Abstract
Lamina-associated polypeptide 1 (LAP1) is an integral protein of the inner nuclear membrane that has been implicated in striated muscle maintenance. Mutations in its gene have been linked to muscular dystrophy and cardiomyopathy. As germline deletion of the gene encoding LAP1 is perinatal lethal, we explored its potential role in myogenic differentiation and development by generating a conditional knockout mouse in which the protein is depleted from muscle progenitors at embryonic day 8.5 (Myf5-Lap1CKO mice). Although cultured myoblasts lacking LAP1 demonstrated defective terminal differentiation and altered expression of muscle regulatory factors, embryonic myogenesis and formation of skeletal muscle occurred in both mice with a Lap1 germline deletion and Myf5-Lap1CKO mice. However, skeletal muscle fibres were hypotrophic and their nuclei were morphologically abnormal with a wider perinuclear space than normal myonuclei. Myf5-Lap1CKO mouse skeletal muscle contained fewer satellite cells than normal and these cells had evidence of reduced myogenic potential. Abnormalities in signalling pathways required for postnatal hypertrophic growth were also observed in skeletal muscles of these mice. Our results demonstrate that early embryonic depletion of LAP1 does not impair myogenesis but that it is necessary for postnatal skeletal muscle growth.
Introduction
Mutations in genes encoding nuclear lamins and other nuclear envelope proteins cause a range of diseases, most commonly affecting tissues in the heart and skeletal muscle (1,2). X-linked Emery-Dreifuss muscular dystrophy (EDMD; OMM #310300), caused by mutations in the EMD gene encoding emerin, an integral protein of the inner nuclear membrane, was the first human disorder to be recognized as a nuclear envelope-linked disease (3–6). EDMD is classically characterized clinically by a triad of slowly progressive muscle weakness and wasting in a scapulo-humeroperoneal distribution, early contractures of the elbows, ankles, and posterior neck and dilated cardiomyopathy with conduction defects (7,8). Cardiomyopathy with variable skeletal muscle involvement, including cases of EDMD and limb girdle muscular dystrophy 1B, is also caused by autosomal mutations in LMNA encoding A-type lamins (9–12). Mutations in LMNA have also been linked to congenital muscular dystrophy with cardiomyopathy (13,14), suggesting that A-type lamins play a role in early striated muscle development. Mutations in genes encoding other nuclear envelope proteins, including nesprin-1/2, LUMA and SUN-1/2, have also been reported in patients with EDMD or EDMD-like phenotypes (15–18). These findings have spurred research on the functions of nuclear envelope proteins in cardiac and skeletal muscle.
Various studies have demonstrated that the elimination of nuclear envelope proteins or expression of disease-causing variants in muscle cells leads to defects in skeletal muscle development and maintenance. Mice lacking A-type lamins display retarded postnatal growth and muscular dystrophy (19). Homozygous Lmna ΔK32 knock-in mice, harbouring a congenital muscular dystrophy-causing mutation, exhibit striated muscle maturation delay (20). Homozygous Lmna knock-in with the H222P amino acid substitution has a muscular dystrophy and cardiomyopathy phenotype (21,22). Emerin null mice do not display gross myopathies, however, defective muscle regeneration is evident and myogenic differentiation is delayed (23). Targeted deletion of the C- terminus of nesprin-1 in mice leads to disorganized myonuclei, central nuclei, and small myofibres, the features of abnormal muscle development and maintenance (16,24). Various in vitro studies using myoblast cell lines, human primary myoblasts or satellite cells have demonstrated that depletion of the endogenous A-type lamins and other nuclear envelope-associated proteins, as well as overexpression of mutant forms of these proteins lead to defective myogenic differentiation (25–29).
Vertebrate skeletal muscle development requires the coordinated events of determination, proliferation and differentiation of myogenic precursors, a fusion of myoblasts, and hypertrophic growth and remodelling of myofibres (30,31). During the early myogenesis period at embryonic day (E) 8, myogenic progenitors are derived from dermomyotome and express the transcription factors paired box protein 3 (Pax3) and 7 (Pax7). These progenitors proliferate and differentiate into multinucleated myofibres through orchestrated activation of myogenic regulatory factors, including myogenic factor 5 (Myf5), myoblast determination protein (MyoD), muscle-specific regulatory factor 4 (MRF4) and myogenin. Myofibre hypertrophy with an increase of myonuclei number occurs during the early postnatal growth period (postnatal day 3 to 21). In addition to the hypertrophic growth of myofibres, the expression of myosin heavy chain isoforms switches from embryonic to adult isoforms, which enhance the maturation to adult skeletal muscle (32,33).
Lamina-associated polypeptide 1 (LAP1) is a protein encoded by the TOR1AIP1 gene (which for simplicity we refer to herein as LAP1 in human or Lap1 in mice) (34). LAP1 was originally discovered as a transmembrane protein of the inner nuclear membrane, with three isoforms, associated with the nuclear lamina (35,36). Subsequently, LAP1 was shown to interact with the endoplasmic reticulum/perinuclear space protein torsinA and with protein phosphatase 1 (37,38). Mice carrying a homozygous gene-trap insertion in the Lap1 locus display late embryonic or perinatal lethality, suggesting that LAP1 has essential roles in early mammalian development (39). We have previously demonstrated that LAP1 interacts with emerin and is essential for adult muscle function or maintenance in mice (40). Furthermore, several case reports have linked mutations in LAP1 to striated muscle disease in humans. A genome-wide homozygosity mapping study led to discovery of a LAP1 mutation in a family with muscular dystrophy with cardiomyopathy (41). In another case report, a homozygous missense mutation in the gene encoding LAP1 was found in a boy with severe dystonia, progressive cerebellar atrophy, and dilated cardiomyopathy (42). Compound heterozygous frame shift and missense mutations in LAP1 have also been linked to heart failure and muscular dystrophy (43). These cases emphasize the essential role of LAP1 in striated muscle function and provide clinical significance to the studies of how LAP1 regulates striated muscle development and maintenance.
We previously generated a mouse line carrying a conditional Lap1 floxed allele and bred it to mice expressing Cre under control of the myosin creatine kinase promoter, which induces Cre expression in differentiating myocytes at E17. The resulting offspring with selective depletion of LAP1 in skeletal muscle (referred as M-CKO mice) develop a muscular dystrophy phenotype starting at 8 weeks of age with progression leading to 100% mortality at 30 weeks of age (40). The phenotypic delay in the M-CKO mice compared to the perinatal lethality of the Lap1 gene-trap knockout mice led us to hypothesize that skeletal muscle selective depletion of LAP1 at an earlier myogenic stage would result in earlier onset phenotypes, as in the germline knockout mice, and display defects during late embryonic or postnatal muscle development. We therefore tested this hypothesis in mice and further analyzed the potential role of LAP1 in skeletal muscle differentiation in cultured cells.
Results
Expression of LAP1 isoforms during skeletal muscle development
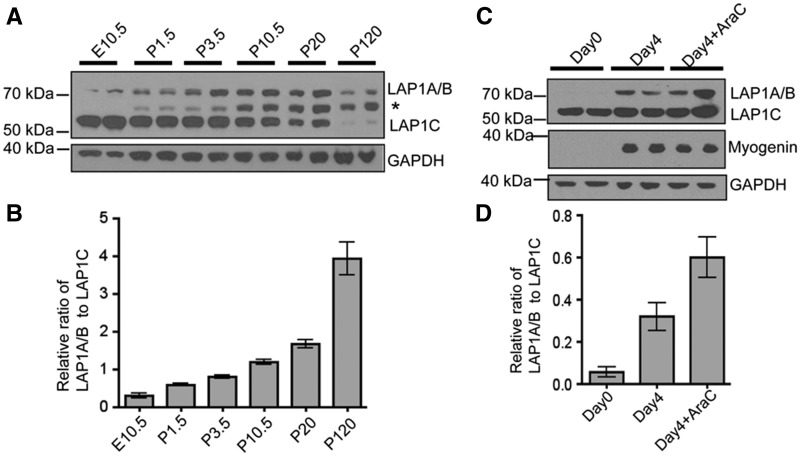
We examined whether isoforms of LAP1 are differentially expressed during mouse skeletal muscle development. There are three LAP1 isoforms that arise by alternative RNA splicing (35,36,44). The smaller isoform LAP1C (∼55 kDa) was predominantly expressed in the embryonic stage at E10.5 and early postnatal (P) day 1.5–10.5, whereas the larger isoforms LAP1A (∼72 kDa) and LAP1B (∼68 kDa) were predominantly expressed in mature skeletal muscle at later stages while LAP1C expression was concurrently diminished (Fig. 1A and B). We next examined the expression of LAP1 isoforms during in vitro differentiation of cultured mouse myoblast. C2C12 myoblasts expressed more LAP1A and LAP1B four days after induced differentiation, as confirmed by myogenin expression, compared to before differentiation; however, the level of LAP1C stayed the same (Fig. 1C and D). These results suggest that LAP1 isoforms may have different roles during skeletal muscle development.
Figure 1.
LAP1 expression is regulated during skeletal muscle development in vivo and myogenic differentiation in vitro. (A) Immunoblots showing differential expression of endogenous LAP1 isoforms during skeletal muscle development in wild type C57BL/6 mice. Whole embryo extract was used for E10.5 samples and hind limb extracts devoid of skin and bones were used for P1.5 and P3.5 samples. The rest of samples were prepared from dissected quadriceps. GAPDH was used as a loading control. The band migrating with an apparent molecular mass of 60 kDa (marked with *) is nonspecific. (B) Graphic representation of relative ratios of band densities of LAP1A and LAP1B to band densities of LAP1C at indicated developmental stages. Band densities were measured from two independent immunoblots using protein extracts from two mice at each stage. Values are means ± standard errors (n = 4). (C) Immunoblots showing increased expression of LAP1A and LAP1B during myogenic differentiation of C2C12 cells. Cells were collected at the indicated time after adding differentiation media. Expressions of GADPH and the differentiation marker myogenin are also shown. Treatment with cytosine arabinoside (AraC) eliminates proliferating myoblasts and enriches myotubes in the culture. LAP1A and LAP1B migrate at the same apparent molecular mass in the immunoblots. (D) Relative ratio of band densities of LAP1A and LAP1B to band densities of LAP1C. Results from immunoblots using four independent cultures were used to measure band densities. Values are means ± standard errors (n = 4).
