplasmic inclusions of ALS cases included RACK1. Double label immunofluorescences for RACK1 and TDP-43 were conducted on spinal cord sections of three controls and three ALS patients. In control sections, RACK1 appeared diffusely in the cytoplasm and TDP-43 was mainly localized in the nucleus. In ALS tissues, even if RACK1 partially maintained a diffuse cytoplasmic localization, it tended to accumulate in cytoplasmic structures labeled by TDP-43. Co-localization analysis (Fig. 8) between RACK1 and TDP-43 indicated that RACK1 overlapped with TDP-43 in about 50% of TDP-43 positive cytoplasmic inclusions (Supplementary MaterialFig. S8A and B and Table S1 in supplementary data). This percentage of co-localization was near to that found for PABPC, another marker of SGs (21,39), and demonstrated that RACK1, as other SG components, was partially mislocalized in patients with ALS.
Figure 8.
RACK1 accumulates in TDP-43 positive inclusions in motor neurons of ALS spinal cord cases. The cytoplasmic localization of RACK1 was examined in motor neurons of three control patients (CTRL, in figure is reported only one case) and three ALS cases. RACK1 appeared diffuse in the cytoplasm of CTRL, while was inclined to (in ALS#1 case) or aggregated with TDP-43 in ALS cases. Scale bar = 10 µm.
Discussion
Although most of studies concerning TDP-43 has focused on its splicing activity, recently it has also been investigated its role in protein synthesis. In Drosophila and in mouse neurons, TDP-43 has been found to repress translation of Futsch/Map1B, Rac1 and GluR1 mRNAs (35,45) and to stimulate the phosphorylation of eIF2α to induce a prolonged state of translational repression (39). Here we report that an increase of cytoplasmic TDP-43 represses global protein synthesis. In this study, considering that TDP-43 is included in cytoplasmic aggregates of motor neurons in ALS/FTLD cases, a mutant form of human TDP-43 lacking the nuclear localization signal (ΔNLS-TDP-43), which main localizes in cytoplasm, has been overexpressed in SH-SY5Y neuroblastoma cells. In previous reports, the overexpression of this TDP-43 form results to be toxic for rat primary cortical neurons (42). Moreover, ΔNLS-TDP-43 overexpressing mice show neurodegeneration, with formation of rare cytoplasmic inclusions in motor neurons (25,46). In our cells, overexpression of ΔNLS-TDP-43 induces a decrease in cell viability, suggesting a putative toxicity of ΔNLS-TDP-43 as reported by (42), and shows a low number of cells containing cytoplasmic inclusions in agreement with mice models. Therefore, the results obtained in our cell model, reproducing identical features seen in vivo (25) and in vitro (42), suggest that a decrease of translational rate may be involved in ALS/FTLD disease.
What mechanism does TDP-43 use to repress translation? In SH-SY5Y cells, the overexpression of ΔNLS-TDP-43 increases 4E-BP1, a translational repressor, and reduces the phosphorylation of eIF4E, which promotes the translation when it is phosphorylated (40). 4E-BP1 represses the translation inhibiting the binding of eIF4E to eIF4F complex, a critical step for association to 5′m7GpppN cap structure of mRNAs to 43S complex and subsequent AUG scanning (41). When phosphorylated by mTOR kinase, 4E-BP1 loss its repressor activity (47). Here, we have not been able to determine neither the level of 4E-BP1 phosphorylation nor its association to eIF4E, however the increase of total 4E-BP1 in ΔNLS-TDP-43 overexpressing cells may partially contribute to reduce protein synthesis. Another contribution to the decrease of the translation in ΔNLS-TDP-43 overexpressing cells may come by the reduction of eIF4E phosphorylation. eIF4E is specifically phosphorylated by the MAPK-interacting kinases (Mnk) to translate specific mRNAs involved in cell growth and cell survival (48). Thus, that may suggest that the ΔNLS-TDP-43 toxicity for the cell, as seen here and in other reports, may be ascribed to translational decrease of eIF4E mRNA targets determined by the affected Mnk-eIF4E axis. However, taken together these results suggest that an increase of cytoplasmic TDP-43 hits the translational initiation phase.
