Abstract
The human anterior cingulate and frontoinsular cortices are distinguished by 2 unique Layer 5 neuronal morphotypes, the von Economo neurons (VENs) and fork cells, whose biological identity remains mysterious. Insights could impact research on diverse neuropsychiatric diseases to which these cells have been linked. Here, we leveraged the Allen Brain Atlas to evaluate mRNA expression of 176 neurotransmitter-related genes and identified vesicular monoamine transporter 2 (VMAT2), gamma-aminobutyric acid (GABA) receptor subunit θ (GABRQ), and adrenoreceptor α-1A (ADRA1A) expression in human VENs, fork cells, and a minority of neighboring Layer 5 neurons. We confirmed these results using immunohistochemistry or in situ hybridization. VMAT2 and GABRQ expression was absent in mouse cerebral cortex. Although VMAT2 is known to package monoamines into synaptic vesicles, in VENs and fork cells its expression occurs in the absence of monoamine-synthesizing enzymes or reuptake transporters. Thus, VENs and fork cells may possess a novel, uncharacterized mode of cortical monoaminergic function that distinguishes them from most other mammalian Layer 5 neurons.
Keywords: ADRA1A, fork cells, GABRQ, VMAT2, von Economo neurons
Introduction
The anterior cingulate (ACC) and frontoinsular (FI) cortices represent key hubs within a large-scale network involved in social-emotional and autonomic functions (Seeley et al. 2007; Jones et al. 2010; Craig 2011). In humans, these regions house an atypical population of Layer 5b projection neurons, the von Economo neurons (VENs), distinguished from neighboring pyramidal neurons by their large spindle-shaped perikarya and thick basal and apical dendrites (von Economo 1926). In FI, VENs are often positioned alongside fork cells, a companion population also characterized by a single large basal dendrite and, in contrast to VENs, a divided apical dendrite (Ngowyang 1936; Seeley et al. 2012). VENs have been identified in humans, great apes, and macaques (Nimchinsky et al. 1999; Allman et al. 2010; Evrard et al. 2012) as well as in distantly related mammals such as cetaceans (Butti et al. 2009), perissodactyls (Raghanti et al. 2014), and elephants (Hakeem et al. 2009). VEN and fork cell functions remain unknown, but the topographic restriction of human VENs and fork cells to ACC and FI suggests involvement in sophisticated social-emotional-autonomic functions (Seeley 2008). Supporting this hypothesis, VENs represent an early target in the behavioral variant of frontotemporal dementia (bvFTD), a neurodegenerative disorder that slowly erodes social behavior and personal conduct (Seeley et al. 2006; Kim et al. 2012; Santillo and Englund 2014). VEN abnormalities have also been linked to other conditions characterized by social-emotional deficits, such as autism (Santos et al. 2011), suicidal psychosis (Brune et al. 2011), agenesis of the corpus callosum (Kaufman et al. 2008), and schizophrenia (Brune et al. 2010). For these reasons, understanding basic VEN and fork cell biology is an important priority for understanding normal and disordered human behavior.
Efforts to determine the VEN molecular profile have thus far consisted of candidate-based studies, mainly using immunohistochemistry. VENs are rich in nonphosphorylated neurofilament (Nimchinsky et al. 1995; Evrard et al. 2012), and some data suggest that VENs are immunopositive for a range of proteins associated with psychiatric disorders, such as DISC1 (Allman et al. 2010; Evrard et al. 2012), serotonin receptor 2B (HTR2B) (Allman et al. 2005; Evrard et al. 2012), and dopamine receptor D3 (DRD3) (Allman et al. 2005; Evrard et al. 2012). VENs also express FEZF2 and CTIP2 (Cobos and Seeley 2013), transcription factors that regulate the fate and differentiation of subcerebral projection neurons (Chen et al. 2008). VEN connections in humans remain unknown and difficult to study, but preliminary evidence suggests that macaque FI VEN projections include ipsilateral midinsula and contralateral FI (Evrard et al. 2012), implying that VENs may have diverse cortical and subcerebral targets (Cobos and Seeley 2013).
To further explore the biochemical phenotype of VENs and fork cells, we adopted a hypothesis-independent approach by leveraging the Allen Brain Atlas (ABA), an online public data resource that integrates extensive gene expression and neuroanatomical data (Hawrylycz et al. 2012). To date, the ABA has provided in situ hybridization (ISH) expression data from 4 neurologically normal subjects of 176 neurotransmitter-related genes across 17–24 brain regions, including ACC and FI. Review of these data uncovered 3 novel VEN and fork cell mRNA markers: vesicular monoamine transporter 2 (VMAT2), gamma-aminobutyric acid (GABA) receptor subunit θ (GABRQ), and adrenoreceptor α-1A (ADRA1A). VMAT2 in VENs and fork cells is not accompanied by monoamine and GABA-synthesizing enzymes or monoamine reuptake transporters, suggesting a new, previously undescribed monoaminergic phenotype among these select cortical projection neurons.
Materials and Methods
ABA ISH Data Mining Approach
Human Subject, Block, and Probe Selection
The ABA is a standardized atlas of gene expression visualized by ISH (© 2015 Allen Institute for Brain Science. Allen Human Brain Atlas [Internet]. Available from: http://human.brain-map.org/) (Hawrylycz et al. 2012). The ABA human neurotransmitter study utilized frozen tissue blocks from 4 control subjects, aged 28–57 years, with 17–24 anatomical structures from one cerebral hemisphere available per donor. Staining for most genes is visualized in multiple sections throughout each tissue block, and every approximately 20th section is Nissl stained with thionin to provide anatomical context for nearby ISH-treated sections. From this ABA study, regions marked “cingulate cortex” (ACC) and “claustrum/insula” (FI) were selected for analysis. The ability to detect VENs and fork cells, which depends on regional position and plane of section, was determined in the Nissl-stained sections. Two blocks per region were selected as those with the highest VEN and fork cell densities: 112 913 273 (donor H0351.1010; sections 1–276) and 121 133 933 (donor H0351.1009; sections 1–848) for ACC; and 113 817 898 (donor H0351.1016; sections 448–1048) and 1 25 512 939 (donor H0351.1012; sections 448–1048) for FI (Figure S1).
Section Rating and Categorization
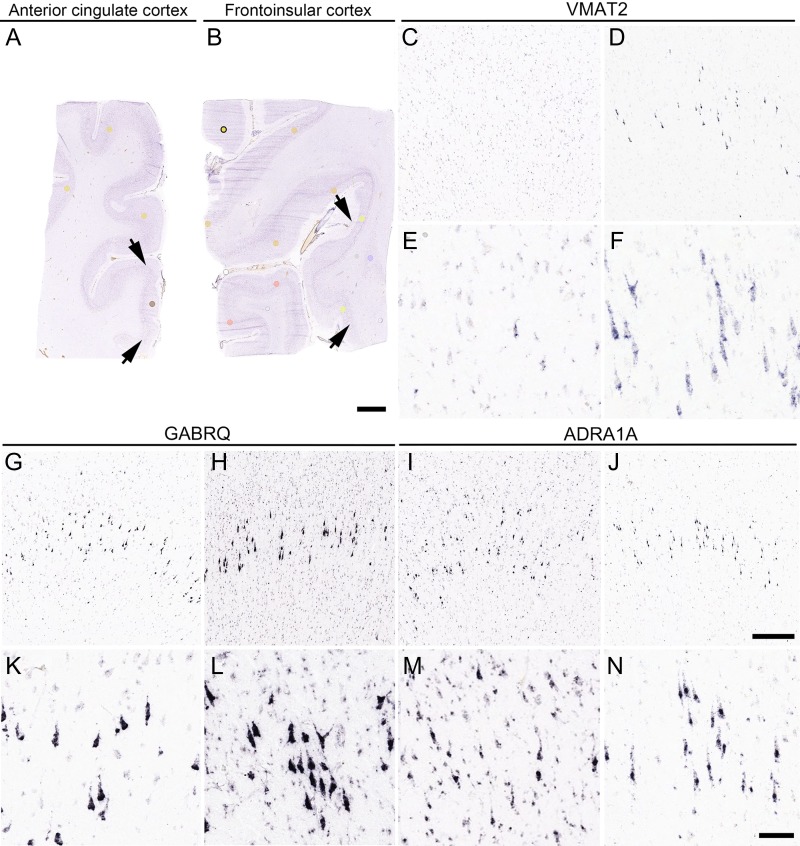
VEN-containing regions in ACC and FI were determined based on the Nissl-stained reference sections. ISH patterns within VEN-containing regions were assessed within each block (details in Figure S1). In brief, rating was initially based on general staining appearance: 1) strong cytoplasmic staining; 2) weak cytoplasmic or nuclear-only staining (precluding neuronal morphotype determination), or 3) absent staining. Positive staining was then assessed by location: 1) Layer 5 predominant; 2) Layer 5 and other layers; or 3) non-Layer 5. If the ISH pattern was strong, cytoplasmic, and present in Layer 5, then the presence of VENs and/or fork cells was evaluated (Figure S2) according to previous guidelines (Kim et al. 2012). Three markers showed Layer 5-predominant staining that prominently included VENs and/or fork cells: VMAT2, GABRQ, and ADRA1A.
VMAT2, GABRQ, and ADRA1A mRNA Expression Across Regions and Species
The presence of VMAT2, GABRQ, and ADRA1A expression was assessed in other publicly available ABA databases. In the BrainSpan Atlas of the Developing Human Brain (© 2015 Allen Institute for Brain Science. BrainSpan Atlas of the Developing Human Brain [Internet], available from: http://brainspan.org/) (Miller et al. 2014), ADRA1A ISH is also available for the insular cortex/putamen in 3-, 5-, and 8-month-old subjects and in 8-, 13-, and 17-year-old subjects. All 3 markers are available in multiple coronally or sagittally sectioned brains in the ABA's extensive mouse ISH data set (© 2015 Allen Institute for Brain Science, Allen Mouse Brain Atlas [Internet]. Available from: http://mouse.brain-map.org/) (Lein et al. 2007). Finally, an ISH atlas for rhesus macaques (© 2015 Allen Institute for Brain Science. NIH Blueprint Non-Human Primate (NHP) Atlas [Internet], available from: http://www.blueprintnhpatlas.org/) currently includes ADRA1A data in a “ventral striatum” block that also contains parts of the subgenual ACC and FI.