Germline Lap1 knockout mice have normal early myogenesis but reduced myofibre size
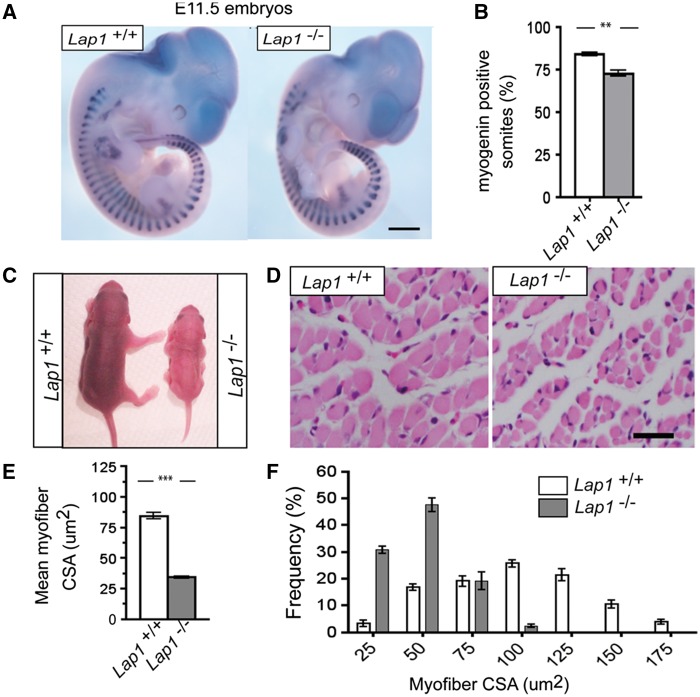
LAP1 expression can be detected in mouse embryos at E10.5 and Lap1 gene trap knockout mice (Lap1-/-) die during the perinatal period (39). We sought to determine if this perinatal lethality is due to defective embryonic myogenesis. The gross morphology and size of Lap1+/+ and Lap1-/- littermate embryos were indistinguishable at E11.5 (Fig. 2A). We next performed whole-mount RNA in situ hybridization to analyze the expression of myogenin, a critical myogenic differentiation gene, in Lap1+/+ and Lap1-/- embryos at E11.5. While the length and shapes of myotomes were not obviously different from wild type embryos, the number of myogenin positive somites was significantly reduced in embryos lacking LAP1 (Fig. 2A and 2B). The few Lap1-/- mice that survived to P2.5 were significantly smaller than Lap+/+ littermates (Fig. 2C). The general skeletal muscle structure was not obviously different in P2.5 Lap1+/+ and Lap1-/- mice; however, the mean myofibre cross sectional area (CSA) was significantly reduced in quadriceps muscle of mice lacking Lap1 (Fig. 2D and 2E) with a shift towards smaller myofibres (Fig. 2F). There was no increase in myofibres with centrally-located nuclei or degenerated myofibres in quadriceps from Lap1-/- mice at P2.5 (data not shown). These data indicate that LAP1 is not essential for early embryonic myogenesis, including muscle progenitor specification, determination and differentiation, but that it is required for late embryonic or postnatal growth of skeletal muscle.
Figure 2.
Lap1 knock out mouse skeletal muscle histology and whole-mount in situ hybridization. (A) Whole-mount in situ hybridization of E11.5 embryos of the indicated genotype with a myogenin probe. Note that myogenin positive signals are detected in somites. Scale bar: 500 µm (B) Percentage of myogenin positive somites among total somites. Values are means ± standard errors (n = 3 per genotype); **P < 0.001. (C) Photographs of Lap1+/+ and Lap1-/- mice at P2.5. (D) Representative hematoxylin and eosin-stained cross sections of quadriceps muscle from Lap1+/+ and Lap1-/- mice at P2.5. Scale bar: 25 µm (E) Quantification of individual quadriceps myofibre CSA from hematoxylin and eosin-stained cross sections from each genotype (n = 2 per genotype). Three different fields from each section from different animals were photographed and measured. A total of 311 myofibres from Lap1+/+ and 355 myofibres from Lap1-/- mice were counted. Values are means ± standard errors; **P < 0.001. (F) Frequency histogram showing the distribution of quadriceps muscle fibre CSA in Lap1+/+ and Lap1-/- mice at P2.5; ***P < 0.0001.
Primary myoblasts lacking LAP1 have defective terminal differentiation and altered expression of skeletal muscle regulatory factors
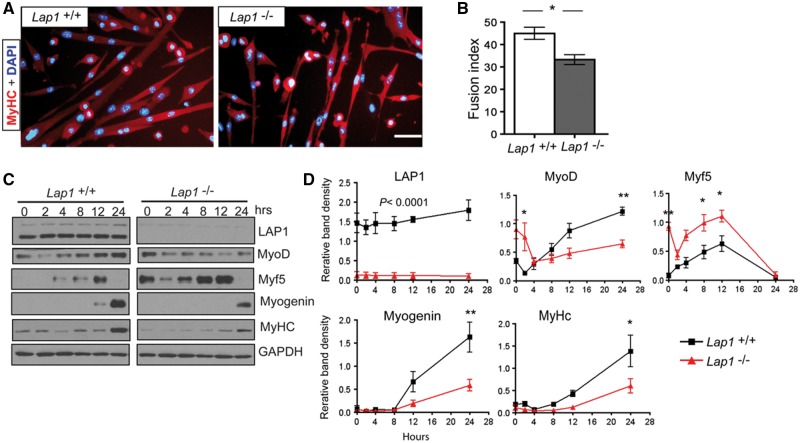
To investigate the requirement for LAP1 in myogenic differentiation in vitro, we isolated primary myoblasts from neonatal Lap1+/+ and Lap1-/-mice. We first confirmed the purity of myoblasts devoid of fibroblasts by staining with anti-Pax7 antibody, which showed that nearly 100% of cells contained Pax7 (Supplementary Material, Fig. S1A). In growth media, Lap+/+ and Lap1-/- primary myoblasts had similar proliferation rates based on 5-bromo-2-deoxyuridine (BrdU) labeling (Supplementary Material, Fig. S1B).
We compared the myogenic differentiation capacity of myoblasts from Lap1+/+ and Lap1-/-mice in differentiation media for 72 h. Myoblasts lacking LAP1 were able to form multi-nucleated myotubes; however, the myotubes were shorter and had smaller diameters than those formed from Lap1+/+ myoblasts (Fig. 3A). The fusion index, which represents the percentage of myotubes with more than two nuclei, was also significantly decreased for Lap1-/- myoblasts (Fig. 3B). During 24 h of differentiation, myoblasts lacking LAP1 had increased expression of Myf5 and delayed expression of myogenin, MyoD and myosin heavy chains (MyHC) (Fig. 3C and D). These results suggest that LAP1 is necessary for the normal expression of myogenic regulatory factors and terminal differentiation in vitro.
Figure 3.
Myogenic differentiation of myoblasts isolated from Lap1+/+ and Lap1-/- mice. (A) Representative immunofluorescence micrographs of myoblasts from Lap1+/+ and Lap1-/- mice after incubation for 72 h in differentiation media. Cells were fixed and stained with antibody against MyHC and nuclei labelled with DAPI. Scale bar: 50 µm. (B) Fusion index indicating percentages of myotubes containing two or more nuclei. Images were taken from 3 different cultures for each genotype and 1,061 nuclei for Lap1+/+ and 916 nuclei for Lap1-/- cells were counted. Values are means ± standard errors; *P < 0.05. (C) Immunoblots showing expression of LAP1, MyoD, Myf5, myogenin, MyHC and GADPH during differentiation of Lap1+/+ and Lap1-/- myoblasts (D) Band densities for indicated proteins normalized to the GAPDH signal at the indicated time points. Results from two independent immunoblots each from two different batches of cultures were used to measure band densities. P < 0.0001 (all time points of LAP1 signals), *P < 0.05, **P < 0.005.
Depletion of LAP1 during early embryonic myogenesis leads to growth retardation and premature death in mice
The germline gene trap Lap1-/- mouse is not an ideal model to determine the specific roles of LAP1 in skeletal muscle development, as non-cell autonomous signals from other tissues can affect the process. To more specifically examine the requirement of LAP1 in early skeletal muscle development, we deleted Lap1 during the early myogenic period by crossing Lap1 floxed mice to Myf5-Cre knock-in mice that have been widely used for this purpose (45–47). We refer this mouse line (Myf5Cre+/-; Lap1f/f) as Myf5-Cre driven Lap1 conditional knockout mice (Myf5-Lap1CKO). Although the Myf5 promoter has been reported to express in non-muscle tissues such as adipose tissue (48,49), the final contribution of the Myf5 lineage to adult myonuclei is significant (50,51). Furthermore, the Myf5 promoter-driven Cre-expressing knock-in mouse has been widely used to assess the roles of genes during myogenesis (47,52–54).