The activity of cytoplasmic TDP-43 may require its localization on ribosomes. Here, we find that TDP-43 associate to translational machinery by the binding to an associated ribosomal protein, RACK1. RACK1, identified three decades ago as a scaffold protein for the activated PKC (49), has been found on 40S ribosome subunits and polyribosomes (29,30,50–52), on which it functions as a docking site for several proteins on the translational machinery (29,31,53). Moreover, given its ability to interact with several kinases, such as PKC, Erk, Src and JNK (54–57), RACK1 is considered a hub for translational control and cell signaling (58). Under stressful conditions, RACK1 is also included in SGs (59) and, recently, it has been found among proteins identified in TDP-43 complex (13,60). Through the binding of TDP-43 to a mutated form of RACK1 (RACK1DE) unable to associate to ribosomes/polyribosomes, we observe a decrease of the level of endogenous TDP-43 on translational machinery and a reduction of ΔNLS-TDP-43 aggregates, which may depend on the exclusion from polyribosomes of ΔNLS-TDP-43. On the basis of these considerations, we would have expected that only the overexpression of RACK1DE would rescue the effects of ΔNLS-TDP-43 on translation mechanisms. Instead we observe that also RACK1WT overexpression represses the ΔNLS-TDP-43 activity. This suggests that RACK1 overexpression, independently on its ribosomal localization, may stimulate a biochemical pathway to inhibit the activity of ΔNLS-TDP-43. However, considering the role of RACK1 on ribosome, we cannot exclude the hypothesis that it may function as a docking site on ribosomes for hypothetical phosphorylation of TDP-43, which may repress its ribosomal activity. In this case, the results obtained by the overexpression of RACK1WT and RACK1DE may be separately interpreted: in RACK1WT overexpression conditions, the hypothetical phosphorylation of TDP-43 on ribosome may inhibit its translational repression, while the exclusion of TDP-43 from ribosomes in RACK1DE overexpression may eliminate its repression activity.
In conclusion, here we propose that an excess of cytoplasmic TDP-43 associates with translational machinery by the binding to RACK1 to repress translation modulating the activity of 4E-BP1 and eIF4E phosphorylation. Moreover, the TDP-43 on polyribosomes may also promote the aggregation of TDP-43 immature cytoplasmic inclusions that, pursuing this state, may mature to cytoplasmic inclusions which may favour the degeneration of motor neurons. Future studies, using ALS relevant cell types, will be required to define by which molecular mechanisms TDP-43 regulates the overall translational rate during ALS/FTLD pathogenesis.
Materials and Methods
Cell culture, transfections and stable clones
Human neuroblastoma SH-SY5Y cells, obtained from American Type Culture Collection (ATCC, Rockville, MD) were cultured in DMEM/F12 medium containing 10% FBS and antibiotics (50 U/ml penicillin and 50 mg/ml streptomycin) at 37 °C in 5% CO2/95% air. Embryonic primary cultures were produced and cultured as reported in (29). SH-SY5Y cells were transfected with Myc, Myc-RACK1WT or RACK1DE, pEGFP, Myc-ΔNLS-TDP-43 or GFP-ΔNLS-TDP43 cDNAs using the manufacturer’s protocol of Lipofectamine 2000 (Thermofisher). Stable SH-SY5Y clone cells for Myc, Myc-RACK1WT and Myc-RACK1DE were produced by selecting transfected cells resistant at 450 mg/ml G418 (Gentamycin, Sigma).
SUnSET
To measure the rate of protein synthesis, cells were treated for 10 min with 5 µg/ml of puromycin (Thermofisher), then lysed and subjected to immunoblotting analysis as described below.