The specificity of the ABA ISH probes was assessed using a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The ABA ISH sequences for VMAT2, GABRQ, and ADRA1A show no significant overlap with other human transcripts.
Validation and Follow-Up Studies
Case Materials
Archival tissue was obtained from the University of California, San Francisco Neurodegenerative Disease Brain Bank (UCSF NDBB). Four control subjects without significant cognitive impairment and having only age-related incidental neuropathological findings were selected for validation of VMAT2 and GABRQ expression using immunohistochemistry. Demographic and pathological details for each donor are provided in Table S1. All brains were freshly cut into coronal slabs that were alternately fixed in 10% neutral buffered formalin or rapidly frozen. After 72 h in formalin, fixed slabs were transferred to phosphate-buffered saline with 0.02% sodium azide (PBS-Az) until dissection into standard tissue blocks. ACC and FI tissue blocks were immersed in graded sucrose solutions, frozen, and cut into 50-µm-thick sections for free-floating immunohistochemistry. Blocks containing the tuberomammilary nucleus (TMN), substantia nigra (SN), dorsal or median raphe nucleus, and locus ceruleus (LC) were similarly sectioned and used as positive controls for histamine-, dopamine-, serotonin-, and noradrenaline-related markers. To perform a more comprehensive regional survey, we performed VMAT2 and GABRQ immunohistochemistry using 8-µm-thick sections from 23 standard diagnostic paraffin-embedded fixed tissue blocks, encompassing samples from 52 distinct brain regions (see Table S3).
For ISH, we used 20-µm-thick frozen sections cut from ACC and FI blocks obtained from 2 control subjects from the NICHD Brain and Tissue Bank for Developmental Disorders, University of Maryland, Baltimore. Sections cut from the left hemi-brainstem of one control subject obtained from Oregon Health & Science University were used as positive controls for reuptake transporter ISH experiments.
Donors or their surrogates provided informed consent to undergo autopsy and brain donation. The UCSF Committee on Human Research and local institutional review boards at contributing brain banks approved all procedures used in the study.
Validation of VMAT2 and GABRQ using Immunohistochemistry
Free-floating 50-μm-thick ACC and FI sections were washed well (6 × 10 min) in PBS, and heat-induced (80 °C) antigen retrieval was performed in TRIS buffer at pH 9.0 for 2 h. After washing with PBS (3 × 10 min), sections were incubated in 0.3% H2O2 diluted in PBS-Az for 30 min to block endogenous peroxidase activity. Next, sections were washed and incubated in 10% normal serum in PBS with 0.25% Triton X (PBT) for 1 h, then in primary antibody (GABRQ 1:750, HPA002063, Sigma-Aldrich; VMAT2 1:1000, AB1598P, EMD Millipore) in PBT for 2 nights at 4 °C. After washing, sections were incubated with biotinylated secondary antibody (BA-1000, Vector Laboratories; 1:500 in PBT, 1 h), washed, then incubated with avidin–biotin–peroxidase complexes (ABC, PK-6100, Vector Laboratories, 1:500 each in PBT). After washing, immunostaining was visualized with chromogen 3,3’-diaminobenzidine (DAB); sections were counterstained with hematoxylin (SH26-500D; Fisher Scientific), dehydrated, and coverslipped. Negative controls for each primary antibody were included by omitting the primary antibody, and showed no immunoreactivity.
Paraffin sections were deparaffinized in xylene for 15 min, washed in 100% and 95% ethanol, and incubated in 3% H2O2 diluted in methanol for 30 min. Next, heat-induced (121 °C autoclave) antigen retrieval was performed in TRIS buffer, pH 9.0, for 5 min. Sections were washed and incubated in 5% nonfat dry milk diluted in PBS for 30 min followed by incubation with primary antibody in 5% milk/PBS (VMAT2: 1:500; GABRQ; 1:250) overnight at room temperature. The next day, the sections were washed in PBS (3 × 5 min) incubated for 30 min with secondary antibody diluted 1:200 in PBS, washed, and then incubated 30 min with ABC diluted 1:200 in PBS. After washing, slides were developed in DAB, counterstained, and coverslipped as above.
The precise epitopes recognized by the VMAT2 and GABRQ polyclonal antibodies are unknown, but a BLAST search on the antigen sequences yielded no other significant hits. Additional validation information for each antibody is provided here.
The specificity of the VMAT2 primary antibody (AB1598P, EMD Millipore) was previously assessed on western blot and immunohistochemistry by preabsorption of the VMAT2 antibody with the peptide antigen used for antibody production ([C]SYPIGDDEESESD) (Liu et al. 1994; Nirenberg et al. 1995; Peter et al. 1995). The specificity has been confirmed by substitution of the primary antibody with nonimmune serum or by preabsorption of the primary antibody with the peptide antigen in rat (Kourtesis et al. 2015). Expression of VMAT2 is present in monoaminergic structures, such as the LC, SN, raphe nucleus, and TMN (Erickson et al. 1996). Our regional survey of VMAT expression throughout the human brain showed a subcortical and brainstem localization strongly convergent with that reported in the literature.
The antibody against GABRQ (HPA002063, Sigma-Aldrich) was previously validated as part of the Human Protein Atlas. Antibody specificity was shown by antigen microarray based on the interaction with 384 different antigens including its own target. GABRQ showed only a single peak corresponding to interaction only with its own antigen. Upon western blotting, the antiserum for GABRQ stains a single band of approximately 70 kD molecular weight on western blot (manufacturer's technical information), and has a predicted molecular weight of 72 kD. To our knowledge, no specificity was tested using antisera. Our regional survey shows that the expression of GABRQ is observed in structures that have been shown to express GABRQ in rodents, such as the LC, raphe nucleus, and periaquiductal gray (Sinkkonen et al. 2000; Moragues et al. 2002; Pape et al. 2009; Masocha 2015).
Each of our experiments included a brainstem section as a positive control. For VMAT2 we used the SN, whereas for GABRQ we used the dorsal raphe nucleus.
Quantification of VMAT2- and GABRQ-Expressing Neurons
The photomicrographs available on the ABA do not lend themselves to rigorous quantification of the proportion of ISH-positive neurons because the ISH sections are not counterstained and the nearest Nissl-stained sections may not be sufficiently close to the ISH sections to provide a meaningful denominator. We therefore quantified VMAT2 and GABRQ expression across cell types in 50-μm-thick immunostained sections that were Nissl counterstained with hematoxylin.
The proportion of VENs, fork cells, and neighboring neurons immunopositive for VMAT2 or GABRQ was assessed in virtual (i.e., digital) sections created from VMAT2- and GABRQ-immunostained sections in 4 controls. For each section, Layer 5 of the FI was traced in StereoInvestigator V11.0.3 (MicroBrightField, Inc). The tracing was exported into ZEN software (Zeiss). Montaged digital, high magnification (×63, 1.4 NA objective) image stacks (z-stack interval: 0.4 µm) of the entire FI Layer 5 were acquired using an AxioObserver Z1 microscope equipped with a motorized x, y, z stage and an ICC1 camera (Zeiss). Stitched montages were initially saved as CZI images, exported as TIFF images, and converted to virtual JPX slides for viewing in StereoInvestigator using a virtual ×40 objective. The total number of VMAT2- or GABRQ-positive and -negative VENs, fork cells, and neighboring Layer 5 neurons with pyramidal morphology was then counted using the optical fractionator probe in StereoInvestigator software to obtain the percentage of immunopositive neurons in each population (West 2002). All slides were counted by a single examiner (A.N.L.) trained to recognize VENs and fork cells following procedures described in our previous work (Kim et al. 2012).
Validation of ADRA1A Expression Using ISH
Due to a lack of specific antibodies to the ADRA1A receptor, some ABA ADRA1A findings were validated in-house using ISH (ViewRNA ISH Tissue 1-Plex Assays; Affymetrix, as per the manufacturer's instructions). Probes were specific to the sequence regions spanning nucleotides 546–1504 for ADRA1A (catalog #VA1-10215) or 63–1177 for the housekeeping gene GAPDH (#VA1-10119). Briefly, fresh frozen ACC and FI tissue blocks (2 donors) and brainstem positive control blocks (one donor) were cut into 20-µm-thick sections on a cryostat; sections were fixed for 16–18 h in 10% neutral buffered formalin, pretreated with 1 h of 60 °C heat and then exposed to 10 min of proteinase K (50 µg/µL) prior to hybridization with the target oligonucleotide probes. Preamplifier, amplifier, and alkaline phosphatase-labeled oligonucleotides were then hybridized sequentially at 40 °C, followed by 30 min of chromogenic precipitate development with Fast Red. Each sample was quality-controlled for RNA integrity with a ViewRNA probe specific to GAPDH RNA, and each target probe was quality-controlled for positive staining signal with brainstem sections including LC, raphe nuclei, and SN. Specific RNA staining signal was identified as red puncta. To visualize the spatial distribution and morphology of VENs and fork cells, samples were counterstained post-ISH with gallocyanin.