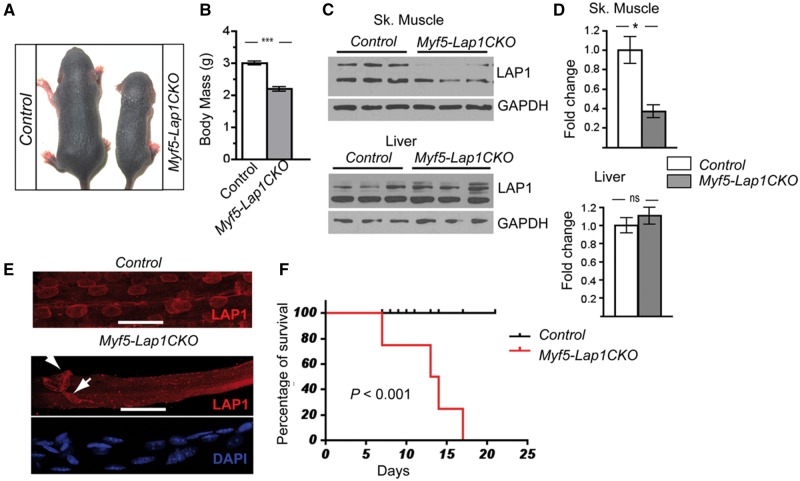
Myf5-Lap1CKO mice were born at the expected Mendelian ratios with reduced body weight (Control [Lap1f/f]): 1.37 ± 0.02 g, n = 6; Myf5-Lap1CKO: 1.27 ± 0.03 g; n = 6, P = 0.03) and drank milk after birth. At P5.5, Myf5-Lap1CKO pups were smaller and had a significantly reduced body mass compared to littermate Control pups (Fig. 4A and B). This was the same for both sexes (data not shown). LAP1 isoforms were reduced by approximately 60% in protein extracts of quadriceps muscle from Myf5-Lap1CKO mice at P5.5 age, while the expression levels of LAP1 were comparable in liver extracts from mice with both genotypes (Fig. 4C and 4D). The presence of LAP1 in protein extracts from the quadriceps muscle of Myf5-Lap1CKO mice is likely due to non-muscle cells and the incomplete expression of Myf5 promoter in skeletal muscle (51). This was confirmed by immunoflurescence microscopy of isolated myofibres from Myf5-Lap1CKO mice, which showed the absence of LAP1 signals in most myonuclei but the expression of LAP1 in some nuclei of either myofibres or possibly connective tissues (Fig. 4E). Most of the nuclei of muscle fibres from both Control and Myf5-Lap1CKO mice at P5.5 expressed lamin A/C, indicating a selective depletion of LAP1 in the myonuclei from Myf5-Lap1CKO mice (Supplementary Material, Fig. S2). Myf5-Lap1CKO mice started to die at P8 and had 100% mortality at P17.5, a significantly shorter median survival than Control mice (Fig. 4F). These data show that skeletal muscle expression of LAP1 after E8 is not essential for embryonic development but is required for early postnatal development of mice.
Figure 4.
Phenotypic analysis of Myf5-Lap1CKO mice. (A) Photos of littermate Control and Myf5-Lap1CKO mice at P5.5. (B) Body mass of Control (n = 11) and Myf5-Lap1CKO (n = 8) mice at P5.5. Values are means ± standard errors; ***P <0.0001. (C) Immunoblots of protein extracts of skeletal muscle (Sk. Muscle) and liver from P5.5 Control and Myf5-Lap1CKO mice. Blots were probed with antibodies against LAP1 and GAPDH. Each lane has protein extracts from a different animal. (D) Quantification of LAP1 expression normalized to GAPDH in protein extracts from skeletal muscle and liver of Control and Myf5-Lap1CKO mice. Values are means ± standard errors (n = 3 for each genotype); *P < 0.05. (E) Confocal immunofluorescence micrographs of quadriceps muscle fibres from Control and Myf5-Lap1CKO mice at P5.5 showing absence of LAP1 signal (red) in myonuclei of Myf5-Lap1CKO mice. Myofibres from a Myf5-Lap1CKO mouse were counterstained with DAPI to show nuclei (blue). Arrows indicate nuclei stained with anti-LAP1 antibody that appear to be outside of myofibres or myonuclei with incomplete deletion of LAP1. Scale bars: 25 µm. (F) Kaplan-Meier survival curves for Control (n = 15) and Myf5-Lap1CKO (n = 16) mice. P < 0.001 for median survival.
Depletion of LAP1 during early embryonic myogenesis results in reduced muscle mass and hypotrophic myofibres
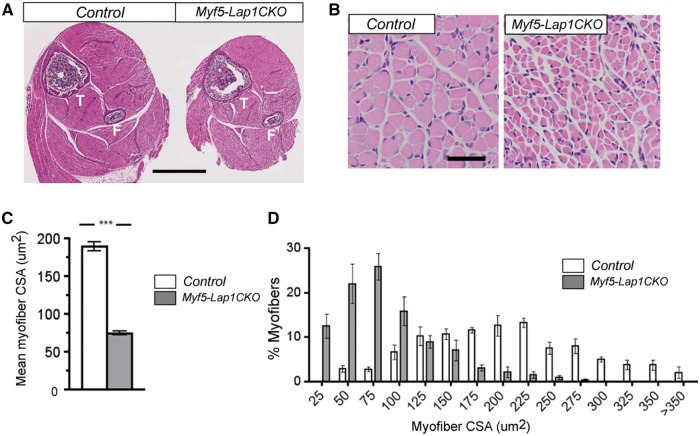
To determine if the neonatal lethality of Myf5-Lap1CKO was caused by skeletal muscle defects, we first performed histopathological analysis on muscles from Myf5-Lap1CKO and littermate Control mice at P5.5 and P10.5. Microscopic examination of hematoxylin and eosin-stained cross sections of hindlimbs demonstrated reduced overall muscle size in Myf5-Lap1CKO mice compared to that of littermate Control mice (Fig. 5A). Higher power examination showed global hypotrophy with markedly reduced myofibre size in Myf5-Lap1CKO mice (Fig. 5B). Laminin and myosin type I-stained cross sections of hind limb muscles from Myf5-Lap1CKO mice showed similar features with significantly reduced myofibre size but type II fibres were not detected, likely because they just start to appear at a relatively low level at this age (Supplementary Material, Fig. S3). However, the general muscle organization and patterning was not changed and there were no myopathic features such as centrally-located myonuclei or myofibre degeneration. There was a significant reduction in mean CSA (Fig. 5C) and a shift towards myofibres with smaller CSAs in Myf5-Lap1CKO mice (Fig. 5D). Collectively, these results indicated that LAP1 is not essential for initial myogenic determination and formation of myofibre but is required for late embryonic myofibre maturation or postnatal hypertrophic muscle growth.
Figure 5.
Histological examination of skeletal muscles from Control and Myf5-Lap1CKO mice at P5.5. (A) Representative micrographs of hematoxylin and eosin-stained cross sections of hindlimbs from Control (Lap1f/f) and Myf5-Lap1CKO (Myf5cre; Lap1f/f) mice at P5.5. The tibia (T) and fibula (F) bones are indicated. Scale bar: 1 mm. (B) Representative hematoxylin and eosin-stained cross sections of quadriceps muscle from Control and Myf5-Lap1CKO mice at P5.5. Scale bar: 50 µm. (C) Mean myofibre size in quadriceps from Control and Myf5-Lap1CKO mice at P5.5. The myofibre cross sectional area (CSA) were measured for three different sections from three mice per genotype. A total of 181 myofibres from Control and 327 myofibres from Myf5-Lap1CKO mice were measured. Values are means ± standard errors; **P <0.001. (D) Frequency histograms showing the distribution of quadriceps muscle fibre CSA in Control and Myf5-Lap1CKO mice at P5.5 (n = 3 per genotype); ***P < 0.0001.
As Myf5 is also expressed in brown adipose precursor cells (48,55), we examined interscapular brown adipose tissues from Control and Myf5-Lap1CKO mice at P5.5. Microscopic examination of hematoxylin and eosin-stained sections of brown adipose tissues from Myf5-Lap1CKO showed cells with eosinophilic cytoplasm containing lipid droplets, a small internal nucleus and no obvious histological abnormalities, which were the same as in Control mice (Supplementary Material, Fig. S4). Hence, the severe skeletal muscle defects observed in Myf5-Lap1CKO mice were not likely caused by defects in brown adipose tissue.
Ultrastructure of myofibres in mice with depletion of LAP1 during early embryonic myogenesis
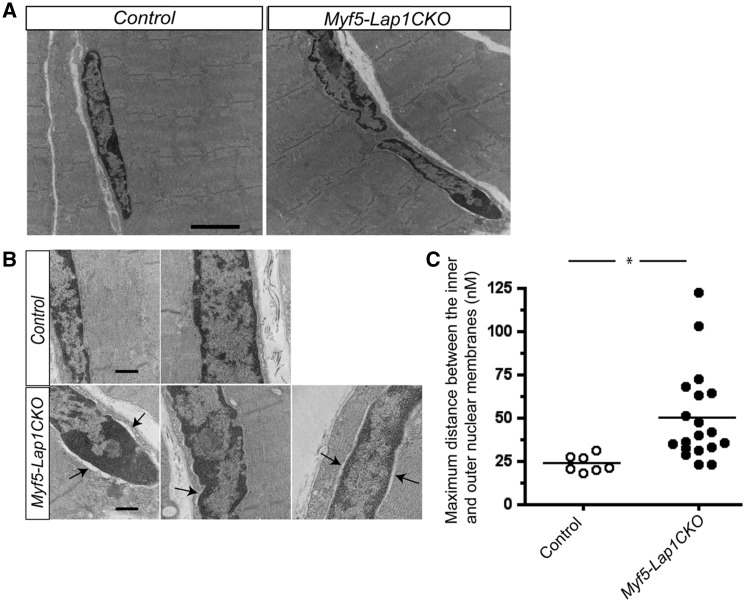
We performed electron microscopy to identify possible abnormalities of sarcomere structure or nuclear envelope architecture in myofibres from Myf5-Lap1CKO mice. Electron micrographic examination of quadriceps muscles from Myf5-Lap1CKO mice did not identify evidence of sarcomere disorganization, fibre degeneration or myopathic features; however, some nuclei at P5.5 showed an abnormal nuclear envelope architecture with ruffled shapes (Fig. 6A). There were moderately hypotropic mitochondria but no abnormal rearrangement of cristae or abnormal proliferation of mitochondria (Supplementary Material, Fig. S5). There was also an enlarged perinuclear space between the inner and outer nuclear membranes (Fig. 6B). To quantify the extent of the enlarged perinuclear space, we measured the maximum distance between the inner and outer nuclear membranes in myonuclei from Control and Myf5-Lap1CKO mice. This revealed that myonuclei from Myf5-Lap1CKO mice had a significantly wider perinuclear space (Fig. 6C). These data suggest that LAP1 is required for proper nuclear envelope architecture formation in myofibres. An enlarged perinuclear space has been observed in cells expressing other abnormal variants of nuclear envelope proteins (15,56,57).