Immunofluorescences and antibodies
Cells were seeded the day before the staining, then rinsed three times with Phosphate Saline Buffer (Na2HPO4 10 mM, KH2PO4 1.8 mM, NaCl 137 mM, KCl 2.7 mM, pH 7.4) (PBS) and fixed with 4% paraformaldehyde for 10 min at room temperature. After three washes with PBS, cells were permeabilized with PBS-Triton-X 0.1% for 10 min at room temperature and then blocked in PBS-BSA 2% at 37 °C for 30 min. The following primary antibodies (dissolved in blocking solution) were incubated for 3 h at room temperature: mouse anti-Myc (Thermofisher, 9E10, 1:1000), mouse anti-RACK1 (BD Biosciences, 610178, 1:200), polyclonal anti-TDP-43 (ProteinTech, 10782-2-AP, 1:200), mouse anti-TIAR (Cell Signaling, 8611, 1:100), polyclonal anti-rpS6 (Cell Signaling, 5610, 1:200) and rabbit polyclonal eIF6/p27BBP (1:200, kindly given by prof. Biffo Stefano, INGM Milan Italy). After rinses, the following secondary antibodies were incubated for 1 h at room temperature in the dark: goat anti-mouse, goat anti-rabbit (Alexa Fluor® secondary antibodies, Molecular Probes), dissolved 1:500 in PBS-DAPI (Molecular Probes NucBlue® Live Ready Probes® Reagent R37605) as manufacturer protocol. After mounting with ProLong Gold (Invitrogen, P36930), cells were examined by confocal microscopy (Leica TCS SP5). For immunofluorescences on tissues, sections from sALS patients were deparaffinized and rehydrated, then treated with Glycin 0.1 M for 30 min. After three washes with PBS, sections were microwaved for 30 min in high citrate buffer. Section were then blocked for 1 h in PBS-0.2% Triton-X with 5% of Normal Donkey Serum. Primary antibodies, dissolved in blocking solution, were incubated overnight at 4 °C. After rinses with PBS, secondary antibodies were incubated for 1 h at room temperature in the dark, together with DAPI (1 µg/ml). Background fluorescence was quenched incubating sections for 3 min with Black Sudan 1%, then sections were washed with PBS and mounted on slides with ProLong Gold, and analysed with confocal microscopy.
SGs induction and quantification
Stress Granules formation was induced by heat, incubating cells for 30 min at 42 °C, 5% CO2/95% air. Then, cells were rapidly fixed for subsequent analysis. For quantification, ten areas of each coverslip were selected, and SGs identified by TIAR immunolabeling. Cells were scored positive when they had at least two TIAR-positive foci. Cell counting was performed on 40× magnification.
Polysome profiles
Growing cells were lysed in polysomal buffer (10 mM Tris–HCl, 150 mM NaCl, 10 mM MgCl2 and 0.1% TritonX-100). MgCl2 was substituted with 5 mM EDTA for polysome profiles in the presence of EDTA. Total lysate was clarified by centrifugation at 15 000g for 5 min at 4 °C and the supernatant was loaded on a continuous sucrose gradient 15–50% in 10 mM Tris–HCl, 150 mM NaCl, 10 mM MgCl2 or 10 mM EDTA. After ultracentrifugation at 4 °C in a SW41Ti Beckman rotor at 37 000 rpm for 2 h, absorbance at 254 nm was recorded by BioLogic LP software (BioRad) and fractions (1 ml each) were collected. The proteins in the collected fractions were precipitated incubating them in 10% trichloroacetic acid (TCA) for 30 min in ice, and then centrifuged at 15 000g for 15 min. The resulting protein pellets were then resuspended for subsequent analysis.
Histidine pull-down
Cells were lysed and protein extracts clarified as seen for polysome profiles. Then, extracts were incubated 2 h with nickel affinity resin (BioRad), pre-equilibrated with polysomal buffer. The resin was then extensively washed with 20 mM Imidazole dissolved in polysomal buffer and finally eluted with 500 mM Imidazole in polysomal buffer. The eluted proteins were then precipitated with 10% TCA, as described for polysomal profiles. For histidine pull down in presence of RNase I, protein extracts were or not treated with 50 µg/ml of RNase I for 30 min at room temperature before histidine purification as previously seen.
Co-immunoprecipitation assay
Cells were lysed and protein extracts clarified as seen for polysome profiles. Protein extracts were pre-cleared for 1 h at 4 °C with Protein G Sepharose Resin (Sigma) pre-equilibrated with polysomal buffer, then anti-Myc antibody (Thermofisher, 9E10, 1:1000) was added to pre-cleared extracts for 16 h at 4 °C. Pre-equilibrated resin was then incubated for 1 h, washed three times with PBS and finally resuspended for immunoblotting.