Follow-Up: Absence of Monoamine-Synthesizing Enzymes and Reuptake Transporters in VENs and Fork Cells
The presence of synthesizing enzymes and reuptake transporters for dopamine, noradrenaline, serotonin, and histamine in ACC and FI sections was examined using immunohistochemistry. The rate-limiting enzyme for dopamine and noradrenaline synthesis is tyrosine hydroxylase (TH; antibody PA1-4605, Thermo Scientific). Serotonin synthesis involves tryptophan hydroxylases 1 and 2 (TPH1 and TPH2); we used a PH8 antibody (MAB5278, Chemicon) to detect these enzymes in fixed tissue. Histamine is synthesized by histidine decarboxylase (HDC; antibody 03-16045, American Research Products). Amino-acid decarboxylase (AADC; antibody D0180, Sigma-Aldrich) catalyzes the conversion of multiple aromatic l-amino acids into neurotransmitters including dopamine, serotonin, and histamine. The presence of monoamine reuptake transporters was explored with antibodies to dopamine (dopamine reuptake transporter [DAT]; AB5990, Abcam), noradrenaline (noradrenaline reuptake transporter [NET]; NET17-1; MAb Technologies), and serotonin (serotonin reuptake transporter [SERT]; sc-1458, Santa Cruz Biotechnology, Inc.) reuptake transporters. Involvement of GABA was tested by immunostaining for GABA-synthesizing enzymes glutamic acid decarboxylase 1 (GAD1) and glutamic acid decarboxylase 2 (GAD2) (GAD1/2; AB1511, Millipore), and aldehyde dehydroxylase 1a1 (ALDH1A1; ab52492, Abcam). Free-floating 50-μm sections of ACC, FI, and positive control tissues (AADC, DAT, and TH: SN; NET: LC; SERT, and PH8: dorsal raphe nuclei in the upper pons at the level of the LC; HDC: TMN; ALDH1A1 and GAD1/2: cerebral cortex) were used for immunohistochemistry as described above. Briefly, heat-induced (80 °C) antigen retrieval was performed in citrate buffer pH 6.0 (TH, PH8, SERT, ALDH1A1) or TRIS buffer pH 9.0 (AADC, DAT, NET, HDC, GAD1/2). Sections were incubated in 5% nonfat dry milk (PH8, TH, SERT, GAD1/2, ALDH1A1) or 5% normal goat (HDC, DAT) or horse (AADC, NET) serum in PBT. Next, we incubated the sections with primary antibody for 2 nights at 4 °C (AADC: 1:30 000; TH: 1:500; PH8: 1:6000; HDC: 1:9000; ALDH1A1: 1:500; GAD1/2: 1:2000; DAT: 1:100; SERT: 1:10 000 and NET: 1:1500). Sections were incubated for 1 h with biotinylated secondary antibody (TH, HDC, ALDH1A1, GAD1/2 BA-1000; AADC, PH8, NET BA-2000; DAT BA-9400; SERT BA-9500; Vector Laboratories) diluted 1:500 in PBT and for 1 h with ABC (1:500 each in PBT). DAB and counterstaining procedures were as described above. Negative controls for each primary antibody were included by omitting the primary antibody and showed no immunoreactivity.
Because it is possible that DAT, NET, and SERT protein are locally synthesized in VEN and fork cell distal dendrites, a proximal somatodendritic signal was needed to rule out the possibility of distal monoamine transporter expression by VENs and fork cells. Therefore, we performed ISH as described above using probes for SERT (nucleotides 1585–2750, #VA1-17005), DAT (801–2899, #VA1-17004), and NET (801–2899, #VA-17003).
Results
VENs and Fork Cells Express VMAT2, GABRQ, and ADRA1A
At the time of this study, the ABA offered human ISH data sets for 176 genes in ACC- or FI-containing sections. Our overall strategy for review of these data is shown in Figure S1 (also see Materials and Methods). In brief, we visually examined over 2000 relevant ISH sections and rated each ISH data set to characterize the intensity, localization, and distribution of hybridization signal (Figure S2). With this approach, we identified 38 genes expressed in VENs and/or fork cells (Table 1). VGLUT1 expression in VENs and fork cells unambiguously confirmed the glutamatergic nature of these cells, as suggested in a previous study (Evrard et al. 2012). ISH for genes coding for HTR2B and DRD3 (Allman et al. 2005; Evrard et al. 2012) did not show a conclusive VEN- or fork cell-like staining pattern because the staining intensity was too weak to distinguish neuronal morphotypes. Three genes—VMAT2, GABRQ, and ADRA1A (Table 1 and Fig. 1)—were expressed predominantly in ACC and FI Layer 5 neurons and conspicuously in VENs and fork cells. A complete listing of genes and ratings in VEN- and fork cell-containing structures is provided in Table S2.
Table 1.
Overview of positive ISH findings in VENs and fork cells in the ACC and FI
| Gene symbol | Gene name | VENs and Layer 5 predominant | VENs and other layers | ||
|---|---|---|---|---|---|
| ACC | FI | ACC | FI | ||
| VMAT2 (SLC18A2) | Vesicular monoamine transporter 2 | n/c | ✓ | ||
| GABRQ | GABA-A receptor, theta | ✓ | ✓ | ||
| ADRA1A | Adrenoceptor alpha 1 A | ✓ | ✓ | ||
| ABAT | 4-Aminobutyrate aminotransferase | ✓ | ✓ | ||
| CBLN2 | Cerebellin 2 precursor | ✓ | ✓ | ||
| CHRM1 | Cholinergic receptor, muscarinic 1 | ✓* | ✓ | ||
| CHRM4 | Cholinergic receptor, muscarinic 4 | n/a | ✓ | ||
| CHRNA4 | Cholinergic receptor, nicotinic, alpha 4 (neuronal) | ✓ | n/c | ||
| CNR1 | Cannabinoid receptor 1 (brain) | n/c | ✓* | ||
| CRYM | Crystallin, mu | ✓ | ✓ | ||
| DLD | Dihydrolipoamide dehydrogenase | ✓ | ✓* | ||
| GABBR1 | Gamma-aminobutyric acid (GABA) B receptor, 1 | ✓ | ✓ | ||
| GABBR2 | Gamma-aminobutyric acid (GABA) B receptor, 2 | n/c | ✓ | ||
| GABRA5 | Gamma-aminobutyric acid (GABA) A receptor, alpha 5 | ✓ | ✓ | ||
| GABRB2 | Gamma-aminobutyric acid (GABA) A receptor, beta 2 | ✓ | ✓ | ||
| GABRB3 | Gamma-aminobutyric acid (GABA) A receptor, beta 3 | n/c | ✓ | ||
| GABRD | Gamma-aminobutyric acid (GABA) A receptor, delta | ✓ | ✓ | ||
| GLRB | Glycine receptor, beta | ✓ | ✓* | ||
| GLS | Glutaminase | ✓ | ✓* | ||
| GOT1 | Glutamic-oxaloacetic transaminase 1, soluble | ✓ | ✓* | ||
| GOT2 | Glutamic-oxaloacetic transaminase 2, mitochondrial | n/c | ✓* | ||
| GRIA1 | Glutamate receptor, ionotropic, AMPA 1 | ✓ | ✓* | ||
| GRIA2 | Glutamate receptor, ionotropic, AMPA 2 | ✓ | n/a | ||
| GRIA3 | Glutamate receptor, ionotropic, AMPA 3 | ✓ | n/a | ||
| GRIK2 | Glutamate receptor, ionotropic, kainate 2 | n/c | ✓* | ||
| GRIN1 | Glutamate receptor, ionotropic, N-methyl D-aspartate 1 | ✓ | ✓* | ||
| GRIN2A | Glutamate receptor, ionotropic, N-methyl D-aspartate 2 A | ✓ | ✓* | ||
| GRIN2B | Glutamate receptor, ionotropic, N-methyl D-aspartate 2B | n/c | ✓* | ||
| GRIN3A | Glutamate receptor, ionotropic, N-methyl-D-aspartate 3 A | n/c | ✓* | ||
| HRH3 | Histamine receptor H3 | n/c | ✓ | ||
| HTR3B | 5-hydroxytryptamine (serotonin) receptor 3B, ionotropic | ✓ | n/a | ||
| NEFH | Neurofilament, heavy polypeptide | ✓ | ✓* | ||
| NNAT | Neuronatin | ✓ | ✓ | ||
| SCG2 | Secretogranin II | ✓ | ✓* | ||
| VGLUT1 (SLC17A7) | Vesicular glutamate transporter 1 | ✓ | ✓* | ||
| SYT2 | Synaptotagmin II | ✓ | n/c | ||
| TAC1 | Tachykinin, precursor 1 | n/c | ✓* | ||
| VGF | VGF nerve growth factor inducible | ✓ | n/c | ||
Note: ✓ = present in both blocks of one region, ✓* = present in one block of one region, but not available or indeterminate in the other block. n/a = not available: no data available in the 2 blocks available for each region. n/c = not consistent: blocks showed inconsistent staining patterns between regions or blocks.
Figure 1.
Human ACC and FI Layer 5 ISH patterns from the ABA. VMAT2, GABRQ, and ADRA1A show Layer 5-predominant expression patterns, including VENs, fork cells, and a subset of neighboring neurons with a pyramidal morphology. Nissl-stained sections capturing the (A) ACC (block ID 112913273) and (B) FI (block ID 113817898) provided anatomical reference. Arrows indicate outer boundaries of each subregion containing VENs and fork cells. VMAT2 ISH in ACC (C, E; weak compared with FI (D, F)). GABRQ ISH in ACC (G, K) and FI (H, L); ADRA1A ISH in ACC (I, M) and FI (J, N). Scale bars represent 50 mm (A, B), 400 μm (C, D, G, H, I, and J), and 100 μm (E, F, K, L, M, and N); (C–N) highlight L5, apical surface up. Image credits: Allen Institute for Brain Science.
VMAT2 and GABRQ Expression Is Present in Other Human Cortical Areas But Absent in Mouse Cerebral Cortex
Having identified VMAT2, GABRQ, and ADRA1A mRNA expression in VENs and fork cells, we sought to determine the human brain-wide distribution of neurons expressing these markers. ISH studies of VMAT2, GABRQ, and ADRA1A are available in the ABA for regions in the frontal, temporal, occipital, and parieto-occipital lobes in 4 adult human controls. In addition, GABRQ is available for adult human hippocampus and striatum. In the human developmental atlas, ADRA1A is available for the FI and putamen. Reviewing these studies, we observed scarce VMAT2 and GABRQ expression in frontal and temporal cortices, with 0–10 positive Layer 5 neurons per section. While no GABRQ expression was observed in primary visual cortex, variable GABRQ expression was noted across subjects in the hippocampal granule cell layer and CA3, as well as parieto-occipital cortex. ADRA1A expression was present in all cortical layers in blocks from parieto-occipital, temporal, and visual cortices, with the most prominent expression in Layers 2 and 5. In the ABA human developmental atlas, Layer 5 cortical neurons were ADRA1A positive in FI of the 8-, 13-, and 17-year-old donors, but it was not possible to determine specific neuronal morphologies. No ADRA1A staining was observed in the 3-, 5-, and 8-month-old donors. Overall, VMAT2 appeared to be more selectively expressed in FI and ACC compared with GABRQ, considering the variable GABRQ expression observed in parieto-occipital cortex. ADRA1A is expressed in Layer 2 and 5 neurons throughout the cerebral cortex.
