Abstract
Clinical trials of tolvaptan showed it to be a promising candidate for the treatment of Autosomal Dominant Polycystic Kidney Disease (ADPKD) but also revealed potential for idiosyncratic drug-induced liver injury (DILI) in this patient population. To identify risk factors and mechanisms underlying tolvaptan DILI, 8 mice in each of 45 strains of the genetically diverse Collaborative Cross (CC) mouse population were treated with a single oral dose of either tolvaptan or vehicle. Significant elevations in plasma alanine aminotransferase (ALT) were observed in tolvaptan-treated animals in 3 of the 45 strains. Genetic mapping coupled with transcriptomic analysis in the liver was used to identify several candidate susceptibility genes including epoxide hydrolase 2, interferon regulatory factor 3, and mitochondrial fission factor. Gene pathway analysis revealed that oxidative stress and immune response pathways were activated in response to tolvaptan treatment across all strains, but genes involved in regulation of bile acid homeostasis were most associated with tolvaptan-induced elevations in ALT. Secretory leukocyte peptidase inhibitor (Slpi) mRNA was also induced in the susceptible strains and was associated with increased plasma levels of Slpi protein, suggesting a potential serum marker for DILI susceptibility. In summary, tolvaptan induced signs of oxidative stress, mitochondrial dysfunction, and innate immune response in all strains, but variation in bile acid homeostasis was most associated with susceptibility to the liver response. This CC study has indicated potential mechanisms underlying tolvaptan DILI and biomarkers of susceptibility that may be useful in managing the risk of DILI in ADPKD patients.
Keywords: Collaborative Cross, drug-induced liver injury, precision medicine, tolvaptan, toxicogenomics.
Tolvaptan is a vasopressin V2-receptor antagonist marketed for the treatment of hyponatremia. Tolvaptan has also been approved for the treatment of autosomal dominant polycystic kidney disease (ADPKD) in some regions (Torres et al., 2012). However, the U.S. Food and Drug Administration approval for this indication has not yet been received, in part due to reported cases of drug-induced liver injury (DILI) associated with tolvaptan use in ADPKD patients.
Hepatotoxicity is relatively rare in tolvaptan-treated ADPKD patients and has not been observed in non-ADPKD patients treated with tolvaptan (Watkins et al., 2015). Furthermore, nonclinical testing of tolvaptan in traditional animal models (mice, rats, and dogs) did not indicate a risk for liver injury (Oi et al., 2011). Studying the mechanisms and identifying risk factors associated with tolvaptan hepatotoxicity has therefore been challenging.
Previous studies have demonstrated the ability of mouse genetic reference populations (GRPs) to model the drug-induced injury observed in humans better than traditional nonclinical animal models (Harrill et al., 2012). GRPs have also been utilized for pharmacogenomic analysis to identify risk factors associated with drug-induced adverse events in liver and kidney (Harrill et al., 2009, 2012; Mosedale et al., 2014). Importantly, findings in GRP mice translate to DILI risk factors in humans (Court et al., 2013; Harrill et al., 2009).
The Collaborative Cross (CC) is a recently developed and highly sophisticated GRP composed of multiparental recombinant inbred lines. This population was strategically designed to overcome limitations of classical inbred GRPs such as population structure and blind spots of genomic variation. The CC strains have high genetic diversity, balanced allele frequencies, and dense, evenly distributed recombination sites (Collaborative Cross Consortium, 2012). Using diverse and genetically defined strains can increase the statistical power to detect the effects of a test chemical (Festing, 2010). Studies with CC strains have demonstrated that the diversity of responses observed in this population is more extensive than standard inbred strain panels (Kelada et al., 2014; Rutledge et al., 2014). Therefore, it was hypothesized that evaluating tolvaptan-induced liver responses within the CC may help to identify strains sensitive to liver injury. These strains could then be used to provide a mechanistic understanding and identify potential risk factors associated with susceptibility to tolvaptan hepatotoxicity in the ADPKD patient population.
Although traditional histological and biochemical endpoints can be used for the identification of a toxic response, recent evidence suggests liver gene expression analysis after a single, maximally tolerated dose may be a more sensitive and translational method to predict an adverse drug response in humans (Laifenfeld et al., 2014; Leone et al., 2014). Furthermore, evaluation of molecular signaling pathways can provide insight into mechanisms of drug toxicity as well as phenotypes for quantitative trait loci (QTL) mapping, even in the absence of overt injury. Based on these findings, traditional biochemical and histological endpoints as well as gene expression profiling were performed in this study.
Finally, it must be noted that clinical tolvaptan DILI cases were typically delayed in onset, presenting between 3 and 18 months of treatment, and exhibited a relatively prompt recurrence upon re-challenge, supporting the involvement of the adaptive immune system in the final critical events underlying the liver injury (Watkins et al., 2015). This CC study was not designed to allow for the evaluation of an adaptive immune response. However, it has been hypothesized that even if an adaptive immune attack is the final insult responsible for eliciting overt liver injury, events that are necessary but not sufficient for the liver injury begin at the hepatocyte level and result in the release of “danger signals” that are important for activating the immune response (Kaplowitz, 2005; Li and Uetrecht, 2010; Mosedale and Watkins, 2016). These early events may require both genetic and nongenetic susceptibility factors that contribute to the idiosyncratic nature of the drug reaction. Therefore, a liver response in sensitive CC strains could have mechanistic similarities to the early hepatocellular events occurring in the ADPKD patients. Thus, to gain insight into potential mechanisms of tolvaptan hepatotoxicity and to generate testable hypotheses regarding risk factors for liver injury in humans, phenotypic findings in combination with genetics and transcriptomics were used to identify variation in specific genes and pathways associated with susceptibility to tolvaptan DILI in the CC mice.
MATERIALS AND METHODS
Animals
The CC is a panel of recombinant inbred lines derived from 8 inbred founder mouse strains (short names in parentheses): 129S1/SvlmJ (129S1), A/J (AJ), C57BL/6J (B6), NOD/ShiLtJ (NOD), NZO/HILtJ (NZO), CAST/EiJ (CAST), PWK/PhJ (PWK), and WSB/EiJ (WSB). The initiation of the CC breeding funnels, description of the CC strains, and animal housing are described elsewhere (Collaborative Cross Consortium, 2012). Eight male mice from each of 45 CC lines (listed on the x-axis of Figure 1) were purchased from the University of North Carolina, Chapel Hill (UNC). All lines came from independent breeding funnels except for 2, CC051/TauUnc and CC059/TauUnc, which descended from the same breeding funnel and were separated at a later generation. Animals were housed in polycarbonate cages, kept on a 12-h light-dark cycle and provided Pelleted PicoLab Rodent Diet 20 (Lab Diet) ad libitum, except during an approximate 18-h period of fasting prior to necropsy. Animals were acclimated for approximately 7 days prior to dosing. At the initiation of dosing, mice were approximately 8–10 weeks of age. Care of mice followed institutional guidelines under a protocol approved by Mispro Biotech Services Institutional Animal Care and Use Committee. Clinical assessments were performed using previously described methods (Burkholder et al., 2012).
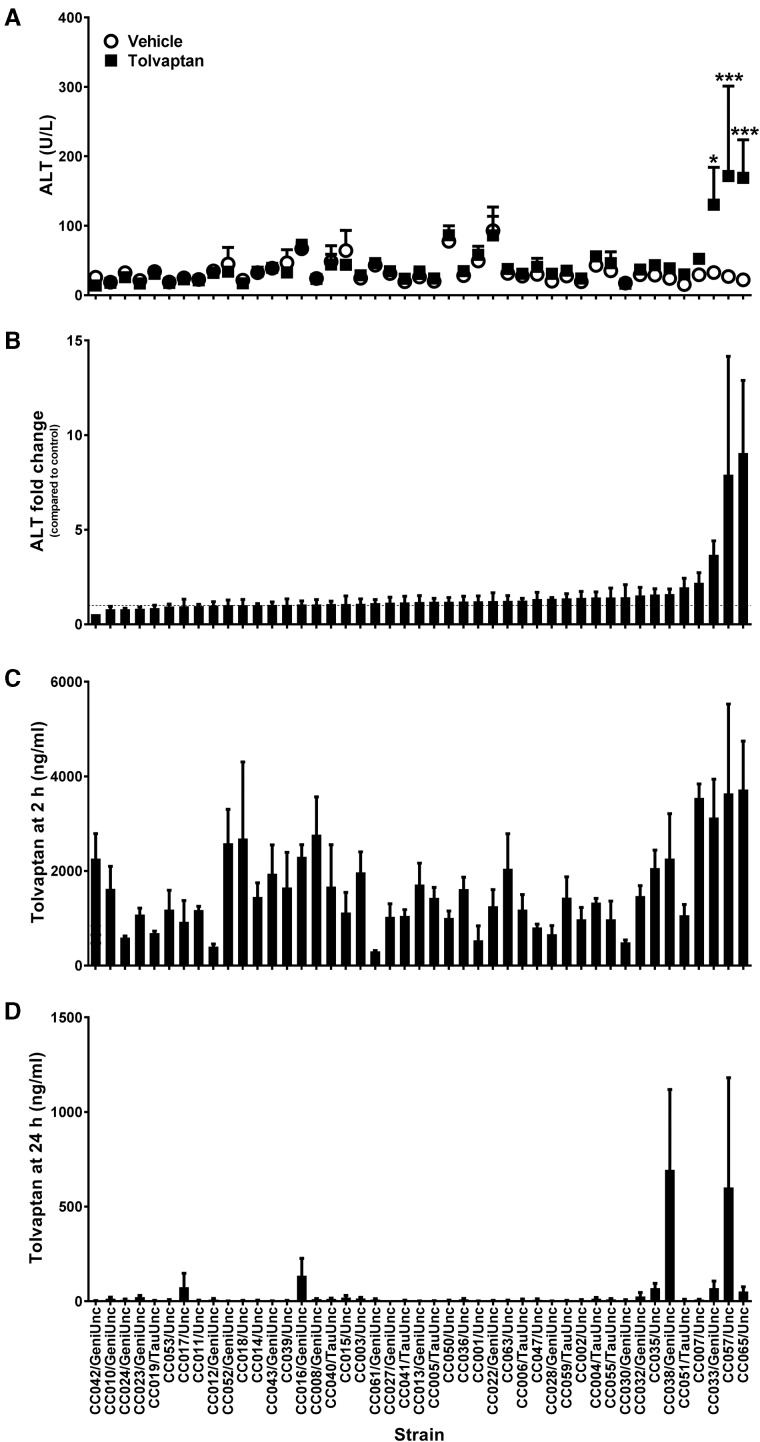
FIG. 1.