Immunoblottings and antibodies
SDS-PAGEs were performed on protein extracts obtained with polysomal buffer as seen for polysomal profiles. Protein concentration was determined with BCA analysis (Thermo Fisher Scientific). Equal amounts of proteins were loaded on each lane and separated on a 10% SDS-PAGEs, then transferred on a PVDF membrane. Membranes were blocked in 5% non-fat milk or 5% Bovine Serum Albumin in PBS1X with Tween (0.01%) for 30 min at 37 °C. The following primary antibodies were used: mouse anti-RACK1 (BD Biosciences, 610178, 1:2000), mouse anti-β-actin (Sigma, A2228, 1:1000), polyclonal anti-TDP43 (ProteinTech, 10782-2-AP, 1:2000), polyclonal anti-rpS6 (Cell Signaling, 5610, 1:1000), polyclonal anti-phospho-eIF2α (Cell Signaling, 9721, 1:1000), mouse anti-puromycin (Millipore, MABE343, 1:10 000) rabbit polyclonal 4E-BP1 (Cell Signaling, 9452, 1:1000), rabbit eIF4E (Cell Signaling, 9742, 1:1000), rabbit polyclonal phospho-eIF4E (Cell Signaling, 9741, 1:1000) and goat polyclonal Lamin B (Santa Cruz, 1:1000). Secondary HRP-conjugated anti-mouse, anti-rabbit or anti-goat antibodies and ECL reagent (1:5000, GE Healthcare) were used. For RACK1, mouse HRP-conjugated anti-IgM (1:5000, Sigma) was used.
Nucleus–cytoplasm fractionation
Cells were lysed in hypotonic buffer (20 mM HEPES pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA) and centrifuged at 700g for 5 min at 4 °C. The supernatant was then centrifuged 10 min at 4 °C at 10 000g, obtaining the cytoplasmic extract. The pellet from the first centrifugation was instead resuspended in nuclear lysis buffer (50mM Tris–HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% Sodium Deoxicolate, 0.1% SDS) and incubated 15 min in ice, then centrifuged 10 min at 4 °C at 10 000×g. The supernatant of this centrifugation is the nuclear extract. Next, nuclear and cytoplasmic proteins were processed as seen in Immunoblotting and antibodies.
MTT assay
A total of 15 000 cells of each genotype were plated in 0.2 ml in 96-well flat bottom plates. At the indicated times, 20 µl of 5 mg/ml MTT solution in PBS were added to each well for 4 h. After removal of the medium, 170 ml of DMSO were added to each well to dissolve the formazan crystals. The absorbance at 540 nm was determined using a Biokinetics plate reader (Bio-Tek Instruments Inc., Winooski, VT, USA). Triplicate wells were assayed for each condition and S.D. determined.
Declarations
All human tissues were obtained through a process that included written informed consent by the subjects’ next of kin. The acquisition process was evaluated by the Institutional Review Boards (Institutional Review Board of the Boston University Medical Campus and the University of Pittsburgh Institutional Review Board/University of Pittsburgh Committee for Oversight of Research Involving the Dead) and determined to be exempt from review by the full committee.
All experiments involving mice were performed in accordance with Italian National Regulations. Experimental protocols were reviewed by local Institutional Animal Care and Use Committees.
Author Contributions
AR performed and analysed experiments, FLR collected and cultured mice primary cells, RS and AC provided data analysis and discussion, MEM, DDW and BW contributed reagents/materials, MC conceived and analysed experiments and wrote the manuscript
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Masato Hasegawa (Tokyo Metropolitan Institute of Medical Science) and Francisco Baralle (ICGEB, International Centre for Genetic Engineering and Biotechnology) for providing vectors encoding mutant forms of TDP-43.
Conflict of Interest statement. None declared.