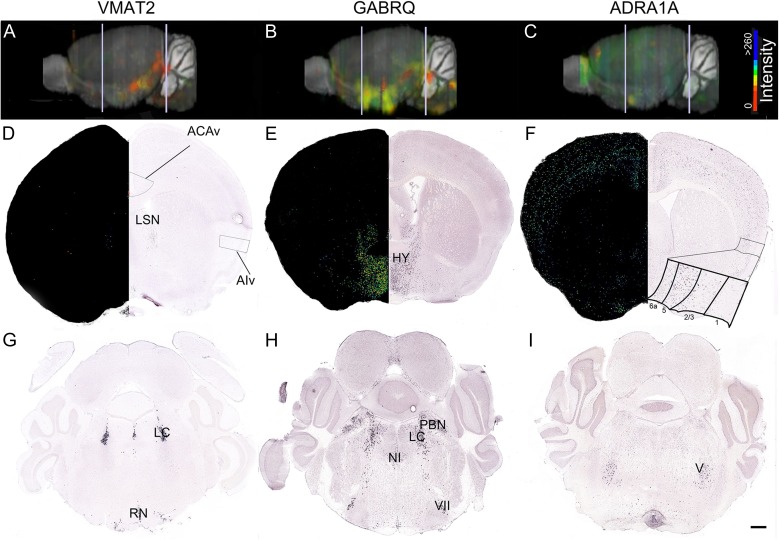
The mouse brain contains cortical regions that are in many ways homologous to human ACC and FI. The nearest mouse homologs for human ACC are infralimbic (IL), prelimbic (PL), and anterior cingulate area (ACA) cortices (Vogt and Paxinos 2014). For human FI, ventral agranular insula (AIv) and dorsal agranular insula (AId) of the mouse pertain (Nieuwenhuys 2012). Despite these homologies, mice lack neuronal morphotypes resembling VENs or fork cells (Nimchinsky et al. 1999). We questioned whether mice might nevertheless harbor a population of Layer 5 neurons expressing VMAT2, GABRQ, and/or ADRA1A. Relevant data from the ABA mouse ISH database are shown in Figure 2. VMAT2 expression was observed in areas including the lateral septal nucleus and monoaminergic nuclei. GABRQ expression was detected in the bed nucleus of the stria terminalis, hypothalamus, amygdala, and several brainstem structures. In contrast to the human brain ABA findings, no VMAT2 or GABRQ staining was observed anywhere throughout the mouse cerebral cortex, including the ACC and FI homologs (Fig. 2). As in humans, mouse ADRA1A showed a more widespread pattern, with expression in regionally distributed Layer 5 cortical neurons and in structures such as the hippocampus, hypothalamus, olfactory bulb, and several brainstem nuclei (Fig. 2).
Figure 2.
Regional distribution of VMAT2, GABRQ, and ADRA1A expression in the mouse (ABA mouse database). A lateral, 3-dimensionally rendered whole-brain view of mRNA expression is shown for VMAT2 (experiment 967) (A), GABRQ (experiment 79591569) (B), and ADRA1A (experiment 75042247) (C). Bar shows ISH staining intensity. White lines in (A–C) indicate level of the higher magnification coronal section views in (D–F) and (G–I). Expression maps and raw ISH data in ACA and agranular ventral insula are shown for VMAT2 (D) GABRQ (E), and ADRA1A (F). ISH patterns in the brainstem are shown for VMAT (G), GABRQ (H), and ADRA1A (I). ACAv, anterior cingulate area, ventral; LSN, lateral septal nucleus; HY, hypothalamus; RN, raphe nucleus; NI, nucleus incertus; PBN, parabrachial nucleus; VII, facial nucleus; V, motor nucleus of the trigeminal nerve. Scale bar represents 500 μm. Image credits: Allen Institute for Brain Science.
Recently, macaque monkeys were added to the list of species in which ACC and FI VENs and fork cells can be identified (Evrard et al. 2012). In the ABA rhesus macaque data set, Layer 5 neurons in the insular and subgenual ACC showed expression of ADRA1A, but VENs and fork cells could not be distinguished due to insufficient staining intensity. To date, no ISH has been performed for VMAT2 or GABRQ in the macaque.
VMAT2, GABRQ, and ADRA1A Protein Is Expressed in VENs, Fork Cells and a Subset of Neighboring Layer 5 Neurons with Pyramidal Morphology
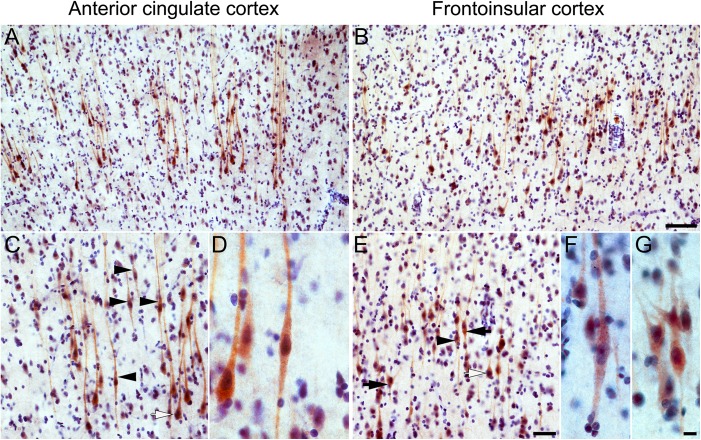
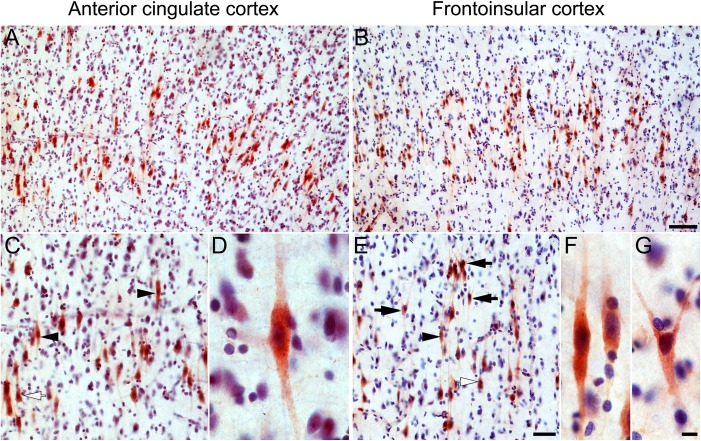
To validate the human ABA ISH findings and estimate the proportion of VENs and fork cells expressing the protein product of the mRNA markers, we performed immunohistochemistry for VMAT2 and GABRQ in human postmortem brain. VENs and fork cells in the ACC and FI were strongly immunopositive for VMAT2 (Fig. 3, Figure S3) and GABRQ (Fig. 4, Figure S3). In addition, we observed VMAT2 and GABRQ immunopositivity in a subset of neighboring large Layer 5 neurons with a pyramidal morphology, in concordance with the human ABA ISH data. Systematic, unbiased counting in the FI revealed that, on average, 92% of VENs (range 88–95%) and 84% (range 63–96%) of fork cells expressed VMAT2 and 82% of VENs (range 78–86%) and 84% of fork cells (range 78–90%) expressed GABRQ. In contrast, 28% (range 19–35%) of Layer 5 pyramidal-shaped neurons were immunopositive for VMAT2 and 35% (range 27–44) for GABRQ.
Figure 3.
VMAT2 immunohistochemistry in human ACC and FI Layer 5. Layer 5-predominant brown VMAT2 immunoreactivity is seen in the ACC (A) and FI (B) and prominently includes VENs and fork cells, as well as a subset of neighboring neurons with a pyramidal morphology (C, E). Higher magnification views of VMAT2-positive VENs in ACC (D) and FI (F) and fork cells in FI (G). Black arrowheads indicate examples of stained VENs, black arrows indicate stained fork cells, and white arrows indicate stained pyramidal-shaped neurons. Scale bars represent 100 μm (A, B), 50 μm (C, E), or 10 μm (D, F, G); apical at top.
Figure 4.
GABRQ immunohistochemistry in human ACC and FI Layer 5. Layer 5-predominant GABRQ immunoreactivity in the ACC (A) and FI (B), including VENs, fork cells, and a subset of neighboring Layer 5 neurons with a pyramidal-shaped morphology (C, E). Higher magnification views of GABRQ-positive VENs in ACC (D) and FI (F) and a fork cell in FI (G). Black arrowheads indicate stained VENs, black arrows indicate stained fork cells, and white arrows indicate stained pyramidal-shaped neurons. Scale bars represent 100 μm (A, B), 50 μm (C, E), 10 μm (D, F, G); apical at top.
To survey VMAT2 and GABRQ expression throughout the human brain, we studied a wide range of brain regions, including cortical, subcortical, limbic, diencephalic, brainstem, and spinal cord regions. This approach revealed only scattered VMAT2- and GABRQ-positive Layer 5 neurons in other cortical regions, including the frontal pole, anterior orbital gyrus, middle and inferior frontal gyri, inferior temporal gyrus, and entorhinal cortex. Among these regions, anterior orbital gyrus showed the most numerous VMAT2-immunopositive Layer 5 neurons, but the number was roughly 5-fold fewer than seen in the VEN-containing regions (Figure S4). For VMAT2, abundant immunopositive neurons were further observed in monoaminergic structures, whereas GABRQ showed a wider range of immunopositive brainstem structures, including monoaminergic nuclei. In contrast to the human ABA ISH findings, IHC did not identify pan-laminar parieto-occipital cortex GABRQ protein expression in any of our 4 donors, suggesting that the ABA findings might have resulted from high background staining in some parieto-occipital cortex blocks. Table S3 provides a complete listing of VMAT2- and GABRQ-stained regions and results.
To validate the expression of ADRA1A in VENS and fork cells, we used ISH (see Materials and Methods) because no specific antibodies were available (Jensen et al. 2009; Bohmer et al. 2014). Our ISH results confirmed ADRA1A mRNA expression in VENs and fork cells (Fig. 5) and in other neurons, including those occupying cortical Layers 1 and 2 of the ACC and FI.