Figure 6.
Ultrastructural analysis of nuclei in quadriceps from Control and Myf5-Lap1CKO mice at P5.5. (A) Electron micrographs of quadriceps from Control and Myf5-Lap1CKO mice at P5.5. Scale bar: 2μm (B) Electron micrographs of nuclei in quadriceps sections from Control and Myf5-Lap1CKO mice. Arrows indicate areas with increased distance between the inner and outer nuclear membranes. Scale bars: 500 nm. (C) Measurement of maximum distance between the inner and outer nuclear membranes in myonuclei of Control mice (n = 7 nuclei) and Myf5-Lap1CKO mice (n = 18 nuclei). Each circle represents the value from an individual myonucleus and horizontal bars the means; *P < 0.05.
LAP1 depletion during early embryonic myogenesis impairs the myogenic potentials of satellite cells
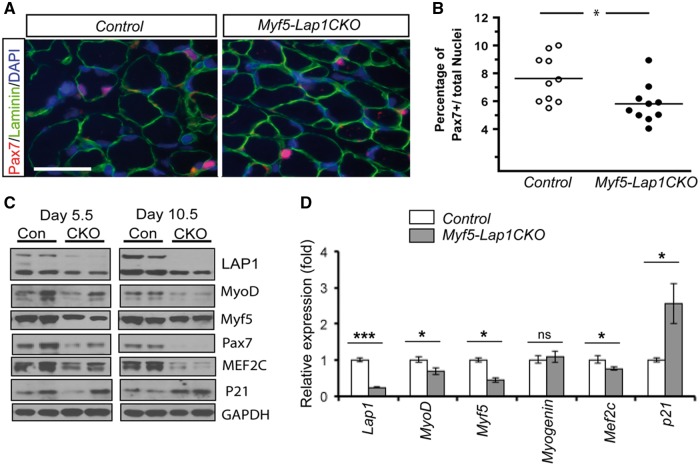
Early postnatal muscle growth from P0 to P21 correlates with an increase in myonuclei resulting from Pax7 positive satellite cell proliferation and fusion (32,58,59) Given the smaller myofibre diameter in skeletal muscle of Myf5-Lap1CKO mice, we suspected that LAP1 depletion at an early postnatal age would prevent committed myoblasts (Pax7/Myf5 positive cells) from fusing into mature myofibres. To test whether the satellite cell population was affected by LAP1 depletion, we performed immuohistochemistry on quadriceps from Myf5-Lap1CKO and control mice at P5.5 to assess Pax7 expression (Fig. 7A). This analysis showed a reduction in the percentage of Pax7 positive cells in sections from Myf5-Lap1CKO mice compared to Control mice (Fig. 7B). We next measured the expression of proteins involved in myogenic cell proliferation and differentiation in skeletal muscle of Control and Myf5-Lap1CKO mice at P5.5 and P10.5. Immunoblotting of protein extracts from quadriceps muscles showed reduced expression of Pax7, MyoD, Myf5 and Mef2C and increased expression of p21, a cell cycle inhibition factor (Fig. 7C). At P5.5, there was also reduced RNAs encoded by MyoD and Myf5 and increased expression of Cdkn1A, which encodes p21 (Fig. 7D). These results indicate that LAP1 depletion during early embryonic myogenesis impairs the myogenic potentials of satellite cells, which can lead to hypotrophic myofibres in Myf5-Lap1CKO mice.
Figure 7.
Reduced myogenic potential of satellite cells in skeletal muscle of Myf5-Lap1CKO mice. (A) Cross sections of quadriceps from Control and Myf5-Lap1CKO mice at P5.5 were stained with antibodies against laminin (green) and Pax7 (red). Images were chosen to show areas where Pax7 signals are frequent. Nuclei were counter stained with DAPI (blue). Scale bar: 25 µm. (B) Percentages of Pax7 positive nuclei among total nuclei in quadriceps from Control and Myf5-Lap1CKO mice. Nuclei in (n = 10) images from five different tissue sections from two animals per genotype were counted. Circles are values for individual images and horizontal bars show means; *P < 0.05. (C) Immunoblots of protein extracts of quadriceps from Control (Con) and Myf5-Lap1CKO (CKO) mice at P5.5 and P10.5. Blots were probed with antibodies against indicated proteins. Each lane contained extracts from different mice. (D). Relative expression of mRNAs encoded by Lap1, MyoD, Myf5, Myog (Myogenin), Mef2c and Cdkn1a (p21) in quadriceps from Control and Myf5-Lap1CKO mice at P5.5. Analysis was performed on total RNA from muscles of Control (n = 4) and Myf5-Lap1CKO (n = 4) mice; *P < 0.05, ***P < 0.001, ns: not significant.
Depletion of LAP1 during early embryonic myogenesis impairs AKT signalling and enhances ubiquitin proteasome pathways
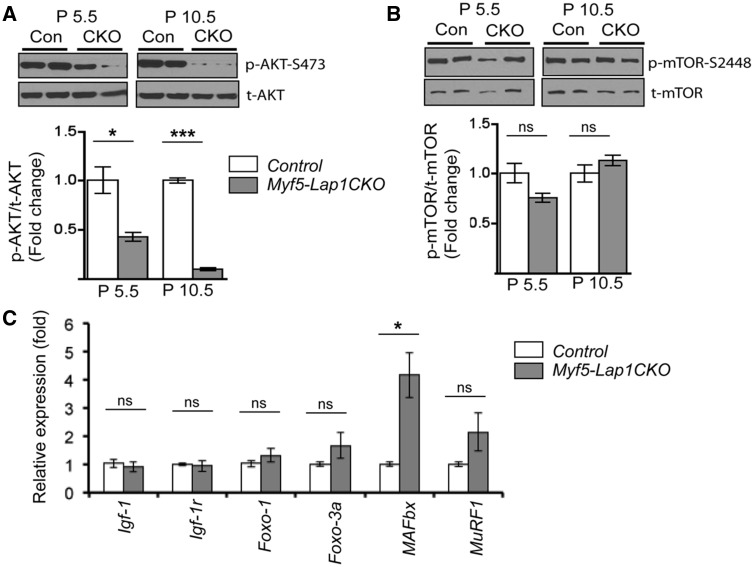
Postnatal skeletal muscle hypertrophic growth occurs through various signalling pathways (60). The proper balance between anabolic and catabolic mechanisms, which control the increased synthesis of muscle proteins or their degradation, determines muscle mass (61,62). One of the key regulators of this is AKT, which stimulates protein synthesis by activating mammalian target of rapamycin (mTOR) and blocking forkhead box O (FoxO) 1 and 3A, thus inhibiting the expression of atrophy-related genes, apoptosis and cell cycle arrest (63–65). We tested if AKT activity was altered in skeletal muscle of Myf5-Lap1CKO mice. There was a significant reduction in Ser-473 phosphorylated AKT in skeletal muscle from Myf5-Lap1CKO mice compared to Control mice at P5.5 and P10.5 (Fig. 8A). AKT regulates macroautophagy through the mTOR pathway (66). However, we did not observe significant changes in Ser-2448 phosphorylated mTOR in skeletal muscle of Myf5-Lap1CKO mice (Fig. 8B). There were also no significant differences in expression of macroautophagy-related genes in skeletal muscle of Control and Myf5-Lap1CKO mice (Supplementary Material, Fig. S6). Next we examined the AKT/FoxO/atrogene pathway responsible for protein degradation by the ubiquitin-proteosome system. AKT phosphorylates FoxO proteins on multiple sites, leading to the exclusion of phosphorylated FoxO proteins from the nucleus and inhibition of their transcriptional activity. Nuclear FoxO proteins induce the transcription of muscle-specific ubiquitin ligases such as muscle RING finer 1 (MuRF1) and muscle atrophy F box (MAFbx, also called atrogin1) during muscle atrophy (64,65,67). There were no significant differences in the levels of mRNAs encoded by Igf-1, Igf-1r, and Foxo-1 and Foxo-3a in skeletal muscle of Myf5-Lap1CKO mice compared to Control mice; however, there was a significant increase of mRNA encoded by MAFbx and a trend towards increased mRNA encoded by MuRF1 (Fig. 8C). Both of these genes are downstream targets of the AKT-FoxO signalling cascade. These data indicated that LAP1 depletion during early embryonic myogenesis lead to decreased AKT activity and increased FoxO-mediated atrogene expression and suggested that accelerated catabolism produces hypotrophic myofibres in Myf5-Lap1CKO mice.
Figure 8.