Tolvaptan induces elevations in plasma ALT in sensitive CC strains. A, Average plasma ALT for vehicle- (white circles) and tolvaptan-treated (black squares) animals. *p<.05 and ***p<.001 indicate the difference between vehicle- and tolvaptan-treated animals within each strain (Bonferroni posttest). B, Average fold change in ALT between pairs of vehicle- and tolvaptan-treated animals within each strain. C, Average plasma tolvaptan concentrations at 2 h postdose and D, 24 h postdose. Data in all panels are represented as mean + SE of N = 4 animals or pairs per strain and/or group, except as noted in the text. Strains are ordered from left to right by increasing average fold change in ALT between pairs of vehicle- and tolvaptan-treated animals within each strain.
Experimental Design
An acute, high-dose exposure study design was selected to enable the use of liver gene expression profiling as an endpoint for the hepatotoxic response (Laifenfeld et al., 2014; Leone et al., 2014). This approach helps to maximize responses at the gene expression level while limiting the opportunity for adaptation. Within each strain, animals were treated with a single oral (gavage) dose of either vehicle (hydroxypropyl methylcellulose; Sigma) or 100 mg/kg tolvaptan (provided by Otsuka Pharmaceutical Co. Ltd) in a dosing volume of 10 ml/kg. The dose selected was based on previous studies of tolvaptan in CD-1 and B6C3F1 mice (Oi et al., 2011), but reduced 3-fold below the maximally tolerated dose (300 mg/kg in B6C3F1 per nonclinical study reports) based on the predicted increased sensitivity of some CC strains. Animals were randomly assigned to treatment groups (N=4 per group, per strain) using a randomization method based on bodyweight. Vehicle- and tolvaptan-treated animals within each strain were treated in pairs, and pairs within each strain were randomized over the course of the study to minimize batch (delivery group) effects.
Whole blood (approximately 100 µl) was collected from all vehicle- and tolvaptan-treated mice at 2 h postdosing (approximate Tmax in mice). Blood was collected from the submandibular (facial) vein or, if necessary, the tail vein or lateral saphenous vein. Blood was transferred to a tube containing sodium heparin and processed into plasma for analysis of tolvaptan and DM-4103 (a primary human tolvaptan metabolite contributing to >10% systemic exposure) concentrations.
Animals were food fasted at 6 h postdosing (to reduce hepatic glycogen levels and improve histopathological assessment) and sacrificed at 24 h postdosing by CO2 inhalation followed by exsanguination. Blood was collected by cardiac puncture. Approximately 100 µl of blood from tolvaptan-treated animals was transferred into a tube containing sodium heparin and processed into plasma for analysis of tolvaptan and DM-4103 concentrations. The remaining blood was transferred into a microcentrifuge tube containing EDTA and processed into plasma for clinical chemistry. The liver of each animal was excised and weighed. A section of the left and median liver lobes was fixed in 10% neutral buffered formalin and processed for histological analysis. A section of liver tissue encompassing all regions of the median lobe was diced, placed in RNAlater (Ambion), and processed for gene expression analysis.
Plasma Drug Concentration
Heparin-treated plasma was shipped to ICON Development Solutions, Whitesboro, NY. Concentrations of parent drug and DM-4103 were determined using reversed-phase liquid chromatography with Turbo IonSpray tandem mass spectrometric detection (LC-MS/MS). Tolvaptan, DM-4103, and an internal standard were extracted from heparinized plasma using solid phase extraction. Calibration standards prepared in plasma and extracted along with samples were used to quantitate the concentrations by weighted (1/x2) linear regression of peak area ratios of analyte-to-internal standard.
Clinical Chemistry
EDTA-treated plasma was subjected to an additional centrifugation step at 16 000 × g for 10 min before freezing and transfer to ANTECH GLP, Research Triangle Park, NC. Clinical chemistry measures were performed on an Olympus AU-640 clinical chemistry analyzer (Beckman Coulter) using standard enzymatic procedures. The following analytes were measured: alanine aminotransferase (ALT), aspartate aminotransferase (AST), and total bilirubin (TBIL).
Circulating microRNA-122
Plasma levels of miR-122 were assayed using previously described methods (Kroh et al., 2010). Briefly, reverse transcription reactions were performed using the TaqMan miRNA Reverse Transcription Kit and miRNA-specific stem-loop primers for miR-122 and miR-39 (Applied Biosystems). Standard curves with known concentrations of miR-122 and miR-39 were generated in parallel using miScript miRNA mimics (Qiagen). qPCR was performed on both samples and standards using the TaqMan Universal PCR Master Mix II with no UNG and miRNA-specific TaqMan primer/probe mixes (Applied Biosystems) on a 7900HT Fast Real-Time PCR System (Applied Biosystems). Cycle threshold (CT) values for miR-122 were normalized using CT values for the spiked-in miR-39 as previously described in Kroh et al. (2010). Absolute values of miR-122 copy number were determined using the synthetic miR-122 standard curve.
Histology
Formalin-fixed liver tissue was paraffin-embedded, cut into 5-µm sections and stained with hematoxylin and eosin (H&E). H&E stained slides were microscopically examined and scored by a Board-Certified Veterinary Pathologist (Charles River Laboratories, Pathology Associates).
Statistical Analysis of Phenotype Data
Descriptive statistics on quantitative values, including single parameter tests, were performed using GraphPad Prism statistical software version 6.07. Mean values from body and organ (relative) weight measurements and clinical laboratory tests were compared with those of control groups using a 2-way analysis of variance. Comparisons between control and treated animals within a strain were performed using a Bonferroni posttest. Fold-change (compared with control) values were calculated for each strain pair, averaged by strain, and used for comparisons across strains. A Spearman’s rank correlation was used when comparing the fold change (for each strain pair) of clinical chemistry analytes to one another or to plasma drug concentration. Group comparisons for clinical observations and gross and microscopic pathology data were made using non-statistical methods.
Six animals died prematurely during the study and were excluded from the analysis of terminal endpoints. All deaths occurred shortly after the interim blood draw. Subdural hemorrhages were noted in 3 of these animals, and it is suspected that these deaths were accidental. No liver lesions were observed during histological examination that could account for the early deaths in any of these animals. Insufficient sample volume, operator error, or values below the limit of quantitation resulted in the inability to obtain terminal endpoint values from 13 animals for ALT and AST, 18 animals for bilirubin, and 12 animals for miR-122. Plasma drug concentration (parent and metabolite) values were obtained from all but 2 animals at the interim 2 h blood draw. One animal (tolvaptan-treated, strain CC057/Unc) was identified as an outlier in several endpoints and was therefore excluded from correlation analysis as well as QTL mapping. At 24 h, plasma tolvaptan concentrations for many animals were below the limit of quantitation (1 ng/ml). These were reported at 1 ng/ml for the purposes of data analysis.
Definitions of Baseline, Postdrug, and Tolvaptan Response Phenotypes for Regression-based Statistical Modeling
The statistical analysis sections below use a set of related regression-based models to infer effects on 3 types of outcome: baseline phenotypes, postdrug phenotypes, and tolvaptan response phenotypes. Unless otherwise specified, baseline phenotypes refer to outcomes measured in vehicle-treated mice (typically after treatment with vehicle), and postdrug phenotypes refer to outcomes measured in tolvaptan-treated mice. Tolvaptan response phenotypes have a special definition based on the experimental pairing of tolvaptan- and vehicle-treated mice described earlier. Formally, we define the ith (matched) pair as comprising 2 mice of CC strain but of opposite treatment assignment, where treatment with vehicle or tolvaptan occurred on the same dosing date . Letting be the (postdrug) phenotype of tolvaptan-treated mouse and be the (baseline) phenotype of its vehicle-treated pair, the tolvaptan response for pair i was defined as
| (1) |
where t is a normalizing transformation; in the case of biochemistry analytes, t is the loge such that measures the log fold change between the 2 paired mice. Tolvaptan response phenotypes have been deposited in the Dryad Digital Repository and can be downloaded at http://dx.doi.org/10.5061/dryad.ch4p5.
Estimating Tolvaptan Response, Strain-Specific Tolvaptan Response, and Heritability in the CC
The response in a given phenotype to tolvaptan treatment across the 45 CC strains was assessed using the linear mixed model,
| (2) |
where the intercept estimates the overall effect of tolvaptan treatment, the random effect estimates the strain-specific deviation from this overall effect, and dosing date and residual error are modeled as random effects and respectively. Although the genetic similarity between most pairs of strains was consistent and low (similarity score of 0.14 on average), similarity between the 2 lines which shared a breeding funnel was high (0.79). Since the strain effect term acts as a proxy for overall effects of genetic background, these lines were modeled as having the same effect, such that the actual number of strain effects modeled was 44. Models were fitted using the R package lmer (Bates et al., 2015) and tests for fixed and random effects were conducted using R package lmerTest (Bates et al., 2015). The heritability of the tolvaptan response for the phenotype was defined as . Heritability estimates are in general imprecise and reported point estimates for h2 in this study should be assumed to have wide CIs. Following identification of a subgroup of sensitive strains, we fitted an additional model that controlled for the subgroup to determine how much strain-specific variation remained, ie,
| (3) |
where is a 0−1 indicator for the sensitive subgroup, and is the additional effect of tolvaptan in this subgroup.
Strain-specific effects and heritabilities for baseline and postdrug outcomes were estimated using linear mixed models similar to those in Equations (2) and (3) but with replaced by the appropriate individual-level measurement, or .
Microarray Processing
Total RNA was isolated from the median lobe liver tissue of all animals using Trizol reagent (Invitrogen) according to manufacturer’s instructions. The isolated RNA was further purified using the QIAcube automated system (Qiagen). Following purification, the quantity of RNA was determined spectrophotometrically and the integrity of the RNA was evaluated with the Agilent 2100 Bioanalyzer. Double-stranded cDNA was synthesized from 50 to 150 ng (dependent on RNA yield) of total RNA, then transcribed to biotin-labeled cRNA using the 3’ Express IVT kit (Affymetrix). Fifteen µg of labeled cRNA was fragmented and prepared for hybridization on HT MG430 PM peg arrays. The Gene Titan robotic instrumentation (Affymetrix) was used to perform the hybridization, washing and scanning of the peg arrays.
Affymetrix CEL files were normalized using the Robust Multi-array Average method with log2 transformation (Irizarry et al., 2003) implemented in Affymetrix Power Tools (APT, v 1.17.0). To avoid confounding from genetic variation (Keane et al., 2011), probes containing any SNPs in the genome of 45 CC strains were masked using the “kill-list” option in the APT module “apt-probeset-summarize”. A second pre-filtering step was then performed to eliminate control probe sets, probe sets without mRNA annotation, and lowly expressed probe sets defined as the maximum of log2 transformed expression level across samples less than an empirically chosen cutoff of 3.66. Principal component analysis (PCA) was used to evaluate the overall performance of the arrays and identify outliers. Hierarchical clustering was then applied using the R function “hclust” with the average link function. Hierarchical clustering and PCA analysis indicated that all arrays were highly correlated and there were no obvious outliers (data not shown). After careful quality control procedures on probe, probe sets, and samples, the dataset for primary analyses consisted of 23 384 probe sets representing 13 878 genes, which is approximately equal to the number of genes expressed in the liver (Fagerberg et al., 2014; Yu et al., 2010).