Funding
This work is supported by PAINCAGE grant (603191, AC) and NIH (ES020395, AG050471, NS089544), Alzheimer Association, Brightfocus, CureAlz, to BW. The founders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. Carri M.T., D′Ambrosi N., Cozzolino M. (2016) Pathways to mitochondrial dysfunction in ALS pathogenesis. Biochem. Biophys. Res. Commun., [DOI] [PubMed] [Google Scholar]
- 2. Lagier-Tourenne C., Cleveland D.W. (2009) Rethinking ALS: the FUS about TDP-43. Cell, 136, 1001–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M.. et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science, 314, 130–133. [DOI] [PubMed] [Google Scholar]
- 4. Rossi S., Cozzolino M., Carri M.T. (2016) Old versus new mechanisms in the pathogenesis of ALS. Brain. Pathol., 26, 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.C., Sun E., Wancewicz E., Mazur C.. et al. (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci., 14, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., Konig J., Hortobagyi T., Nishimura A.L., Zupunski V.. et al. (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci., 14, 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Colombrita C., Onesto E., Megiorni F., Pizzuti A., Baralle F.E., Buratti E., Silani V., Ratti A. (2012) TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J. Biol. Chem., 287, 15635–15647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lagier-Tourenne C., Polymenidou M., Cleveland D.W. (2010) TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet., 19, R46–R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buratti E., Baralle F.E. (2008) Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci., 13, 867–878. [DOI] [PubMed] [Google Scholar]
- 10. Fiesel F.C., Voigt A., Weber S.S., Van den Haute C., Waldenmaier A., Gorner K., Walter M., Anderson M.L., Kern J.V., Rasse T.M.. et al. (2010) Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J., 29, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang I.F., Wu L.S., Chang H.Y., Shen C.K. (2008) TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J. Neurochem., 105, 797–806. [DOI] [PubMed] [Google Scholar]
- 12. Fallini C., Bassell G.J., Rossoll W. (2012) The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum. Mol. Genet., 21, 3703–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Freibaum B.D., Chitta R.K., High A.A., Taylor J.P. (2010) Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome. Res., 9, 1104–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alami N.H., Smith R.B., Carrasco M.A., Williams L.A., Winborn C.S., Han S.S., Kiskinis E., Winborn B., Freibaum B.D., Kanagaraj A.. et al. (2014) Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron, 81, 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu-Yesucevitz L., Lin A.Y., Ebata A., Boon J.Y., Reid W., Xu Y.F., Kobrin K., Murphy G.J., Petrucelli L., Wolozin B. (2014) ALS-linked mutations enlarge TDP-43-enriched neuronal RNA granules in the dendritic arbor. J. Neurosci., 34, 4167–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aulas A., Vande Velde C. (2015) Alterations in stress granule dynamics driven by TDP-43 and FUS: a link to pathological inclusions in ALS? Front. Cell. Neurosci., 9, 423.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Higashi S., Kabuta T., Nagai Y., Tsuchiya Y., Akiyama H., Wada K. (2013) TDP-43 associates with stalled ribosomes and contributes to cell survival during cellular stress. J. Neurochem., 126, 288–300. [DOI] [PubMed] [Google Scholar]
- 18. Ayala V., Granado-Serrano A.B., Cacabelos D., Naudi A., Ilieva E.V., Boada J., Caraballo-Miralles V., Llado J., Ferrer I., Pamplona R.. et al. (2011) Cell stress induces TDP-43 pathological changes associated with ERK1/2 dysfunction: implications in ALS. Acta Neuropathol., 122, 259–270. [DOI] [PubMed] [Google Scholar]
- 19. Meyerowitz J., Parker S.J., Vella L.J., Ng D., Price K.A., Liddell J.R., Caragounis A., Li Q.X., Masters C.L., Nonaka T.. et al. (2011) C-Jun N-terminal kinase controls TDP-43 accumulation in stress granules induced by oxidative stress. Mol. Neurodegener., 6, 57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y.. et al. (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun., 351, 602–611. [DOI] [PubMed] [Google Scholar]