Depletion of LAP1 in skeletal muscle leads to down-regulation of AKT activity and enhanced expression of atrophy-related genes. (A) Immunoblots of protein extracts of quadriceps from Con (Control) and CKO (Myf5-Lap1CKO animals at P5.5. Blots were probed with antibodies against total AKT and Ser-473 phosphorylated AKT. In the lower panel, bar graphs show quantification of the ratio of the phosphorylated protein signals to their respective total protein signals. Values are means ± standard errors (n = 4 per genotype). *P < 0.05, ***P <0.0001. Graphs are presented as fold change over controls. (B) Immunoblot analysis of total mTOR and Ser-2448 phosphorylated mTOR from protein extracts of quadriceps from Con (Control) and CKO (Myf5-Lap1CKO animals at P5.5. Bar graphs show the ratio of phosphorylated protein signals to their respective total protein signals. Values are means ± standard errors (n = 4 per genotype). ns: not significant. (C) Means ± standard errors of relative expression of mRNA encoded by Igf-1, Igf-1r, Foxo-1, Foxo-3a, MAFbx and MuRF1 in quadriceps of Control (n = 4) and Myf5-Lap1CKO (n = 4) mice at P5.5. *P < 0.05, ns: not significant.
Discussion
We have shown that early embryonic expression of LAP1 isoforms, integral polypeptides of the inner nuclear membrane, is required for normal postnatal skeletal muscle growth. Embryos of Lap1-/- mice had fewer myotomes at E11.5 compared to Lap1+/+ mice but the general size and structure of myotomes were not changed. Although we observed impairment of myoblast differentiation in cultured primary myoblasts from Lap1-/- mice, early myogenesis was not ostensibly altered. Myofibres of Lap1-/- mice at P2.5 appeared to be normally formed and the general skeletal muscle organization was not altered. However, fibre CSA was significantly reduced in skeletal muscle of Lap1-/- mice, indicating hypotrophic growth. From these results, we conclude that LAP1 is not essential for myogenic progenitor determination and initial skeletal muscle formation but is required for late embryonic and postnatal muscle growth.
We previously reported that conditional skeletal muscle deletion of LAP1 starting at E17.5 lead to myopathic features typically seen in muscular dystrophy starting at P55 to P60 (40). There are also case reports of human subjects with genetic mutations affecting single isoforms of LAP1 or leading to amino acid substitutions that cause limb-girdle or Emery-Dreifuss-like muscular dystrophy (41,43). In contrast, our findings in mice with conditional skeletal muscle deletion of all LAP1 isoforms starting at E8.5 did not include typical myopathic features such as disorganized myofibres, degenerative myofibres or internal myonuclei. Instead, this earlier embryonic depletion of LAP1 caused dramatic skeletal muscle hypotrophy by P2.5 leading to 100% mortality by P17.5. Hence, some LAP1 expression during early embryogenesis appears to be necessary for normal postnatal skeletal muscle development and prolonged postnatal survival. However, depletion later in embryonic development or germline mutations leading to more-subtle alterations in isoform expression or protein structure can lead to later-onset muscular dystrophy.
We used the Myf5 promoter to drive Cre expression to deplete LAP1 in early embryonic myogenesis in mice. While Myf5 lineage cells contribute to a substantial percentage of adult myofibres (51), they can also develop into non-muscle tissues including adipose, bone, cartilage and dermis (48,68,69). Therefore, the phenotype of the Myf5-Lap1CKO mice we generated could be caused by alterations in tissues other than skeletal muscle. However, we did not observe any gross abnormalities in bone, cartilage or skin in Myf5-Lap1CKO mice. We also examined interscapular brown adipose tissue at P5.5, as it shares Myf5 lineage progenitors with skeletal muscle, and did not find any histological abnormalities. Given the profound skeletal muscle defects with lack of obvious gross defects in other tissues of Myf5-Lap1CKO mice, we are confident that their phenotype results from early embryonic deletion of LAP1 from muscle precursors.
Proliferation and fusion of satellite cells contribute to postnatal muscle growth (32,60). Myf5-Lap1CKO mice had a small but significant reduction in the number of Pax7 positive satellite cells at P5.5. Postnatal skeletal muscle also had decreased expression of myogenic factors such as MyoD, Myf5 and Mef2C and increased expression of a cell cycle inhibition factor. These data suggest that LAP1 is necessary for postnatal satellite cells proliferation and activation. As some Pax7 positive satellite cells do not express Myf5 (68), LAP1 may not have been depleted in all satellite cells in the Myf5-Lap1CKO mice population. Mice with floxed Lap1 alleles and Pax7-driven expression of Cre can be used to better elucidate the contribution of satellite cell depletion of LAP1 on skeletal muscle.
Postnatal skeletal muscle growth is controlled by various signalling pathways (31,60). We found reduced AKT activity and increased downstream FoxO-mediated atrophy-related gene expression in skeletal muscle of postnatal Myf5-Lap1CKO mice. This suggests that LAP1 is required for the suppression of protein catabolic pathways in postnatal muscle. A more comprehensive approach using transcriptome or proteomic profiling may lead to the identification of other signalling pathways altered in mice lacking LAP1 in skeletal muscle. This could lead to the identification of candidate targets for novel therapeutics to treat muscular dystrophies and skeletal muscle hypotrophy associated with physiological gaining and debilitating medical conditions.
Materials and methods
Mice
The Institutional Animal Care and Use Committee of Columbia University Medical Center approved all protocols. Mice were kept at room temperature and fed normal chow. Lap1-/- mice have been described previously (39) and were maintained in a 129S6/SvEv background. The generation and maintenance of floxed alleles of Lap1 (Lap1f/f) mice were previously described (40,70). The Myf5-Cre knock-in mice (stock number 007893) were purchased and maintained according to the protocol from the Jackson Laboratory. Both mouse lines were maintained in a C57BL/6 background. To generate Myf5-Cre;Lap1flf mice (referred to as Myf5-Lap1CKO mice), Myf5-Cre+/- mice were bred with Lap1f/f mice to obtain Myf5-Cre+/-; Lap1fl+ mice. These animals were fertile and produced at the expected Mendelian frequency. They were subsequently backcrossed with Lap1f/f mice to obtain Myf5-Lap1CKO mice. To genotype Myf5-Cre mice, PCR was performed from tail biopsies using the following primers (Forward: 5’ CGTAGACGCCTGAAGAAGGTCAACCA Reverse: 5’ ACGAAGTTATTAAGGTCCCTCGAC). When newborn pups were found in the morning, we considered their age to be P0.5.
Whole mount RNA in situ hybridization
The plasmid containing myogenin cDNA has been described (71). For whole-mount RNA in situ hybridization, digoxigenin labelled riboprobe and embryos were prepared as described previously (72,73). In brief, the plasmid was linearized and digoxigenin-labelled antisense RNA was transcribed in vitro using a DIG RNA labeling mix with T7 RNA polymerase (Roche). Labelled riboprobes were purified using ProbeQuant G-50 micro columns (Amersham) before hybridization to embryos.
E11.5 embryos were prepared essentially as described previously (72), except that they were treated with 10 μg/ml proteinase K (Qiagen) in phosphate-buffered saline (PBS) containing 0.1% polysorbate 20 for one hr. Embryos were rinsed, post-fixed, and hybridized with digoxigenin-labelled probe in the hybridization mix [50% formamide, 1.3 x saline-sodium citrate buffer, 5 mM ethylenediaminetetraacetic acid, 50 μg/ml yeast RNA, 0.2% polysorbate 20, 0.5% 3-[(3-cholamidopropyl) dimethylammonio] propanesulfonate and 100 μg/ml heparin] overnight at 65°C. After washing and blocking, embryos were incubated overnight with alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche) in 2% Blocking Reagent (Roche) containing 20% heat-inactivated lamb serum in 100 mM maleic acid (pH 7.5), 150 mM NaCl and 0.1% polysorbate 20. After washing with buffer containing 100 mM NaCl, 100 mM Tris-HCl (pH 9.5), 50 mM MgCl2, and 0.1% polysorbate 20, signals were developed using BM Purple (Roche).
Histology and myofibre quantification
For light microscopy, mouse hindlimbs or dissected quadriceps muscles were placed in 10% neutral-buffered formalin for 48 hr, embedded in paraffin and sectioned at 5 µm. Sections were stained with hematoxylin and eosin for histological analysis. To analyze myofibre size, representative images of stained cross sections of hindlimbs from two Lap1+/+ and two Lap1-/- mice at P2.5 were photographed with a DP72 digital camera attached to a BX53 upright light microscope (Olympus). Three different areas of each section were photographed to assess general features of myofibres. Individual myofibres from each image were processed using Adobe Photoshop CS (Adobe Systems). CSAs of individual myofibres were measured with Image J software (https://imagej.nih.gov/ij/; date last accessed November 4, 2016) and graphically represented with GraphPad Prism 4 software. The same procedures were applied to the measurement of CSAs of myofibres in the sections of the quadriceps from three control and three Myf5C-Lap1CKO mice at P5.5 of age.
C2C12 and primary myoblast cultures
The methods for isolation and culturing primary myoblasts and C2C12 cells were adapted from previous reports (73–75). C2C12 cells were obtained from the American Type Culture Collection and cultured in Dulbecco's Modified Eagle Medium supplemented with 10% fetal bovine serum. To induce differentiation of C2C12 cells, cultures grown to approximately 90% confluency were transferred to Dulbecco's Modified Eagle Medium with 2% horse serum. Some cultures were treated in differentiation media containing 10 mM cytosine arabinoside to eliminate proliferating myoblasts and enrich myotubes.