Statistical Analysis of Gene Expression Data
Two complementary approaches were used on the gene expression data. First, to identify transcripts correlated ALT fold change, a simple linear regression analysis was performed to regress ALT fold change values on fold change in gene expression. Second, to identify transcripts differentially expressed with treatment (tolvaptan vs vehicle) and response to tolvaptan (sensitive vs resistant strains), a linear mixed model was used to analyze expression differences between tolvaptan- and vehicle-treated pairs of mice (explained below), while accounting for dependencies among biological replicates of each strain.
The contribution of CC strains, scan date, dosing date, and error to variability in expression data was examined first. The variabilities explained by each term were similar in the 2 treatment groups across probe sets. In tolvaptan-treated mice, the mean variabilities across probe sets were explained by error (61%), followed by strain (29%), scan date (9%), and dosing date (0.9%). Similarly, in vehicle-treated mice, the mean variabilities in expression values across the probe sets were explained by error (66%), followed by strain (24%), scan date (9%), and dosing date (1%). This result suggests that scan date and dosing date together contributed ∼10% of the variability in expression levels and hence needed to be adjusted accordingly.
Next, batch effects from scan date and dosing date were regressed out using a linear model, and the residuals were used to test for expression differences between tolvaptan- and vehicle-treated pairs. For a given probe set, letting be the expression difference for the th pair, we fitted the linear mixed model
| (4) |
with other terms as described by Equation (2). The effect of tolvaptan on expression was estimated as intercept and p-values for this parameter were calculated using the t-test with Satterthwaite approximations to the degrees of freedom using R package lmerTest (Bates et al., 2015).
A tolvaptan-induced difference in expression is not necessarily relevant to liver toxicity. To assess this relevance for each probe set, we compared the fit of Equation (4) with that of a model including an indicator for the sensitive strains (as in from Equation 3), ie,
| (5) |
whereby significant improvement in fit supports relevance. The p-values for this comparison were based on an F-test with Kenward-Roger approximation using R package pbkrtest (Halekoh and Højsgaard, 2014). To correct for multiple statistical comparisons, we applied the Benjamini-Hochberg false discovery rate (FDR)-controlling procedure (Benjamini and Hochberg, 1995).
Pathways enriched among statistically significant, differentially expressed genes in the data were identified using the Tox Analysis module in Ingenuity Pathway Analysis (Ingenuity Systems; Build version 366632M; Content version: 24718999). Gene expression data generated for this manuscript can be downloaded in its entirety from the Gene Expression Omnibus repository under the accession number GSE83739. All data are MIAME compliant.
Genotyping and Haplotype Reconstruction of the CC Mice
The genome of each CC strain comprises a mosaic of segments, or haplotypes, inherited from the 8 CC founders. Genotyping of the strains is followed by inference of the underlying haplotype mosaic, which is then used in the detection of QTL. Genotyping was performed using MegaMUGA, a custom 78K-marker array built on the Illumina Infinium genotyping platform; it has been designed to support the CC, and is optimized to identify founder contributions, and to detect any residual heterozygosity (Morgan and Welsh, 2015). Probe intensities from MegaMUGA were then converted using a hidden Markov model (HMM) (Fu et al., 2012) into a probabilistic representation of the haplotype mosaic: for each strain at each genotyped locus, the HMM calculates the probability of having inherited each of the possible haplotype pairings (diplotype), giving rise to 36 diplotype probabilities. The diplotype probabilities for 45 CC lines used in these experiments had been calculated previously and were downloaded from the CC status website (http://csbio.unc.edu/CCstatus/index.py?run=AvailableLines; last accessed January 4, 2017). The 77K markers on the genome were reduced to the 7767 genome segments for faster analyses and reduction of genotyping errors by averaging the probability matrices of 10 markers to create a new matrix for each segment. Genome scans were comparable using 77K markers versus 7767 segments, supporting the averaging across markers. All genomic locations were reported using NCBI mouse genome build 37.
General Statistical Model for QTL and eQTL Mapping
QTL mapping was performed by testing the association between tolvaptan response and the inherited founder haplotypes at each point in the genome. Genetic association was tested at each genomic locus by first fitting a null model for tolvaptan response that included effects of strain and dosing date, and then fitting an alternative model that included a QTL additive effect based on the haplotype reconstruction at the locus. Comparison of these 2 models produced scores of association across the genome (genome scans) for which genome-wide significance thresholds were then calculated. These 2 models are described in detail below.
The null model included random effects of strain and dosing date, and was identical to that used to estimate heritability (Equation 2). Note that our use of the strain random effect is equivalent to a polygenic (or kinship) effect that models all CC strains as equally related, which they approximately are. Note also that a covariate for body weight was not included since weights between each pair were similar by design. The alternative model for locus m on the th pair’s tolvaptan response was equivalent to the null model with an additional term,
| (6) |
where the QTL contribution represents the sum of the effects of the founder haplotypes, weighted by haplotype count for individual i at locus m. For strain s this quantity is defined as
| (7) |
where, eg, is the effect of the AJ haplotype on tolvaptan response, and is the number of founder AJ haplotypes present at locus m in CC strain . However, since the haplotype counts are not observed directly, being approximated only via the diplotype probabilities and therefore subject to uncertainty, we modeled using 1 of 2 approximations: for QTL mapping, that is, mapping of nontranscript phenotypes, we used a new, computationally demanding method that accounts for haplotype uncertainty; for eQTL mapping, we used an existing approach that does not account for uncertainty but is computationally efficient for large scale analyses. These are described below.
QTL Mapping Using Multiple Imputation and Genome Permutation
For QTL mapping of nontranscript phenotypes, the QTL model in Equations (2) and (6) was fitted using a new method based on multiple imputations (MIs). This new method is closely related to previous methods used for multiparent populations (Mott et al., 2000) but incorporates additional steps to take into account that the haplotype counts in Equation (7) are subject to uncertainty following Zhang et al. (2014) and references therein and also Sen and Churchill (2001). At each locus m, the diplotype probabilities were used to generate I=50 imputed datasets, that is, datasets in which all of the diplotype assignments, and therefore the haplotype counts, were treated as known. For example, if the 36 diplotype probabilities for strain s were , then a dataset with the haplotype count , which corresponds to the homozygote AJ diplotype, would be imputed with probability . Each imputed dataset was then used to fit the QTL model in Equations (2) and (6). This fitted model was then compared against the null model using a likelihood ratio test (LRT), leading to a −log10(p) score (the negative log10 of the uncorrected p-value from the LRT). These imputed −log10(p)’s were then aggregated, with their median being used as the final reported −log10(p) for locus m. This was repeated for all loci to produce a genome scan.
Genome-wide significance thresholds were calculated using a permutation procedure designed to control family-wide error rate, also developed specifically for this study. At each permutation, the genome scanning procedure described above was repeated but with the genome information of strains 44 having been reassigned to a random reordering of those strains, eg, ; from this genome scan, the maximum -log10(p) was then recorded. The maximum −log10(p) for all permutations was used to fit a generalized extreme value distribution (Dudbridge and Koeleman, 2004), and the upper fifth percentile of this fitted distribution was taken as the 0.05 genome-wide significance threshold.
eQTL Mapping Using Regression on Probabilities and FDR
QTL mapping of transcript abundance (eQTL mapping) was performed using an approximation for the QTL contribution described previously in Mott et al. (2000) and referred to here as regression on probabilities (ROPs) after Zhang et al. (2014). Briefly, in ROP, the haplotype counts in Equation (7) are substituted by their expectations based on the diplotype probabilities, such that, for example, where f indexes the founders. Unlike MI, this approximation does not account for uncertainty, however, it also does not require I model fittings per locus, and thus is considerably faster and scalable. The −log10(p) for each locus was calculated from an LRT of the null versus alternative model, and for scalability, thresholds were based on the FDR (Benjamini and Hochberg, 1995). For “targeted” eQTL mapping where we examined all eQTLs within the region of ± 10 Mb around 80% CI of the QTL peak, we calculated FDR from p-values of the most significant eQTL for each probe set within the targeted region.
As reported in many eQTL studies, there are 2 distinct types of expression associations: local eQTL (cis) and distal eQTL (trans). Following Rockman and Kruglyak (2006), the terms local eQTL and distal eQTL were used. Local eQTL was defined as segment-transcript pairs in the range of 10 Mb upstream and downstream of the physical location of the target transcript. Distal eQTLs included all the segment-transcript pairs outside the local region.
Haplotype Effects at Detected QTL and CIs for QTL Location
For detected QTL we estimated the effect of alternate haplotype substitutions on drug response using the Diploffect model of Zhang et al. (2014). This is a Bayesian linear mixed model that provides CIs (as highest posterior density intervals) both for additive (haplotype) effects and dominance (heterozygous diplotype) effects, incorporating any uncertainty in the haplotype assignments from the HMM.
CIs for the location of detected QTL were defined using positional bootstrapping after Visscher et al. (1996), as previously described by Phillippi et al. (2014) and Woods et al. (2012). To provide estimates that are computationally stable, 1000 bootstraps were performed for each QTL and the 80% CI was reported.
Testing of Candidate SNPs via Merge Analysis
To perform a merge analysis (Yalcin et al., 2005) on segregating SNPs within the 80% CI of QTL on chromosome 14, whole-genome sequencing data (release 1410) was downloaded from the Wellcome Trust Sanger Institute’s Mouse Genomes Project (http://www.sanger.ac.uk/resources/mouse/genomes/; last accessed January 4, 2017). Prior to the merge analysis, filtering was performed to remove SNPs with poor variant calling, more than 1 alternative allele, and noninformative strain distribution patterns across the 8 CC founder strains (eg, all 8 founder strains have the same allele). At each variant within the 80% CI, p-values from the 2-allele merged model were compared with those from the 8-allele haplotype model to evaluate how well genotypic variation at each SNP represented by strain distribution pattern correlates with the phenotypic distribution and whether the correlation is stronger than that seen in the 8-allele model.
Circulating Secretory Leukocyte Peptidase Inhibitor
Levels of circulating secretory leukocyte peptidase inhibitor were assayed in the remaining plasma of the 3 sensitive strains (CC033/GeniUnc, CC057/Unc, CC065/Unc) and 2 resistant strains (CC012/GeniUnc and CC027/GeniUnc) using an ELISA (Cloud-Clone Corp.). No plasma was remaining for 1 tolvaptan-treated animal in strain CC033/GeniUnc. One animal (tolvaptan-treated, strain CC065/Unc) was identified as an outlier (Grubbs’ test, α=0.05) and was therefore excluded from statistical analyses. Statistical analyses were performed using GraphPad Prism statistical software version 6.07. Fold-change (compared with control) values were calculated for each strain pair, averaged by strain, log2 transformed, and used for comparisons across strains. Fold-change values were also log2 transformed, averaged by the responses, and compared using a Student’s t-test. A Spearman’s correlation was used when comparing the fold change (for each strain pair) of analytes (without transformation) to one another.