- 21. Bentmann E., Neumann M., Tahirovic S., Rodde R., Dormann D., Haass C. (2012) Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43). J. Biol. Chem., 287, 23079–23094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bosque P.J., Boyer P.J., Mishra P. (2013) A 43-kDa TDP-43 species is present in aggregates associated with frontotemporal lobar degeneration. PLoS One, 8, e62301.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Igaz L.M., Kwong L.K., Xu Y., Truax A.C., Uryu K., Neumann M., Clark C.M., Elman L.B., Miller B.L., Grossman M.. et al. (2008) Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am. J. Pathol., 173, 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arnold E.S., Ling S.C., Huelga S.C., Lagier-Tourenne C., Polymenidou M., Ditsworth D., Kordasiewicz H.B., McAlonis-Downes M., Platoshyn O., Parone P.A.. et al. (2013) ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl. Acad. Sci. USA, 110, E736–7E745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Igaz L.M., Kwong L.K., Lee E.B., Chen-Plotkin A., Swanson E., Unger T., Malunda J., Xu Y., Winton M.J., Trojanowski J.Q.. et al. (2011) Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J. Clin. Invest., 121, 726–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang C., Tong J., Bi F., Zhou H., Xia X.G. (2012) Mutant TDP-43 in motor neurons promotes the onset and progression of ALS in rats. J. Clin. Invest., 122, 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu-Yesucevitz L., Bilgutay A., Zhang Y.J., Vanderweyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M.. et al. (2010) Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One, 5, e13250.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Graber T.E., Hebert-Seropian S., Khoutorsky A., David A., Yewdell J.W., Lacaille J.C., Sossin W.S. (2013) Reactivation of stalled polyribosomes in synaptic plasticity. Proc. Natl. Acad. Sci. USA, 110, 16205–16210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ceci M., Welshhans K., Ciotti M.T., Brandi R., Parisi C., Paoletti F., Pistillo L., Bassell G.J., Cattaneo A. (2012) RACK1 is a ribosome scaffold protein for beta-actin mRNA/ZBP1 complex. PLoS One, 7, e35034.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ceci M., Gaviraghi C., Gorrini C., Sala L.A., Offenhauser N., Marchisio P.C., Biffo S. (2003) Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature, 426, 579–584. [DOI] [PubMed] [Google Scholar]
- 31. Jannot G., Bajan S., Giguere N.J., Bouasker S., Banville I.H., Piquet S., Hutvagner G., Simard M.J. (2011) The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Rep., 12, 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Coyle S.M., Gilbert W.V., Doudna J.A. (2009) Direct link between RACK1 function and localization at the ribosome in vivo. Mol. Cell. Biol., 29, 1626–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruan Y., Sun L., Hao Y., Wang L., Xu J., Zhang W., Xie J., Guo L., Zhou L., Yun X.. et al. (2012) Ribosomal RACK1 promotes chemoresistance and growth in human hepatocellular carcinoma. J. Clin. Invest., 122, 2554–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kedersha N.L., Gupta M., Li W., Miller I., Anderson P. (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol., 147, 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Coyne A.N., Siddegowda B.B., Estes P.S., Johannesmeyer J., Kovalik T., Daniel S.G., Pearson A., Bowser R., Zarnescu D.C. (2014) Futsch/MAP1B mRNA is a translational target of TDP-43 and is neuroprotective in a Drosophila model of amyotrophic lateral sclerosis. J. Neurosci., 34, 15962–15974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Coyne A.N., Yamada S.B., Siddegowda B.B., Estes P.S., Zaepfel B.L., Johannesmeyer J.S., Lockwood D.B., Pham L.T., Hart M.P., Cassel J.A.. et al. (2015) Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum. Mol. Genet., 24, 6886–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Winton M.J., Igaz L.M., Wong M.M., Kwong L.K., Trojanowski J.Q., Lee V.M. (2008) Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem., 283, 13302–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ayala Y.M., Zago P., D′Ambrogio A., Xu Y.F., Petrucelli L., Buratti E., Baralle F.E. (2008) Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell. Sci., 121, 3778–3785. [DOI] [PubMed] [Google Scholar]