Primary myoblasts were isolated from dissected quadriceps muscles from the hindlimbs of Lap1+/+ and Lap1-/- mice at P0.5 or P1.5. The dissected muscles were incubated with a collagenase and dispase mixture solution and further dissociated by gentle mechanical trituration. Dissociated cells were filtered and centrifuged before being plated on non-coated tissue culture dishes with growth media (Ham’s F-10 media with 20% fetal bovine serum, 2.5 ng/ml basic fibroblast growth factor). To remove fibroblasts, floating cells were collected to a collagen-coated culture dish after 12 h of initial plating. Growth media were replaced every 48 h and only floating cells were replated to leave fibroblast behind. The purity of myoblasts was confirmed by Pax7 staining. To differentiate myoblasts, cells at 70–80% confluence in growth media were switched to differentiation media (Ham’s F-10 media with 5% horse serum). To maintain the cell’s primary characteristics, all experiments were performed using cultures that had undergone between four and seven passages.
Immnofluorescence microscopy on cultured cells
Primary myoblasts were cultured on collagen-coated coverslips and fixed with 4% paraformaldehyde in PBS at room temperature. Cells were blocked and permeabilized with 0.2% Triton X-100 in PBS for 20 min at room temperature and then incubated with anti-Pax7 (Developmental Studies Hybridoma Bank) and anti-MyHC (Santa Cruz Biotechnologies #SC-20641) antibodies in blocking buffer (PBS with 5% normal goat or donkey serum [Jackson Laboratory]) overnight at 4°C. After washing four times with 0.2% polysorbate 20 in PBS, cells were incubated with secondary antibodies (Molecular Probes) in blocking buffer for 1 h at room temperature. After washing again with 0.2% polysorbate 20 in PBS, coverslips were mounted using ProLong Gold Anti-fade Kit (Life Technologies) with 4',6-diamidino-2-phenylindole (DAPI; Life Technologies). Stained cells were photographed using a DP72 digital camera attached to a BX53 upright light microscope (Olympus). The myoblast fusion index was calculated as the percentage of myotubes containing two or more nuclei. Three independent experiments were performed in duplicate and at least three random fields were imaged per genotype for quantification.
Myofibre staining and immunohistochemistry
For myofibre staining, dissected hindlimb muscles from Control and Myf5-Lap1CKO mice at P5.5 were fixed with 4% paraformaldehyde for 30 min at room temperature. Muscles were washed with PBS three times, finely teased with a forceps with a sharp #5 tip and then incubated with blocking solution (1% bovine serum albumin/5% normal goat serum/0.5% Triton X-100 in PBS) overnight at 4°C. Myofibres were then incubated with anti-LAP1 (37) or anti-lamin A/C (Santa Cruz Biotechnologies #SC-20681) antibodies in the same blocking solution overnight at 4°C. After three washes with PBS containing 0.2% polysorbate 20, myofibres were incubated with Texas Red conjugated secondary antibodies (Life Technologies) in blocking buffer for 1 h at room temperature to visualize the primary antibody labeling. After three additional washes, myofibres were stained with DAPI before mounting on coverslips. Microscopy was performed on a AIR MP multiphoton confocal microscope with imaging software NIS-Elements (Nikon).
For immunohistochemistry, mouse quadriceps muscles were dissected and rapidly frozen in liquid nitrogen-cooled isopentane for sectioning on a cryostat and cryosections were cut at 5 µm thickness. For Pax7 and laminin staining, frozen muscle sections were fixed with 4% paraformaldehyde for 20 min at room temperature and then permeabilized in methanol for 6 min at −20°C. Antigen retrieval was performed by boiling the sections in 0.01M citric acid buffer (pH 6.0) for 10 min in a microwave oven. Sections were cooled to room temperature and washed three times with PBS. They were then incubated with anti-Pax7 and anti-laminin (Sigma-Aldrich, #L9393) antibodies in blocking solution (PBS with 5% normal goat serum [Jackson Laboratory]) overnight at 4°C. After washing four times with 0.2% polysorbate 20 in PBS, sections were incubated with Alexa Fluor 488 and Texas Red conjugated secondary antibodies (Life Technologies) in blocking buffer for 1 h at room temperature and sections were stained with DAPI before mounting on coverslips. To quantify Pax7 positive myonuclei, a total of five sections from two mice from Control and Myf5-Lap1CKO mice were imaged using a 40X objective and numbers of total nuclei (DAPI labelled) and Pax7 positive nuclei from two different areas of each section were averaged and used for plotting. The hind limb sections from Control and Myf5-Lap1CKO were also stained with anti-laminin and myosin heavy chain type I (Sigma, clone NOQ7.5.4D) and type II (Sigma, clone My-32) antibodies.
Electron microscopy
Electron microscopy was performed as previously described (39,40). Briefly, quadriceps from P5.5 Control and Myf5-Lap1CKO mice were drop fixed with 1% paraformaldehyde/2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4), postfixed with 1% osmium tetroxide in 0.1 M cacodylate buffer (pH 7.4) and incubated with uranyl acetate and dehydrated with ethanol. Tissues were subsequently rinsed with propylene oxide and embedded. Sections 60 nm thick were counterstained with uranyl acetate and lead citrate and examined on a JEM-1200EX electron microscope (JEOL). Measurements of perinuclear spacing were taken with ImageJ software with the nanometer/pixel ratio set according to the scale in electron micrograph. For each myonucleus, three different perinuclear areas showing increased distances between inner nuclear membrane and outer nuclear membrane were selected and measured and mean values plotted (57). A total of 7 nuclei from control mice and 19 nuclei from Myf5-Lap1CKO mice were analyzed.
Protein extraction and immunoblotting
Lysates of cultured cells and dissected skeletal muscles were homogenized in radioimmunoprecipitation assay buffer (RIPA Buffer, Cell Signaling) containing Protease Inhibitor Cocktail (Roche) plus 1 mM phenylmethanesulfonyl fluoride (Sigma). Proteins in the samples were denatured by boiling in Laemmli sample buffer (76) containing β-mercaptoethanol for 5 min, separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes for immunoblotting.
For immunoblotting, the membranes containing proteins transferred from SDS-polyacrylamide gels were washed with blocking buffer (5% bovine serum albumin and 0.2% polysorbate 20 in PBS for 30 min and probed with primary antibodies in blocking buffer overnight at 4°C. The primary antibodies used for immunoblotting were against LAP1 (37) GAPDH (Ambion #AM4300), MyoD (BD Parmingen #554130), Myf5 (Santa Cruz Biotechnologies #SC-302), myogenin (Santa Cruz Biotechnologies #SC-12732), MyHC, MEF2C (Santa Cruz Biotechnologies #SC-17785), Pax7, P21 (Santa Cruz Biotechnologies #SC-391), total AKT (Cell Signaling, #4691), phosphorylated AKT S473 (Cell Signaling #4969), total mTOR (Cell Signaling, #2972) and phosphorylated mTOR S2448 (Cell Signaling #2971). Blots were washed with 0.2% polysorbate 20 in PBS and then incubated in blocking buffer with horseradish peroxidase-conjugated secondary antibodies (Amersham) for 1 h at room temperature. Recognized proteins were visualized by enhanced chemiluminescence (Thermo Fisher Scientific) and detected by exposure on X-ray films (Kodak). To quantify signals, immunoblots were scanned and the densities of the bands were quantified using ImageJ software. For kinases, band intensity of phosphorylated protein was normalized to the band intensity of respective total protein.
BrdU labeling
The BrdU stock solution was purchased from Abcam (#ab142567) and labeling performed according to the manufacturer’s instruction. Briefly, final concentration of 10 μM BrdU labeling solution was added on culture media and incubated for 2 h at 37°C in a 5% CO2 incubator. After removing labeling solution, cells were fixed with 4% paraformaldehyde. For DNA hydrolysis, fixed cells were incubated in 1 N HCl for 10 mins followed by 2 N HCl for 10 mins. After neutralization with 0.1 M sodium borate, standard immunocytochemistry procedures were performed with anti-BrdU antibody (Abcam, #ab6326) and DAPI.
Quantitative real-time qPCR
Total RNA was extracted from dissected quadriceps muscle from Control and Myf5-Lap1CKO mice at P5.5 age using the RNeasy isolation Kit (Qiagen) as described previously (77). Quality and concentrations of RNA were measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific). The cDNA was synthesized using Superscript First Strand Synthesis System according to the manufacturer’s instructions (Life Technologies). For each replicate in each experiment, RNAs from skeletal muscles of different animals were used. The sequences of qPCR primers were either designed using Primer3 software or used with validated primers from the PrimerBank (https://pga.mgh.harvard.edu/primerbank; date last accessed November 4, 2016) (78). The primer sequences used for the study are listed in Supplementary Material, Table S1. qPCR was performed on an ABI 7300 Real-Time PCR System (Applied Biosystems) using HotStart-IT SYBR green qPCR Master Mix (Affymetrix). Relative levels of mRNA expression were calculated using the ΔΔCT method (79). Individual expression values were normalized by comparison to GAPDH mRNA.
Statistical analysis
Statistical analyses were carried out using GraphPad Prism 4. Comparisons between two groups were tested using a two-tailed t-test. Two way ANOVA was used for comparisons in Fig. 2F, Fig. 3D and Fig. 5D. Mouse survival was analyzed using the Kaplan-Meier estimator (80) and differences in median survival were compared using a log-rank test. Differences were considered significant at P-values less than 0.05.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Kristy Brown (Department of Pathology and Cell Biology, Columbia University) for advice and assistance with electron microscopy. We thank Cecilia Östlund for helpful comments on the manuscript.
Conflict of Interest statement. None declared.