RESULTS
Tolvaptan Induces Plasma ALT Elevations in Sensitive CC Strains
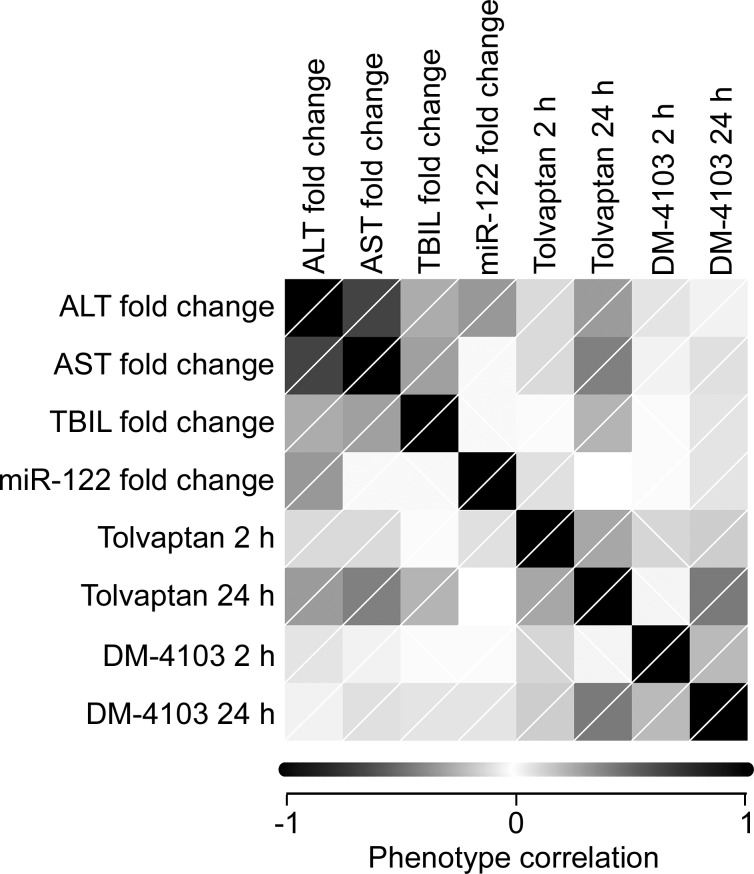
Overall, the single dose (100 mg/kg) of tolvaptan was well tolerated across all CC strains. Tolvaptan-related clinical findings were minimal and consisted mainly of moderate dehydration, an expected pharmacological effect of the drug (Miyazaki et al., 2007). No gross findings were observed consistently in the livers of tolvaptan-treated animals in any strain, and no statistically significant differences in the liver-to-body weight ratio were observed between tolvaptan- and vehicle-treated animals at necropsy (p>.05, Bonferroni posttest). No histological changes in the liver were associated with tolvaptan treatment in any strain. However, statistically significant elevations in serum ALT levels in tolvaptan-treated animals relative to vehicle-treated controls were observed in 3 of the 45 strains (Figure 1A). These strains had the highest mean fold change in ALT between tolvaptan-treated animals and their paired, vehicle-treated controls (Figure 1B). ALT fold change was significantly correlated with changes in plasma AST (Spearman r=0.6528, p<.0001), TBIL (Spearman r=0.2626, p=.0009), and miR-122 (Spearman r=0.3202, p<.0001) supporting that the ALT elevations reflect subclinical liver toxicity. A correlation matrix for all phenotypic variables is shown in Figure 2.
FIG. 2.
Correlation matrix representing relationships between phenotypic variables analyzed in tolvaptan-treated CC mice. Phenotypic variables are plotted against themselves and other variables on the x- and y-axes. Shading intensity of the corresponding squares reflect the Spearman correlation coefficient, r. Diagonal lines within the squares reflect the directionality of the correlation (ie, lines going from bottom left corner to top right corner indicate positive correlation; lines from top left corner to bottom right corner indicate negative correlation).
For each of ALT, AST, TBIL, and miR-122, linear mixed modeling was used to examine how tolvaptan response, the log fold change between matched vehicle- and drug-treated mice, varied with genetic background. Tolvaptan response was partitioned into an overall effect, constant across strains, and a set of strain-specific effects, collectively representing the heritable component of vulnerability to DILI. Next, the amount of heritability that remained after controlling for the group of 3 sensitive strains and the magnitude of this sensitive group effect was examined. Last, heritability of drug response was compared with heritability of the phenotype at baseline, as estimated from the vehicle-treated mice. These comparisons are summarized below, with full results in Supplementary Tables 1–3.
For ALT, the overall effect of tolvaptan on fold change was only borderline significant (p=.06), but the effect of CC strain was highly significant (p=2.3E-4), explaining 27% of the variance. Most of this heritability of tolvaptan response could be explained by the grouping of 3 sensitive and 42 nonsensitive strains (where 2 of the nonsensitive strains were modeled as having the same effect owing to their high genetic similarity, as described in Materials and Methods section); after controlling for the sensitive group, the effect of CC strain was insignificant (p=.49). For AST, the overall effect of tolvaptan on fold change was a little stronger (p=.01) with slightly less explained by CC strain (18%, p=.02); but as with ALT, this heritability was insignificant after controlling for the sensitive strains (p=.49). Notably, although baseline levels of ALT and AST were heritable (ALT 38%, p=9.2E-8; AST 34%, p=4.2E-6), they were not predictive of tolvaptan response in their matched pairs.
The effect of tolvaptan on TBIL fold change was highly significant overall (p=4.7E-5), but the effect of CC strain was not significant (p=.12), explaining 12% of the variance; baseline TBIL was also not significantly heritable across the CC strains (2.6%, p=.72). In contrast, the effect of tolvaptan on miR-122 fold change was insignificant, both overall and across CC strains; this was despite baseline miR-122 being significantly heritable (20%; p=5E-3). In general, heritable effects on baseline measures were not predictive of heritable effects on drug response, nor vice versa.
Plasma Tolvaptan Concentration Varies by Strain but Is Correlated With ALT
To determine whether the strain-dependent variability in plasma ALT levels was related to drug exposure, plasma tolvaptan levels were measured at 2 and 24 h postdose. Mean plasma tolvaptan concentrations at 2 h ranged from 301 to 3719 ng/ml (Figure 1C). Tolvaptan plasma concentrations at 2 h were not significantly correlated with ALT fold change in the corresponding pairs (Spearman r=0.1060, p=.1809). By 24 h, the concentration had decreased in all strains (range: 1–694 ng/ml), with many strains having values below the limit of quantitation (Figure 1D); nonetheless, correlation of these values with ALT fold change in the corresponding pairs was significant (Spearman r=0.3053, p<.0001). At both 2 and 24 h, tolvaptan concentration showed strain-specific variation, giving heritability estimates of 37% (p=3.1E-7) and 27% (p=1.9E-4) respectively, these being only slightly reduced when controlling for the sensitive strain group to 33 and 21%, respectively (Supplementary Table 3).
Plasma concentrations of a significant human metabolite of tolvaptan, DM-4103, were also measured. Plasma DM-4103 concentrations at 2 h were not significantly correlated with plasma tolvaptan concentrations at 2 h (Spearman r=−0.1204, p=.1106) or with ALT fold change values in the corresponding pairs (Spearman r=0.07457, p=.3472). Plasma DM-4103 concentrations at 24 h were weakly correlated with plasma tolvaptan concentrations at 2 h (Spearman r=0.1514, p=.0481) and strongly correlated with plasma tolvaptan concentrations at 24 h (Spearman r=0.4267, p<.0001). However, plasma levels of DM-4103 at 24 h were not correlated with ALT fold change in the corresponding pairs (Spearman r=0.03979, p=.6152). Plasma drug concentrations are included among the variables represented in the correlation matrix shown in Figure 2. The heritability of DM-4103 concentration among the CC strains was estimated at 24% for both time points and was not meaningfully changed by controlling for the sensitive strain group (Supplementary Tables 1–3).
Tolvaptan-Induced ALT Elevation Is Most Associated With Genetic Variation on Chromosome 14
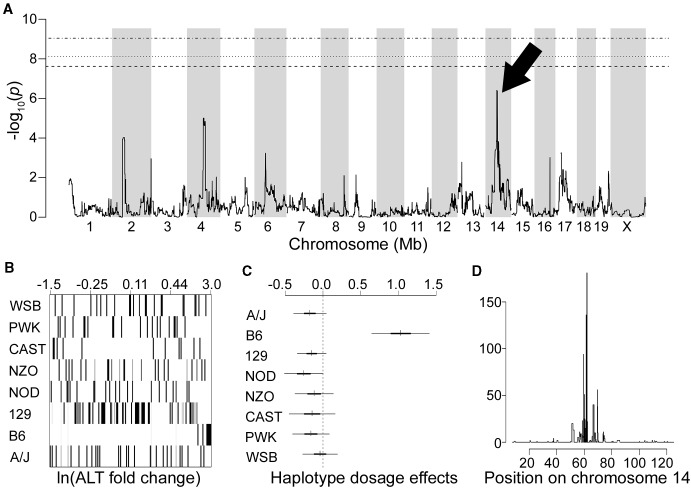
Genetic mapping was used to identify genomic regions whose variation might underlie the strain-dependent variability in ALT fold change, and more generally tolvaptan-induced liver response. Several potential QTL were found to be associated with ALT fold change at varying levels of significance (Figure 3A). Although none reached the strict permutation-based threshold of 8.11 for a genome-wide error rate of α=0.05, the strongest association (termed ALT1), and the one that best explained the strain-specificity, was found on chromosome 14. Founder strain probabilities at the ALT1 peak (chromosome 14 at 59.47 Mb, nominal −log10(p)=6.41) show an overrepresentation of C57BL/6J DNA among the pairs of mice with the greatest ALT elevations in response to tolvaptan (Figure 3B). Bayesian modeling of the QTL estimated a strong positive effect of the C57BL/6J haplotype on ALT elevation (Figure 3C). ALT1 was mapped to an 80% CI of 12 Mb (58.27–69.85 Mb) (Figure 3D). This region was interrogated further to identify quantitative trait genes (QTGs) that may explain the liver response.
FIG. 3.
Genetic mapping using ALT fold change identifies significantly associated locus on chromosome 14. A, Genome scan plot illustrating QTL associated with tolvaptan-induced ALT fold change. Dashed horizontal lines indicate genome wide significance at the 0.01, 0.05, and 0.1 levels. An arrow points to the genomic interval with the highest LOD score (Marker: JAX00381082, Chr14: 59.47 Mb). B, Founder strain probabilities at the locus peak. The x-axis plots paired animals in order from left to right by increasing ln(ALT fold change). Each horizontal row indicates the probability of a given CC founder haplotype being present at the locus, with darker intensities indicating higher probabilities (0–1.0). C, Estimates and CIs for haplotype substitution effects at the peak locus. D, CI plot for the QTL on chromosome 14. The 80% CI (Chr14: 58.27–69.85 Mb) is spanned by black bars.