Figure 5.
ADRA1A mRNA expression in ACC and FI confirmed by ISH. Red punctate ADRA1A ISH in VENs, fork cells, and some neighboring Layer 5 neurons with a pyramidal morphology (A, C). Higher magnification views of ADRA1A-expressing VENs in ACC (B) and FI (D) and a fork cell in FI (E). Purple gallocyanin counterstain. Arrowheads indicate VENs. Scale bars represent 50 μm (A, C) or 10 μm (B, D, E).
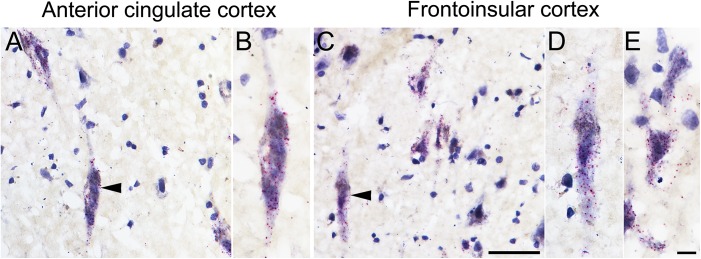
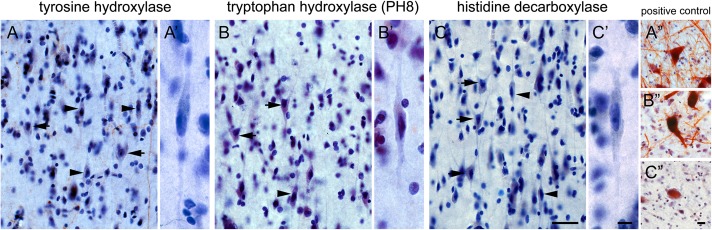
VMAT2-Expressing VENs and Fork Cells Do Not Utilize a Classical Monoaminergic Output
VMAT2 is a transporter protein that packages monoamines into synaptic vesicles (Schafer et al. 2013). Therefore, observing VMAT2 expression in human VENs and fork cells led us to the unconventional hypothesis that these cells utilize, in addition to glutamate, a classical monoamine output transmitter. To explore this possibility, we evaluated the synthesizing enzymes and reuptake transporters for the monoamines dopamine, serotonin, noradrenaline, and histamine, to the extent possible, in ACC and FI using immunohistochemistry and ISH. TH is the rate-limiting enzyme involved in dopamine and noradrenaline synthesis. In the human ABA, ACC and FI TH showed a “weak or nuclear” staining pattern, predominantly in Layer 5/6, without evidence of VEN or fork cell expression. Previous immunohistochemical studies of multiple regions, including ACC, have shown that TH is expressed in interneurons and a small minority of cortical pyramidal-like neurons, chiefly in Layers 5 and 6 (Trottier et al. 1989; Ikemoto et al. 1999). Here, we replicated these findings in the ACC and extended them to FI, but found no TH in VENs or fork cells in either region (Fig. 6), consistent with the ABA ISH data. To date, no human data from the literature or ABA ISH are available concerning ACC or FI expression of the serotonin-synthesizing enzymes TPH1 and TPH2 or the histamine-synthesizing enzyme, HDC. Using immunohistochemistry, we observed no expression of TPH1, TPH2, or HDC in VENs, fork cells, or other cells in the ACC and FI (Fig. 6). Moreover, AADC, which catalyzes the final step in dopamine, serotonin, and noradrenaline synthesis, was not detectable in VENs and fork cells. We did identify small populations of TH- and AADC-expressing Layer 5 and 6 pyramidal-shaped neurons in ACC and FI, corroborating a previous report on AADC-positive pyramidal neurons in the ACC (Figure S5) (Ikemoto et al. 1999).
Figure 6.
Absence of monoamine-synthesizing enzymes in FI VENs and fork cells. Immunohistochemistry for the synthesizing enzymes for norepinephrine and dopamine (A; TH), serotonin (B; TPH, PH8), or histamine (C; HDC) in FI. Immunohistochemistry showed a pattern consistent with arriving TH axons, but no immunostaining was observed in VEN, fork cell, or neighboring pyramidal-shaped neuron somata. Higher magnification views are shown for VENs stained for TH (A’), PH8 (B’), and HDC (C’). Right panel shows positive controls for TH (SN, A”), PH8 (raphe nucleus, B”), and HDC (TMN, C”). Arrowheads indicate VENs; arrows indicate fork cells. Scale bars represent 50 μm (A, B, C) and 10 μm (A’, A”, B’, B”, C’, C”)
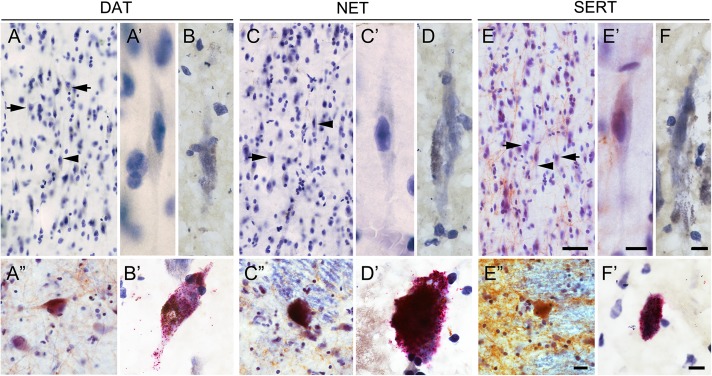
Having ruled out synthesis of the major monoamines in VENs and fork cells, we next questioned whether these neurons might internalize, via reuptake transporters, monoamines arriving from brainstem or hypothalamic projections (Lebrand et al. 1998). Using immunohistochemistry, we assessed the presence of the DAT, NET, and SERT in adult human ACC and FI. Mechanisms of neuronal histamine reuptake remain unknown. It has been postulated that the plasma membrane monoamine transporter (PMAT) and the extracellular monoamine transporter (EMT) are transporters for histamine in human astrocytes (Grundemann et al. 1999; Yoshikawa et al. 2013), but the relevance of PMAT and EMT in cortical neurons remains to be explored. As expected, arriving monoaminergic axons showed immunoreactivity for DAT, NET, and SERT, but these proteins were not detected in VEN and fork cell perikarya (Fig. 7). Immunohistochemistry would fail to detect these proteins in VENs and fork cells, however, if the mRNA is locally translated in distal dendrites. Accordingly, we performed ISH for DAT, NET, and SERT in the ACC and FI. Using this method, we found no DAT, NET, or SERT mRNA expression in VENs and fork cells (Fig. 7).
Figure 7.
Absence of monoamine reuptake transporters in VENs and fork cells. Immunohistochemistry (A, C, E) and ISH (B, D, F) for reuptake transporters in ACC and FI. Reuptake transporters for dopamine (A, B; DAT), norepinephrine (C, D; NET), and serotonin (E, F; SERT) were absent in VENs and fork cells. The FI is strongly innervated by arriving SERT axons (E). Lower panel shows the positive controls for DAT IHC (A”) and ISH (B’) in SN, NET IHC (C”) and ISH (D’) in LC, and SERT IHC (E”) and ISH (F’) in raphe nucleus. Arrowheads indicate VENs and arrows indicate fork cells. Scale bars represent 50 μm (A, C, E), 10 μm (A’, B, C’, D, E’, F, A”, C”, E”), and 20 μm (B’, D’, F’).
Another possible substrate for VMAT2 is GABA. In dopaminergic neurons, VMAT2 is known to use both dopamine and GABA as a substrate, even in the absence of the vesicular GABA transporter (Tritsch et al. 2012). Using the ABA, we observed neither evidence for expression of the GABA-synthesizing enzymes glutamate decarboxylase (GAD), GAD1 and GAD2, nor evidence for the main neuronal GABA reuptake transporter, GAT-1, in VENs and fork cells (Figure S6). In addition, immunohistochemistry for GAD1 and GAD2 revealed only punctate, likely synaptic, staining in ACC and FI, with no staining of VEN or fork cell perikarya (Figure S7). Recently, it was shown that ALDH1A1 is also responsible for GABA synthesis in VMAT2-positive nigral dopaminergic neurons (Kim et al. 2015). Here, we found that ALDH1A1 mRNA is expressed in Layer 5 in ACC and FI. Using immunohistochemistry, we detected ALDH1A1 protein expression in Layer 5 pyramidal-shaped neurons and bipolar interneurons. No immunopositive VENs of fork cells were found (Figure S8). Taken together, these findings suggest that VMAT2 expression in most VENs and fork cells and a minority of neighboring Layer 5 pyramidal-shaped neurons relates to a novel and yet to be characterized mode of monoaminergic function or some other cellular process that remains to be identified.
Discussion
The VENs and fork cells represent a clinically relevant yet mysterious neuronal class embedded in the heart of the social brain. We utilized the ABA, an online public database, to discover a novel molecular identity of VENs and fork cells in humans. Using a data-driven approach, we derived a neurochemical profile of VENs and fork cells that confirmed the glutamatergic identity of these neurons. Remarkably, within this profile we discovered VMAT2, GABRQ, and ADRA1A as a previously unrecognized set of human Layer 5-predominant markers expressed in VENs, fork cells, and a subpopulation of neighboring Layer 5 neurons. Among the 3 markers identified here, VMAT2 expression was the most selective for human ACC and FI and labeled nearly all VENs and fork cells and a subpopulation of neurons with a pyramidal morphology but was not observed in any neuron within homologous regions of the mouse cerebral cortex. These findings suggest that VENs, fork cells, and the neighboring pyramidal-shaped neurons may share a specialized and as yet unknown function. In addition, these findings suggest that not only the VENs and fork cells but also Layer 5 more generally has undergone evolutionary changes since the last common ancestor shared by mice and humans. VMAT2 and GABRQ may also be expressed in other highly social, large-brained mammals in which VENs and fork cells are found, but additional work is needed to detail the comparative neuroanatomy of the anterior cingulate and anterior insula in light of our findings.