Funding
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health [grant number AR048997] and the Muscular Dystrophy Association [grant number 294537]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1. Dauer W.T., Worman H.J. (2009) The nuclear envelope as a signaling node in development and disease. Dev. Cell, 17, 626–638. [DOI] [PubMed] [Google Scholar]
- 2. Chatzifrangkeskou M., Bonne G., Muchir A. (2015) Nuclear envelope and striated muscle diseases. Curr. Opin. Cell Biol., 32, 1–6. [DOI] [PubMed] [Google Scholar]
- 3. Bione S., Maestrini E., Rivella S., Mancini M., Regis S., Romeo G., Toniolo D. (1994) Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet., 8, 323–327. [DOI] [PubMed] [Google Scholar]
- 4. Nagano A., Koga R., Ogawa M., Kurano Y., Kawada J., Okada R., Hayashi Y.K., Tsukahara T., Arahata K. (1996) Emerin deficiency at the nuclear membrane in patients with Emery-Dreifuss muscular dystrophy. Nat. Genet., 12, 254–259. [DOI] [PubMed] [Google Scholar]
- 5. Manilal S., Recan D., Sewry C.A., Hoeltzenbein M., Llense S., Leturcq F., Deburgrave N., Barbot J., Man N., Muntoni F., et al. (1998) Mutations in emery-dreifuss muscular dystrophy and their effects on emerin protein expression. Hum. Mol. Genet., 7, 855–864. [DOI] [PubMed] [Google Scholar]
- 6. Manilal S., Nguyen T.M., Sewry C.A., Morris G.E. (1996) The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum. Mol. Genet., 5, 801–808. [DOI] [PubMed] [Google Scholar]
- 7. Emery A.E., Dreifuss F.E. (1966) Unusual type of benign X-linked muscular dystrophy. J. Neurol. Neurosurg. Psychiatry, 29, 338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Emery A.E. (1987) X-linked muscular dystrophy with early contractures and cardiomyopathy (Emery-Dreifuss type). Clin. Genet., 32, 360–367. [DOI] [PubMed] [Google Scholar]
- 9. Bonne G., Di Barletta M.R., Varnous S., Becane H.M., Hammouda E.H., Merlini L., Muntoni F., Greenberg C.R., Gary F., Urtizberea J.A., et al. (1999) Mutations in the gene encoding lamin a/c cause autosomal dominant emery-dreifuss muscular dystrophy. Nat. Genet., 21, 285–288. [DOI] [PubMed] [Google Scholar]
- 10. Fatkin D., MacRae C., Sasaki T., Wolff M.R., Porcu M., Frenneaux M., Atherton J., Vidaillet H.J. Jr., Spudich S., De Girolami U., et al. (1999) Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med., 341, 1715–1724. [DOI] [PubMed] [Google Scholar]
- 11. Bonne G., Mercuri E., Muchir A., Urtizberea A., Becane H.M., Recan D., Merlini L., Wehnert M., Boor R., Reuner U., et al. (2000) Clinical and molecular genetic spectrum of autosomal dominant Emery-Dreifuss muscular dystrophy due to mutations of the lamin A/C gene. Ann. Neurol., 48, 170–180. [PubMed] [Google Scholar]
- 12. Muchir A., Bonne G., van der Kooi A.J., van Meegen M., Baas F., Bolhuis P.A., de Visser M., Schwartz K. (2000) Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B). Hum. Mol. Genet., 9, 1453–1459. [DOI] [PubMed] [Google Scholar]
- 13. D'Amico A., Haliloglu G., Richard P., Talim B., Maugenre S., Ferreiro A., Guicheney P., Menditto I., Benedetti S., Bertini E., et al. (2005) Two patients with ‘dropped head syndrome’ due to mutations in LMNA or SEPN1 genes. Neuromuscul. Disord., 15, 521–524. [DOI] [PubMed] [Google Scholar]
- 14. Quijano-Roy S., Mbieleu B., Bonnemann C.G., Jeannet P.Y., Colomer J., Clarke N.F., Cuisset J.M., Roper H., De Meirleir L., D'Amico A., et al. (2008) De novo LMNA mutations cause a new form of congenital muscular dystrophy. Ann. Neurol., 64, 177–186. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Q., Bethmann C., Worth N.F., Davies J.D., Wasner C., Feuer A., Ragnauth C.D., Yi Q., Mellad J.A., Warren D.T., et al. (2007) Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet., 16, 2816–2833. [DOI] [PubMed] [Google Scholar]
- 16. Puckelwartz M.J., Kessler E.J., Kim G., Dewitt M.M., Zhang Y., Earley J.U., Depreux F.F., Holaska J., Mewborn S.K., Pytel P., et al. (2010) Nesprin-1 mutations in human and murine cardiomyopathy. J. Mol. Cell. Cardiol., 48, 600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liang W.C., Mitsuhashi H., Keduka E., Nonaka I., Noguchi S., Nishino I., Hayashi Y.K. (2011) Tmem43 mutations in Emery-Dreifuss muscular dystrophy-related myopathy. Ann. Neurol., 69, 1005–1013. [DOI] [PubMed] [Google Scholar]
- 18. Meinke P., Mattioli E., Haque F., Antoku S., Columbaro M., Straatman K.R., Worman H.J., Gundersen G.G., Lattanzi G., Wehnert M., et al. (2014) Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet., 10, e1004605.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B. (1999) Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol., 147, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bertrand A.T., Renou L., Papadopoulos A., Beuvin M., Lacene E., Massart C., Ottolenghi C., Decostre V., Maron S., Schlossarek S., et al. (2012) Delk32-lamin A/C has abnormal location and induces incomplete tissue maturation and severe metabolic defects leading to premature death. Hum. Mol. Genet., 21, 1037–1048. [DOI] [PubMed] [Google Scholar]
- 21. Arimura T., Helbling-Leclerc A., Massart C., Varnous S., Niel F., Lacene E., Fromes Y., Toussaint M., Mura A.M., Keller D.I., et al. (2005) Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum. Mol. Genet., 14, 155–169. [DOI] [PubMed] [Google Scholar]
- 22. Muchir A., Kim Y.J., Reilly S.A., Wu W., Choi J.C., Worman H.J. (2013) Inhibition of extracellular signal-regulated kinase 1/2 signaling has beneficial effects on skeletal muscle in a mouse model of Emery-Dreifuss muscular dystrophy caused by lamin A/C gene mutation. Skelet. Muscle, 3, 17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Melcon G., Kozlov S., Cutler D.A., Sullivan T., Hernandez L., Zhao P., Mitchell S., Nader G., Bakay M., Rottman J.N., et al. (2006) Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum. Mol. Genet., 15, 637–651. [DOI] [PubMed] [Google Scholar]
- 24. Zhang X., Xu R., Zhu B., Yang X., Ding X., Duan S., Xu T., Zhuang Y., Han M. (2007) Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development, 134, 901–908. [DOI] [PubMed] [Google Scholar]
- 25. Favreau C., Higuet D., Courvalin J.C., Buendia B. (2004) Expression of a mutant lamin A that causes Emery-Dreifuss muscular dystrophy inhibits in vitro differentiation of C2C12 myoblasts. Mol. Cell. Biol., 24, 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frock R.L., Kudlow B.A., Evans A.M., Jameson S.A., Hauschka S.D., Kennedy B.K. (2006) Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes Dev., 20, 486–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kandert S., Wehnert M., Muller C.R., Buendia B., Dabauvalle M.C. (2009) Impaired nuclear functions lead to increased senescence and inefficient differentiation in human myoblasts with a dominant p.R545C mutation in the LMNA gene. Eur. J. Cell Biol., 88, 593–608. [DOI] [PubMed] [Google Scholar]
- 28. Huber M.D., Guan T., Gerace L. (2009) Overlapping functions of nuclear envelope proteins NET25 (lem2) and emerin in regulation of ERK signaling in myoblast differentiation. Mol. Cell. Biol., 29, 5718–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gotic I., Schmidt W.M., Biadasiewicz K., Leschnik M., Spilka R., Braun J., Stewart C.L., Foisner R. (2010) Loss of LAP2 alpha delays satellite cell differentiation and affects postnatal fiber-type determination. Stem Cells, 28, 480–488. [DOI] [PubMed] [Google Scholar]
- 30. Buckingham M. (2006) Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr. Opin. Genet. Dev., 16, 525–532. [DOI] [PubMed] [Google Scholar]
- 31. Braun T., Gautel M. (2011) Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell. Biol., 12, 349–361. [DOI] [PubMed] [Google Scholar]
- 32. White R.B., Bierinx A.S., Gnocchi V.F., Zammit P.S. (2010) Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev. Biol., 10, 21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gokhin D.S., Ward S.R., Bremner S.N., Lieber R.L. (2008) Quantitative analysis of neonatal skeletal muscle functional improvement in the mouse. J. Exp. Biol., 211, 837–843. [DOI] [PubMed] [Google Scholar]
- 34. Shin J.Y., Dauer W.T., Worman H.J. (2014) Lamina-associated polypeptide 1: Protein interactions and tissue-selective functions. Semin. Cell Dev. Biol., 29C, 164–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Senior A., Gerace L. (1988) Integral membrane proteins specific to the inner nuclear membrane and associated with the nuclear lamina. J. Cell Biol., 107, 2029–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Foisner R., Gerace L. (1993) Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell, 73, 1267–1279. [DOI] [PubMed] [Google Scholar]
- 37. Goodchild R.E., Dauer W.T. (2005) The AAA+ protein torsinA interacts with a conserved domain present in lap1 and a novel ER protein. J. Cell Biol., 168, 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Santos M., Rebelo S., Van Kleeff P.J., Kim C.E., Dauer W.T., Fardilha M., da Cruz E.S.O.A., da Cruz E.S.E.F. (2013) The nuclear envelope protein, LAP1B, is a novel protein phosphatase 1 substrate. PLoS One, 8, e76788.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim C.E., Perez A., Perkins G., Ellisman M.H., Dauer W.T. (2010) A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc. Natl. Acad. Sci. U S A, 107, 9861–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shin J.Y., Mendez-Lopez I., Wang Y., Hays A.P., Tanji K., Lefkowitch J.H., Schulze P.C., Worman H.J., Dauer W.T. (2013) Lamina-associated polypeptide-1 interacts with the muscular dystrophy protein emerin and is essential for skeletal muscle maintenance. Dev. Cell, 26, 591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kayman-Kurekci G., Talim B., Korkusuz P., Sayar N., Sarioglu T., Oncel I., Sharafi P., Gundesli H., Balci-Hayta B., Purali N., et al. (2014) Mutation in TOR1AIP1 encoding LAP1B in a form of muscular dystrophy: A novel gene related to nuclear envelopathies. Neuromuscul. Disord., 24, 624–633. [DOI] [PubMed] [Google Scholar]