Merge Analysis Identifies Candidate QTGs in QTL on Chromosome 14
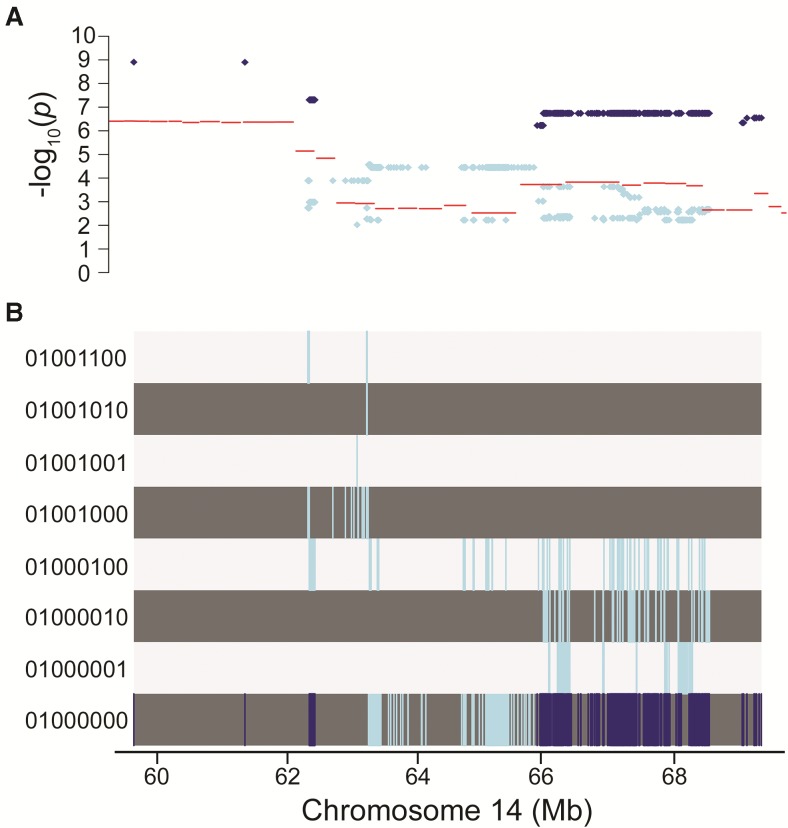
Initially, candidate causal variants were identified using merge analysis (Phillippi et al., 2014; Vered et al., 2014; Yalcin et al., 2005). Merge analysis compares the significance of association for individual variants obtained from the haplotype model used for genome-wide analysis to those obtained using a biallelic model, where the founder strain haplotypes are “merged” into 2 groups for each biallelic SNP. For potentially causative variants, both models will explain a significant portion of the variation, but the biallelic model will do so with far fewer parameters, thus providing a more parsimonious fit. In the ALT1 region on chromosome 14 (58.27–69.85 Mb), 753 variants were identified having a −log10(p) from merge analysis greater than −log10(p) from MIs and a −log10(p) from merge analysis >5 (Figure 4A). All 753 variants meeting these criteria reflected only 1 strain distribution pattern: C57BL/6J discordant from all other founder strains (Figure 4B). In total, 33 unique genes were represented among the 753 variants identified by merge analysis (Table 1). Three genes have human orthologs, are expressed in the liver, and have been associated with liver injury and are thus considered priority candidate QTGs for further investigation: Ephx2 (soluble epoxide hydrolayse), Scara3 (scavenger receptor class A, member 3), and Zfp395 (zinc finger protein 395).
FIG. 4.
Merge analysis of chromosome 14 QTL identifies candidate causal variants. A, Comparison of −log10(p) values from 8-allele haplotype model (lines) to −log10(p) values from 2-allele merge analysis (dots) for variants in Chr14: 58.27–69.85 Mb with a −log10(p) from merge analysis >2. Dark dots indicates variants with a −log10(p) from merge analysis > −log10(p) from 8-allele haplotype model and a −log10(p) from merge analysis >5. B, Corresponding strain distribution pattern for variants represented in A. Each track corresponds to strain distribution pattern indicated in the left margin by a string of 0's and 1's representing the alleles for the 8 founder strains, in the order: A/J, C57BL/6J, 129S1Sv/ImJ, NOD/ShiLtJ, NZO/H1LtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ. Within each SDP track, the vertical lines indicate the locations of the variants within the strain distribution pattern.
TABLE 1.
Candidate Quantitative Trait Genes Identified by Merge Analysis
| Gene Symbol | Gene Name | Human Orthologue | Liver Expression | Biological Relevance |
|---|---|---|---|---|
| 4930438E09Rik | RIKEN cDNA 4930438E09 gene | − | NA | NA |
| Adam2 | A disintegrin and metallopeptidase domain 2 | + | − | NA |
| Adam28 | A disintegrin and metallopeptidase domain 28 | + | − | NA |
| Adam7 | A disintegrin and metallopeptidase domain 7 | + | − | NA |
| Adra1a | Adrenoceptor alpha 1A | + | + | − |
| Bnip3l | BCL2/adenovirus E1B 19kDa interacting protein 3-like | + | + | − |
| Cdca2 | Cell division cycle associated 2 | + | − | NA |
| Dock5 | Dedicator of cytokinesis 5 | + | + | − |
| Dpysl2 | Dihydropyrimidinase-like 2 | + | + | − |
| Ebf2 | Early B-cell factor 2 | + | − | NA |
| Elp3 | Elongator acetyltransferase complex subunit 3 | + | + | − |
| Ephx2a | Epoxide hydrolase 2, cytoplasmic | + | + | + |
| Fbxo16 | F-box protein 16 | + | + | − |
| Fzd3 | frizzled homolog 3 | + | + | − |
| Gm10032 | Predicted gene 10032 | − | NA | NA |
| Gm10860 | Predicted gene 10860 | − | NA | NA |
| Gm20667 | Predicted gene 20667 | − | NA | NA |
| Gm23899 | Predicted gene 23899 | − | NA | NA |
| Gm24258 | Predicted gene 24258 | − | NA | NA |
| Gm27647 | Predicted gene 27647 | − | NA | NA |
| Gm6878 | Predicted gene 6878 | − | NA | NA |
| Kctd9 | Potassium channel tetramerization domain containing 9 | + | + | − |
| Mir6539 | MicroRNA 6539 | – | NA | NA |
| Nuggc | Nuclear GTPase, germinal center associated | + | + | – |
| Pnma2 | Paraneoplastic antigen MA2 | + | − | NA |
| Pnoc | Prepronociceptin | + | − | NA |
| Ppp2r2a | Protein phosphatase 2, regulatory subunit B, alpha | + | + | − |
| Ptk2b | PTK2 protein tyrosine kinase 2 beta | + | + | − |
| Scara3a | Scavenger receptor class A, member 3 | + | + | + |
| Scara5 | Scavenger receptor class A, member 5 | + | + | − |
| Spata13 | Spermatogenesis associated 13 | + | + | − |
| Stmn4 | Stathmin-like 4 | + | − | NA |
| Zfp395a | Zinc finger protein 395 | + | + | + |
Indicates priority candidate QTGs based on meeting all 3 criteria: human ortholog, liver expression, and biological relevance.
Fold change in the other measured plasma chemistries were also used for QTL mapping to see if additional risk factors could be identified using biomarkers with different mechanisms of release and/or kinetics. Not surprisingly, the highly correlated AST also produced a suggestive QTL (−log10(p)>4) on chromosome 14 (peak at 61.81 Mb, −log10(p)=4.70), but no suggestive QTLs were observed with TBIL or miR-122 (data not shown).
Expression QTL Mapping Is Used to Identify Additional QTGs Associated With Sensitivity to the Tolvaptan-Induced Liver Response
The identification of candidate QTGs using phenotype-based QTL mapping alone is founded on the assumption that a gene variant (ie, a SNP) causes a change in the protein-coding region of a gene that ultimately affects the drug response. However, recent studies have highlighted the importance of variants that influence responses by regulating gene expression. To explore these relationships, phenotypic data can be evaluated in the context of gene expression data from the target organ. This approach has been used successfully in CC mice to identify new candidate genes for asthma (Kelada et al., 2014; Rutledge et al., 2014). A similar approach was applied here to identify additional QTGs. Microarray profiling was performed using liver tissue of all vehicle- and tolvaptan-treated animals included this study. Whole genome expression QTL (eQTL) mapping was performed using expression values from vehicle- and tolvaptan-treated mice separately. More importantly, gene-by-environment, or in the case of this study, gene-by-treatment (GxT) eQTLs were identified using the fold change (delta) in expression between strain pairs of vehicle- and tolvaptan-treated mice. A complete description of the overall eQTL mapping findings is described in the Supplementary material. Findings, as they relate to QTGs for susceptibility to the tolvaptan-induced liver response, are described below.
Initially, a “targeted approach” was used to identify additional QTGs associated with the tolvaptan liver response. Similar to Kelada et al. (2014) and Rutledge et al. (2014), inquiry focused on local eQTLs (segment-probe set pairs) within the region of the phenotype QTL (in this case, the QTL termed ALT1). Unlike those approaches, however, which focused on eQTLs present in treated mice only (Kelada et al., 2014; Rutledge et al., 2014), our analysis was performed using GxT eQTLs, which are more directly relevant for identifying gene variants that influence treatment-induced responses. As described in the Supplementary material, the most significant GxT eQTLs were distal. Even with a targeted approach designed to increase statistical power, no local GxT eQTLs in the region of ALT1 passed the threshold for statistical significance (FDR p<.001).
As mentioned earlier, liver gene expression analysis after an acute, high-dose exposure can also be used to identify activation of molecular phenotypes that may indicate sensitivity toward an adverse drug response in humans (Laifenfeld et al., 2014; Leone et al., 2014). In the absence of overt injury, molecular phenotypes can be used to identify variants influencing individual treatment-induced responses. To investigate this possibility, genome-wide GxT interaction eQTLs (both local and distal) were examined to identify new molecular phenotypes as well as variants influencing their response. No local GxT interaction eQTLs were observed. However, 385 distal eQTLs were identified (FDR p<.001). These eQTLs comprise 14 unique probe sets comprising 11 genes (Table 2). Four of these genes have human orthologs expressed in the liver with biological relevance to DILI and thus are considered candidate QTGs for further investigation: Angptl4 (angiopoietin-like 4), Irf3 (interferon regulatory factor 3, Itgb5 (integrin, beta 5), and Mff (mitochondrial fission factor).
TABLE 2.
Candidate Quantitative Trait Genes Identified by Whole Genome Gene by Treatment eQTL Mapping
| Gene Symbol | Gene Name | Human Orthologue | Liver Expression | Biological Relevance |
|---|---|---|---|---|
| Angptl4a | Angiopoetin-like 4 | + | + | + |
| Cpne1 | Copine 1 | + | + | − |
| Hist1h2be | Histone Cluster 1, H2be | + | − | NA |
| Htra1 | HtrA Serine Peptidase 1 | + | + | − |
| Irf3a | Interferon regulatory factor 3 | + | + | + |
| Itgb5a | Integrin, Beta 5 | + | + | + |
| Lrrtm3 | Leucine rich repeat transmembrane neuronal 3 | + | − | NA |
| Mffa | Mitochondrial fission factor | + | + | + |
| Rarres1 | Retinoic acid receptor responder 1 | + | + | − |
| Slc7a6 | Solute Carrier Family 7, member 6 | + | − | NA |
| Sox9 | Sex determining region Y box 9 | + | + | − |
Indicates priority candidate QTGs based on meeting all 3 criteria: human ortholog, liver expression, and biological relevance.