An Uncharacterized Mode of Monoaminergic Function in VENs and Fork Cells
Classical VMAT2 expression takes place in monoaminergic neurons of brainstem and hypothalamic projection nuclei (Erickson et al. 1996). Within these neurons, VMAT2 transports monoamines across the synaptic vesicular membrane by exchanging 2 protons for a monoamine, facilitating neurotransmission and limiting the toxicity of cytosolic monoamines via sequestration (Parsons 2000; Schafer et al. 2013). VMAT2 expression in these nuclei is accompanied by expression of monoamine-synthesizing enzymes and high affinity transporter proteins DAT, NET, and SERT, which reclaim serotonin, noradrenaline, and dopamine from the synaptic cleft after vesicle fusion with the presynaptic membrane.
Nearly all cortical Layer 5 projection neurons, including VENs and fork cells, use glutamate as their major excitatory output neurotransmitter (Evrard et al. 2012). Reports describing monoamine release by cortical neurons are limited (Lebrand et al. 1998). Corelease of monoamines and glutamate by VMAT2-expressing neurons has been described in monoaminergic neurons of the brainstem and hypothalamus (Hnasko et al. 2010; Hnasko and Edwards 2012), but not in cortical neurons. For these reasons, we first questioned whether VMAT2 expression in VENs and fork cells indicated an unprecedented cortical monoaminergic neuronal phenotype. We found no evidence, however, that VENs or fork cells express monoamine-synthesizing enzymes (AADC, TH, TPH1, TPH2, or HDC), suggesting that VMAT2 might serve a different, nonclassical function in these neurons.
Nonclassical VMAT2 function has been implicated in 2 neuronal contexts in which monoamines may be used as the VMAT2 substrate. First, neuron populations in several rodent brain regions express VMAT2 during a brief period of development. In some of these neurons, VMAT2 and SERT are transiently co-expressed in the absence of synthesizing enzymes for serotonin or other monoamines (Lebrand et al. 1998). This observation suggests that such VMAT2/SERT neurons take up serotonin from the extracellular space. The absence of monoamine transporter proteins in VENs and fork cells argues against this possibility. In the second nonclassical context, monoaminergic neurons constitutively express one of the monoamine-synthesizing enzymes but no other enzymes in the monoamine synthesizing pathways. The expression of dopamine-related synthesizing enzymes, such as TH and AADC, has been thoroughly explored in rats and humans (Ikemoto et al. 1999; Balan et al. 2000; Ugrumov et al. 2004; Ugrumov 2009, 2013). Several neuronal populations in human ACC express TH but lack AADC, or express AADC but lack TH (Ikemoto et al. 1999). These “mono-enzymatic neurons” are thought to synthesize dopamine in cooperation: TH-expressing neurons produce l-dopa and may transfer it using an unknown mechanism to mono-enzymatic neurons that lack TH but express AADC (Ugrumov et al. 2014). Notably, these neuron populations have not been evaluated for VMAT2 expression in the human brain. We found expression of AADC or TH in ACC and FI Layers 3, 5, and 6 in scattered fusiform and pyramidal-shaped neurons, consistent with the literature (Ikemoto et al. 1999). In contrast, VENs and fork cells expressed neither AADC nor TH, ruling out this second nonclassical form of VMAT2 expression.
A noncanonical role for VMAT2 itself has been described in dopaminergic neurons, where VMAT2 transports dopamine, as well as GABA, into vesicles (Tritsch et al. 2012). These dopaminergic neurons express TH and the GABA-synthesizing enzyme GAD, suggesting that both substrates are synthesized within the cell. Another GABA-synthesizing pathway has been described in which ALDH1A1 mediates GABA synthesis in nigral dopaminergic neurons of the mouse (Kim et al. 2015). We found no expression of GAD or ALDH1A1 in VENs and fork cells. The absence of monoamine- and GABA-synthesizing enzymes and reuptake transporters suggests a novel role for VMAT2 in VENs, fork cells, and some nearby pyramidal-shaped neurons. One possibility is that VMAT2 transports another as yet unidentified substrate into vesicles. Another possibility is that histamine is taken up by VENs and fork cells via mechanisms that remain unknown. Finally, VMAT2 could play a role unrelated to neurotransmission, such as sequestration of some toxic metabolite or other substrate.
Presence of GABRQ and ADRA1A Receptors Suggests a Specialized Role for VENs and Fork Cells in Monoaminergic Neurotransmission and Autonomic Functioning
Functional GABA-A receptors represent a heteropentameric assembly (Nayeem et al. 1994), most often consisting of 2 α, 2 β, and 1 γ subunits (McKernan and Whiting 1996). Nineteen distinct receptor subunits have been described, giving rise to an enormous diversity of potential subunit combinations. The GABA-A receptor subunit assembly state influences the receptor's functional properties and binding affinities for GABA and GABA-ergic drugs (Olsen and Sieghart 2009). Using the ABA, we observed VEN and fork cell mRNA expression of GABA-A receptor subunits α-5, β-2, β-3, δ, and, most specifically, θ (encoded by GABRQ). Other GABA-A subunits showed staining patterns inadequate to identify VENs and fork cells. For example, γ-1 and ε subunits showed Layer 5-predominant nuclear and weak cytoplasmic mRNA expression in ACC and FI (Table S2), but VENs and fork cells, though possibly included among cells expressing these markers, could not be identified by morphology. The θ subunit is expressed only in mammals. In rat LC neurons, θ is co-expressed with α-2, α-3, β-1, β-3, and ε subunits, but not with others (Sinkkonen et al. 2000; Moragues et al. 2002), but the precise co-assembly subunit partners of θ in rats and humans remain unknown. Additional work is needed to determine which subunits combine with θ to form functional GABA-A receptors in VENs and fork cells and how these assemblies impact phasic and tonic inhibitory neurotransmission onto these neurons.
Regarding topography, we found GABRQ expression in selected human brainstem and subcortical/limbic regions, predominantly those linked to monoaminergic neurotransmission (raphe nuclei and LC) and autonomic functioning (hypothalamus, parabrachial nucleus, and others, Table S3) (Bonnert et al. 1999), consistent with GABRQ findings from the mouse ABA (Fig. 2) and GABRQ protein and mRNA expression in previous studies of rodents and monkeys (Bonnert et al. 1999; Moragues et al. 2002; Pape et al. 2009). Focal GABRQ binding has also been observed with autoradiography in monkey ACC and piriform cortex, in a region near or including the agranular anterior insula (Bonnert et al. 1999), but no cortical expression has been observed across methodologically diverse rodent studies (Sinkkonen et al. 2000; Moragues et al. 2002; Pape et al. 2009; Masocha 2015). In the human brain, we found prominent GABRQ expression in ACC and FI Layer 5 VENs, fork cells, and some pyramidal-shaped neurons as well as scattered Layer 5 pyramidal-shaped neurons in selected frontal and temporal cortices. The divergent DNA sequences of rodent and human GABRQ homologs suggest that the θ subunit has evolved more rapidly than other GABA-A receptor subunits (Sinkkonen et al. 2000; Martyniuk et al. 2007). Therefore, as for VMAT2, GABRQ expression in human ACC and FI Layer 5 neurons may represent an evolutionary specialization within large-brained mammals, the significance of which should be further explored.
The expression of ADRA1A in VENs and fork cells indicates that these neurons receive noradrenergic input. The ascending adrenergic system responds to salient internal and external stimuli by increasing the gain within forebrain systems for attention, alerting, and other cognitive functions (Sirvio and MacDonald 1999). Adrenergic receptors are G protein-coupled receptors, including α-1, α-2, and β receptor subtypes. α-1 Receptors are further divided into α-1a (ADRA1A), α-1b (ADRA1B), and α-1d (ADRA1D) subtypes. Adrenergic α-1 and α-2 receptor mRNAs other than ADRA1A were included in the ABA human ISH study, but due to weak staining patterns these materials could not be used to distinguish cellular morphotypes. In transgenic mice expressing a constitutively active mutant α-1a receptor, increases in α-1a stimulation are linked to improved learning and memory, decreased anxiety and depression, and increased lifespan (Doze et al. 2011; Collette et al. 2014). ADRA1A knockout mice, in contrast, display poor learning and memory (Doze et al. 2011). In the rat brain, ADRA1A is involved in the mechanism of action of antidepressant drugs (Nalepa et al. 2002; Kreiner et al. 2011). Two selective α-1a receptor antagonists are known, phentolamine and WB4101 (Zhao et al. 1996), and could be used to manipulate receptor function in model systems.
In rodents, ADRA1A is expressed across a wide range of brain regions, including hippocampus, hypothalamus, and brainstem structures such as the nucleus ambiguus, parabrachial nucleus, and reticular formation (Day et al. 1997; Papay et al. 2006). In rats, the strongest telencephalic ADRA1A mRNA expression was observed in piriform cortex using ISH (Domyancic and Morilak 1997). The human and mouse ABA data reveal a more widespread ADRA1A mRNA expression pattern than VMAT2 and GABRQ, being present throughout the cortex, especially in Layers 2 and 5, and in brainstem structures including the nucleus ambiguus, parabrachial nucleus, and the reticular formation. Collectively, many regions characterized by ADRA1A expression can be considered part of a central homeostatic-autonomic-alertness core. In humans, the ACC and FI are important hubs within a large-scale network that processes homeostatically salient information (Seeley et al. 2007; Touroutoglou et al. 2012; Zhou and Seeley 2014). Engagement of these hubs increases with noradrenergic (Hermans et al. 2011) and autonomic nervous system activation (Critchley and Harrison 2013), and focal vascular or degenerative lesions to ACC and FI are associated with social, emotional, and autonomic deficits (Bush et al. 2000; Zhou et al. 2010; Cho et al. 2012). Further studies are needed to determine how ADRA1A expression relates to VENs and fork cell functioning within the network hubs where these cells reside.