- 39. Kim H.J., Raphael A.R., LaDow E.S., McGurk L., Weber R.A., Trojanowski J.Q., Lee V.M., Finkbeiner S., Gitler A.D., Bonini N.M. (2014) Therapeutic modulation of eIF2alpha phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat. Genet., 46, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Waskiewicz A.J., Flynn A., Proud C.G., Cooper J.A. (1997) Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J., 16, 1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hinnebusch A.G., Ivanov I.P., Sonenberg N. (2016) Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science, 352, 1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barmada S.J., Skibinski G., Korb E., Rao E.J., Wu J.Y., Finkbeiner S. (2010) Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J. Neurosci., 30, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vanderweyde T., Yu H., Varnum M., Liu-Yesucevitz L., Citro A., Ikezu T., Duff K., Wolozin B. (2012) Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J. Neurosci., 32, 8270–8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Colombrita C., Zennaro E., Fallini C., Weber M., Sommacal A., Buratti E., Silani V., Ratti A. (2009) TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem., 111, 1051–1061. [DOI] [PubMed] [Google Scholar]
- 45. Majumder P., Chu J.F., Chatterjee B., Swamy K.B., Shen C.J. (2016) Co-regulation of mRNA translation by TDP-43 and Fragile X Syndrome protein FMRP. Acta Neuropathol., 132(5):721–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walker A.K., Spiller K.J., Ge G., Zheng A., Xu Y., Zhou M., Tripathy K., Kwong L.K., Trojanowski J.Q., Lee V.M. (2015) Functional recovery in new mouse models of ALS/FTLD after clearance of pathological cytoplasmic TDP-43. Acta Neuropathol., 130, 643–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gingras A.C., Gygi S.P., Raught B., Polakiewicz R.D., Abraham R.T., Hoekstra M.F., Aebersold R., Sonenberg N. (1999) Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev., 13, 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bramham C.R., Jensen K.B., Proud C.G. (2016) Tuning specific translation in cancer metastasis and synaptic memory: control at the MNK-eIF4E axis. Trends. Biochem. Sci., 41, 847–858. [DOI] [PubMed] [Google Scholar]
- 49. Ron D., Chen C.H., Caldwell J., Jamieson L., Orr E., Mochly-Rosen D. (1994) Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc. Natl. Acad. Sci. USA, 91, 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Angenstein F., Staak S. (1997) Receptor-mediated activation of protein kinase C in hippocampal long-term potentiation: facts, problems and implications. Prog. Neuropsychopharmacol. Biol. Psychiatry, 21, 427–454. [DOI] [PubMed] [Google Scholar]
- 51. Sengupta J., Nilsson J., Gursky R., Spahn C.M., Nissen P., Frank J. (2004) Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat. Struct. Mol. Biol, 11, 957–962. [DOI] [PubMed] [Google Scholar]
- 52. Rabl J., Leibundgut M., Ataide S.F., Haag A., Ban N. (2011) Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science, 331, 730–736. [DOI] [PubMed] [Google Scholar]
- 53. Gallo S., Manfrini N. (2015) Working hard at the nexus between cell signaling and the ribosomal machinery: an insight into the roles of RACK1 in translational regulation. Translation (Austin), 3, e1120382.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vomastek T., Iwanicki M.P., Schaeffer H.J., Tarcsafalvi A., Parsons J.T., Weber M.J. (2007) RACK1 targets the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway to link integrin engagement with focal adhesion disassembly and cell motility. Mol. Cell. Biol., 27, 8296–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chang B.Y., Harte R.A., Cartwright C.A. (2002) RACK1: a novel substrate for the Src protein-tyrosine kinase. Oncogene, 21, 7619–7629. [DOI] [PubMed] [Google Scholar]
- 56. Lopez-Bergami P., Habelhah H., Bhoumik A., Zhang W., Wang L.H., Ronai Z. (2005) RACK1 mediates activation of JNK by protein kinase C [corrected]. Mol. Cell., 19, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gandin V., Gutierrez G.J., Brill L.M., Varsano T., Feng Y., Aza-Blanc P., Au Q., McLaughlan S., Ferreira T.A., Alain T.. et al. (2013) Degradation of newly synthesized polypeptides by ribosome-associated RACK1/c-Jun N-terminal kinase/eukaryotic elongation factor 1A2 complex. Mol. Cell. Biol., 33, 2510–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ron D., Adams D.R., Baillie G.S., Long A., O′Connor R., Kiely P.A. (2013) RACK1 to the future—a historical perspective. Cell Commun. Signal., 11, 53.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., Takekawa M. (2008) Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell. Biol., 10, 1324–1332. [DOI] [PubMed] [Google Scholar]
- 60. Blokhuis A.M., Koppers M., Groen E.J., van den Heuvel D.M., Dini Modigliani S., Anink J.J., Fumoto K., van Diggelen F., Snelting A., Sodaar P.. et al. (2016) Comparative interactomics analysis of different ALS-associated proteins identifies converging molecular pathways. Acta Neuropathol., 132, 175–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.