Gene Expression Profiling Also Identifies Mechanisms Associated With the Tolvaptan-Induced Liver Response
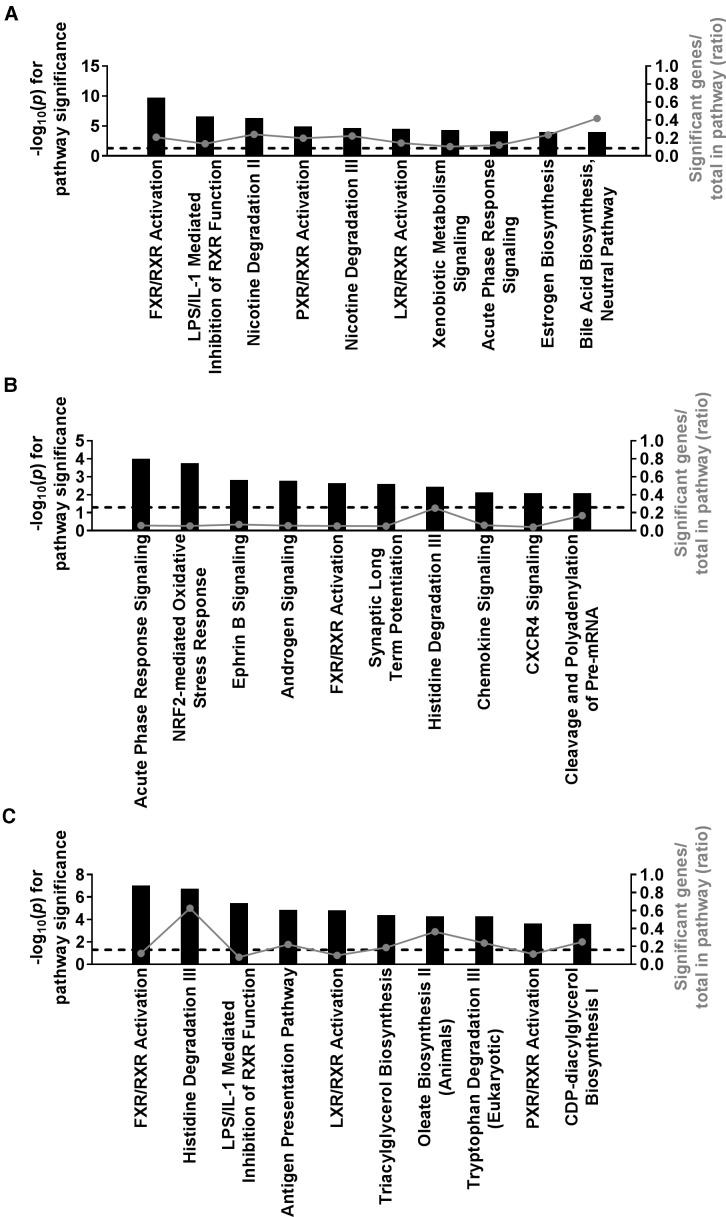
Finally, gene expression data were analyzed to provide insight into mechanisms of toxicity associated with the tolvaptan-induced liver response. Three different analytical approaches were used. First, a regression analysis was performed using the fold change in gene expression for each probe set on the array and fold change in ALT. Tolvaptan-induced changes in 1020 probe sets were significantly correlated with tolvaptan-induced changes in ALT (FDR p<.2). The top pathway enriched among these genes, and a second pathway in the top 10, are associated with the regulation of bile acid homeostasis: “FXR/RXR Activation” and “Bile Acid Biosynthesis” (Figure 5A). Second, transcripts differentially expressed with treatment (tolvaptan vs vehicle) were identified using a linear mixed effect model as described in the methods. A total of 250 probe sets were differentially expressed with tolvaptan treatment across all strains (FDR p<.2). Included in the top 10 pathways enriched among these genes are pathways associated with immune response: “Acute Phase Response Signaling”, “Chemokine Signaling”, and “CXCR4 Signaling” and oxidative stress: “Nrf2-meidated Oxidative Stress Response” (Figure 5B). Last, all strains were categorized as “sensitive” or “resistant” based on tolvaptan-induced elevations in ALT, and transcripts differentially expressed with response (sensitive vs resistant strains) were identified using a linear mixed effect model as described in the methods. This approach identified 449 tolvaptan-responsive probe sets differentially expressed between sensitive and resistant strains (FDR p<.2). The top pathway enriched among these genes is associated with the regulation of bile acid levels within the hepatocytes: “FXR/RXR Activation” (Figure 5C).
FIG. 5.
Ingenuity pathway enrichment analysis of liver gene expression identifies general mechanisms associated with the tolvaptan-induced liver response. Top 10 pathways enriched among gene expression changes associated with A, ALT fold change B, tolvaptan treatment, and C, tolvaptan response (sensitive vs resistant strains). Significant probe sets (FDR p<.2) were identified by a regression analysis in A or linear mixed model in B and C. Pathway significance is plotted by −log10(p) on the left y-axis and represented by the black bars on the graphs. The dashed line indicates a −log10(p) >1.3 threshold for significance. The ratio of significant genes relative to the total genes in the pathway is plotted on the right y-axis and represented by the gray dots (connected by a gray line) on the graphs.
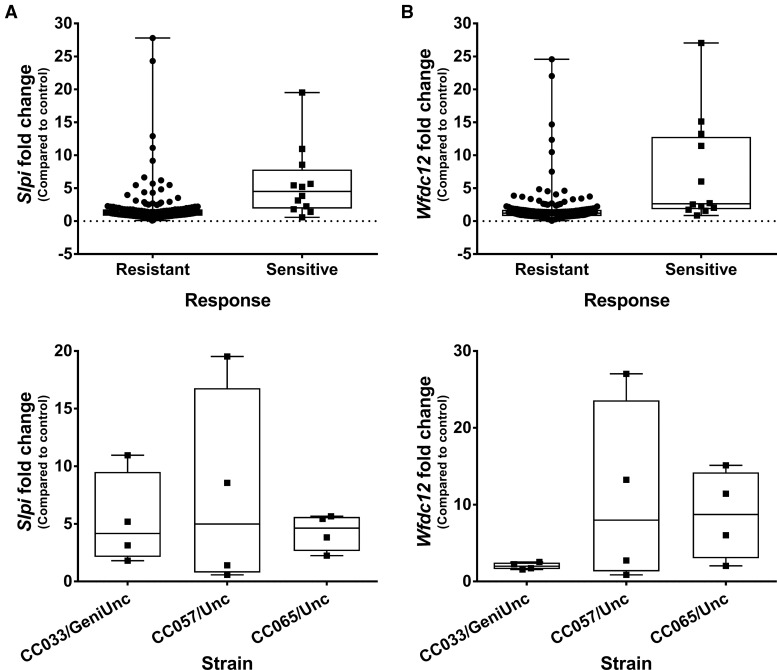
Overlap of Treatment and Response Associated Genes Identifies Individual Transcripts That May Contribute to the Tolvaptan-Induced Liver Response in CC Mice
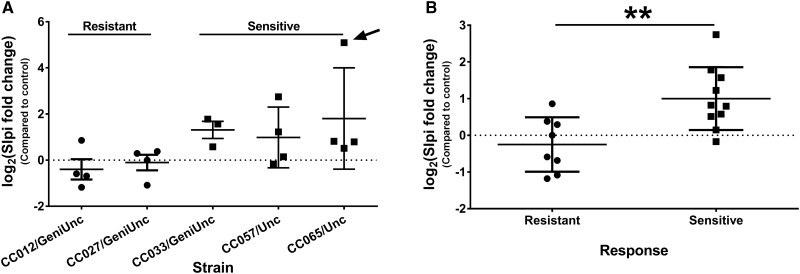
Two probe sets were found to be significantly differentially expressed with both treatment and response (FDR p<.2) and have a fold change>|2| associated with both factors. These probe sets corresponded to the genes: Slpi (secretory leukocyte peptidase inhibitor) and Wfdc12 (whey acidic protein 4-disulfide-core 12) (Figure 6). Both genes play a similar role in modulating inflammation (Glasgow et al., 2015; Odaka et al., 2003; Reeves et al., 2013; Williams et al., 2006) and the more extensively studied Slpi has been shown recently to be elevated in livers of patients with acetaminophen-induced acute liver failure (AALF) (Antoniades et al., 2014). In these patients, elevated transcript levels corresponded to elevations in Slpi protein, both in liver tissue and in circulation. To see if this was also true in the CC mice, Slpi levels were measured in plasma from the 3 sensitive strains (demonstrating elevated Slpi transcript levels) as well as 2 resistant strains (with no increase in Slpi transcript). Circulating Slpi was consistently elevated in the sensitive strains (Figure 7A) and the average fold change in sensitive strains was significantly higher than the average fold change in resistant strains (Figure 7B). Fold change in circulating Slpi was also correlated with fold change in ALT (Spearman r=0.6883, p=.0016), and fold change in Slpi transcript levels in the liver (Spearman r=0.6526, p=.0025).
FIG. 6.
Gene expression changes in Slpi and Wfdc12 are associated with tolvaptan treatment and liver response. Average fold change for each strain pair is plotted by response on the top and strain (for the 3 sensitive strains) on the bottom for A, Slpi and B, Wfdc12. Both probe sets were significantly associated with treatment and response (FDR p<.2, FC > 2).
FIG. 7.
Slpi protein is elevated in plasma of sensitive strains. A, Average log2(fold change) by strain for strain pairs from 2 resistant and 3 sensitive strains. Arrow points to outlier pair in sensitive strain CC065/Unc (Grubbs’ test, α = 0.05) that was removed prior to statistical analysis in B. B, Average log2(fold change) by response (with outlier from A removed). **p<.01 indicates the difference between sensitive and resistant strains (Student’s t-test).
DISCUSSION
The design of this study exploits 2 key features of the CC population: (1) the ability to repeatedly measure phenotypes under drug- and vehicle-treatments that were randomized between genetically identical, matched mouse pairs; and (2) the ability to observe these phenotypes across a diverse and equally related set of genetic backgrounds. This combination of features allows rigorous estimation of the causal effect of drug treatment, where the tolvaptan response defined by each pair is confounded by neither baseline outcomes nor genetic background. Additionally, these features provide a stable basis for detecting GxT effects and replicating them in follow-up studies. By comparison, the use of a single inbred strain provides replicability and rigor but no genetic diversity and hence no power. On the other end of the spectrum, the use of outbred resources such as the Diversity Outbred popul-ation provides genetic diversity and high mapping resolution (Svenson et al., 2012), but cannot support such a rigorous measurement of treatment response nor provide a stable basis for replication studies, follow-up mechanistic studies, or testing next-in-class compounds.