Future Directions
Our findings provide new avenues for understanding the phenomenology and pathogenesis of prevalent neuropsychiatric illnesses, such as bvFTD, autism, and schizophrenia, in which VENs and fork cells have been implicated (Seeley et al. 2006, 2008; Brune et al. 2010; Santos et al. 2011; Kim et al. 2012; Santillo and Englund 2014). As neurochemical mediators, the proteins encoded by VMAT2, GABRQ, and ADRA1A may represent new drug targets for symptomatic therapy. In human genetics studies, variants in VMAT2 have been associated with altered cognition in patients with psychosis (Simons et al. 2013), and GABRQ truncating mutations characterize a family with autism spectrum disorder (Piton et al. 2013). Our findings further underscore the importance of these potential therapeutic targets and provide novel opportunities to refine the biological identity of VENs and fork cells. For example, these markers provide a potential means for identifying VENs and fork cells in human-induced pluripotent stem cell-derived neuron cultures and in relevant animal model and human postmortem tissue research. Comparative anatomists can now investigate whether the human VEN- and fork cell signature is seen across the diverse VEN- and fork cell-containing mammalian species. As cell surface receptors, the GABRQ and α-1a adrenergic receptor provide potential targets not only for drugs but also for molecular imaging ligands that could be used for diagnosis and treatment monitoring. Mapping the density and distribution of these and other receptors along the unique VEN and fork cell dendritic arbors may help clarify the information processed by these specialized neurons.
Supplementary Material
Notes
We thank Dr Randy Woltjer (Oregon Health and Science University) for assistance and Drs. Robert Edwards, Samuel Shelton, and Michael Oldham at the University of California, San Francisco for helpful discussion. Finally, we thank the Allen Institute for Brain Sciences for providing an invaluable data resource. Conflict of Interest: None declared.
Funding
We thank the participant donors from the University of California, San Francisco Neurodegenerative Disease Brain Bank, National institute of Child Health and Human Development (NICHD) Brain and Tissue Bank for Developmental Disorders (funded by National Institutes of Health (NIH), contract no. #HHSN275200900011C and ref. no.NO1-HD-9-0011) at the University of Maryland, and Oregon Alzheimer's Disease Center (funded by NIH P30AG8017) at Oregon Health and Science University.
Supplementary Material
References
- Allman JM, Tetreault NA, Hakeem AY, Manaye KF, Semendeferi K, Erwin JM, Park S, Goubert V, Hof PR. 2010. The von Economo neurons in frontoinsular and anterior cingulate cortex in great apes and humans. Brain Struct Funct. 214:495–517. [DOI] [PubMed] [Google Scholar]
- Allman JM, Watson KK, Tetreault NA, Hakeem AY. 2005. Intuition and autism: a possible role for Von Economo neurons. Trends Cogn Sci. 9:367–373. [DOI] [PubMed] [Google Scholar]
- Balan IS, Ugrumov MV, Calas A, Mailly P, Krieger M, Thibault J. 2000. Tyrosine hydroxylase-expressing and/or aromatic L-amino acid decarboxylase-expressing neurons in the mediobasal hypothalamus of perinatal rats: differentiation and sexual dimorphism. J Comp Neurol. 425:167–176. [DOI] [PubMed] [Google Scholar]
- Bohmer T, Pfeiffer N, Gericke A. 2014. Three commercial antibodies against alpha1-adrenergic receptor subtypes lack specificity in paraffin-embedded sections of murine tissues. Naunyn Schmiedebergs Arch Pharmacol. 387:703–706. [DOI] [PubMed] [Google Scholar]
- Bonnert TP, McKernan RM, Farrar S, le Bourdelles B, Heavens RP, Smith DW, Hewson L, Rigby MR, Sirinathsinghji DJ, Brown N, et al. 1999. theta, a novel gamma-aminobutyric acid type A receptor subunit. Proc Natl Acad Sci U S A. 96:9891–9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune M, Schobel A, Karau R, Benali A, Faustmann PM, Juckel G, Petrasch-Parwez E. 2010. Von Economo neuron density in the anterior cingulate cortex is reduced in early onset schizophrenia. Acta Neuropathol. 119:771–778. [DOI] [PubMed] [Google Scholar]
- Brune M, Schobel A, Karau R, Faustmann PM, Dermietzel R, Juckel G, Petrasch-Parwez E. 2011. Neuroanatomical correlates of suicide in psychosis: the possible role of von Economo neurons. PLoS ONE. 6:e20936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush G, Luu P, Posner MI. 2000. Cognitive and emotional influences in anterior cingulate cortex. Trends Cogn Sci. 4:215–222. [DOI] [PubMed] [Google Scholar]
- Butti C, Sherwood CC, Hakeem AY, Allman JM, Hof PR. 2009. Total number and volume of Von Economo neurons in the cerebral cortex of cetaceans. J Comp Neurol. 515:243–259. [DOI] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. 2008. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci U S A. 105:11382–11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Kim SJ, Hwang SJ, Jo MK, Kim HJ, Seeley WW, Kim EJ. 2012. Social-emotional dysfunction after isolated right anterior insular infarction. J Neurol. 259:764–767. [DOI] [PubMed] [Google Scholar]
- Cobos I, Seeley WW. 2013. Human von Economo neurons express transcription factors associated with layer V subcerebral projection neurons. Cereb Cortex. 25(1):213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collette KM, Zhou XD, Amoth HM, Lyons MJ, Papay RS, Sens DA, Perez DM, Doze VA. 2014. Long-term alpha1B-adrenergic receptor activation shortens lifespan, while alpha1A-adrenergic receptor stimulation prolongs lifespan in association with decreased cancer incidence. Age. 36:9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AD. 2011. Significance of the insula for the evolution of human awareness of feelings from the body. Ann N Y Acad Sci. 1225:72–82. [DOI] [PubMed] [Google Scholar]
- Critchley HD, Harrison NA. 2013. Visceral influences on brain and behavior. Neuron. 77:624–638. [DOI] [PubMed] [Google Scholar]
- Day HE, Campeau S, Watson SJ Jr, Akil H. 1997. Distribution of alpha 1a-, alpha 1b- and alpha 1d-adrenergic receptor mRNA in the rat brain and spinal cord. J Chem Neuroanat. 13:115–139. [DOI] [PubMed] [Google Scholar]
- Domyancic AV, Morilak DA. 1997. Distribution of alpha1A adrenergic receptor mRNA in the rat brain visualized by in situ hybridization. J Comp Neurol. 386:358–378. [DOI] [PubMed] [Google Scholar]
- Doze VA, Papay RS, Goldenstein BL, Gupta MK, Collette KM, Nelson BW, Lyons MJ, Davis BA, Luger EJ, Wood SG, et al. 2011. Long-term alpha1A-adrenergic receptor stimulation improves synaptic plasticity, cognitive function, mood, and longevity. Mol Pharmacol. 80:747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JD, Schafer MK, Bonner TI, Eiden LE, Weihe E. 1996. Distinct pharmacological properties and distribution in neurons and endocrine cells of two isoforms of the human vesicular monoamine transporter. Proc Natl Acad Sci U S A. 93:5166–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard HC, Forro T, Logothetis NK. 2012. Von Economo neurons in the anterior insula of the macaque monkey. Neuron. 74:482–489. [DOI] [PubMed] [Google Scholar]
- Grundemann D, Liebich G, Kiefer N, Koster S, Schomig E. 1999. Selective substrates for non-neuronal monoamine transporters. Mol Pharmacol. 56:1–10. [DOI] [PubMed] [Google Scholar]
- Hakeem AY, Sherwood CC, Bonar CJ, Butti C, Hof PR, Allman JM. 2009. Von Economo neurons in the elephant brain. Anat Rec. 292:242–248. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 489:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans EJ, van Marle HJ, Ossewaarde L, Henckens MJ, Qin S, van Kesteren MT, Schoots VC, Cousijn H, Rijpkema M, Oostenveld R, et al. 2011. Stress-related noradrenergic activity prompts large-scale neural network reconfiguration. Science. 334:1151–1153. [DOI] [PubMed] [Google Scholar]
- Hnasko TS, Chuhma N, Zhang H, Goh GY, Sulzer D, Palmiter RD, Rayport S, Edwards RH. 2010. Vesicular glutamate transport promotes dopamine storage and glutamate corelease in vivo. Neuron. 65:643–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnasko TS, Edwards RH. 2012. Neurotransmitter corelease: mechanism and physiological role. Annu Rev Physiol. 74:225–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemoto K, Kitahama K, Nishimura A, Jouvet A, Nishi K, Arai R, Jouvet M, Nagatsu I. 1999. Tyrosine hydroxylase and aromatic L-amino acid decarboxylase do not coexist in neurons in the human anterior cingulate cortex. Neurosci Lett. 269:37–40. [DOI] [PubMed] [Google Scholar]
- Jensen BC, Swigart PM, Simpson PC. 2009. Ten commercial antibodies for alpha-1-adrenergic receptor subtypes are nonspecific. Naunyn Schmiedebergs Arch Pharmacol. 379:409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CL, Ward J, Critchley HD. 2010. The neuropsychological impact of insular cortex lesions. J Neurol Neurosurg Psychiatry. 81:611–618. [DOI] [PubMed] [Google Scholar]
- Kaufman JA, Paul LK, Manaye KF, Granstedt AE, Hof PR, Hakeem AY, Allman JM. 2008. Selective reduction of Von Economo neuron number in agenesis of the corpus callosum. Acta Neuropathol. 116:479–489. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Sidhu M, Gaus SE, Huang EJ, Hof PR, Miller BL, DeArmond SJ, Seeley WW. 2012. Selective frontoinsular von Economo neuron and fork cell loss in early behavioral variant frontotemporal dementia. Cereb Cortex. 22:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Ganesan S, Luo SX, Wu YW, Park E, Huang EJ, Chen L, Ding JB. 2015. Aldehyde dehydrogenase 1a1 mediates a GABA synthesis pathway in midbrain dopaminergic neurons. Science. 350(6256):102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtesis I, Kasparov S, Verkade P, Teschemacher AG. 2015. Ultrastructural correlates of enhanced norepinephrine and neuropeptide Y cotransmission in the spontaneously hypertensive rat brain, ASN Neuro. 7(5):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiner G, Zelek-Molik A, Kowalska M, Bielawski A, Antkiewicz-Michaluk L, Nalepa I. 2011. Effects of the noradrenergic neurotoxin DSP-4 on the expression of alpha1-adrenoceptor subtypes after antidepressant treatment. Pharmacol Rep. 63:1349–1358. [DOI] [PubMed] [Google Scholar]