- 42. Dorboz I., Coutelier M., Bertrand A.T., Caberg J.H., Elmaleh-Berges M., Laine J., Stevanin G., Bonne G., Boespflug-Tanguy O., Servais L. (2014) Severe dystonia, cerebellar atrophy, and cardiomyopathy likely caused by a missense mutation in TOR1AIP1. Orphanet. J. Rare Dis., 9, 174.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ghaoui R., Benavides T., Lek M., Waddell L.B., Kaur S., North K.N., MacArthur D.G., Clarke N.F., Cooper S.T. (2016) TOR1AIP1 as a cause of cardiac failure and recessive limb-girdle uscular dystrophy. Neuromuscul. Disord., 26, 500–503. [DOI] [PubMed] [Google Scholar]
- 44. Santos M., Domingues S.C., Costa P., Muller T., Galozzi S., Marcus K., da Cruz e., Silva E.F., da Cruz e., Silva O.A., Rebelo S. (2014) Identification of a novel human lap1 isoform that is regulated by protein phosphorylation. PLoS One, 9, e113732.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tallquist M.D., Weismann K.E., Hellstrom M., Soriano P. (2000) Early myotome specification regulates PDGFA expression and axial skeleton development. Development, 127, 5059–5070. [DOI] [PubMed] [Google Scholar]
- 46. Hallock P.T., Xu C.F., Park T.J., Neubert T.A., Curran T., Burden S.J. (2010) Dok-7 regulates neuromuscular synapse formation by recruiting Crk and Crk-l. Genes Dev., 24, 2451–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huh M.S., Parker M.H., Scime A., Parks R., Rudnicki M.A. (2004) Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J. Cell Biol., 166, 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scime A., Devarakonda S., Conroe H.M., Erdjument-Bromage H., et al. (2008) Prdm16 controls a brown fat/skeletal muscle switch. Nature, 454, 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kajimura S., Seale P., Tomaru T., Erdjument-Bromage H., Cooper M.P., Ruas J.L., Chin S., Tempst P., Lazar M.A., Spiegelman B.M. (2008) Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev., 22, 1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haldar M., Karan G., Tvrdik P., Capecchi M.R. (2008) Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev. Cell, 14, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Comai G., Sambasivan R., Gopalakrishnan S., Tajbakhsh S. (2014) Variations in the efficiency of lineage marking and ablation confound distinctions between myogenic cell populations. Dev. Cell, 31, 654–667. [DOI] [PubMed] [Google Scholar]
- 52. Gruenbaum-Cohen Y., Harel I., Umansky K.B., Tzahor E., Snapper S.B., Shilo B.Z., Schejter E.D. (2012) The actin regulator N-WASp is required for muscle-cell fusion in mice. Proc. Natl. Acad. Sci. U S A, 109, 11211–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Beedle A.M., Turner A.J., Saito Y., Lueck J.D., Foltz S.J., Fortunato M.J., Nienaber P.M., Campbell K.P. (2012) Mouse fukutin deletion impairs dystroglycan processing and recapitulates muscular dystrophy. J. Clin. Invest., 122, 3330–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matheny R.W. Jr., Riddle-Kottke M.A., Leandry L.A., Lynch C.M., Abdalla M.N., Geddis A.V., Piper D.R., Zhao J.J. (2015) Role of phosphoinositide 3-OH kinase p110beta in skeletal myogenesis. Mol. Cell. Biol., 35, 1182–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martinez-Lopez N., Athonvarangkul D., Sahu S., Coletto L., Zong H., Bastie C.C., Pessin J.E., Schwartz G.J., Singh R. (2013) Autophagy in Myf5+ progenitors regulates energy and glucose homeostasis through control of brown fat and skeletal muscle development. EMBO Rep., 14, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zwerger M., Kolb T., Richter K., Karakesisoglou I., Herrmann H. (2010) Induction of a massive endoplasmic reticulum and perinuclear space expansion by expression of lamin B receptor mutants and the related sterol reductases TM7SF2 and DHCR7. Mol. Biol. Cell, 21, 354–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cain N.E., Tapley E.C., McDonald K.L., Cain B.M., Starr D.A. (2014) The SUN protein UNC-84 is required only in force-bearing cells to maintain nuclear envelope architecture. J. Cell Biol., 206, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pallafacchina G., Blaauw B., Schiaffino S. (2013) Role of satellite cells in muscle growth and maintenance of muscle mass. Nutr. Metab. Cardiovasc. Dis., 23 Suppl 1, S12–S18. [DOI] [PubMed] [Google Scholar]
- 59. Serrano A.L., Baeza-Raja B., Perdiguero E., Jardi M., Munoz-Canoves P. (2008) Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab., 7, 33–44. [DOI] [PubMed] [Google Scholar]
- 60. Schiaffino S., Dyar K.A., Ciciliot S., Blaauw B., Sandri M. (2013) Mechanisms regulating skeletal muscle growth and atrophy. Febs J., 280, 4294–4314. [DOI] [PubMed] [Google Scholar]
- 61. Glass D.J. (2005) Skeletal muscle hypertrophy and atrophy signaling pathways. Int. J. Biochem. Cell Biol., 37, 1974–1984. [DOI] [PubMed] [Google Scholar]
- 62. Sandri M. (2013) Protein breakdown in muscle wasting: Role of autophagy-lysosome and ubiquitin-proteasome. Int. J. Biochem. Cell Biol., 45, 2121–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Laplante M., Sabatini D.M. (2012) mTOR signaling in growth control and disease. Cell, 149, 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S.H., Goldberg A.L. (2004) Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell, 117, 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stitt T.N., Drujan D., Clarke B.A., Panaro F., Timofeyva Y., Kline W.O., Gonzalez M., Yancopoulos G.D., Glass D.J. (2004) The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell, 14, 395–403. [DOI] [PubMed] [Google Scholar]
- 66. Bodine S.C., Stitt T.N., Gonzalez M., Kline W.O., Stover G.L., Bauerlein R., Zlotchenko E., Scrimgeour A., Lawrence J.C., Glass D.J., et al. (2001) Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell. Biol., 3, 1014–1019. [DOI] [PubMed] [Google Scholar]
- 67. Bodine S.C., Latres E., Baumhueter S., Lai V.K., Nunez L., Clarke B.A., Poueymirou W.T., Panaro F.J., Na E., Dharmarajan K., et al. (2001) Identification of ubiquitin ligases required for skeletal muscle atrophy. Science, 294, 1704–1708. [DOI] [PubMed] [Google Scholar]
- 68. Kuang S., Kuroda K., Le Grand F., Rudnicki M.A. (2007) Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell, 129, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gensch N., Borchardt T., Schneider A., Riethmacher D., Braun T. (2008) Different autonomous myogenic cell populations revealed by ablation of myf5-expressing cells during mouse embryogenesis. Development, 135, 1597–1604. [DOI] [PubMed] [Google Scholar]
- 70. Shin J.Y., Le Dour C., Sera F., Iwata S., Homma S., Joseph L.C., Morrow J.P., Dauer W.T., Worman H.J. (2014) Depletion of lamina-associated polypeptide 1 from cardiomyocytes causes cardiac dysfunction in mice. Nucleus, 5, 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sassoon D., Lyons G., Wright W.E., Lin V., Lassar A., Weintraub H., Buckingham M. (1989) Expression of two myogenic regulatory factors myogenin and myoD1 during mouse embryogenesis. Nature, 341, 303–307. [DOI] [PubMed] [Google Scholar]
- 72. Mulieri P.J., Kang J.S., Sassoon D.A., Krauss R.S. (2002) Expression of the boc gene during murine embryogenesis. Dev. Dyn., 223, 379–388. [DOI] [PubMed] [Google Scholar]
- 73. Cole F., Zhang W., Geyra A., Kang J.S., Krauss R.S. (2004) Positive regulation of myogenic bHLH factors and skeletal muscle development by the cell surface receptor CDO. Dev. Cell, 7, 843–854. [DOI] [PubMed] [Google Scholar]
- 74. Rando T.A., Blau H.M. (1994) Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol., 125, 1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kang J.S., Mulieri P.J., Miller C., Sassoon D.A., Krauss R.S. (1998) CDO, a robo-related cell surface protein that mediates myogenic differentiation. J. Cell Biol., 143, 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage t4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 77. Muchir A., Shan J., Bonne G., Lehnart S.E., Worman H.J. (2009) Inhibition of extracellular signal-regulated kinase signaling to prevent cardiomyopathy caused by mutation in the gene encoding A-type lamins. Hum. Mol. Genet., 18, 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang X., Spandidos A., Wang H., Seed B. (2012) Primerbank: A PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res., 40, D1144–D1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ponchel F., Toomes C., Bransfield K., Leong F.T., Douglas S.H., Field S.L., Bell S.M., Combaret V., Puisieux A., Mighell A.J., et al. (2003) Real-time PCR based on SYBR-green I fluorescence: An alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol., 3, 18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kaplan E.L., Meier P. (1958) Nonparametric estimation from incomplete observations. J. Amer. Statist. Assoc., 53, 457–481. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.