After a single dose of tolvaptan only 4× the daily human equivalent dose used in the ADPKD clinical trials (Watkins et al., 2015), evidence for liver stress was observed in 3 CC strains. This finding was unexpected, given that no liver injury was observed in traditional nonclinical models after multiple treatments with higher doses (Oi et al., 2011). This rapid response at a relatively low dose suggests that these 3 CC strains may be more genetically sensitive to a tolvaptan-induced liver response than other CC strains and traditional mouse models.
Variation in plasma tolvaptan exposure explained <10% of variability in ALT values observed across animals in this study. This is consistent with the lack of an obvious relationship between tolvaptan exposure and hepatotoxicity in subjects from the tolvaptan clinical trials (Watkins et al., 2015). As a result, genetic variants that may contribute to this sensitivity in mice were explored further using ALT fold change as a marker of the liver response and a QTL was identified on chromosome 14. To further ensure this QTL was not simply the result of exposure, genetic mapping was also performed using plasma concentrations of tolvaptan and DM-4103 at 2 h and no suggestive peaks (−log10(p)>4) were observed (data not shown). Furthermore, mapping was also performed using ALT fold change with plasma concentration of tolvaptan or DM-4103 as a covariate, and the most significant QTL was still on chromosome 14.
Three priority candidate QTGs identified via merge analysis have biological relevance to liver injury. Ephx2 is a liver enzyme that catalyzes the conversion of protective epoxyeicosatrienoic acid to dihydroxyeicosatrienoic acid, which is associated with increased hepatic inflammation and injury (Schuck et al., 2014). Scara3 is a cellular stress response protein that has been shown to protect cells, including hepatocytes, from oxidative stress by scavenging reactive oxygen species (Oguro et al., 2014). Zfp395, the mouse ortholog of human zinc finger protein 395 (ZNF395), is a hypoxia-inducible transcription factor that contributes to inflammation by superactivating proinflammatory cytokines (Herwartz et al., 2015). Associations of DILI risk with these genes may suggest a role for oxidative stress and/or inflammation in the tolvaptan liver response.
In addition to using traditional phenotypes for QTL mapping, whole-genome eQTL mapping was performed to identify drug-induced molecular phenotypes that are influenced by genetic variation. Genes with drug-induced eQTLs may influence the drug response and can inform potential mechanisms of toxicity as well as genetic risk factors for toxicity susceptibility. Four genes that may mediate or modulate a DILI response were identified using drug-induced fold change in gene expression as the primary phenotype. Angptl4 controls inflammation and apoptosis in the liver (Guo et al., 2015). Irf3 regulates cytokine levels and plays a role in liver diseases such as steatosis, inflammation, and alcoholic liver injury (Loi et al., 2013; Ni et al., 2015). Itgb5 is a subunit of an integrin receptor involved in cell adhesion, viral entry, and antigen processing (Lyle and McCormick, 2010). Mff2 controls mitochondrial and peroxisomal fission, an important process affecting mitochondrial adaptation and apoptosis (Gandre-Babbe and van der Bliek, 2008). Alterations in these genes may suggest a role for immunotoxicity and/or mitochondrial dysfunction in the tolvaptan liver response.
Gene expression profiling was also used to gain mechanistic insight into the tolvaptan liver response. Pathways most enriched among genes associated with tolvaptan-induced ALT elevations included “FXR/RXR Activation” and “Bile Acid Biosynthesis”. These pathways are made up of genes involved in the regulation of bile acid levels within hepatocytes and support recent studies implicating alterations in bile acid homeostasis as a possible mechanism underlying tolvaptan DILI (Slizgi et al., 2016; Woodhead et al., 2016).
The levels of 2 individual genes, Slpi and Wfdc12, were also found to change not only with tolvaptan treatment but also to differentiate sensitive and resistant strains. Both genes appear to play a similar role in regulating immune response. Slpi inhibits nuclear factor kappa B (NF-kB)-dependent proinflammatory signaling (Reeves et al., 2013; Williams et al., 2006), and macrophages secrete Slpi in response to phagocytosis of apoptotic cells (Odaka et al., 2003). Therefore, the elevations of Slpi observed in the present study may indicate an initial inflammatory response, apoptosis, and subsequent macrophage activation in the livers of sensitive tolvaptan-treated animals. A similar mechanism has been proposed to explain the elevation in SLPI mRNA and Slpi protein observed in the livers of patients with AALF (Antoniades et al., 2014). Liver levels corresponded to Slpi elevations in plasma samples of AALF patients—a finding that translated to this study. As susceptible mice in this study had minimal elevations in serum ALT and no histological evidence of liver injury, blood levels of Slpi levels may be elevated in the plasma of patients prior to clinical evidence of tolvaptan DILI.
More importantly, Slpi provides a potential connection between the initial drug-induced cellular stress occurring at the hepatocyte level and a delayed adaptive immune response. In addition to inhibiting the proinflammatory response, elevated Slpi levels also suppress secondary immune response (immune tolerance) by inhibiting regulatory T-cell differentiation (Muller et al., 2012). There is strong support that the loss of immune tolerance is a critical step in the pathogenesis of IDILI (Mosedale and Watkins, Accepted). Taken together, this suggests that tolvaptan-induced elevations in Slpi in sensitive strains may ultimately result in loss of immune tolerance and precede the onset of an adaptive immune attack.
A proposed alternative explanation for the delay in liver injury observed in ADPKD patients is the contribution of disease progression to DILI susceptibility (Watkins et al., 2015; Woodhead et al., 2016). Interestingly, both rodent models of ADPKD and ADPKD patients have increased bile acid concentrations and inflammatory cytokines in liver cyst fluid (Li et al., 2008; Munoz-Garrido et al., 2015), and in this study genes and pathways implicated in altering bile acid homeostasis and immune response were found to be associated with tolvaptan-induced liver stress in the sensitive CC strains. It is therefore conceivable that these findings reflect specific DILI risk factors associated with disease progression among ADPKD patients.
Finally, pathways associated with tolvaptan treatment, independent of response suggested tolvaptan-induced alterations in immune signaling and oxidative stress in all animals, regardless of susceptibility to liver stress. These findings are supported by a recently published study demonstrating oxidative stress and apoptosis in HepG2 (hepatocellular carcinoma) cells exposed to tolvaptan (Wu et al., 2015). However, given that oxidative stress was not associated with ALT elevations in the mice (and that oxidative stress was observed in HepG2 cells), it may not underlie susceptibility to tolvaptan DILI.
In conclusion, 3 of the 45 CC strains evaluated in this study were found to be sensitive to tolvaptan-induced liver stress. Plasma drug concentration levels were only minimally correlated with the degree of liver response. Genetic mapping identified a locus associated with tolvaptan-induced liver stress on chromosome 14 that contained 3 candidate genes previously implicated in liver injury. eQTL mapping also identified 4 new molecular phenotypes as well as variants influencing their response. Finally, gene expression profiling was used to gain a mechanistic understanding of the liver response and suggests alterations in bile acid homeostasis are most likely to underlie susceptibility to the stress in sensitive strains. Together with mitochondrial dysfunction and oxidative stress, which were observed in all tolvaptan-treated CC mice, these findings may reflect early hepatocyte stress, even in the absence of overt injury. In sensitive strains, these events were associated with the release of Slpi, consistent with an innate immune response and suppression of immune tolerance. Eventually, these events may promote an adaptive immune attack and overt liver injury. Taken together, the findings from this CC study may now guide a targeted, hypothesis-based approach to biomarker discovery in the DNA and sequential samples of plasma and urine that were collected and archived from clinical trials. The ultimate goal of this research is to inform a precision medicine approach to manage the risk of DILI in tolvaptan-treated patients.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
ACKNOWLEDGMENTS
We wish to acknowledge Darla Miller and Fernando Pardo Manuel de Villena of University of North Carolina, Chapel Hill and Michael Black, Darol Dodd, and Linda Pluta of The Hamner Institutes for their technical guidance and support of these studies.
FUNDING
Otsuka Pharmaceutical Development and Commercialization, Inc. R.W.C., G.R.K., W.V., and X.Y. were supported in part by a grant from the National Institute for General Medical Sciences under award R01-GM104125 (W.V.).