- Lebrand C, Cases O, Wehrle R, Blakely RD, Edwards RH, Gaspar P. 1998. Transient developmental expression of monoamine transporters in the rodent forebrain. J Comp Neurol. 401:506–524. [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445:168–176. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schweitzer ES, Nirenberg MJ, Pickel VM, Evans CJ, Edwards RH. 1994. Preferential localization of a vesicular monoamine transporter to dense core vesicles in PC12 cells. J Cell Biol. 127(5):1419–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyniuk CJ, Aris-Brosou S, Drouin G, Cahn J, Trudeau VL. 2007. Early evolution of ionotropic GABA receptors and selective regimes acting on the mammalian-specific theta and epsilon subunits. PLoS ONE. 2:e894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masocha W. 2015. Comprehensive analysis of the GABAergic system gene expression profile in the anterior cingulate cortex of mice with Paclitaxel-induced neuropathic pain. Gene Expr. 16:145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKernan RM, Whiting PJ. 1996. Which GABAA-receptor subtypes really occur in the brain? Trends Neurosci. 19:139–143. [DOI] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. 2014. Transcriptional landscape of the prenatal human brain. Nature. 508:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moragues N, Ciofi P, Tramu G, Garret M. 2002. Localisation of GABA(A) receptor epsilon-subunit in cholinergic and aminergic neurones and evidence for co-distribution with the theta-subunit in rat brain. Neuroscience. 111:657–669. [DOI] [PubMed] [Google Scholar]
- Nalepa I, Kreiner G, Kowalska M, Sanak M, Zelek-Molik A, Vetulani J. 2002. Repeated imipramine and electroconvulsive shock increase alpha 1A-adrenoceptor mRNA level in rat prefrontal cortex. Eur J Pharmacol. 444:151–159. [DOI] [PubMed] [Google Scholar]
- Nayeem N, Green TP, Martin IL, Barnard EA. 1994. Quaternary structure of the native GABAA receptor determined by electron microscopic image analysis. J Neurochem. 62:815–818. [DOI] [PubMed] [Google Scholar]
- Ngowyang G. 1936. Neuere Befunde über die Gabelzellen. Z Zellforsch Mikrosk Anat. 25:236. [Google Scholar]
- Nieuwenhuys R. 2012. The insular cortex: a review. Prog Brain Res. 195:123–163. [DOI] [PubMed] [Google Scholar]
- Nimchinsky EA, Gilissen E, Allman JM, Perl DP, Erwin JM, Hof PR. 1999. A neuronal morphologic type unique to humans and great apes. Proc Natl Acad Sci U S A. 96:5268–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchinsky EA, Vogt BA, Morrison JH, Hof PR. 1995. Spindle neurons of the human anterior cingulate cortex. J Comp Neurol. 355:27–37. [DOI] [PubMed] [Google Scholar]
- Nirenberg MJ, Liu Y, Peter D, Edwards RH, Pickel VM. 1995. The vesicular monoamine transporter 2 is present in small synaptic vesicles and preferentially localizes to large dense core vesicles in rat solitary tract nuclei. Proc Natl Acad Sci U S A. 92(19):8773–8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen RW, Sieghart W. 2009. GABA A receptors: subtypes provide diversity of function and pharmacology. Neuropharmacology. 56:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papay R, Gaivin R, Jha A, McCune DF, McGrath JC, Rodrigo MC, Simpson PC, Doze VA, Perez DM. 2006. Localization of the mouse alpha1A-adrenergic receptor (AR) in the brain: alpha1AAR is expressed in neurons, GABAergic interneurons, and NG2 oligodendrocyte progenitors. J Comp Neurol. 497:209–222. [DOI] [PubMed] [Google Scholar]
- Pape JR, Bertrand SS, Lafon P, Odessa MF, Chaigniau M, Stiles JK, Garret M. 2009. Expression of GABA(A) receptor alpha3-, theta-, and epsilon-subunit mRNAs during rat CNS development and immunolocalization of the epsilon subunit in developing postnatal spinal cord. Neuroscience. 160:85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons SM. 2000. Transport mechanisms in acetylcholine and monoamine storage. FASEB J. 14:2423–2434. [DOI] [PubMed] [Google Scholar]
- Peter D, Liu Y, Sternini C, de Giorgio R, Brecha N, Edwards RH. 1995. Differential expression of two vesicular monoamine transporters. J Neurosci. 15(9):6179–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton A, Jouan L, Rochefort D, Dobrzeniecka S, Lachapelle K, Dion PA, Gauthier J, Rouleau GA. 2013. Analysis of the effects of rare variants on splicing identifies alterations in GABAA receptor genes in autism spectrum disorder individuals. Eur J Hum Genet. 21:749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghanti MA, Spurlock LB, Robert Treichler F, Weigel SE, Stimmelmayr R, Butti C, Hans Thewissen JG, Hof PR. 2014. An analysis of von Economo neurons in the cerebral cortex of cetaceans, artiodactyls, and perissodactyls. Brain Struct Funct. 220(4):2303–2314. [DOI] [PubMed] [Google Scholar]
- Santillo AF, Englund E. 2014. Greater loss of von Economo neurons than loss of layer II and III neurons in behavioral variant frontotemporal dementia. Am J Neurodegener Dis. 3:64–71. [PMC free article] [PubMed] [Google Scholar]
- Santos M, Uppal N, Butti C, Wicinski B, Schmeidler J, Giannakopoulos P, Heinsen H, Schmitz C, Hof PR. 2011. Von Economo neurons in autism: a stereologic study of the frontoinsular cortex in children. Brain Res. 1380:206–217. [DOI] [PubMed] [Google Scholar]
- Schafer MK, Weihe E, Eiden LE. 2013. Localization and expression of VMAT2 aross mammalian species: a translational guide for its visualization and targeting in health and disease. Adv Pharmacol. 68:319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW. 2008. Selective functional, regional, and neuronal vulnerability in frontotemporal dementia. Curr Opin Neurol. 21:701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Carlin DA, Allman JM, Macedo MN, Bush C, Miller BL, Dearmond SJ. 2006. Early frontotemporal dementia targets neurons unique to apes and humans. Ann Neurol. 60:660–667. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. 2007. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 27:2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Merkle FT, Gaus SE, Craig AD, Allman JM, Hof PR. 2012. Distinctive neurons of the anterior cingulate and frontoinsular cortex: a historical perspective. Cereb Cortex. 22:245–250. [DOI] [PubMed] [Google Scholar]
- Simons CJ, van Winkel R, Group . 2013. Intermediate phenotype analysis of patients, unaffected siblings, and healthy controls identifies VMAT2 as a candidate gene for psychotic disorder and neurocognition. Schizophr Bull. 39:848–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen ST, Hanna MC, Kirkness EF, Korpi ER. 2000. GABA(A) receptor epsilon and theta subunits display unusual structural variation between species and are enriched in the rat locus ceruleus. J Neurosci. 20:3588–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirvio J, MacDonald E. 1999. Central alpha1-adrenoceptors: their role in the modulation of attention and memory formation. Pharmacol Ther. 83:49–65. [DOI] [PubMed] [Google Scholar]
- Touroutoglou A, Hollenbeck M, Dickerson BC, Feldman Barrett L. 2012. Dissociable large-scale networks anchored in the right anterior insula subserve affective experience and attention. Neuroimage. 60:1947–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritsch NX, Ding JB, Sabatini BL. 2012. Dopaminergic neurons inhibit striatal output through non-canonical release of GABA. Nature. 490:262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier S, Geffard M, Evrard B. 1989. Co-localization of tyrosine hydroxylase and GABA immunoreactivities in human cortical neurons. Neurosci Lett. 106:76–82. [DOI] [PubMed] [Google Scholar]
- Ugrumov M, Taxi J, Pronina T, Kurina A, Sorokin A, Sapronova A, Calas A. 2014. Neurons expressing individual enzymes of dopamine synthesis in the mediobasal hypothalamus of adult rats: functional significance and topographic interrelations. Neuroscience. 277:45–54. [DOI] [PubMed] [Google Scholar]
- Ugrumov MV. 2009. Non-dopaminergic neurons partly expressing dopaminergic phenotype: distribution in the brain, development and functional significance. J Chem Neuroanat. 38:241–256. [DOI] [PubMed] [Google Scholar]
- Ugrumov MV. 2013. Brain neurons partly expressing dopaminergic phenotype: location, development, functional significance, and regulation. Adv Pharmacol. 68:37–91. [DOI] [PubMed] [Google Scholar]
- Ugrumov MV, Melnikova VI, Lavrentyeva AV, Kudrin VS, Rayevsky KS. 2004. Dopamine synthesis by non-dopaminergic neurons expressing individual complementary enzymes of the dopamine synthetic pathway in the arcuate nucleus of fetal rats. Neuroscience. 124:629–635. [DOI] [PubMed] [Google Scholar]
- Vogt BA, Paxinos G. 2014. Cytoarchitecture of mouse and rat cingulate cortex with human homologies. Brain Struct Funct. 219:185–192. [DOI] [PubMed] [Google Scholar]
- von Economo C. 1926. Eine neue art spezialzellen des lobus cinguli und lobus insulae. Zeitschrift für die gesamte Neurologie und Psychiatrie. 100:706–712. [Google Scholar]
- West MJ. 2002. Design-based stereological methods for counting neurons. Prog Brain Res. 135:43–51. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Naganuma F, Iida T, Nakamura T, Harada R, Mohsen AS, Kasajima A, Sasano H, Yanai K. 2013. Molecular mechanism of histamine clearance by primary human astrocytes. Glia. 61:905–916. [DOI] [PubMed] [Google Scholar]
- Zhao MM, Hwa J, Perez DM. 1996. Identification of critical extracellular loop residues involved in alpha 1-adrenergic receptor subtype-selective antagonist binding. Mol Pharmacol. 50:1118–1126. [PubMed] [Google Scholar]
- Zhou J, Greicius MD, Gennatas ED, Growdon ME, Jang JY, Rabinovici GD, Kramer JH, Weiner M, Miller BL, Seeley WW. 2010. Divergent network connectivity changes in behavioural variant frontotemporal dementia and Alzheimer's disease. Brain. 133:1352–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Seeley WW. 2014. Network dysfunction in Alzheimer's disease and frontotemporal dementia: implications for psychiatry. Biol Psychiatry. 75:565–573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.