Supplementary Material
REFERENCES
- Antoniades C. G., Khamri W., Abeles R. D., Taams L. S., Triantafyllou E., Possamai L. A., Bernsmeier C., Mitry R. R., O'Brien A., Gilroy D., et al. (2014). Secretory leukocyte protease inhibitor: A pivotal mediator of anti-inflammatory responses in acetaminophen-induced acute liver failure. Hepatology 59, 1564–1576. [DOI] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B., Walker S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 671, 1–48. [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R Stat. Soc. 57, 289–300. [Google Scholar]
- Burkholder T., Foltz C., Karlsson E., Linton C. G., Smith J. M. (2012). Health evaluation of experimental laboratory mice. Curr. Protoc. Mouse Biol. 2, 145–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Cross Consortium. (2012). The genome architecture of the collaborative cross mouse genetic reference population. Genetics 190, 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court M. H., Peter I., Hazarika S., Vasiadi M., Greenblatt D. J., Lee W. (2013). Candidate gene polymorphisms in patients with acetaminophen-induced acute liver failure. Drug Metabolism and Disposition [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F., Koeleman B. P. C. (2004). Efficient Computation of significance levels for multiple associations in large studies of correlated data, including genomewide association studies. Am. J. Hum. Genet. 75, 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L., Hallström B. M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., et al. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festing M. F. W. (2010). Inbred strains should replace outbred stocks in toxicology, safety testing, and drug development. Toxicol. Pathol. 38, 681–690. [DOI] [PubMed] [Google Scholar]
- Fu C. P., Welsh C. E., Villena FPMd., McMillan L. (2012). Inferring ancestry in admixed populations using microarray probe intensities In Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine, Orlando, Florida, ACM, pp. 105–112. [Google Scholar]
- Gandre-Babbe S., van der Bliek A. M. (2008). The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 19, 2402–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow A. M. A., Small D. M., Scott A., McLean D. T., Camper N., Hamid U., Hegarty S., Parekh D., O'Kane C., Lundy F. T., et al. (2015). A role for whey acidic protein four-disulfide-core 12 (WFDC12) in the regulation of the inflammatory response in the lung. Thorax 70, 426–432. [DOI] [PubMed] [Google Scholar]
- Guo L., Li S., Zhao Y., Qian P., Ji F., Qian L., Wu X., Qian G. (2015). silencing angiopoietin-like protein 4 (ANGPTL4) protects against lipopolysaccharide-induced acute lung injury via regulating SIRT1/NF-kB pathway. J. Cell. Physiol. 230, 2390–2402. [DOI] [PubMed] [Google Scholar]
- Halekoh U., Højsgaard S. (2014). A Kenward-Roger approximation and parametric bootstrap methods for tests in linear mixed models – The R package pbkrtest. J. Stat. Softw. 599, 1–32.26917999 [Google Scholar]
- Harrill A. H., DeSmet K. D., Wolf K. K., Bridges A. S., Eaddy J. S., Kurtz C. L., Hall J. E., Paine M. F., Tidwell R. R., Watkins P. B. (2012). A mouse diversity panel approach reveals the potential for clinical kidney injury due to DB289 not predicted by classical rodent models. Toxicol. Sci. 130, 416–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill A. H., Watkins P. B., Su S., Ross P. K., Harbourt D. E., Stylianou I. M., Boorman G. A., Russo M. W., Sackler R. S., Harris S. C., et al. (2009). Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res. 19, 1507–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwartz C., Castillo-Juárez P., Schröder L., Barron B. L., Steger G. (2015). The transcription factor ZNF395 is required for the maximal hypoxic induction of proinflammatory cytokines in U87-MG cells. Mediators Inflamm. 2015, 804264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer‐Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Kaplowitz N. (2005). Idiosyncratic drug hepatotoxicity. Nat. Rev. Drug Discov. 4, 489–499. [DOI] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., Yalcin B., Heger A., Agam A., Slater G., Goodson M., et al. (2011). Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelada S. N. P., Carpenter D. E., Aylor D. L., Chines P., Rutledge H., Chesler E. J., Churchill G. A., Pardo-Manuel de Villena F., Schwartz D. A., Collins F. S. (2014). Integrative genetic analysis of allergic inflammation in the murine lung. Am. J. Respir. Cell Mol. Biol. 51, 436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroh E. M., Parkin R. K., Mitchell P. S., Tewari M. (2010). Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 50, 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laifenfeld D., Qiu L., Swiss R., Park J., Macoritto M., Will Y., Younis H. S., Lawton M. (2014). Utilization of causal reasoning of hepatic gene expression in rats to identify molecular pathways of idiosyncratic drug-induced liver injury. Toxicol. Sci. 137, 234–248. [DOI] [PubMed] [Google Scholar]
- Leone A., Nie A., Brandon Parker J., Sawant S., Piechta L. A., Kelley M. F., Mark Kao L., Jim Proctor S., Verheyen G., Johnson M. D., et al. (2014). Oxidative stress/reactive metabolite gene expression signature in rat liver detects idiosyncratic hepatotoxicants. Toxicol. Appl. Pharmacol. 275, 189–197. [DOI] [PubMed] [Google Scholar]
- Li J., Uetrecht J. P. (2010). The danger hypothesis applied to idiosyncratic drug reactions. Handb Exp Pharmacol 493–509. [DOI] [PubMed] [Google Scholar]
- Li X., Magenheimer B. S., Xia S., Johnson T., Wallace D. P., Calvet J. P., Li R. (2008). A tumor necrosis factor-[alpha]-mediated pathway promoting autosomal dominant polycystic kidney disease. Nat Med 14, 863–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi P., Yuan Q., Torres D., Delbauve S., Laute M. A., Lalmand M. C., Pétein M., Goriely S., Goldman M., Flamand V. (2013). Interferon regulatory factor 3 deficiency leads to interleukin-17-mediated liver ischemia-reperfusion injury. Hepatology 57, 351–361. [DOI] [PubMed] [Google Scholar]
- Lyle C., McCormick F. (2010). Integrin αvβ5 is a primary receptor for adenovirus in CAR-negative cells. Virol. J. 7, 148-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T., Fujiki H., Yamamura Y., Nakamura S., Mori T. (2007). Tolvaptan, an orally active vasopressin V2-receptor antagonist—pharmacology and clinical trials. Cardiovasc. Drug Rev. 25, 1–13. [DOI] [PubMed] [Google Scholar]
- Morgan A. P., Welsh C. E. (2015). Informatics resources for the Collaborative Cross and related mouse populations. Mamm. Genome 26, 521–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale M., Watkins P. B. (2016). Drug-induced liver injury: Advances in mechanistic understanding that will inform risk management. Clin. Pharmacol. Ther. doi:10.1002/cpt.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale M., Wu H., Kurtz C. L., Schmidt S. P., Adkins K., Harrill A. H. (2014). Dysregulation of protein degradation pathways may mediate the liver injury and phospholipidosis associated with a cationic amphiphilic antibiotic drug. Toxicol. Appl. Pharmacol. [DOI] [PubMed] [Google Scholar]
- Mott R., Talbot C. J., Turri M. G., Collins A. C., Flint J. (2000). A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. U.S.A. 97, 12649–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A. M., Jun E., Conlon H., Sadiq S. A. (2012). Inhibition of SLPI ameliorates disease activity in experimental autoimmune encephalomyelitis. BMC Neurosci. 13, 30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Garrido P., Marin J. J. G., Perugorria M. J., Urribarri A. D., Erice O., Sáez E., Uriz M., Sarvide S., Portu A., Concepcion A. R., et al. (2015). Ursodeoxycholic acid inhibits hepatic cystogenesis in experimental models of polycystic liver disease. J. Hepatol. 63, 952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Xu T., Wang Y., He Y., Zhou Q., Huang C., Meng X., Li J. (2015). Inhibition of IRF3 expression reduces TGF-β1-induced proliferation of hepatic stellate cells. J. Physiol. Biochem. 72, 9–23. [DOI] [PubMed] [Google Scholar]
- Odaka C., Mizuochi T., Yang J., Ding A. (2003). Murine Macrophages produce secretory leukocyte protease inhibitor during clearance of apoptotic cells: Implications for resolution of the inflammatory response. J. Immunol. 171, 1507–1514. [DOI] [PubMed] [Google Scholar]
- Oguro A., Koyama C., Xu J., Imaoka S. (2014). A cellular stress response (CSR) that interacts with NADPH-P450 reductase (NPR) is a new regulator of hypoxic response. Biochem. Biophys. Res. Commun. 445, 43–47. [DOI] [PubMed] [Google Scholar]
- Oi A., Morishita K., Awogi T., Ozaki A., Umezato M., Fujita S., Hosoki E., Morimoto H., Ishiharada N., Ishiyama H., et al. (2011). Nonclinical safety profile of tolvaptan. Cardiovasc. Drugs Ther. 25(Suppl 1), S91–S99. [DOI] [PubMed] [Google Scholar]
- Phillippi J., Xie Y., Miller D. R., Bell T. A., Zhang Z., Lenarcic A. B., Aylor D. L., Krovi S. H., Threadgill D. W., Pardo-Manuel de Villena F., et al. (2014). Using the emerging Collaborative Cross to probe the immune system. Genes Immun. 15, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves E. P., Banville N., Ryan D. M., O'Reilly N., Bergin D. A., Pohl K., Molloy K., McElvaney O. J., Alsaleh K., Aljorfi A., et al. (2013). Intracellular secretory leukoprotease inhibitor modulates inositol 1,4,5-triphosphate generation and exerts an anti-inflammatory effect on neutrophils of individuals with cystic fibrosis and chronic obstructive pulmonary disease. BioMed Res. Int. 2013, 560141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Kruglyak L. (2006). Genetics of global gene expression. Nat. Rev. Genet. 7, 862–872. [DOI] [PubMed] [Google Scholar]
- Rutledge H., Aylor D. L., Carpenter D. E., Peck B. C., Chines P., Ostrowski L. E., Chesler E. J., Churchill G. A., de Villena F. P. M., Kelada S. N. P. (2014). Genetic regulation of Zfp30, CXCL1, and neutrophilic inflammation in murine lung. Genetics 198, 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck R. N., Zha W., Edin M. L., Gruzdev A., Vendrov K. C., Miller T. M., Xu Z., Lih F. B., DeGraff L. M., Tomer K. B., et al. (2014). The cytochrome P450 epoxygenase pathway regulates the hepatic inflammatory response in fatty liver disease. PLoS One 9, e110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Ś., Churchill G. A. (2001). A statistical framework for quantitative trait mapping. Genetics 159, 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slizgi J. R., Lu Y., Brouwer K. R., St. Claire R. L., Freeman K. M., Pan M., Brock W. J., Brouwer K. L. R. (2016). Inhibition of human hepatic bile acid transporters by tolvaptan and metabolites: contributing factors to drug-induced liver injury? Toxicol. Sci. 1491, 237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson K. L., Gatti D. M., Valdar W., Welsh C. E., Cheng R., Chesler E. J., Palmer A. A., McMillan L., Churchill G. A. (2012). High-resolution genetic mapping using the mouse diversity outbred population. Genetics 190, 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres V. E., Chapman A. B., Devuyst O., Gansevoort R. T., Grantham J. J., Higashihara E., Perrone R. D., Krasa H. B., Ouyang J., Czerwiec F. S. (2012). Tolvaptan in patients with autosomal dominant polycystic kidney disease. N. Engl. J. Med. 367, 2407–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vered K., Durrant C., Mott R., Iraqi F. A. (2014). Susceptibility to klebsiella pneumonaie infection in collaborative cross mice is a complex trait controlled by at least three loci acting at different time points. BMC Genomics 15, 865.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., Thompson R., Haley C. S. (1996). Confidence intervals in QTL mapping by bootstrapping. Genetics 143, 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins P.B., Lewis J.H., Kaplowitz N., Alpers D., Blais J., Smotzer D., Krasa H., Ouyang J., Torres V., Czerwiec F., et al. (2015). Clinical pattern of tolvaptan-associated liver injury in subjects with autosomal dominant polycystic kidney disease: Analysis of clinical trials database. Drug Saf. 38, 1103–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. E., Brown T. I., Roghanian A., Sallenave J. M. (2006). SLPI and elafin: one glove, many fingers. Clin. Sci. (Lond) 110, 21–35. [DOI] [PubMed] [Google Scholar]
- Woodhead J. L., Brock W. J., Roth S. E., Shoaf S. E., Brouwer K. L. R., Church R., Grammatopoulos T. N., Stiles L., Siler S. Q., Howell B. A., et al. (2016). Application of a mechanistic model to evaluate putative mechanisms of tolvaptan drug-induced liver injury and identify patient susceptibility factors. Toxicol. Sci. doi:10.1093/toxsci/kfw193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods L. C. S., Holl K. L., Oreper D., Xie Y., Tsaih S. W., Valdar W. (2012). Fine-mapping diabetes-related traits, including insulin resistance, in heterogeneous stock rats. Physiol. Genomics 44, 1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Beland F. A., Chen S., Liu F., Guo L., Fang J. L. (2015). Mechanisms of tolvaptan-induced toxicity in HepG2 cells. Biochem. Pharmacol. 95, 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Flint J., Mott R. (2005). Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics 171, 673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Ping J., Chen H., Jiao L., Zheng S., Han Z. G., Hao P., Huang J. (2010). A comparative analysis of liver transcriptome suggests divergent liver function among human, mouse and rat. Genomics 96, 281–289. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wang W., Valdar W. (2014). Bayesian Modeling of Haplotype Effects in Multiparent Populations. Genetics 198, 139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.